Abstract
The incidence and development of colorectal cancer (CRC) is a process with multiple gene interactions. We have previously demonstrated that ATP synthase-coupling factor 6, mitochondrial (ATP5J) is associated with CRC migration and 5-fluorouracil resistance; nevertheless, the exact molecular mechanism remains unclear. The following study uses microarray and bioinformatics methods to identify candidate genes and long non-coding RNAs (lncRNAs) in CRC cells (two pairs) with upregulated and downregulated ATP5J. Briefly, a total of 2,190 differentially expressed mRNAs (DEmRNAs) were sorted. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed for 4 DEmRNAs to validate the results of microarray analysis. Functional annotation and pathway enrichment were analyzed for DEmRNAs using the Database for Annotation, Visualization and Integrated Discovery. Significantly enriched pathways included the regulation of gene expression and cell growth. The protein-protein interaction network was constructed, and AKT serine/threonine kinase 2 (AKT2) was considered as one of the hub genes. For further analysis, 51 DEmRNAs and 30 DElncRNAs were selected that were positively or negatively associated with the expression of ATP5J in the two cell pairs. X-inactive specific transcript (XIST), premature ovarian failure 1B (POF1B) and calmin (CLMN) were identified in the DEmRNA-DElncRNA co-expression network. The expression of AKT2 and XIST in CRC cells was confirmed by RT-qPCR. To sum up, the candidate genes and lncRNAs, as well as potential signaling pathways, which were identified using integrated bioinformatics analysis, could improve the understanding of molecular events involved in the function of ATP5J in CRC.
Keywords: colorectal cancer, ATP5J, microarray, long non-coding RNA, bioinformatics analysis
Introduction
For patients with colorectal cancer (CRC), the overall survival benefits of systemic treatments, including tumor resection, 5-fluorouracil (5-FU)-based chemotherapy and targeted therapies, have been firmly established (1,2). Nevertheless, recurrence and metastasis caused by the drug resistance of tumor cells continue to obstruct significant therapeutic efficacy.
The development of CRC is a multi-gene, multi-step and multi-factor interactive process. The improvement of therapeutic efficacy significantly relies on improved studies of gene functions and mechanisms (3). It has been widely recognized that abnormal bioenergetics is one of the most common phenotypes of a number of tumor cells, including CRC (4). Changes in ATP synthetase are found in tumor cells and are considered to be associated with the energy metabolism and drug resistance within the tumor (5,6).
ATP synthase-coupling factor 6, mitochondrial (ATP5J) is a protein connecting F0 and F1, which are two components of ATP synthetase (7). ATP5J has been hypothesized to recover the activity of ATPase inhibited by oligomycin, as well as the exchange between ATP and inorganic phosphorus (8). Our previous study reported that ATP5J may be a tumor biomarker. Higher expression of ATP5J was found in CRC tissue compared with that in normal tissue, and in metastatic lymph nodes compared with that in primary tumors (9). In vitro experiments demonstrated that upregulation of ATP5J expression in DLD1 cells enhances cell migration and 5-FU resistance, while downregulation reverses this reaction. Investigation of ATP5J has been mainly focused on cardiovascular disease. Only a few studies have discussed the role of ATP5J in cancer, including renal carcinoma and hepatocellular carcinoma (10,11). The molecular mechanism of ATP5J in CRC cells remains unknown.
Based on previous studies, the following study investigated differentially expressed mRNAs (DEmRNAs) and lncRNAs (DElncRNAs) using microarray and bioinformatics methods. The obtained results may provide valuable information on the function and mechanism of ATP5J in CRC, and a better understanding may be beneficial for the improvement of CRC management.
Materials and methods
Cell lines
In our previous study (9), the human colon cancer DLD1 cell line was used for cell transfection and stable colony selection. The expression of ATP5J was highly downregulated in clone 4, while it was highly upregulated in clone 2. Consequently, 4 DLD1 cells that were stably transfected with pcDNA3.1(+)/ATP5J plasmid (DLD1/A2), ATP5J short hairpin (sh)RNA plasmid (DLD1/SA4) and the corresponding control vectors (DLD1/C6 and DLD1/CA6), respectively, were selected and used in the present study.
RNA extraction and microarray
Total RNA was extracted from the 4 cell types by TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocols. RNA quality was detected by an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). The microarray analysis was performed by CapitalBio Corporation (Beijing, China). Total RNA was extracted from the cells and the cDNA that was then produced by reverse transcription served as the template to synthesize the fluorescently labeled (Cy5 or Cy3) cDNA using Klenow fragment polymerase. The labeled cDNA was hybridized on the lncRNA + mRNA Human Gene Expression Microarray (4×180K; Agilent Technologies, Inc.). The microarrays were washed three times with wash solution (0.2% SDS, 2X SSC at 42°C for 120 sec) and scanned using an Agilent G2565CA Microarray Scanner (Agilent Technologies, Inc.), according to the manu facturer' s protocols. The raw data were analyzed and then normalized using percentile normalization. Only the mRNAs or lncRNAs with a fold-change ≥2.0 and a P-value cutoff of <0.05 were considered as DEmRNAs or DElncRNAs.
Pathway enrichment analysis of DEmRNAs
The Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/) provides a comprehensive set of functional annotation tools for investigators to extract biological meaning from genes or proteins. The Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/) is a database resource for functional interpretation and annotation of enriched pathways using large-scale gene datasets. The Gene Ontology (GO; http://www.geneontology.org/) database is commonly used to unify the representation of gene and gene product attributes. GO and KEGG pathway enrichment analyses were conducted for differentially expressed genes using DAVID. A P-value of <0.05 was selected as the cut-off criterion.
