Abstract
SUMOylation is a reversible post-translational modification which has emerged as a crucial molecular regulatory mechanism, involved in the regulation of DNA damage repair, immune responses, carcinogenesis, cell cycle progression and apoptosis. Four SUMO isoforms have been identified, which are SUMO1, SUMO2/3 and SUMO4. The small ubiquitin-like modifier (SUMO) pathway is conserved in all eukaryotes and plays pivotal roles in the regulation of gene expression, cellular signaling and the maintenance of genomic integrity. The SUMO catalytic cycle includes maturation, activation, conjugation, ligation and de-modification. The dysregulation of the SUMO system is associated with a number of diseases, particularly cancer. SUMOylation is widely involved in carcinogenesis, DNA damage response, cancer cell proliferation, metastasis and apoptosis. SUMO can be used as a potential therapeutic target for cancer. In this review, we briefly outline the basic concepts of the SUMO system and summarize the involvement of SUMO proteins in cancer cells in order to better understand the role of SUMO in human disease.
Keywords: SUMO pathway, cancer, post-translational modification, SUMO protease, Ubc9
1. Introduction
Post-translational protein modification (also known as PTM) is one of the important regulatory mechanisms of cellular proteins with a number of biological functions. Any protein in the proteome can be modified following translation or during translation. Different types of modifications alter the charge state, hydrophobicity, conformation and stability of the protein, and ultimately affect its function. Protein modification is reversible and has different functions in different organelles (1). Specific protein modification controls almost all physiological processes, including immune function (2), and the position, duration and intensity of exact physiological process to ensure the rapid and dynamic response of the cells on the extracellular and intracellular stimulation (3). To date, >450 unique protein modifications have been identified, including phosphorylation, acetylation, ubiquitination and SUMOylation, which can alter the activity of target proteins, intracellular distribution, protein interactions and protein longevity through post-translational modification (4). Phosphorylation is one of the most common and most extensively studied protein modifications and there are >500 different kinases in mammals to catalyze protein phos-phorylation (5). Phosphorylation occurs mainly in the serine, threonine and tyrosine residues of target substrate proteins (6). The stability of proteins, protein interactions, cellular localization of proteins and enzyme activity are determined according to the different substrates and phosphorylation sites (7). Ubiquitination is also a widely studied method of post-translational protein modification which regulates many biological processes, including immune responses (8), apoptosis (9) and cancer (10). It is mainly catalyzed by three enzymes, the ubiquitin activating enzyme E1, the ubiquitin conjugating enzyme E2 and the ubiquitin ligase E3, in which the E3 ligase determine the substrate specificity. Ubiquitin molecules bind to target proteins to form poly-ubiquitin chains, which can be identified by the 26S proteasome, causing protein degradation. The specific regulatory mechanism mediated by ubiquitination varies with the different structure of the poly-ubiquitin chain (11). Competing with ubiquitination, acetylation is a modification of the lysine residues of the target protein (12). Both serine and threonine are substrates of acetylation and phosphorylation.
SUMOylation has attracted increasing attention as a widely used post-translational protein modification. As this pathway exists in almost all eukaryotes and is essential for the maintenance of genomic integrity, transcriptional regulation, gene expression and the regulation of intracellular signal transduction. The association between SUMOylation and other post-translational protein modifications, such as phosphorylation (13), ubiquitination (14,15), methylation (16) and acetylation (17-20) is currently one of the most important issues. SUMOylation regulates a number of biological processes, including DNA damage repair, immune responses, carcinogenesis, cell cycle progression and apoptosis. Small ubiquitin-like modifier (SUMO) is also involved in the regulation of mitochondrial division, ion channels and biological rhythms. Therefore, multiple layers of regulation or SUMOylation may play important role in the complex protein regulatory networks. The disorder of SUMOylation can lead to the development of certain diseases and tumors. SUMO can thus be used as a potential therapeutic target for cancer (21-25).
2. SUMO protein and SUMO catalytic cycle
SUMO protein is a type of protein which is similar to ubiq-uitin in molecular structure. However, the amino sequence and surface charge distribution of SUMO differ from those of ubiquitin, and thus they have different functions. The amino acid sequence homology of SUMO and ubiquitin molecules is only 18%, but the three-dimensional structure of the two is very similar which contains a typical ββαββαβ fold and a double glycerin C terminal (26). SUMO molecules are highly conserved in evolution, widely found in protozoa, metazoan, plants and fungi. SUMO protein was first discovered in 1996 and there are three types of SUMO isoforms in mammals, which are SUMO-1, SUMO-2 and SUMO-3 (27,28). SUMO-2 and SUMO-3 are very similar as regards their amino acid sequence and are often written as SUMO-2/3. SUMO-1 mainly modifies the physiological state proteins, while SUMO-2/3 mainly modifies stress proteins (29). SUMO4 belongs to another SUMO protein family. SUMO4 mRNA is only found in the kidneys, spleen and lymph nodes, while SUMO 1-3 is widely expressed in human tissues. SUMO4-related research is limited and its protein expression in vivo has not yet been fully determined (30,31). Eight types of SUMO protein isoforms have been found in Arabidopsis thaliana (32). Similar to ubiquitin, SUMO protein can modify one or more lysine residues of the target protein, and the poly-SUMO protein chain is formed on the substrate molecule (33).
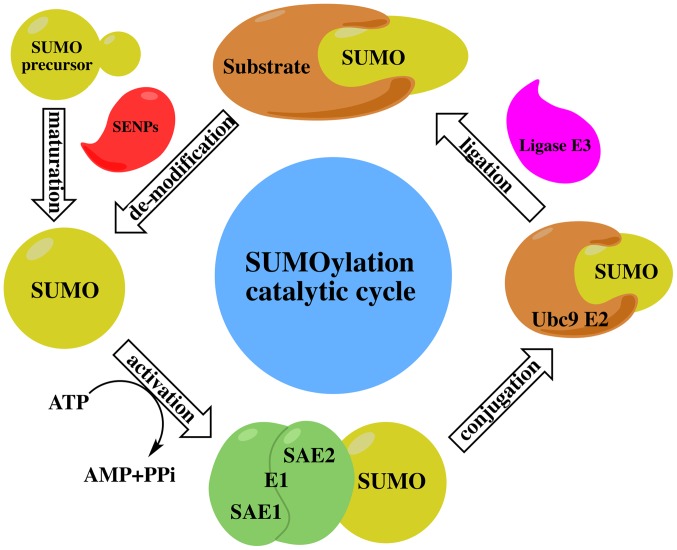
The SUMO cycle and the ubiquitin cycle are similar, but distinct in function. The ubiquitin-modified target protein mainly makes the target protein recognized and is degraded by the proteasome, while the SUMO-modified protein is more stable and SUMOylation modulates protein-protein interactions in order to mediate the localization and functional regulation of target proteins. All SUMO proteins undergo the same enzyme catalytic mechanism with substrate protein attachment or dissociation. The SUMO cycle consists of maturation, activation, conjugation, ligation and de-modification (Fig. 1) as described below:
Figure 1.
The catalytic cycle of SUMOylation. Maturation: Small ubiquitin-like modifier (SUMO) precursors are cleaved by members of the SUMO1/sentrin specific peptidase (SENP) family to expose a C-terminal di-glycine motif. Activation: The mature form of SUMO is then activated by the E1 enzyme SAE1/SAE2 which is ATP-dependent. Conjugation: The activated SUMO is then passed to the active site cysteine of the E2 conjugating enzyme, Ubc9. Ligation: SUMO is then attached to specific lysine residue in the substrate which usually requires E3 ligases. De-modification: SUMO proteins are removed from substrates by SENP, and free SUMO proteins are available for another catalytic cycle.
Maturation
The SUMO protein has a molecular weight of about 11 kDa and is an inactive precursor molecule at the beginning. It is activated by ubiquitin-like specific protease 1 (Ulp1) or sentrin-specific protease 1 (SENP; in humans) to cut the four amino acids at the C terminal and exposed a terminal diglycine GG motif (34).
Activation
SUMO E1-activating enzyme is a 110 kDa protein that contains two subunits, which are SUMO-activating enzyme E1 (SAE1 or Aos1) and SUMO-activating enzymeE2 (SAE2 or Uba2). These two subunits usually form a heterodimer of SAE1-SAE2 or Aos1-Uba2. The heterodimer is mediated by ATP-dependent SUMO adenine nucleotide intermediates to active the SUMO protein by connecting its C terminal carboxyl group with the cysteine residue of SAE2/Uba2 through the thioester bond (35).
Conjugation
There is only one SUMO-conjugating enzyme E2 (SUMO E2 or Ubc9) which can be located at the cytoplasmic side of the nuclear pore complex (NPC) or the nucleoplasm side of the NPC. The activated SUMO protein is transferred to the conserved number 93 cysteine residue of Ubc9 via transesterification reaction to form an E2-SUMO thioester compound. Finally, Ubc9 completes the SUMOylation by connecting the SUMO molecules to the lysine residues of target proteins via the isopeptide bond (36).
Ligation
Although experiments have indicated that SUMO E1 and Ubc9 are sufficient to SUMOylate the substrates, SUMO E3 ligase is essential in the process of SUMO protein targeting the substrate (37). SUMO E3 ligase recognizes substrates and participates in the promotion of SUMOylation through two different mechanisms. SUMO E3 ligase promotes SUMO protein dissociation from Ubc9 by stabilizing the substrate and SUMO-E2 complex contact, and then connecting the SUMO protein to the substrate (38). Through the accurate positioning of the SUMO-E2 complex, SUMO E3 ligase decreases the distance between the thioester bond of SUMO-E2 complex and the substrate lysine residue, making it close enough to enhance the specificity of the substrate (39).
There are approximately three classes of SUMO E3 ligase. Protein inhibitor of activated STAT (PIAS 1-4) (40) and MMS21 (NSE2) (41) are one class of SUMO E3 ligase existing in mammalian cells. Ran binding protein 2 (RanBP2 or NUP358) is kind of long cytoplasmic fragments of the nuclear pore complex. The activity of the RanBP2 SUMO E3 ligase is located in approximately 100 residues of the internal repeat domain (termed R1-M-IR2). Which are related to SUMO-modified RanGAP1, as well as Ubc9 (38). This ternary complex indicates the multi-subunit function of RanBP2 SUMO E3 ligase to stimulate SUMO E2 dissociation with SUMO protein (42). Another type of SUMO E3 ligase is poly-comb protein PC2 or chromobox4 (CBX4), which can promote the ligation of SUMO protein and C-terminal binding protein 1 (CtBP1). It also contributes to the localization of activated Ubc9 and target proteins (43,44). Tripartite motif (TRIM) is the fourth class of SUMO E3 ligase and includes at least eight different TRIM family members (45). TRIM enzyme requires both the RING domain and the B-boxes zinc binding domain to stimulate the SUMO complex and target protein binding (46).
De-modification
SUMOylation is a reversible process in which de-modification involves the SUMO terminal glycine being removed from the lysine residues of the target protein by specific proteases. Thus far, the SUMO protease family has been found to have six SENPs (SENP-1,2,3,5,6,7) in mammals or humans (47). SENP1 and SENP2 can dissociate SUMO1 and SUMO2/3 proteins, while SENP3 and SENP5 are mainly dissociated by SUMO2/3 protein. SUMO2/3 poly chain is dissociated by SENP6 and SENP7 (48). SENPs are mainly located in nuclear or nuclear-related structures. It has recently been found that SUMO protease includes desumoylating isopeptidase 1 (DESI1), desumoylating isopeptidase 2 (DESI2) and ubiquitin-specific protease-like 1 (USPL1) (49). DESI1 and DESI2 are also expressed in the cytoplasm, and USPL1 dissociation enzymes are found in Cajal bodies in the nucleus. The localization of these enzymes in the cell differs and is substrate-specific. Nuclear SENP does not catalyze the deSUMOylation of the cytoplasmic protein. There are two deSUMOylation isomerases, Ulp1 and Ulp2, in yeast and prokaryotes. The two enzymes have different substrate specificity and distribution in cells. Ulp1 is mainly distributed around the nucleus. Experiments have indicated that Ulp1 is connected with the nuclear pore, while Ulp2 is unevenly distributed in the cytoplasm. One of the basic functions of Ulp1 is to process the C end of SUMO precursor and Ulp2 is mainly mediated deSUMOylation (50-52). SENP8 is a novel SUMO protease which mainly processes the full length NEDD8 to the mature form or neddylation (53).
