Abstract
This study examined the effects of long non-coding RNA (lncRNA) BC032020 on the development of human pancreatic ductal adenocarcinoma (PDAC), and the potential molecular mechanisms responsible for these effects. The expression of BC032020 was assessed in 20 pairs of PDAC tumor tissues and adjacent normal tissues. The overexpression of BC032020 was enforced in the AsPC-1 and PANC-1 cells, and the effects on cell proliferation, cell cycle distribution, cell migration and apoptosis were determined. We also analyzed the functions of zinc finger protein 451 (ZNF451), which shares a gene sequence with two exons of BC032020 and a non-coding region with another two exons, in PDAC cells. The AsPC-1 and PANC-1 cells that overexpressed BC032020 were used to establish a subcutaneous tumor xenograft model in order to examine the effects of BC032020 on tumor growth in vivo. The results revealed that the BC032020 levels in the PDAC tumor tissues were lower than those in the adjacent normal tissues, and ZNF451 expression inversely correlated with the BC032020 levels in the PDAC tumor tissues and cell lines. BC032020 overexpression led to a decrease in ZNF451 expression; it also suppressed the proliferation and migration of the AsPC-1 and PANC-1 cells, and induced G1 phase arrest and cell apoptosis. The results of in vivo experiments revealed that BC032020 suppressed tumor growth in a xenograft model by inhibiting ZNF451 expression. Taken together, the findings of this study indicate that BC032020 suppresses the survival of PDAC cells by inhibiting ZNF451 expression.
Keywords: pancreatic ductal adenocarcinoma, long non-coding RNA, cell proliferation, cell cycle, cell migration
Introduction
Pancreatic cancer is a very lethal disease with a very poor prognosis, and is the fourth leading cause of cancer-related mortality in the developed world. The standard therapy for patients with metastatic disease has provided only minimal improvements in prognosis, and the incidence and mortality rate associated with pancreatic cancer are almost parallel (1). From a histological viewpoint, almost 95% of pancreatic cancers in humans are pancreatic ductal adenocarcinomas (PDACs), which have the worst prognosis among the major types of cancer (2). The incidence and mortality rates associated with PDAC have remained stable and have not improved over the past three decades. The average lifetime risk of pancreatic cancer for both males and females is 1 in 65 (1.5%), and the 5-year survival rate is 7% (3). As the symptoms of PDAC are difficult to detect at an early stage, the disease is usually diagnosed during its later stages (4,5).
Thus, there is a desperate need for an effective screening program that can detect PDAC and aid in establishing a prognosis during the early stages of the disease. Even though a family history (6,7), cigarette smoking history (7), chronic pancreatitis (8), and diabetes mellitus (9) have been identified as important risk factors for pancreatic cancer, a standard screening protocol is still lacking. When viewed as a genetic disease, pancreatic cancer may be induced by an accumulation of somatic mutations in oncogenes and tumor suppressor genes (10). KRAS, CDKN2A, TP53, and SMAD4 have been identified as major driver genes in PDAC (5,11). The identification of biomarkers that can aid in the prediction and detection of PDAC may prove to be of great clinical significance. Increasing evidence indicates that genetic and modifiable risk factors contribute to the development of PDAC (12,13). As regards genetic conditions, hereditary breast and ovarian cancer syndrome, Lynch syndrome, familial adenomatous polyposis, Peutz-Jeghers Syndrome, familial atypical multiple mole melanoma syndrome, hereditary pancreatitis, cystic fibrosis and ataxia-telangiectasia have been shown to increase the risk of developing pancreatic cancer (14). As regards modifiable risk factors, tobacco exposure, alcohol use, chronic pancreatitis, diet, obesity and diabetes mellitus, as well as certain abdominal surgeries and infections have also been confirmed as important risk factors for the development of PDAC (15,16).
Long non-coding RNAs (lncRNAs) comprise a group of non-coding RNA molecules that have 200 nt- to 100 kb-long transcripts, and lack an open-reading frame and the capability to code for proteins (17–19). The dysregulation of lncRNAs occurs in numerous diseases, including cancers, and affects tumor development and progression. Numerous studies have indicated that lncRNAs may serve as novel biomarkers for the early diagnosis and prognosis of cancer (18,20,21). Even though the function of most lncRNAs remains to be fully elucidated, a number of lncRNAs are now known to act as important regulators in diverse biological processes (22–25). Increasing evidence indicates that lncRNAs are involved in chromosome dosage compensation, epigenetic regulation, nuclear and cytoplasmic trafficking, splicing, transcription, translation, cell cycle control, and cell differentiation (26–28). Furthermore, lncRNAs can regulate the expression of downstream genes by mediating histone modification, chromatin remodeling or serving as precursors for microRNAs (miRNAs or miRs) or small interfering RNAs (siRNAs) (20).
In pancreatic cancer, some lncRNAs have been shown to play important roles in cell proliferation (29), cell cycle, cell apoptosis (30), cell migration (31), epithelial-mesenchymal transition (32) and drug resistance (33). Although a number of lncRNAs have been found to be dysregulated, little is known about the overall biological functions of lncRNAs in pancreatic cancer. With the avalanche of biological sequences generated in the post-genomic age, much research is required to computationally analyze their structures and functions. Typically, predictors based on machine learning techniques contain three main steps: Feature extraction, predictor construction and performance evaluation. Currently, several web servers and stand-alone tools have been developed to facilitate the biological sequence analysis (34,35).
In this study, we obtained two datasets of lncRNAs in pancreatic cancer tissues from the Cancer RNA-Seq Nexus (CRN) database, and from the intersection of the two datasets we found 13 lncRNAs that were differently expressed in the PDAC tissues when compared with the adjacent non-tumor tissues. Furthermore, we verified that the expression levels of lncRNA BC032020 differed significantly between the two types of tissue (tumor and non-tumor tissue). BC032020 was only slightly expressed in the PDAC tissues and cell lines, and exhibited an inverse correlation with zinc finger protein 451 (ZNF451) expression. The overexpression of BC032020 suppressed cell proliferation and migration, and induced G1 phase arrest and the apoptosis of PDAC cells by inhibiting ZNF451.
Materials and methods
Cell culture and reagents
The PDAC cell lines, AsPC-1 (CRL-1682), BxPC-3 (CRL-1687), PANC-1 (CRL-1469), CFPAC-1 (CRL-1918) and MIA PaCa-2 (CRL-1420) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Human pancreatic ductal epithelial (HPDE) cells were purchased from Otwo Biotech Inc. (Shenzhen, China). The PDAC cell lines were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The HPDE cells were cultured in M3 medium containing 10% heat-inactivated fetal bovine serum plus epithelial growth factor (EGF), penicillin and streptomycin (Invitrogen, Carlsbad, CA, USA).
Clinical samples
PDAC tumor tissues and adjacent tissues were collected from 20 patients with pancreatic cancer during surgery at Xiangya Hospital (Changsha, China). The details of the patient characteristics were shown in Table I. The collected tumor tissues were identified as PDAC by pathological analysis. The tumor tissues were immediately frozen and stored at −80°C. The protocol for this study was approved by the Institutional Ethics Committee of Xiangya Hospital, and all enrolled patients signed a written informed consent document.
Table I.
