Abstract
Improved in vitro models are needed to better understand cancer progression and bridge the gap between in vitro proof-of-concept studies, in vivo validation, and clinical application. Multicellular tumor spheroids (MCTS) are a popular method for three-dimensional (3D) cell culture, because they capture some aspects of the dimensionality, cell–cell contact, and cell–matrix interactions seen in vivo. Many approaches exist to create MCTS from cell lines, and they have been used to study tumor cell invasion, growth, and how cells respond to drugs in physiologically relevant 3D microenvironments. However, there are several discrepancies in the observations made of cell behaviors when comparing between MCTS formation methods. To resolve these inconsistencies, we created and compared the behavior of breast, prostate, and ovarian cancer cells across three MCTS formation methods: in polyNIPAAM gels, in microwells, or in suspension culture. These methods formed MCTS via proliferation from single cells or passive aggregation, and therefore showed differential reliance on genes important for cell–cell or cell–matrix interactions. We also found that the MCTS formation method dictated drug sensitivity, where MCTS formed over longer periods of time via clonal growth were more resistant to treatment. Toward clinical application, we compared an ovarian cancer cell line MCTS formed in polyNIPAAM with cells from patient-derived malignant ascites. The method that relied on clonal growth (PolyNIPAAM gel) was more time and cost intensive, but yielded MCTS that were uniformly spherical, and exhibited the most reproducible drug responses. Conversely, MCTS methods that relied on aggregation were faster, but yielded MCTS with grapelike, lobular structures. These three MCTS formation methods differed in culture time requirements and complexity, and had distinct drug response profiles, suggesting the choice of MCTS formation method should be carefully chosen based on the application required.
Keywords: polyNIPAAM, 3D, ovarian cancer, breast cancer, prostate cancer, mafosfamide
Graphical abstract
INTRODUCTION
Two-dimensional (2D) cell culture monolayers are traditionally used to study cancer biology, gain insight into mechanisms of cancer progression, and screen for novel anticancer treatments. However, traditional 2D cell culture platforms used in drug screening do not accurately model cancer tumors,1–3 as they lack cell heterogeneity, differentiated phenotypes, extracellular matrix (ECM) architecture, and drug resistance seen in vivo.4–7 In vitro model systems that can better mimic the in vivo microenvironment could improve therapeutics discovery by reducing false positives that later fail in preclinical or clinical trials. Three-dimensional (3D) cell culture models can recapitulate some of the physiological behavior of in vivo tumors,5,8 including nonuniform distribution of oxygen and nutrients1 and multicellular drug resistance.9 Moreover, gene expression profiles of 3D cell culture models are often closer to gene expression profiles of tumors than 2D models.4,10,11 Therefore, 3D models can better reflect some of the complexity of tumors that impact biological responses, such as drug resistance, angiogenesis, cell migration, cell invasion, and metastasis.5,12
The most common approach for 3D cell culture models is the multicellular tumor spheroids (MCTS) model,13,14 which can be applied to established cancer cell lines or samples isolated from patients.15 MCTS are characterized by a 3D structure with strong cell–cell and cell-ECM interactions, which make them more similar to in vivo tumors over 2D cell monolayers.2,7 Unlike 2D models, MCTS models can also recapitulate the oxygen and nutrient gradients seen in vivo.16–18 Because of these features, MCTS have been used to study tumor biology, and for high-throughput drug screening.9,12,15,19 MCTS are used in development and testing of several therapeutic approaches like chemotherapy, radiotherapy, and immunotherapy.19–21 Additionally, many researchers have found increased resistance in MCTS compared to cells grown on tissue culture polystyrene (TCPS).22–27 Despite these attributes, several factors limit the use of MCTS in drug discovery and development, including the need to (1) produce high quantities of uniform MCTS,28,29 (2) predictably control MCTS size for consistent results,16 and (3) technical adaptation of existing screening assays to 3D MCTS models.28,29 Addressing these limitations could make 3D high-throughput drug screening methods more suitable to commercial development.
Current MCTS formation techniques include stationary or rotating culture incubation systems. Stationary formation systems include the liquid-overlay technique, the hanging drop method, and the suspension method in nonadherent plates.1,30 The advantages of these systems are low cost, easy operation, and production of reproducible MCTS.31 However, they yield very few MCTS, which limits their adaptation to large scale studies. Rotating formation systems include the spinner flask method, the gyratory rotation system, and the roller tube system.1,30 The advantages of these techniques include massive production and control of culture conditions,31 while the biggest limitations are the infrastructure, and high quantities of medium and drugs required.30 Although these methods each have their advantages, the existence of multiple MCTS formation methods introduces variability in MCTS size, shape, and formation ability.14,32,33 To the best of our knowledge, this is the first work that compares MCTS formation methods in terms of the implications in gene expression and drug response, which may improve future studies across many applications, such as high-throughput drug screening.
In this study, we explored how three MCTS formation methods changed gene expression and drug response in breast, prostate, and ovarian cancer cells. These methods required either aggregation followed by compaction or clonal proliferation, probably leading to different subpopulations of cells in the resulting MCTS. Although each method produced similar size MCTS, different methods lead to different expression profiles of cell–cell and cell–matrix interaction genes. Moreover, the MCTS formation methods used dictated drug sensitivity, suggesting significant implications to improve future drug screening studies.
MATERIALS AND METHODS
Cell Culture
All cell lines were cultured at 37 °C and with 5% CO2 unless otherwise noted. Cell culture supplies were purchased from Thermo Fisher Scientific (Waltham, MA) with the exception of bovine insulin (Sigma-Aldrich, St. Louis, MO). The cell lines AU565, BT549, BT474, HCC 1395, HCC 1419, HCC 1428, HCC 1806, HCC 1954, HCC 202, HCC 38, LNCaPcol, HCC 70, PC-3, SKOV-3, and ZR-75-1 were cultured in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/ streptomycin (Pen/Strep). OVCAR-3 cells were cultured in RPMI with 20% FBS, 1% Pen/Strep, and 0.01 mg/mL bovine insulin. Hs578T, MCF7, MDA-MB-468, MDA-MB-231, MDA-MB-231 BoM (bone tropic), MDA-MB-231 BrM2a (brain tropic), MDA-MB-231 LM2 (lung tropic), and SkBr3 were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS, 1% Pen/Strep. MDA-MB-175 was cultured in Leibovitz’s L-15 medium with 10% FBS and 1% Pen/Strep without supplemental CO2, and MDA-MB-134 and MDA-MB-361 were cultured in Leibovitz’s L-15 medium with 20% FBS and 1% Pen/Strep without supplemental CO2. The media described for their respective cell lines will be referred to as “routine culture medium” and any modifications will be noted where applicable. Highly metastatic MDA-MB-231 variants isolated from in vivo selection (BoM, BrM2a, and LM2) were kindly provided by Joan Massagué,34–36 MDA-MB-231 by Sallie Smith Schneider, BT549, MCF7, and SkBr3 by Shannon Hughes, PC3 by Evan Keller, and LNCaPcol derived through serial passage on collagen type I from LNCaP by Michael Long.37 SKOV-3 and OVCAR-3 were purchased from American Type Culture Collection (ATCC), and all others were kindly provided by Mario Niepel.
PolyNIPAAM MCTS
Single cells were suspended at 100 cells/ µL in polyNIPAAM (Cosmo Bio USA, Carlsbad, CA) on ice and gelled as 150 µL volumes at 37 °C for 5 min. Routine culture medium or routine culture medium +100 ng/mL epidermal growth factor (EGF, R&D Systems, Minneapolis, MN) was added and medium was changed every 2–3 days until MCTS were collected at Day 14. MCTS were recovered from polyNIPAAM by replacing the cell culture medium with serum free medium at 4 °C and placing them on ice for 5 min. The dissolved gel was diluted in additional serum free medium and put in a conical tube to concentrate the MCTS via gravity sedimentation on ice for 30 min. Medium was removed, and the MCTS pellet was lysed for RT-PCR, or used for encapsulation in 3D hydrogels. MCTS were handled using cut pipet tips to minimize shear stress.
