Abstract
Mutations in MYBPC3, the gene encoding cardiac myosin binding protein C (cMyBP-C), are a major cause of hypertrophic cardiomyopathy (HCM). While most mutations encode premature stop codons, missense mutations causing single amino acid substitutions are also common. Here we investigated effects of a single proline for alanine substitution at amino acid 31 (A31P) in the C0 domain of cMyBP-C, which was identified as a natural cause of HCM in cats. Results using recombinant proteins showed that the mutation disrupted C0 structure, altered sensitivity to trypsin digestion, and reduced recognition by an antibody that preferentially recognizes N-terminal domains of cMyBP-C. Western blots detecting A31P cMyBP-C in myocardium of cats heterozygous for the mutation showed a reduced amount of A31P mutant protein relative to wild-type cMyBP-C, but the total amount of cMyBP-C was not different in myocardium from cats with or without the A31P mutation indicating altered rates of synthesis/degradation of A31P cMyBP-C. Also, the mutant A31P cMyBP-C was properly localized in cardiac sarcomeres. These results indicate that reduced protein expression (haploinsufficiency) cannot account for effects of the A31P cMyBP-C mutation and instead suggest that the A31P mutation causes HCM through a poison polypeptide mechanism that disrupts cMyBP-C or myocyte function.
Keywords: cMyBP-C, hypertrophic cardiomyopathy, missense mutation, animal models of cardiac disease
Introduction
Hypertrophic cardiomyopathy (HCM) is the most common genetic cause of cardiomyopathy and is estimated to affect 1 in 500 people [1-3]. Hundreds of mutations in genes encoding sarcomeric proteins are thought to cause HCM [3]. However, the majority of mutations occur in two genes, MYH7 and MYBPC3. MYH7 encodes the β-myosin heavy chain, the primary force generating protein of cardiac muscle, while MYBPC3 encodes cardiac myosin binding protein C (cMyBP-C), an essential regulatory protein that modulates the force and speed of cardiac contraction. Although the majority of mutations in MYH7 cause single amino acid substitutions, most MYBPC3 mutations are non-sense or frameshift mutations that encode premature termination codons and are thus predicted to cause early termination of cMyBP-C. However, truncated cMyBP-C proteins have not been detected in myocardium from HCM patients [4-6], most likely because cell quality control mechanisms either efficiently degrade mRNAs that contain premature termination codons or because truncated or misfolded proteins are rapidly degraded thus preventing their accumulation [4, 7, 8]. The apparent lack of expression of mutant truncated proteins combined with observations that the total amount of cMyBP-C protein is reduced in some patients with HCM has led to the hypothesis that haploinsufficiency causes sub-stoichiometric amounts of cMyBP-C within the sarcomere which impair contractile function and cause disease. Because reduced cMyBP-C protein expression was also found in myocardium of some patients with cMyBP-C missense mutations that cause single amino acid substitutions [5], both truncation and missense mutations could cause disease through a common mechanism of reduced cMyBP-C expression. In support of this idea, partial extraction of MyBP-C from cardiac or skeletal myocytes increased calcium sensitivity of force [9] and increased shortening velocity in a dose dependent matter [10], suggesting that insufficient amounts of cMyBP-C could disrupt muscle function and thereby trigger disease. However, the idea that haploinsufficiency can account for effects of cMyBP-C mutations has been challenged [6] and other mechanisms are possible that can account for deleterious effects of cMyBP-C missense mutations. For instance, missense mutations in cMyBP-C could directly disrupt the function of cMyBP-C by acting as poison polypeptides or by causing protein misfolding that impairs protein function. Protein misfolding could also affect cMyBP-C protein stability and abundance and/or impact overall cell homeostasis by increasing the burden on protein quality control clearance mechanisms [7, 8]. Given the complex basis of HCM where hundreds of different cMyBP-C sequence variants are believed causative for the disease, it is likely that both haploinsufficiency and poison polypeptide mechanisms will be relevant depending on the individual mutation.
To investigate the cellular mechanisms by which a missense mutation in cMyBP-C leads to HCM, we used a naturally occurring feline model of HCM caused by a single amino acid substitution in cMyBP-C. The missense mutation, a proline for alanine substitution at amino acid 31 in the C0 domain of cMyBP-C (A31P, Figure 1A,B), was first identified as a cause of HCM in a research colony of domestic cats (Maine Coon/mixed breed) with inherited HCM [11]. HCM is the most common cause of heart failure in cats and, similar to HCM in people, is characterized by abnormal regional or global thickening of the left ventricle, left ventricle outflow tract obstruction including systolic anterior motion of the mitral valve (SAM), and sudden cardiac death [12-14]. Since its original description, the A31P allele has been identified with high frequency in outbred Maine Coon cat populations throughout the world where homozygous inheritance of the A31P allele is associated with an increased odds ratio for HCM in cats [15-17]. Because inheritance of the A31P allele was associated with reduced cMyBP-C protein in the original report of this mutation [11], we sought to determine whether cMyBP-C protein structure was altered by the A31P mutation, whether the A31P mutation affects localization of cMyBP-C within the sarcomere, and to quantify cMyBP-C expression in a larger population of cats heterozygous and homozygous for the A31P allele. Circular dichroism analysis demonstrated that the mutation disrupted the structure of C0. Consistent with altered structure, recombinant proteins with the A31P mutation showed altered susceptibility to trypsin digestion. Western blots using an antibody that preferentially recognizes N-terminal domains of cMyBP-C including C0 demonstrated diminished epitope recognition for proteins containing the A31P mutation in vitro, as well as in cardiomyocytes from cats with the A31P mutation. The total amount of cMyBP-C was found similar in cats with or without the A31P mutation and immunohistochemistry revealed proper incorporation of the mutant A31P protein into sarcomeres. However, the amount of A31P mutant protein was relatively low compared to normal cMyBP-C in the myocardium of cats heterozygous for the A31P mutation, suggesting altered protein synthesis or stability of A31P cMyBP-C. Therefore, our results suggest that the A31P mutation alters cMyBP-C structure, potentially leading to altered protein stability and function, rather than causing disease through haploinsufficiency.