Protein-protein interaction (PPI) network construction
The Search Tool for the Retrieval of Interacting Genes (STRING; http://www.string-db.org/) database is an online database containing direct and indirect associations of proteins. DEmRNAs were mapped to STRING to assess the interactions. Next, PPI networks were established using Cytoscape software (version 3.6.0; http://www.cytoscape.org/). The Molecular Complex Detection (MCODE) plug-in was utilized to filter the modules of the PPI network in Cytoscape. An MCODE score of >5 was the selection criteria. Hub genes were exported. Function and pathway enrichment analyses were performed for DEmRNAs in the modules. A P-value of <0.05 was considered statistically significant.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted as aforementioned. RNA was quantified using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Inc.). cDNA was generated using an RNeasy Mini kit (Takara Bio, Inc., Otsu, Japan). qPCR analysis was performed with SYBR-Green Master mix (Takara Bio, Inc.) for lncRNA and mRNA detection. The qPCR was performed at 95°C for 2 min, 40 cycles of 95°C for 5 sec and 60°C for 30 sec, one cycle of 95°C for 5 sec, 60°C for 1 min and 95°C for 15 sec, and finally, 50°C for 30 sec. Relative expression was analyzed using the 2−ΔΔCq method (12). Human GAPDH was used as an endogenous reference gene. The sequences of the primers are shown in Table I.
Table I.
Sequences of primers used for reverse transcription-quantitative polymerase chain reaction.
| mRNAs/lncRNAs | Primer sequences |
|---|---|
| ATP5J | F: TCAGCCGTCTCAGTCCATTT |
| R: CCAAACATTTGCTTGAGCTT | |
| CRTAM | F: CCAAATACCAGCTTCTTCATCA |
| R: CTTCAAACCGGAAGGGTGCT | |
| CD44 | F: AGTCACAGACCTGCCCAATGCCTTT |
| R: TTTGCTCCACCTTCTTGACTCCCATG | |
| CST1 | F: AGGAGACCATGGCCCAGTAT |
| R: GCAGCGGACGTCTGTAGTAG | |
| EEF1A2 | F: TCTCCAAGAATGGGCAGACG |
| R: TTGACGATCTCGTCGTAGCG | |
| XIST | F: CTCTCCATTGGGTTCAC |
| R: GCGGCAGGTCTTAAGAGATGAG | |
| AKT2 | F: GCCACCATGAATGAGGTGAATA |
| R: CCTTGTACCCAATGAAGGAGC | |
| GAPDH | F: ACCACAGTCCATGCCATCAC |
| R: TCCACCACCCTGTTGCTGTA |
ATP5J, ATP synthase-coupling factor 6, mitochondrial; CRTAM, cytotoxic and regulatory T cell molecule; CST1, cystatin SN; CD44, cluster of differentiation 44; EEF1A2, eukaryotic translation elongation factor 1α2; AKT2, AKT serine/threonine kinase 2; XIST, X-inactive specific transcript; lnCRNA, long non-coding RNA; F, forward; R, reverse.
Construction of DEmRNA-DElncRNA co-expression network
A DElncRNA-DEmRNA co-expression network was constructed to explore the association between the RNAs. Pearson's correlation coefficient (PCC) was calculated between DElncRNAs and DEmRNAs according to corresponding expression levels. The criteria of |PCC| ≥0.90 was used to determine DElncRNA-DEmRNA pairs for network construction. The DElncRNA-DEmRNA network was constructed using Cytoscape 3.5.1 (http://cytoscape.org/).
Statistical analysis
The results of the qPCR were presented using Graph Pad Prism (version 6.0; GraphPad Software, Inc., La Jolla, CA, USA). Data are presented as the mean ± standard error of the mean. The independent samples t-test was performed for data comparison. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DEmRNAs
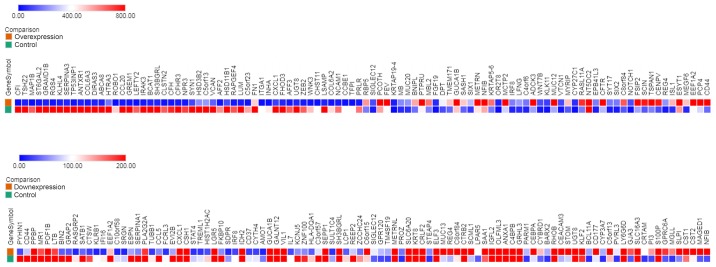
According to microarray analysis, 32,206 mRNAs were detected in two pairs of cells, including 2,190 DEmRNAs (503 upregulated and 1,687 downregulated), which were significantly differentially expressed with a fold-change of ≥2.0. In addition, the DEmRNAs were divided into two sets: Set A contained all DEmRNAs from cell pair DLD1/A2 vs. DLD1/C6, and set B contained all DEmRNAs from DLD1/SA4 vs. DLD1/CA6. Heat map representation showed the top 100 differentially DEmRNAs (Fig. 1).
Figure 1.
Heat map of the top 100 differentially expressed DEmRNAs in two pairs of cells. Red and blue represent high expression and low expression, respectively.
RT-qPCR validation of DEmRNAs
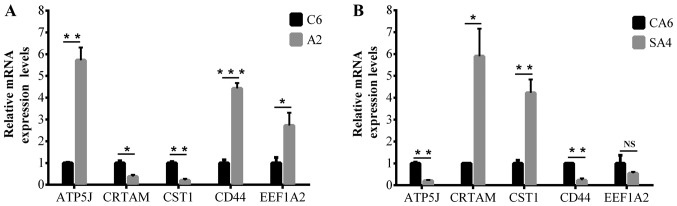
To validate the results of microarray analysis, RT-qPCR was performed in ATP5J and four randomly selected mRNAs. As shown in Fig. 2, cytotoxic and regulatory T cell molecule and cystatin SN were downregulated in DLD1/A2 vs. DLD1/C6 and upregulated in DLD1/SA4 vs. DLD1/CA6, while opposite results were obtained for ATP5J, cluster of differentiation 44 and eukaryotic translation elongation factor 1α2. The PCR results were basically concurrent with the microarray data.