Patient information.
| Clinical parameter | No. of cases (%) |
|---|---|
| Age (years) | |
| ≤55 | 6 (30.0) |
| >55 | 14 (70.0) |
| Sex | |
| Male | 11 (55.0) |
| Female | 9 (45.0) |
| Tumor location | |
| Distal | 9 (45.0) |
| Middle | 4 (20.0) |
| Proximal | 7 (35.0) |
| Tumor size | |
| ≤5 cm | 14 (70.0) |
| >5 cm | 6 (30.0) |
| TNM stage | |
| I | 6 (30.0) |
| II | 8 (40.0) |
| III | 4 (20.0) |
| IV | 2 (10.0) |
| Lymphatic metastasis | |
| Yes | 10 (50.0) |
| No | 10 (50.0) |
| Distant metastasis | |
| Yes | 2 (10.0) |
| No | 18 (90.0) |
In addition two datasets of lncRNAs in pancreatic cancer tissues datasets (GSE28735 and GSE15471) were obtained from the CRN database (http://syslab4.nchu.edu.tw/CRN).
CCK-8 assay
A Cell Counting kit-8 (CCK-8; Dojido, Kumamoto, Japan) was used to assess cell proliferation according to the manufacturer's instructions. In brief, aliquots containing 3,000 cells in 100 µl of medium were seeded into the individual wells of a 96-well plate. The cells were then transfected with the indicated plasmids or small interfering RNA (siRNA; pcDNA3.1-Vector, pcDNA3.1-BC032020, pcDNA3.1-ZNF451, si-Control or si-ZNF451; details of the plasmids are provided below) for 24 h; after which, 10 µl of CCK-8 were added to each well. The cells were then further incubated for 1–3 h at 37°C in a water-saturated atmosphere containing 5% CO2. A microplate reader (PerkinElmer, Waltham, MA, USA) was used to detect the absorbance values at 450 nm.
Wound healing assay
Wound healing assays were performed by measuring the movement of cells in a scraped cellular area to assess their motility. The AsPC-1 or PANC-1 cells were seeded onto a 24-well plate at a density of 80–90%. A scratch was then gently and slowly created on the monolayer using a 1 ml pipette tip across the center of the well. After scratching, the wells were gently washed with medium to remove the detached cells and the well was replenishedwith fresh medium. Wound closure was observed and images were acquired using an optical microscope (Olympus-IX51; Olympus, Tokyo, Japan) to assess the levels of migration after 0 and 48 h, respectively.
Cell cycle analysis
The AsPC-1 and PANC-1 cells were transfected with the plasmids (pcDNA3.1-Vector, pcDNA3.1-BC032020 or pcDNA3.1-ZNF451+ pcDNA3.1-BC032020) or siRNAs (NC siRNA or ZNF451 siRNA) for 12 h, and the cells were then fixed with 80% cold ethanol, and incubated with 0.5% Triton X-100 solution containing 1 mg/ml RNase A at 37°C for 30 min. The cells were then stained with 50 µg/ml propidium iodide (PI; Sigma, St. Louis, MO, USA) for 30 min in the dark. Cell cycle analysis was performed using a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the PDAC tumor tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). A 1 µg sample of RNA was used as a template for single strand cDNA synthesis performed using random primers and Primescript reverse transcriptase (M-MLV, Takara, Shiga, Japan) according to the manufacturer's instructions. qPCR was performed for GAPDH, BC032020, AX748062, DHRS4-AS1, AK127787 and ZNF451. qPCR was performed as follows: pre-denaturation at 95°C for 10 sec, followed by 40 cycles of denaturation at 95°C for 10 sec and elongation at 60°C for 30 sec. The results were normalized to GAPDH expression. The 2−ΔΔCq method was used for calculating the relative expression levels. The primers used in this study are listed in Table II. The relative expression levels were detected with an Applied Biosystems step-one plus sequence detection system (Applied Biosystems, Foster City, CA, USA), and fold changes in gene expression were calculated using the 2−ΔΔCq method (36).
Table II.
Sequences of the primers used for RT-qPCR.
| Name | Sequence (5′→3′) |
|---|---|
| H-GAPDH-F | TGTTCGTCATGGGTGTGAAC |
| H-GAPDH-R | ATGGCATGGACTGTGGTCAT |
| BC032020-F | CACTGAAGCCTCAACCTCC |
| BC032020-R | ACTTGCTGGGAGTGTTGGT |
| AX748062-F | GTTTGTTGCCTACCGATGAAG |
| AX748062-R | AGAAGAGTTACGAGTTGCCTGAT |
| DHRS4-AS1-F | GATTCGGAGTGGCTTCAGG |
| DHRS4-AS1-R | GCTCGCAAATAAGGTGGC |
| AK127787-F | CTCTGGGGAATAGACTTGGAT |
| AK127787-R | CAGGAAGACTTATGATGTGGG |
Cell transfection
Different plasmids (pcDNA3.1-Vector, pcDNA3.1-BC032020 and pcDNA3.1-ZNF451) were created and used to enforce the overexpression of BC032020 and ZNF451 in the AsPC-1 and PANC-1 cells. The BC032020 full-length sequence was synthesized and subcloned into the pCDNA3.1 vector (Invitrogen, Shanghai, China). A siRNA that targets human ZNF451 was used to knockdown ZNF451 expression. The sequence of siRNA used against ZNF451 is shown in Table III. The siRNA was purchased from RiboBio (Guangzhou, China). The cells were transfected with the plasmids or siRNA for 48 h using Lipofectamine 3000 (Invitrogen, Shanghai, China) following the manufacturer's instructions.
Table III.
The sequence of the siRNA used against zinc finger protein 451 (ZNF451).
| Name | Sequence (5′→3′) |
|---|---|
| si-ZNF451 | GCAGAATTCAGGACAGCAGAATTCAGGACA |
| siRNA-Control | TGCGCTGCTGGTGCCAACCCTATTCT |
Xenograft model of pancreatic cancer
All procedures involving animals conformed to the national guidelines of and were approved by the Animal Care Ethics Committee of Xiangya Hospital of Central South University. Male BALB/c nude mice (age, 5–6 weeks; weight, 20–25 g; n=20) were purchased from the Laboratory Animal Center of Shanghai Academy of Science. The mice were randomly divided into 4 groups (AsPC-1-NC, AsPC-BC032020, PANC-1-NC and PANC-1-BC032020; n=5 per group). A human BC032020 lentiviral vector was constructed (GenePharma, Shanghai, China) and used to establish stable BC032020-overexpressing AsPC-1 and PANC-1 cells. Subsequently, 1×106 BC032020-overexpressing AsPC-1 or PANC-1 cells were injected into the right flanks of the BALB/c nude mice. Tumor volumes were measured using a caliper every 3 days, and calculated using the formula, V = (ab2)/2, where 'a' is the long axis, and 'b' is the short axis. The mice were sacrificed when the mean tumor long axis exceeded 10 mm. The maximum diameter of a single tumor found was 12 mm and no mouse developed multiple tumors. The maximum tumor volume was 672 mm3 in our study. All BALB/c nude mice were sacrificed on the same day (body weight, 30–38 g).
