Abstract
Purpose
To measure the apparent diffusion coefficients (ADC) of five main metabolites in the human brain at 3 T with PRESS and STEAM, avoiding measurement biases due to cross-terms. Cross-terms arise from interactions between slice-selection and spoiler gradients in the localized spectroscopy sequence and the diffusion gradients.
Methods
Diffusion-weighted spectra were acquired from the prefrontal cortex in 5 healthy subjects using STEAM (TE/TM/TR=21.22/105/3000 ms, b-values=0 and 3172 s/mm2) and PRESS (TE/TR=54.2/3000 ms, b-values=0 and 2204 s/mm2). Diffusion weighting was applied using bipolar gradients in three orthogonal directions. Post-processed spectra were analyzed with LCModel, and the trace/3 ADC values were calculated.
Results
Comparable trace/3 ADC values (0.14–0.18 µm2/ms) were obtained for five main metabolites with both methods. These metabolites were quantified with Cramer-Rao lower bounds below 15%.
Conclusion
The ADC values of the five main metabolites were successfully measured in the human brain at 3 T with eliminated directional dependence. Both STEAM and PRESS can be used to probe the diffusivity of metabolites in normal brain and various pathologies on the clinical scanner with slightly higher precision achieved with STEAM for glutamate and myo-inositol.
Keywords: STEAM, PRESS, ADC, glutamate, myo-inositol
Introduction
Diffusion-weighted magnetic resonance spectroscopy (DW-MRS) is a unique, noninvasive tool which allows the investigation of the intracellular microenvironment in the brain in vivo. By probing the diffusivity of metabolites, specific information on compartmentation can be obtained at both cellular and subcellular levels (1–4). The most commonly measured brain metabolites with spectroscopy are total N-acetyl aspartate (tNAA, N-acetylaspartate (NAA) plus N-acetylaspartylglutamate (NAAG)), total creatine (tCr, creatine plus phosphocreatine), choline containing compounds (tCho, phosphorylcholine plus glycerophosphorylcholine), glutamate (Glu), and myo-inositol (mIns). NAA and Glu are essentially located in neurons, mIns is thought to be preferentially compartmentalized in astrocytes, whereas tCr and tCho are located in both neuronal and glial cells.
The MRS sequences most commonly used in diffusion studies are STEAM (5) and PRESS (6). STEAM is advantageous in DW-MRS since the mixing period (TM) can be used to increase the diffusion time without compromising the signal-to-noise ratio (SNR). In this way, relatively short echo-time (TE) can be achieved in STEAM however only half the signal is observed compared to spin-echo sequences. PRESS, on the other hand, allows shorter diffusion time at relatively long TE although at high fields chemical shift displacement (CSD) error might be large due to the limited bandwidth of the refocusing pulses.
DW-MRS studies have been used to study both healthy and diseased brains (7–14). The apparent diffusion coefficients (ADC) of NAA, tCr and tCho have been commonly reported in human and animal brains at several field strengths due to ease in measuring these singlets (7–13). By taking advantage of the high SNR and spectral resolution, the ADC of glutamate (Glu) was first measured in the human brain at 7 T (12). In contrast, the ADC of several J-coupled metabolites such as Glu, mIns, glucose, glutamine, lactate, and taurine previously have only been measured in animal brains at 4.7 T (15) and 9.4 T (10). These measurements were possible due to narrower spectral linewidth in ppm achievable in animals compared to humans and higher spectral dispersion at high fields. The ADC of Glu and mIns were recently determined in the human brain at 3 T (16,17), however in these studies the cross-term effects were not taken into account and this could lead to inaccurate ADC estimations (13). Cross-terms result from interactions between the DW gradients and the slice-selection and spoiler gradients in the spectroscopic sequence. These effects are different based on the directions and polarities of the applied DW gradients.
The aims of this study were: 1) to measure the ADC of tCr, tCho, NAA, Glu and mIns in the human brain on a clinical 3 T scanner using both PRESS and STEAM sequences where DW spectra were acquired with positive and negative diffusion gradient polarity to allow for the removal of cross-terms, and 2) to compare the sensitivity of both acquisition techniques in terms of metabolite quantification precision (reflected by Cramér-Rao Lower bounds, CRLB) obtained during LCModel analysis and spectral SNR.