Figure 2.
Reverse transcription-quantitative polymerase chain reaction results for ATP5J and four randomly selected mRNAs. (A) The expression level in DLD1/A2 and DLD1/C6; (B) The expression level in DLD1/SA4 and DLD1/CA6. *P<0.05, **P<0.01 and ***P<0.001. NS, not significant; ATP5J, ATP synthase-coupling factor 6, mitochondrial; CRTAM, cytotoxic and regulatory T cell molecule; CST1, cystatin SN; CD44, cluster of differentiation 44; EEF1A2, eukaryotic translation elongation factor 1α2.
Functional and pathway enrichment analysis
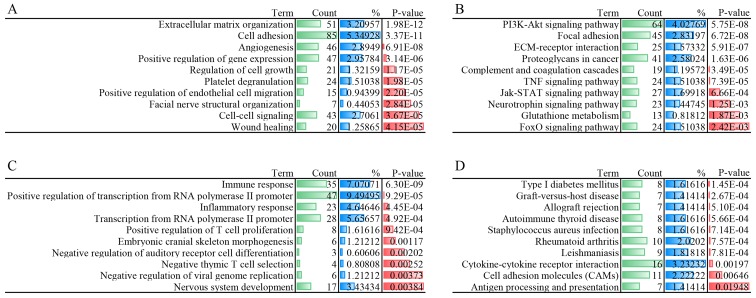
In order to investigate potential gene and gene products, GO functional and KEGG pathway enrichment analyses were performed for sets A and B, respectively. The DEmRNAs from set A were mainly enriched in biological processes associated with 'positive regulation of gene expression', 'cell-cell signaling' and 'regulation of cell growth'. The 10 most significantly enriched GO terms for DEmRNAs from sets A and B are shown in Fig. 3A and C, while the 10 most significantly enriched KEGG pathways in sets A and B are presented in Fig. 3B and D.
Figure 3.
Gene Ontology functional and KEGG pathway enrichment analysis. The top 10 terms were selected according to P-value. (A) Biological process of set A. (B) Enriched KEGG pathways of set A. (C) Biological process of set B. (D) Enriched KEGG pathways of set B. KEGG, Kyoto Encyclopedia of Genes and Genomes.
PPI network analysis
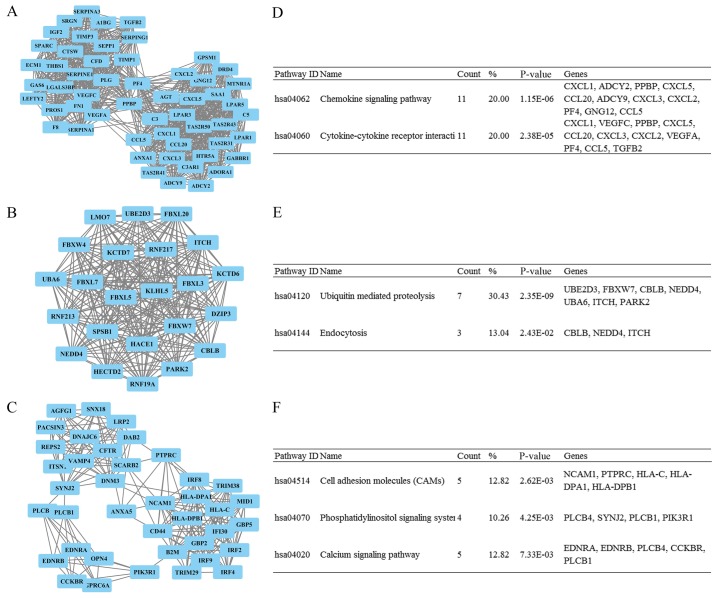
By submitting all DEmRNAs from sets A and B into STRING, the PPI network was obtained (data not shown). Next, the MCODE plug-in was used to analyze the results of STRING in Cytoscape software. The 3 most significant modules were acquired; the functions were mainly associated with 'cytokine-cytokine receptor interaction', 'ubiquitin-mediated proteolysis' and 'cell adhesion molecules (CAMs)' (Fig. 4). Moreover, a total of 20 genes were considered as hub genes (Table II), among which AKT serine/threonine kinase 2 (AKT2) obtained the highest value for neighborhood connectivity, outdegree and closeness centrality. This meant that AKT2 was the most important gene within the PPI network and represented the closest connection with other genes in the module (13).
Figure 4.
Top three modules of the protein-protein interaction network. (A) Module 1, (B) module 2, (C) module 3 and their genes, combined with enriched pathways, are shown in excel format in (D, E and F).
Table II.
Hub genes from Molecular Complex Detection analysis.
| Gene symbol | Neighborhood connectivity | Outdegree | Closeness centrality |
|---|---|---|---|
| MTNR1A | 42.20 | 12 | 3.23×10−1 |
| SPSB1 | 30.65 | 8 | 2.93×10−1 |
| CCKBR | 47.25 | 8 | 2.94×10−1 |
| AKT2a | 57.50 | 34 | 3.54×10−1 |
| GPX3 | 16.18 | 15 | 2.62×10−1 |
| PTPN6 | 41.71 | 31 | 3.38×10−1 |
| CLIC3 | 17.20 | 5 | 2.28×10−1 |
| TNFRSF19 | 10.00 | 6 | 2.56×10−1 |
| IFT80 | 45.83 | 2 | 2.03×10−1 |
| AMHR2 | 34.69 | 1 | 2.93×10−1 |
| MAMLD1 | 41.00 | 8 | 2.96×10−1 |
| DHRSX | 30.63 | 3 | 1.93×10−1 |
| SLC47A1 | 5.00 | 1 | 1.00 |
| SLC30A4 | 3.75 | 0 | 0.00 |
| CYBRD1 | 19.33 | 1 | 1.00 |
| INTS6 | 14.67 | 0 | 0.00 |
| CLDN7 | 9.50 | 4 | 2.04×10−1 |
| TNFSF12 | 56.67 | 9 | 2.93×10−1 |
| DGKA | 21.50 | 3 | 3.13×10−1 |
| HOMER2 | 27.13 | 2 | 2.49×10−1 |
Gene showing the closest connection with other genes.
