Abstract
The assessment of B-cell clonality is a critical component of the evaluation of suspected lymphoproliferative disorders, but analysis from formalin fixed paraffin embedded tissues can be challenging if fresh tissue is not available for flow cytometry. Immunohistochemical and conventional bright field in situ hybridization stains for kappa and lambda are effective for evaluation of plasma cells, but are often insufficiently sensitive to detect the much lower abundance of light chains present in B cells. We describe an ultrasensitive RNA in situ hybridization assay which has been adapted for use on an automated immunohistochemistry platform and compare results with flow cytometry in 203 consecutive tissues and 104 consecutive bone marrows. Overall, in 203 tissue biopsies, RNA in situ hybridization identified light chain restricted B-cells in 85 (42%) vs. 58 (29%) by flow cytometry. Within 83 B-cell non-Hodgkin lymphomas, RNA in situ hybridization identified a restricted B-cells in 74 (89%) vs. 56 (67%) by flow cytometry. B-cell clonality could be evaluated in only 23/104 (22%) bone marrow cases due to poor RNA preservation, but evaluable cases showed 91% concordance with flow cytometry. RNA in situ hybridization allowed for recognition of biclonal/composite lymphomas not identified by flow cytometry, and highlighted unexpected findings, such as coexpression of kappa and lambda RNA in 2 cases and the presence of lambda light chain RNA in a T lymphoblastic lymphoma. Automated RNA in situ hybridization showed excellent interobserver reproducibility for manual evaluation (average K=0.92), and an automated image analysis system showed high concordance (97%) with manual evaluation. Automated RNA in situ hybridization staining, which can be adopted on commonly utilized immunohistochemistry instruments, allows for the interpretation of clonality in the context of the morphologic features in formalin fixed, paraffin embedded tissues with a clinical sensitivity similar or superior to flow cytometry.
Keywords: Light chain restriction, B-cell lymphoma, CISH, RNA in situ hybridization, RNAscope
An evaluation of B-cell clonality through detection of light chain restriction is a mainstay of pathologic workup of suspected lymphoproliferative disorders.1,2 Currently, flow cytometric analysis serves as the gold standard for evaluation of kappa and lambda light chain protein expression due to a high sensitivity over a broad dynamic range of protein expression.3–5 Flow cytometry, however, requires fresh tissue for analysis, which is not always available in routine practice. Other methods may therefore be used in routine clinical diagnosis. PCR studies for IGH and IGK rearrangements can be used to document clonality from formalin fixed paraffin embedded tissue.6,7 PCR studies however are time consuming and are performed only in specialized molecular laboratories. PCR studies also separate detection of clonality from the morphologic findings, which can lead to challenges integrating unexpected test results. The detection of clonality by PCR also provides no information regarding which light chain is expressed as clonal IGH and IGK rearrangements are found in both kappa-restricted and lambda-restricted B-cells.6,7 Immunohistochemical stains and conventional colorimetric RNA in situ hybridization are widely used in routine clinical practice. These techniques, however, suffer from a limited dynamic range such that they are capable of detecting light chain restriction in plasma cells or other B-cells with very abundant amounts of light chain protein, but they are limited for detection of the much lower amounts of surface immunoglobulin present on most resting B-cells.8–11
In the last several years, advances in techniques for in situ hybridization have allowed for highly sensitive detection of RNA down to the single molecule level.12–18 In a prior study, we described a novel, ultrasensitive bright field RNA in situ hybridization for detection of light chain expression in B-cells.19 This technique employed manual staining procedures with probes to kappa light chain, lambda light chain, and IGLL5. IGLL5, an immunoglobulin-like protein that is expressed in lymphoid and non-lymphoid cells, shares the same sequences with the immunoglobulin light chain constant region and is also recognized by the lambda RNA in situ hybridization probe.19–21 The use of kappa, lambda, and IGLL5 probes in algorithmic fashion were previously shown to accurately detect B-cell clonality in multiple subtypes of B-cell lymphoma.19,22
In the current study, we have adapted this technique to an automated procedure performed on a commonly used immunohistochemistry platform available in routine clinical diagnostic laboratories. We further assessed the utility of this assay by analyzing 203 consecutive tissue biopsies and 104 consecutive bone marrow samples submitted for flow cytometric evaluation with a suspicion for lymphoma.
MATERIALS AND METHODS
Case Selection
This study was conducted following waiver of written informed consent and approval of the study by the Cleveland Clinic institutional review board. We identified 203 consecutive tissue samples and 104 consecutive bone marrow aspirate samples submitted to the Cleveland Clinic flow cytometry laboratory for workup of a suspected lymphoproliferative disorder with a corresponding formalin fixed paraffin embedded tissue block available for further analysis. Cases were excluded if nonviable cell suspensions had been derived from the submitted tissue or no lymphoid tissues identified in formalin fixed paraffin embedded specimens. All tissue biopsies were fixed in 10% neutral buffered formalin prior to paraffin embedding. Trephine bone marrow core biopsies were fixed in zinc formalin (Poly Scientific R&D Corp, Bay Shore, NY) and decalcified in an EDTA plus dilute hydrochloric acid decalcifying solution (Thermo Scientific, Waltham MA) for 30 minutes prior to embedding.