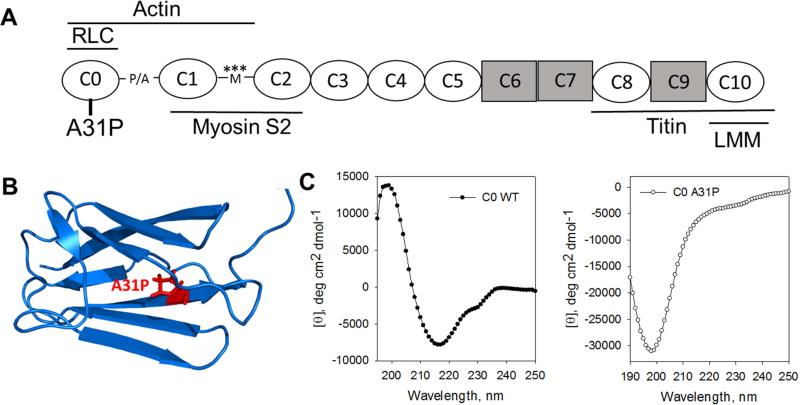
Figure 1.
A. Cartoon depicting the domain structure of cMyBP-C. cMyBP-C consists of 8 immunoglobulin-like domains (ovals) and 3 fibronectin type III-like domains (rectangles) along with an unstructured proline-alanine rich linker sequence (P/A) between C0 and C1 and the regulatory M-domain (M) between C1 and C2 with protein kinase phosphorylation sites indicated by asterisks. The A31P mutation is located in C0, the first immunoglobulin domain unique to the cardiac isoform of MyBP-C. The positions of binding sites for other sarcomeric proteins are indicated by lines above or below the domains. B. Ribbon diagram of C0 domain structure (PDB 2K1M) showing the location of the A31P mutation. C. Circular dichroism spectra (mean residue ellipticity versus wavelength) of C0-WT (left) and C0-A31P (right) showing that C0-WT is high in β-sheet content while C0-A31P showed a spectrum consistent with disordered proteins.
Materials and methods
Recombinant protein expression and purification
Recombinant proteins encoding the C0 domain of feline cMyBP-C or domains C0-C2 (inclusive of C0, the proline-alanine rich region, C1, the M-domain and C2 (Figure 1A)) were expressed with or without the A31P mutation (C0-WT, C0-A31P, C0C2-WT and C0C2-A31P, respectively). Cat cDNAs were cloned by RT-PCR using mRNA isolated from cat myocardium. Cloned cDNA sequences were inserted into the pQE2 expression vector (Qiagen) immediately downstream of a His6-tag affinity tag that was appended to the N-terminus of coding sequences to facilitate purification of recombinant proteins. The A31P mutation was introduced into cloned cDNA sequences using site-directed mutagenesis and the substitution confirmed by DNA sequencing. Recombinant proteins were expressed using a bacterial expression system in M15 cells as described previously [18]. Briefly, proteins were purified under non-denaturing conditions using a Ni-NTA affinity column (Qiagen) according to the manufacturer's directions and eluted in a buffer containing (in mM): 250 imidazole, 25 Tris-HCl, 200 NaCl, 1 2-Mercaptoethanol, 0.1 PMSF and 1 μg/ml pepstatin A, pH 7.5. Fractions that contained protein were pooled and supplemented with 1:200 EDTA-free Halt protease inhibitor cocktail (Thermo Scientific). C0C2 was exchanged into buffer ‘A’ (in mM: 50 Tris-HCl, 50 NaCl, 25 imidazole, 0.001 pepstatin A, 0.1 PMSF, 1 EDTA. pH 8.0 and further purified using a SO3− cation exchange column. C0C2 was eluted using a NaCl gradient increasing from 230-365 mM. Halt protease inhibitor cocktail (Thermo Scientific) was added to fractions that contained proteins. Coomassie-stained SDS-PAGE was used to determine the appropriate fractions to pool.
Circular dichroism (CD)
WT and A31P C0 proteins used for circular dichroism (CD) were eluted from a Ni-NTA affinity column as described above. Halt protease cocktail was replaced by 1 mM Pefabloc and 0.05% NaN3. Subsequently, the recombinant proteins were transferred to CD buffer (in mM: 20 Na-phosphate, pH 7.4, 100 (NH4)2SO4, 0.2 EGTA, 0.2 DTT) using PD10 desalting columns (GE Healthcare). C0 concentration was determined by measuring their difference spectra in 6M guanidine-HCl as described [19]. The CD spectra of C0 at 0.1 mg/ml were measured between 250-190 nm at 10°C using an Aviv model 400 spectropolarimeter (Lakewood, NJ) in 1 mm cuvettes.
Trypsin digestion
The influence of the A31P mutation on the susceptibility of the N-terminal domains of cMyBP-C to digestion by trypsin was determined by incubating recombinant C0 or C0C2 proteins with and without the A31P mutation with trypsin for increasing amounts of time (0 min - 3 hr). Trypsin was dissolved in phosphate buffer saline (PBS) (3.3 μg/ml; porcine pancreas, Sigma-Aldrich) and incubated with recombinant proteins at a 1:500 trypsin:protein mass ratio at 30°C for each desired time, when reactions were stopped by addition of Laemmli sample buffer after which samples were heated to 95°C for 5 min. Proteins were separated by SDS-PAGE (15% polyacrylamide) and visualized with Coomassie stain.
Animals
Care and handling of all animals was in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals using protocols approved by the Institutional Animal Care and Use Committee (IACUC) at University of California, Davis. Cats were obtained from a research colony of Maine Coon/mixed breed cats with heritable hypertrophic cardiomyopathy (HCM) [11, 14]. All cats were genotyped for the A31P cMyBP-C mutation using PCR amplification of the coding sequence containing the mutation (forward primer 5’-AAGAAGCCAGTCTCAGCCT-3’ and reverse primer 5’-CTTGCCCTTGAACCACTT-3’) followed by direct sequencing of PCR product [11]. HCM disease status was determined based on echocardiographic images [14], where HCM affected status was defined by diastolic regional or global wall thickness >6 mm in the left ventricular free wall and/or the interventricular septum [13, 14, 20]. Cardiac tissue was obtained from cats that were euthanized due to heart failure, systemic disease such as renal failure, or as a study end point. A summary describing genotype and phenotype of all animals used in this study is provided in Table 1.
