Abstract
Small-cell lung cancer (SCLC) is an aggressive malignancy associated with a poor prognosis. First-line treatment has remained unchanged for decades, and a paucity of effective treatment options exists for recurrent disease. Nonetheless, advances in our understanding of SCLC biology have led to the development of novel experimental therapies. Poly [ADP-ribose] polymerase (PARP) inhibitors have shown promise in preclinical models, and are being clinically tested in combination with cytotoxic therapies and inhibitors of cell-cycle checkpoints. Preclinical data indicate that targeting of histone-lysine N-methyltransferase EZH2, a regulator of chromatin remodelling implicated in acquired therapeutic resistance, might augment and prolong chemotherapy responses. High expression of the inhibitory Notch ligand Delta-like protein 3 (DLL3) in most SCLCs has been linked to expression of Achaetescute homologue 1 (ASCL1; also known as ASH-1), a key transcription factor driving SCLC oncogenesis; encouraging preclinical and clinical activity has been demonstrated for an anti-DLL3-antibody–drug conjugate. The immune microenvironment of SCLC seems to be distinct from that of other solid tumours, with few tumour-infiltrating lymphocytes and low levels of the immune-checkpoint protein programmed cell death 1 ligand 1 (PD-L1). Nonetheless, immunotherapy with immune-checkpoint inhibitors holds promise for patients with this disease, independent of PD-L1 status. Herein, we review the progress made in uncovering aspects of the biology of SCLC and its microenvironment that are defining new therapeutic strategies and offering renewed hope for patients.
Graphical Abstract
For three decades, the treatment of small-cell lung cancer (SCLC) has remained essentially unchanged, and patient outcomes remain dismal. In the past 5 years, however, advances in our understanding of the disease, at the molecular level, have resulted in the development of new therapeutic strategies, encompassing immunotherapies and novel molecularly targeted agents. Herein, authors review the breakthroughs that hold the promise to improve SCLC outcomes.
Lung cancer is the most common cancer worldwide, with an estimated 1.8 million cases diagnosed each year and an estimated 1.6 million lung-cancer-related deaths annually1,2. Globally, small-cell lung cancer (SCLC) accounts for 13–15% of all lung cancers, with approximately 250,000 cases diagnosed annually, and is the sixth most common cause of cancer-related mortality3–7. SCLC is an aggressive high-grade neuroendocrine tumour associated with a short doubling time, a high growth fraction, and early development of widespread metastases, which contribute to the extremely poor prognosis of patients with the disease. The typical life expectancy for a patient diagnosed with SCLC, and the standard options for therapy, have not changed over the past three decades (FIG. 1); the median overall survival duration of patients with extensive-stage (ES)-SCLC is stalled, frustratingly, at <10 months, with a discouraging 5-year overall survival of 1–5%4. The incidence of SCLC in the developed world has decreased in parallel with the declining rates of smoking, although SCLC remains a substantial cause of cancer-related mortality worldwide2,8. Among the major lung-cancer subtypes, SCLC has the strongest association with smoking, with only 2% of cases occurring in never-smokers9,10. Consequently, SCLCs have a high load of somatic mutations induced by tobacco carcinogens11–13. The most common genetic alterations in SCLC include inactivation of the tumour-suppressor genes TP53 and RB1, as well as copy-number gains of genes encoding MYC family members, enzymes involved in chromatin remodelling, receptor tyrosine kinases and their downstream effectors, and Notch family proteins12,13. Importantly, the high mutational burden of SCLC might provide opportunities for therapeutic intervention. In this Review, we explore the progress made in defining the molecular aetiology of SCLC and discuss the development of rational therapeutic strategies based on the disease biology.
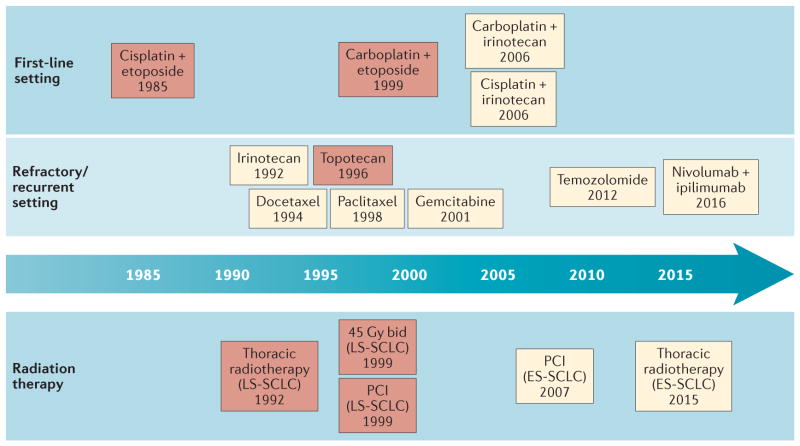
Figure 1. Timeline of therapeutic advances for small-cell lung cancer (SCLC).
This timeline illustrates the paucity of new treatment options for patients with SCLC over the past three decades. The red-shaded boxes represent standard-of-care therapies that have been approved by the FDA; the yellow-shaded boxes represent therapies that have been recommended by the National Comprehensive Cancer Network (NCCN)20, but are not currently approved by the FDA. Since 1985, the cisplatin and etoposide chemotherapy regimen has remained the standard-of-care first-line systemic treatment for patients with extensive-stage (ES)-SCLC. Subsequent regimens, in which carboplatin or irinotecan substitute for cisplatin or etoposide, respectively, have comparable effectiveness, but differing toxicity profiles. Second-line therapies that are recommended in the NCCN guidelines include topoisomerase inhibitors, taxols, alkylating agents, and, since 2016, immunotherapy, although only topotecan is approved by the FDA for use in this setting. For limited-stage (LS)-SCLC, radiation treatment early in the course of chemotherapy is recommended, classically at a total dose of 45 Gy delivered in 30 twice-daily (b.i.d.) fractions of 1.5 Gy (over the course of 3 weeks), with additional prophylactic cranial irradiation (PCI). More recently, thoracic irradiation has been shown to be of benefit for some patients with ES-SCLC; however, the role of thoracic radiation and PCI in the treatment of ES-SCLC remains controversial.
Clinical overview
Pathology
SCLC is one constituent of a group of neuroendocrine lung tumours that also includes large-cell neuroendocrine carcinoma, and typical and atypical carcinoid tumours. The diagnosis of SCLC is based primarily on histological appearance by light microscopy, demonstrating dense sheets of small cells with neuroendocrine differentiation (characterized by scant cytoplasm; poorly defined cell borders; dispersed, finely granular nuclear chromatin; absent or inconspicuous nucleoli; and prominent nuclear moulding). Necrosis is typically extensive and the mitotic count is exceptionally high (>10 mitoses per 10 high-power fields), with a high Ki67 labelling index (using the MIB-1 antibody) of around 90–100% also indicating rapid cell proliferation14. Current classifications of SCLC subtypes include ‘small-cell carcinoma’ and ‘combined small-cell carcinoma’, with the latter comprising small-cell carcinomas harbouring an additional component of non-small-cell carcinoma (NSCLC), such as adenocarcinoma, squamous-cell carcinoma, or large-cell carcinoma14. Combined small-cell carcinoma accounts for approximately 10–25% of SCLC cases. Most SCLCs express the neuroendocrine markers CD45, CD56, chromogranin, and synaptophysin; fewer than 10% of SCLCs are negative for all neuroendocrine markers14.
SCLC can be staged according to the conventional ‘TNM’ criteria, as defined by the Union for International Cancer Control and the American Joint Committee on Cancer4. Surgery can have a role in the treatment of patients with early, TNM stage I disease (tumours <5 cm in diameter with no lymph-node involvement or metastasis); however, disease presentation at such early stage is the exception to the norm. More commonly, SCLC is staged as limited-stage or extensive-stage disease; these distinctions are both prognostic and guide the use of the available treatment options.
Limited-stage disease
Limited-stage (LS)-SCLC is defined as disease confined to a single radiation port (that is, to a tolerable treatment field), with or without mediastinal lymph-node involvement. Only around one-third of patients diagnosed with SCLC present with LS-SCLC. In contrast to NSCLC, low-dose CT screening has not been shown to improve the survival of patients with SCLC, or to increase the number of patients diagnosed with early stage disease15. Treatment advances involving thoracic and cranial irradiation have led to improved outcomes for patients with LS-SCLC6 (FIG. 1). The rapid proliferation rate of this malignancy confers an exquisite sensitivity to DNA-damaging therapies; thus, the standard of care for LS-SCLC is concurrent chemoradiotherapy with cisplatin and etoposide, which results in an objective response rate (ORR) of 70–90%16, and has been associated with 44% survival at 2 years and 23% survival at 5 years17,18. The radiotherapy regimen best supported by randomized clinical trial data is 45 Gy delivered in 30 twice-daily (b.i.d.) 1.5-Gy fractions (over 3 weeks)17. In this practice-defining study17, however, the b.i.d. fractionation schema was compared with a regimen comprising the same nominal dose of 45 Gy delivered in 25 once-daily 1.8-Gy fractions (over 5 weeks), which is not a biologically equivalent dose. Accordingly, in the ongoing EORTC 08072 CONVERT trial (NCT00433563) and the CALGB 30610/RTOG 0538 trial (NCT00632853), the standard b.i.d. regimen is being compared to biologically equivalent doses: once-daily 2-Gy fractions to a total dose of 66 Gy and 70 Gy, respectively (over 6.5–7 weeks). Early results of the CONVERT trial, presented in abstract form at the ASCO 2016 meeting19, indicate equivalent overall survival and toxicity between the two arms, providing some evidence to support the use of either regimen.