Microwell MCTS
Square pyramidal microwells (400 µm side-wall dimension) were fabricated as described previously38,39 or purchased (AggreWell, Stem Cell Technologies, Canada). For fabrication, master molds containing square-pyramidal pits were generated by anisotropic etching of 100 crystalline silicon in potassium hydroxide (KOH). Microwell surfaces for tissue culture were then generated from poly(dimethylsiloxane) (PDMS) using a two-stage replica molding process of the master mold as described previously.38,39 Microwells were arranged in a square array with no space between adjacent wells and placed in 6 or 12-well plates. To prepare microwells for cell seeding, microwell surfaces were UV sterilized and pretreated with 5% Pluronic F-127 (Sigma-Aldrich) for 30 min at room temperature and then washed twice with sterile water. Cells were distributed over microwell surfaces at concentrations of 1.03 × 105 cells/cm2 or 1.00 × 104 cells/cm2. After 24 h, MCTS were collected by shaking the plate gently to dislodge most of them, and gently aspirating medium and MCTS. MCTS solution was spun down at 400 rpm for 5 min. Medium was removed, and the MCTS pellet was lysed for RT-PCR, or encapsulated in 3D hydrogels. MCTS were handled using cut pipet tips to minimize shear stress.
Suspension MCTS
Single cells were seeded at 1.05 × 104 cells/ cm2, 1.05 × 103 cells/cm2, or 1.05 × 102 cells/cm2 in a 6-well flat ultralow attachment plate (Corning, Tewksbury, MA). After 3 days, MCTS were collected by aspiration of medium and MCTS. MCTS solution was spun down at 400 rpm for 5 min. Medium was removed, and the MCTS pellet was lysed for RT-PCR or encapsulated in 3D hydrogels. MCTS were handled using cut pipet tips to minimize shear stress.
Characterization of Gene Expression by RT-PCR
The expression of cell–cell adhesion molecules and ECM mRNA transcripts was measured by quantitative RT-PCR. Total RNA was isolated using the GenElute mammalian total RNA kit (Sigma-Aldrich) and 0.5 µg total RNA was reverse transcribed using the RevertAid reverse transcription system (Thermo). Ten nanograms cDNA was then amplified using 10 pmol of integrin-specific primers (Table S1) and the Maxima SYBR green master mix (Thermo) on a Rotor-Gene Q thermocycler (Qiagen, Valencia, CA) as follows: 50 °C for 2 min, 95 °C for 10 min followed by 45 cycles at 95 °C for 10 s, 58 °C for 30 s, and 72 °C for 30 s. Both β-actin and ribosomal protein S13 were included as reference genes to permit gene expression analysis using the 2−ΔΔCt method.40
PEG-Maleimide (PEG-MAL) Hydrogels
3D hydrogels were prepared with a 20 kDa 4-arm PEG-maleimide (PEG-MAL, Jenkem Technology, Plano, TX) at 10 wt % solution with 2 mM of cell adhesion peptide RGD (see the Supporting Information) and cross-linked at a 1:1 ratio with 1 kDa linear PEG-dithiol (Sigma-Aldrich) in sterile 2 mM triethanolamine (pH 7.4). Briefly, the MCTS pellet obtained from each method was resuspended in the PEG-RGD solution, and casting of hydrogels was done by mixing PEG-RGD-MCTS with the cross-linker at a ratio of 10:1. One microliter of the cross-linking solution was placed on the bottom of the plate, and then 9 µL of PEG-RGD-MCTS solution was added with vigorous mixing. Volumes of the hydrogels were limited to 10 µL to avoid oxygen and nutrient diffusion limitations. Gelation proceeded for 5 min41 at 37 °C to ensure complete polymerization before the addition of culture medium. MCTS from the three MCTS formation methods were transferred to 3D PEG-MAL hydrogels, by using similar seeding densities. PolyNIPAAM MCTS were encapsulated at a ratio of one 150 µL polyNIPAAM gel to nine 10 µL PEG-MAL hydrogels, microwell MCTS at a ratio of 4 cm2, 1 mL to nine 10 µL PEG-MAL hydrogels, and suspension MCTS at a ratio of 9 cm2, 3 mL to nine 10 µL PEG-MAL hydrogels. MCTS created via either polyNIPAAM, microwells or suspension were transferred with cut pipet tips to minimize shear stress.
Primary Ovarian Cancer Ascites Culture
Ascites samples were received from patients undergoing paracentesis at UMass Medical School (Worcester, MA), were transported to UMass Amherst (Amherst, MA) on the day of collection, and used immediately upon receipt. Samples were deidentified and were IRB exempt. Pathology reports are provided in Table S2. Either single cells or ovarian carcinoma ascites spheroids (OCAS) were recovered from patient samples. For single cells, the ascites fluid was centrifuged at 1,000 rpm for 10 min at 4 °C, and the supernatant was removed. Red blood cells were removed by resuspending the cell pellet in cold red blood cell lysis buffer (0.83% ammonium chloride, 0.1% potassium bicarbonate, and 0.0037% EDTA).42 The tube was rotated at room temperature for 10 min, cells spun down at 1,000 rpm for 10 min at 4 °C, and washed with PBS once prior to seeding on TCPS or in polyNIPAAM. For collection of OCAS directly from the patient sample, ascites fluid was filtered through a 40 µm mesh cell strainer. The retained OCAS were collected by using a cut pipet tip on the inverted cell strainer and encapsulated directly into polyNIPAAM or 3D PEG-MAL hydrogels. Supernatant from ascites pellets was stored at −80 °C and filtered through a 0.45 µm syringe filter prior to use as a culture medium without any additional supplements for single cell and OCAS polyNIPAAM culture.
Proliferation and Drug Screening
AU565, BT549, SKOV-3, and primary ovarian single cancer cells were seeded in RPMI with 5% FBS at 6250 cell/cm2. MCTS from the same cell lines formed in polyNIPAAM, microwells, and suspension were recovered and encapsulated in 3D PEG-MAL hydrogels. OCAS from primary ovarian cancer samples were collected and also encapsulated in 3D PEG-MAL hydrogels. Drugs were added after 24 h, and cells were incubated with drugs for 48 h. Cisplatin (Tocris Bioscience, United Kingdom), paclitaxel (MP Biomedicals, Santa Ana, CA), sorafenib (LC Laboratories, Woburn, MA), or mafosfamide (Niomech, Germany) were added in 10-fold serial dilutions at concentrations of 1 × 10−5 to 1 × 102 µM, and dimethyl sulfoxide (DMSO, Sigma-Aldrich) was used as a vehicle control. Viability was assayed after 48 h of incubation using the CellTiter Glo luminescent viability assay (Promega, Madison, WI). Luminescence values were read in a BioTek Synergy H1 plate reader (Winooski, VT), and GR5043 was calculated using GraphPad Prism v6.0h (La Jolla, CA) for each cell line and drug. Traditionally, the IC50, which is the concentration of drug required to kill half the population of living cells has been used to quantify drug response, but can lead to misleading results between studies44 because it does not take the number of cell divisions during the assay into consideration. We applied a new parameter for calculating drug response known as the GR50, which takes into account the growth rate of the cells, with GR50 being the dose where the cell growth rate is reduced by 50%.43 GR50 and proliferation results for the three MCTS formation methods are presented and discussed as relative to TCPS (fold change from the TCPS results).
MCTS Staining
MCTS in microwells plates at days 0 and 1, and from polyNIPAAM, microwells, and suspension encapsulated in 3D PEG-MAL hydrogels for 24 h or 3 days, were assessed for viability with live/dead staining (L3224, Thermo) according to the manufacturer’s instructions, and for proliferation via Ki67 immunofluorescence. For the Ki67 staining, samples were rinsed three times with PBS, fixed with 4% formaldehyde, permeabilized with Trisbuffered saline (TBS) containing 0.5% Triton X-100 (Promega), and blocked with AbDil (2 wt % bovine serum albumin (BSA) in TBS with 0.1% Triton X-100, TBS-T). Samples were incubated for 2 h at room temperature with the primary antibody (ab16667, 1:200-Abcam, UK), washed, and incubated with goat anti-Rabbit IgG (H+L) secondary antibody for 2 h (Alexa Fluor 647, 1:500, Promega). Cell nuclei were labeled with DAPI at 1:10000 (Thermo) for 5 min. Brightfield imaging was performed on a Zeiss Axio Observer Z1 (Carl Zeiss AG, Oberkochen, Germany), and fluorescence imaging on a Zeiss Spinning Disc Cell Observer SD (Zeiss).