Detection of ZNF451 by immunofluorescence
The transfected cells were fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 for 15 min, after which they were washed with PBS, blocked with 5% BSA in PBS, and incubated with anti-ZNF451 antibody ZNF451 (SAB2102880, Sigma) overnight at 4°C. After washing with PBS, the cells were incubated with a secondary antibody conjugated with Alexa Fluorescence 568 (1:1,000, Invitrogen, Carlsbad, CA, USA) at 37°C for 1 h. DAPI (1:5,000; Sigma) was used to stain the cell nuclei. The immunostained cells were visualized with using Leica TCS-SP2 confocal microscope (Leica, Wetzlar, Germany).
Hoechst fluorescent staining
Hoechst fluorescent staining was performed to detect apoptosis at the nuclear stage. In brief, the cells were fixed in ice-cold methanol for 5 min and then incubated with 1 µg/ml Hoechst 33258 (Life Technologies, Carlsbad, CA, USA) solution for 10 min at room temperature. The cells were then washed and examined using a Leica TCS-SP2 confocal microscope (Leica).
Immunohistochemistry
The PDAC tumor tissues and adjacent non-tumor tissues were cut at a thickness of 4 µm, after which the sections were fixed in 10% formalin, dehydrated and embedded in paraffin. The slides with the tissue sections were baked, deparaffinized in xylene, passed through a graded alcohol series, and then subjeted to antigen retrieval with 1 mM EDTA, pH 8.0 (Invitrogen, Carlsbad, CA, USA). Staining was performed with a primary antibody against ZNF451 (SAB2102880, Sigma) used at a 1:500 dilution. Following immunostaining, the slides were washed and treated with Signal Stain Boost IHC detection reagent (HRP, rabbit; Cell Signaling Technology, Danvers, MA, USA). Areas of tissue staining were developed by incubation with DAB (ab64261) and DAB Enhancer (ab675) (both from Abcam, Cambridge, MA, USA).
Western blot analysis
The cells were lysed in ice-cold radioimmunoprecipitation assay buffer (150 mm NaCl, 10 mm Tris, pH 8.0, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS and 5 mm EDTA). The protein extracts from the tumor tissues were prepared by homogenizing the tissues in ice-cold DPBS containing protease and phosphatase inhibitors. Protein concentrations of the collected lysates were measured using a BCA Protein Assay kit (Pierce, Rockford, IL, USA). A 50 µg aliquot of protein from each sample was separated by electrophoresis on a 12% SDS-PAGE gel. The separated protein bands were transferred onto PVDF membranes (Millipore, Billerica, MA, USA) which were then incubated overnight at 4°C with the following primary antibodies specific for ZNF451 (Cat. no. SAB2102880; Sigma), p21 (Cat. no. 2946; Cell Signaling Technology), p15 (Cat. no. sc-612, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and GAPDH (Cat. no. 5174, Cell Signaling Technology). The stained protein bands were then detected using an Alpha Innotech Flour Chem-FC2 imaging system (Alpha Innotech, San Leandro, CA, USA).
Sequencing analysis
Sequencing data was illustrated using IGV2.3 software. The following website was used to predict the targets of lncRNA: http://www.herbbol.org:8001/lrt/.
Statistical analysis
All results are presented as the means ± SD of values obtained in least 3 independent experiments. The Student's t-test, Mann-Whitney U test, and log-rank test were used to assess differences between 2 groups. For multiple group comparisons ANOVA followed by Tukey's post hoc test was performed. Individual statistics of paired or unpaired samples was performed by paired or unpaired t-test and if not normally distributed by Mann-Whitney test. Karl Pearson's co-efficient of correlation was used to explore the correlation between the expression of BC032020 and ZNF451. A P-value <0.05 was considered to indicate a statistically significant difference.
Results
Decreased expression of BC032020 in pancreatic cancer tissues
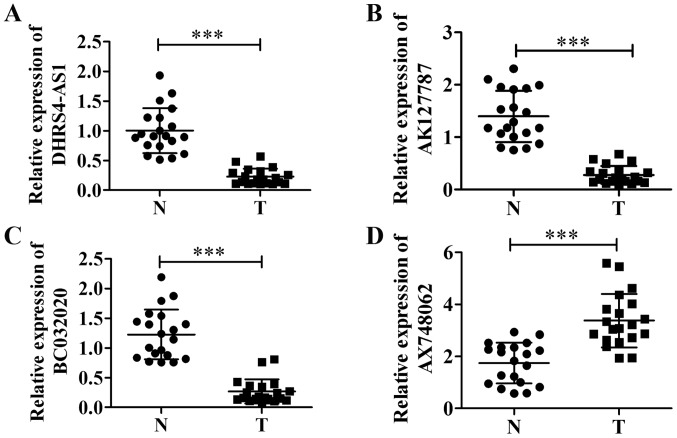
We analyzed two datasets from the CRN database (http://syslab4.nchu.edu.tw/CRN) that listed lncRNAs which exhibited an abnormal expression in patients with pancreatic cancer, and found that 13 lncRNAs were present in both datasets (Table IV). After excluding lncRNAs which had different transcripts in both datasets, we identified 4 lncRNAs that had the same transcript in PDAC tumor tissues and adjacent tissues (Fig. 1). The data indicated that the levels of lncRNAs DHRS4-AS1, AK127787 and BC032020 (Fig. 1A–C) were decreased in the PDAC tumor tissues compared with the adjacent non-tumor tissues, whereas AX748062 (Fig. 1D) was highly expressed in the tumor tissues. As little is known about BC032020 in PDAC and other tumors, we selected BC032020 in order to explore its roles in the development and progression of PDAC.
Table IV.
Differentially expressed lncRNAs as shown in the two pancreatic cancer datasets from the CRN database.
| Isoform ID (CRN) | Gene name | Isoform ID (UCSC) |
|---|---|---|
| uc010vwj.1 | C17orf76-AS1 | uc010vwj.2 |
| uc003pdq.1 | BC032020 | uc003pdq.1 |
| uc001kfo.1 | AX748062 | uc001kfo.1 |
| uc001kbj.3 | TMEM254-AS1 | uc001kbj.4 |
| uc001wkz.2 | DHRS4-AS1 | uc001wkz.3 |
| uc004bgo.2 | FAM225A | uc004bgo.3 |
| uc002eqh.2 | AF086125 | uc002eqh.3 |
| uc002mdn.2 | BC033124 | uc002mdn.3 |
| uc003dpv.2 | LOC401074 | uc003dpv.3 |
| uc002lgd.1 | AK127787 | uc002lgd.1 |
| uc003ihi.1 | SLC7A11-AS1 | uc003ihi.2 |
| uc010lea.1 | MAGI2-AS3 | uc010lea.2 |
| uc001deb.1 | GNG12-AS1 | uc001deb.2 |
CRN, Cancer RNA-Seq Nexus; UCSC, University of California Santa Cruz.
Figure 1.
Expression of long non-coding RNAs (lncRNAs) in pancreatic ductal adenocarcinoma (PDAC) tumor tissues and adjacent non-tumor tissues. The expression levels of (A) DHRS4-AS1, (B) AK127787, (C) BC032020, and (D) AX748062 in samples of pancreatic cancer tissue were analyzed by RT-qPCR. ***P<0.001. N, adjacent non-tumor (normal) tissues; T, tumor tissues.