The blood–brain barrier can restrict access of systemically delivered therapies into the brain and, therefore, most patients with LC-SCLC experience recurrent disease in the central nervous system (CNS). Thus, prophylactic cranial irradiation (PCI) is recommended for patients with LS-SCLC who respond to upfront concurrent chemoradiotherapy and have no evidence of brain metastasis on MRI, in order to eliminate undetectable micrometastatic disease that might be present20. Results of a large meta-analysis have confirmed the survival benefit of this approach21. In summary, the preferred therapeutic strategy for patients with LS-SCLC is concurrent chemoradiotherapy (with cisplatin and etoposide)16, followed by PCI in those who achieve complete remission21; the associated 5-year survival ranges from 20–40% depending on disease stage and degree of nodal involvement22.
Extensive-stage disease
ES-SCLC, defined as disease that has spread beyond a single radiation port — generally synonymous with distant metastasis — accounts for two-thirds of all SCLC diagnoses6,23. As described previously, the outcomes of patients with ES-SCLC are dismal, with median overall survival durations of <10 months and 5-year survival <5%4. First-line treatment for such patients, which has remained unchanged for more than three decades (FIG. 1), generally consists of 4–6 cycles of chemotherapy with a platinum-based agent (either cisplatin or carboplatin) and etoposide. ORRs associated with this regimen approach 70%; however, most patients suffer rapid disease relapse within 6 months24–30. The use of a platinum-based drug in combination with irinotecan is also an acceptable first-line treatment, and this regimen is commonly used in Japan28,29. Results of the largest North American study reported to date31, however, did not confirm the clinical benefit of cisplatin and irinotecan versus cisplatin and etoposide previously reported in Japanese cohorts28,29.
Despite the inevitable and rapid disease relapse after first-line treatment, topotecan is the only systemic therapy approved by the FDA for the treatment of recurrent ES-SCLC. Furthermore, the activity of this agent is, in general, limited to patients with chemosensitive relapse, defined as those with an objective response to first-line chemotherapy that persisted for at least 3 months after completion of therapy32. Unfortunately, the mechanisms of acquired chemoresistance in SCLC are poorly understood.
The roles of thoracic radiation therapy and PCI in the treatment of ES-SCLC remain controversial. A randomized phase III trial of post-chemotherapy consolidative chest radiation was negative for its primary end point of an improvement in overall survival at 1 year compared with that observed without thoracic radiotherapy (33% versus 28%; P = 0.066), but results of a post-hoc analysis indicated an overall survival benefit at 2 years (13% versus 3%; P= 0.004)33. A survival benefit from consolidative thoracic radiotherapy was also later reported for the subgroup of patients with residual intrathoracic disease (HR 0.81,95% CI 0.66–1.00; P= 0.044)34. The low-dose radiation regimen used (30 Gy in 10 fractions) was well tolerated, with no marked differences in toxicities between the control and experimental groups33. Further work is necessary to confirm these results, and to identify additional subsets of patients with ES-SCLC who are most likely to benefit from consolidative thoracic radiotherapy. In this study33, all patients received PCI, which has been reported to improve overall survival in patients with ES-SCLC (27.1% at 1 year versus 13.3% without PCI)35; however, radiological assessment before PCI was not mandated in this study35, and thus the inclusion of patients with overt brain metastases might have contributed to the apparent clinical benefit. In a more recent Japanese phase III trial36, in which pretreatment brain imaging was mandated, PCI did not improve the overall survival of patients with ES-SCLC. Of note, both patients and physicians have concerns regarding the potential for delayed PCI-related neurotoxicity, which must be balanced with the need to achieve disease control37,38.
Essentially, the management of ES-SCLC has remained unchanged for the past 20 years — since the approval of topotecan for recurrent and/or refractory disease in 1996 (FIG. 1) — and effectiveness of the few available treatment options is limited; therefore, novel, targeted, and biomarker-guided therapies an important unmet need. Advances in genomic, epigenetic, and proteomic profiling, tumour immunology, and tumour biology have led to exciting new experimental therapies, some of which have been shown to provide a meaningful clinical benefit in early phase clinical trials. In the following sections, we discuss the breakthroughs in our understanding of the molecular and cellular biology of SCLC, and highlight the emerging targets for novel treatments.
SCLC developmental regulatory pathways
SCLCs display poorly differentiated neuroendocrine features14; three distinct molecular subtypes have been defined by gene-expression profiles determined by differential expression of the neuronal basic helix–loop–helix transcription factors achaetescute homologue 1 (ASCL1, also known as ASH-1) and neurogenic differentiation factor 1 (NEUROD1)39–41. A subgroup defined by expression of ASCL1 has been described as the ‘classic’ subtype, while a subgroup defined by high expression levels of NEUROD1 has been described as the ‘variant’ subtype41–43. The third, minor subgroup lacks expression of either of these neuroendocrine markers. These subgroups are clearly distinct disease entities based on gene-expression profiling, although the clinical implications of this molecular classification have not been defined — for example, whether these subtypes have differential capacities for invasion or metastasis, or differential responses to standard therapies is unknown.
In a genetically engineered mouse model, conditional Cre-mediated excision of the Tp53 and Rb1 gene in lung epithelial cells gives rise to lung tumours resembling the classic subtype of SCLC44. Interestingly, tumours resembling variant subtype SCLC are not observed in this model45. The results of subsequent experiments have revealed that ASCL1 activates pulmonary neuroendocrine differentiation46; regulates the expression of multiple Notch-pathway genes, including that encoding the inhibitory Notch ligand Delta-like protein 3 (DLL3)39; and is required for tumour initiation in this model, whereas NeuroD1 is dispensable47,48. In 2017, however, Mollaoglu et al.49 demonstrated that overexpression of an oncogenic MycT58A allele in this context promotes the development of ‘neuroendocrine-low’ tumours resembling variant subtype SCLC, characterized by a high level of NeuroD1 expression and low ASCL1 expression. Importantly, a targeted drug screen in this model revealed the therapeutic potential of aurora kinase inhibitors, in combination with cisplatin and etoposide chemotherapy, in MYC-driven variant subtype SCLC49. ASCL1 drives the expression of many proto-oncogenes implicated in SCLC progression and cell survival, including MYCL1, RET, SOX2, NF-IB, and BCL2 (REFS 39,50). Although NEUROD1 does not seem to be required for the tumorigenesis of classic subtype SCLC, this transcription factor has been shown to induce migratory signalling pathways in human SCLC cells51, and can activate a neuroendocrine differentiation programme in a mouse lung epithelial (non-neuroendocrine) cell line52.
The SCLC genome, epigenome, and proteome
Genetic landscape
SCLC cells are typically aneuploid, with a high incidence of chromosomal deletions of 3p, 4q, 5q, 10q, 13q, and 17p, and copy-number gains of 3q, 5p, 6p, 8q, 17q, 19, and 20q (REFS 53–55). The genetic mutational landscape of SCLC is complex and varied, but functional inactivation of both TP53 and RB1 is essentially universal12,13. Other molecular abnormalities detected in SCLCs include amplification and overexpression of MYC family oncogenes56, overexpression of BCL2 (REF. 57) and KIT58, as well as activation of autocrine loops through bombesin-like peptides59. Gene-expression profiling data suggest that several neuroendocrine genes are expressed in SCLCs, including those encoding chromogranins A, B and C (CHGA, CHGB, and CHGC (SCG2))60, insulinoma-associated gene 1 (INSM1)61, as well as ASCL1 (REF. 62); however, the relevance of many of these markers, with respect to improving the diagnosis or directing therapy for the disease, remains unclear.
In the past 5 years, comprehensive genomic studies, including exome, whole-genome, transcriptome, and copy-number alteration analyses of primary human SCLC samples, have provided the first overview of genomic landscape of this disease12,13,63,64. SCLCs have a mean mutation rate of 7.4 nonsynonymous mutations per million base pairs — similar to that of other tobacco-associated lung cancers12,65. As noted, biallelic inactivation of TP53 and RB1 is near-ubiquitous in SCLC12,13,63,64. Recurrent mutations in CREBBP, EP300, MLL (also known as KMT2A), PTEN, SLIT2, EPHA7, and Notch genes, and amplification of FGFR1 and SOX2, have also been reported12,13,63. In particular, mutations affecting Notch receptors have been detected in 25% of SCLCs, and these receptors have been validated as tumour suppressors in mouse models of SCLC12. Indeed, alterations in tumour-suppressor genes were the most-frequent finding in SCLC samples. Lastly, RNA-sequencing data have identified multiple fusion transcripts in primary SCLC specimens, including a recurrent RLF–MYCL1 fusion13. Further work is needed to better understand how individual genomic alterations associated with SCLC relate to patient response to therapy and outcome.
Epigenetic landscape
Epigenetic processes have a central role in carcinogenesis, and tumour maintenance, progression, and responses to treatment66. Most early cancer epigenetics studies focused on DNA methylation at CpG-dinucleotide islands67,68. DNA-methylation patterns are associated with gene expression, and promoter hyper-methylation has been shown to lead to a heterochromatin state and repressed gene expression69. Repression of tumour-suppressor genes and genes required for programmed cell death in this manner, as well as de-repression of oncogenes, can provide a fitness advantage for cancer cells67,68. Evidence for this paradigm in SCLC was provided by the finding that several SCLC cell lines were insensitive to TNF-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis owing to epigenetic silencing of genes encoding caspase-8, FAS, and TRAIL-R1 via promoter CpG-island methylation70. Interestingly, treatment with the DNA methyltransferase (DNMT) inhibitor decitabine, in combination with IFNγ (which has been shown to upregulate expression of caspase-8), partially restored caspase-8 expression and increased the sensitivity of the SCLC cell lines to TRAIL-induced apoptosis70. Subsequently, the combination of DNMT inhibition using decitabine and histone deacetylase (HDAC) inhibition using either valproic acid or CI-994 could restore expression of caspase-8, sensitizing the cell lines to TRAIL-mediated cell death71. These findings provide insight into the mechanisms by which DNA-methylation events can impair the extrinsic apoptosis pathway and provided early evidence of the potential for pharmacological reversibility of such events in SCLC cells.