Statistical Analysis and Correlations
Statistical analysis was performed with GraphPad Prism v6.0h. Data shown are the averages of the means from three separate biological replicates, and the error bars represent standard error (95% confidence level). Patient samples had only one biological replicate each. Statistical significance was evaluated by two-way analysis of variance (ANOVA), followed by Tukey’s post-test for pairwise comparisons. Spearman rank correlation is reported as ρ with significance (p) determined by a two-tailed t test. For both tests, p < 0.05 was considered statistically significant. p < 0.05 is marked with *, ≤ 0.01 with **, ≤ 0.001 with ***, and ≤0.0001 with ****, p ≥ 0.05 was considered not significant (“ns”).
Image Processing
ImageJ (NIH, Bethesda, MD) was used for diameter and circularity measurements of MCTS and cells, as well as compiling individual staining images. Most nonspherical MCTS had a 4-pointed star shape. In these cases, diameter was measured as the distance between two opposing points. In other cases, such as MCTS that were ellipsoid shaped rather than spherical, the longest dimension of the spheroid was taken as the diameter.
Information for “RGD synthesis” and “Gene Expression Clustering and PCA” can be found in Supporting Information and Methods.
RESULTS
PolyNIPAAM, Microwells, and Suspension Methods Generated Diverse MCTS across Cell Lines
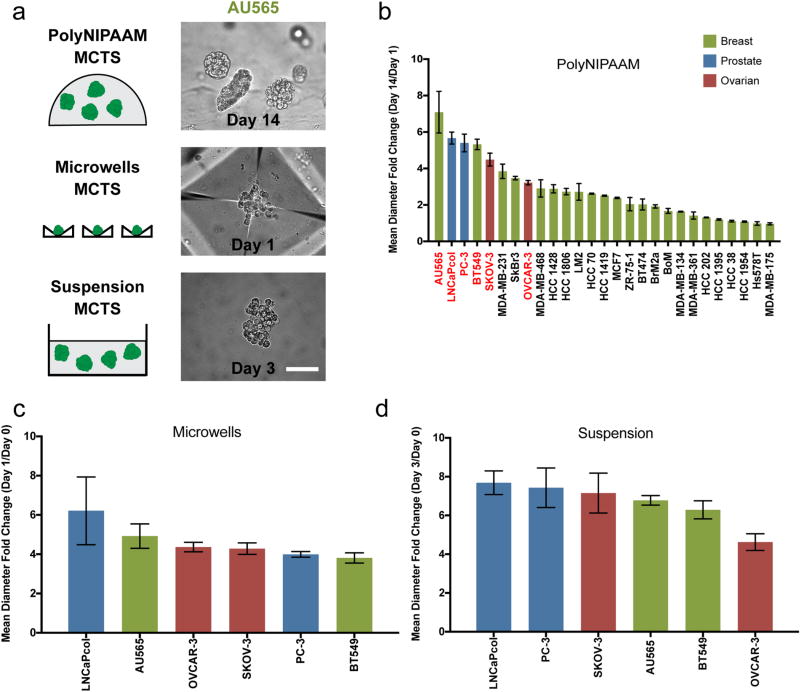
We screened the MCTS formation abilities of 23 breast, 2 ovarian, and 2 prostate cancer cell lines across three different MCTS methods: polyNIPAAM gels, microwells, and suspension, in nonadherent plates (Figure 1a). For all three culture methods, single cells were seeded, and after a certain culture time (14 days for the polyNIPAAM, 3 days for the suspension, and 1 day for the microwells), MCTS reached approximately 100 µm. In the polyNIPAAM method, many cell lines formed uniformly sized MCTS that had at least a 2-fold increase in mean diameter, with many reaching approximately 100 µm in diameter in 14 days (i.e., AU565, BT549, LNCaPcol, PC-3, SKOV-3, OVCAR-3, HCC 1419, HCC 1428, MCF7, MDA-MB-231, SkBr3, ZR-75-1, BT474). Some cell lines (i.e., HCC 1954, Hs578T) were incapable of growing into MCTS, and most of these cells died within the 14-day period (data not shown). Other cell lines formed few MCTS (i.e., HCC 1806, MDA-MB-468, MDA-MB-231 LM2, HCC 70), suggesting that only a small percentage of cells were capable of forming MCTS via this method (Figure 1b, Figure S1).
Figure 1.
PolyNIPAAM, microwells, and suspension methods generate diverse MCTS. (a) Schematic of methods used to form MCTS and representative images of AU565 MCTS formed by each method. Scale bar: 100 µm. (b) Mean diameter fold change of 23 breast (green), 2 prostate (blue), and 2 ovarian (red) cancer cell lines into MCTS in polyNIPAAM (cell lines highlighted in red were used in c and d) N ≥ 2. (c, d) Mean diameter fold change of 2 breast (green), 2 prostate (blue), and 2 ovarian (red) cancer cell lines into MCTS in microwells (c) and suspension (d) N ≥ 3. Mean diameter fold change shows how MCTS diameters changed over time. ANOVA followed by Tukey post-test was used to compare statistically significant differences across methods for each cell line: SKOV-3 (polyNIPAAM vs suspension*) and (microwells vs suspension*), and PC3 (microwells vs suspension**).
We observed many different morphologies across these MCTS, and they were reminiscent of work reported from the Bissell group that described four classes of spheroids that formed in 3D laminin-rich ECM (3D lrECM): round, mass, grape-like, and stellate.45 In polyNIPAAM, BT474 and MCF7 formed very compact MCTS, whereas SkBr3 and MDA-MB-468 formed grapelike MCTS. As for the cell lines that were stellate in 3D lrECM, BT549 formed compact MCTS, MDA-MB-231 formed large, loose MCTS, and Hs578T did not form MCTS in polyNIPAAM. Invasion or cell spreading reminiscent of the stellate morphology were never observed because there were no cell–matrix adhesion sites in the polyNIPAAM gel, and it was not cell-degradable. We did not examine any cell lines that were categorized by Bissell as round.
The polyNIPAAM method allowed growth of MCTS in more than half of the cell lines tested, independent of breast cancer clinical subtype (data not shown). A negative correlation was found between doubling times and MCTS size for all the 17 cancer cell lines for which doubling times are known46 (Figure S2a), indicating that faster growing cells might produce bigger MCTS in polyNIPAAM (ρ = −0.78, p = ***). EGF has been shown to increase spheroid size in various tumor cell lines,47–49 and many of the breast cancer cells lines used here overexpress EGFR.50 For this reason, we attempted to increase spheroid growth with supplemental EGF. We found that most cell lines formed bigger MCTS when grown with supplemental EGF, although some did not change (i.e., HCC 1428, HCC 1806, BrM2a, BoM, MDA-MB-134, MDA-MB-361, HC 1954, and Hs578T) and a few decreased in size, such as AU565, MDA-MB-231, and MDA-MB-468, which already make large MCTS (Figure S3). The data showed that EGF caused only a small size increase for cell lines that formed small MCTS, while it caused larger size increases in the majority of the cell lines that made larger MCTS even without EGF.
All cell lines that formed MCTS in the polyNIPAAM method were collected after 14 days of culture, and encapsulated into 3D PEG-MAL hydrogels. This was done to demonstrate that the MCTS were easily handled, and remained intact when transferred to another system. Viability was assessed after 3 days, and the majority of the MCTS were viable (Figure S4). There was no evidence of a necrotic core, but this was likely because the MCTS diameters were relatively small.