Flow cytometry
Six color flow cytometry was performed using CD45 and side scatter gating. While the complete panel of antibodies employed varied depending on the available number of cells, B-cell clonality was evaluated in all cases using the following six color combination: CD5-perdinin chlorophyll protein complex-Cy5.5 (PerCP-Cy5.5), CD19-allophycocyanin (APC), CD20-APC-Cy7, CD45-phycoerythrin (PE)-Cy7, and κ-fluorescein isothiocyanate and λ-PE immunoglobulin light chains (Becton Dickenson, San Jose, CA). Analysis was completed using an FACSCanto flow cytometer (Becton Dickinson). Flow cytometry data were analyzed using FCS Express (v3.0, De Novo Software, Los Angeles, CA).
Fully Automated Ultrasensitive RNA in situ Hybridization Assay
A two-color duplex RNAscope assay (Advanced Cell Diagnostics, Newark, CA) for the simultaneous detection of kappa and lambda Ig mRNA in lymphoma and bone marrow samples was performed using the Dual Color Open Probe software on a Ventana Benchmark XT (Benchmark XT, Roche Ventana Medical Systems, Tucson, AZ). The RNAscope technology, probe design, and amplification system have been previously described.19 In brief, sections were baked (32 min at 60°C) and deparaffinized on the instrument, followed by target retrieval (24 min at 97°C for tissues) and protease treatment (16 min at 37°C). Probes were then hybridized for 2 h at 43°C followed by RNAscope amplification and chromogenic detection using VS detection reagents. The following RNAscope probes were used in this study: dapB (negative control), Hs-IGLL5-C2, Hs-IgK, and Hs-IgL-C2. On one section kappa (IgK) and lambda (IgL) probes were hybridized together while on the consecutive section IGLL5 and dapB probes were hybridized together.
RNA in situ hybridization slides and corresponding H&E sections were reviewed (LG, JRC) without knowledge of the flow cytometry results according to the previously published algorithm (Figure 1)19. Clonal restriction was defined as either only signals of one light chain mRNA type or an excess of at least 5:1 signals for one light chain mRNA versus the alternate light chain.
Figure 1.
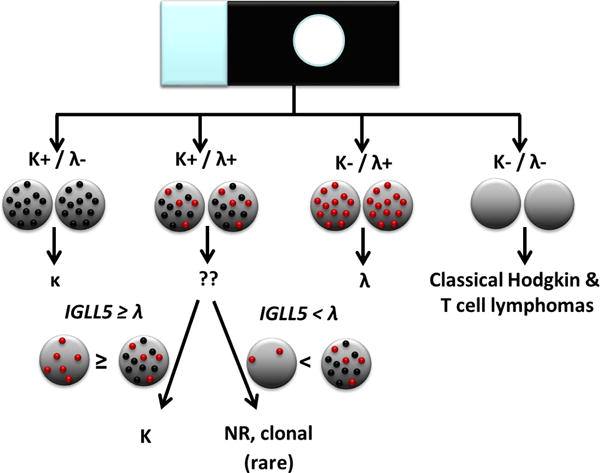
Algorithm for interpretation of kappa/lambda RNA in situ hybridization staining.19 Cells displaying only kappa or lambda signal are interpreted as kappa- or lambda-restricted, respectively. The neoplastic cells in classical Hodgkin and T-cell neoplasms are expected to lack staining for both light chains. When cells appear to express both kappa and lambda chains, the IGLL5 stain is reviewed. If IGLL5 signal is greater than or equal to the lambda signal, then the observed lambda signal is presumed to represent IGLL5 and the RNA in situ hybridization results are interpreted as kappa-restricted. If IGLL5 staining represents less than the lambda signal, then the neoplastic cell is interpreted as showing coexpression of kappa and lambda RNA.
Interobserver Reproducibility
The inter-observer reproducibility of manual interpretation of RNA in situ hybridization was determined by independent review of a subset of cases by two additional hematopathologists (CC and SO). Each pathologist first reviewed a teaching set of 10 cases, chosen to represent typical examples of RNA in situ hybridization clonality patterns, together with the corresponding flow cytometric data. Each pathologist then individually reviewed a test set of 30 cases, blinded to flow cytometry results and to the results of other scorers. The average Cohen’s Kappa statistic for each pairwise comparison was calculated (GraphPad Prism software, Graph Pad, La Jolla, CA).
Digital Image Analysis
Images of RNA in situ hybridization stained slides were acquired using a Leica Biosystems Aperio AT2 Digital Pathology scanner, and images were evaluated using the Halo image analysis software (Indica Labs, Albuquerque, New Mexico). The digital image analysis was first developed using a subset of lymphoma cases (n=25). A region of interest (ROI) was identified by a hematopathologist (JC) for each case. The software detected the number of red and black dots per ROI, indicating either kappa (black)/lambda (red) or dapB (black)/IGLL5 (red) signals per ROI. A classifier was used to identify and exclude plasma cells from the analysis. Kappa and Lambda probe copies per ROI were normalized to dapB and IGLL5 probe copies per ROI, respectively. The ratio between Kappa:Lambda was then calculated, and the following image analysis ratios were designated for interpretation: <0.3 Lambda-restricted, ≥0.3 but ≤3 non-restricted, and >3 Kappa-restricted. Using this algorithm, there was only one case that was discordant between the digital image analysis interpretation and the flow cytometry phenotype, and between the digital image analysis interpretation and the manual interpretation by a hematopathologist (JC). This algorithm was then used on the remaining lymphoma cases with no changes.