In the first genome-scale analysis of methylation changes in SCLC72, 73 genes that were methylated in >77% of SCLC tumours were identified. Methylated gene promoters were enriched in binding sites for the neurogenic transcription factors NEUROD1, heart and neural crest derivatives-expressed protein 1 (HAND1), zinc finger protein 423 (ZNF423), and RE1-silencing transcription factor (REST), which the authors interpreted as being indicative of a defect in neuroendocrine differentiation72. Results of a subsequent genome-wide study, performed at single-nucleotide resolution, demonstrated overall DNA hypomethylation in primary SCLC samples compared with non-neoplastic lung specimens; however, CpG-island-containing promoters were found to be hypermethylated in SCLC to a greater degree than most tumour types included in The Cancer Genome Atlas40. Hypermethylated sites were concentrated focally at transcription start sites, whereas hypomethylated sites were distributed diffusely throughout promoter regions, suggesting a functional role for hypermethylation in cancer-specific gene silencing40. The authors further demonstrated that a major subgroup of SCLC has increased promoter methylation in a manner similar to what has been described in other tumour types as the ‘CpG-island methylator phenotype’ (CIMP)40. Of note, the CIMP has been associated with an unfavourable prognosis across multiple tumour types73,74. Neither initial genome-scale study in SCLC reported relationships of methylation profiles with patient outcome data40,72. In an ensuing study, however, SCLC samples could be similarly stratified according to CIMP status, and patients with CIMP-positive tumours had a poorer prognosis than those with CIMP-negative disease75. These data suggest that clinically relevant subtypes of SCLC are defined by DNA-methylation patterns, consistent with observations among other lung carcinoma epitypes74.
Proteomic landscape
Proteomic and transcriptomic analyses of 34 SCLC and 74 NSCLC cell lines revealed a number of proteins of potential therapeutic interest that are differently expressed between these tumour types76. In comparison to NSCLCs, SCLCs had substantially increased levels of the growth-factor receptor KIT; the antiapoptotic protein Bcl-2 and the pro-apoptotic Bcl-2 family members BIM (Bcl-2-like protein 11) and BAX (Bcl-2-like protein 4); Upregulated histone-lysine N-methyltransferase EZH2 (also known as enhancer of zeste homologue 2), a chromatin-remodelling factor; thymidylate synthase; and DNA-repair proteins, including poly [ADP-ribose] polymerase (PARP) enzymes76.
Potential therapeutic targets in SCLC
Many of the new insights into the biology of SCLC are now being actively translated into clinical trials of novel treatments for patients with this disease. In the following sections of this Review, we highlight some of the particularly attractive targets for clinical investigation.
PARP inhibitors
PARP enzymes were first identified in 1963 (REF. 77). To date, 17 structural PARP enzymes have been described78. PARP1 — the most abundantly expressed isoform in humans — and PARP2 function to detect and mark DNA single-strand breaks (SSB) by binding to the site of DNA damage and synthesizing poly [ADP-ribose] chains, which recruit a host of scaffold proteins and DNA-repair enzymes to mend the break79. As alluded to, PARP protein levels are upregulated in SCLC relative to other lung cancers76. In particular, PARP1 has been found to be highly expressed at both the mRNA and protein levels in SCLC samples76. In addition to DNA repair, this protein is a co-activator of the transcription factor E2F1 (FIG. 2), and has been implicated in various cellular processes involved in tumorigenesis — including cell differentiation, proliferation, and transformation80. The results of initial in vitro studies indicated that SCLC cell lines are sensitive to PARP inhibitors, and provided preclinical validation that PARP inhibition enhances the anticancer activity of chemotherapy (and perhaps other DNA-damaging therapies) by downregulating key DNA-repair mechanisms76. These preclinical data supported the inclusion of a cohort of patients with SCLC in a phase I study of monotherapy with the PARP inhibitor talazoparib; the reported ORR was 9% and the clinical benefit rate at ≥16 weeks was 26%81. Indeed, clinical trials of a number of PARP inhibitors, in a range of treatment settings, are ongoing in patients with SCLC (TABLE 1).
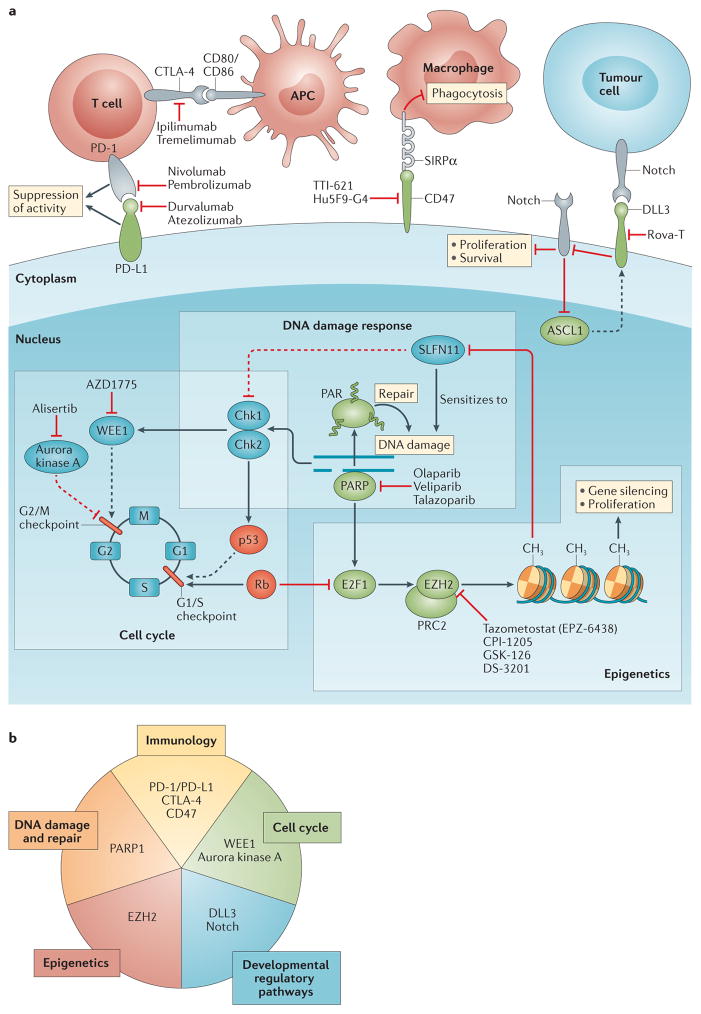
Figure 2. Signalling pathways and physiological domains that are the focus of experimental targeted therapies for small-cell lung cancer (SCLC).
a | Dashed and solid lines indicate indirect and direct interactions, respectively. Proteins in green are typically upregulated in SCLCs compared with nonmalignant lung tissue, while those in red are downregulated or absent. Examples of the investigational molecularly targeted agents or antibody-based treatments targeting each signalling node are provided. b | The novel, investigational, targeted therapeutics for SCLC are predicated on five aspects of cancer biology. Immune-checkpoint blockade with antibodies targeting programmed cell death protein 1 (PD-1), programmed cell death 1 ligand 1 (PD-L1), and/or cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) can prime the adaptive immune response to SCLC cells, whereas antibody-mediated blockade of the ‘don’t eat me’ protein CD47 can enable phagocytosis of tumour cells by macrophages. The use of small-molecule inhibitors of key regulators of the cell cycle, such as the protein kinases WEE1 and aurora kinase A, exploits the inherent lack of the G1/S-checkpoint activity resulting from loss of the tumour suppressors p53 and retinoblastoma-associated protein (Rb) in most SCLC cells. WEE1 regulates the G2/M cell-cycle checkpoint that is essential for ensuring the integrity of the genome in such cancer cells and, thus, inhibition of this kinase can lead to mitotic catastrophe and apoptosis, particularly if combined with DNA-damaging therapies. By contrast, aurora kinase A has essential roles in mitosis, and inhibitors of this protein results in cell-cycle arrest, preventing cell proliferation. Inhibitors of Notch have demonstrated antitumour effects in preclinical studies. Moreover, Delta-like protein 3 (DLL3), an inhibitory Notch ligand, is specifically upregulated in SCLC and can, therefore, be leveraged for selective tumour targeting with the antibody–drug conjugate rovalpituzumab tesirine (Rova-T). Inhibitors of the histone-lysine N-methyltransferase EZH2 (also known as enhancer of zeste homologue 2), the enzymatic histone-lysine N-methyltransferase subunit of the polycomb repressive complex 2 (PRC2) chromatin-remodelling machinery, can prevent chemoresistance and cell proliferation by counteracting epigenetic gene silencing, in particular, of schlafen family member 11 (SLFN11) — a protein that negatively regulates homologous recombination DNA repair. Poly [ADP-ribose] polymerase (PARP) inhibitors prevent the activation of DNA-repair proteins by PARP1 and trap this enzyme on DNA, which causes further DNA damage that can eventually result in cell death. APC, antigen-presenting cell; ASCL1, achaetescute homologue 1 (also known as ASH-1); Chk1/2, checkpoint kinase 1/2; E2F1, transcription factor E2F1 (also known as retinoblastoma-associated protein 1); PAR, poly-ADP-ribosylation.
Table 1.