Methods
Healthy subjects (N = 5, 24 ± 3 years old, 4 males) were scanned using a 3 T whole-body Siemens Prismafit scanner (Siemens Medical Solutions, Erlangen, Germany). The scanner was equipped with a gradient coil capable of reaching 80 mT/m on each of the three orthogonal axes simultaneously. Informed consent approved by the Institutional Review Board at the University of Minnesota was obtained from all subjects prior to the scan. The standard body coil was used for excitation and the 32-channel receive-only head coil for reception.
Whole brain T1-weighted MPRAGE (18) images (0.8 mm3 isotropic resolution, repetition time, TR = 2400 ms, TE = 2.24 ms, inversion time, TI = 1060 ms, flip angle = 8°, GRAPPA acceleration factor = 2 (19)) were acquired and a volume-of-interest (VOI) of 25×25×25 mm3 was positioned in the prefrontal cortex (anterior to the genu of corpus callosum and centered on the interhemispheric fissure). First and second-order shims in the VOI were automatically adjusted using the system 3D gradient-echo shim, operated in the “brain” mode.
Localized DW spectra were acquired in the same MR session using STEAM (TE/TM = 21.22/105 ms) and PRESS (TE = 54.2 ms) sequences in each subject. Water suppression was achieved using VAPOR interleaved with OVS pulses in both sequences (20). For optimal spectral SNR, B1 field for the 90° was calibrated inside the VOI while the 180° pulses in PRESS were set relative to the 90° pulse (21). A similar calibration was performed to optimize the water suppression flip angle. All data were obtained with a TR of 3 s, a spectral width of 6 kHz and with 2048 complex points and saved as individual free induction decays (FIDs) for further post-processing offline. The carrier frequency of the localization radio frequency (RF) pulses was set to 3 ppm.
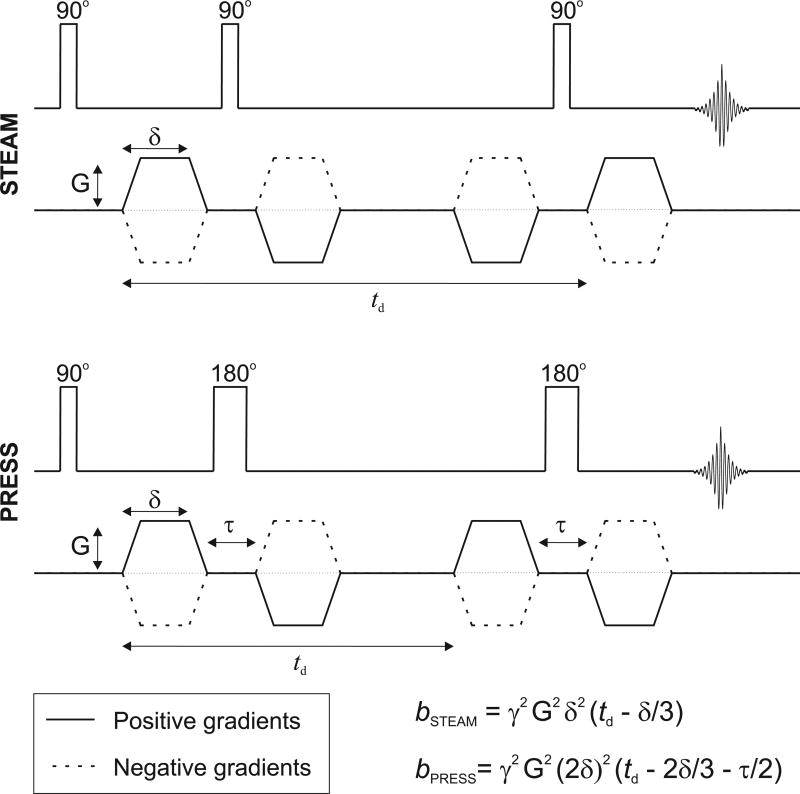
Diffusion weighting was applied in three orthogonal directions ([1,1,−0.5], [1,−0.5,1], and [−0.5,1,1]) using bipolar gradients (13) to reduce the effect of eddy currents generated by the DW gradients (22). In STEAM, the diffusion gradient duration (δ) was 5.85 ms operating at 70 mT/m (maximum strength applied per axis) with a diffusion time (td) of 118 ms resulting in a nominal b-value of 3172 s/mm2 (Figure 1A). The TM period was set to 105 ms and was optimized using density matrix simulation such that Glu and mIns signals were maximized. In PRESS, δ was 6.4 ms at 70 mT/m with td of 27 ms such that the nominal b-value was 2204 s/mm2; TE1 was 28.14 ms and TE2 was 26.06 ms (Figure 1B). These b-values were determined using the chronograms of the actual pulse sequences as applied in vivo (23). The equations given in Figure 1 provide approximate b-values.