Low expression of BC032020 and high expression of ZNF451 in PDAC
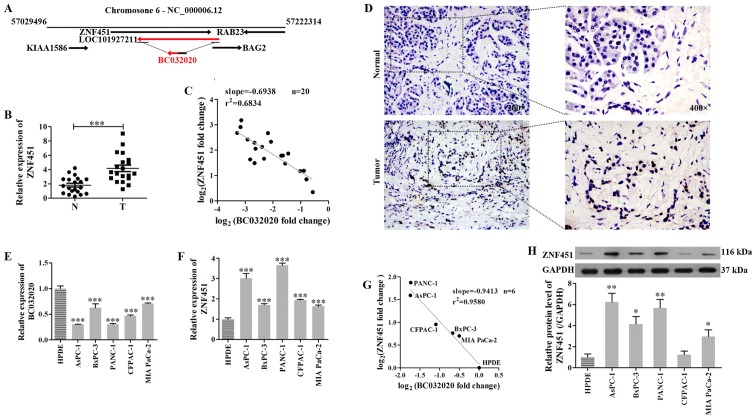
LncRNAs can mediate epigenetic alterations by recruiting chromatin remodeling complexes to specific genomic loci (37). In addition, they can regulate RNA polymerase II activity by interacting with the initiation complex to influence promoter choice (38). Sequence analysis found that ZNF451 shares a gene sequence (456 bp) with two exons of BC032020 and a 3′-non-coding region (234 bp) with another two exons of BC032020 (Fig. 2A), which suggests that an intron sequence or 3′-non-coding region of ZNF451 is very closely related to the BC032020 sequence, and ZNF451 may thus be a potential gene regulated by BC032020. Subsequently, we found that ZNF451 expression was upregulated in the PDAC tumor tissues when compared with the adjacent non-tumor tissues (Fig. 2B and D). Furthermore, there was an inverse correlation between the levels of BC032020 expression and ZNF451 expression in the specimens of pancreatic cancer tissue (Fig. 2C). We then assessed the levels of BC032020 and ZNF451 expression in the PDAC cells and HPDE cells, and found that the expression levels of BC032020 in the PDAC cells (AsPC-1, BxPC-3, PANC-1, CFPAC-1 and MIA PaCa-2) were lower than those in the HPDE cells (Fig. 2E). The mRNA and protein levels of ZNF451 in the PDAC cells were both significantly increased when compared with the levels in the HPDE cells (Fig. 2F and H). Similarly, ZNF451 expression inversely correlated with BC032020 expression in these cell lines (Fig. 2G).
Figure 2.
Zinc finger protein 451 (ZNF451) expression in pancreatic ductal adenocarcinoma (PDAC) tumor tissues and cell lines inversely correlates with BC032020 expression. (A) The genomic context and locations of BC032020 and ZNF451 are shown. (B) ZNF451 mRNA levels in PDAC tumor tissues and adjacent tissues. (C) Correlation between BC032020 and ZNF451 expression in PDAC tumor tissues. (D) ZNF451 expression in PDAC tumor tissues and adjacent non-tumor tissues was detected by immunohistochemistry (×200 magnification). (E and F) Expression levels of BC032020 and ZNF451 in PDAC cells (AsPC-1, BxPC-3, PANC-1, CFPAC-1 and MIA PaCa-2) and human pancreatic ductal epithelial (HPDE) cells were evaluated by RT-qPCR. (G) Correlation between BC032020 and ZNF451 expression in the PDAC cell lines. (H) Protein levels of ZNF451 in PDAC cells and HPDE cells were evaluated by western blot analysis. The relative expression levels were normalized to those of GAPDH. Data represent the means ± SD of values obtained from triplicate samples. *P<0.05, **P<0.01, ***P<0.001 vs. HPDE cells.
Overexpression of BC032020 suppresses ZNF451 expression in PDAC cells
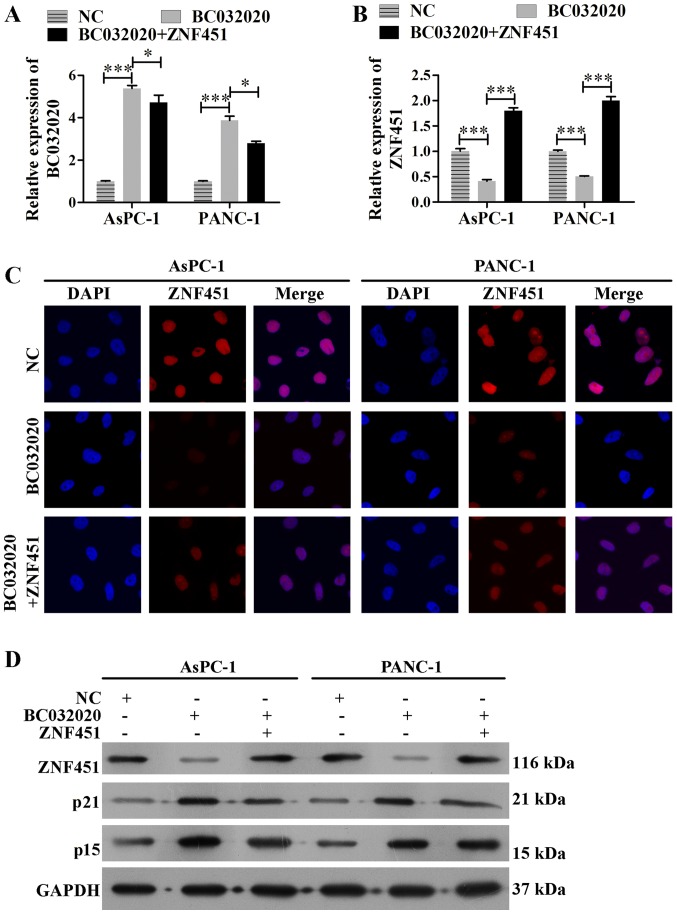
In order to investigate the assocation between BC032020 and ZNF451, we enforced the overexpression of BC032020 in the AsPC-1 and PANC-1 cells. The transfection efficiency is shown in Fig. 3A. We found that the mRNA levels of ZNF451 in the AsPC-1 and PANC-1 cells were significantly decreased following BC032020 overexpression, but were restored by ZNF451 overexpression (Fig. 3B). Immunofluorescence assay and western blot analysis also revealed that ZNF451 expression in the AsPC-1 and PANC-1 cells was decreased following BC032020 overexpression (Fig. 3C and D). As ZNF451 knockdown has been reported to enhance transforming growth factor (TGF)-β-induced p15 and p21 expression and lead to growth arrest in HaCaT cells (39), we also analyzed the levels of p15 and p21 expression. As shown in Fig. 3D, the protein levels of p15 and p21 were enhanced following BC032020 overexpression.
Figure 3.
BC032020 overexpression decreases zinc finger protein 451 (ZNF451) expression in pancreatic cancer cells. (A) AsPC-1 and PANC-1 cells were transfected with different plasmids (pcDNA3.1-Vector, pcDNA3.1-BC032020 and pcDNA3.1-ZNF451) to enforce the overexpression of BC032020 or ZNF451. The levels of BC032020 in the pancreatic cancer cells were evaluated by RT-qPCR. Data represent the means ± SD of values obtained from triplicate samples, *P<0.05, ***P<0.001. (B) mRNA expression levels of ZNF451 in pancreatic cancer cells were evaluated by RT-qPCR. (C) Immunofluorescence analysis of ZNF451 expression in pancreatic cancer cells (scale bar, 10 µm). (D) Protein levels of ZNF451n in human pancreatic epithelial cells were examined by western blot analysis.