RESULTS
RNA in situ hybridization staining of normal tonsil and hyperplastic lymph node controls (Figure 2) revealed results similar to those previously described for manual staining. Plasma cells exhibited very strong cytoplasmic light chain expression. Lymphocytes, including germinal center and mantle zone lymphocytes, showed weaker cytoplasmic staining. Discrete dot-like nuclear lambda staining was frequently observed in lymphoid and non-lymphoid cells. This nuclear staining corresponded to IGLL5 expression, as shown by comparison of the two color K/L stained slides to those stained with an IGLL5 specific probe.
Figure 2.
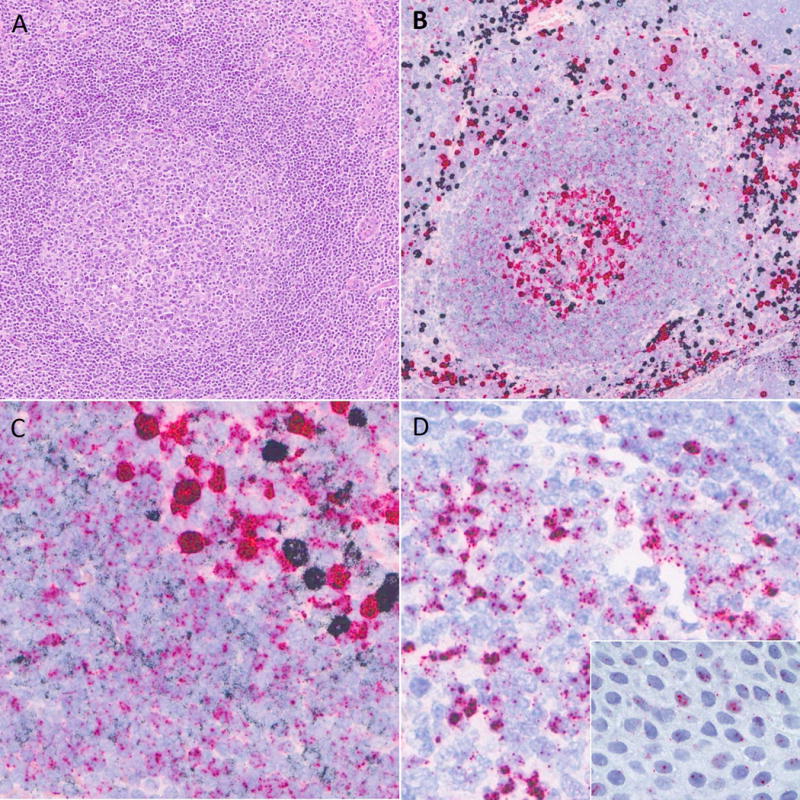
RNA in situ hybridization staining in normal lymphoid tissues. A) Reactive germinal center in routine H&E. B) Kappa/lambda RNA in situ hybridization shows strongly positive kappa (black) and lambda (red) plasma cells at low power. C) At higher power, polytypic expression is seen in mantle zone lymphocytes (lower left) and germinal center lymphocytes (upper right). D) IGLL5 signal contains nuclear dots and, especially within germinal center lymphocytes, cytoplasmic staining. Inset, IGLL5 nuclear staining in tonsil epithelium.
In 203 tissue biopsies submitted for flow cytometric analysis, RNA in situ hybridization studies identified a light chain restricted B-cell population in 85 (42%) compared to 58 (29%) by flow cytometry. Representative examples of RNA in situ hybridization staining are illustrated in Figures 3. Review of the IGLL5 stain was necessary in 16% of cases. RNA in situ hybridization staining showed excellent interobserver agreement between 3 expert hematopathologists (average K=0.92). The incidence of light chain restriction detected by RNA in situ hybridization and flow cytometry for specific diagnostic categories is illustrated in Figure 4. In every diagnostic category, light chain restriction was identified at a similar or greater incidence by RNA in situ hybridization compared to flow cytometry. Cases of B-cell lymphomas that failed to show light chain restriction by RNA in situ hybridization included cases with numerous admixed benign B-cells, limited involvement of the tissue block stained by RNA in situ hybridization, little to no staining of light chains in neoplastic cells, and/or necrotic tumor.
Figure 3.