Figure 1.
Schematic of the diffusion-weighted STEAM and PRESS sequences as used in the present study. To reduce eddy current effects, bipolar gradients were employed in both acquisition methods. Approximate b-values can be calculated with the provided equations, where δ represents the duration of the diffusion gradient, td is the diffusion time, and τ is the time separation between the bipolar gradients. These equations assume infinitely short gradient rise time. Chronograms were used to determine the exact b-value.
Single-shot STEAM and PRESS metabolite spectra were acquired at two b-values to allow further post-processing; a null b-value with 16 averages and a high nominal b-value with 48 averages. To remove the cross-term effects between all applied gradients, additional spectra were acquired with diffusion gradients of opposite polarity for each gradient direction such that a total of 6 DW directions were utilized. Water reference scans at null and high b-values (6 directions) were also acquired for eddy-current correction and for measuring the ADC of tissue water. The total acquisition time per sequence was approximately 16 minutes for one complete diffusion dataset.
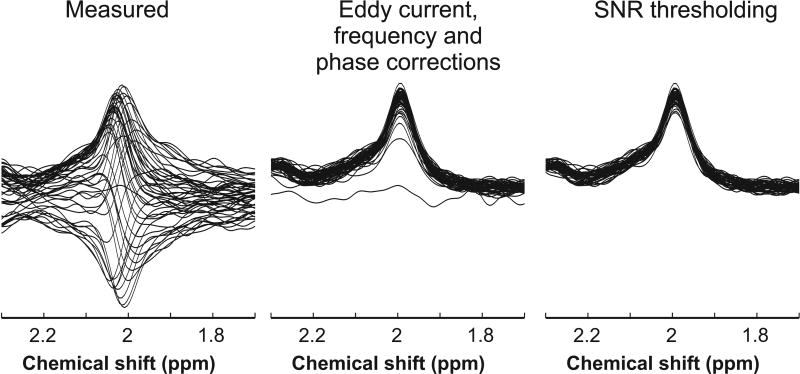
All spectra were processed using Matlab (MathWorks Inc., Natick, MA). Spectra were first corrected for eddy current effects. To obtain the best quality data, each single-shot spectrum was then frequency corrected using a cross-correlation algorithm and zero-order phase corrected using a least-squares algorithm. Diffusion-weighted spectra which showed evidence of low SNR (Figure 2) were eliminated if the spectrum SNR was lower than ~90% of the mean SNR of all spectra in each b-value dataset. Remaining high SNR spectra, at null b-value and at high b-value for all six gradient diffusion directions, were then averaged separately and analyzed with LCModel (24) version 6.3-0G (Stephen Provencher Inc., Ontario, Canada). No baseline correction, water-removal, zero-filling, or apodization functions were applied, and the spectra were fitted from 0.5 to 4.1 ppm.
Figure 2.
Stacked plot of NAA singlet at 2 ppm measured in one diffusion direction with STEAM from one subject; 48 individually measured spectra (left panel), spectra after eddy current, frequency, and phase corrections (middle panel), and the resulting individual spectra after removing low SNR spectra as described in the text (right panel).