BC032020 suppresses the survival of PDAC cells by targeting ZNF451
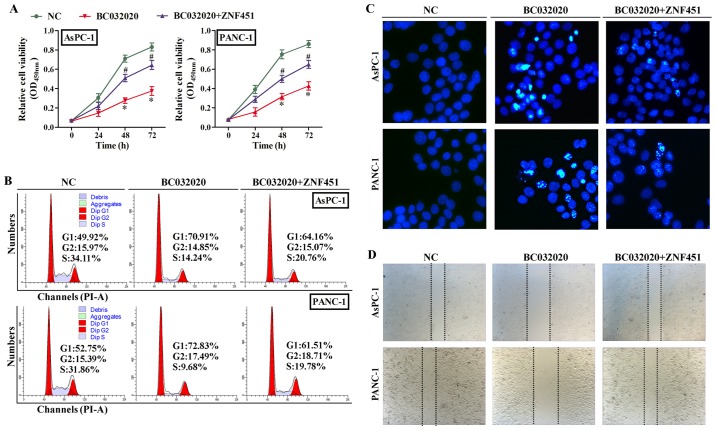
In order to explore the role of BC032020 in PDAC cells, we examined the proliferation, cell cycle distribution, migration and apoptotic rate of the AsPC-1 and PANC-1 cells in which the overexpression of BC032020 was enforced by transfection with the expression plasmid. As shown in Fig. 4A, the overexpression of BC032020 inhibited the proliferation of the AsPC-1 and PANC-1 cells, whereas the overexpression of ZNF451 restored their proliferation. Moreover, the overexpression of BC032020 markedly decreased the number of AsPC-1 cells in the S and G2/M phases, and increased the percentage of cells undergoing G0/G1 cell cycle arrest from 49.92 to 70.91% (Fig. 4B). Similar results were observed in the PANC-1 cells, where once again, ZNF451 overexpression caused a decrease in the percentage of arrested G1 phase cells. As the overexpression of BC032020 inhibited cell proliferation and induced G1 phase arrest, we also examined whether cell apoptosis was affected by BC032020. Our results revealed that the rate of apoptosis of both the AsPC-1 and PANC-1 cells was increased by the overexpression of BC032020, whereas the overexpression of ZNF451 decreased the rate of apoptosis of both cell lines (Fig. 4C). Finally, the results from wound healing assay revealed that cell migration was suppressed by BC032020 overexpression (Fig. 4D).
Figure 4.
BC032020 suppresses the proliferation and migration, and induces G1 phase arrest and the apoptosis of pancreatic cancer cells. The AsPC-1 and PANC-1 cells were transfected with different plasmids (NC, pcDNA3.1-Vector; BC032020, pcDNA3.1-BC032020; BC032020 + zinc finger protein 451 (ZNF451), pcDNA3.1-BC032020 and pcDNA3.1-ZNF451) to enforce the overexpression of BC032020 or ZNF451. (A) AsPC-1 and PANC-1 cell proliferation was assessed by CCK-8 assay. NC vs. BC032020 (*P<0.05); BC032020 vs. BC032020 + ZNF451 (#P<0.05). (B) The cell cycle distributions of AsPC-1 and PANC-1 cells were detected by flow cytometry. (C) Hoechst staining for AsPC-1 and PANC-1 was performed after 48 h of plasmid transfection (scale bar, 10 µm) (D) Wound healing assay was used to detect cell migration. Data represent the means ± SD of values obtained from triplicate samples.
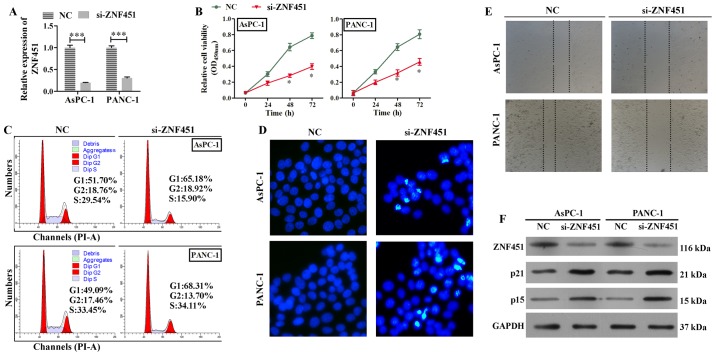
Knockdown of ZNF451 suppresses the survival of PDAC cells
We found that the overexpression of ZNF451 partially reversed the effects of BC032020 on the survival of AsPC-1 and PANC-1 cells. However, the roles played by ZNF451 in the proliferation, cell cycle, migration and apoptosis of the AsPC-1 and PANC-1 cells required further investigation. Therefore, we silenced ZNF451 expression in the AsPC-1 and PANC-1 cells (Fig. 5A), and found that the knockdown of ZNF451 significantly inhibited the proliferation of both cell lines (Fig. 5B). We also found that ZNF451 knockdown induced G1 phase arrest (Fig. 5C). In addition, greater numbers of apoptotic cells were induced by ZNF451 knockdown when compared with the negative control (NC siRNA) group (Fig. 5D). Moreover, ZNF451 knockdown inhibited the migratory ability of both the AsPC-1 and PANC-1 cells (Fig. 5E). The results from western blot analysis also revealed that the protein levels of p21 and p15 were significantly increased by the silencing of ZNF451 (Fig. 5F). Taken together, our results demonstrated that the silencing of ZNF451 in the AsPC-1 and PANC-1 cells, suppressed cell proliferation and migration, but induced G1 phase arrest and apoptosis.
Figure 5.
Zinc finger protein 451 (ZNF451) knockdown suppresses cell proliferation/migration, and induces G1 phase arrest and the apoptosis of pancreatic cancer cells. The AsPC-1 and PANC-1 cells were transfected with small interfering RNA (siRNA; NC, siRNA without any target; si-ZNF451, siRNA targeting ZNF451). (A) The efficiency of ZNF451 knockdown was confirmed by RT-qPCR. NC vs. si-ZNF451, ***P<0.001. (B) The proliferation of transfected AsPC-1 and PANC-1 cells was assessed by CCK-8 assay. NC vs. si-ZNF451, *P<0.05. (C) The cell cycle distributions of transfected AsPC-1 and PANC-1 cells were detected by flow cytometry. (D) Hoechst staining foci in AsPC-1 and PANC-1 cells were detected after 48 h of siRNA transfection (scale bar,10 µm). (E) Wound healing assay was used to detect cell migration. (F) Western blot analyses were performed to detect the protein levels of ZNF451, p15, and p21 in the AsPC-1 and PANC-1 cells. Data represent the means ± SD of results obtained from triplicate samples.