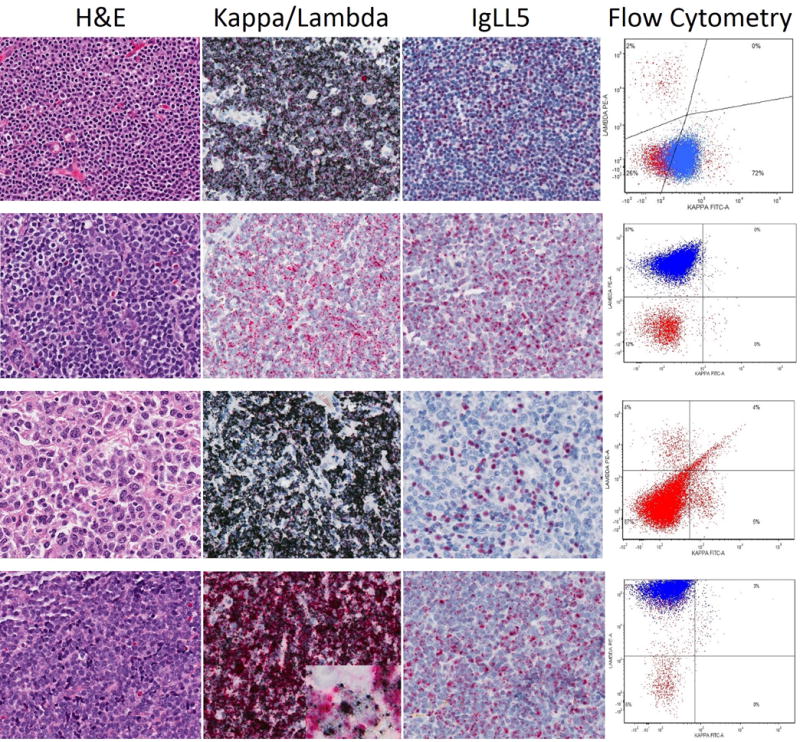
Representative RNA in situ hybridization staining in non-Hodgkin lymphomas, including routine H&E, Kappa/Lambda RNA in situ hybridization, IGLL5 pattern and flow cytometry (CD5 positive B-cells gated in blue). First line: a case of CLL/SLL exhibits kappa light chain restriction by RNA in situ hybridization and by flow cytometry. IGLL5 nuclear signal is present. Second line: a case of MCL shows lambda light chain restriction by RNA in situ hybridization and flow cytometry. Third line: a case of DLBCL with kappa light chain restriction by RNA in situ hybridization. Flow cytometry showed only few, polytypic B-cells. Fourth line: a case of MCL with coexpression of kappa and lambda mRNA by RNA in situ hybridization. IGLL5 shows less staining than with lambda probe. Flow cytometry shows lambda light chain protein only.
Figure 4.
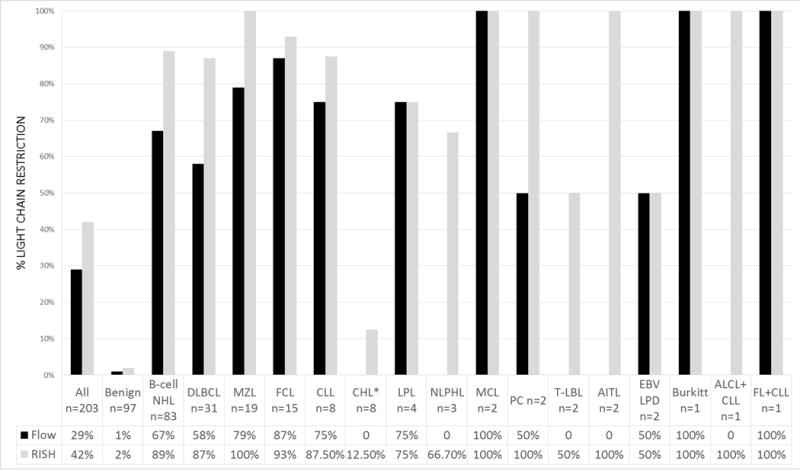
Incidence of light chain restriction as identified in tissue biopsies by RNA in situ hybridization or flow cytometry in various diagnostic categories. *Light chain restricted small B-cells present; Reed-Sternberg cells negative for kappa and lambda.
The correlation between light chain usage results by RNA in situ hybridization versus flow cytometry in tissue biopsy samples is detailed in Table 1 for all 203 cases and Table 2 for the 83 cases of B-cell lymphoma. Overall, identical light chain usage results were obtained in 166/203 (82%) cases. Light chain restricted B-cells were identified by RNA in situ hybridization in 16/128 (12%) cases with only polytypic B-cells or no B-cells identified by flow cytometry, including 6 diffuse large B-cell lymphoma (DLBCL), 2 marginal zone lymphomas (MZL), 2 nodular lymphocyte predominant Hodgkin lymphomas with clonal “popcorn” cells, 1 grade 3B follicular lymphoma (FL) with DLBCL, 1 grade 1–2 FL, 1 angioimmunoblastic T-cell lymphoma (AITL) with clonal B-cells, 1 anaplastic large cell lymphoma (ALCL) with admixed chronic lymphocytic leukemia, 1 classical Hodgkin lymphoma with negative Reed-Sternberg cells but aggregates of light chain restricted small B-cells, and 1 salivary gland benign cystic lymphoepithelial lesion. Light chain restriction was also identified by RNA in situ hybridization in 11/17 (65%) cases demonstrating surface Ig negative B-cells, including 5 DLBCL, 1 MZL, 1 AITL with clonal B-cells, 1 CLL, 1 mantle cell lymphoma (MCL), 1 pediatric type FL, and 1 T-lymphoblastic leukemia with lambda light chain mRNA expression (described further below). One case of MCL was found to show coexpression of both light chains by RNA in situ hybridization while only lambda light chain was detected by flow cytometry (Figure 3, bottom row).