Basis sets were simulated using custom software in Matlab based on density matrix formalism (25) using published chemical shift and J-coupling values (26–28). Due to minimal CSD error (3.5%/ppm) in STEAM, non-localized simulation with actual RF shapes and timings was used. The large CSD error in PRESS (11.1%/ppm for the refocusing pulse) was accounted for by performing a 2D localized simulation using 40×40 spatial points (29) along the direction of the refocusing pulses while utilizing all RF shapes and timings used in vivo. Localization in PRESS was achieved by frequency sweeping the refocusing pulses from BWb + 0 ppm to BWb + 4.5 ppm where BWb is the bandwidth of the pulse at the base (30). The basis set for both sequences contained 19 metabolites: alanine, ascorbate, aspartate, creatine, γ-aminobutyric acid, glucose, Glu, glutamine, glutathione, glycerophosphorylcholine, mIns, scyllo-inositol, lactate, NAA, NAAG, phosphocreatine, phosphorylcholine, phosphorylethanolamine, and taurine. Separate basis spectra were generated for the singlet and multiplet (i.e., CH3 and CH2 groups) resonances of NAA (denoted as sNAA and mNAA, respectively). Macromolecule spectra were measured using the metabolite-null technique (31) (TR/TI = 2500/740 ms, 128 averages) where an extra inversion pulse was added in both STEAM and PRESS without the diffusion gradient applied. These macromolecule spectra were included in the basis sets for each sequence.
Metabolites with CRLB < 15% at all b-values were selected for further analysis. ADC was determined in each gradient diffusion direction by taking the logarithmic geometric mean of the LCModel signal amplitude measured with both gradient polarities divided by the signal at null b-value (13). The final trace/3 ADC was obtained by taking the average of the ADC values in all three directions. Trace/3 ADCs were calculated for tNAA, tCr, tCho, Glu, mIns, and water. The fractions of gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) in the VOI were determined by segmenting the MPRAGE images using FSL and using an in-house written Matlab script. Reported SNR was measured in the frequency domain and is defined as peak height of sNAA divided by root mean square noise.
Differences in ADC values between STEAM and PRESS sequences were analyzed using a two-tailed paired Student’s t-test. Differences were considered statistically significant if P < 0.05, with Bonferroni correction for multiple comparisons.
Results
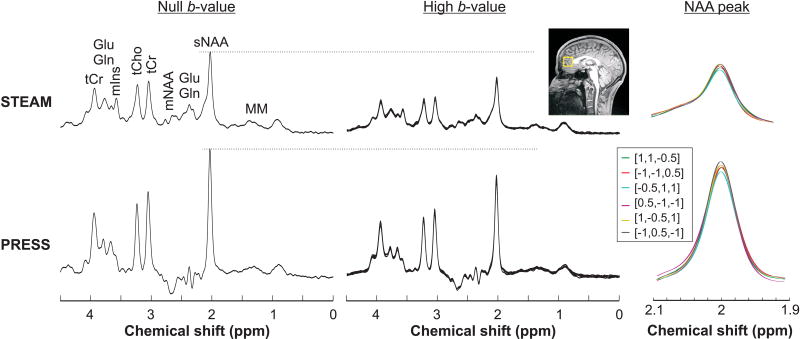
Proton spectra acquired at null and high b-values from one representative subject using both PRESS and STEAM sequences in one session are shown in Figure 3. High quality spectra with no lipid contamination were obtained with an average spectral linewidth of 6.4 Hz (tCr at 3.03 ppm). No huge baseline artifact was observed from the residual water due to ~99% suppression efficiency using VAPOR. The resonance of the five major metabolites, i.e., tNAA, tCr, tCho, Glu, and mIns, could be easily distinguished in all spectra in addition to other low concentration metabolites. The variation in the amplitude of the sNAA peak across all six diffusion directions showed the effect of cross-terms, as observed in both sequences (Figure 3). The GM fraction was 0.64 ± 0.01, WM fraction was 0.23 ± 0.02, and CSF fraction was 0.13 ± 0.01 in the VOI.
Figure 3.