3. SUMO and cancer
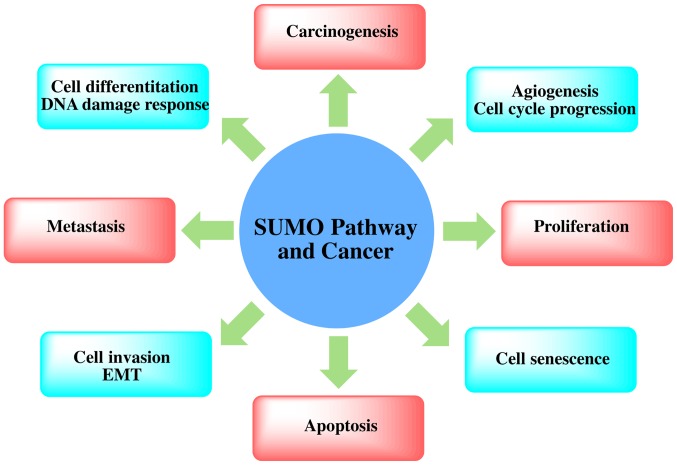
Sumoylation is an important post-translational modification that fine-tunes virtually all cell function and pathological processes. The important role of SUMOylation in human tumorigenesis has gradually emerged. Alterations in the expression or activity of different components in the SUMO signaling pathway may alter the nature of the cell completely. The SUMO pathway can induce cell proliferation, apoptosis resistance and the potential of metastasis by regulating proteins involved in the carcinogenesis (54-58) (Fig. 2). Abnormal SUMOylation can lead to the development of a number of diseases, including cancer. Although the association between the expression of various components in the SUMO signaling pathway and cancer progression or metastasis is not yet fully understood, an increasing number of studies have shown that SUMOylation plays an important role in cancer (25, and refs. therein).
Figure 2.
Association of the SUMO pathway and cancer. SUMOylation has an impact on cancer cell signaling and gene networks that regulate inflammation, immunity and DNA damage, which provides link with carcinogenesis, proliferation, metastasis and apoptosis.
SUMOylation is widely involved in DNA damage response (DDR) and regulates DNA damage sensing and repair protein, which is mainly found in the nucleus (59), particularly in chromatin (60) and nuclear bodies (61). SUMO-modified proteins are ultimately required to perform specific targeting functions. SUMOylation can block the binding sites of substrate proteins and cell interactions, and can influence the function of the proteins by blocking protein-interaction domains. For example, SUMOylation blocks the dimerization of FoxM1 and enhances its transcriptional activity. As the SUMOylation of FoxM1 peaks during the G2 and M phase, these findings contribute to the understanding of the role of SUMOylation during cell-cycle progression (62). SUMO attachment severely impairs E2-25K (UBE2K) ubiquitin thioester and then disrupts the activity of ubiquitin-conjugating enzyme E2-25K (Hip2). It has been found that the protein secondary structure elements are also part of SUMO attachment signals (63). SUMOylation can also produce new docking sites to facilitate the interaction with other proteins. RAD51 interacts non-covalently with SUMO and it interacts more efficiently with SUMO-modified bloom syndrome protein (BLM) at damaged replication forks in vitro. BLM SUMOylation can promote RAD51 function (64). The covalent binding of SUMO protein to the substrate can lead to the conformational change of the substrate, which affects the protein interaction, enzyme activity and cell localization.
Some proteins contain SUMO-interaction motifs (SIMs) and these functional domains usually contain a number of hydrophobic residues, some acidic residues or regions nearby. SIMs can facilitate the non-covalent interaction between the target protein and SUMO protein (65). They can also promote the interaction between SIM-containing proteins and covalent SUMOylation proteins (66). For example, SIMs can enhance the function of SUMO E3 ligase chromebox homologue 4 (CBX4) (67). Protein complexes, nucleosomes, chromatin and other nuclear structures can become more robust through multiple SUMO-SIM interactions. This spatial SUMO regulation through substrate binding and dissociation may be an important factor in the specificity of dynamic SUMO systems, since the enzyme structure of the SUMO system is not as complex as the ubiquitin system (68).
Based on these mechanisms, SUMO can rapidly regulate a number of cellular processes, such as gene expression, transcriptional regulation (69), DDR (70), nucleocytoplasmic transport (71), cell signaling, mRNA maturation, meiosis, mitosis, chromatin remodeling, ion channel activity, cell cycle regulation (72) and cell growth and apoptosis (73). Since SUMO affects the process and function of most intracellular pathways, the SUMO proteins play an important role in certain diseases, particularly cancer.
Recent proteomics data have confirmed that there are at least 1,000 types of human-derived proteins, and as many as 3,000 sites will be modified by SUMO (74). Although the function of these SUMO-modified proteins is not yet clear, many functions are found in all normal cells, including some cancer hallmark functions. SUMOylated proteins are directly or indirectly related to cell apoptosis, inflammatory reactions, immune regulation, DNA damage signaling pathways and gene network regulation, angiogenesis and metastasis-related regulation, the replication of DNA, cell division and cell cycle regulation (22) (Fig. 3). The mechanism of SUMOylation in telomere maintenance and chromosome replication has a further connection with carcinogenesis (75).
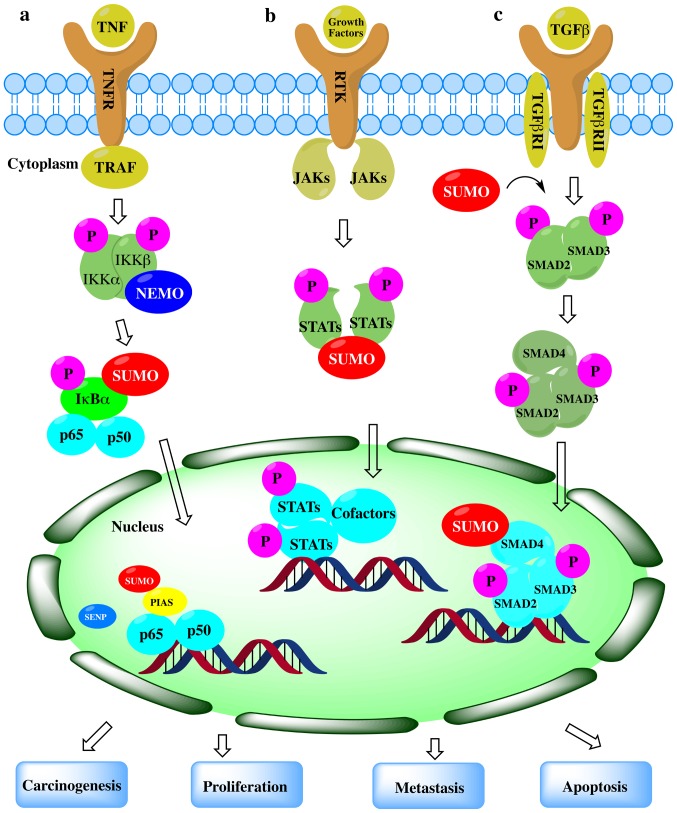
Figure 3.
The role of SUMOylation in representative cancer pathways. (a) SUMOylation in the NF-кB signaling pathway. SUMOylation and de-modification of NF-кB essential modifier (NEMO) proteins are observed both in the cytoplasm and nucleus at steps in this pathway. SUMOylation can positively or negatively regulate NF-кB activation. (b) SUMOylation in the JAK-STAT pathway. SUMOylation of STATs can inhibit phosphorylation and repress target gene expression activated by growth factors. (c) SUMOylation in the TGF-β pathway. SUMOylation exerts both promoting and suppressive effects on target gene expression through the regulation of SMADs.
Role of SUMOylation in cell differentiation and carcinogenesis
The key to cell differentiation is the synthesis of specific proteins. The essence of synthetic-specific proteins is the differential expression of tissue-specific genes (or luxury genes) in time and space due to the combinatorial regulation of gene expression. Thus, cell differentiation is the result of gene expression. Cell carcinogenesis is the performance of normal cell differentiation which is out of control (76). However, the little is known about the mechanisms through which SUMOylation controls gene activity.
Metal toxicants, such as chromium and arsenic are closely related to carcinogenesis through the upregulation of the overall modification of many cellular proteins by SUMO2/3. Androgen receptor (AR) has an established a role in prostate carcinogenesis (77). SUMOylation has diverse effects on AR transcriptional activity via the direct modification of the AR or AR co-regulators (78,79). DNA endonuclease Mus81 is involved in homologous recombination repair, and is among the identified proteins whose SUMOylation is increased following treatment with arsenic trioxide (As2O3). Mus81 SUMOylation is important for normal mitotic chromosome congression, and cells expressing SUMO-resistant Mus81 mutants exhibit compromised DNA damage responses related to tumorigenesis (80). SENP1 exhibits carcinogenic properties associated with the promotion of androgen receptor-dependent and -independent cell proliferation, the stabilization of hypoxia-inducible factor (HIF)1α, an increase in vascular endothelial growth factor (VEGF) expression and the support of angiogenesis (81). Ubc9, PIAS1 and SENP1 are highly expressed in human prostate cancer specimens and are associated with hypoxia-inducing factor 1alpha (HIF1α) expression, and SENP1 enhances prostate epithelial cell proliferation. SENP1 overexpression induces the transformation of the normal prostate gland to high-grade prostatic intraepithelial neoplasia both in vitro and in vivo (82). Increased SUMOylation and deSUMOylation levels at the same time perhaps indicate that cancer cells need to 'speed up' the SUMO cycle, to increase SUMO-modified proteins and to modify the frequency and increase the turnover of the enzyme.
SUMOylation can stabilize PES1 by inhibiting its ubiquitination, which is stimulated by estrogen (83). PES1 is a component of the PeBoW complex which is required for the formation of the 60S ribosomal subunits. The control of ribosome biogenesis is a critical cellular nodal point and the deregulation of ribosome may cause carcinogenesis (84). DNA replication is highly conserved and controlled, the dysregulation of which can lead to genomic instability and even carcinogenesis. USP7 as a replisome-enriched SUMO deubiquitinase which can lead to the deubiquitination of SUMOylated proteins at the replication fork and plays a pivotal role in the control of DNA replication (85,86). Epithelial-mesenchymal transition (EMT) plays an essential role in organogenesis and contributes to a host of pathologies, including carcinogenesis. SIRT1 plays an important role in tumorigenesis and opposed ovarian cancer metastasis by impeding EMT both in vitro and in vivo. SUMO E3 ligase PIAS4 is induced by hypoxia and prevents Sp1 from binding to the SIRT1 promoter in cancer cells (87). Amplified in breast cancer 1 (AIB1) is a transcriptional co-activator of nuclear receptors, which is implicated in breast carcinogenesis. AIB1 is covalently modified by SUMO-1 and PIAS1 may play a crucial role in the regulation of AIB1 transcriptional activity through SUMOylation (88). The extracellular signal-regulated kinase (ERK) and mitogen-activated protein kinase (MAPK) cascade (Raf-MEK-ERK) mediates mitogenic signaling, and is frequently hyperactivated by Ras oncogenes in human cancer and is related to carcinogenesis (89). Oncogenic Ras efficiently activates the ERK pathway both by activating Raf and by inhibiting MEK SUMOylation, thereby inducing carcinogenesis (90). The SMC protein complexes play important roles in chromosome dynamics and the MAGEG1 protein is part of this complex. MAGE proteins play important roles in carcinogenesis and apoptosis (91).