Table 1.
Results of two-color automated RNA in situ hybridization in entire cohort of 203 consecutive samples submitted for flow cytometry.
| RISH | Flow cytometry | |||
|---|---|---|---|---|
| Polytypic or no B cells | Kappa | Lambda | sIg negative | |
| Polytypic or no B cells | 112 | 0 | 1 | 6 |
| Kappa | 9 | 25 | 0 | 3 |
| Lambda | 7 | 1 | 29 | 8 |
| Coexpression of kappa and Lambda | 0 | 0 | 1 | |
| Both Kappa and Lambda | 0 | 1 | 0 | 0 |
Table 2.
Results of two-color automated RNA in situ hybridization and flow cytometry in 83 cases diagnosed as B-cell non-Hodgkin lymphoma.
| Flow cytometry | ||||
|---|---|---|---|---|
| RISH | Polytypic or no B cells | Kappa | Lambda | sIg negative |
| Polytypic or no B cells | 4 | 0 | 1 | 4 |
| Kappa | 7 | 24 | 0 | 3 |
| Lambda | 2 | 1 | 28 | 7 |
| Coexpression of kappa and Lambda | 0 | 0 | 1 | |
| Both Kappa and Lambda | 0 | 1 | 0 | 0 |
The use of RNA in situ hybridization also highlighted unusual or unexpected findings in a minority of cases. Light chain restricted B-cells were found by RNA in situ hybridization in 2/97 cases diagnosed as benign lesions, including a salivary gland benign lymphoepithelial lesion with polyclonal flow results and one case diagnosed as atypical marginal zone hyperplasia with lambda light chain restriction by flow cytometry but polyclonal PCR results. One case diagnosed as a FL with kappa light chain restriction by flow cytometry was shown by RNA in situ hybridization to represent a composite of two FL (one kappa light chain restricted and one lambda light chain restricted) with only subtle morphologic differences identified between the two areas in retrospective review of the routine H&E sections. Seven cases of classical Hodgkin lymphoma (CHL) each demonstrated absence of light chain restriction in R-S cells, although one case demonstrated light chain restriction in background small B cells. In contrast, 2/3 (67%) nodular lymphocyte predominant Hodgkin lymphomas (NLPHL) demonstrated light chain restriction in the large neoplastic cells. T-cell lymphomas were infrequent in this consecutive series and the majority of these lacked restricted light chain mRNA signals. The exceptions were 2 cases of AITL demonstrating light chain restriction in B-cells by RNA in situ hybridization, but not by flow cytometry. In one case, lambda light chain mRNA expression was found in a T lymphoblastic lymphoma that lacked surface immunoglobulin light chains or other B-cell markers by flow cytometry.
One hundred and four consecutive bone marrow cases examined included 63 benign samples and 41 cases involved by B-cell lymphoproliferative disorders. While at least focal plasma cells with light chain staining by RNA in situ hybridization were present in 87 (84%) cases, lymphoid cells with interpretable RNA in situ hybridization signals were present in only 23 (22%), including 2 benign samples and 21 involved by lymphoproliferative disorders. RNA in situ hybridization and flow cytometry were concordant in 21 of these 23 (91%) evaluable cases. (Figure 5). The 2 discordant cases included one CLL with lambda light chain restriction by flow cytometry and polytypic B-cells by RNA in situ hybridization, and one case of HCL with lambda light chain restriction by flow cytometry and aberrant expression of both kappa and lambda mRNA by RNA in situ hybridization.
Figure 5.
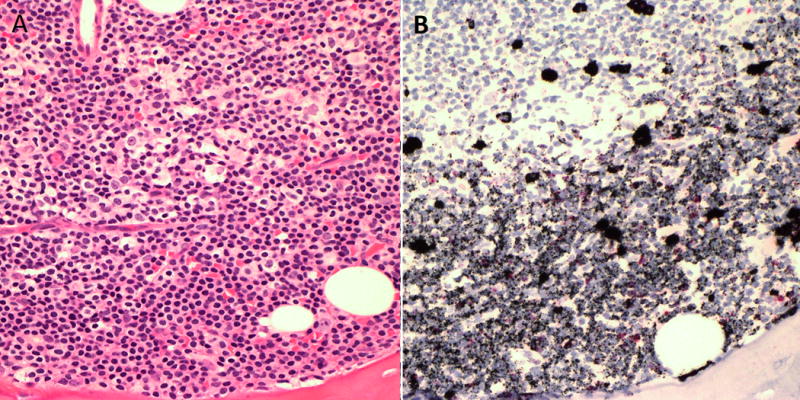
Automated RNA in situ hybridization staining in bone marrow. A) A paratrabecular aggregate of small lymphocytes and plasma cells in a case of kappa light chain restricted lymphoplasmacytic lymphoma. B) RNA in situ hybridization shows kappa light chain restriction in small lymphocytes and in scattered plasma cells.