Table 1.
Clinical characteristics of cats
| A31P Genotype | HCM Phenotype | N | Age (yr) | M/F | IVSd (mm) | LVPWd (mm) |
|---|---|---|---|---|---|---|
| WT | Unaffected | 6 | 10.0 ± 0.6 | 4/2 | 4.9 ± 0.2 | 5.3 ± 0.2 |
| Affected | 4 | 10.6 ± 0.8 | 2/2 | 6.1 ± 0.3 | 6.7 ± 0.2 | |
| HT | Unaffected | 4 | 9.8 ± 0.8 | 1/4 | 4.5 ± 0.1 | 4.6 ± 0.3 |
| Affected | 8 | 12.1 ± 1.1 | 3/5 | 6.7 ± 0.3 | 6.5 ± 0.3 | |
| HO | Unaffected | 1 | 3.0 ±0 | 1/0 | 5.8 ±0 | 5.2 ±0 |
| Affected | 4 | 5.3 ± 2.3 | 1/3 | 7.8 ± 1.0 | 6.5 ± 1.0 | |
WT, wild-type; HT, heterozygous, HO, homozygous; IVSd, interventricular septal diameter in diastole; LVPWd, diastolic left ventricular posterior wall diameter in diastole.
cMyBP-C protein expression in feline myocardium
Hearts were explanted from euthanized cats, rinsed in ice-cold Ringer's solution (in mM: 100 NaCl, 24 NaHCO3, 2.5 KCl, 1 MgSO4, 1 Na2HPO4, 1 CaCl2), and dissected into sections of left ventricle (LV), right ventricle (RV), interventricular septum (IVS), and right atrium (RA). All samples were subsequently snap frozen in liquid nitrogen and stored at −80°C until used.
For analysis of cMyBP-C protein content, myocardial tissue was homogenized using a Polytron homogenizer in an ice-cold relaxing solution containing (in mM) 5.95 Na2ATP, 6.04 MgCl2, 2.0 EGTA, 139.6 KCl, 10.0 imidazole. Subsequently, homogenates were treated with 10% trichloroacetic acid [21] and dissolved in a 2x urea-thiourea sample buffer containing (in M): 8 urea, 2 thiourea, 0.05 trizma base, 0.075 DTT, 3% SDS, 0.05% Bromophenol Blue [22]. Proteins were separated on SDS-PAGE (10% polyacrylamide) and transferred to nitrocellulose membranes (Biorad) for detection of proteins by western blot. Two antibodies were used to quantify total amounts of cMyBP-C: a rabbit polyclonal antibody that preferentially recognizes epitopes near the N-terminus of cMyBP-C (C-pro, [23, 24] 1:10,000) and a commercially available goat polyclonal antibody raised against internal domains of cMyBP-C (K-16, 1:200, Santa Cruz Biotechnology). An additional rabbit polyclonal antibody was raised against a short peptide sequence containing the A31P substitution to specifically detect mutant A31P cMyBP-C (rabbit polyclonal, 1:1,000, Invitrogen). All blots were probed for α-actinin (Sigma-Aldrich A7811, 1:2,000 in combination with the C-pro and A31P antibody or Santa Cruz Biotechnologies sc-15335, 1:2,000 in combination with the K16 antibody) to correct for protein loading. Bands were visualized using enhanced chemiluminescence (ECL,Thermo Scientific) and imaged with a G:Box gel doc system (Syngene). ECL signal intensity was analyzed with GeneTools software (Syngene). Samples from each cat were analyzed on 3-5 separate western blots for each antibody, after which cMyBP-C content was determined by averaging the obtained cMyBP-C or A31P signals corrected to α-actinin for each animal.
Immunohistochemistry
Cardiomyocyte homogenates were obtained by mechanical disruption of frozen cardiac tissue in relaxing buffer as described above and then fixed in 4.44% formaldehyde in phosphate buffered saline (PBS) for a minimum of 2 hr at 4°C with agitation. Fixed cardiomyocytes were pelleted by centrifugation at 3000 × g and incubated with buffer containing 20 mM glycine to quench the fixative. Cardiomyocytes were then incubated with 5% normal goat serum (NGS) to block non-specific antibody binding. Primary antibodies against cMyBP-C (C-pro, 1:6,000 [23]) or the A31P mutation (A31P, 1:200) were diluted in Tris buffered saline plus 0.05% Tween 20 (TBS-T) and 5% NGS and added to myocytes for incubation overnight at 4°C with agitation. Each of the primary antibodies was incubated simultaneously with an antibody against α-actinin (Sigma-Aldrich A7811, 1:500). After washing cardiomyocytes with TBS-T, myocytes were incubated with species appropriate secondary antibodies (anti-rabbit AlexaFluor 488 for C-pro and A31P and anti-mouse AlexaFluor 568 for α-actinin, both diluted 1:10,000, Invitrogen) for 1 hr at room temperature with agitation. Following additional rinses in fresh TBS-T, myocytes were pelleted and mounted in a drop of antifade (Prolong® Gold antifade with DAPI, Invitrogen) on a microscope slide. A coverslip was applied and the antifade was allowed to harden. Cells were imaged using a DeltaVision Deconvolution microscope and an Olympus 100X/ 1.40, Plan Apo, IX70 objective. Image stacks were deconvolved using softWoRx® version 1.1.0 (GE Healthcare). The images were false colored in ImageJ [25] with C-pro or A31P cMyBP-C shown in green and α-actinin in red.
Results
The A31P mutation alters structure of cMyBP-C N-terminal domains
Meurs et al. [11] identified the A31P mutation in cardiac myosin binding protein C (cMyBP-C) as a cause of HCM in a research colony of Maine Coon domestic cats. Computational algorithms performed in their study predicted that the A31P mutation disrupts the structure of the N-terminus of cMyBP-C [11]. Here we directly tested that prediction by expressing the A31P mutation in recombinant feline C0 protein as well as in a longer protein, C0C2, consisting of the C0 through C2 domains of feline cMyBP-C (Figure 1A,B). We assessed structure of the wild-type and mutant proteins using three independent techniques: circular dichroism (CD), antibody epitope recognition on western blots, and susceptibility of protein degradation by trypsin digestion.