Heat shock proteins (HSPs) constitute a large family of proteins involved in protein folding and maturation, which play a significant role in cellular proliferation, differentiation and carcinogenesis (92). A recent study found that HSPgp96 promoted hepatocellular carcinogenesis (93). Heat shock (HS) is type of stress response of cells and can increase the levels of SUMO conjugates (94). HS-triggered SUMOylation targets promoters and enhancers of actively transcribed genes where it restricts the transcriptional activity of the HS-induced genes using ChIP-seq in cells. The silencing of SUMOylation machinery either by the depletion of UBC9 or PIAS1 enhances the expression of HS-induced genes (95). The deSUMOylation of lens epithelium-derived growth factor (LEDGF) by Sumo-specific protease-1 regulates its transcriptional activation of small heat shock protein and the cellular response (96). Heat shock can cause the accumulation of SUMO-2/3 conjugates which is blocked by proteasome inhibition (97). HSP27 increases the number of cell proteins modified by SUMO-2/3 and blocks heat shock factor 1 (HSF1) transactivation capacity (98). SUMO-2 and SUMO-3 are required for cells to survive heat shock. In summary, SUMO is polymerized into polySUMO chains in response to heat shock and is redistributed to a regulated wide range of protein functions, including folding, transcription, translation, cell cycle regulation, DNA replication and apoptosis (99). SUMO-1 localization on chromatin is dynamic throughout the cell cycle by using chromatin affinity purification coupled with next-generation sequencing, which is consistent with the reversible nature of SUMOylation (100). SUMOylation occurs on many target promoters to regulate transcriptional activation which is consistent with the rules of group modification (101).
Myc (c-Myc) is a regulator gene that codes for a transcription factor. Myc mutation causes Myc protein to be constitutively expressed, which leads to the unregulated expression of a number of genes, some of which are involved in cell proliferation, and results in the development of cancer. Myc protein activates the SUMO-activating enzyme subunit 1 (SAE1) transcription by directly binding to canonical E-Box sequences located close to the SAE1 transcription start site (102). The loss of SAE1/2 enzymatic activity drives synthetic lethality with Myc. SUMOylation-dependent Myc switchers (SMS genes) is required for Myc-driven tumorigenesis. Patients with breast cancer with Myc overexpression and a low expression of SAE1/2 exhibit significantly reduced cancer cell metastasis and an improved survival compared with those with a high SAE1/2 expression. The detection of SAE1 and SAE2 expression may be a useful tool for evaluating and predicting the malignancy of patients with breast cancer (103). The inhibition of SUMOylation disables Myc-induced proliferation and triggers G2/M cell-cycle arrest in mouse and human Myc-driven lymphomas (104). SUMOylated c-Myc is subsequently ubiquitylated and degraded by the proteasome, which is regulated by PIAS1, SENP7 and RNF4 (105).
Ubc9 is overexpressed in ovarian cancer cell lines and tissues and is associated with the downregulation of Bcl-2 in tumorigenesis (106). HPV oncoproteins induce the upregulation of UBC9 during the very early steps of head and neck tumorigenesis via the autophagic process (107). Akt SUMOylation has been found to regulate cell proliferation and tumorigenesis through the direct phosphorylation of Ubc9 at Thr35 and the phosphorylation of SUMO1 at Thr76 (108). Akt SUMOylation is promoted by SUMO E3 ligase PIAS1 and is reversed by SENP1. The expression of PIAS1 and SUMO1 increases Akt activity, whereas the expression of SENP1 reduces Akt1 activity. The loss of SUMOylation markedly reduces Akt1 E17K-mediated cell proliferation and tumorigenesis (109). Tissue microarray analysis has revealed that the expression of Ubc9 is increased during the transformation process of normal colonic epithelial cells into the early stages of cancer and subsequently into advanced colon cancer. Ubc9 is also upregulated in advanced melanomas, prostate intraepithelial neoplasia and primary prostate cancer. The level of Ubc9 is five-fold higher in tumor tissue than in matched normal breast tissue. However, the expression of Ubc9 is downregulated in metastatic breast cancer, prostate cancer and lung cancer, compared with corresponding normal tissues and primary tumors (110). MicroRNAs (miRNAs or miRs) miR-30e and miR-214 can negatively regulate UBC9 expression, and more importantly, the expression of these two factors in some tumors is downregulated (111).
The RANBP2 protein is related to cancer cells, and the point mutation of the RANBP2 gene and gene translocation is related to tumorigenesis. Single point mutations in the RANBP2 gene have been found in human colorectal cancer (112). Insulin-like growth factor-1 receptor (IGF-1R) SUMOylation requires for interaction with RanBP2 and plays critical roles in cancer cell growth. IGF-1R is SUMOylated and translocated into the cell nucleus where it activates gene transcription (113). A recent study found that RANBP2 functions in a complex with SUMO1, namely the as RanBP2/RanGAP1*SUMO1/Ubc9 complex (114).
TRIM family proteins have both SUMO E3 ligase and ubiquitin E3 ligase activities, and are involved in multiple cellular processes including carcinogenesis (45,115,116). TRIM contains a common domain structure composed of a RING finger, one or two B-box motifs and a coiled-coil motif. TRIM29 overexpression enhances cell growth and transforming activity and promotes tumor growth by reducing the acetylation of p53 (117). TRIM40 promotes the neddylation of inhibitor of nuclear factor κB kinase subunit γ and consequently causes the inhibition of NF-κB activity. Nuclear factor-κB (NF-κB) is an important transcription factor for carcinogenesis in chronic inflammatory diseases and plays a key role in promoting inflammation-associated carcinoma in the gastrointestinal tract (118). TRIM45 overexpression also suppresses cell growth by inhibiting the NF-κB signal (119). TRIM24 functions as an oncogene in colorectal carcinogenesis by using a lentivirus-mediated RNA interference system (120). The TRIM66 expression level is higher in osteosarcoma tissues than in normal tissues and it acts as an oncogene by suppressing the apoptotic pathway and promoting TGF-β signaling in osteosarcoma carcinogenesis (121). TRIM59 is upregulated in bladder cancer tissues and is oncogenically active via the TGF-β/Smad2/3 signaling pathway (122).
The SENPs deconjugate modified proteins and are important for maintaining SUMO homeostasis. Alterations in SENP1 levels can transform normal prostate epithelia to a dysplastic state. The enhanced SUMO conjugation of cellular substrates in breast cancer cells promotes tumorigenesis (123). The catalytic activity of SENP is inhibited in hypoxic cell extracts and oxygen controls SENP activity to hypoxic reprogramming of metabolism (124). The Pin1 prolyl isomerase regulates phosphorylation signaling and its upregulation promotes oncogenesis. SENP1-mediated deSUMOylation increases both Pin1 protein stability and Pin1 levels in human breast cancer specimens (125). SENP2 overexpression suppresses the growth and colony-forming ability of hepatocellular carcinoma cells by modulating the stability of β-catenin (126). The upregulation of SENP3/SMT3IP1 promotes epithelial ovarian cancer progression and may serve as a potential biomarker for prognosis (127). The overexpression of SENP5 is associated with the differentiation of oral squamous cell carcinoma (128). SENP5 also enhances cell growth in osteosarcoma cells (129) and promotes tumorigenesis in hepatocellular carcinoma (130). SENP6 promotes gastric cancer cell growth via the deSUMOylation of the transcription factor fork head box protein M1 (FoxM1). The expression of two SENP7 genes in breast cancer is opposite. The short splice variant SENP7S gene is highly expressed in normal breast tissue, while the long splice variant SENP7L gene is highly expressed in breast cancer tissues. SENP7L promotes gene expression and favors aberrant proliferation and initiates EMT in breast cancer (131).
SUMOylation plays significant role in mammalian DNA damage response
Proliferating cell nuclear antigen (PCNA) is an essential factor for DNA replication and repair. Ubiquitin and SUMO compete for the modification of PCNA and regulate the accuracy of replication and repair, contributing to overall genomic stability (132). It is interesting to determine the mechanisms through which the SUMO modification of human PCNA maintains genomic stability. One route is preventing replication fork collapse to DNA double-strand breaks (DSBs) (133). SUMO-PCNA signals for the recruitment of the anti-recombinogenic DNA helicase Srs2 to sites of replication, which need the Srs2 carboxy-terminal domain which harbors tandem receptor motifs (134).
BRCA1 participates in the DNA damage response and mutations in BRCA1 are associated with a high risk of breast and ovarian cancer. SUMO modification of the BRCA1/BARD1 heterodimer greatly increases its ligase activity in vitro (135). Hybrid SUMO-ubiquitin chains are synthesized by SUMO-targeted ubiquitin E3 ligase RNF4 which are recognized by RAP80 to promote BRCA1 recruitment and DNA DSB repair (136). PIAS1 and PIAS4 are required for effective ubiquitin-adduct formation mediated by RNF8, RNF168 and BRCA1 at sites of DNA damage (137). Deubiquitylation enzyme (DUB) ataxin-3 counteracts RNF4 activity during DSBs and is essential for a mediator of DNA damage checkpoint 1 (MDC1)-dependent signaling and repair of DSBs (138). The recruitment of RNF4 to DSBs requires its RING and SUMO interaction motif (SIM) domains and DNA damage factors such as NBS1, MDC1, RNF8, 53BP1 and BRCA1. RNF4 plays a role in the integration of SUMO modification and ubiquitin signaling in the cellular response to DSBs (139). RNF4 targets proteins to the proteasome and regulates the turnover of the DSB-responsive factors MDC1 and replication protein A (RPA) at DNA damage sites (140).