RNA in situ hybridization stained slides were also evaluated by image analysis software using an algorithm that excluded intensely staining plasma cells and scored only the fainter staining present in lymphocytes. Manual evaluation and image analysis results were concordant in 197/203 (97%) cases. In three discrepant cases, the clonal cells were plasmacytic or lymphoplasmacytic with very intense cytoplasmic staining that was excluded by the software algorithm. The remaining 3 discrepant cases included one benign spleen interpreted as lambda light chain by image analysis (possibly due to the presence of numerous T-cells with nuclear IGLL5 staining in the field analyzed), one FL with faint cytoplasmic kappa light restriction and admixed polytypic B-cells interpreted as polytypic by image analysis, and the MCL case with coexpression of kappa and lambda by RNA in situ hybridization interpreted as lambda restricted by the image analysis algorithm.
DISCUSSION
In this study, we have expanded on our prior report19 of a manually processed, ultrasensitive RNA in situ hybridization assay for kappa and lambda expression by adapting this technology to a commercial automated immunohistochemistry platform and evaluating the feasibility and utility of this method in routine clinical practice by staining of 203 consecutive tissue biopsies and 104 consecutive bone marrow samples submitted for flow cytometric analysis. In tissue biopsies, RNA in situ hybridization staining revealed light chain restriction with a sensitivity equivalent or greater than that of flow cytometry across a broad range of lymphoproliferative disorders. The detection rate by RNA in situ hybridization in bone marrow samples was much lower than tissue biopsies due to failure to detect any light chain mRNA in lymphocytes, likely due to decalcification procedures which are known to degrade RNA.20,21 Within bone marrow samples where light chain signal was evaluable, however, RNA in situ hybridization showed excellent concordance with flow cytometry. Other investigators have recently described other sensitive in situ hybridization techniques which show good correlation with flow cytometry in smaller series of selected cases.20,21 The current study represents the largest series to date of the methodology, and for the first time explores the diagnostic yield in a consecutive case series designed to be representative of routine clinical practice.
The use of RNA in situ hybridization to assess light chain expression in FFPE tissues offers several advantages over flow cytometry. First, RNA in situ hybridization offers the ability to examine light chain expression in the context of tissue morphology. For example, one case of a benign salivary gland lymphoepithelial lesion displayed polytypic B-cells by flow cytometry, and indeed polytypic reactive germinal centers were present by RNA in situ hybridization, but RNA in situ hybridization also clearly demonstrated light chain restriction in the intra-epithelial B-cell population. Secondly, RNA in situ hybridization identified light chain restricted B-cell proliferations that were missed by flow cytometry due to loss of surface immunoglobulin expression, selective loss of the neoplastic cell populations, or admixed polytypic B-cells. Finally, RNA in situ hybridization offers the ability to assess clonality in formalin fixed, paraffin embedded tissue cases where flow cytometric analysis is not possible due to the lack of fresh tissue. PCR analysis can be used to document clonality in such cases, but PCR is a time consuming technique not available in many routine laboratories, while the RNA in situ hybridization method described in this study can be adapted in any laboratory using a commercial immunostainer. RNA in situ hybridization techniques, however, are unlikely to completely replace the need for flow cytometry in the assessment of suspected lymphomas. The current routine use of 6 or 8 color flow cytometry allows for documentation of a thorough phenotypic profile and selective analysis of light chain expression only within specific phenotypic subsets (e.g., selective gating on dimly CD20 positive B-cells, or gating selectively on CD5 positive or CD5 negative B-cells). Flow cytometry may also remain helpful in cases where poor RNA preservation precludes the use of RNA in situ hybridization, particularly in decalcified bone marrow specimens. However, the latter obstacle for RNA in situ hybridization may be overcome by developing decalcification procedures that better preserve RNA.
One potential challenge in the interpretation of light chain RNA in situ hybridization is the expression of IGLL5. IGLL5 RNA expression, which does not require somatic recombination, is found in both lymphoid and non-lymphoid cells.19–21 Unfortunately, it has not been possible to design lambda specific probes that do not recognize IGLL5 because IGLL5 shares two exons with the IGL constant region, and analysis of light chain staining by RNA in situ hybridization must therefore account for IGLL5 expression. Rimsza et al23 noted that IGLL5 staining is predominantly nuclear, and suggested that nuclear signals may be disregarded when evaluating sensitive kappa and lambda stains. Arora et al,24 in a small series of 30 cases, reported that IGLL5 signal was observed only in kappa restricted B-cells. Our prior study and the current report, however, show that IGLL5 is not limited to dot-like nuclear staining, but substantial cytoplasmic expression may also be seen, especially within germinal center B-cells. Moreover, IGLL5 expression is clearly present in both kappa-restricted and lambda-restricted B-cell lymphomas. The use of sequential kappa/lambda and IGLL5-specific probes as indicated in our algorithmic approach clarifies potential confusion due to IGLL5 expression.
This report also describes an image analysis software algorithm intended to help facilitate interpretation of staining. This image analysis approach shows an excellent correlation with manual interpretation, and could be used to provide a more objective evaluation of RNA in situ hybridization staining. It should be noted, however, that image analysis still requires a trained pathologist to identify the areas of interest for evaluation. Moreover, manual review of RNA in situ hybridization slides showed excellent inter-observer reproducibility between hematopathologists. These results suggest that while image analysis protocols may be useful for specific research applications, manual interpretation of RNA in situ hybridization staining can be readily interpreted by experienced pathologists.