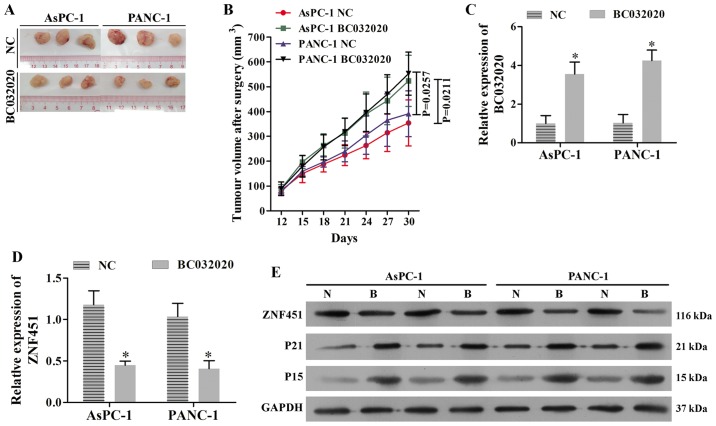
BC032020 suppresses tumor growth in a xenograft model of PDAC
To explore the role of BC032020 in vivo, we established two subcutaneous xenograft models in nude mice using the AsPC-1 and PANC-1 cells in which the stable overexpression of BC032020 was induced. Our experimental results revealed that tumor growth was significantly suppressed by BC032020 overexpression in both the the AsPC-1 and PANC-1 tumor-derived models when compared with the tumor growth in the control mice (Fig. 6A and B). The increased levels of BC032020 expression in the tumor tissues were confirmed by RT-qPCR (Fig. 6C). We found that the ZNF451 levels were significantly decreased in the tumor tissues of mice derived from BC032020-overexpressing cells (Fig. 6D). Moreover, the protein levels of p21 and p15 increased in conjunction with the decreased levels of ZNF451 in the BC032020-overexpressing tumor tissues (Fig. 6E).
Figure 6.
BC032020 suppresses tumor growth in a xenograft model of pancreatic cancer by inhibiting zinc finger protein 451 (ZNF451) expression. A human BC032020 lentiviral vector was constructed and used to transfect the AsPC-1 and PANC-1 cells to induce the stable overexpression of BC032020. The BC032020-overexpressing AsPC-1 and PANC-1 cells were used to establish a subcutaneous xenograft model. (A) Representative images of tumors in the subcutaneous xenograft model. NC, transfected with empty lentiviral vector; BC032020, transfected with the BC032020 overexpressing lentiviral vector. (B) Tumor growth curves were calculated (n=5 mice per group). (C) BC032020 levels in tumor tissues were evaluated by RT-qPCR. (D) mRNA levels of ZNF451 in tumor tissues were evaluated by RT-qPCR. Data represent the means ± SD of data obtained from triplicate samples. *P<0.05, vs. NC. (E) Western blot analyses were performed to assess the protein levels of ZNF451, p15 and p21 in tumor tissues. N, NC cells; B, overexpressing BC032020 cells.
Discussion
ZNF451, a poorly characterized vertebrate zinc-finger protein, acts as a transcriptional co-regulator, partially localizing to promyelocytic leukemia bodies (40–42). It is sumoylated itself, and this sumoylation is required for its promyelocytic leukemia body localization. As products of the promyelocytic leukemia gene, promyelocytic leukemia bodies are stress-responsive nuclear structures which are involved in cell differentiation, cell senescence, cell apoptosis, immune responses, and DNA repair (43,44). ZNF451 is a SUMO substrate containing two predicted N-terminal SIMs followed by twelve C2H2 zinc finger domains and an ubiquitin-interacting motif, and interacts with proteins of the SUMOylation machinery (41,42). A number of non-coding RNAs can use RNA-RNA interactions to target specific pre-mRNAs, and achieve their diverse functions (45). lncRNAs can regulate the level of a target gene by mediating histone modification, chromatin remodeling, or acting as an miRNA or small interfering RNA precursor (20). A bioinformatics analysis identified ZNF451 as a gene potentially regulated by BC032020, which share a gene sequences (456 bp) with two exons of BC032020 and a 3′-non-coding region (234 bp) with another two exons of BC032020. The present study also confirmed that ZNF451 has an inverse correlation with BC032020 expression in PDAC cell lines and pancreatic cancer samples. However, the specific mechanisms underlying the regulation of ZNF451 expression by BC032020 warrant further investigation.
To investigate the association between BC032020 and ZNF451, we enforced the overexpression of BC032020 in the AsPC-1 and PANC-1 cells. We found that BC032020 over-expression decreased the levels of ZNF451 expression in the pancreatic cancer cells. ZNF451 knockdown has been reported to enhance TGF-induced p15 and p21 expression, and lead to growth arrest in HaCaT cells (39). Our analysis also revelaed that BC032020 overexpression enhanced the levels of p15 and p21. ZNF451 has been described as a Smad3/4-binding protein, and has been reported to inhibit TGF-β-induced cell growth inhibition. The overexpression of ZNF451 has been shown to attenuate TGF-β-induced growth inhibition and gene transcription responses (39). In HaCaT cells, p15 and p21 mRNA and protein levels induced by TGF-β were enhanced by ZNF451 knockdown (39). In this study, BC032020 overexpression enhanced the protein levels of p15 and p21 in pancreatic cancer cells. Furthermore, ZNF451 knockdown also enhanced p15 and p21 expression, while the overexpression of ZNF451 decreased these expression levels.
As a tumor suppressor gene involved in cell cycle regulation, p15 plays an important role in negatively regulating cell proliferation by inhibiting cyclin-dependent kinase 4/6 activity (46–48). A decreased p15 protein expression can lead to retinoblastoma phosphorylation, the subsequent progression of G1/S phase transition, and an acceleration of uncontrolled cell proliferation (49–51). p21 is a potent universal CDK inhibitor that regulates the cell cycle, cell proliferation, senescence and apoptosis (52–56). The lost expression of p21 and p15 mutations is involved in the occurrence of pancreatic cancer (57–60). The p21 protein can prevent Rb phosphorylation by suppressing cyclin D1-CDK4 and cyclin E-CDK2 activity, which leads to cell cycle arrest. The activated tumor suppressor p53 can induce either G1 phase cell cycle arrest or a chronic state of senescence or apoptosis by enhancing the transient expression of p21 (61–64).
In this study, BC032020 overexpression decreased the levels of ZNF451 expression in pancreatic cancer cells, and the knockdown of ZNF451 enhanced the levels of p21 and p15 expression. In view of the roles played by p21 and p15 in cell cycle regulation, cell proliferation, senescence and apoptosis, our data suggest that BC032020 suppresses the proliferation and migration, and induces G1 phase arrest and the apoptosis of pancreatic cancer cells. The results of our in vivo experiments indicated that BC032020 suppressed tumor growth in a xenograft model of pancreatic cancer by inhibiting ZNF451 expression. In conclusion, the findings of the present study BC032020 indicate that suppresses the survival of human PADC cells by targeting ZNF451.
Acknowledgments
Not applicable.
Footnotes
Funding
This study was supported by grants from the Key Projects of Hunan Province Science and Technology Plan (no. 2016JC2040).