We next selected a subset of the cell lines that had at least a 2-fold increase in diameter in the polyNIPAAM method (red text in Figure 1c) to compare MCTS formation in microwells (Figure 1d) and suspension (Figure 1f) methods. In these aggregation methods, MCTS size increased with seeding cell density (Figure S5), and densities of 1.00 × 104 cells/cm2 (microwells) and 1.05 × 104 cells/cm2 (suspension) were used to achieve MCTS sizes between 80 and 150 µm (Figure S6). In the microwells, the cells slowly coalesced into MCTS for the first ~10 h, then the MCTS further compacted over the first day of culture (Figure S7). MCTS from microwells were both viable and proliferative (Figure S8). Among the six cell lines used for the microwells and suspension methods, there was no correlation between MCTS size and growth rate, likely because these methods are dependent upon cell aggregation rather than growth (Figure S2b, c). No correlation was found between MCTS size and doubling time with the polyNIPAAM method when only the 6 cell lines that were used in the other methods were taken into account (data not shown). Since MCTS in microwells compacted over time, circularity was found to increase, which eventually yielded more uniform MCTS. In contrast, circularity was preserved in polyNIPAAM throughout the growth process, and decreased in suspension (Figure S9). It is likely that MCTS in suspension became less compact over time because there were no solid surfaces that cells could contact. By contrast, cells in polyNIPAAM are confined in the gel, and cells in the microwells are in contact with the microwell surfaces. Together, these results demonstrated that MCTS can be formed with three different methods. Furthermore, the differences in culture times suggest that in the polyNIPAAM method single cells grew into MCTS (Figure 1 and Figure S2a), whereas in the microwells and suspension methods, MCTS formed as a result of aggregation, independent of proliferation (Figure S2b, c).
Fibronectin and Claudin 4 Expression Depend on MCTS Formation Method
Gene expression was quantified in breast, prostate, and ovarian cancer cell lines in the three MCTS formation methods and compared to basal gene levels in 2D TCPS culture. To select the relevant genes for this study, RNA-Seq data was analyzed from breast cancer cell lines (MDA-MB-231 and SkBr3) that were grown on TCPS, in polyNIPAAM for 14 days, or grown in polyNIPAAM and then dissociated and plated back onto TCPS (Figure S10a; GSE93562). Using Gene Set Enrichment Analysis (GSEA), we observed that cell surface receptor-linked signal transduction genes, including several integrins, were enriched on TCPS when compared to MCTS formed in polyNIPAAM for 14 days, whereas cell–cell adhesion genes, such as claudin 4, were enriched in cells grown into MCTS in polyNIPAAM over 14 days compared to TCPS (Figure S10b, c). From this data set, we selected a subset of cell adhesion genes including integrin subunits (α2 (ITGA2), β1 (ITGB1) and β4 (ITGB4)), cell–cell junction proteins (cadherins 3 (CDH3) and 5 (CDH5) and claudin 4 (CLDN4)), and the ECM protein, fibronectin (FN1), to examine in each MCTS formation method (abbreviations used in Figure 2a).
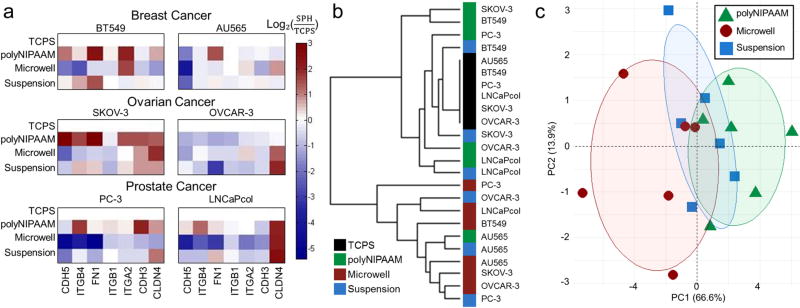
Figure 2.
MCTS method determines expression of cell–ECM and cell–cell adhesion genes. (a) RT-PCR of genes collected from 2 breast (AU565 and BT549), 2 prostate (PC3 and LNCaPcol), and 2 ovarian (SKOV-3 and OVCAR-3) cell lines reported as log2(fold change from TCPS). Genes are ordered based on expression clustering. Shades of red indicate gene upregulation compared to TCPS and shades of blue indicate gene downregulation compared to TCPS. (b) Dendrogram of RT-PCR data by platform in TCPS (black), in polyNIPAAM (green), in microwells (red), and in suspension (blue). (c) Principal component analysis (PCA) of gene expression by MCTS formation method. Ovals represent 0.5 probability for each group of polyNIPAAM (green triangle), microwells (red circle), and suspension (blue square), N ≥ 3.
We found that either claudin 4 or fibronectin were upregulated in all MCTS methods compared to TCPS (Figure 2a) with the few exceptions of OVCAR-3 in polyNIPAAM, and BT549 and PC-3 in microwells. Gene expression patterns varied with the time required for MCTS formation across the three methods. Fibronectin was downregulated for all the cell lines in microwells, while claudin 4 was upregulated in polyNIPAAM method for all the cells lines, except for AU565 and OVCAR-3. These gene expression changes were dependent on the cell line as well as the MCTS formation method. Variations in gene expression with MCTS formation method was confirmed by dendrogram clustering for all combinations of examined cell lines and methods, which revealed that TCPS and polyNIPAAM primarily clustered together, and the shorter methods of microwells and suspension also clustered together (Figure 2b). This was further confirmed by principal component analysis (PCA), which revealed that samples did not cluster by cell line or cancer type (data not shown), but rather that PC1 separated samples by method, with those that had the shortest and longest times of culture being the most distinct from one another (Figure 2c). Although all of these systems formed MCTS of similar size, the expression of cell contact genes varied across methods, which may have affected the compactness of MCTS. We thus hypothesized that MCTS that upregulated cell–cell contact genes and took longer times to form MCTS may be more resistant to first line therapies than 2D TCPS because of their compact morphology.
MCTS Growth Method Dictated Drug Response
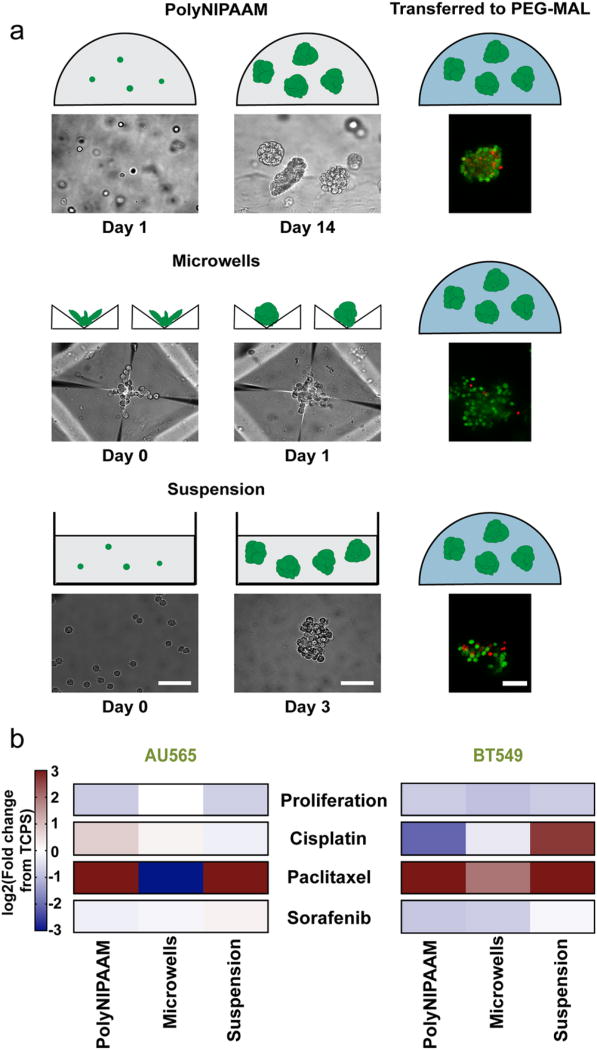
To test drug resistance, we selected two breast cancer cell lines, one triple negative (BT549) and one HER2-enriched (AU565) for drug screening experiments. These subtypes have poor prognosis and they are clinically treated with chemotherapeutic drugs.51 Breast cancer MCTS were created with the three methods, encapsulated in 3D PEG-MAL hydrogels, and their viability was verified with live/dead staining (Figure 3a). They were then treated with either the chemotherapeutic drugs cisplatin or paclitaxel, or the targeted drug sorafenib, which is a Raf kinase inhibitor.52 Transferring the MCTS to the 3D PEG-MAL hydrogels allowed us to test cell response to the drugs as a function of MCTS formation method, independent of the effects of the MCTS formation platform. The maximum MCTS diameter for these two cell lines was kept at 150 µm (Figure S6). In all cases, the MCTS were intact and viable 24 h after encapsulation in 3D PEG-MAL (Figure S11).