SUMOylation in cancer cell proliferation
The knockdown of the SAE2 and UBC9 genes by lentivirus-based short hairpin RNAs (shRNA) reduces SUMO conjugation activity and inhibits the proliferation of human cancer cells and xenograft tumor growth (141). SUMO-1 can modify IGF-1R, which increases the proliferation of leukemia cell lines (142). These results suggest that IGF-1R and its SUMOylation may be a novel therapeutic target for acute myeloid leukemia (143). Zinc finger protein 131 (ZNF131) is a target for SUMO modification and a substrate for the SUMO E3 ligase human polycomb protein 2 (hPc2), and SUMOylation attenuates estrogen-induced cell growth in a breast cancer cell line (144). USPL1, as a type of SUMO protease, has a low abundance and co-localizes with coilin in Cajal bodies. Cajal bodies are nuclear structures that are involved in the biogenesis of snRNPs and snoRNPs, the maintenance of telomeres and the processing of histone mRNA. The depletion of USPL1 causes striking coilin mislocalization and impairs cell proliferation as a potent SUMO isopeptidase (145). SENP1 is overexpressed and plays a key role in the IL-6 induced proliferation and survival of multiple myeloma cells via NF-кB signaling (146). β-catenin SUMOylation is part of the mechanisms involved in the dysregulated proliferation of myeloma cells. SUMOylation inhibition downregulates β-catenin at the protein level via the promotion of ubiquitin-proteasomal-mediated degradation (147). PIAS1 enhances VCaP prostate cancer cell proliferation by interacting with AR at chromatin sites and also with the pioneer factor FOXA1 (148). The tumor suppressor phosphatase and tensin homolog (PTEN) plays a critical role in the regulation of multiple cellular processes that include survival, cell cycle, proliferation and apoptosis. PIASxα is a SUMO E3 ligase for PTEN which physically interacts with PTEN both in vitro and in vivo. The overexpression of PIASxα leads to G0/G1 cell cycle arrest, thus triggering cell proliferation inhibition and tumor suppression by negatively regulating the PI3K-Akt pathway through the stabilization of PTEN protein (149). The SUMOylation of spliceosome factors USP39 decreases the proliferation-enhancing effect of USP39 on prostate cancer cells (150). Celastrol and triptolide were identified as natural 26s proteasome inhibitors, which promote cell apoptosis and inhibit prostate cancer cell growth. SENP1 expression is downregulated by triptolide at both the mRNA and protein level in a dose-dependent and time-dependent manner, resulting in an enhanced cellular SUMOylation in prostate cancer cells (151). SAE2 protein is significantly associated with the higher expression of c-MYC in primary gastric cancer tissues and the knockdown of SAE2 expression inhibits the proliferation of gastric cancer cells (152). Glucocorticoid receptor (GR) is regulated by SUMOylation which affects genes which are significantly associated with pathways of cellular proliferation and survival. SUMOylation modulates the chromatin occupancy of GR on several loci associated with cellular growth (153). In mechanism study the formation of a GR-SUMO-SMRT/NCoR1-HDAC3-repressing complex is indispensable for NF-κB/AP1-mediated glucocorticoid-induced transrepression in vitro (154). SUMO proteins have been shown to suppress the transcriptional activity of c-Myb. The interplay between the phosphorylation and SUMOylation of c-Myb is essential for the regulation of properly balanced hematopoiesis through the transcriptional regulation of genes directly controlling cellular processes, such as proliferation, differentiation and apoptosis (155). The zinc finger transcription factor of Krüppel-like factor 4 (KLF4) interacts with SUMO-1 to enhance cell proliferation (156). Low dose H2O2 induces an increase in the protein levels of SENP3, which removes SUMO2/3 from promyelocytic leukemia (PML), resulting in an enhanced cell proliferation (157). SUMOylation can regulate the strength of the p53-MDM2 interaction, inhibiting proliferation or inducing the death of potential tumor cells (158). Protein-tyrosine phosphatase 1B (PTP1B) negatively regulates growth factor signaling and cell proliferation by binding to and dephosphorylating key receptor tyrosine kinases, such as the insulin receptor. PIAS1 catalyzes the SUMOylation of PTP1B, reduces its catalytic activity and inhibits the negative effect of PTP1B on insulin receptor signaling in mammalian fibroblasts (159). SUMO-1 gene silencing by transfection with shRNA-SUMO-1 decreases the proliferation and promote the apoptosis of gastric cancer cells (160). The overexpression of E3 ligase PC2 (CBX4) leads to the proliferation of hepatocellular carcinoma. The immunohistochemical staining of PC2 in tumor cells is much stronger than that in normal tissues, and its high expression in the cytoplasm is associated with the poor prognosis of patients with hepatocellular carcinoma (161). Cyclin-dependent kinase 6 (CDK6) is modified by SUMO1 and stabilizes the protein which drives the cell cycle for the cancer development and progression in glioblastoma (162).
SUMOylation in cancer cell invasion, migration and metastasis
Hypoxia (low oxygen supply) aids tumor metastasis, in part by promoting EMT in cancer cells. SENP1 has been shown to enhance the invasion and lung metastasis of triple-negative breast cancer (TNBC) cells in a xenograft tumor model with nude mice (163). SENP1 promotes prostate cancer progression and bone metastasis by regulating two bone remodeling proteins, matrix metalloproteinase (MMP)-2 and MMP-9, through the HIF1α signaling pathway (164). SENP2 inhibits bladder cancer cell invasion and metastasis through the deactivation of the promotor of MMP13 through deSUMOylation of TBL1/TBLR1 (165).
CBX4 participates in the polycomb repressive complex (PRC1) which suppresses metastasis via the recruitment of histone deacetylase 3 (HDAC3) to the key transcription factor Runx2 promoter in colorectal carcinoma (166). CBX4 also enhances hypoxia-induced VEGF expression and angiogenesis in hepatocellular carcinoma cells (167) by enhancing the SUMOylation of HIF-1α. Cbx4 expression promotes the progression and metastasis of orthotopically transplanted tumors in nude mice (168). Smad nuclear interacting protein 1 (SNIP1) is a transcription repressor for the TGF-β and NF-κB signaling pathways and its SUMOylation is enhanced by SUMO E3 ligase PIAS proteins and inhibited by SUMO proteases SENP1/2. The SUMOylation of SNIP1 enhances TGF-β-regulated cell migration and invasion (169). Hepatocyte growth factor (HGF)/c-Met signaling is implicated in the process of EMT in hepatocellular carcinoma. SENP1 silencing via lentivirus-mediated small hairpin RNA (shRNA) transduction has been shown to inhibit EMT and to reduce the HGF-induced proliferation and migration of hepatocellular carcinoma cells (170). N-Myc downstream-regulated gene 2 (NDRG2) protein (171) is covalently modified by SUMO1 and inhibits the growth, metastasis and invasion of human lung adenocarcinomas cells; RNF4 increases the efficiency of this process (172). SUMO and SUMO E3 ligases play a critical role in the spatial and temporal regulation of homologous recombination repair during DSB in heterochromatin (173). SAE2 is highly expressed and the downregulation of SAE2 expression inhibits the migration and invasion of small cell lung cancer (174). SENP1 enhances the progression and metastasis of pancreatic ductal adenocarcinoma via the upregulation of MMP-9, and its expression positively correlates with the lymph node metastasis of cancer cells (175). Bioinformatics analysis has revealed that the expression level of SENP5 positively correlates with the prognosis and survival rate of patients with breast cancer. Patients with a low expression of SENP5 have a good prognosis and a higher survival rate. SENP5 silencing inhibits the anchorage-independence growth, proliferation, migration and invasion of breast cancer cell lines through the regulation of the TGFβRI and MMP-9 levels (176). The mRNA expression of SENP6 in breast cancer tissues is lower than that in normal tissues (177).
SENP 2 suppresses cell migration and invasion partly through inhibiting the expression of MMP13 in bladder cancer cells (178). The Rho GDP dissociation inhibitor (RhoGDI) can affect actin polymerization and cell motility by binding to small GTPases and maintaining them in a biologically inactive state in the cytoplasm (179). RhoGDI interacts with X-linked inhibitor of apoptosis protein (XIAP) and negatively modulates RhoGDI SUMOylation and cancer cell migration, invasion and metastasis (180). The Rho-like GTPase, Rac1, induces cytoskeletal rearrangements and it interacts with PIAS3, which is required for increased Rac activation and optimal cell migration and invasion (181). The expression of galectin-1 belongs to the lectin family, participating in malignant tumor development through the regulation of HIF for the cellular response to hypoxia. PHD3-SUMO conjugation represses HIF1 transcriptional activity (182). Galectin-1 mediates the HIF-1-induced migration and invasion of colorectal cancer cells during hypoxia (183). Ubc9 promotes breast cancer cell invasion and metastasis by interacting with metastasis-related genes, such as CDC42/CXCR4 and regulating the expression of several miRNAs, such as for example, the downregulation of miR-224 (184). The upregulation of Ubc9 expression promotes the invasion and metastasis of lung cancer cells (185).
In a previous study, the whole genome shRNA library screened out a potent synthetic lethal effect between encoding SUMO E1 subunits SAE1/SAE2 genes and K-ras genes. The SAE1 and SAE2 genes were able to selectively inhibit the K-Ras mutant following shRNA interference or silencing, but not to the non-carcinogenic K-Ras wild-type. The SAE gene is critical for K-Ras-induced tumor growth. Therefore, it can be inferred that SAE1/2 can be used to evaluate the invasive and metastastic ability of the mutated K-ras gene. It was also suggested that the SUMO activating enzyme E1 may be a good target for developing new methods for the treatment of human malignancies in a specific genetic context (186).
However, it is of interest to determine the reasons why advanced-stage cancer is refractory to conventional chemotherapy and radiation therapy. As previously demonstrated, Ubc9 is expressed in high levels in melanoma-positive lymph nodes and plays an important role in the progression and metastasis of melanoma. In that study, Ubc9 exerted protective effects, which prevented advanced-stage melanomas from undergoing chemotherapy-induced apoptosis (187). The upregulation of Ubc9 mRNA and the downregulation of miR-214 are greatly associated with the enhanced invasion of glioma. miR-214 overexpression suppresses the endogenous Ubc9 protein and inhibits glioma cell proliferation (188). Ubc9 protein is overexpressed in some breast cancer cell lines and tissues. Ubc9 is associated with alterations in the tumor cellular response to anticancer drugs, as well as tumor growth by regulating Bcl-2 expression through the ER signaling pathway (189).
Metastasis and not the primary tumor is the cause of the majority of most cancer-related deaths (190). A large proportion of solid tumors are derived from epithelial cells. Metastasis involves the physical translocation of a cancer cell to a distant organ and the ability of the cancer cell to develop into a metastatic lesion via EMT. The TGF-β family signaling is essential for EMT and angiogenesis (191). The function of TGF-β is paradoxical, which inhibits tumor growth at the early stage and promote EMT and metastasis at the late stage. TGF-β was modified by SUMO and amplifies TGFβ signaling at multiple levels under various situations (192).
As regards aerobic respiration in cells, the hypoxic signal is a key signaling pathway in regulating gene expression and metabolic process (such as the Warburg effect) and promotes angiogenesis and metastasis in cancer cells (193). HIF controls the hypoxia-signaling cascade and has a direct association with SUMOylation (194). Hypoxia can profoundly affect SUMOylation and it has been shown that the protein expression of the SUMO1 (195) is increased under hypoxic conditions. SUMO-1 promotes glycolysis during periods of hypoxic stress (196). RWD-domain-containing SUMOylation enhancer (RSUME) enhances SUMO conjugation by interacting with the SUMO conjugase Ubc9, increases Ubc9 thioester formation and is overexpressed under conditions of hypoxic stress. Hypoxia-induced RSUME protein SUMOylation can enhance the stability and activity of the HIF1α protein (197,198). SUMO E3 ligase PIAS4 (87) and SENP1 (199) are induced by hypoxia, which is positively associated with cancer aggressiveness. In hepatocellular carcinoma, the SUMOylation of CBX4-dependent HIF1α protein increases the transcriptional activity of HIF1, thereby increasing the expression of vascular endothelial growth factor and angiogenesis (167). Hypoxia aids tumor metastasis, in part by promoting EMT in cancer cells. These results can correspond with the NF-κB and TGF-β pathway observed above.
Role of SUMOylation in cancer cell senescence and apoptosis
The present research suggests that SUMOylation is neither a tumor promoting factor nor a tumor suppressor factor, and that it plays a key role in all cells. Although an increasing number of studies have shown that SUMOylation in tumor cells is greater than that in normal cells (104,152,157,164,175), the drugs or factors which have an effect on the SUMOylation pathway in cancer cells also have the same effect on normal cells.