DW spectra acquired from one subject using STEAM (TE/TM = 21.2/105 ms) and PRESS (TE = 54.2 ms) sequences. Spectra acquired at null b-value (left panel) and at high b-value in all six directions (middle panel) are shown. Excellent spectral quality, with no lipid contaminations and flat baseline were obtained in the prefrontal lobe VOI (yellow box on the T1-weighted image) with both techniques. The effects of cross-terms from the DW gradients are visible as the variation of the NAA amplitude at 2 ppm (right panel).
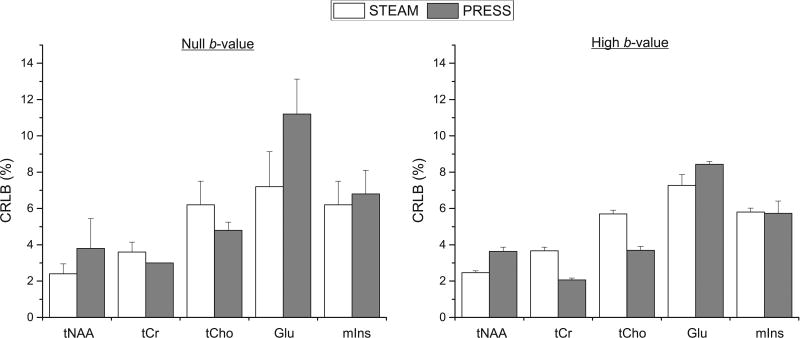
High spectral SNR (> 50) was achieved at both b-values with both protocols. Although the SNR was almost two times higher with PRESS compared to STEAM (105 ± 20 vs. 64 ± 12 at null b-value, and 128 ± 4 vs. 67 ± 2 at high b-values), this difference in SNR did not lead to differences in quantification of metabolites since the CRLBs for the five major metabolites considered in this study were below 12% at null b-value and below 9% at high b-values due to having more averages (Figure 4). The CRLBs of Glu and mIns were slightly lower with STEAM than PRESS (7.2 ± 1.9% vs. 11.2 ± 1.9% for Glu, and 6.2 ± 1.3% vs. 6.8 ± 1.3% for mIns at null b-value) and could be explained by the relatively short TE used in STEAM where J-evolution is minimized.
Figure 4.
Average CRLBs of the 5 main metabolites for STEAM (TE/TM = 21.2/105 ms) and PRESS (TE = 54.2 ms) sequences at different b-values (16 averages for null b-value and 48 averages for high b-value). At high b-value, the average CRLBs from all 6 directions are shown. Error bars represent standard deviation.
No statistically significant difference in trace/3 ADC values (0.11–0.16 µm2/ms, Table 1) were obtained for the five major metabolites between the two techniques, although the ADC values measured with STEAM had a tendency to be lower compared to PRESS. Interestingly, the standard deviation was much smaller for STEAM than PRESS for all metabolites except for tCr and Glu where the standard deviations were close with both methods. As expected, the trace/3 ADC of water was higher (0.70 – 0.78 µm2/ms) compared to metabolites. If low SNR spectra were not excluded during the post-processing steps, ADC values of metabolites would have been 10 to 25% higher than those reported in Table 1.
Table 1.
ADC obtained with positive (ADC+) vs. negative (ADC−) polarity diffusion gradients, and trace/3 ADC values (µm2/ms) of metabolites and water measured in the prefrontal lobe in 5 healthy subjects at 3 T using STEAM and PRESS sequences. ADC+ and ADC− contain cross-term effects which were higher in STEAM than PRESS. No significant difference in trace/3 ADC values were observed between the two acquisition techniques.