Co-expression network of DEmRNA-DElncRNA
Based on microarray analysis, 39,304 lncRNAs were detected. A total of 3,002 DElncRNAs (2,319 in DLD1/A2 vs. DLD1/C6; 683 in DLD1/SA4 vs. DLD1/CA6) were significantly differentially expressed with a fold-change of ≥2.0. For further analysis, DElncRNAs upregulated in DLD1/A2 vs. DLD1/C6 and downregulated in DLD1/SA4 vs. DLD1/CA6 were selected, which meant that the expression of DElncRNAs was positively associated with the expression of ATP5J in the two cell pairs. The DElncRNAs that were negatively associated with ATP5J in the two cell pairs were also included. Certain DEmRNAs were selected according to the aforementioned criteria. Therefore, 51 DEmRNAs and 30 DElncRNAs were identified and listed in Tables III and IV.
Table III.
Selected differentially expressed mRNAs for co-expression network construction (n=51).
| Ensemble ID | Gene symbol | Log2FC (A2 vs. C6) | log2FC (SA4 vs. CA6) |
|---|---|---|---|
| Upregulation | |||
| ENST00000463422 | ATOH8 | 2.3664742 | −1.5749975 |
| ENST00000308108 | CCNE2 | 1.0017490 | −1.0507374 |
| ENST00000278385 | CD44 | 3.8303200 | −3.9564400 |
| ENST00000298912 | CLMN | 1.2790971 | −1.2013025 |
| ENST00000329882 | CSH1 | 1.2013025 | −2.2145548 |
| ENST00000249749 | DLL4 | 1.4630564 | −1.3910798 |
| ENST00000217182 | EEF1A2 | 3.5102847 | −2.7776937 |
| ENST00000371021 | FRAT1 | 1.1014051 | −1.1662812 |
| ENST00000435292 | GUCA1B | 1.7285424 | −1.9108304 |
| ENST00000377791 | HIST1H2AC | 1.0301589 | −2.0924091 |
| ENST00000330062 | IDH2 | 1.3220385 | −2.0191135 |
| ENST00000518982 | IL7 | 1.3051137 | −1.8685079 |
| ENST00000268638 | IRF8 | 1.8297353 | −2.0302430 |
| ENST00000378111 | KCNAB2 | 1.1377705 | −1.4946451 |
| ENST00000394683 | KIAA1199 | 1.0515032 | −1.0550995 |
| ENST00000420981 | MGC39372 | 1.2202988 | −1.6457891 |
| ENST00000396217 | MYRIP | 1.9900737 | −1.2685270 |
| ENST00000277541 | NOTCH1 | 2.4109862 | −1.5415106 |
| ENST00000276124 | POF1B | 1.1683197 | −3.3742200 |
| ENST00000313755 | PRODH | 1.1842122 | −1.4703089 |
| ENST00000341901 | SBK1 | 1.1663734 | −1.3532438 |
| ENST00000247182 | SIX1 | 1.7563076 | −1.2851509 |
| ENST00000393062 | SPIRE2 | 1.3516588 | −1.1577921 |
| ENST00000328526 | XKRX | 1.1844625 | −1.1355352 |
| Downregulation | |||
| ENST00000376726 | ACSS1 | −1.4264563 | 1.0032806 |
| ENST00000374111 | C10orf53 | −3.0494199 | 2.7619016 |
| ENST00000397899 | C2orf55 | −1.0406728 | 1.8891068 |
| ENST00000492730 | C4BPB | −1.8177981 | 1.7743891 |
| ENST00000509979 | C5orf13 | −4.4611087 | 2.1796174 |
| ENST00000503763 | CRHBP | −2.4869404 | 1.2709899 |
| ENST00000227348 | CRTAM | −2.1076543 | 2.4206290 |
| ENST00000450719 | CSHL1 | −1.2476245 | 1.3876114 |
| ENST00000398402 | CST1 | −1.7663736 | 3.7468631 |
| ENST00000304725 | CST2 | −3.1294575 | 4.1763420 |
| ENST00000292414 | CYP3A7 | −1.5009923 | 2.0561150 |
| ENST00000317811 | FJX1 | −1.1050119 | 1.2775639 |
| ENST00000310357 | GPRC6A | −1.3399930 | 2.4899235 |
| ENST00000393584 | GSTA5 | −1.7383062 | 1.0165440 |
| ENST00000309446 | KLF7 | −1.5819120 | 1.1883107 |
| ENST00000375827 | LY6G6D | −1.1885548 | 2.3284416 |
| ENST00000427231 | NEB | −2.4703722 | 1.1157570 |
| ENST00000319792 | PVRL3 | −4.1147676 | 2.2300897 |
| ENST00000216392 | PYGL | −3.4774067 | 1.2769729 |
| ENST00000405158 | SAA1 | −3.2550786 | 1.6204370 |
| ENST00000414546 | SAA2 | −1.4848727 | 1.1160498 |
| ENST00000051659 | SCML1 | −2.6339436 | 1.5956994 |
| ENST00000486749 | SLC2A3 | −2.8174708 | 1.1550741 |
| ENST00000358525 | SLC6A20 | −1.2244166 | 1.4633511 |
| ENST00000338380 | SLPI | −1.9716522 | 3.2052798 |
| ENST00000394511 | UGT8 | −4.2315920 | 1.8874178 |
FC, fold-change.