Figure 3.
MCTS method formation dictates drug response. (a) Schematic of MCTS formation by polyNIPAAM, microwells, and suspension followed by encapsulation in 10 wt % 3D PEG-MAL hydrogels. Representative brightfield and live/dead images of MCTS (green, live cells; red, dead cells) are shown under each. Scale bar: 100 µm. (b) Heatmap of proliferation and GR50 values of cisplatin, paclitaxel, and sorafenib in AU565 and BT549 cell lines. Values reported as log2(fold change from TCPS). Shades of red indicate higher drug resistance compared to TCPS, and shades of blue indicate higher drug sensitivity compared to TCPS, N ≥ 3.
MCTS proliferation was measured relative to TCPS as a baseline for cell growth across platforms. AU565 and BT549 cells proliferated less in all the three methods used to form MCTS compared to TCPS (Figure 3b). Next, drug response was determined by calculating the GR50 for MCTS formed by all three methods, as well as for each cell line grown on TCPS. Drug responses were reported as fold changes between GR50 values of each drug in individual 3D MCTS models and TCPS; a fold change of >1 means that the 3D MCTS model displayed greater drug resistance than TCPS. We found that the MCTS formation method dictated drug sensitivity. For example, AU565 cells were less sensitive to cisplatin in MCTS formed in polyNIPAAM compared to suspension and microwells (Figure 3b). However, the effect of formation method on drug response also varied between cell lines. For example, BT549 were more sensitive to cisplatin when they were formed in the polyNIPAAM method, and less sensitive when they were formed in the suspension method, while the opposite was true for AU565. We also found that drug response varied across drugs: AU565 MCTS cultured in microwells were significantly more sensitive to paclitaxel, while they were less sensitive to cisplatin and sorafenib. Interestingly, MCTS formed in all methods were slightly more sensitive to sorafenib than on TCPS, with the exception of AU565 MCTS formed in suspension. This may be because sorafenib works as a targeted agent rather than interfering with cell division. Overall, the microwell method sensitized MCTS to drugs, with the exception of the BT549 response to paclitaxel. However, MCTS created with the polyNIPAAM and suspension methods were more resistant to drug treatments, although the response was slightly more heterogeneous across conditions. We then hypothesized that MCTS formed in polyNIPAAM could show similar response to drugs as seen in in vivo tumors. To test this hypothesis, we compared drug responses of SKOV-3 MCTS and patient-derived ovarian carcinoma ascites.
SKOV-3 MCTS Modeled Primary Ovarian Cancer Drug Response
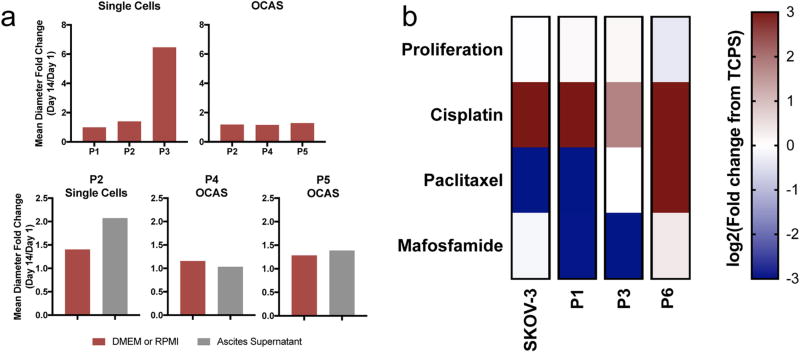
With the goal of discovering whether MCTS were able to recapitulate features of primary patient samples, we compared the drug responses of SKOV-3 MCTS with that of cells gathered from ascites of ovarian cancer patients. We seeded single cells and OCAS collected from patients in polyNIPAAM to determine if single cells could form MCTS and if OCAS seeding would increase in MCTS size. There was variation in growth for single cells between patients (P1, P2, and P3) and culture medium (Figure 4a). However, OCAS seeded in polyNIPAAM did not appreciably increase in size compared to single cells (Figure 4a). Because most samples were not able to form MCTS from single cells in our polyNIPAAM method, and ascites samples are often rich in OCAS, we collected the spheroids and encapsulated them directly into our 3D PEG-MAL hydrogel to capture native architecture, cell-cell contacts, and cell type heterogeneity for drug screening. These samples were compared to SKOV-3 MCTS, which were formed in polyNIPAAM before being transferred to 3D PEG-MAL hydrogels.
Figure 4.
MCTS formed in polyNIPAAM as a model for ovarian cancer drug discovery. (a) Formation of single cells (Patients 1–3) into MCTS and OCAS (Patients 2, 4, and 5) in polyNIPAAM under different medium conditions, N = 1. (b) Heatmap of proliferation and GR50 values of cisplatin, paclitaxel, and mafosfamide in SKOV-3 and patient samples (Patients 1, 3, and 6). SKOV-3 MCTS were grown in polyNIPAAM and encapsulated in 10 wt % 3D PEG-MAL hydrogels with RGD, N ≥ 3. Patients OCAS were encapsulated in 10 wt % 3D PEG-MAL hydrogels with RGD upon receipt, N = 1. Values reported as log2(fold change from TCPS). Shades of red indicate higher drug resistance compared to TCPS, and shades of blue indicate higher drug sensitivity compared to TCPS.
We chose to examine the response to cisplatin and paclitaxel, which are first line clinical therapeutics for ovarian cancer, and mafosfamide, a drug that has never been in clinical trials for ovarian cancer, but had promising results on TCPS.53,54 First, we found that the SKOV-3 MCTS grown in polyNIPAAM had similar responses to cisplatin and paclitaxel compared to the patient-derived OCAS (Figure 4b). To corroborate that patient-derived OCAS response was the same as what was seen in the actual clinical response, we compared our results with pathology reports of the patient samples (Table S2). Interestingly, OCAS collected from patients P1, P3 and P6, previously treated with platinum-based therapy (cisplatin or carboplatin), predicted cisplatin resistance where TCPS models did not (Figure 4b). The exception was high resistance to paclitaxel observed for P6, who had been previously treated with this drug, which potentially explains the observed resistance (Table S2). This suggests that OCAS from patients transferred directly into 3D PEG-MAL hydrogels may model drug response more accurately than TCPS. Surprisingly, mafosfamide, a drug that has never been used to treat ovarian cancer, was more effective relative to TCPS against P1 and P3 OCAS (Figure 4b). These data demonstrate that MCTS in polyNIPAAM could be a good model for the discovery of new drugs for the treatment of primary ovarian cancer.