Selected ongoing studies of targeted therapy for extensive stage small-cell lung cancer
| Therapy | Molecular target | Study name | Study phase | ClinicalTrials.gov study identifier | Estimated primary completion date |
|---|---|---|---|---|---|
| First line | |||||
| Veliparib (plus carboplatin and etoposide) | PARP | M14-361/2014-001764-35 | I/II | NCT02289690 | August 2017 |
| Rova-T (± cisplatin and etoposide) | DLL3 | SCRX001-004 | I | NCT02819999 | October 2020 |
| Maintenance | |||||
| Olaparib plus cediranib or no maintenance treatment (after cis/carboplatin and etoposide ± cediranib) | PARP; VEGF-RTKs | SUKSES-B | II | NCT02899728 | December 2018 |
| Rova-T plus dexamethasone versus placebo (after cis/carboplatin and etoposide) | DLL3 | MERU | III | NCT03033511 | August 2019 |
| Second line and beyond | |||||
| Olaparib plus CRLX101 (nanoparticle camptothecin) | PARP | 160107/16-C-0107 | I/II | NCT02769962 | June 2018 |
| Olaparib plus cediranib | PARP; VEGF-RTKs | NCI-2015-01097/9881 | II | NCT02498613 | May 2018 |
| Olaparib plus durvalumab | PARP; PD-L1 | MEDIOLA | I/II | NCT02734004 | October 2018 |
| Olaparib plus AZD1775 | PARP; WEE1 | D6010C00005/REFMAL 384 | I | NCT02511795 | January 2018 |
| AZD1775 | WEE1 | 2015-10-178 | II | NCT02593019 | March 2017 |
| AZD1775 plus carboplatin | WEE1 | D419QC00002/2016-001202-42 | II | NCT02937818 | May 2020 |
| Rova-T | DLL3 | TRINITY | II | NCT02674568 | March 2017 |
| Rova-T | DLL3 | SCRX001-007 | I | NCT02874664 | June 2017 |
Cis/carboplatin, cisplatin or carboplatin; DLL3, Delta-like protein 3; PARP, poly [ADP-ribose] polymerase; PD-L1, programmed cell death 1 ligand 1; Rova-T, rovalpituzumab tesirine; VEGF-RTKs, vascular endothelial growth factor receptor tyrosine kinases; WEE1, Wee1-like protein kinase.
Various PARP inhibitors have received FDA approval or a ‘breakthrough therapy’ designation for the treatment of patients with ovarian cancer harbouring deleterious BRCA1 or BRCA2 mutations82,83. BRCA1/2 are critical mediators of the DNA double-strand break repair pathway involving homologous recombination (HR). HR-deficient (HRD) tumour cells depend on PARP-mediated SSB-repair pathway84,85, as well as other back-up pathways involving RAD52 (REFS 86,87) and DNA polymerase θ, for survival88,89. This synthetic lethal dependency can be exploited therapeutically; thus, PARP inhibition leads to selective lethality of BRCA1/2-mutated tumour cells. However, BRCA1/2 mutations are notably rare in primary human SCLCs, occurring in <2% of cases12,13. Beyond BRCA1/2 mutations, HRD can be assessed using allele specific copy-number analysis of data generated from single-nucleotide polymorphism (SNP) microarrays and now next-generation sequencing (NGS) approaches that quantify the resulting characteristic large-scale chromosomal aberrations90, and which have been shown to predict PARP inhibitor sensitivity in patients with BRCA1/2-wild-type ovarian cancer91. Surprisingly, however, HRD-assay scores do not seem to correlate with sensitivity to PARP inhibitors in SCLC cell lines92.
A distinct mechanism, high expression levels of schlafen family member 11 (SLFN11), has been identified as a critical determinant of PARP-inhibitor sensitivity in SCLC cell lines and patient-derived xenografts92,93. SLFN11 is actively recruited to sites of DNA damage, inhibits HR94, and activates a cellular replication-stress response93. Notably, SLFN11 expression correlates with sensitivity to DNA-damaging agents (such as irinotecan, etoposide, and cisplatin) in other malignancies95–98. In line with the preclinical evidence92, high levels of SLFN11 expression (H-score ≥1) was associated with favourable tumour responses, progression-free survival (PFS), and overall survival in patients with SCLC who were treated with temozolomide and the PARP inhibitor veliparib, but not temozolomide plus placebo, in a randomized phase II clinical trial99. Of note, SLFN11 expression levels were defined using tumour samples obtained at initial diagnosis, and all patients enrolled had received at least one prior treatment99, which could have led to downregulation of SLFN11 at the time of study treatment; thus, this population might not have had ideal responses to PARP inhibition. Indeed, mechanisms to upregulate SLFN11 in chemoresistant patients are currently being considered. The utility of SLFN11 expression as a predictive biomarker for PARP-inhibitor therapy in SCLC will require validation in prospective biomarker-stratified trials.
EZH2 inhibition
The polycomb repressor complex 2 (PRC2) is a multiprotein chromatin-modifying complex that inhibits gene expression by promoting local histone methylation. EZH2 is the enzymatic histone-lysine N-methyltransferase subunit of PRC2, and mediates histone H3 lysine 27 dimethylation and trimethylation (H3K27me2 and H3K27me3)100 (FIG. 2). EZH2 is mutated in some human cancers at gain of function hotspots that increase its enzymatic activity and thereby promote H3K27me3 (REF. 101). EZH2 is not commonly mutated in SCLC but the level of EZH2 expression is higher in SCLCs than in any tumour type included in The Cancer Genome Atlas40,76. Expression of the EZH2 gene is under the direct control of E2F family of transcription factors, including E2F1 (REF. 102) (for which PARP1, itself overexpressed in SCLC cells76, acts a co-activator). E2F transcriptional activity is negatively regulated by product of the RB1 tumour-suppressor gene (Rb); the nearly universal loss of RB1 — and thus functional Rb — in SCLC cells results in a high level of E2F transcriptional activity, and consequent high EZH2 expression levels103. These observations define a model in which EZH2 expression is primarily promoted by one of the pathognomonic genetic alterations of SCLC (FIG. 2).
In 2017, findings from multiple patient-derived xenografts linked the upregulation of EZH2 with H3K27me3-associated SLFN11 gene silencing as a frequent mechanism of acquired chemoresistance in SCLC104. EZH2-mediated suppression of SLFN11 was observed in 40% of SCLC models selected in vivo for acquired chemotherapeutic resistance104. Mechanistically, loss of SLFN11 expression increases HR efficiency and, therefore, augments repair of DNA damage induced by cytotoxic chemotherapy. Importantly, EZH2 inhibition was found to prevent SLFN11 silencing and maintain the sensitivity of SCLC xenografts to chemotherapy104, suggesting a potential combinatorial strategy to enhance the effectiveness of current standard therapies for this recalcitrant disease. Various EZH2 inhibitors are undergoing clinical testing in the treatment of a range of malignancies; although clinical trials enrolling patients with SCLC are a research priority, at present, no such studies are underway.
WEE1-targeted cell-cycle vulnerabilities
The protein product of TP53, p53, has a critical role in the DNA-damage-response network, inducing cell-cycle arrest and initiation of apoptosis in cells exposed to genotoxic stress. Accordingly, TP53 deficiency leads to defective cell-cycle arrest at the G1/S checkpoint (FIG. 2), blunts the DNA-damage response, and contributes to replication stress105–107. In SCLC cells, combined loss of RB1 and TP53 result in markedly defective G1/S cell-cycle checkpoint capacity and, consequently, increased dependency on the G2/M checkpoint for adequate DNA repair and ultimately cell survival; rational targeting of the G2/M checkpoint might exploit this tumour-specific vulnerability107.
The tyrosine kinase WEE1 is an important gatekeeper of G2/M checkpoint (FIG. 2), and induces G2 arrest via inhibitory phosphorylation of cyclin-dependent kinases 1 and 2 (REF. 108). The combination of a WEE1 inhibitor with any of several classes of DNA-damaging agents, including antimetabolites (gemcitabine or 5-fluorouracil), topoisomerase inhibitors (camptothecin or doxorubicin), DNA-crosslinking agents (cisplatin or carboplatin), and PARP inhibitors, results in synergistic efficacy in TP53-deficient cervical, colon, pancreatic, and NSCLC cell lines109–111. As such, WEE1 is a promising target for SCLC therapy. Single-agent and combination studies of WEE1 inhibitors are now active in the clinic112,113, and include studies in patients with relapsed and/or refractory SCLC (TABLE 1). The role of TP53/RB1 status (that is, context dependency) and WEE1 expression as predictive biomarkers of response to WEE1-inhibitor therapy are of great interest, and remain an area of substantial controversy in preclinical studies109,111,114,115.
The inhibitory Notch ligand DLL3
DLL3 is normally expressed in the developing CNS and has a key role in somitogenesis, the process by which somites (bilaterally paired blocks of mesoderm tissue) form along the anterior–posterior axis of the developing embryo116,117. The Notch pathway has been implicated in regulating neuroendocrine versus epithelial-cell differentiation in embryonic lung development118 and, more recently, in SCLC oncogenesis12. Notch activation is oncogenic in some tumour types; however, in neuroendocrine tumours, Notch signalling suppresses oncogenesis and tumour growth12,119. Unlike other mammalian Notch family members, DLL3 is predominantly located in the Golgi apparatus and inhibits Notch 1 signalling in cis120. In high-grade neuroendocrine tumours, including SCLC, DLL3 is highly upregulated and aberrantly expressed on the cell surface, making it a potential therapeutic target121.
Rovalpituzumab tesirine (Rova-T) is a novel, first-in-class, antibody–drug conjugate with high specificity for DLL3. Rova-T binds to DLL3 expressed on the cell surface, is internalized, and subsequent cleavage of a linker moiety releases the pyrrolobenzodiazepine (PBD) dimer cytotoxic payload from the anti-DLL3 antibody, resulting in tumour-specific DNA damage and cell death. PBDs, originally discovered in Streptomyces species of bacteria, are a class of sequence-selective, DNA-minor-groove-binding agents that form covalent crosslinks between the N2 of guanine and the C11 position of the PBD122,123. The resulting PBD–DNA adduct leads to replication-fork stalling, preventing DNA replication and causing cell-cycle arrest. PBDs are not inherently tumour specific; thus, conjugation of PBDs to an anti-DLL3 antibody enables a potential targeted therapeutic approach in SCLC. Preclinical findings demonstrated in vivo efficacy of Rova-T in patient-derived xenograft models of SCLC, with a strong correlation between the level of DLL3 expression and therapeutic activity121.