Table IV.
Selected differentially expressed long non-coding RNAs for co-expression network construction (n=30).
| Ensemble gene ID | Gene symbol | log2FC (A2 vs. C6) | log2FC (SA4 vs. CA6) |
|---|---|---|---|
| Upregulation | |||
| ENSG00000026508 | CD44 | 5.7112436 | −4.6520596 |
| ENSG00000248690 | HAS2-AS1 | 1.5613460 | −1.3604736 |
| ENSG00000104432 | IL7 | 1.2327710 | −2.0651786 |
| ENSG00000242887 | IGHJ3 | 1.1636353 | −1.1661916 |
| ENSG00000227372 | TP73-AS1 | 1.1705691 | −1.1062335 |
| ENSG00000237807 | RP11-400K9.4 | 1.0098063 | −1.4481292 |
| ENSG00000227357 | HLA-DRB4 | 1.0012778 | −1.1985005 |
| ENSG00000212999 | AC117834.1 | 1.2642217 | −1.2010332 |
| ENSG00000170011 | MYRIP | 1.9657596 | −1.1791620 |
| ENSG00000174562 | KLK15 | 1.7544044 | −1.0959578 |
| ENSG00000140968 | IRF8 | 2.0519360 | −1.8488283 |
| Downregulation | |||
| ENSG00000100181 | TPTEP1 | −1.1547785 | 1.0562963 |
| ENSG00000256124 | RP11-84E24.3 | −1.0987811 | 1.4957023 |
| ENSG00000255071 | SAA2-SAA4 | −2.9303002 | 1.6360683 |
| ENSG00000080823 | MOK | −1.2133837 | 1.0177355 |
| ENSG00000253668 | RP11-463C14.1 | −3.7030535 | 2.1768303 |
| ENSG00000151612 | ZNF827 | −1.9309711 | 1.2170410 |
| ENSG00000225383 | SFTA1P | −1.1355629 | 1.3586316 |
| ENSG00000229791 | LINC00420 | −1.6585293 | 1.3905687 |
| ENSG00000236654 | AC079780.3 | −4.6469680 | 1.8889217 |
| ENSG00000254148 | RP11-84E24.2 | −2.1362443 | 1.6749586 |
| ENSG00000102098 | SCML2 | −1.3423405 | 1.0036592 |
| ENSG00000229807 | XIST | −2.4771900 | 1.0491910 |
| ENSG00000173432 | SAA1 | −1.9162921 | 1.2718056 |
| ENSG00000212232 | SNORD17 | −1.2717161 | 1.0156708 |
| ENSG00000206402 | LY6G6D | −1.5264271 | 2.9315690 |
| ENSG00000071575 | TRIB2 | −1.3852354 | 1.0461107 |
| ENSG00000138735 | PDE5A | −1.5525875 | 1.1479750 |
| ENSG00000183531 | Z98256.1 | −2.9538560 | 2.0470590 |
| ENSG00000174607 | UGT8 | −3.3867934 | 2.0097785 |
FC, fold-change.
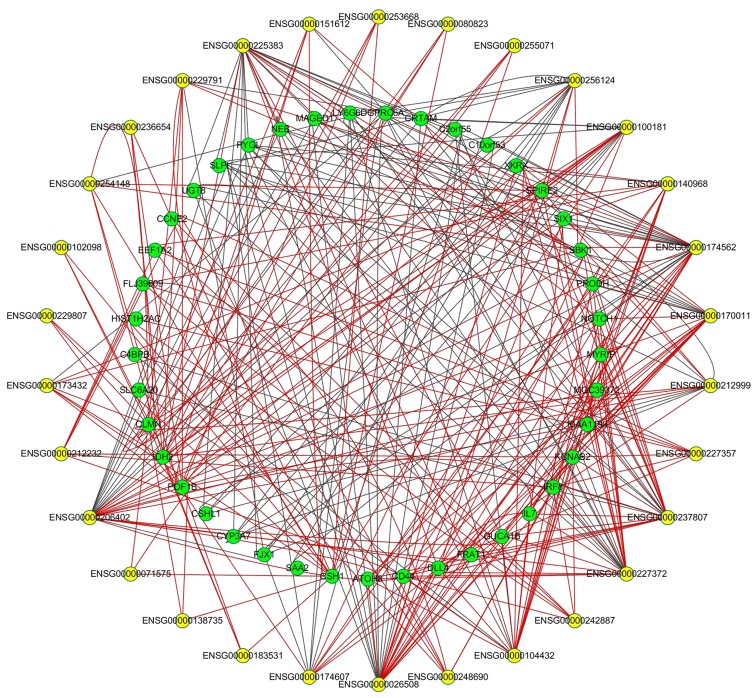
To further investigate the potential association between DEmRNAs and DElncRNAs, PCC was calculated according to the expression levels of 51 DEmRNAs and 30 DElncRNAs. Using |PCC| ≥0.90, 343 DEmRNA/DElncRNA pairs were used to construct the co-expression network, including 239 pairs exhibiting positive correlation. The network is shown in Fig. 5. The pair of premature ovarian failure 1B (POF1B) and ENSG00000236654 presented the most significant positive correlation coefficiency, while the most marked negative correlation coefficiency was found in the pair of glycogen phosphorylase L and ENSG00000170011. One DElncRNA may target multiple DEmRNAs; for example, XIST was associated with POF1B, chorionic somatomammotropin hormone 1 (CSH1) and calmin (CLMN), leading to multiple signaling pathways or mechanisms.
Figure 5.
DElncRNA-DEmRNA co-expression network constructed based on the correlation analysis. Yellow nodes represent DElncRNAs and green nodes represent the target DEmRNAs. Red lines represent a positive association and black lines represent a negative association. DElncRNA, differentially-expressed long non-coding RNA.