DISCUSSION
To the best of our knowledge, a comparative analysis between MCTS formation methods has never been reported in a single study. Traditional models generate very large MCTS, generally created through the hanging drop method or suspension in nonadherent plates.22,25,55 These techniques often use just one large MCTS within a well of a 96-well plate, and are limited by the presence of a necrotic core,56 which affects drug response and assay readout. A system that can generate many smaller MCTS (to limit diffusion and hypoxia effects and allow encapsulation in many hydrogels), would be an improvement over current 3D drug screening models. Although diffusion limitations and hypoxia effects are an important part of recapitulating in vivo conditions, we eliminated this confounding aspect to isolate cell–cell and cell–ECM regulators of MCTS formation. Hypoxia causes changes in gene expression and metabolic pathways, and induces the expression of a class of proteins known as “hypoxia induced factors” (HIFs).57 In a study like this one, a hypoxic core would complicate interpretation of results by introducing variability in gene expression due to HIFs instead of culture method. The methods described here can generate a large number of small MCTS (between 50 and 150 µm) (Figure S6), which were easily recovered and transferred to other materials while maintaining high viability (Figure S11). Additionally, these MCTS provided many features of tumors, such as oxygen and nutrient diffusion limitations (but not to the extent that a necrotic core is present). Incorporation of the RGD peptide into the PEG-MAL hydrogel into which MCTS were transferred is also relevant to mimicking tumor features. It is required for the survival of adherent cells in a 3D environment, but it also allows adhesion of cells to the hydrogel, mimicking cell–ECM interactions, which is an important factor in cancer cell migration and invasion. We used 2 mM RGD in our PEG-MAL gel, which has been shown to promote 3D cell adhesion and spreading.41 The PEG-MAL hydrogel we have used in this study is low-cost, and it has the practical advantage of achieving gelation in 5 min,41 but it is not cell-degradable, which means that it does not mimic the breakdown of ECM by cancer cells. To recapitulate this effect, cell-degradable hydrogels could be used.58
MCTS have previously been used to study tumor biology and drug resistance. However, MCTS size, shape, and even the ability to form MCTS changes with the method and conditions. Piggott et al. formed MDA-MB-231 MCTS with ultralow adherence plates, whereas Iglesias et al. failed to do so with the same method,32,33 likely because of differences in seeding density. Casey et al. found that SKOV-3 cells did not form MCTS with the liquid overlay technique,59 whereas in this work we present SKOV-3 MCTS formation with all three methods (Figure 1). SkBr3s did not form MCTS when seeded onto soft agar,60 but formed MCTS in our polyNIPAAM method. Moreover, SkBr3s, MDA-MB-231s, and MDA-MB-468s did not form MCTS when seeded in suspension,61 but formed large, loose MCTS in our polyNIPAAM method (Figure 1b). Our polyNIPAAM results are in good agreement with work from the Bissell group using lrECM. For example, both works observe a mass phenotype in BT474 and MCF7, a grapelike appearance in AU565, SkBr3, and MDA-MB-468.45 These results stress the need to compare the behavior of cell lines across multiple MCTS formation methods.
Our data suggested that cells employ one of two possible mechanisms for MCTS formation: they either secrete their own local matrix to provide binding sites and structure, or they rely on cell–cell contacts. In MCTS formed in the microwells (briefest culture time), claudin 4 was upregulated, whereas those formed via polyNIPAAM or suspension (longer culture time), both claudin 4 and fibronectin were upregulated (Figure 2a). Immunofluorescence staining has shown that in free MCTS (a MCTS configuration with significant spacing between cells) fibronectin is distributed within the intercell space throughout the spheroid, while in compact MCTS fibronectin is found on the outer edge of the MCTS.62 This can be compared with our suspension and polyNIPAAM MCTS, respectively. On the other hand, knocking down claudin 4 reduces in vitro MCTS formation63 because it is essential for tight junctions. Interesting, claudin 4 is expressed in the majority of ovarian cancers,64 and also in our SKOV-3 and OVCAR-3 MCTS. Additionally, breast cancer cells grown into MCTS using the overlay method, similar to our suspension method, upregulated claudin 4 and several other cell–cell adhesion genes.65 Our results are largely cell-line dependent, without any correlation to cancer type.
We examined how drug response changed with MCTS formation methods. Changes in drug resistance compared to 2D TCPS were greater in MCTS formed in polyNIPAAM or suspension than the microwells method (Figure 3b). Decreased sensitivity of MCTS to drug treatment in a 3D model versus 2D screening agrees with other studies that have looked at the effect of dimensionality on drug response.22–27 Both breast cancer cell lines tested were slightly more sensitive to sorafenib in all the three methods compared to TCPS, with the exception of AU565 MCTS grown in suspension. This may be because sorafenib works as a targeted agent rather than interfering with cell divisions, as chemotherapeutic drugs do. We believe that the drug response observed in 3D drug screening is a result of the properties of the MCTS culture time and method. For example, MCTS formed in polyNIPAAM were more resistant to first line therapies because of their high proliferation and more compact morphology, obtained with longer culture times.
Finally, we compared our polyNIPAAM MCTS to patient-derived ovarian cancer ascites, because drug approval for ovarian cancer therapy has begun to stagnate. Only two ovarian cancer drugs have been approved by the U.S Food and Drug Administration (FDA) in the past 10 years.66 Therefore, there is a pressing need to more accurately model ovarian cancer in vitro to better identify effective treatments. In this study, OCAS from patient-derived samples were compared to an established ovarian cancer cell line formed in MCTS in polyNIPAAM. In most cases, the patient-derived and cancer cell line MCTS grown in polyNIPAAM responded similarly (Figure 4b). Our results suggest that the 3D methods presented here may show drug efficacy and drug resistance for an individual patient more accurately than TCPS. Interestingly, patient samples were more sensitive to mafosfamide compared to TCPS, a drug that has never been in clinical trials for ovarian cancer. This shows that selection of an MCTS method that mimics an in vivo environment can facilitate future drug discovery.
In ovarian cancer, the presence of ascites indicates disease progression and poor prognosis.67 The disseminated cells and OCAS in the peritoneum are those that become metastatic, and contribute to drug resistance and recurrence.68,69 Ascites fluid is enriched in cancer stem cells, which can form tumors in vivo.70 Therefore, patient-specific drug screening of the cells isolated from ascites would be extremely beneficial to make treatment decisions that could result in better patient outcomes. The clinical application of drug screening to ascites would be relatively easy because the fluid is in great excess in many patients. However, only few research groups have performed drug screening in ascites samples,56,71 and they have demonstrated that ascites responses mimic cell line data. Sensitivity to carboplatin and paclitaxel in ascites-derived cells treated in vitro mimicked the clinical chemosensitive or chemoresistant phenotype in each patient.71 Therefore, MCTS formation methods that can grow MCTS from patient derived ascites can be useful clinical tools.
Selection of a suitable 3D MCTS model is not straightforward. Drug response in 3D MCTS models depends on the cell line and drug of interest, as well as the MCTS formation method. The most accurate 3D MCTS model would produce MCTS similar to the in vivo MCTS, as well as exhibit similar gene expression. The choice of MCTS formation method may also be based on whether a clonal growth method (polyNIPAAM) or an aggregation method (microwells, suspension) is desired. Finally, the time requirements of each method are different, which is a practical factor to be considered for drug screening assays.
CONCLUSIONS
We applied three different MCTS formation methods: polyNIPAAM gels, microwells, and suspension culture across 27 cancer cell lines (breast, ovarian, and prostate) to investigate the implications of these methods on gene expression and drug response. The MCTS formation methods depend on either proliferation from single cells, which requires longer culture times or passive aggregation, which requires shorter culture times. The cell proliferation method (PolyNIPAAM gel) produced MCTS with a uniform spherical shape, although the aggregation methods (microwells and suspension) produced MCTS with grapelike structure. To form MCTS with these methods, cells rely on either the production of ECM (fibronectin) or the robust expression of cell–cell contact genes (claudin 4). MCTS that were formed with the three methods were then used for drug screening in a 3D hydrogel platform. We found that drug sensitivity was dependent on MCTS formation technique. To address the need for improved patient treatments, we compared the drug response of MCTS of an ovarian cancer cell line grown in polyNIPAAM to OCAS obtained directly from patients and we demonstrated that our 3D drug testing platform is a good model for patient-derived samples.
Supplementary Material
Acknowledgments
The authors thank Lauren Jansen and Thomas McCarthy for peptide synthesis and Dr. Sarah Perry and Dr. Lila Gierasch for use of equipment. Also, Hong Bing (Amy) Chen, Chien-I (Mike) Chang, and Cristian Fraioli at the Cancer Center Tissue and Tumor Bank at UMass Medical School for primary ovarian cancer ascites. S.R.P. was supported by the Pew Charitable Trusts and by a faculty development award from Barry and Afsaneh Siadat. This work was also funded by the National Institute of Health (NIH) New Innovator award (DP2CA186573-01) and a National Science Foundation (NSF) CAREER grant (DMR-1454806) to S.R.P. L.E.B. was partially supported by National Research Service Award (T32 GM008515) from NIH. A.D.S. was supported by a NSF Graduate Research Fellowship (1451512). E.A.B. was partially supported by National Research Service Award T32 GM008515 from the National Institutes of Health.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsbiomaterials.7b00069.
The authors declare no competing financial interest.