Availability of data and materials
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
ZZ and HC designed and performed cell culture experiment and interpreted the data. YL and TF performed the clinical and animal experiments and interpreted the data. WS conceived the study and wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
For the use of human samples, the protocol for this study was approved by the Institutional Ethics Committee of Xiangya Hospital of Central South University (Changsha, China) and all enrolled patients signed a written informed consent document. In addition, all procedures involving animals conformed to the national guidelines of and were approved by the Animal Care Ethics Committee of Xiangya Hospital of Central South University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein AP. Identifying people at a high risk of developing pancreatic cancer. Nat Rev Cancer. 2013;13:66–74. doi: 10.1038/nrc3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillen S, Schuster T, Meyer Zum Buschenfelde C, Friess H, Kleeff J. Preoperative/neoadjuvant therapy in pancreatic cancer: A systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010;7:e1000267. doi: 10.1371/journal.pmed.1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bapat AA, Hostetter G, Von Hoff DD, Han H. Perineural invasion and associated pain in pancreatic cancer. Nat Rev Cancer. 2011;11:695–707. doi: 10.1038/nrc3131. [DOI] [PubMed] [Google Scholar]
- 5.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 6.Hruban RH, Canto MI, Goggins M, Schulick R, Klein AP. Update on familial pancreatic cancer. Adv Surg. 2010;44:293–311. doi: 10.1016/j.yasu.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iodice S, Gandini S, Maisonneuve P, Lowenfels AB. Tobacco and the risk of pancreatic cancer: A review and meta-analysis. Langenbecks Arch Surg. 2008;393:535–545. doi: 10.1007/s00423-007-0266-2. [DOI] [PubMed] [Google Scholar]
- 8.Raimondi S, Lowenfels AB, Morselli-Labate AM, Maisonneuve P, Pezzilli R. Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection. Best Pract Res Clin Gastroenterol. 2010;24:349–358. doi: 10.1016/j.bpg.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Bosetti C, Rosato V, Li D, Silverman D, Petersen GM, Bracci PM, Neale RE, Muscat J, Anderson K, Gallinger S, et al. Diabetes, antidiabetic medications, and pancreatic cancer risk: An analysis from the International Pancreatic Cancer Case-Control Consortium. Ann Oncol. 2014;25:2065–2072. doi: 10.1093/annonc/mdu276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood LD, Hruban RH. Pathology and molecular genetics of pancreatic neoplasms. Cancer J. 2012;18:492–501. doi: 10.1097/PPO.0b013e31827459b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 13.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker AE, Hernandez YG, Frucht H, Lucas AL. Pancreatic ductal adenocarcinoma: Risk factors, screening, and early detection. World J Gastroenterol. 2014;20:11182–11198. doi: 10.3748/wjg.v20.i32.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falzone L, Marconi A, Loreto C, Franco S, Spandidos DA, Libra M. Occupational exposure to carcinogens: Benzene, pesticides and fibers. Mol Med Rep. 2016;14:4467–4474. doi: 10.3892/mmr.2016.5791. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decker GA, Batheja MJ, Collins JM, et al. Risk factors for pancreatic adenocarcinoma and prospects for screening. Gastroenterol Hepatol (N Y) 2010;6:246–254. [PMC free article] [PubMed] [Google Scholar]
- 17.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Chen Z, Xu H, He A, Liu Y, Huang W. Long noncoding RNA CCAT2 as a novel biomaker of metastasis and prognosis in human cancer: A meta-analysis. Oncotarget. 2017;8:75664–75674. doi: 10.18632/oncotarget.18161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aftab MN, Dinger ME, Perera RJ. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch Biochem Biophys. 2014;563:60–70. doi: 10.1016/j.abb.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng H, Wang JM, Li M, Tang R, Tang K, Su Y, Hou Y, Zhang J. Long non-coding RNAs: New biomarkers for prognosis and diagnosis of colon cancer. Tumour Biol. 2017;39 doi: 10.1177/1010428317706332. 1010428317706332. [DOI] [PubMed] [Google Scholar]
- 21.Ding L, Ren J, Zhang D, Li Y, Huang X, Ji J, Hu Q, Wang H, Ni Y, Hou Y. The TLR3 agonist inhibit drug efflux and sequentially consolidates low-dose cisplatin-based chemoimmunotherapy while reducing side effects. Mol Cancer Ther. 2017;16:1068–1079. doi: 10.1158/1535-7163.MCT-16-0454. [DOI] [PubMed] [Google Scholar]
- 22.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Deng K, Guo X, Wang H, Xia J. The lncRNA-MYC regulatory network in cancer. Tumour Biol. 2014;35:9497–9503. doi: 10.1007/s13277-014-2511-y. [DOI] [PubMed] [Google Scholar]
- 24.Ye N, Wang B, Quan ZF, et al. Functional roles of long non-coding RNA in human breast cancer. Asian Pac J Cancer Prev. 2014;15:5993–5997. doi: 10.7314/APJCP.2014.15.15.5993. [DOI] [PubMed] [Google Scholar]
- 25.Min F, Xinying LI, Wei J, Yun H, Jingdong LI, Wang Z. A long non-coding RNA, TCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. 2013;5:1143–1146. doi: 10.3892/etm.2013.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwok ZH, Tay Y. Long noncoding RNAs: Lincs between human health and disease. Biochem Soc Trans. 2017;45:805–812. doi: 10.1042/BST20160376. [DOI] [PubMed] [Google Scholar]
- 27.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J, et al. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 29.Wei W, Liu Y, Lu Y, Yang B, Tang L. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J Cell Biochem. 2017;118:3349–3358. doi: 10.1002/jcb.25988. [DOI] [PubMed] [Google Scholar]
- 30.Gu L, Zhang J, Shi M, Zhan Q, Shen B, Peng C. lncRNA MEG3 had anti-cancer effects to suppress pancreatic cancer activity. Biomed Pharmacother. 2017;89:1269–1276. doi: 10.1016/j.biopha.2017.02.041. [DOI] [PubMed] [Google Scholar]