RT-qPCR validation of hub genes and DElncRNA
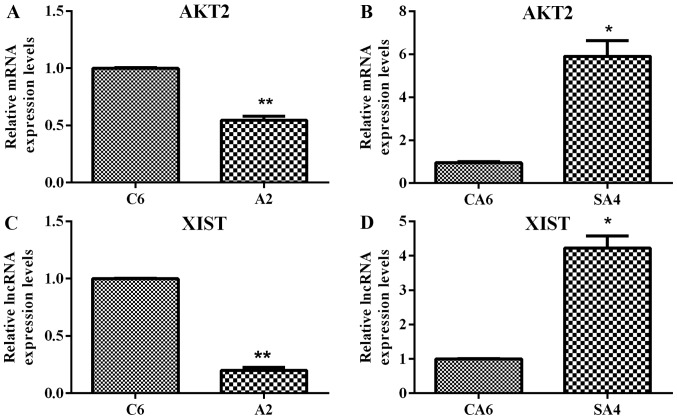
RT-qPCR was used to validate AKT2 and XIST. As shown in Fig. 6, AKT2 and XIST were downregulated in DLD1/A2 vs. DLD1/C6 (P<0.01) and upregulated in DLD1/SA4 vs. DLD1/CA6 (P<0.05). In general, the validation results were consistent with the microarray data.
Figure 6.
Reverse transcription-quantitative polymerase chain reaction validation of AKT2 and XIST. (A) The expression level of AKT2 in DLD1/A2 and DLD1/C6. (B) The expression level of AKT in DLD1/SA4 and DLD1/CA6. (C) The expression level of XIST in DLD1/A2 and DLD1/C6. (D) The expression level of XIST in DLD1/SA4 and DLD1/CA6. *P<0.05 and **P<0.01. lncRNA, long non-coding RNA; AKT2, AKT serine/threonine kinase 2; XIST, X-inactive specific transcript.
Discussion
ATP5J has been associated with several cancer types, including diffuse large B-cell lymphoma renal carcinoma and hepatocellular carcinoma (10,11,14). Furthermore, our previous study confirmed that ATP5J is associated with CRC cell migration and 5-fluorouracil resistance. The present study further investigated the molecular mechanism and potential key genes interacting with ATP5J in CRC via microarray and bioinformatics analyses.
The mRNAs from set A, which were differentially expressed in the upregulation of ATP5J, were mainly enriched in the GO terms of 'extracellular matrix organization', 'cell adhesion', 'positive regulation of gene expression' and 'regulation of cell growth'. These biological processes are essential for tumor cell survival and growth. Emerging data have confirmed the crucial role of the extracellular matrix in tumor metastasis (15,16). Changes in the content of the extracellular matrix may influence tumor cell properties, including proliferation and motility (17). Moreover, the primary enriched GO terms of set B contained enzyme regulation ('regulation of transcription from RNA polymerase II promoter') and biological responses ('immune response' and 'inflammatory response'). It is widely accepted that a causal association exists between inflammation, immunity and cancer (18). Human immunity responses, such as immune surveillance, are part of an innate and efficient antineoplastic system. Therefore, it could be concluded that the upregulation of ATP5J may affect cell migration and 5-FU sensitivity by influencing the biological processes of the intracellular and extracellular environment, thus promoting cell growth; its downregulation may operate by regulating immune and inflammatory responses. The KEGG pathways of set A were enriched primarily in tumor-related processes, including the phosphoinositide 3-kinase-protein kinase B signaling pathway, the tumor necrosis factor (TNF) signaling pathway and the Janus kinase-signal transducer and activator of transcription signaling pathway (19,20).
According to PPI network analysis, several biomarkers were identified. Some of these genes are associated with CRC biological behavior, including AKT2, TNF receptor superfamily member 19, claudin 7, cholecystokinin B receptor, glutathione peroxidase 3 and homer scaffolding protein 2. AKT2, one of the most significantly meaningful genes, has been shown to be essential in several cellular pathways involved in cell proliferation, metastasis and drug resistance (21). Overexpression of AKT2 has been detected in breast, ovarian and colon cancer (22,23). Previous studies have investigated the role of AKT2 in CRC via the construction of cell and animal models. The results showed that AKT2 deficiency may decrease the metastatic ability of CRC cells in mice (24). In addition, an shRNA-mediated AKT2-knockdown system has been shown to decrease cell survival and proliferation in primary tumors (25). Furthermore, low AKT2 expression in HCT116 cells has been shown to be correlated with increased chemosensitivity to paclitaxel (26). Taken together, the regulation of AKT2 may be a critical process for ATP5J exerting an effect on CRC cell biological functions.
Numerous studies have confirmed the association between lncRNAs and tumors. Previous studies have indicated that lncRNAs could have an important regulatory role in gene expression and are associated with cancer development (27). In the present study, microarray analysis was performed for lncRNAs. From the lncRNA expression profiles, DElncRNAs meeting the criteria were selected for further analysis. According to the results, TP73-AS1, HAS2-AS1, SFTA1P and XIST were highlighted and have been previously associated with the development and progression of certain cancer types, including hepatocellular carcinoma and oral squamous cell carcinoma (28–30). Among them, XIST was the only lncRNA that has been previously investigated in CRC (31). XIST, located in the X chromosome, was one of the first lncRNAs to be found that was involved in the progression of cancer, including non-small cell lung cancer and human glioblastoma (32,33). Certain studies have found the mechanism of XIST in several cancer types; for example, the lncRNA XIST has been shown to promote the malignancy of esophageal squamous cell carcinoma through the modulation of miR-101/enhancer of zeste homolog 2 (34). In addition, by regulating miR-186-5p, XIST controls cell proliferation and invasion in non-small cell lung cancer (35). Moreover, significantly high XIST expression has been found in CRC tumor tissues compared with that in paired adjacent normal tissues (36). High-level expression of XIST could predict poor disease-free survival times in CRC patients. Moreover, XIST is considered to be an oncogene that could promote cell proliferation through the miR-132-3p/mitogen-activated protein kinase 1 axis (31).