Results of the first-in-human phase I clinical trial of Rova-T in patients with recurrent metastatic SCLC and large-cell neuroendocrine lung cancers were published in 2017 (REF. 124). The study investigators defined a recommended phase II dose and schedule (0.3 mg/kg every 6 weeks), and identified dose-limiting toxicities, including thrombocytopenia, liver-test abnormalities, and serosal effusions. Among the evaluable patients, 17% (11 of 65) had a confirmed objective response and 54% (35 of 65) had stable disease. The median duration of response was 5.6 months (95% CI 2.5–8.3 months), median PFS was 3.1 months (95% CI 2.7–4.1 months), and the median overall survival was 4.6 months (95% CI 3.9–7.1 months). In patients with a high level of DLL3 expression (defined immunohistochemically as detectable protein expression in at least 50% of tumour cells), the ORR was 39% (10 of 26) and the disease-control rate (stable disease or objective response) was 89% (22 of 26). Most notably, among those patients with tissue available for protein analysis, responses were observed exclusively in those with a high DLL3 expression level, further supporting the preclinical observation that the efficacy of Rova-T correlates with the level of DLL3 expression. In an exploratory analysis, median PFS was 4.5 months (95% CI 3.0–5.4 months) in the DLL3-high patient subgroup compared with 2.3 months (95% CI 1.3–3.3 months) in the DLL3-low subgroup; overall survival differences between the DLL3-high and DLL3-low subgroups were not reported124. These data support DLL3 as a candidate predictive bio-marker for this therapy — potentially the first such bio-marker in SCLC. These results, in a heavily pretreated patient population, seem promising; however, additional studies are needed to determine the clinical benefits of Rova-T treatment and multiple trials of this agent are ongoing (TABLE 1), including an open-label, multicentre, phase II study of the efficacy of Rova-T in the third-line and later-line treatment of patients with DLL3-positive ES-SCLC (NCT02674568). Further proposed studies of Rova-T in patients with ES-SCLC encompass frontline treatment of DLL3-high disease (NCT02819999), maintenance therapy following first-line platinum-based chemotherapy (NCT03033511), and combination treatment with nivolumab — with or without ipilimumab — in the second-line setting (NCT03026166).
Aurora kinase
The aurora kinase family proteins have key roles in mitosis. Aurora kinase A is essential for centrosome function, spindle assembly, chromosome alignment, and mitotic entry125. Knockdown of Aurora A expression induces G2/M-phase arrest and thereby inhibits the proliferation of human SCLC cells126. Moreover, targeted drug screens have indicated that the neuroendocrine-low, variant subtype of SCLC with high MYC and NEUROD1 expression is vulnerable to aurora kinase inhibition, which strongly suppressed tumour progression when combined with chemotherapy in the aforementioned mouse model of this disease subtype49. Alisertib, an investigational, orally administered, selective inhibitor of aurora kinase A, has preclinical therapeutic activity across multiple tumour types127. In a multicentre phase I/II study of this agent in patients with various solid tumours128, a recommended phase II dose and schedule was defined, and dose-limiting toxicities, including neutropenia, leukopenia, and anaemia, were noted. In a phase II study expansion cohort comprising patients with relapsed and/or refractory SCLC, single-agent alisertib therapy resulted in an ORR of 21% (10 of 48 patients; 95% CI 10–35)128. Furthermore, in a randomized phase II study of paclitaxel plus either alisertib or placebo in the second-line treatment of patients with SCLC, the ORR was 22% in the experimental arm (20 of 89) and 18% in the control arm (16 of 89); the median PFS was 101 days versus 66 days (HR 0.71,95% CI 0.51–0.99; P= 0.04)129. Further clinical investigations are needed to better study and optimize the therapeutic utility of this compound in SCLC.
Immunotherapy for SCLC
Escape from immune surveillance is a well-recognized feature of cancer130. The development of therapies to enhance antitumour immune responses — particularly antagonistic antibodies targeting the inhibitory immune-checkpoint proteins cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1), or its ligand PD-L1 (FIG. 2) — has led to exciting new treatment options for patients, across multiple tumour types131,132. The high mutational burden of SCLCs, resulting in a large number of potential tumour-specific antigens, has raised hope that immunotherapy might be effective in this disease. At present, only limited data have been reported on immune-checkpoint blockade in patients with SCLC, although a number of clinical trials of this promising therapeutic approach are underway (TABLE 2).
Table 2.
Ongoing studies of immune-checkpoint blockade in small-cell lung cancer
| Immunotherapy | Study name | Study phase | ClinicalTrials.gov study identifier | Estimated primary completion date |
|---|---|---|---|---|
| LS-SCLC | ||||
| Nivolumab plus ipilimumab | STIMULI | II | NCT02046733 | October 2019 |
| Pembrolizumab and concurrent radiotherapy ± chemotherapy (cis/carboplatin and etoposide) | NA | I | NCT02402920 | July 2023 |
| ES-SCLC: first line | ||||
| Pembrolizumab with cis/carboplatin and etoposide | KEYNOTE-011 | I | NCT01840579 | June 2019 |
| Cis/carboplatin and etoposide ± pembrolizumab | REACTION | II | NCT02580994 | June 2020 |
| ES-SCLC: maintenance | ||||
| Nivolumab ± ipilimumab versus placebo | CheckMate 451 | III | NCT02538666 | September 2018 |
| ES-SCLC: second line and beyond | ||||
| Nivolumab versus topotecan or amrubicin | CheckMate 331 | III | NCT02481830 | March 2018 |
| Carboplatin and etoposide plus atezolizumab or placebo | IMpower133 | I/III | NCT02763579 | June 2019 |
| Tremelimumab and durvalumab ± radiation | NCI-2016-00026/Winship3112-15/ESR-14-10531 | II | NCT02701400 | April 2019 |
| Pembrolizumab versus topotecan | AFT-17 | II | NCT02963090 | May 2019 |
| BMS-986012 ± nivolumab | CA001-030 | I/II | NCT02247349 | October 2018 |
| Pembrolizumab | KEYNOTE-158 | II | NCT02628067 | September 2017 |
| Pembrolizumab plus irinotecan | PembroPlus | Ib/II | NCT02331251 | December 2016 |
| Durvalumab plus olaparib | MEDIOLA | I/II | NCT02734004 | October 2018 |
Cis/carboplatin, cisplatin or carboplatin; ES-SCLC, extensive-stage SCLC; LS-SCLC, limited-stage SCLC; NA, not applicable.
CTLA-4
Ipilimumab, a fully human monoclonal IgG1, binds to CTLA-4 expressed by T cells and blocks the interaction of this receptor with its ligands CD80 and CD86 on antigen-presenting cells (FIG. 2). Upon ligand binding, CTLA-4 transmits signals that suppress T-cell priming, and thus blockade of this interaction using ipilimumab can promote T-cell activation and an anticancer immune response133. In a randomized phase II study, investigators evaluated the activity of ipilimumab in combination with paclitaxel and carboplatin versus paclitaxel and carboplatin alone in patients with ES-SCLC134; 133 patients with previously untreated ES-SCLC were randomly assigned (1:1:1) to receive either concurrent ipilimumab and chemotherapy, phased-ipilimumab and chemotherapy, or the control regimen of chemotherapy alone. This study was not stratified by PD-L1 expression status, and the immune-related response criteria (irRC) were used to assess response to therapy. The irRC are modified RECIST criteria that capture the unique tumour response patterns observed with immunotherapy, including regression of index lesions coincident with the appearance of new lesions, and initial progression followed by tumour stabilization or a decrease in tumour burden135. The phased administration of ipilimumab in combination with paclitaxel and carboplatin, after induction with the same chemotherapy regimen plus placebo, resulted in an improvement in immune-related PFS (as per the iRC) compared with that observed with induction and maintenance chemotherapy plus placebo (HR 0.64; P= 0.03); however, no significant improvement in PFS (HR 0.93; P= 0.37) or overall survival (HR 0.75; P= 0.13) was demonstrated134. Moreover, induction therapy with concurrent ipilimumab plus chemotherapy with maintenance chemotherapy plus placebo did not improve patient outcomes. In the same setting, a follow-up phase III trial of phased ipilimumab in combination with more-conventional first-line chemotherapy (comprising etoposide and a platinum-based agent) versus the same chemotherapy regimen plus placebo was negative for its primary end point of overall survival: median 11.0 months versus 10.9 months (HR 0.94, 95% CI 0.81–1.09; P= 0.38)136. Furthermore, ipilimumab was associated with a high frequency of some toxicities, including diarrhoea, rash, and colitis136.
PD-1 and PD-L1 blockade
PD-L1 is expressed on a range of cell types, including some neoplastic and non-neoplastic cells within tumours, and interaction of this protein with PD-1 on T cells results in local suppression of T-cell activation and cytotoxicity, and promotes T-cell exhaustion (FIG. 2). Nivolumab, a fully human IgG4, binds to PD-1 and blocks its interaction with PD-L1, which can reinvigorate T-cell activity and potentially unleash suppressed antitumour immunity. Although ipilimumab alone did not improve chemotherapy responsiveness of patients with SCLC136, preclinical data from the melanoma literature suggests that combined PD-1 and CTLA-4 blockade can synergistically enhance activation of tumour-specific T cells and antitumour activity through complementary mechanisms137. Outcome data from CheckMate 032, a multicentre, open-label, phase I/II trial of nivolumab with or without ipilimumab in patients with recurrent ES-SCLC138, was reported in 2016. The study investigators enrolled 216 patients, including 98 patients treated with nivolumab alone and the remainder with alternative schedules incorporating both nivolumab and ipilimumab. The toxicity profiles were similar to those observed in prior studies of these agents, including grade 3–4 events in 13% of patients receiving nivolumab alone, and between 19% and 30% of patients receiving nivolumab and ipilimumab. The ORRs were 10% in the nivolumab arm and 19–23% in the combination arms, translating into an encouraging 1-year survival of 33% and 35–43%, respectively138. These results have led to the incorporation of the nivolumab and ipilimumab combination as a National Comprehensive Cancer Network (NCCN) guideline recommendation for the second-line treatment of ES-SCLC20. However, the efficacy of checkpoint inhibitors in patients with SCLC needs to be confirmed in randomized trials, and these treatments have not been formally approved for the treatment of SCLC in the USA or elsewhere and, therefore, remain experimental. Potential toxicity remains an ongoing concern with combination immunotherapy, and physicians should have a substantive discussion with their patients regarding potential risks and benefits of treatment. Close monitoring of the endocrine axis and early detection of immune-related adverse events, such as colitis and pneumonitis, are critical because patients can benefit from early intervention with high-dose steroids and/or drug discontinuation.