| Metabolite | ADC+ | STEAM ADC− |
ADC | ADC+ | PRESS ADC− |
ADC |
|---|---|---|---|---|---|---|
| tNAA | 0.14 ± 0.02 | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.16 ± 0.03 | 0.16 ± 0.02 | 0.16 ± 0.02 |
| tCr | 0.15 ± 0.03 | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.14 ± 0.01 | 0.14 ± 0.01 | 0.14 ± 0.01 |
| tCho | 0.14 ± 0.03 | 0.11 ± 0.03 | 0.12 ± 0.01 | 0.15 ± 0.05 | 0.14 ± 0.04 | 0.14 ± 0.04 |
| Glu | 0.17 ± 0.05 | 0.13 ± 0.03 | 0.15 ± 0.03 | 0.16 ± 0.05 | 0.15 ± 0.04 | 0.15 ± 0.04 |
| mIns | 0.12 ± 0.04 | 0.11 ± 0.02 | 0.11 ± 0.02 | 0.15 ± 0.07 | 0.17 ± 0.07 | 0.16 ± 0.07 |
| Water | 0.74 ± 0.08 | 0.68 ± 0.10 | 0.70 ± 0.04 | 0.84 ± 0.06 | 0.89 ± 0.13 | 0.78 ± 0.03 |
To better understand the effects of cross-terms, the ADC of metabolites and water were also evaluated separately using signals from positive (ADC+) and negative (ADC−) polarity of the DW gradients (Table 1). ADC+ values were significantly lower compared to ADC− with STEAM while no significant cross-term effects were observed in PRESS.
The CRLB of the ADC were also determined as previously described (32). The lowest CRLB of the ADC was observed for tNAA and was comparable between both sequences (3.5 ± 0.8% with STEAM versus 3.3 ± 0.6% with PRESS). The highest CRLB of the ADC were observed for tCho (19.5 ± 5.3% with STEAM versus 24.3 ± 10.4% with PRESS). For the other reported metabolites, the CRLB of the ADC ranged from 3.8 to 7.2% and were comparable between both sequences.
Discussion
This study shows the feasibility of measuring the trace/3 ADC of the five major metabolites and water on a clinical scanner. The high spectral SNR at high b-value has successfully enabled the measurement of the diffusivity of Glu and mIns for the first time at 3 T with biases from cross-terms removed. Although the ADC values were comparable between PRESS and STEAM techniques, the quantification precision for these two J-coupled metabolites was better with STEAM.
The effect of cross-terms was more pronounced in STEAM than in PRESS data (Table 1). This observation is consistent with two previous studies utilizing STEAM (13,33) where the variation in ADC values was apparent between positive and negative DW gradients. On the other hand, small cross-terms were observed in PRESS most likely due to the bipolar DW scheme used and the symmetric spoiler gradients applied between the two refocusing pulses which minimize the effect (12).
The measured ADC values of the 3 singlets (tNAA, tCr and tCho) in this study were in agreement with some studies (12,13,34) while being different from other studies (7,35,36) in the human brain. This disagreement could be related to the different proportion of WM and GM in the selected VOI between various reported studies since it is known that ADC values are different between GM and WM (12). Another reason for this discrepancy could be related to uncorrected cross-terms effects (13) or the duration of diffusion times used in the MRS sequences (13,37). Our results are consistent with the published ADC values at 7 T which ranged from 0.10 to 0.16 µm2/ms where cross-terms effects were suppressed in addition to having comparable GM, WM and CSF tissue composition in the VOI. The ADC of water also agrees with literature values ranging from 0.65 – 0.78 in healthy subjects (13,35).
Glu and mIns are the two most highly concentrated (> 6 mM) J-coupled metabolites in the brain next to the three major singlets. Owing to the high b-value achieved at relatively short TE in both STEAM and PRESS, it was possible to successfully measure the ADC of both Glu and mIns at 3 T without any cross-terms. The measured ADC of Glu was comparable to that previously reported at a much higher field of 7 T from the occipital gray matter. Recently the diffusivity of Glu was assessed by two groups at 3 T (16,17) however the reported ADC values were higher than in the present study most likely due to absence of cross-term correction and difference in gray and white matter content in the selected VOI (17).
To the best of our knowledge, the ADC of mIns has only recently been reported in the human brain at 3 T (16,17) although several animal studies have successfully measured its diffusivity (9–11). In rat and monkey brains (9,10), the ADC values for mIns were found to be comparable between species and were also similar to the ADC of NAA although the tissue content was different; rat brain consists mostly of gray matter while in the monkey study the VOI was positioned in both the GM and WM. The fact that the ADC of mIns was comparable to that of NAA in the current study suggests that the ADC value of mIns was accurately measured. Consequently, the previously reported ADC value for mIns seems to be overestimated (16,17), similarly to Glu, due to uncorrected cross-terms.