Figure 1C shows circular dichroism (CD) spectra of the C0 domain with and without the A31P mutation after normalization for protein concentration. The C0-WT spectrum has a minimum at 217 nm and maximum at 199 nm typical for β-structural proteins, while the spectrum for C0-A31P has very low ellipticity between 190 and 210 nm with a minimum at 199 nm that is typical for disordered proteins [26]. The difference between the CD spectra for C0-WT and mutant C0-A31P thus showed that the A31P mutation reduced β-sheet content of the protein and caused disruption of the structure of the C0 domain.
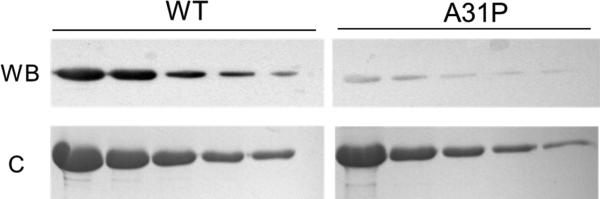
To determine if the A31P mutation altered antibody recognition of epitopes in C0 we made use of a polyclonal antibody raised against full-length cMyBP-C that preferentially recognizes epitopes within the C0 domain (C-pro [23, 24]). Figure 2 shows western blots (WB) and Coomassie (C) stained gels loaded with increasing amounts of C0C2-WT and C0C2-A31P. Results showed that despite comparable loading of C0C2-WT and C0C2-A31P protein, the C-pro antibody signal was reduced on blots of C0C2-A31P compared to C0C2-WT. That is, the C-pro antibody readily detected as little as 0.03 μg of C0C2-WT, while reduced signal was obtained from a much greater amount (0.5 μg) of C0C2-A31P loaded on the same blot. These results indicate that the A31P mutation alters antibody epitope sequence recognition.
Figure 2.
The A31P mutation reduces antibody epitope recognition of C0. Top: Western blot analysis (WB) of recombinant WT and A31P C0C2 protein (0.5 μg to 0.03 μg per lane) were probed with a polyclonal antibody against cMyBP-C (C-pro). Less signal was detected from the C0C2-A31P protein. Western blot compared to C0C2-WT despite comparable amounts of protein loaded as demonstrated by Coomassie blue staining of duplicate gels (bottom).
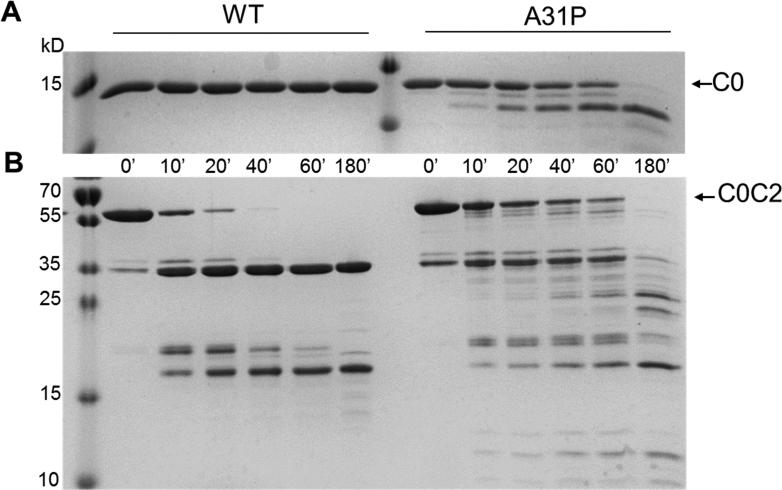
Because misfolded cMyBP-C may be subject to enhanced degradation and clearance by cell quality control mechanisms [7, 8] we evaluated whether recombinant proteins containing the A31P substitution showed increased susceptibility to limited trypsin proteolysis. Figure 3A shows a comparison of C0-WT and C0-A31P after trypsin digestion. Whereas C0-WT was completely intact even after 3 hr of incubation with trypsin, the mutant C0-A31P protein was rapidly degraded with cleavage products appearing as soon as 10 min after trypsin incubation. The major cleavage product was an approximately 12 kD fragment. Trypsin digestion of the longer C0C2 protein (Figure 3B) also showed an altered pattern of degradation. Whereas C0C2-WT was rapidly cleaved to an approximately 35 kD fragment that was stable and persisted for up to 3 hr (the length of the experiment), C0C2-A31P was degraded into multiple cleavage products and the 35 kD fragment was completely degraded by 180 min. Together these altered proteolysis patterns suggest that the A31P mutation not only alters the conformation of the C0 domain, but that the A31P mutation also affects stability of adjacent downstream domains of cMyBP-C.
Figure 3.
Limited trypsin digestion of WT or A31P C0 and C0C2. A. C0-WT was resistant to cleavage by trypsin for up to 180 min whereas C0-A31P was readily cleaved by trypsin after only 10 min as evident by the appearance of an ~12 kD cleavage fragment. Time of protein incubation with trypsin is indicated below each lane. B. Trypsin digestion of C0C2-WT or C0C2-A31P. The ~12 kD product was again apparent after 10 min of incubation with trypsin. However, whereas the full length C0C2-WT band disappeared between 40-60 min post incubation with trypsin, full length C0C2-A31P was still present at 60 min. In addition, several digestion products sized ~18-32 KD appeared when C0C2-A31P was digested but not during the digestion of C0C2-WT.