A high mutational burden has been identified as a potential predictor of effective immunotherapy139; however, despite the fact that SCLCs have among the highest mutational burdens of all tumour types11, the clinical efficacy of checkpoint inhibitors in this disease seems to be far less pronounced than would be expected based on the experiences in other highly mutated cancers, such as melanoma and NSCLC. In stark contrast to other tumour types, SCLCs rarely express PD-L1, which might at least partially explain this disparity. In an immunohistochemical analysis of archival formalin-fixed paraffin-embedded SCLC specimens using two different assays (VENTANA PD-L1 (SP142) and 28–8 pharmDx)140, the overall prevalence of PD-L1 expression in tumour cells was found to be low (16.5%) and was not markedly different between LS-SCLC and ES-SCLC samples. Importantly, PD-L1 positivity does not seem to be predictive of immunotherapy response in this setting138. Work to define predictive biomarkers of immunotherapy response and the characteristics of the SCLC immune microenvironment is ongoing. In addition, multiple clinical trials of combination strategies to bolster the efficacy of immune-checkpoint inhibition are underway in patients with SCLC (TABLE 2).
CD47
CD47 is a cell-surface molecule that inhibits activation and phagocytic activity of macrophages by engaging signal-regulatory protein α141 (SIRPα; FIG. 2). In fact, CD47 is involved in regulating a wide variety of physiological processes, including platelet and neutrophil activation, T-cell function, vascular signalling by nitric oxide, suppression of dendritic cell activity, and inhibition of monocyte activation142. This protein is expressed on many normal cells, but is highly upregulated on the surface of human SCLC cells and has been implicated in immune escape by tumours143. In particular, disruption of the interaction of CD47 and SIRPα using anti-CD47 antibodies induced macrophage-mediated phagocytosis of human SCLC cell lines in vitro and in mouse xenograft models143. Moreover, CD47 blockade has been shown to trigger T-cell-mediated destruction of immunogenic tumours144, and thus combination strategies with immune-checkpoint blockade might enhance the antitumour effects of anti-CD47 antibodies. Clinical exploration of CD47/SIRPα inhibition as a therapeutic strategy for SCLC is expected to begin in 2017.
Conclusions
Large-scale genomic, proteomic, and transcriptomic analyses have led to the identification of new druggable targets in SCLC (FIG. 2). PARP1, EZH2, WEE1, and DLL3 are all examples of novel targets implicated as vulnerabilities, and tractable therapeutic opportunities, in SCLC; drugs for each of these targets are under active clinical investigation. Rapid progress in the field of immuno-oncology has similarly opened a door to new treatment options. Treatment with the combination of ipilimumab and nivolumab is now NCCN-recommended for patients with recurrent SCLC after platinum-based therapy. Nevertheless, the immune microenvironment of SCLC seems to be distinct from that of other solid tumours, with alternative targets, such as CD47, coming to the fore. Defining predictive biomarkers for targeted therapies and optimizing activation of antitumour immune response in SCLC are areas of intensive ongoing investigation. The progress made in defining novel therapeutic targets in SCLC has renewed hope for advances in combatting this recalcitrant disease. Several ongoing and upcoming clinical trials will test whether these new insights into tumour biology can be successfully translated into major therapeutic breakthroughs in what has been a singularly challenging and deadly disease.
Key points.
Small-cell lung cancer (SCLC) is a high-grade neuroendocrine tumour associated with a poor overall survival, and limited progress has been made in the treatment of this disease over the past three decades
Over the past 5 years, advances in our understanding of multiple aspects of the biology of SCLC have led to the development of new therapies that are currently under clinical investigation
Poly [ADP-ribose] polymerase (PARP) is abundantly expressed in SCLC and is involved in DNA-damage repair; clinical trials of the PARP inhibitors veliparib, olaparib, and talazoparib are ongoing in patients with SCLC
Enhancer of zeste homologue 2 (EZH2) is a regulator of chromatin remodelling that can drive acquired chemoresistance; therapeutic targeting of EZH2 might augment and extend the durability of chemotherapy responses
Delta-like protein 3 (DLL3) is an inhibitory Notch ligand that is overexpressed in many SCLCs; rovalpituzumab tesirine (Rova-T), an anti-DLL3-antibody drug conjugate, has shown promising activity in preclinical and early phase clinical studies
SCLC has a high mutational burden, raising hopes regarding immunotherapy, and immune-checkpoint blockade has shown encouraging clinical activity in patients with this disease, despite typically low tumoural expression of immune-checkpoint proteins
Acknowledgments
The work of the authors is supported by funding from the National Cancer Institute (grants T32 CA009207 to J.K.S. and R01 CA197936, P30 CA008748 to C.M.R.), and from the Conquer Cancer Foundation of ASCO, the Lung Cancer Research Foundation, and the Radiological Society of North America (to B.H.L.).
Biographies
Joshua K. Sabari, MD, is a medical oncology fellow at the Memorial Sloan Kettering Cancer Center (MSKCC), New York, New York, USA.
Benjamin H. Lok, MD, is a radiation oncology resident at MSKCC.
James H. Laird is a medical student at New York University School of Medicine, USA.
John T. Poirier, PhD, is an Assistant Member in the Department of Medicine and the Molecular Pharmacology Program at MSKCC.
Charles M. Rudin, MD, PhD, is a Member in the Department of Medicine and the Molecular Pharmacology Program at the MSKCC, and a professor of medicine at Weill Cornell Medical College, New York, New York, USA.
Footnotes
Author contributions
J.K.S. and B.H.L. made equal contributions to the manuscript and should be considered co-first authors. J.K.S., B.H.L., J.H.L., and J.T.P. researched data for article. All authors made substantial contributions to discussion of content, writing of the Review, and review/editing of the manuscript.
Competing interests statement
C.M.R. has been a paid consultant regarding oncology drug development for Bristol Myers Squibb, Celgene, G1 Therapeutics, Harpoon Therapeutics, Medivation, and Novartis. The other authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Subject ontology terms
Health sciences / Diseases / Cancer / Lung cancer / Small-cell lung cancer
[URI/692/699/67/1612/2143]
Health sciences / Diseases / Cancer / Oncogenes
[URI/692/699/67/395]
Health sciences / Diseases / Cancer / Cancer therapy / Cancer immunotherapy
[URI/692/699/67/1059/2325]
Health sciences / Diseases / Cancer / Cancer therapy / Targeted therapies
[URI/692/699/67/1059/602]
Health sciences / Diseases / Cancer / Cancer therapy / Cancer therapeutic resistance
[URI/692/699/67/1059/2326]
References
- 1.Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson AG, et al. The International Association for the Study of Lung Cancer Lung Cancer staging project: proposals for the revision of the clinical and pathologic staging of small cell lung cancer in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:300–311. doi: 10.1016/j.jtho.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, et al. Cancer Statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 6.Govindan R, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the Surveillance, Epidemiologic, and End Results database. J Clin Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 7.American Cancer Society. Cancer Facts & Figures 2016. American Cancer Society; 2016. [Google Scholar]
- 8.Fitzmaurice C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pesch B, et al. Cigarette smoking and lung cancer — relative risk estimates for the major histological types from a pooled analysis of case-control studies. Int J Cancer. 2012;131:1210–1219. doi: 10.1002/ijc.27339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varghese AM, et al. Small-cell lung cancers in patients who never smoked cigarettes. J Thorac Oncol. 2014;9:892–896. doi: 10.1097/JTO.0000000000000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George J, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudin CM, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brambilla E, BM, Auerbach O, Kuschner M. In: WHO Classification of Tumors: Tumors of the Lung, Pleura, Thymus and Heart — Pathology and Genetics. Travis WD, Brambilla E, Burke AP, Marx A, Nicholson GA, editors. IARC; 2015. [DOI] [PubMed] [Google Scholar]
- 15.Silva M, et al. Screening with low-dose computed tomography does not improve survival of small cell lung cancer. J Thorac Oncol. 2016;11:187–193. doi: 10.1016/j.jtho.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Pignon JP, et al. A meta-analysis of thoracic radiotherapy for small-cell lung cancer. N Engl J Med. 1992;327:1618–1624. doi: 10.1056/NEJM199212033272302. [DOI] [PubMed] [Google Scholar]
- 17.Turrisi AT, III , et al. Twice-daily compared with once-daily thoracic radiotherapy in limited small-cell lung cancer treated concurrently with cisplatin and etoposide. N Engl J Med. 1999;340:265–271. doi: 10.1056/NEJM199901283400403. [DOI] [PubMed] [Google Scholar]
- 18.Turrisi AT, III, Glover DJ, Mason BA. A preliminary report: concurrent twice-daily radiotherapy plus platinum-etoposide chemotherapy for limited small cell lung cancer. Int J Radiat Oncol Biol Phys. 1988;15:183–187. doi: 10.1016/0360-3016(88)90364-1. [DOI] [PubMed] [Google Scholar]
- 19.Faivre-Finn C, et al. CONVERT: an international randomised trial of concurrent chemo-radiotherapy (cCTRT) comparing twice-daily (BD) and once-daily (OD) radiotherapy schedules in patients with limited stage small cell lung cancer (LS-SCLC) and good performance status (PS) [abstract] J Clin Oncol. 2016;34:8504. doi: 10.1136/bmjopen-2015-009849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Comprehensive Cancer Network. National Comprehensive Cancer Network Guidelines: Small Cell Lung Cancer Version 2.2017. NCCN; 2016. [DOI] [PubMed] [Google Scholar]
- 21.Aupérin A, et al. Prophylactic cranial irradiation for patients with small-cell lung cancer in complete remission. N Engl J Med. 1999;341:476–484. doi: 10.1056/NEJM199908123410703. [DOI] [PubMed] [Google Scholar]
- 22.Vallières E, et al. The IASLC lung cancer staging project: proposals regarding the relevance of TNM in the pathologic staging of small cell lung cancer in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol. 2009;4:1049–1059. doi: 10.1097/JTO.0b013e3181b27799. [DOI] [PubMed] [Google Scholar]