Measured ADC values were slightly, but not significantly, lower with STEAM than PRESS, although cross-term effects were taken into account in the current study (Table 1). This discrepancy in ADC might be explained by difference in duration and amplitude of DW gradients, however these parameters were comparable between both sequences. One possible explanation for the observed lower ADC values with STEAM could be related to the difference in diffusion times; td was approximately 4 times longer with STEAM compared to PRESS in this study. Previous animal studies have shown that ADCs are dependent on the diffusion time (13,37) and this seems to be consistent with our findings: at shorter td ADC was higher with PRESS while at longer td ADC was lower with STEAM. The high CSD error in PRESS might also play a part in the slightly higher ADC values; NAA and mIns signals are displaced in a VOI containing mostly WM and it is known that ADC values are higher in WM.
DW spectra are very sensitive to motion (bulk or physiological) which occurs during the application of the diffusion gradients, and this will result in phase and signal amplitude variations of the spectra (7,35,38). To partially mitigate these effects, several studies have used cardiac gating in addition to the phase information from the residual water peak during post-processing (7,14,33,38). In the current study, it was possible to correct for phase variations of the single-shot spectrum due to the high SNR of the diffusion data without any gating as previously reported (35). To avoid overestimating the ADC values, low SNR data (i.e., spectra with low amplitude fluctuations) were discarded (Figure 2). This resulted in ADCs consistent with literature values as discussed above. Hence, cardiac gating might not be essential if the single-shot spectral SNR is high enough to correct for phase fluctuations between shots, and post-processing is important to avoid bias in the determined ADC value.
Conclusion
This study shows the feasibility of successfully measuring the ADC values of the five major metabolites (tNAA, tCr, tCho, Glu and mIns) in the human prefrontal gray matter at 3 T, a particularly challenging region for B0 shimming due to the proximity to the sinus cavities. By reporting the mean diffusivity, i.e., trace/3 ADC values, the directional dependence of the measured ADC was eliminated. In conclusion, both STEAM and PRESS sequences can be used to probe changes in the diffusivity of metabolites in normal brain and various pathologies on the clinical scanner.
Acknowledgments
This work was supported by NIH grants: R21AG045606, P41 EB015894, P30 NS076408. We would like to thank Drs. Julien Valette and Francesca Branzoli for helpful discussion.
References
- 1.Nicolay K, Braun KPJ, Graaf RA, Dijkhuizen RM, Kruiskamp MJ. Diffusion NMR spectroscopy. NMR Biomed. 2001;14(2):94–111. doi: 10.1002/nbm.686. [DOI] [PubMed] [Google Scholar]
- 2.Bihan DL. Molecular diffusion, tissue microdynamics and microstructure. NMR Biomed. 1995;8(7):375–386. doi: 10.1002/nbm.1940080711. [DOI] [PubMed] [Google Scholar]
- 3.Ronen I, Valette J. eMagRes. Vol. 4. John Wiley & Sons, Ltd; 2015. Diffusion-Weighted Magnetic Resonance Spectroscopy; pp. 733–750. [Google Scholar]
- 4.Cao P, Wu EX. In vivo diffusion MRS investigation of non-water molecules in biological tissues. NMR in Biomedicine. 2017;30(3):e3481-n/a. doi: 10.1002/nbm.3481. [DOI] [PubMed] [Google Scholar]
- 5.Frahm J, Merboldt K-D, Hänicke W. Localized proton spectroscopy using stimulated echoes. J Magn Reson. 1987;72(3):502–508. doi: 10.1002/mrm.1910170113. [DOI] [PubMed] [Google Scholar]