Cellular senescence is usually irreversible, and promotes cell cycle arrest and limits cell proliferation. Senescence is generally due to the shortening of telomeres or cellular stress responses, such as DNA damage, oxidative stress and the expression of some oncogenes (200). Emerging evidence indicates that cellular senescence plays a key role in tumor suppression; however, the molecular signaling pathway involved has not yet been determined. During the process of cellular senescence, cellular protein secretion is also altered, which is termed the senescence-associated secretory phenotype (SASP). SASP in cells promotes the secretion of a series of inflammatory cytokines, chemokines, growth factors and matrix remodeling factors or matrix metalloproteinases, which alter the local tissue environment and contributes to chronic inflammation and cancer (201). p53 promotes cellular senescence, as well as programmed cell death (apoptosis) as a tumor suppressor (202). The SUMO-1 conjugation with p53 results in p53 stabilization and activation, which causes the induction of senescence (203). By inhibiting or consuming UBC9 protein, hypoSUMOylation leads to the cessation of senescence, such as the growth of human fibroblasts. SUMO protein is selectively retained at the histone maintenance of a repressive environment at histone and tRNA loci is a distinguishing feature of the senescent state. Senescence is related to the widespread increase of SUMO protein. A large number of SUMO chromatin proteins are retained in some selected genomic loci, such as histone or tRNA genes in senescent cells (204). It can be seen that there is a reciprocal association between SUMO and senescence. Oncogenic mutations or DNA damage signals induce SUMO disorder that ultimately leads to cell senescence, which is the current study on the effects of tumor or tumor microenvironment. In turn, senescence phenotype has an important influence on the SUMO balance.
The silencing of the SUMO gene can markedly decrease tumor growth in vivo. In a previous study, it was demonstrated that the gastric cancer cell apoptotic rate was significantly increased following SUMO-1 gene silencing with shRNA-SUMO-1. The expression levels of apoptosis-related genes, such as Bcl-2, c-Myc and p53 were decreased following transfection with shRNA-SUMO plasmid (204). The transcription factor X box-binding protein 1 (XBP1) is a key component of the endoplasmic reticulum (ER) stress response, the transcriptional activity of which is increased by SENP1. ER stress-induced apoptosis is significantly increased by SENP1 deficiency through the accumulation of XBP1 SUMOylation (205). The knockdown of endogenous SENP1 promotes cell apoptosis by inhibiting the phosphorylation of IκBα and Akt and the expression of its downstream regulation factors, Bcl-xL and cyclin D1 (206). PIAS1 regulates ultraviolet (UV)-induced apoptosis through its N-terminal and C-terminal domains (207).
Synthetic lethality interactions are genetic interactions of two mutations whereby the presence of either mutation alone has no effect on cell viability, but the combination of the two mutations results in cell death. The presence of one of these mutations in cancer cells, but not in normal cells can therefore create opportunities to selectively kill cancer cells and spare normal cells.
4. Conclusions and therapeutic perspectives
From the above-mentioned studies, SUMO modification may improve the stability of complex signaling pathways through a wide range of regulatory mechanisms. Studies on the role of SUMO modification in NF-B, TGF-β and JAK/STAT signaling pathways have shown that SUMO proteins play a role in promoting or antagonizing the output of the pathway by targeting not only one, but often more proteins in the signaling pathway components. SUMO modification is an important post-translational modification. It is not only the key factor regulating cell activities, but also plays a role in the pathological processes, which has been widely accepted and confirmed by extensive research. SUMO modification is closely associated with carcinogenesis, and the proliferation and metastasis of tumors; however, the underlying molecular mechanisms remain poorly understood. SUMOylation is upregulated significantly in the majority of cancers, and may thus be a potential target for cancer therapy. The SUMO pathways have profound impacts on protein-protein interactions and some SUMO inhibitors have been developed (208,209). However, a large number of clinical trials are warranted in order to verify these findings and provide useful information for the diagnosis and prognosis of cancer.
Acknowledgments
Not applicable.
Footnotes
Funding
The authors are grateful to the National Natural Science Foundation of China (grant nos. 81670594 and 31270532) and the Cuiying Scientific and Technological Innovation Program of Lanzhou University Second Hospital for their financial support.
Availability of data and materials
Not applicable.
Authors' contributions
ZJH and YHF participated in the writing of the manuscript. BHG contributed to the editing of the manuscript. HC and YML served as scientific advisors. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zamaraev AV, Kopeina GS, Prokhorova EA, Zhivotovsky B, Lavrik IN. Post-translational modification of caspases: The other side of apoptosis regulation. Trends Cell Biol. 2017;27:322–339. doi: 10.1016/j.tcb.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Qian C, Cao X. Post-translational modification control of innate immunity. Immunity. 2016;45:15–30. doi: 10.1016/j.immuni.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 4.Venne AS, Kollipara L, Zahedi RP. The next level of complexity: Crosstalk of posttranslational modifications. Proteomics. 2014;14:513–524. doi: 10.1002/pmic.201300344. [DOI] [PubMed] [Google Scholar]
- 5.Woolfrey KM, Dell'Acqua ML. Coordination of protein phosphorylation and dephosphorylation in synaptic plasticity. J Biol Chem. 2015;290:28604–28612. doi: 10.1074/jbc.R115.657262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 7.Johnson LN. The regulation of protein phosphorylation. Biochem Soc Trans. 2009;37:627–641. doi: 10.1042/BST0370627. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2011;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: A post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- 10.Fulda S, Rajalingam K, Dikic I. Ubiquitylation in immune disorders and cancer: From molecular mechanisms to therapeutic implications. EMBO Mol Med. 2012;4:545–556. doi: 10.1002/emmm.201100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 12.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 13.Guo M, Huang BX. Integration of phosphoproteomic, chemical, and biological strategies for the functional analysis of targeted protein phosphorylation. Proteomics. 2013;13:424–437. doi: 10.1002/pmic.201200274. [DOI] [PubMed] [Google Scholar]
- 14.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamoliatte F, McManus FP, Maarifi G, Chelbi-Alix MK, Thibault P. Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat Commun. 2017;8:14109. doi: 10.1038/ncomms14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16:5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 17.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 18.Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta. 2016;1864:1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Menzies KJ, Zhang H, Katsyuba E, Auwerx J. Protein acetylation in metabolism - metabolites and cofactors. Nat Rev Endocrinol. 2016;12:43–60. doi: 10.1038/nrendo.2015.181. [DOI] [PubMed] [Google Scholar]
- 20.Verdin E, Ott M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 21.Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett. 2012;316:113–125. doi: 10.1016/j.canlet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 22.Eifler K, Vertegaal AC. SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci. 2015;40:779–793. doi: 10.1016/j.tibs.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flotho A, Melchior F. Sumoylation: A regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 24.Rabellino A, Andreani C, Scaglioni PP. The role of PIAS SUMO E3-ligases in cancer. Cancer Res. 2017;77:1542–1547. doi: 10.1158/0008-5472.CAN-16-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17:184–197. doi: 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Dasso M. SUMOylation and deSUMOylation at a glance. J Cell Sci. 2009;122:4249–4252. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/S0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 28.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 30.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 31.Wang CY, Yang P, Li M, Gong F. Characterization of a negative feedback network between SUMO4 expression and NFkappaB transcriptional activity. Biochem Biophys Res Commun. 2009;381:477–481. doi: 10.1016/j.bbrc.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 32.Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol. 2007;10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 34.Nayak A, Müller S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014;15:422. doi: 10.1186/s13059-014-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 36.Tatham MH, Kim S, Jaffray E, Song J, Chen Y, Hay RT. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat Struct Mol Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- 37.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/S0092-8674(02)00630-X. [DOI] [PubMed] [Google Scholar]
- 38.Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell. 2012;46:287–298. doi: 10.1016/j.molcel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 39.Cappadocia L, Pichler A, Lima CD. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat Struct Mol Biol. 2015;22:968–975. doi: 10.1038/nsmb.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rytinki MM, Kaikkonen S, Pehkonen P, Jääskeläinen T, Palvimo JJ. PIAS proteins: Pleiotropic interactors associated with SUMO. Cell Mol Life Sci. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephan AK, Kliszczak M, Morrison CG. The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett. 2011;585:2907–2913. doi: 10.1016/j.febslet.2011.04.067. [DOI] [PubMed] [Google Scholar]
- 42.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang SH, Sharrocks AD. The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol Cell Biol. 2010;30:2193–2205. doi: 10.1128/MCB.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/S0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 45.Hatakeyama S. TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42:297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Koliopoulos MG, Esposito D, Christodoulou E, Taylor IA, Rittinger K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 2016;35:1204–1218. doi: 10.15252/embj.201593741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendes AV, Grou CP, Azevedo JE, Pinto MP. Evaluation of the activity and substrate specificity of the human SENP family of SUMO proteases. Biochim Biophys Acta. 2016;1863:139–147. doi: 10.1016/j.bbamcr.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 49.Shin EJ, Shin HM, Nam E, Kim WS, Kim JH, Oh BH, Yun Y. DeSUMOylating isopeptidase: A second class of SUMO protease. EMBO Rep. 2012;13:339–346. doi: 10.1038/embor.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeh ET. SUMOylation and De-SUMOylation: Wrestling with life's processes. J Biol Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, Baek SH. Emerging roles of desumoylating enzymes. Biochim Biophys Acta. 2009;1792:155–162. doi: 10.1016/j.bbadis.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 52.Huang CJ, Wu D, Khan FA, Huo LJ. DeSUMOylation: An important therapeutic target and protein regulatory event. DNA Cell Biol. 2015;34:652–660. doi: 10.1089/dna.2015.2933. [DOI] [PubMed] [Google Scholar]