Mutant A31P-cMyBP-C is expressed in cardiac myocytes and is properly incorporated into cardiac sarcomeres
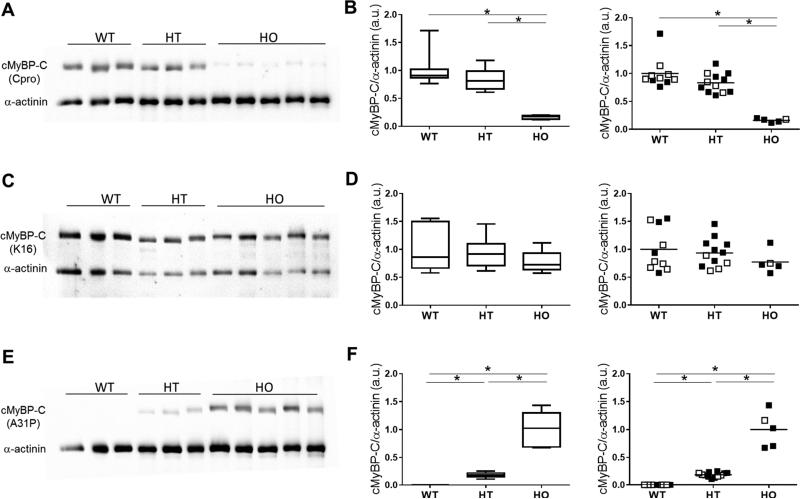
To determine in vivo consequences of altered protein folding due to the A31P mutation, we next investigated native cMyBP-C content in myocardium from cats where the A31P mutation occurs naturally [11, 14]. Cardiac tissue from cats heterozygous or homozygous for the A31P mutation was analyzed for cMyBP-C protein content and compared to cMyBP-C protein in WT cats (i.e., in control cats without the A31P mutation) using western blots probed with antibodies to different epitopes on cMyBP-C (Figure 4). As shown in Figure 4A,B, western blots probed with the polyclonal C-pro antibody showed an apparent reduction of cMyBP-C in cats homozygous for the A31P mutation compared to wild-type cats, but no difference in cMyBP-C content was detected in cats heterozygous for A31P cMyBP-C compared to wild-type cats. However, because the C-pro polyclonal antibody showed a diminished ability to recognize recombinant cMyBP-C with the A31P mutation (Figure 2), we next probed duplicate blots with the K16 antibody (Santa Cruz) that recognizes epitopes within the central domains of cMyBP-C, which are distant from epitopes affected by the A31P mutation in the C0 domain. In this case, the K16 antibody (Figure 4C,D) revealed similar amounts of cMyBP-C content in homozygous A31P myocardium compared to both WT and heterozygous myocardium, indicating that the total amount of cMyBP-C protein content was not significantly different in cats with or without the A31P cMyBP-C. A comparison of cMyBP-C expression in cats affected by HCM and unaffected cats (Figure 4D) showed no correlation between amount of cMyBP-C expressed and disease status.
Figure 4.
cMyBP-C protein expression in myocardium from wild-type (WT), heterozygous (HT) and homozygous (HO) cats determined using western blot analysis. A. Representative western blots probed with the C-pro antibody that primarily recognizes the C0 domain of cMyBP-C. The positions of cMyBP-C and α-actinin (loading control) are indicated. B. Left panel: Box-and-whiskers plot (median, 25-75 percentile box and min-max whiskers) summarizing data from western blots probed using the C-pro antibody with signal normalized to α-actinin. Right panel: The same data shown at left but plotted to show the mean of each group and disease status. Filled symbols, HCM affected cats; Open symbols, unaffected cats. C. Representative western blots probed with the K16 antibody (Santa Cruz) that recognizes internal regions of cMyBP-C. D. Left panel: Summary data from western blots probed using the K16 antibody with signal normalized to α-actinin. Right panel: The same data shown at left but plotted to show the mean of each group and disease status. E. Representative western blots probed with an antibody specific for the A31P substitution. F. Left panel: Summary data from western blots probed using the A31P antibody with signal normalized to α-actinin. Right panel: The same data shown at left but plotted to show the mean of each group and disease status.
We next measured amounts of mutant A31P cMyBP-C in myocardium from cats heterozygous and homozygous for the A31P mutation by using an antibody specific for A31P cMyBP-C. Representative western blots and their summary quantification are shown in Figure 4E,F. As expected, the antibody raised against the A31P sequence did not detect cMyBP-C in wild-type cats that do not express A31P cMyBP-C. However, the mutant A31P protein was readily detected in cardiac samples from cats heterozygous and homozygous for the A31P cMyBP-C mutation. As expected based on allelic dose, i.e., protein expression from one mutant allele in heterozygous cats versus two mutant alleles in homozygous cats, A31P protein content was lower in heterozygous myocardial samples than in homozygous samples. However, instead of the expected relative ~2 fold decrease in heterozygous compared to homozygous myocardium, A31P cMyBP-C was about 5 fold lower in heterozygous cats (Figure 4E,F). Because the total amount of cMyBP-C in heterozygous or homozygous cats was comparable to wild-type cats (Figure 4C,D), these results suggest that the mutant A31P protein makes up significantly less than the expected 50% (1:1 ratio) of the total cMyBP-C protein pool in heterozygous cats. This conclusion is also supported by results from western blots (Figure 4A,B) using the C-pro antibody which showed similar cMyBP-C protein content in heterozygous myocardium compared to wild-type myocardium, indicating that most of the cMyBP-C in heterozygous hearts is the wild-type cMyBP-C that is efficiently recognized by the C-pro antibody. Thus, the mutant A31P cMyBP-C contributes only a small proportion to the total cellular pool of cMyBP-C in myocardium from heterozygous cats.