To further elucidate the potential functions of lncRNAs and find out their target genes in the cell pairs of the present study, a co-expression network of DEmRNA/DElncRNA was constructed. XIST was associated with 3 mRNAs (POF1B, CSH1 and CLMN). The gene encoding POF1B was critical for ovarian function (37). However, its impact was also confirmed in the regulation of cell adhesion in human intestinal cell lines (38). CLMN has been shown to regulate the cell cycle in a report involving neuroblastoma cells (39). Therefore, we hypothesized that the co-repression of XIST and POF1B and CLMN was involved in the molecular mechanisms of ATP5J in CRC cells, which will be confirmed by further cell line experiments in future studies.
In conclusion, the present study determined the expression profiles of mRNAs and lncRNAs in CRC cells overexpressing/underexpressing ATP5J. According to bioinformatics analyses, AKT2 and XIST expression was identified as a potential biomarker participating in the effect of ATP5J in CRC, which in turn was associated with cell migration and 5-FU resistance. Further research is necessary to confirm this hypothesis.
Acknowledgments
This study represents partial fulfillment of the requirements for a Master degree for Dr Bingjun Bai.
Footnotes
Funding
This study was supported by the National Natural Science Foundation of China (grant no. 81272681) and the Medical and Health Science and Technology Foundation of Zhejiang Province (grant no. 2017 KY593).
Availability of data and materials
The datasets analyzed during the current study are not publicly available as they will be used in our further studies, but they are available from the corresponding author on reasonable request.
Authors' contributions
Guarantor of integrity of entire study: BJB, BBX, ZYP and HBZ. Study concepts: BJB, BBX, ZYP and HBZ. Study design: BJB, BBX, ZYP and HBZ. Data acquisition: LNS and ZJP. Data analysis/interpretation: LNS and ZJP. Statistical analysis: BJB, BBX, ZYP and HBZ. Manuscript preparation: LNS and ZJP. Manuscript revision/review: BJB, BBX, ZYP and HBZ. Manuscript final version approval: BJB, BBX, ZYP and HBZ.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflicts of interest
All authors listed in the manuscript agree on the content of the and this submission. The authors declare no conflict of interest.
References
- 1.Chau I, Cunningham D. Chemotherapy in colorectal cancer: New options and new challenges. Br Med Bull. 2002;64:159–180. doi: 10.1093/bmb/64.1.159. [DOI] [PubMed] [Google Scholar]
- 2.Shao YC, Chang YY, Lin JK, Lin CC, Wang HS, Yang SH, Jiang JK, Lan YT, Lin TC, Li AF, et al. Neoadjuvant chemotherapy can improve outcome of colorectal cancer patients with unresectable metastasis. Int J Colorectal Dis. 2013;28:1359–1365. doi: 10.1007/s00384-013-1713-x. [DOI] [PubMed] [Google Scholar]
- 3.Liu HY, Zhang CJ. Identification of differentially expressed genes and their upstream regulators in colorectal cancer. Cancer Gene Ther. 2017;24:244–250. doi: 10.1038/cgt.2017.8. [DOI] [PubMed] [Google Scholar]
- 4.Pedersen PL. Bioenergetics of cancer cells - A brief orientation to this minireview series. J Bioenerg Biomembr. 1997;29:301–302. doi: 10.1023/A:1022417911796. [DOI] [Google Scholar]
- 5.Cuezva JM, Krajewska M, de Heredia ML, Krajewski S, Santamaría G, Kim H, Zapata JM, Marusawa H, Chamorro M, Reed JC. The bioenergetic signature of cancer: A marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- 6.Shin YK, Yoo BC, Chang HJ, Jeon E, Hong SH, Jung MS, Lim SJ, Park JG. Down-regulation of mitochondrial F1F0-ATP synthase in human colon cancer cells with induced 5-fluorouracil resistance. Cancer Res. 2005;65:3162–3170. doi: 10.1158/0008-5472.CAN-04-3300. [DOI] [PubMed] [Google Scholar]
- 7.Collinson IR, van Raaij MJ, Runswick MJ, Fearnley IM, Skehel JM, Orriss GL, Miroux B, Walker JE. ATP synthase from bovine heart mitochondria. In vitro assembly of a stalk complex in the presence of F1-ATPase and in its absence. J Mol Biol. 1994;242:408–421. doi: 10.1006/jmbi.1994.1591. [DOI] [PubMed] [Google Scholar]
- 8.Joshi S, Pringle MJ. ATP synthase complex from bovine heart mitochondria. Passive H+ conduction through mitochondrial coupling factor 6-depleted F0 complexes. J Biol Chem. 1989;264:15548–15551. [PubMed] [Google Scholar]
- 9.Zhu H, Chen L, Zhou W, Huang Z, Hu J, Dai S, Wang X, Huang X, He C. Over-expression of the ATP5J gene correlates with cell migration and 5-fluorouracil sensitivity in colorectal cancer. PLoS One. 2013;8:e76846. doi: 10.1371/journal.pone.0076846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, Lu Y, Xu Y, Xu L, Zheng W, Wu Y, Li L, Shen P. Estrogen represses hepatocellular carcinoma (HCC) growth via inhibiting alternative activation of tumor-associated macrophages (TAMs) J Biol Chem. 2012;287:40140–40149. doi: 10.1074/jbc.M112.348763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjerregaard H, Pedersen S, Kristensen SR, Marcussen N. Reference genes for gene expression analysis by real-time reverse transcription polymerase chain reaction of renal cell carcinoma. Diagn Mol Pathol. 2011;20:212–217. doi: 10.1097/PDM.0b013e318212e0a9. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Carson MB, Lu H. Network-based prediction and knowledge mining of disease genes. BMC Med Genomics. 2015;8(Suppl 2):S9. doi: 10.1186/1755-8794-8-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamani-Ahmadmahmudi M, Najafi A, Nassiri SM. Reconstruction of canine diffuse large B-cell lymphoma gene regulatory network: Detection of functional modules and hub genes. J Comp Pathol. 2015;152:119–130. doi: 10.1016/j.jcpa.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynes RO. The extracellular matrix: Not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: An updated review. Ann Med. 2014;46:372–383. doi: 10.3109/07853890.2014.912836. [DOI] [PubMed] [Google Scholar]