Several cases in this series displayed notable findings by RNA in situ hybridization. Multiple composite lymphoproliferative disorders were identified including B-cell and T-cell lymphomas with admixed CLL/SLL, T-cell lymphomas with clonal B-cells, and a composite of two follicular lymphomas (one kappa restricted, one lambda restricted). The ability to analyze light chain restriction in the context of tissue morphology facilitates the recognition of such cases which might be missed by flow cytometry alone. Two cases in this series showed coexpression of both kappa and lambda mRNA. Rare examples of coexpression of kappa and lambda light chain proteins have been reported,25,26 but there is little published data regarding coexpression of light chain mRNAs. During B-cell development, the IGK locus rearranges first, with rearrangement of the IGL locus occurring only if a productive IGK rearrangement is not produced.25,26 The coexpression of kappa and lambda mRNA may therefore represent an IGK rearrangement that is capable of being expressed at the mRNA level but not capable of yielding a functional protein product. Both of the cases coexpressing kappa and lambda in this report expressed lambda protein by flow cytometry, consistent with this hypothesis. Finally, we also noted one case of a T lymphoblastic lymphoma with expression of lambda RNA. This case displayed a typical T-ALL phenotype without immunoglobulin light chain proteins by flow cytometry. It is well known that immunoglobulin gene rearrangements may be detected by PCR or Southern blotting in T-cell neoplasms including T-ALL,27–29 but this is the first demonstration of light chain RNA expression to our knowledge. This phenomenon should be further investigated to determine if it may show biological or clinical significance.
In conclusion, this report describes an automated method for ultrasensitive RNA in situ hybridization detection of light chain restriction in suspected lymphomas with a clinical sensitivity that meets or exceeds that of flow cytometry. Additional studies of this technique from other laboratories should be performed to validate the automated RNA in situ hybridization assay. If confirmed, ultrasensitive automated RNA in situ hybridization offers the potential to become a new gold standard for the evaluation of light chain restriction. Sensitive detection of light chain restriction in the context of morphologic features by an automated platform provides further insight into lymphoproliferative disorders in both research applications and in clinical diagnostic practice.
Acknowledgments
This work was funded by National Cancer Institute grant 2R44CA168019-02A1
Footnotes
DISCLOSURE: Courtney Anderson, Emerald Doolittle, Siobhan Kernag, and Xiao-Jun Ma are employees of Advanced Cell Diagnostics. Zhen Wang and James Cook are eligible to receive royalties for reagents employed in this study. Ling Guo, Claudiu Cotta and Sarah Ondrejka have no duality of interest to declare.
References
- 1.NCCN. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Non-Hodgkin’s Lymphomas. Version 1.2016. 2015:1–466. Available at: www.NCCN.com.
- 2.Swerdlow SH. Lymphoma classification and the tools of our trade: an introduction to the 2012 USCAP Long Course. Mod Pathol. 2013;26:S1–S14. doi: 10.1038/modpathol.2012.177. [DOI] [PubMed] [Google Scholar]
- 3.de Tute RM. Flow cytometry and its use in the diagnosis and management of mature lymphoid malignancies. Histopathology. 2011;58:90–105. doi: 10.1111/j.1365-2559.2010.03703.x. [DOI] [PubMed] [Google Scholar]
- 4.Craig FE, Foon KA. Flow cytometric immunophenotyping for hematologic neoplasms. Blood. 2008;111:3941–3967. doi: 10.1182/blood-2007-11-120535. [DOI] [PubMed] [Google Scholar]
- 5.Virgo PF, Gibbs GJ. Flow cytometry in clinical pathology. Ann Clin Biochem. 2012;49:17–28. doi: 10.1258/acb.2011.011128. [DOI] [PubMed] [Google Scholar]
- 6.Langerak AW, et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leuk. 2012;26:2159–71. doi: 10.1038/leu.2012.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans PAS, et al. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leuk Off J Leuk Soc Am Leuk Res Fund, U K. 2007;21:207–214. doi: 10.1038/sj.leu.2404479. [DOI] [PubMed] [Google Scholar]