- 23.Gaspar LE, et al. Small-cell lung cancer: prognostic factors and changing treatment over 15 years. Clin Lung Cancer. 2012;13:115–122. doi: 10.1016/j.cllc.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Fukuoka M, et al. Randomized trial of cyclophosphamide, doxorubicin, and vincristine versus cisplatin and etoposide versus alternation of these regimens in small-cell lung cancer. J Natl Cancer Inst. 1991;83:855–861. doi: 10.1093/jnci/83.12.855. [DOI] [PubMed] [Google Scholar]
- 25.Roth BJ, et al. Randomized study of cyclophosphamide, doxorubicin, and vincristine versus etoposide and cisplatin versus alternation of these two regimens in extensive small-cell lung cancer: a phase III trial of the Southeastern Cancer Study Group. J Clin Oncol. 1992;10:282–291. doi: 10.1200/JCO.1992.10.2.282. [DOI] [PubMed] [Google Scholar]
- 26.Niell HB, et al. Randomized phase III intergroup trial of etoposide and cisplatin with or without paclitaxel and granulocyte colony-stimulating factor in patients with extensive-stage small-cell lung cancer: Cancer and Leukemia Group B Trial 9732. J Clin Oncol. 2005;23:3752–3759. doi: 10.1200/JCO.2005.09.071. [DOI] [PubMed] [Google Scholar]
- 27.Schmittel A, et al. A randomized phase II trial of irinotecan plus carboplatin versus etoposide plus carboplatin treatment in patients with extended disease small-cell lung cancer. Ann Oncol. 2006;17:663–667. doi: 10.1093/annonc/mdj137. [DOI] [PubMed] [Google Scholar]
- 28.Noda K, et al. Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N Engl J Med. 2002;346:85–91. doi: 10.1056/NEJMoa003034. [DOI] [PubMed] [Google Scholar]
- 29.Hanna N, et al. Randomized phase III trial comparing irinotecan/cisplatin with etoposide/cisplatin in patients with previously untreated extensive-stage disease small-cell lung cancer. J Clin Oncol. 2006;24:2038–2043. doi: 10.1200/JCO.2005.04.8595. [DOI] [PubMed] [Google Scholar]
- 30.Rossi A, et al. Carboplatin- or cisplatin-based chemotherapy in first-line treatment of small-cell lung cancer: the COCIS meta-analysis of individual patient data. J Clin Oncol. 2012;30:1692–1698. doi: 10.1200/JCO.2011.40.4905. [DOI] [PubMed] [Google Scholar]
- 31.Primo NL, et al. Phase III trial of irinotecan/cisplatin compared with etoposide/cisplatin in extensive-stage small-cell lung cancer: clinical and pharmacogenomic results from SWOG S0124. J Clin Oncol. 2009;27:2530–2535. doi: 10.1200/JCO.2008.20.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ardizzoni A, et al. Topotecan, a new active drug in the second-line treatment of small-cell lung cancer: a phase II study in patients with refractory and sensitive disease. The European Organization for Research and Treatment of Cancer Early Clinical Studies Group and New Drug Development Office, and the Lung Cancer Cooperative Group. J Clin Oncol. 1997;15:2090–2096. doi: 10.1200/JCO.1997.15.5.2090. [DOI] [PubMed] [Google Scholar]
- 33.Slotman BJ, et al. Use of thoracic radiotherapy for extensive stage small-cell lung cancer: a phase 3 randomised controlled trial. Lancet. 2015;385:36–42. doi: 10.1016/S0140-6736(14)61085-0. [DOI] [PubMed] [Google Scholar]
- 34.Slotman BJ, et al. Radiotherapy for extensive stage small-cell lung cancer — authors’ reply. Lancet. 2015;385:1292–1293. doi: 10.1016/S0140-6736(15)60679-1. [DOI] [PubMed] [Google Scholar]
- 35.Slotman B, et al. Prophylactic cranial irradiation in extensive small-cell lung cancer. N Engl J Med. 2007;357:664–672. doi: 10.1056/NEJMoa071780. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi T, et al. Prophylactic cranial irradiation versus observation in patients with extensive-disease small-cell lung cancer: a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2017;18:663–671. doi: 10.1016/S1470-2045(17)30230-9. [DOI] [PubMed] [Google Scholar]
- 37.Giuliani M, et al. Utilization of prophylactic cranial irradiation in patients with limited stage small cell lung carcinoma. Cancer. 2010;116:5694–5699. doi: 10.1002/cncr.25341. [DOI] [PubMed] [Google Scholar]
- 38.Lok BH, et al. The factors influencing the utilization of prophylactic cranial irradiation in patients with limited-stage small cell lung cancer. Int J Radiat Oncol Biol Phys. 2015;93:E420–E421. doi: 10.1016/j.adro.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borromeo MD, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poirier JT, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene. 2015;34:5869–5878. doi: 10.1038/onc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poirier JT, et al. Selective tropism of Seneca Valley virus for variant subtype small cell lung cancer. J Natl Cancer Inst. 2013;105:1059–1065. doi: 10.1093/jnci/djt130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gazdar AF, Carney DN, Nau MM, Minna JD. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res. 1985;45:2924–2930. [PubMed] [Google Scholar]
- 43.Carney DN, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–2923. [PubMed] [Google Scholar]
- 44.Meuwissen R, et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 45.Gazdar AF, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol. 2015;10:553–564. doi: 10.1097/JTO.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito T, et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127:3913–3921. doi: 10.1242/dev.127.18.3913. [DOI] [PubMed] [Google Scholar]
- 47.Jiang T, et al. Achaetescute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res. 2009;69:845–854. doi: 10.1158/0008-5472.CAN-08-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osada H, Tatematsu Y, Yatabe Y, Horio Y, Takahashi T. ASH1 gene is a specific therapeutic target for lung cancers with neuroendocrine features. Cancer Res. 2005;65:10680–10685. doi: 10.1158/0008-5472.CAN-05-1404. [DOI] [PubMed] [Google Scholar]
- 49.Mollaoglu G, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell. 2017;31:270–285. doi: 10.1016/j.ccell.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Augustyn A, et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci USA. 2014;111:14788–14793. doi: 10.1073/pnas.1410419111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osborne JK, et al. NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas via signaling molecules TrkB and NCAM. Proc Natl Acad Sci USA. 2013;110:6524–6529. doi: 10.1073/pnas.1303932110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neptune ER, et al. Targeted disruption of NeuroD, a proneural basic helix-loop-helix factor, impairs distal lung formation and neuroendocrine morphology in the neonatal lung. J Biol Chem. 2008;283:21160–21169. doi: 10.1074/jbc.M708692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Girard L, Zöchbauer-Müller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- 54.Shivapurkar N, et al. Deletions of chromosome 4 at multiple sites are frequent in malignant mesothelioma and small cell lung carcinoma. Clin Cancer Res. 1999;5:17–23. [PubMed] [Google Scholar]
- 55.Petersen I, et al. Small-cell lung cancer is characterized by a high incidence of deletions on chromosomes 3p, 4q, 5q, 10q, 13q and 17p. Br J Cancer. 1997;75:79–86. doi: 10.1038/bjc.1997.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson BE, et al. MYC family DNA amplification in small cell lung cancer patients’ tumors and corresponding cell lines. Cancer Res. 1988;48:5163–5166. [PubMed] [Google Scholar]
- 57.Brambilla E, et al. Apoptosis-related factors p53, Bcl2, and Bax in neuroendocrine lung tumors. Am J Pathol. 1996;149:1941–1952. [PMC free article] [PubMed] [Google Scholar]
- 58.Tamborini E, et al. Detection of overexpressed and phosphorylated wild-type kit receptor in surgical specimens of small cell lung cancer. Clin Cancer Res. 2004;10:8214–8219. doi: 10.1158/1078-0432.CCR-04-1013. [DOI] [PubMed] [Google Scholar]
- 59.Carney DN, Cuttitta F, Moody TW, Minna JD. Selective stimulation of small cell lung cancer clonal growth by bombesin and gastrin-releasing peptide. Cancer Res. 1987;47:821–825. [PubMed] [Google Scholar]
- 60.Pedersen N, et al. Transcriptional gene expression profiling of small cell lung cancer cells. Cancer Res. 2003;63:1943–1953. [PubMed] [Google Scholar]
- 61.Fujino K, et al. Insulinoma-associated protein 1 is a crucial regulator of neuroendocrine differentiation in lung cancer. Am J Pathol. 2015;185:3164–3177. doi: 10.1016/j.ajpath.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 62.Borges M, et al. An achaetescute homologue essential for neuroendocrine differentiation in the lung. Nature. 1997;386:852–855. doi: 10.1038/386852a0. [DOI] [PubMed] [Google Scholar]
- 63.Peifer M, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dowlati A, et al. Clinical correlation of extensive-stage small-cell lung cancer genomics. Ann Oncol. 2016;27:642–647. doi: 10.1093/annonc/mdw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baylin SB, Jones PA. A decade of exploring the cancer epigenome — biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 68.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 69.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 70.Hopkins-Donaldson S, et al. Silencing of death receptor and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ. 2003;10:356–364. doi: 10.1038/sj.cdd.4401157. [DOI] [PubMed] [Google Scholar]
- 71.Kaminskyy VO, Surova OV, Vaculova A, Zhivotovsky B. Combined inhibition of DNA methyltransferase and histone deacetylase restores caspase-8 expression and sensitizes SCLC cells to TRAIL. Carcinogenesis. 2011;32:1450–1458. doi: 10.1093/carcin/bgr135. [DOI] [PubMed] [Google Scholar]
- 72.Kalari S, Jung M, Kernstine KH, Takahashi T, Pfeifer GP. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene. 2013;32:3559–3568. doi: 10.1038/onc.2012.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, et al. Prognostic and predictive value of CpG island methylator phenotype in patients with locally advanced nonmetastatic sporadic colorectal cancer. Gastroenterol Res Pract. 2014;2014:436985. doi: 10.1155/2014/436985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karlsson A, et al. Genome-wide DNA methylation analysis of lung carcinoma reveals one neuroendocrine and four adenocarcinoma epitypes associated with patient outcome. Clin Cancer Res. 2014;20:6127–6140. doi: 10.1158/1078-0432.CCR-14-1087. [DOI] [PubMed] [Google Scholar]