References
- 1.Santini MT, Rainaldi G. Three-dimensional spheroid model in tumor biology. Pathobiology. 1999;67(3):148–157. doi: 10.1159/000028065. [DOI] [PubMed] [Google Scholar]
- 2.Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014;12(4):207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smalley KS, Lioni M, Herlyn M. Life isn’t flat: Taking cancer biology to the next dimension. In Vitro Cell. Dev. Biol. Anim. 2006;42(8–9):242–247. doi: 10.1290/0604027.1. [DOI] [PubMed] [Google Scholar]
- 4.Shiras A, Bhosale A, Patekar A, Shepal V, Shastry P. Differential expression of CD44 (S) and variant isoforms v3, v10 in three-dimensional cultures of mouse melanoma cell lines. Clin. Exp. Metastasis. 2002;19(5):445–455. doi: 10.1023/a:1016305611858. [DOI] [PubMed] [Google Scholar]
- 5.Baker BM, Chen CS. Deconstructing the third dimension-how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012;125(13):3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perche F, Torchilin VP. Cancer cell spheroids as a model to evaluate chemotherapy protocols. Cancer Biol. Ther. 2012;13(12):1205–1213. doi: 10.4161/cbt.21353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa EC, Moreira AF, de Melo-Diogo D, Gaspar VM, Carvalho MP, Correia IJ. 3D tumor spheroids: an overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016;34(8):1427–1441. doi: 10.1016/j.biotechadv.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Sutherland RM, McCredie JA, Inch WR. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J. Natl. Cancer Inst. 1971;46(1):113–120. [PubMed] [Google Scholar]
- 9.Desoize B, Jardillier J-C. Multicellular resistance: a paradigm for clinical resistance? Crit. Rev. Oncol. Hematol. 2000;36(2):193–207. doi: 10.1016/s1040-8428(00)00086-x. [DOI] [PubMed] [Google Scholar]
- 10.L’Esperance S, Bachvarova M, Tetu B, Mes-Masson A-M, Bachvarov D. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC Genomics. 2008;9(1):99. doi: 10.1186/1471-2164-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zietarska M, Maugard CM, Filali-Mouhim A, Alam-Fahmy M, Tonin PN, Provencher DM, Mes-Masson AM. Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC) Mol. Carcinog. 2007;46(10):872–885. doi: 10.1002/mc.20315. [DOI] [PubMed] [Google Scholar]
- 12.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130(4):601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Kunz-Schughart LA, Freyer JP, Hofstaedter F, Ebner R. The use of 3-D cultures for high-throughput screening: the multicellular spheroid model. J. Biomol. Screening. 2004;9(4):273–285. doi: 10.1177/1087057104265040. [DOI] [PubMed] [Google Scholar]
- 14.Weiswald L-B, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17(1):1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyga A, Cheema U, Loizidou M. 3D tumour models: novel in vitro approaches to cancer studies. J. Cell Commun. Signal. 2011;5(3):239–248. doi: 10.1007/s12079-011-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 2010;148(1):3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Ham SL, Joshi R, Luker GD, Tavana H. Engineered Breast Cancer Cell Spheroids Reproduce Biologic Properties of Solid Tumors. Adv. Healthcare Mater. 2016;5(21):2788–2798. doi: 10.1002/adhm.201600644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinci M, Box C, Zimmermann M, Eccles SA. Tumor spheroid-based migration assays for evaluation of therapeutic agents. In: Jurge M, Colombo R, editors. Target Identification and Validation in Drug Discovery. Vol. 986. Methods in Molecular Biology; Springer; New York: 2013. pp. 253–266. [DOI] [PubMed] [Google Scholar]
- 19.Mueller-Klieser W. Tumor biology and experimental therapeutics. Crit. Rev. Oncol. Hematol. 2000;36(2):123–139. doi: 10.1016/s1040-8428(00)00082-2. [DOI] [PubMed] [Google Scholar]
- 20.Lambert B, De Ridder L, Slegers G, de Gelder V, Dierckx R, Thierens H. Screening for supra-additive effects of cytotoxic drugs and gamma irradiation in an in vitro model for hepatocellular carcinoma. Can. J. Physiol. Pharmacol. 2004;82(2):146–152. doi: 10.1139/y04-008. [DOI] [PubMed] [Google Scholar]
- 21.Fehlauer F, Muench M, Rades D, Stalpers LJ, Leenstra S, van der Valk P, Slotman B, Smid EJ, Sminia P. Effects of irradiation and cisplatin on human glioma spheroids: inhibition of cell proliferation and cell migration. J. Cancer Res. Clin. Oncol. 2005;131(11):723–732. doi: 10.1007/s00432-005-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst. 2011;136(3):473–8. doi: 10.1039/c0an00609b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinci GS, Boxall F, Patterson L, Zimmermann M, Court W, Lomas C, Mendiola M, Hardisson D, Eccles SA. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10(29) doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Wang W, Yu W, Xie Y, Zhang X, Zhang Y, Ma X. Development of an in Vitro Multicellular Tumor Spheroid Model Using Microencapsulation and Its Application in Anticancer Drug Screening and Testing. Biotechnol. Prog. 2005;21(4):1289–96. doi: 10.1021/bp050003l. [DOI] [PubMed] [Google Scholar]
- 25.Vorsmann H, Groeber F, Walles H, Busch S, Beissert S, Walczak H, Kulms D. Development of a human three-dimensional organotypic skin-melanoma spheroid model for in vitro drug testing. Cell Death Dis. 2013;4:e719. doi: 10.1038/cddis.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshikata A, Matsushita T, Ueoka R. Enhancement of drug efflux activity via MDR1 protein by spheroid culture of human hepatic cancer cells. J. Biosci. Bioeng. 2011;111(5):590–3. doi: 10.1016/j.jbiosc.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Ho WY, Yeap SK, Ho CL, Rahim RA, Alitheen NB. Development of multicellular tumor spheroid (MCTS) culture from breast cancer cell and a high throughput screening method using the MTT assay. PLoS One. 2012;7(9):e44640. doi: 10.1371/journal.pone.0044640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zanoni M, Piccinini F, Arienti C, Zamagni A, Santi S, Polico R, Bevilacqua A, Tesei A. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016;6(19103) doi: 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta G, Hsiao AY, Ingram M, Luker GD, Takayama S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Controlled Release. 2012;164(2):192–204. doi: 10.1016/j.jconrel.2012.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedrich J, Ebner R, Kunz-Schughart LA. Experimental anti-tumor therapy in 3-D: spheroids-old hat or new challenge? Int. J. Radiat. Biol. 2007;83(11–12):849–871. doi: 10.1080/09553000701727531. [DOI] [PubMed] [Google Scholar]
- 31.Lin RZ, Chang HY. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008;3(9–10):1172–1184. doi: 10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]
- 32.Iglesias JM, Beloqui I, Garcia-Garcia F, Leis O, Vazquez-Martin A, Eguiara A, Cufi S, Pavon A, Menendez JA, Dopazo J, Martin AG. Mammosphere formation in breast carcinoma cell lines depends upon expression of E-cadherin. PLoS One. 2013;8(10):e77281. doi: 10.1371/journal.pone.0077281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piggott L, Omidvar N, Pérez SM, Eberl M, Clarkson RW. Suppression of apoptosis inhibitor c-FLIP selectively eliminates breast cancer stem cell activity in response to the anti-cancer agent, TRAIL. Breast Cancer Res. 2011;13(5):1. doi: 10.1186/bcr2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 35.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bos PD, Zhang XH-F, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall CL, Dai J, van Golen KL, Keller ET, Long MW. Type I collagen receptor (α2β1) signaling promotes the growth of human prostate cancer cells within the bone. Cancer Res. 2006;66(17):8648–8654. doi: 10.1158/0008-5472.CAN-06-1544. [DOI] [PubMed] [Google Scholar]