- 31.Wu BQ, Jiang Y, Zhu F, Sun DL, He XZ. Long noncoding RNA VT1 promotes EMT and cell proliferation and migration through downregulating p21 in pancreatic cancer cells. Technol Cancer Res Treat. 2017;16:819–827. doi: 10.1177/1533034617700559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S, Zhang JQ, Chen JZ, Chen H-X, Qiu F-N, Yan M-L, Chen Y-L, Peng C-H, Tian Y-F, Wang Y-D. The over expression of long non-coding RNA ANRIL promotes epithelial-mesenchymal transition by activating the ATM-E2F1 signaling pathway in pancreatic cancer: An in vivo and in vitro study. Int J Biol Macromol. 2017;102:718–728. doi: 10.1016/j.ijbiomac.2017.03.123. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Guan Z, He K, Qian J, Cao J, Teng L. LncRNA UCA1 in anti-cancer drug resistance. Oncotarget. 2017;8:64638–64650. doi: 10.18632/oncotarget.18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu B. BioSeq-Analysis: A platform for DNA, RNA and protein sequence analysis based on machine learning approaches. Brief Bioinform. 2017 Dec 19; doi: 10.1093/bib/bbx165. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 35.Lin Y, Qian F, Shen L, Chen F, Chen J, Shen B. Computer-aided biomarker discovery for precision medicine: Data resources, models and applications. Brief Bioinform. 2017 Nov 29; doi: 10.1093/bib/bbx158. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) methods. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015;12:1094–1098. doi: 10.1080/15476286.2015.1063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 39.Feng Y, Wu H, Xu Y, Zhang Z, Liu T, Lin X, Feng X-H. Zinc finger protein 451 is a novel Smad corepressor in transforming growth factor-beta signaling. J Biol Chem. 2014;289:2072–2083. doi: 10.1074/jbc.M113.526905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karvonen U, Jaaskelainen T, Rytinki M, Kaikkonen S, Palvimo JJ. ZNF451 is a novel PML body- and SUMO-associated transcriptional coregulator. J Mol Biol. 2008;382:585–600. doi: 10.1016/j.jmb.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 41.Abascal F, Tress ML, Valencia A. Alternative splicing and co-option of transposable elements: The case of TMPO/LAP2alpha and ZNF451 in mammals. Bioinformatics. 2015;31:2257–2261. doi: 10.1093/bioinformatics/btv132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisenhardt N, Chaugule VK, Koidl S, Droescher M, Dogan E, Rettich J, Sutinen P, Imanishi SY, Hofmann K, Palvimo JJ, et al. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat Struct Mol Biol. 2015;22:959–967. doi: 10.1038/nsmb.3114. [DOI] [PubMed] [Google Scholar]
- 43.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 44.Droescher M, Chaugule VK, Pichler A. SUMO rules: Regulatory concepts and their implication in neurologic functions. Neuromolecular Med. 2013;15:639–660. doi: 10.1007/s12017-013-8258-6. [DOI] [PubMed] [Google Scholar]
- 45.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Cheng J, Xu C, Liu S, Jiang S, Xu Q, Chen X, Zhuang H, Lu F. Quantitative methylation analysis reveals gender and age differences in p16INK4a hypermethylation in hepatitis B virus-related hepatocellular carcinoma. Liver Int. 2012;32:420–428. doi: 10.1111/j.1478-3231.2011.02696.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhang JC, Gao B, Yu ZT, Liu X-B, Lu J, Xie F, Luo H-J, Li H-P. Promoter hypermethylation of p14 (ARF), RB, and INK4 gene family in hepatocellular carcinoma with hepatitis B virus infection. Tumour Biol. 2014;35:2795–2802. doi: 10.1007/s13277-013-1372-0. [DOI] [PubMed] [Google Scholar]
- 48.Fukai K, Yokosuka O, Imazeki F, Tada M, Mikata R, Miyazaki M, Ochiai T, Saisho H. Methylation status of p14ARF, p15INK4b, and p16INK4a genes in human hepatocellular carcinoma. Liver Int. 2005;25:1209–1216. doi: 10.1111/j.1478-3231.2005.01162.x. [DOI] [PubMed] [Google Scholar]
- 49.Shcherba M, Liang Y, Fernandes D, Perez-Soler R, Cheng H. Cell cycle inhibitors for the treatment of NSCLC. Expert Opin Pharmacother. 2014;15:991–1004. doi: 10.1517/14656566.2014.902935. [DOI] [PubMed] [Google Scholar]
- 50.Gong H, Wen H, Zhu X, Lian Y, Yang X, Qian Z, Zhu J. High expression of long non-coding RNA ZEB1-AS1 promotes colorectal cancer cell proliferation partially by suppressing p15 expression. Tumour Biol. 2017;39 doi: 10.1177/1010428317705336. 1010428317705336. [DOI] [PubMed] [Google Scholar]
- 51.Tian XP, Jin XH, Li M, Huang WJ, Xie D, Zhang JX. The depletion of PinX1 involved in the tumorigenesis of non-small cell lung cancer promotes cell proliferation via p15/cyclin D1 pathway. Mol Cancer. 2017;16:74. doi: 10.1186/s12943-017-0637-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 53.Ning P, Hu C, Li X, Zhou Y, Hu A, Zhang Y, Gao L, Gong C, Guo K, Zhang X, et al. Classical swine fever virus Shimen infection increases p53 signaling to promote cell cycle arrest in porcine alveolar macrophages. Oncotarget. 2017;8:55938–55949. doi: 10.18632/oncotarget.18997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang C, Liu G, Luckhardt T, Antony V, Zhou Y, Carter AB, Thannickal VJ, Liu R-M. Serpine 1 induces alveolar type II cell senescence through activating p53-p21-Rb pathway in fibrotic lung disease. Aging Cell. 2017;16:1114–1124. doi: 10.1111/acel.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato F, Kohsaka A, Takahashi K, Otao S, Kitada Y, Iwasaki Y, Muragaki Y. Smad3 and Bmal1 regulate p21 and S100A4 expression in myocardial stromal fibroblasts via TNF-alpha. Histochem Cell Biol. 2017;148:617–624. doi: 10.1007/s00418-017-1597-x. [DOI] [PubMed] [Google Scholar]
- 56.Ren J, Ding L, Xu Q, Shi G, Li X, Li X, Ji J, Zhang D, Wang Y, Wang T, et al. LF-MF inhibits iron metabolism and suppresses lung cancer through activation of P53-miR-34a-E2F1/E2F3 pathway. Sci Rep. 2017;7:749. doi: 10.1038/s41598-017-00913-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kis A, Tatar TZ, Gall T, et al. Frequency of genetic and epigenetic alterations of p14ARF and p16INK4A in head and neck cancer in a Hungarian population. Pathology oncology research. POR. 2014;20:923–929. doi: 10.1007/s12253-014-9775-9. [DOI] [PubMed] [Google Scholar]
- 58.Imamura T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Ohashi T, Kiuchi J, Nishibeppu K, Konishi H, Shiozaki A, et al. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci Rep. 2017;7:5708. doi: 10.1038/s41598-017-06137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villarino N, Signaevskaia L, van Niekerk J, Medal R, Kim H, Lahmy R, Scully K, Pinkerton A, Kim SW, Lowy A, et al. A screen for inducers of bHLH activity identifies pitavastatin as a regulator of p21, Rb phosphorylation and E2F target gene expression in pancreatic cancer. Oncotarget. 2017;8:53154–53167. doi: 10.18632/oncotarget.18587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohammad RM, Li Y, Muqbil I, Aboukameel A, Senapedis W, Baloglu E, Landesman Y, Philip PA, Azmi AS. Targeting Rho GTPase effector p21 activated kinase 4 (PAK4) suppresses p-Bad-microRNA drug resistance axis leading to inhibition of pancreatic ductal adenocarcinoma proliferation. Small GTPases. 2017 Jun 22; doi: 10.1080/21541248.2017.1329694. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zubair H, Bhardwaj A, Ahmad A, Srivastava SK, Khan MA, Patel GK, Singh S, Singh AP. Hydroxytyrosol induces apoptosis and cell cycle arrest and suppresses multiple oncogenic signaling pathways in prostate cancer cells. Nutr Cancer. 2017;69:932–942. doi: 10.1080/01635581.2017.1339818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He L, Chen Y, Feng J, Sun W, Li S, Ou M, Tang L. Cellular senescence regulated by SWI/SNF complex subunits through p53/p21 and p16/pRB pathway. Int J Biochem Cell Biol. 2017;90:29–37. doi: 10.1016/j.biocel.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 63.Ren J, Nie Y, Lv M, Shen S, Tang R, Xu Y, Hou Y, Zhao S, Wang T. Estrogen upregulates MICA/B expression in human non-small cell lung cancer through the regulation of ADAM17. Cell Mol Immunol. 2015;12:768–776. doi: 10.1038/cmi.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]