- 75.Saito Y, et al. Prognostic significance of CpG island methylator phenotype in surgically resected small cell lung carcinoma. Cancer Sci. 2016;107:320–325. doi: 10.1111/cas.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Byers LA, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2:798–811. doi: 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 78.Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat Commun. 2013;4:2240. doi: 10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simbulan-Rosenthal CM, et al. PARP-1 binds E2F-1 independently of its DNA binding and catalytic domains, and acts as a novel coactivator of E2F-1-mediated transcription during re-entry of quiescent cells into S phase. Oncogene. 2003;22:8460–8471. doi: 10.1038/sj.onc.1206897. [DOI] [PubMed] [Google Scholar]
- 81.de Bono J, et al. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-16-1250. http://dx.doi.org/10.1158/2159-8290.CD-16-1250. [DOI] [PMC free article] [PubMed]
- 82.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 83.Swisher EM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 84.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 85.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 86.Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32:3552–3558. doi: 10.1038/onc.2012.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lok BH, Powell SN. Molecular pathways: understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin Cancer Res. 2012;18:6400–6406. doi: 10.1158/1078-0432.CCR-11-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ceccaldi R, et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mateos-Gomez PA, et al. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16:211. doi: 10.1186/bcr3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mirza MR, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 92.Lok BH, et al. PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin Cancer Res. 2017;23:523–535. doi: 10.1158/1078-0432.CCR-16-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Murai J, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget. 2016;7:76534–76550. doi: 10.18632/oncotarget.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mu Y, et al. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 2016;17:94–109. doi: 10.15252/embr.201540964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Deng Y, et al. High SLFN11 expression predicts better survival for patients with KRAS exon 2 wild type colorectal cancer after treated with adjuvant oxaliplatin-based treatment. BMC Cancer. 2015;15:833. doi: 10.1186/s12885-015-1840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang SW, et al. SLFN11 is a transcriptional target of EWS-FLI1 and a determinant of drug response in Ewing sarcoma. Clin Cancer Res. 2015;21:4184–4193. doi: 10.1158/1078-0432.CCR-14-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nogales V, et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget. 2016;7:3084–3097. doi: 10.18632/oncotarget.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Byers LA, et al. Improved small cell lung cancer response rates with veliparib and temozolomide: results from a phase II trial [abstract MA11.07] J Thorac Oncol. 2016;12:S406–S407. [Google Scholar]
- 100.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Coe BP, et al. Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer. PLoS ONE. 2013;8:e71670. doi: 10.1371/journal.pone.0071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gardner EE, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell. 2017;31:286–299. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 106.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 107.Dobbelstein M, Sorensen CS. Exploiting replicative stress to treat cancer. Nat Rev Drug Discov. 2015;14:405–423. doi: 10.1038/nrd4553. [DOI] [PubMed] [Google Scholar]
- 108.Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 109.Hirai H, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8:2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- 110.Hirai H, et al. MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol Ther. 2010;9:514–522. doi: 10.4161/cbt.9.7.11115. [DOI] [PubMed] [Google Scholar]
- 111.Rajeshkumar NV, et al. MK-1775, a potent Wee1 inhibitor, synergizes with gemcitabine to achieve tumor regressions, selectively in p53-deficient pancreatic cancer xenografts. Clin Cancer Res. 2011;17:2799–2806. doi: 10.1158/1078-0432.CCR-10-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Matheson CJ, Backos DS, Reigan P. Targeting WEE1 kinase in cancer. Trends Pharmacol Sci. 2016;37:872–881. doi: 10.1016/j.tips.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 113.Do K, et al. Phase I study of single-agent AZD1775 (MK-1775), a Wee1 kinase inhibitor, in patients with refractory solid tumors. J Clin Oncol. 2015;33:3409–3415. doi: 10.1200/JCO.2014.60.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guertin AD, et al. Preclinical evaluation of the WEE1 inhibitor MK-1775 as single-agent anticancer therapy. Mol Cancer Ther. 2013;12:1442–1452. doi: 10.1158/1535-7163.MCT-13-0025. [DOI] [PubMed] [Google Scholar]
- 115.Van Linden AA, et al. Inhibition of Wee1 sensitizes cancer cells to antimetabolite chemotherapeutics in vitro and in vivo, independent of p53 functionality. Mol Cancer Ther. 2013;12:2675–2684. doi: 10.1158/1535-7163.MCT-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ladi E, et al. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol. 2005;170:983–992. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Loomes KM, et al. Dll3 and Notch1 genetic interactions model axial segmental and craniofacial malformations of human birth defects. Dev Dyn. 2007;236:2943–2951. doi: 10.1002/dvdy.21296. [DOI] [PubMed] [Google Scholar]
- 118.Morimoto M, Nishinakamura R, Saga Y, Kopan R. Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development. 2012;139:4365–4373. doi: 10.1242/dev.083840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kunnimalaiyaan M, Chen H. Tumor suppressor role of Notch-1 signaling in neuroendocrine tumors. Oncologist. 2007;12:535–542. doi: 10.1634/theoncologist.12-5-535. [DOI] [PubMed] [Google Scholar]
- 120.Chapman G, Sparrow DB, Kremmer E, Dunwoodie SL. Notch inhibition by the ligand DELTA-LIKE 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum Mol Genet. 2011;20:905–916. doi: 10.1093/hmg/ddq529. [DOI] [PubMed] [Google Scholar]
- 121.Saunders LR, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med. 2015;7:302ra136. doi: 10.1126/scitranslmed.aac9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Antonow D, Thurston DE. Synthesis of DNA-interactive pyrrolo[2,1–c][1,4]benzodiazepines (PBDs) Chem Rev. 2011;111:2815–2864. doi: 10.1021/cr100120f. [DOI] [PubMed] [Google Scholar]
- 123.Gerratana B. Biosynthesis, synthesis and biological activities of pyrrolobenzodiazepines. Med Res Rev. 2012;32:254–293. doi: 10.1002/med.20212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rudin CM, et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017;18:42–51. doi: 10.1016/S1470-2045(16)30565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL, Golemis EA. Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci. 2013;70:661–687. doi: 10.1007/s00018-012-1073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lu Y, et al. Knocking down the expression of Aurora A gene inhibited cell proliferation and induced G2/M phase arrest in human small cell lung cancer cells. Oncol Rep. 2014;32:243–249. doi: 10.3892/or.2014.3194. [DOI] [PubMed] [Google Scholar]
- 127.Manfredi MG, et al. Characterization of alisertib (MLN8237), an investigational small-molecule inhibitor of aurora A kinase using novel in vivo pharmacodynamic assays. Clin Cancer Res. 2011;17:7614–7624. doi: 10.1158/1078-0432.CCR-11-1536. [DOI] [PubMed] [Google Scholar]
- 128.Melichar B, et al. Safety and activity of alisertib, an investigational aurora kinase A inhibitor, in patients with breast cancer, small-cell lung cancer, non-small-cell lung cancer, head and neck squamous-cell carcinoma, and gastro-oesophageal adenocarcinoma: a five-arm phase 2 study. Lancet Oncol. 2015;16:395–405. doi: 10.1016/S1470-2045(15)70051-3. [DOI] [PubMed] [Google Scholar]
- 129.Owonikoko T, et al. Randomized phase 2 study: alisertib (MLN8237) or placebo + paclitaxel as second-line therapy for small-cell lung cancer (SCLC) [abstract OA05.05] J Thorac Oncol. 2017;12:S261–S262. [Google Scholar]
- 130.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 131.Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol. 2016;13:273–290. doi: 10.1038/nrclinonc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 133.Hoos A, et al. Development of ipilimumab: contribution to a new paradigm for cancer immunotherapy. Semin Oncol. 2010;37:533–546. doi: 10.1053/j.seminoncol.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 134.Reck M, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line therapy in extensive-disease-small-cell lung cancer: results from a randomized, double-blind, multicenter phase 2 trial. Ann Oncol. 2012;24:75–83. doi: 10.1093/annonc/mds213. [DOI] [PubMed] [Google Scholar]
- 135.Wolchok JD, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15:7412–7420. doi: 10.1158/1078-0432.CCR-09-1624. [DOI] [PubMed] [Google Scholar]
- 136.Reck M, et al. Phase III randomized trial of ipilimumab plus etoposide and platinum versus placebo plus etoposide and platinum in extensive-stage small-cell lung cancer. J Clin Oncol. 2016;34:3740–3748. doi: 10.1200/JCO.2016.67.6601. [DOI] [PubMed] [Google Scholar]
- 137.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci USA. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Antonia SJ, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol. 2016;17:883–895. doi: 10.1016/S1470-2045(16)30098-5. [DOI] [PubMed] [Google Scholar]
- 139.Rizvi NA, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu H, et al. PD-L1 expression by two complementary diagnostic assays and mRNA in situ hybridization in small cell lung cancer. J Thorac Oncol. 2017;12:110–120. doi: 10.1016/j.jtho.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Weiskopf K, Weissman IL. Macrophages are critical effectors of antibody therapies for cancer. MAbs. 2015;7:303–310. doi: 10.1080/19420862.2015.1011450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matozaki T, Murata Y, Okazawa H, Ohnishi H. Functions and molecular mechanisms of the CD47–SIRPα signalling pathway. Trends Cell Biol. 2009;19:72–80. doi: 10.1016/j.tcb.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 143.Weiskopf K, et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J Clin Invest. 2016;126:2610–2620. doi: 10.1172/JCI81603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu X, et al. CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat Med. 2015;21:1209–1215. doi: 10.1038/nm.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]