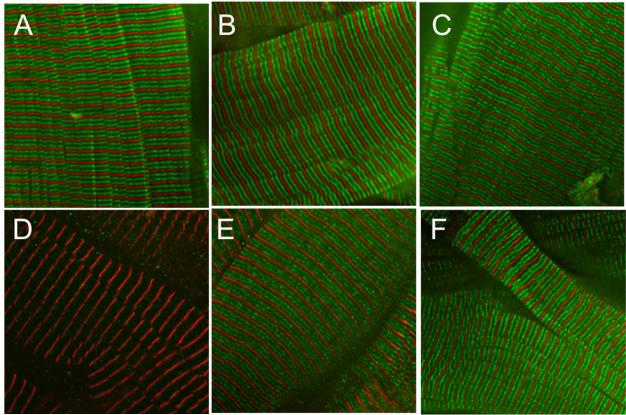
To determine whether the mutant A31P cMyBP-C is properly localized in cardiac sarcomeres, we imaged myocytes that were immunofluorescently labeled with antibodies directed against cMyBP-C to compare the pattern of cMyBP-C localization in wild-type cats and cats with the A31P mutation. Immunofluorescence staining with the C-pro antibody (Figure 5A-C) revealed the normal doublet pattern of cMyBP-C localization in myocytes that arises from the characteristic position of cMyBP-C in 7-9 transverse stripes within the C-zone of each half sarcomere [27-29]. At this magnification the individual cMyBP-C stripes cannot be resolved and instead appear merged as two green fluorescent stripes within each sarcomere. As shown in Figure 5A-C, cMyBP-C doublets were readily apparent in myocytes from all cats indicating proper localization of cMyBP-C, including in myocytes from homozygous cats that exclusively express the mutant A31P cMyBP-C. The ability of the C-pro polyclonal antibody to reveal cMyBP-C localization in A31P homozygous myocytes despite its diminished recognition of mutant epitopes in C0 in western blots presumably reflects the persistence of other antigenic epitopes not affected by the A31P mutation, such as those that result from the three dimensional structure of the native protein as it occurs within the sarcomere. To determine whether mutant A31P cMyBP-C was also incorporated along with wild-type cMyBP-C into sarcomeres of heterozygous cats that express both wild-type cMyBP-C and A31P cMyBP-C, we labeled myocytes with the antibody specific for A31P cMyBP-C. Figure 5D-F shows that staining with the A31P antibody revealed the doublet pattern of localization in both A31P heterozygous and A31P homozygous myocytes but not in wild-type myocytes that do not express the mutant A31P cMyBP-C. Collectively these results suggest that both mutant A31P protein and WT cMyBP-C are properly incorporated into cardiac sarcomeres from A31P mutant cats.
Figure 5.
Immunofluorescence localization of cMyBP-C in cat myocardium. Top: Immunofluorescence staining using the C-pro antibody showed the expected doublet pattern of cMyBP-C localization (green) in myocardium from wild-type (A), A31P heterozygous (B), and A31P homozygous (C) cats. Staining for α-actinin (red) was used to label sarcomere Z-lines as a reference. Bottom: Immunofluorescence localization using the A31P antibody showed that mutant A31P cMyBP-C is also properly incorporated into sarcomeres as demonstrated by the characteristic doublet pattern of cMyBP-C localization (green) in myocardium from heterozygous (E) and homozygous (F) cats but not wild-type (D) cats that lack A31P cMyBP-C. α-actinin localization (red) was used to label sarcomere Z-lines as a reference.
Discussion
Results from this study establish that the A31P missense mutation in cMyBP-C disrupts the structure of C0, the first immunoglobulin-like domain of cMyBP-C, causing altered protein folding, altered epitope recognition, and altered proteolytic susceptibility of recombinant proteins containing the A31P mutation. Because mutant A31P cMyBP-C expressed in cat myocardium also showed reduced epitope recognition by the C-pro antibody and the mutant A31P cMyBP-C was decreased in abundance relative to wild-type cMyBP-C in cats heterozygous for the A31P mutation, it is likely that the structure and stability of the native protein is similarly altered in vivo. However, despite altered or misfolded protein structure, we show that total cMyBP-C protein content was not different in cats with or without the A31P mutation and that the mutant protein is properly incorporated into cardiac sarcomeres along with wild-type cMyBP-C protein. Proper incorporation into sarcomeres was also true for cats homozygous for the A31P allele that exclusively express the mutant protein. Thus, haploinsufficiency, i.e. reduced expression of cMyBP-C, is not a likely mechanism whereby the A31P mutation causes HCM in A31P homozygous cats [5, 11]. Instead, our results provide new evidence for an alternative hypothesis that the A31P mutation generates a poison polypeptide whereby the normal function of cMyBP-C is either directly impaired or whereby the mutant A31P protein interferes with myocyte homeostasis, e.g. by placing a burden on protein quality control systems in the cell.
Our finding that A31P cMyBP-C does not reduce total cMyBP-C expression differs from the initial findings of Meurs et al. [11] who first identified the A31P cMyBP-C substitution in the cats from the founding research colony of Maine Coon/mixed breed domestic cats studied here. However, we show that the C-pro antibody used by Meurs et al. [11] unexpectedly exhibits reduced ability to recognize the mutant A31P cMyBP-C. The sensitivity of the C-pro antibody to a single epitope is surprising because this polyclonal antibody was raised against full-length native cMyBP-C purified from rat heart [23] and is thus expected to react with numerous epitopes throughout the entire length of cMyBP-C. However, recent epitope mapping experiments revealed that the C-pro polyclonal antibody preferentially reacts with epitopes in C0 and to a lesser extent with other N-terminal domains of cMyBP-C such as the regulatory M-domain [24]. The significance of the strong antigenic nature of C0 is currently unknown, but the N-terminus of cMyBP-C has been identified in the development of immunogenic myocarditis suggesting that it is especially provocative to immune cells [30, 31]. N-terminal proteolytic cleavage products of cMyBP-C that include C0 and that are released during cardiac stress have also been shown to be cardio-toxic [30, 32]. Regardless, reduced epitope recognition by the C-pro antibody for the A31P C0 domain as shown here (Figures 2 and 4) could cause cMyBP-C content to appear lower than actual protein content in cats carrying the A31P mutation as initially reported by Meurs et al. [11].