- 20.Groner B, von Manstein V. Jak Stat signaling and cancer: Opportunities, benefits and side effects of targeted inhibition. Mol Cell Endocrinol. 2017;451:1–14. doi: 10.1016/j.mce.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 21.Cheung M, Testa JR. Diverse mechanisms of AKT pathway activation in human malignancy. Curr Cancer Drug Targets. 2013;13:234–244. doi: 10.2174/1568009611313030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarwal E, Brattain MG, Chowdhury S. Cell survival and metastasis regulation by Akt signaling in colorectal cancer. Cell Signal. 2013;25:1711–1719. doi: 10.1016/j.cellsig.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chau NM, Ashcroft M. Akt2: A role in breast cancer metastasis. Breast Cancer Res. 2004;6:55–57. doi: 10.1186/bcr739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rychahou PG, Kang J, Gulhati P, Doan HQ, Chen LA, Xiao SY, Chung DH, Evers BM. Akt2 overexpression plays a critical role in the establishment of colorectal cancer metastasis. Proc Natl Acad Sci USA. 2008;105:20315–20320. doi: 10.1073/pnas.0810715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal E, Robb CM, Smith LM, Brattain MG, Wang J, Black JD, Chowdhury S. Role of Akt2 in regulation of metastasis suppressor 1 expression and colorectal cancer metastasis. Oncogene. 2017;36:3104–3118. doi: 10.1038/onc.2016.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding Z, Xu F, Li G, Tang J, Tang Z, Jiang P, Wu H. Knockdown of Akt2 expression by shRNA inhibits proliferation, enhances apoptosis, and increases chemosensitivity to paclitaxel in human colorectal cancer cells. Cell Biochem Biophys. 2015;71:383–388. doi: 10.1007/s12013-014-0209-9. [DOI] [PubMed] [Google Scholar]
- 27.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Huang Y, Huang Y, Fu Y, Tang D, Kang R, Zhou R, Fan XG. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36:51. doi: 10.1186/s13046-017-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu G, Wang S, Chen J, Wang Z, Liang X, Wang X, Jiang J, Lang J, Li L. Long noncoding RNA HAS2-AS1 mediates hypoxia-induced invasiveness of oral squamous cell carcinoma. Mol Carcinog. 2017;56:2210–2222. doi: 10.1002/mc.22674. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Xiong Y, Xia R, Wei C, Shi X, Nie F. The pseudogene-derived long noncoding RNA SFTA1P is downregulated and suppresses cell migration and invasion in lung adenocarcinoma. Tumour Biol. 2017;39:1010428317691418. doi: 10.1177/1010428317691418. [DOI] [PubMed] [Google Scholar]
- 31.Song H, He P, Shao T, Li Y, Li J, Zhang Y. Long non-coding RNA XIST functions as an oncogene in human colorectal cancer by targeting miR-132-3p. J BUON. 2017;22:696–703. [PubMed] [Google Scholar]
- 32.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 33.Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, Chen L, Xi Z, Teng H, Wang Z, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 34.Wu X, Dinglin X, Wang X, Luo W, Shen Q, Li Y, Gu L, Zhou Q, Zhu H, Li Y, et al. Long noncoding RNA XIST promotes malignancies of esophageal squamous cell carcinoma via regulation of miR-101/EZH2. Oncotarget. 2017;8:76015–76028. doi: 10.18632/oncotarget.18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Shen Q, Zhang X, Yang C, Cui S, Sun Y, Wang L, Fan X, Xu S. The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem. 2017;41:2221–2229. doi: 10.1159/000475637. [DOI] [PubMed] [Google Scholar]
- 36.Kara M, Yumrutas O, Ozcan O, Celik OI, Bozgeyik E, Bozgeyik I, Tasdemir S. Differential expressions of cancer-associated genes and their regulatory miRNAs in colorectal carcinoma. Gene. 2015;567:81–86. doi: 10.1016/j.gene.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 37.Bione S, Rizzolio F, Sala C, Ricotti R, Goegan M, Manzini MC, Battaglia R, Marozzi A, Vegetti W, Dalprà L, et al. Mutation analysis of two candidate genes for premature ovarian failure, DACH2 and POF1B. Hum Reprod. 2004;19:2759–2766. doi: 10.1093/humrep/deh502. [DOI] [PubMed] [Google Scholar]
- 38.Crespi A, Bertoni A, Ferrari I, Padovano V, Della Mina P, Berti E, Villa A, Pietrini G. POF1B localizes to desmosomes and regulates cell adhesion in human intestinal and keratinocyte cell lines. J Invest Dermatol. 2015;135:192–201. doi: 10.1038/jid.2014.327. [DOI] [PubMed] [Google Scholar]
- 39.Marzinke MA, Clagett-Dame M. The all-trans retinoic acid (atRA)-regulated gene Calmin (Clmn) regulates cell cycle exit and neurite outgrowth in murine neuroblastoma (Neuro2a) cells. Exp Cell Res. 2012;318:85–93. doi: 10.1016/j.yexcr.2011.10.002. [DOI] [PubMed] [Google Scholar]