- 53.Enchev RI, Schulman BA, Peter M. Protein neddylation: Beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 55.Thomson TM, Guerra-Rebollo M. Ubiquitin and SUMO signalling in DNA repair. Biochem Soc Trans. 2010;38:116–131. doi: 10.1042/BST0380116. [DOI] [PubMed] [Google Scholar]
- 56.Ulrich HD. Ubiquitin and SUMO in DNA repair at a glance. J Cell Sci. 2012;125:249–254. doi: 10.1242/jcs.091801. [DOI] [PubMed] [Google Scholar]
- 57.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 58.Sarangi P, Zhao X. SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem Sci. 2015;40:233–242. doi: 10.1016/j.tibs.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 60.Stielow B, Sapetschnig A, Krüger I, Kunert N, Brehm A, Boutros M, Suske G. Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol Cell. 2008;29:742–754. doi: 10.1016/j.molcel.2007.12.032. [DOI] [PubMed] [Google Scholar]
- 61.Zhong S, Müller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- 62.Schimmel J, Eifler K, Sigurðsson JO, Cuijpers SA, Hendriks IA, Verlaan-de Vries M, Kelstrup CD, Francavilla C, Medema RH, Olsen JV, et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell. 2014;53:1053–1066. doi: 10.1016/j.molcel.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 63.Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Körner R, Olsen JV, Jentsch S, Melchior F, Sixma TK. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol. 2005;12:264–269. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- 64.Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 2009;7:e1000252. doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keusekotten K, Bade VN, Meyer-Teschendorf K, Sriramachandran AM, Fischer-Schrader K, Krause A, Horst C, Schwarz G, Hofmann K, Dohmen RJ, et al. Multivalent interactions of the SUMO-interaction motifs in RING finger protein 4 determine the specificity for chains of the SUMO. Biochem J. 2014;457:207–214. doi: 10.1042/BJ20130753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merrill JC, Melhuish TA, Kagey MH, Yang SH, Sharrocks AD, Wotton D. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS One. 2010;5:e8794. doi: 10.1371/journal.pone.0008794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodríguez JA. Interplay between nuclear transport and ubiquitin/SUMO modifications in the regulation of cancer-related proteins. Semin Cancer Biol. 2014;27:11–19. doi: 10.1016/j.semcancer.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 69.Müller S, Ledl A, Schmidt D. SUMO: A regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 70.Nie M, Boddy MN. Cooperativity of the SUMO and ubiquitin pathways in genome stability. Biomolecules. 2016;6:14. doi: 10.3390/biom6010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Melchior F, Schergaut M, Pichler A. SUMO: Ligases, isopeptidases and nuclear pores. Trends Biochem Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 72.Eifler K, Vertegaal AC. Mapping the SUMOylated landscape. FEBS J. 2015;282:3669–3680. doi: 10.1111/febs.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi SG, Kim H, Jeong EI, Lee HJ, Park S, Lee SY, Lee HJ, Lee SW, Chung CH, Jung YK. SUMO-Modified FADD recruits cytosolic Drp1 and caspase-10 to mitochondria for regulated necrosis. Mol Cell Biol. 2017;37:37. doi: 10.1128/MCB.00254-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- 75.Peuscher MH, Jacobs JJ. Posttranslational control of telomere maintenance and the telomere damage response. Cell Cycle. 2012;11:1524–1534. doi: 10.4161/cc.19847. [DOI] [PubMed] [Google Scholar]
- 76.von Wangenheim KH, Peterson HP. The role of cell differentiation in controlling cell multiplication and cancer. J Cancer Res Clin Oncol. 2008;134:725–741. doi: 10.1007/s00432-008-0381-7. [DOI] [PubMed] [Google Scholar]
- 77.Vlachostergios PJ, Papandreou CN. The role of the small ubiquitin-related modifier (SUMO) pathway in prostate cancer. Biomolecules. 2012;2:240–255. doi: 10.3390/biom2020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng Z, Cai C, Omwancha J, Chen SY, Baslan T, Shemshedini L. SUMO-3 enhances androgen receptor transcriptional activity through a sumoylation-independent mechanism in prostate cancer cells. J Biol Chem. 2006;281:4002–4012. doi: 10.1074/jbc.M509260200. [DOI] [PubMed] [Google Scholar]
- 79.Bawa-Khalfe T, Cheng J, Wang Z, Yeh ET. Induction of the SUMO-specific protease 1 transcription by the androgen receptor in prostate cancer cells. J Biol Chem. 2007;282:37341–37349. doi: 10.1074/jbc.M706978200. [DOI] [PubMed] [Google Scholar]
- 80.Hu L, Yang F, Lu L, Dai W. Arsenic-induced sumoylation of Mus81 is involved in regulating genomic stability. Cell Cycle. 2017;16:802–811. doi: 10.1080/15384101.2017.1302628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bawa-Khalfe T, Yang FM, Ritho J, Lin HK, Cheng J, Yeh ET. SENP1 regulates PTEN stability to dictate prostate cancer development. Oncotarget. 2017;8:17651–17664. doi: 10.18632/oncotarget.13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bawa-Khalfe T, Cheng J, Lin SH, Ittmann MM, Yeh ET. SENP1 induces prostatic intraepithelial neoplasia through multiple mechanisms. J Biol Chem. 2010;285:25859–25866. doi: 10.1074/jbc.M110.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li S, Wang M, Qu X, Xu Z, Yang Y, Su Q, Wu H. SUMOylation of PES1 upregulates its stability and function via inhibiting its ubiquitination. Oncotarget. 2016;7:50522–50534. doi: 10.18632/oncotarget.10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Finkbeiner E, Haindl M, Raman N, Muller S. SUMO routes ribosome maturation. Nucleus. 2011;2:527–532. doi: 10.4161/nucl.2.6.17604. [DOI] [PubMed] [Google Scholar]
- 85.Lecona E, Rodriguez-Acebes S, Specks J, Lopez-Contreras AJ, Ruppen I, Murga M, Muñoz J, Mendez J, Fernandez-Capetillo O. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat Struct Mol Biol. 2016;23:270–277. doi: 10.1038/nsmb.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smits VA, Freire R. USP7/HAUSP: A SUMO deubiquitinase at the heart of DNA replication. BioEssays. 2016;38:863–868. doi: 10.1002/bies.201600096. [DOI] [PubMed] [Google Scholar]
- 87.Sun L, Li H, Chen J, Iwasaki Y, Kubota T, Matsuoka M, Shen A, Chen Q, Xu Y. PIASy mediates hypoxia-induced SIRT1 transcriptional repression and epithelial-to-mesenchymal transition in ovarian cancer cells. J Cell Sci. 2013;126:3939–3947. doi: 10.1242/jcs.127381. [DOI] [PubMed] [Google Scholar]
- 88.Li S, Yang C, Hong Y, Bi H, Zhao F, Liu Y, Ao X, Pang P, Xing X, Chang AK, et al. The transcriptional activity of co-activator AIB1 is regulated by the SUMO E3 ligase PIAS1. Biol Cell. 2012;104:287–296. doi: 10.1111/boc.201100116. [DOI] [PubMed] [Google Scholar]
- 89.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kubota Y, O'Grady P, Saito H, Takekawa M. Oncogenic Ras abrogates MEK SUMOylation that suppresses the ERK pathway and cell transformation. Nat Cell Biol. 2011;13:282–291. doi: 10.1038/ncb2169. [DOI] [PubMed] [Google Scholar]
- 91.Taylor EM, Copsey AC, Hudson JJ, Vidot S, Lehmann AR. Identification of the proteins, including MAGEG1, that make up the human SMC5-6 protein complex. Mol Cell Biol. 2008;28:1197–1206. doi: 10.1128/MCB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu J, Liu T, Rios Z, Mei Q, Lin X, Cao S. Heat shock proteins and cancer. Trends Pharmacol Sci. 2017;38:226–256. doi: 10.1016/j.tips.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 93.Rachidi S, Sun S, Wu BX, Jones E, Drake RR, Ogretmen B, Cowart LA, Clarke CJ, Hannun YA, Chiosis G, et al. Endoplasmic reticulum heat shock protein gp96 maintains liver homeostasis and promotes hepatocellular carcinogenesis. J Hepatol. 2015;62:879–888. doi: 10.1016/j.jhep.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pinto MP, Carvalho AF, Grou CP, Rodríguez-Borges JE, Sá-Miranda C, Azevedo JE. Heat shock induces a massive but differential inactivation of SUMO-specific proteases. Biochim Biophys Acta. 2012;1823:1958–1966. doi: 10.1016/j.bbamcr.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 95.Niskanen EA, Malinen M, Sutinen P, Toropainen S, Paakinaho V, Vihervaara A, Joutsen J, Kaikkonen MU, Sistonen L, Palvimo JJ. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 2015;16:153. doi: 10.1186/s13059-015-0717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ishihara K, Fatma N, Bhargavan B, Chhunchha B, Kubo E, Dey S, Takamura Y, Kumar A, Singh DP. Lens epithelium-derived growth factor deSumoylation by Sumo-specific protease-1 regulates its transcriptional activation of small heat shock protein and the cellular response. FEBS J. 2012;279:3048–3070. doi: 10.1111/j.1742-4658.2012.08686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Castorálová M, Březinová D, Svéda M, Lipov J, Ruml T, Knejzlík Z. SUMO-2/3 conjugates accumulating under heat shock or MG132 treatment result largely from new protein synthesis. Biochim Biophys Acta. 2012;1823:911–919. doi: 10.1016/j.bbamcr.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 98.Brunet Simioni M, De Thonel A, Hammann A, Joly AL, Bossis G, Fourmaux E, Bouchot A, Landry J, Piechaczyk M, Garrido C. Heat shock protein 27 is involved in SUMO-2/3 modification of heat shock factor 1 and thereby modulates the transcription factor activity. Oncogene. 2009;28:3332–3344. doi: 10.1038/onc.2009.188. [DOI] [PubMed] [Google Scholar]
- 99.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 100.Liu HW, Zhang J, Heine GF, Arora M, Gulcin Ozer H, Onti-Srinivasan R, Huang K, Parvin JD. Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res. 2012;40:10172–10186. doi: 10.1093/nar/gks819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aguilar-Martinez E, Chen X, Webber A, Mould AP, Seifert A, Hay RT, Sharrocks AD. Screen for multi-SUMO-binding proteins reveals a multi-SIM-binding mechanism for recruitment of the transcriptional regulator ZMYM2 to chromatin. Proc Natl Acad Sci USA. 2015;112:E4854–E4863. doi: 10.1073/pnas.1509716112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Amente S, Lavadera ML, Palo GD, Majello B. SUMO-activating SAE1 transcription is positively regulated by Myc. Am J Cancer Res. 2012;2:330–334. [PMC free article] [PubMed] [Google Scholar]
- 103.Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hoellein A, Fallahi M, Schoeffmann S, Steidle S, Schaub FX, Rudelius M, Laitinen I, Nilsson L, Goga A, Peschel C, et al. Myc-induced SUMOylation is a therapeutic vulnerability for B-cell lymphoma. Blood. 2014;124:2081–2090. doi: 10.1182/blood-2014-06-584524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.González-Prieto R, Cuijpers SA, Kumar R, Hendriks IA, Vertegaal AC. c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle. 2015;14:1859–1872. doi: 10.1080/15384101.2015.1040965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mo YY, Yu Y, Theodosiou E, Ee PL, Beck WT. A role for Ubc9 in tumorigenesis. Oncogene. 2005;24:2677–2683. doi: 10.1038/sj.onc.1208210. [DOI] [PubMed] [Google Scholar]