Our data provide new evidence that strongly argues against haploinsufficiency as being the mechanism whereby the A31P mutation causes disease in homozygous cats. Instead our results provide support for the alternative hypothesis that the A31P mutation generates a poison polypeptide whose presence disrupts myocyte function. While the precise function of the C0 domain of cMyBP-C is not yet known, C0 is unique to cardiac isoforms of MyBP-C suggesting a specialized function in heart muscle contraction. In vitro binding experiments showed that C0 can interact with the regulatory light chains (RLC) of myosin [33] and with actin of thin filaments [34-36]. Disruption of C0 structure by the A31P substitution as described here (Figure 1) could impact these interactions potentially leading to abnormal regulation of contraction by cMyBP-C. Alternatively, because the A31P mutation affects the folded structure of C0, increased degradation of misfolded A31P cMyBP-C could lead to secondary cellular stress effects that contribute to or are causative in disease pathogenesis. For instance, recombinant truncated cMyBP-C proteins were preferred substrates that overwhelmed the ubiquitin-proteasome system (UPS) when they were overexpressed in neonatal rat cardiomyocytes [7] and mice with a truncation mutation in cMyBP-C showed UPS impairment while cMyBP-C knockout mice did not [8]. The relative reduction (less than the expected 50% protein content) of A31P cMyBP-C shown here for cats heterozygous for the A31P mutation (Figure 4E,F) suggests that the synthesis and/or stability and degradation of the mutant A31P cMyBP-C protein differs from the wild-type protein, potentially increasing burden on cell quality control mechanisms especially in cats homozygous for the A31P mutation. Allelic imbalances that presumably reflect differences in protein synthesis or stability have also recently been reported in human myocardial samples where the fraction of mutant protein compared to wild-type protein for mutations in several different sarcomeric proteins ranged from 30-84% [6].
The finding that the A31P mutant protein makes up only a small percentage of the total pool of cMyBP-C in cats heterozygous for the mutation provides an explanation for why heterozygous A31P cats are mostly unaffected whereas A31P homozygous cats are at elevated risk for HCM in the general cat population [15-17]. That is, heterozygous cats have near normal amounts of wild-type cMyBP-C (Figure 4A,B) and express only a small amount of A31P mutant cMyBP-C (Figure 4E,F), which is apparently not sufficient to significantly elevate risk for HCM. To determine if variability in A31P mutant protein content could potentially account for some of the variability in disease penetrance, for instance if cats that expressed a higher percentage of the mutant A31P protein were HCM affected while those that expressed lower amounts of the mutant A31P protein were not, we assessed whether A31P mutant protein content correlated to disease status. However, we observed little variability in A31P mutant protein content in heterozygous cats and detected no correlation between A31P mutant protein content and affected status (Figure 4F).
The apparent relative up-regulation of protein production from the wild-type allele (Figure 4) to compensate for reduced A31P mutant protein in heterozygous cats is consistent with findings from cMyBP-C knockout mouse models showing that heterozygous knockout mice have near normal or only modest reductions in cMyBP-C protein content despite complete loss of protein production from the knockout allele [23, 37]. Thus, it is only when the entire cellular pool of cMyBP-C consists of the mutant A31P protein that homozygous cats are at increased risk for HCM. However, the susceptibility of A31P homozygous cats to HCM [15-17] contrasts with the more typical autosomal dominant pattern of inheritance for HCM mutations in human patients where inheritance of a single variant allele (heterozygous genotype) is generally sufficient to cause disease [3]. Homozygous inheritance of the A31P substitution in cats may be more similar to homozygous inheritance of mutant alleles or compound inheritance patterns in people where gene dosage effects have been reported with disease being more severe or occurring with an earlier onset in homozygous patients or in patients that carry 2 or more sequence variants in different genes [3, 38]. In addition, disease penetrance in humans is highly variable, i.e. identical mutations cause severe HCM in some individuals while others remain symptom free (genotype positive/phenotype negative individuals), implying that genetic or environmental modifiers greatly influence disease development. The relatively high prevalence of HCM affected cats in our research colony that either do not have the A31P mutation (WT cats) or that are heterozygous for A31P suggests that additional genetic modifiers or causative mutations are also likely present in this research population of cats and is consistent with observations that the A31P mutation is not the sole cause of HCM in outbred Maine Coon cat populations [15-17, 39] and with the multiple genetic causes of HCM in people.
In summary, our results demonstrate that the naturally occurring cMyBP-C A31P missense mutation does not significantly reduce total cMyBP-C protein content even in homozygous cats that exclusively express the A31P mutant protein. However, in heterozygous cats that express both the wild-type and mutant proteins the ratio of mutant A31P protein to wild-type protein is less than 1:1. These results indicate that either the mutant protein is degraded more rapidly than the wild-type protein or that its synthesis is reduced. In either event, relatively low expressed amounts of A31P cMyBP-C combined with overtly normal total cMyBP-C content provides an explanation for the low incidence of HCM reported in A31P heterozygous cats [15-17]. By contrast, homozygous A31P cats express only the mutant A31P protein which correlates with higher disease penetrance in homozygous cats [15-17]. Because we demonstrate that the A31P mutation alters structure, epitope recognition, and proteolytic susceptibility of the C0 domain, our results provide strong support for the hypothesis that the A31P protein functions as a poison polypeptide which impairs function of cMyBP-C and/or otherwise affects protein quality control pathways in A31P homozygous cats.