- 8.Marshall-Taylor CE, Cartun RW, Mandich D, DiGiuseppe JA. Immunohistochemical detection of immunoglobulin light chain expression in B-cell non-Hodgkin lymphomas using formalin-fixed, paraffin-embedded tissues and a heat-induced epitope retrieval technique. Appl Immunohistochem Mol Morphol AIMM. 2002;10:258–62. doi: 10.1097/00129039-200209000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Weiss LM, Loera S, Bacchi CE. Immunoglobulin Light Chain Immunohistochemistry Revisited, With Emphasis on Reactive Follicular Hyperplasia Versus Follicular Lymphoma. Appl Immunohistochem Mol Morphol. 2010;18:199–205. doi: 10.1097/PAI.0b013e3181c59a81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck RC, Tubbs RR, Hussein M, Pettay J, Hsi ED. Automated colorimetric in situ hybridization (CISH) detection of immunoglobulin (Ig) light chain mRNA expression in plasma cell (PC) dyscrasias and non-Hodgkin lymphoma. Diagn Mol Pathol. 2003;12:14–20. doi: 10.1097/00019606-200303000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Magro C, Crowson AN, Porcu P, Nuovo GJ. Automated kappa and lambda light chain mRNA expression for the assessment of B-cell clonality in cutaneous B-cell infiltrates: its utility and diagnostic application. J Cutan Pathol. 2003;30:504–11. doi: 10.1034/j.1600-0560.2003.00102.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, et al. Automated Quantitative RNA in Situ Hybridization for Resolution of Equivocal and Heterogeneous ERBB2 (HER2) Status in Invasive Breast Carcinoma. J Mol Diagnostics. 2013;15:210–219. doi: 10.1016/j.jmoldx.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, et al. RNAscope. J Mol Diagnostics. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ukpo OC, et al. High-Risk Human Papillomavirus E6/E7 mRNA Detection by a Novel In Situ Hybridization Assay Strongly Correlates With p16 Expression and Patient Outcomes in Oropharyngeal Squamous Cell Carcinoma. Am J Surg Pathol. 2011;35:1343–1350. doi: 10.1097/PAS.0b013e318220e59d. [DOI] [PubMed] [Google Scholar]
- 15.Schache AG, et al. Validation of a novel diagnostic standard in HPV-positive oropharyngeal squamous cell carcinoma. Br J Cancer. 2013;108:1332–1339. doi: 10.1038/bjc.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MA, Jung JE, Lee HE, Yang HK, Kim WH. In situ analysis of HER2 mRNA in gastric carcinoma: comparison with fluorescence in situ hybridization, dual-color silver in situ hybridization, and immunohistochemistry. Hum Pathol. 2013;44:487–94. doi: 10.1016/j.humpath.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 17.Bordeaux JM, et al. Quantitative In Situ Measurement of Estrogen Receptor mRNA Predicts Response to Tamoxifen. PLoS One. 2012;7:e36559. doi: 10.1371/journal.pone.0036559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop JA, et al. Detection of Transcriptionally Active High-risk HPV in Patients With Head and Neck Squamous Cell Carcinoma as Visualized by a Novel E6/E7 mRNA In Situ Hybridization Method. Am J Surg Pathol. 2012;36:1874–1882. doi: 10.1097/PAS.0b013e318265fb2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tubbs RR, et al. Ultrasensitive RNA in situ hybridization for detection of restricted clonal expression of low-abundance immunoglobulin light chain mRNA in B-cell lymphoproliferative disorders. Am J Clin Pathol. 2013;140:736–46. doi: 10.1309/AJCPJTWK07FSABRJ. [DOI] [PubMed] [Google Scholar]
- 20.Guglielmi P, Davi F. Expression of a novel type of immunoglobulin Cλ transcripts in human mature B lymphocytes producing χ light chains. Eur J Immunol. 1991;21:501–508. doi: 10.1002/eji.1830210237. [DOI] [PubMed] [Google Scholar]
- 21.Evans RJ, Hollis GF. Genomic structure of the human Ig lambda 1 gene suggests that it may be expressed as an Ig lambda 14.1-like protein or as a canonical B cell Ig lambda light chain: implications for Ig lambda gene evolution. J Exp Med. 1991;173:305–11. doi: 10.1084/jem.173.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minca EC, et al. Detection of immunoglobulin light-chain restriction in cutaneous B-cell lymphomas by ultrasensitive bright-field mRNA in situ hybridization. J Cutan Pathol. 2015;42:82–89. doi: 10.1111/cup.12415. [DOI] [PubMed] [Google Scholar]
- 23.Rimsza LM, et al. Kappa and lambda light chain mRNA in situ hybridization compared to flow cytometry and immunohistochemistry in B cell lymphomas. Diagn Pathol. 2014;9:144. doi: 10.1186/1746-1596-9-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arora K, et al. Branched-chain in situ hybridization for κ and λ light chains: A powerful ancillary technique for determining B-cell clonality in cytology samples. Cancer Cytopathol. 2016;124:203–12. doi: 10.1002/cncy.21629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D. Dual surface immunoglobulin light-chain expression in B-cell lymphoproliferative disorders. Arch Pathol Lab Med. 2006;130:853–6. doi: 10.5858/2006-130-853-DSILEI. [DOI] [PubMed] [Google Scholar]
- 26.Jiwani S, Bornhost J, Alapat D. Biphenotypic plasma cell myeloma: two cases of plasma cell neoplasm with a coexpression of kappa and lambda light chains. Int J Clin Exp Pathol. 2015;8:8536–44. [PMC free article] [PubMed] [Google Scholar]
- 27.Zaki MAA, et al. Presence of B-cell clones in T-cell lymphoma. Eur J Haematol. 2011;86:412–419. doi: 10.1111/j.1600-0609.2011.01597.x. [DOI] [PubMed] [Google Scholar]
- 28.van Dongen JJM, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leuk Off J Leuk Soc Am Leuk Res Fund, U K. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 29.Brüggemann M, et al. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leuk Off J Leuk Soc Am Leuk Res Fund, UK. 2007;21:215–221. doi: 10.1038/sj.leu.2404481. [DOI] [PubMed] [Google Scholar]