- 107.Mattoscio D, Casadio C, Miccolo C, Maffini F, Raimondi A, Tacchetti C, Gheit T, Tagliabue M, Galimberti VE, De Lorenzi F, et al. Autophagy regulates UBC9 levels during viral-mediated tumorigenesis. PLoS Pathog. 2017;13:e1006262. doi: 10.1371/journal.ppat.1006262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin CH, Liu SY, Lee EH. SUMO modification of Akt regulates global SUMOylation and substrate SUMOylation specificity through Akt phosphorylation of Ubc9 and SUMO1. Oncogene. 2016;35:595–607. doi: 10.1038/onc.2015.115. [DOI] [PubMed] [Google Scholar]
- 109.Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K, Wang P. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 2013;73:5742–5753. doi: 10.1158/0008-5472.CAN-13-0538. [DOI] [PubMed] [Google Scholar]
- 110.Moschos SJ, Jukic DM, Athanassiou C, Bhargava R, Dacic S, Wang X, Kuan SF, Fayewicz SL, Galambos C, Acquafondata M, et al. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol. 2010;41:1286–1298. doi: 10.1016/j.humpath.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 111.Wu F, Zhu S, Ding Y, Beck WT, Mo YY. MicroRNA-mediated regulation of Ubc9 expression in cancer cells. Clin Cancer Res. 2009;15:1550–1557. doi: 10.1158/1078-0432.CCR-08-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gylfe AE, Kondelin J, Turunen M, Ristolainen H, Katainen R, Pitkänen E, Kaasinen E, Rantanen V, Tanskanen T, Varjosalo M, et al. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology. 2013;145:540–3.e22. doi: 10.1053/j.gastro.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 113.Packham S, Warsito D, Lin Y, Sadi S, Karlsson R, Sehat B, Larsson O. Nuclear translocation of IGF-1R via p150 (Glued) and an importin-β/RanBP2-dependent pathway in cancer cells. Oncogene. 2015;34:2227–2238. doi: 10.1038/onc.2014.165. [DOI] [PubMed] [Google Scholar]
- 114.Ritterhoff T, Das H, Hofhaus G, Schröder RR, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3 ligase is a disassembly machine for Crm1-dependent nuclear export complexes. Nat Commun. 2016;7:11482. doi: 10.1038/ncomms11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 116.Watanabe M, Hatakeyama S. TRIM proteins and diseases. J Biochem. 2017;161:135–144. doi: 10.1093/jb/mvw087. [DOI] [PubMed] [Google Scholar]
- 117.Sho T, Tsukiyama T, Sato T, Kondo T, Cheng J, Saku T, Asaka M, Hatakeyama S. TRIM29 negatively regulates p53 via inhibition of Tip60. Biochim Biophys Acta. 2011;1813:1245–1253. doi: 10.1016/j.bbamcr.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 118.Noguchi K, Okumura F, Takahashi N, Kataoka A, Kamiyama T, Todo S, Hatakeyama S. TRIM40 promotes neddylation of IKKγ and is downregulated in gastrointestinal cancers. Carcinogenesis. 2011;32:995–1004. doi: 10.1093/carcin/bgr068. [DOI] [PubMed] [Google Scholar]
- 119.Shibata M, Sato T, Nukiwa R, Ariga T, Hatakeyama S. TRIM45 negatively regulates NF-κB-mediated transcription and suppresses cell proliferation. Biochem Biophys Res Commun. 2012;423:104–109. doi: 10.1016/j.bbrc.2012.05.090. [DOI] [PubMed] [Google Scholar]
- 120.Wang J, Zhu J, Dong M, Yu H, Dai X, Li K. Knockdown of tripartite motif containing 24 by lentivirus suppresses cell growth and induces apoptosis in human colorectal cancer cells. Oncol Res. 2014;22:39–45. doi: 10.3727/096504014X14078436005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Y, Guo Y, Yang H, Shi G, Xu G, Shi J, Yin N, Chen D. TRIM66 overexpresssion contributes to osteosarcoma carcinogenesis and indicates poor survival outcome. Oncotarget. 2015;6:23708–23719. doi: 10.18632/oncotarget.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen W, Zhao K, Miao C, Xu A, Zhang J, Zhu J, Su S, Wang Z. Silencing Trim59 inhibits invasion/migration and epithelial-to-mesenchymal transition via TGF-β/Smad2/3 signaling pathway in bladder cancer cells. Onco Targets Ther. 2017;10:1503–1512. doi: 10.2147/OTT.S130139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bawa-Khalfe T, Yeh ET. SUMO losing balance: SUMO proteases disrupt SUMO homeostasis to facilitate cancer development and progression. Genes Cancer. 2010;1:748–752. doi: 10.1177/1947601910382555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kunz K, Wagner K, Mendler L, Hölper S, Dehne N, Müller S. SUMO signaling by hypoxic inactivation of SUMO-specific isopeptidases. Cell Reports. 2016;16:3075–3086. doi: 10.1016/j.celrep.2016.08.031. [DOI] [PubMed] [Google Scholar]
- 125.Chen CH, Chang CC, Lee TH, Luo M, Huang P, Liao PH, Wei S, Li FA, Chen RH, Zhou XZ, et al. SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function. Cancer Res. 2013;73:3951–3962. doi: 10.1158/0008-5472.CAN-12-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen HJ, Zhu HY, Yang C, Ji F. SENP2 regulates hepatocellular carcinoma cell growth by modulating the stability of β-catenin. Asian Pac J Cancer Prev. 2012;13:3583–3587. doi: 10.7314/APJCP.2012.13.8.3583. [DOI] [PubMed] [Google Scholar]
- 127.Cheng J, Su M, Jin Y, Xi Q, Deng Y, Chen J, Wang W, Chen Y, Chen L, Shi N, et al. Upregulation of SENP3/SMT3IP1 promotes epithelial ovarian cancer progression and forecasts poor prognosis. Tumour Biol. 2017;39:1010428317694543. doi: 10.1177/1010428317694543. [DOI] [PubMed] [Google Scholar]
- 128.Ding X, Sun J, Wang L, Li G, Shen Y, Zhou X, Chen W. Overexpression of SENP5 in oral squamous cell carcinoma and its association with differentiation. Oncol Rep. 2008;20:1041–1045. [PubMed] [Google Scholar]
- 129.Wang K, Zhang XC. Inhibition of SENP5 suppresses cell growth and promotes apoptosis in osteosarcoma cells. Exp Ther Med. 2014;7:1691–1695. doi: 10.3892/etm.2014.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jin ZL, Pei H, Xu YH, Yu J, Deng T. The SUMO-specific protease SENP5 controls DNA damage response and promotes tumorigenesis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20:3566–3573. [PubMed] [Google Scholar]
- 131.Bawa-Khalfe T, Lu LS, Zuo Y, Huang C, Dere R, Lin FM, Yeh ET. Differential expression of SUMO-specific protease 7 variants regulates epithelial-mesenchymal transition. Proc Natl Acad Sci USA. 2012;109:17466–17471. doi: 10.1073/pnas.1209378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 133.Gali H, Juhasz S, Morocz M, Hajdu I, Fatyol K, Szukacsov V, Burkovics P, Haracska L. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 2012;40:6049–6059. doi: 10.1093/nar/gks256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012;483:59–63. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 136.Guzzo CM, Berndsen CE, Zhu J, Gupta V, Datta A, Greenberg RA, Wolberger C, Matunis MJ. RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci Signal. 2012;5:ra88. doi: 10.1126/scisignal.2003485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pfeiffer A, Luijsterburg MS, Acs K, Wiegant WW, Helfricht A, Herzog LK, Minoia M, Böttcher C, Salomons FA, van Attikum H, et al. Ataxin-3 consolidates the MDC1-dependent DNA double-strand break response by counteracting the SUMO-targeted ubiquitin ligase RNF4. EMBO J. 2017;36:1066–1083. doi: 10.15252/embj.201695151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012;26:1196–1208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012;26:1179–1195. doi: 10.1101/gad.188284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.He X, Riceberg J, Pulukuri SM, Grossman S, Shinde V, Shah P, Brownell JE, Dick L, Newcomb J, Bence N. Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PLoS One. 2015;10:e0123882. doi: 10.1371/journal.pone.0123882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang J, Huang FF, Wu DS, Li WJ, Zhan HE, Peng MY, Fang P, Cao PF, Zhang MM, Zeng H, et al. SUMOylation of insulin-like growth factor 1 receptor, promotes proliferation in acute myeloid leukemia. Cancer Lett. 2015;357:297–306. doi: 10.1016/j.canlet.2014.11.052. [DOI] [PubMed] [Google Scholar]
- 143.You L, Liu C, Tang H, Liao Y, Fu S. Advances in targeting insulin-like growth factor signaling pathway in cancer treatment. Curr Pharm Des. 2014;20:2899–2911. doi: 10.2174/13816128113199990595. [DOI] [PubMed] [Google Scholar]
- 144.Oh Y, Chung KC. Small ubiquitin-like modifier (SUMO) modification of zinc finger protein 131 potentiates its negative effect on estrogen signaling. J Biol Chem. 2012;287:17517–17529. doi: 10.1074/jbc.M111.336354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schulz S, Chachami G, Kozaczkiewicz L, Winter U, Stankovic-Valentin N, Haas P, Hofmann K, Urlaub H, Ovaa H, Wittbrodt J, et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012;13:930–938. doi: 10.1038/embor.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xu J, Sun HY, Xiao FJ, Wang H, Yang Y, Wang L, Gao CJ, Guo ZK, Wu CT, Wang LS. SENP1 inhibition induces apoptosis and growth arrest of multiple myeloma cells through modulation of NF-κB signaling. Biochem Biophys Res Commun. 2015;460:409–415. doi: 10.1016/j.bbrc.2015.03.047. [DOI] [PubMed] [Google Scholar]
- 147.Huang HJ, Zhou LL, Fu WJ, Zhang CY, Jiang H, Du J, Hou J. β-catenin SUMOylation is involved in the dysregulated proliferation of myeloma cells. Am J Cancer Res. 2014;5:309–320. [PMC free article] [PubMed] [Google Scholar]
- 148.Toropainen S, Malinen M, Kaikkonen S, Rytinki M, Jääskeläinen T, Sahu B, Jänne OA, Palvimo JJ. SUMO ligase PIAS1 functions as a target gene selective androgen receptor coregulator on prostate cancer cell chromatin. Nucleic Acids Res. 2015;43:848–861. doi: 10.1093/nar/gku1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang W, Chen Y, Wang S, Hu N, Cao Z, Wang W, Tong T, Zhang X. PIASxα ligase enhances SUMO1 modification of PTEN protein as a SUMO E3 ligase. J Biol Chem. 2014;289:3217–3230. doi: 10.1074/jbc.M113.508515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wen D, Xu Z, Xia L, Liu X, Tu Y, Lei H, Wang W, Wang T, Song L, Ma C, et al. Important role of SUMOylation of Spliceosome factors in prostate cancer cells. J Proteome Res. 2014;13:3571–3582. doi: 10.1021/pr4012848. [DOI] [PubMed] [Google Scholar]
- 151.Huang W, He T, Chai C, Yang Y, Zheng Y, Zhou P, Qiao X, Zhang B, Liu Z, Wang J, et al. Triptolide inhibits the proliferation of prostate cancer cells and down-regulates SUMO-specific protease 1 expression. PLoS One. 2012;7:e37693. doi: 10.1371/journal.pone.0037693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shao DF, Wang XH, Li ZY, Xing XF, Cheng XJ, Guo T, Du H, Hu Y, Dong B, Ding N, et al. High-level SAE2 promotes malignant phenotype and predicts outcome in gastric cancer. Am J Cancer Res. 2014;5:140–154. [PMC free article] [PubMed] [Google Scholar]
- 153.Paakinaho V, Kaikkonen S, Makkonen H, Benes V, Palvimo JJ. SUMOylation regulates the chromatin occupancy and anti-proliferative gene programs of glucocorticoid receptor. Nucleic Acids Res. 2014;42:1575–1592. doi: 10.1093/nar/gkt1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hua G, Ganti KP, Chambon P. Glucocorticoid-induced tethered transrepression requires SUMOylation of GR and formation of a SUMO-SMRT/NCoR1-HDAC3 repressing complex. Proc Natl Acad Sci USA. 2016;113:E635–E643. doi: 10.1073/pnas.1522826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bies J, Sramko M, Wolff L. Stress-induced phosphorylation of Thr486 in c-Myb by p38 mitogenactivated protein kinases attenuates conjugation of SUMO-2/3. J Biol Chem. 2013;288:36983–36993. doi: 10.1074/jbc.M113.500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Du JX, McConnell BB, Yang VW. A small ubiquitin-related modifier-interacting motif functions as the transcriptional activation domain of Krüppel-like factor 4. J Biol Chem. 2010;285:28298–28308. doi: 10.1074/jbc.M110.101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Han Y, Huang C, Sun X, Xiang B, Wang M, Yeh ET, Chen Y, Li H, Shi G, Cang H, et al. SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. J Biol Chem. 2010;285:12906–12915. doi: 10.1074/jbc.M109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 159.Dadke S, Cotteret S, Yip SC, Jaffer ZM, Haj F, Ivanov A, Rauscher F, III, Shuai K, Ng T, Neel BG, et al. Regulation of protein tyrosine phosphatase 1B by sumoylation. Nat Cell Biol. 2007;9:80–85. doi: 10.1038/ncb1522. [DOI] [PubMed] [Google Scholar]