Acknowledgments
This work was supported in part by NIH R21 HL093603 and R01 HL080367 to SPH. SJvD is supported in part by a postdoctoral fellowship from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 2.Elliott P, McKenna WJ. Lancet. 2004;363:1881–1891. doi: 10.1016/S0140-6736(04)16358-7. [DOI] [PubMed] [Google Scholar]
- 3.Maron BJ, Maron MS. The Lancet. 2013;381:242–255. doi: 10.1016/S0140-6736(12)60397-3. [DOI] [PubMed] [Google Scholar]
- 4.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ, van der Velden J. Circulation. 2009;119:1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 5.Marston S, Copeland O, Jacques A, Livesey K, Tsang V, McKenna WJ, Jalilzadeh S, Carballo S, Redwood C, Watkins H. Circ. Res. 2009;105:219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 6.Helms AS, Davis FM, Coleman D, Bartolone SN, Glazier AA, Pagani F, Yob JM, Sadayappan S, Pedersen E, Lyons R, Westfall MV, Jones R, Russell MW, Day SM. Circ. Cardiovasc. Genet. 2014;7:434–443. doi: 10.1161/CIRCGENETICS.113.000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarikas A, Carrier L, Schenke C, Doll D, Flavigny J, Lindenberg KS, Eschenhagen T, Zolk O. Cardiovasc. Res. 2005;66:33–44. doi: 10.1016/j.cardiores.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Schlossarek S, Englmann DR, Sultan KR, Sauer M, Eschenhagen T, Carrier L. Basic Res. Cardiol. 2012;107:235. doi: 10.1007/s00395-011-0235-3. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann PA, Hartzell HC, Moss RL. J. Gen. Physiol. 1991;97:1141–1163. doi: 10.1085/jgp.97.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann PA, Greaser ML, Moss RL, Physiol J. 1991;439:701–715. doi: 10.1113/jphysiol.1991.sp018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meurs KM, Sanchez X, David RM, Bowles NE, Towbin JA, Reiser PJ, Kittleson JA, Munro MJ, Dryburgh K, Macdonald KA, Kittleson MD. Hum. Mol. Genet. 2005;14:3587–3593. doi: 10.1093/hmg/ddi386. [DOI] [PubMed] [Google Scholar]
- 12.Payne J, Luis Fuentes V, Boswood A, Connolly D, Koffas H, Brodbelt D, Small Anim J. Pract. 2010;51:540–547. doi: 10.1111/j.1748-5827.2010.00989.x. [DOI] [PubMed] [Google Scholar]
- 13.Fox PR, Liu SK, Maron BJ. Circulation. 1995;92:2645–2651. doi: 10.1161/01.cir.92.9.2645. [DOI] [PubMed] [Google Scholar]
- 14.Kittleson MD, Meurs KM, Munro MJ, Kittleson JA, Liu SK, Pion PD, Towbin JA. Circulation. 1999;99:3172–3180. doi: 10.1161/01.cir.99.24.3172. [DOI] [PubMed] [Google Scholar]
- 15.Godiksen MT, Granstrom S, Koch J, Christiansen M. Acta Vet. Scand. 2011;53:7. doi: 10.1186/1751-0147-53-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longeri M, Ferrari P, Knafelz P, Mezzelani A, Marabotti A, Milanesi L, Pertica G, Polli M, Brambilla PG, Kittleson M, Lyons LA, Porciello F. J. Vet. Intern. Med. 2013;27:275–285. doi: 10.1111/jvim.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mary J, Chetboul V, Sampedrano CC, Abitbol M, Gouni V, Trehiou-Sechi E, Tissier R, Queney G, Pouchelon JL, Thomas A. J. Vet. Cardiol. 2010;12:155–161. doi: 10.1016/j.jvc.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Razumova MV, Bezold KL, Tu AY, Regnier M, Harris SP. J. Gen. Physiol. 2008;132:575–585. doi: 10.1085/jgp.200810013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostyukova AS, Hitchcock-Degregori SE, Greenfield NJ. J. Mol. Biol. 2007;372:608–618. doi: 10.1016/j.jmb.2007.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norsworthy GD, Foshee Grace S, Crystal MA, Tilley LP. Wiley-Blackwell. 2010:1072. [Google Scholar]
- 21.Zaremba R, Merkus D, Hamdani N, Lamers JM, Paulus WJ, Dos Remedios C, Duncker DJ, Stienen GJ, van der Velden J. Proteomics Clin. Appl. 2007;1:1285–1290. doi: 10.1002/prca.200600891. [DOI] [PubMed] [Google Scholar]
- 22.Fritz JD, Swartz DR, Greaser ML. Anal. Biochem. 1989;180:205–210. doi: 10.1016/0003-2697(89)90116-4. [DOI] [PubMed] [Google Scholar]
- 23.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL. Circ. Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 24.Lee K, Harris SP, Sadayappan S, Craig R. J. Mol. Biol. 2015;427:274–286. doi: 10.1016/j.jmb.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasband WS, ImageJ US. National Institutes of Health; Bethesda, Maryland, USA: 1997-2015. http://imagej.nih.gov/ij/ [Google Scholar]
- 26.Greenfield NJ. Nat. Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Offer G, Moos C, Starr R. J. Mol. Biol. 1973;74:653–376. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 28.Bennett PM, Furst DO, Gautel M. Rev. Physiol. Biochem. Pharmacol. 1999;138:203–234. doi: 10.1007/BFb0119628. [DOI] [PubMed] [Google Scholar]
- 29.Luther PK, Winkler H, T. K., Zoghbi ME, Craig R, Padrón R, Squire J.m., Liu J. Proc Natl. Acad. Sci. 2011;108:11423–11428. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Razzaque MA, Gupta M, Osinska H, Gulick J, Blaxall BC, Robbins J. Circ. Res. 2013;113:553–561. doi: 10.1161/CIRCRESAHA.113.301225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch TL, Sadayappan S. Proteomics Clin. Appl. 2014;8:569–577. doi: 10.1002/prca.201400011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witayavanitkul N, Ait Mou Y, Kuster DW, Khairallah RJ, Sarkey J, Govindan S, Chen X, Ge Y, Rajan S, Wieczorek DF, Irving T, Westfall MV, de Tombe PP, Sadayappan S. J. Biol. Chem. 2014;289:8818–8827. doi: 10.1074/jbc.M113.541128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratti J, Rostkova E, Gautel M, Pfuhl M. J. Biol. Chem. 2011;286:12650–12658. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y, Kwan AH, Trewhella J, Jeffries CM. J. Mol. Biol. 2011;413:908–913. doi: 10.1016/j.jmb.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 35.Orlova A, Galkin VE, Jeffries CM, Egelman EH, Trewhella J. J. Mol. Biol. 2011;412:379–386. doi: 10.1016/j.jmb.2011.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitten AE, Jeffries CM, Harris SP, Trewhella J. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrier L, Knoll R, Vignier N, Keller DI, Bausero P, Prudhon B, Isnard R, Ambroisine ML, Fiszman M, Ross J, Jr., Schwartz K, Chien KR. Cardiovasc. Res. 2004;63:293–304. doi: 10.1016/j.cardiores.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Carrier L, Mearini G, Stathopoulou K, Cuello F. Gene. 2015;573:188–197. doi: 10.1016/j.gene.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kittleson MD, Meurs K, Munro M. J. Vet. Intern. Med. 2010;24:1242–1243. doi: 10.1111/j.1939-1676.2010.0614.x. author reply 1244. [DOI] [PubMed] [Google Scholar]