- 38.Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One. 2008;3(2):e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens K, Ungrin M, Schwartz R, Ng S, Carvalho B, Christine K, Chaturvedi R, Li C, Zandstra P, Chen C, Bhatia SN. InVERT molding for scalable control of tissue microarchitecture. Nat. Commun. 2013;4:1847. doi: 10.1038/ncomms2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Phelps EA, Enemchukwu NO, Fiore VF, Sy JC, Murthy N, Sulchek TA, Barker TH, García AJ. Maleimide cross-linked bioactive peg hydrogel exhibits improved reaction kinetics and cross-linking for cell encapsulation and in situ delivery. Adv. Mater. 2012;24(1):64–70. doi: 10.1002/adma.201103574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park K-Y, Li G, Platt MO. Monocyte-derived macrophage assisted breast cancer cell invasion as a personalized, predictive metric to score metastatic risk. Sci. Rep. 2015;5(13855) doi: 10.1038/srep13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat. Methods. 2016;13(6):521–527. doi: 10.1038/nmeth.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ, Quackenbush J. Inconsistency in large pharmacogenomic studies. Nature. 2013;504(7480):389–393. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenny PA, L G, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Peterson OW, Gray JW, Bissell MJ. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007;1(1):84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heiser LM, Sadanandam A, Kuo W-L, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, Bayani N, Hu Z, Billig JI, Dueregger A, Lewis S, Jakkula L, Korkola JE, Durinck S, Pepin F, Guan Y, Purdom E, Neuvial P, Bengtsson H, Wood KW, Smith PG, Vassilev LT, Hennessy BT, Greshock J, Bachman KE, Hardwicke MA, Park JW, Marton LJ, Wolf DM, Collisson EA, Neve RM, Mills GB, Speed TP, Feiler HS, Wooster RF, Haussler D, Stuart JM, Gray JW, Spellman PT. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2012;109(8):2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dufau I, Frongia C, Sicard F, Dedieu L, Cordelier P, Ausseil F, Ducommun B, Valette A. Multicellular tumor spheroid model to evaluate spatio-temporal dynamics effect of chemo-therapeutics: application to the gemcitabine/CHK1 inhibitor combination in pancreatic cancer. BMC Cancer. 2012;12(1):15. doi: 10.1186/1471-2407-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engebraaten O, Bjerkvig R, Pedersen P-H, Laerum OD. Effects of EGF, BFGF, NGF and PDGF(bb) on cell proliferative, migratory and invasive capacities of human brain-tumour biopsies In Vitro. Int. J. Cancer. 1993;53(2):209–214. doi: 10.1002/ijc.2910530206. [DOI] [PubMed] [Google Scholar]
- 49.Mueller-Klieser W. Three-dimensional cell cultures: from molecular mechanisms to clinical applications. Am. J. Physiol. - Cell Physiol. 1997;273(4):C1109–C1123. doi: 10.1152/ajpcell.1997.273.4.C1109. [DOI] [PubMed] [Google Scholar]
- 50.Subik K, Lee J-F, Baxter L, Strzepek T, Costello D, Crowley P, Xing L, Hung M-C, Bonfiglio T, Hicks DG. The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer. 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- 51.Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM. The Triple Negative Paradox: Primary Tumor Chemosensitivity of Breast Cancer Subtypes. Clin. Cancer Res. 2007;13(8):2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 52.Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, Wilhelm S, Lynch M, Carter C. Sorafenib blocks the RAF/MEK/ ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66(24):11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 53.Mueck A, Seeger H, Wallwiener D, Huober J. Is the combination with 2-methoxyestradiol able to reduce the dosages of chemotherapeutices in the treatment of human ovarian cancer? Preliminary in vitro investigations. Eur. J. Gynaecol. Oncol. 2003;25(6):699–701. [PubMed] [Google Scholar]
- 54.Alberts DS, Einspahr JG, Struck R, Bignami G, Young L, Surwit EA, Salmon SE. Comparative in vitro cytotoxicity of cyclophosphamide, its major active metabolites and the new oxazaphosphorine ASTA Z 7557 (INN mafosfamide) Invest. New Drugs. 1984;2(2):141–148. doi: 10.1007/BF00232343. [DOI] [PubMed] [Google Scholar]
- 55.Yu L, Chen MC, Cheung KC. Droplet-based microfluidic system for multicellular tumor spheroid formation and anticancer drug testing. Lab Chip. 2010;10(18):2424–32. doi: 10.1039/c004590j. [DOI] [PubMed] [Google Scholar]
- 56.Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, Gao S, Mills GB, Brugge JS. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21(2):227–39. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keith B, Simon MC. Hypoxia-Inducible Factors, Stem Cells, and Cancer. Cell. 2007;129(3):465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zustiak SP, Leach JB. Hydrolytically degradable poly- (ethylene glycol) hydrogel scaffolds with tunable degradation and mechanical properties. Biomacromolecules. 2010;11(5):1348–1357. doi: 10.1021/bm100137q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casey RC, Burleson KM, Skubitz KM, Pambuccian SE, Oegema TR, Ruff LE, Skubitz AP. β1-integrins regulate the formation and adhesion of ovarian carcinoma multicellular spheroids. Am. J. Pathol. 2001;159(6):2071–2080. doi: 10.1016/s0002-9440(10)63058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuhas JM, Tarleton AE, Molzen KB. Multicellular Tumor Spheroid Formation by Breast Cancer Cells Isolated from Different Sites. Cancer Res. 1978;38(8):2486–2491. [PubMed] [Google Scholar]
- 61.Manuel Iglesias J, Beloqui I, Garcia-Garcia F, Leis O, Vazquez-Martin A, Eguiara A, Cufi S, Pavon A, Menendez JA, Dopazo J, Martin AG. Mammosphere formation in breast carcinoma cell lines depends upon expression of E-cadherin. PLoS One. 2013;8(10):e77281. doi: 10.1371/journal.pone.0077281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alessandri K, Sarangi BR, Gurchenkov VV, Sinha B, Kieβling TR, Fetler L, Rico F, Scheuring S, Lamaze C, Simon A, et al. Cellular capsules as a tool for multicellular spheroid production and for investigating the mechanics of tumor progression in vitro. Proc. Natl. Acad. Sci. U. S. A. 2013;110(37):14843–14848. doi: 10.1073/pnas.1309482110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boylan KL, Misemer B, De Rycke MS, Andersen JD, Harrington KM, Kalloger SE, Gilks CB, Pambuccian SE, Skubitz AP. Claudin 4 Is differentially expressed between ovarian cancer subtypes and plays a role in spheroid formation. Int. J. Mol. Sci. 2011;12(2):1334–58. doi: 10.3390/ijms12021334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shang X, Lin X, Alvarez E, Manorek G, Howell SB. Tight Junction Proteins Claudin-3 and Claudin-4 Control Tumor Growth and Metastases. Neoplasia. 2012;14(10):974–IN22. doi: 10.1593/neo.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pacheco-Marin R, Melendez-Zajgla J, Castillo-Rojas G, Mandujano-Tinoco E, Garcia-Venzor A, Uribe-Carvajal S, Cabrera-Orefice A, Gonzalez-Torres C, Gaytan-Cervantes J, Mitre-Aguilar IB, Maldonado V. Transcriptome profile of the early stages of breast cancer tumoral spheroids. Sci. Rep. 2016;6:23373. doi: 10.1038/srep23373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Narod S. Have we given up on a cure for ovarian cancer? Curr. Oncol. 2015;22(3):139. doi: 10.3747/co.22.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmed N, Stenvers KL. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front. Oncol. 2013;3:256. doi: 10.3389/fonc.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kipps E, Tan DS, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat. Rev. Cancer. 2013;13(4):273–82. doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ayantunde AA, Parsons SL. Pattern and prognostic factors in patients with malignant ascites: a retrospective study. Ann. Oncol. 2007;18(5):945–9. doi: 10.1093/annonc/mdl499. [DOI] [PubMed] [Google Scholar]
- 70.Alvero AB, Chen R, Fu HH, Montagna M, Schwartz PE, Rutherford T, Silasi DA, Steffensen KD, Waldstrom M, Visintin I, Mor G. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle. 2009;8(1):158–66. doi: 10.4161/cc.8.1.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Flick MB, O'Malley D, Rutherford T, Rodov S, Kamsteeg M, Hao XY, Schwartz P, Kacinski BM, Mor G. Apoptosis-Based Evaluation of Chemosensitivity in Ovarian Cancer Patients. J. Soc. Gynecol. Invest. 2004;11(4):252–259. doi: 10.1016/j.jsgi.2003.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.