- 160.Jin L, Shen K, Chen T, Zhang H, Yu W. SUMO-1 gene silencing inhibits proliferation and promotes apoptosis of human gastric cancer SGC-7901 Cells. Cell Physiol Biochem. 2017;41:987–998. doi: 10.1159/000460836. [DOI] [PubMed] [Google Scholar]
- 161.Wang B, Tang J, Liao D, Wang G, Zhang M, Sang Y, Cao J, Wu Y, Zhang R, Li S, et al. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S684–S692. doi: 10.1245/s10434-013-3171-7. [DOI] [PubMed] [Google Scholar]
- 162.Bellail AC, Olson JJ, Hao C. SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat Commun. 2014;5:4234. doi: 10.1038/ncomms5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang Z, Jin J, Zhang J, Wang L, Cao J. Depletion of SENP1 suppresses the proliferation and invasion of triple-negative breast cancer cells. Oncol Rep. 2016;36:2071–2078. doi: 10.3892/or.2016.5036. [DOI] [PubMed] [Google Scholar]
- 164.Wang Q, Xia N, Li T, Xu Y, Zou Y, Zuo Y, Fan Q, Bawa-Khalfe T, Yeh ET, Cheng J. SUMO-specific protease-1 promotes prostate cancer progression and metastasis. Oncogene. 2013;32:2493–2498. doi: 10.1038/onc.2012.250. [DOI] [PubMed] [Google Scholar]
- 165.Tan M, Gong H, Wang J, Tao L, Xu D, Bao E, Liu Z, Qiu J. SENP2 regulates MMP13 expression in a bladder cancer cell line through SUMOylation of TBL1/TBLR1. Sci Rep. 2015;5:13996. doi: 10.1038/srep13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao D, Wang G, Qin G, Xu RH, Kang T. CBX4 suppresses metastasis via recruitment of HDAC3 to the Runx2 promoter in colorectal carcinoma. Cancer Res. 2016;76:7277–7289. doi: 10.1158/0008-5472.CAN-16-2100. [DOI] [PubMed] [Google Scholar]
- 167.Li J, Xu Y, Long XD, Wang W, Jiao HK, Mei Z, Yin QQ, Ma LN, Zhou AW, Wang LS, et al. Cbx4 governs HIF-1α to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25:118–131. doi: 10.1016/j.ccr.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 168.Mei Z, Jiao H, Wang W, Li J, Chen G, Xu Y. Polycomb chromobox 4 enhances migration and pulmonary metastasis of hepatocellular carcinoma cell line MHCC97L. Sci China Life Sci. 2014;57:610–617. doi: 10.1007/s11427-014-4663-9. [DOI] [PubMed] [Google Scholar]
- 169.Liu S, Long J, Yuan B, Zheng M, Xiao M, Xu J, Lin X, Feng XH. SUMO modification reverses inhibitory effects of Smad nuclear interacting protein-1 in TGF-β responses. J Biol Chem. 2016;291:24418–24430. doi: 10.1074/jbc.M116.755850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang W, Sun H, Shi X, Wang H, Cui C, Xiao F, Wu C, Guo X, Wang L. SENP1 regulates hepatocyte growth factor-induced migration and epithelial-mesenchymal transition of hepatocellular carcinoma. Tumour Biol. 2016;37:7741–7748. doi: 10.1007/s13277-015-4406-y. [DOI] [PubMed] [Google Scholar]
- 171.Hu W, Fan C, Jiang P, Ma Z, Yan X, Di S, Jiang S, Li T, Cheng Y, Yang Y. Emerging role of N-myc downstream-regulated gene 2 (NDRG2) in cancer. Oncotarget. 2016;7:209–223. doi: 10.18632/oncotarget.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tantai J, Pan X, Hu D. RNF4-mediated SUMOylation is essential for NDRG2 suppression of lung adenocarcinoma. Oncotarget. 2016;7:26837–26843. doi: 10.18632/oncotarget.8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, Karpen GH, Chiolo I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol. 2015;17:1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu X, Xu Y, Pang Z, Guo F, Qin Q, Yin T, Sang Y, Feng C, Li X, Jiang L, et al. Knockdown of SUMO-activating enzyme subunit 2 (SAE2) suppresses cancer malignancy and enhances chemotherapy sensitivity in small cell lung cancer. J Hematol Oncol. 2015;8:67. doi: 10.1186/s13045-015-0164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ma C, Wu B, Huang X, Yuan Z, Nong K, Dong B, Bai Y, Zhu H, Wang W, Ai K. SUMO-specific protease 1 regulates pancreatic cancer cell proliferation and invasion by targeting MMP-9. Tumour Biol. 2014;35:12729–12735. doi: 10.1007/s13277-014-2598-1. [DOI] [PubMed] [Google Scholar]
- 176.Cashman R, Cohen H, Ben-Hamo R, Zilberberg A, Efroni S. SENP5 mediates breast cancer invasion via a TGFβRI SUMOylation cascade. Oncotarget. 2014;5:1071–1082. doi: 10.18632/oncotarget.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mooney SM, Grande JP, Salisbury JL, Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- 178.Tan MY, Mu XY, Liu B, Wang Y, Bao ED, Qiu JX, Fan Y. SUMO-specific protease 2 suppresses cell migration and invasion through inhibiting the expression of MMP13 in bladder cancer cells. Cell Physiol Biochem. 2013;32:542–548. doi: 10.1159/000354458. [DOI] [PubMed] [Google Scholar]
- 179.Garcia-Mata R, Boulter E, Burridge K. The 'invisible hand': Regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yu J, Zhang D, Liu J, Li J, Yu Y, Wu XR, Huang C. RhoGDI SUMOylation at Lys-138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility. J Biol Chem. 2012;287:13752–13760. doi: 10.1074/jbc.M111.337469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Castillo-Lluva S, Tatham MH, Jones RC, Jaffray EG, Edmondson RD, Hay RT, Malliri A. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat Cell Biol. 2010;12:1078–1085. doi: 10.1038/ncb2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Núñez-O'Mara A, Gerpe-Pita A, Pozo S, Carlevaris O, Urzelai B, Lopitz-Otsoa F, Rodríguez MS, Berra E. PHD3-SUMO conjugation represses HIF1 transcriptional activity independently of PHD3 catalytic activity. J Cell Sci. 2015;128:40–49. doi: 10.1242/jcs.151514. [DOI] [PubMed] [Google Scholar]
- 183.Zhao XY, Chen TT, Xia L, Guo M, Xu Y, Yue F, Jiang Y, Chen GQ, Zhao KW. Hypoxia inducible factor-1 mediates expression of galectin-1: The potential role in migration/invasion of colorectal cancer cells. Carcinogenesis. 2010;31:1367–1375. doi: 10.1093/carcin/bgq116. [DOI] [PubMed] [Google Scholar]
- 184.Zhu S, Sachdeva M, Wu F, Lu Z, Mo YY. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene. 2010;29:1763–1772. doi: 10.1038/onc.2009.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li H, Niu H, Peng Y, Wang J, He P. Ubc9 promotes invasion and metastasis of lung cancer cells. Oncol Rep. 2013;29:1588–1594. doi: 10.3892/or.2013.2268. [DOI] [PubMed] [Google Scholar]
- 186.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Moschos SJ, Smith AP, Mandic M, Athanassiou C, Watson-Hurst K, Jukic DM, Edington HD, Kirkwood JM, Becker D. SAGE and antibody array analysis of melanoma-infiltrated lymph nodes: Identification of Ubc9 as an important molecule in advanced-stage melanomas. Oncogene. 2007;26:4216–4225. doi: 10.1038/sj.onc.1210216. [DOI] [PubMed] [Google Scholar]
- 188.Zhao Z, Tan X, Zhao A, Zhu L, Yin B, Yuan J, Qiang B, Peng X. microRNA-214-mediated UBC9 expression in glioma. BMB Rep. 2012;45:641–646. doi: 10.5483/BMBRep.2012.45.11.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lu Z, Wu H, Mo YY. Regulation of bcl-2 expression by Ubc9. Exp Cell Res. 2006;312:1865–1875. doi: 10.1016/j.yexcr.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 190.Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 191.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Morrison CD, Parvani JG, Schiemann WP. The relevance of the TGF-β Paradox to EMT-MET programs. Cancer Lett. 2013;341:30–40. doi: 10.1016/j.canlet.2013.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Núñez-O'Mara A, Berra E. Deciphering the emerging role of SUMO conjugation in the hypoxia-signaling cascade. Biol Chem. 2013;394:459–469. doi: 10.1515/hsz-2012-0319. [DOI] [PubMed] [Google Scholar]
- 195.Shao R, Zhang FP, Tian F, Anders Friberg P, Wang X, Sjöland H, Billig H. Increase of SUMO-1 expression in response to hypoxia: Direct interaction with HIF-1alpha in adult mouse brain and heart in vivo. FEBS Lett. 2004;569:293–300. doi: 10.1016/j.febslet.2004.05.079. [DOI] [PubMed] [Google Scholar]
- 196.Agbor TA, Cheong A, Comerford KM, Scholz CC, Bruning U, Clarke A, Cummins EP, Cagney G, Taylor CT. Small ubiquitin-related modifier (SUMO)-1 promotes glycolysis in hypoxia. J Biol Chem. 2011;286:4718–4726. doi: 10.1074/jbc.M110.115931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Antico Arciuch VG, Tedesco L, Fuertes M, Arzt E. Role of RSUME in inflammation and cancer. FEBS Lett. 2015;589:3330–3335. doi: 10.1016/j.febslet.2015.07.048. [DOI] [PubMed] [Google Scholar]
- 198.Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, Holsboer F, Arzt E. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell. 2007;131:309–323. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 199.Xu Y, Zuo Y, Zhang H, Kang X, Yue F, Yi Z, Liu M, Yeh ET, Chen G, Cheng J. Induction of SENP1 in endothelial cells contributes to hypoxia-driven VEGF expression and angio-genesis. J Biol Chem. 2010;285:36682–36688. doi: 10.1074/jbc.M110.164236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Childs BG, Baker DJ, Kirkland JL, Campisi J, van Deursen JM. Senescence and apoptosis: Dueling or complementary cell fates. EMBO Rep. 2014;15:1139–1153. doi: 10.15252/embr.201439245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Davalos AR, Coppe JP, Campisi J, Desprez PY. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 2010;29:273–283. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, Zhao Z, Thapar V, Joyce JA, Krizhanovsky V, et al. Non-cell-autonomous tumor suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ivanschitz L, Takahashi Y, Jollivet F, Ayrault O, Le Bras M, de Thé H. PML IV/ARF interaction enhances p53 SUMO-1 conjugation, activation, and senescence. Proc Natl Acad Sci USA. 2015;112:14278–14283. doi: 10.1073/pnas.1507540112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Neyret-Kahn H, Benhamed M, Ye T, Le Gras S, Cossec JC, Lapaquette P, Bischof O, Ouspenskaia M, Dasso M, Seeler J, et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013;23:1563–1579. doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jiang Z, Fan Q, Zhang Z, Zou Y, Cai R, Wang Q, Zuo Y, Cheng J. SENP1 deficiency promotes ER stress-induced apoptosis by increasing XBP1 SUMOylation. Cell Cycle. 2012;11:1118–1122. doi: 10.4161/cc.11.6.19529. [DOI] [PubMed] [Google Scholar]
- 206.Xia W, Tian H, Cai X, Kong H, Fu W, Xing W, Wang Y, Zou M, Hu Y, Xu D. Inhibition of SUMO-specific protease 1 induces apoptosis of astroglioma cells by regulating NF-κB/Akt pathways. Gene. 2016;595:175–179. doi: 10.1016/j.gene.2016.09.040. [DOI] [PubMed] [Google Scholar]
- 207.Sudharsan R, Azuma Y. The SUMO ligase PIAS1 regulates UV-induced apoptosis by recruiting Daxx to SUMOylated foci. J Cell Sci. 2012;125:5819–5829. doi: 10.1242/jcs.110825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhou Y, Ji C, Cao M, Guo M, Huang W, Ni W, Meng L, Yang H, Wei JF. Inhibitors targeting the SUMOylation pathway: A patent review 2012–2015 (Review) Int J Mol Med. 2018;41:3–12. doi: 10.3892/ijmm.2017.3231. [DOI] [PubMed] [Google Scholar]
- 209.Scott DE, Bayly AR, Abell C, Skidmore J. Small molecules, big targets: Drug discovery faces the protein-protein interaction challenge. Nat Rev Drug Discov. 2016;15:533–550. doi: 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]