- 6.Bottomley PA. Spatial Localization in NMR Spectroscopy in Vivo. Ann NY Acad Sci. 1987;508(1):333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 7.Posse S, Cuenod CA, Le Bihan D. Human brain: proton diffusion MR spectroscopy. Radiology. 1993;188(3):719–725. doi: 10.1148/radiology.188.3.8351339. [DOI] [PubMed] [Google Scholar]
- 8.Ronen I, Ercan E, Webb A. Axonal and glial microstructural information obtained with diffusion-weighted magnetic resonance spectroscopy at 7T. Front Integr Neurosci. 2013;7:13. doi: 10.3389/fnint.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valette J, Guillermier M, Besret L, Boumezbeur F, Hantraye P, Lebon V. Optimized diffusion-weighted spectroscopy for measuring brain glutamate apparent diffusion coefficient on a whole-body MR system. NMR Biomed. 2005;18(8):527–533. doi: 10.1002/nbm.985. [DOI] [PubMed] [Google Scholar]
- 10.Pfeuffer J, Tkáč I, Gruetter R. Extracellular-intracellular distribution of glucose and lactate in the rat brain assessed noninvasively by diffusion-weighted 1H nuclear magnetic resonance spectroscopy in vivo. J Cereb Blood Flow Metab. 2000;20(4):736–746. doi: 10.1097/00004647-200004000-00011. [DOI] [PubMed] [Google Scholar]
- 11.Dreher W, Busch E, Leibfritz D. Changes in apparent diffusion coefficients of metabolites in rat brain after middle cerebral artery occlusion measured by proton magnetic resonance spectroscopy. Magn Reson Med. 2001;45(3):383–389. doi: 10.1002/1522-2594(200103)45:3<383::aid-mrm1050>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 12.Kan HE, Techawiboonwong A, van Osch MJP, Versluis MJ, Deelchand DK, Henry P-G, Marjańska M, van Buchem MA, Webb AG, Ronen I. Differences in apparent diffusion coefficients of brain metabolites between grey and white matter in the human brain measured at 7 T. Magn Reson Med. 2012;67(5):1203–1209. doi: 10.1002/mrm.23129. [DOI] [PubMed] [Google Scholar]
- 13.Najac C, Branzoli F, Ronen I, Valette J. Brain intracellular metabolites are freely diffusing along cell fibers in grey and white matter, as measured by diffusion-weighted MR spectroscopy in the human brain at 7 T. Brain Struct Funct. 2016;221(3):1245–1254. doi: 10.1007/s00429-014-0968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harada M, Uno M, Hong F, Hisaoka S, Nishitani H, Matsuda T. Diffusion-weighted in vivo localized proton MR spectroscopy of human cerebral ischemia and tumor. NMR Biomed. 2002;15(1):69–74. doi: 10.1002/nbm.759. [DOI] [PubMed] [Google Scholar]
- 15.Mayer D, Dreher W, Leibfritz D, Spielman DM. RF refocused echoes of J-coupled spin systems: Effects on RARE-based spectroscopic imaging. Magn Reson Med. 2007;57(5):967–971. doi: 10.1002/mrm.21206. [DOI] [PubMed] [Google Scholar]
- 16.Döring A, Adalid V, Brandejsky V, Boesch C, Kreis R. ISMRM Workshop on MR Spectroscopy: From Current Best Practice to Latest Frontiers. Lake Constance; Germany: 2016. Optimized Estimation of Apparent Diffusion Coefficients of Metabolites. [Google Scholar]
- 17.Landheer K, Schulte R, Geraghty B, Hanstock C, Chen AP, Cunningham CH, Graham SJ. Diffusion-weighted J-resolved spectroscopy. Magn Reson Med. doi: 10.1002/mrm.26514. [DOI] [PubMed] [Google Scholar]
- 18.Brant-Zawadzki M, Gillan GD, Nitz WR. MP RAGE: a three-dimensional, T1-weighted, gradient-echo sequence--initial experience in the brain. Radiology. 1992;182(3):769–775. doi: 10.1148/radiology.182.3.1535892. [DOI] [PubMed] [Google Scholar]
- 19.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 20.Tkáč I, Starčuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41(4):649–656. doi: 10.1002/(sici)1522-2594(199904)41:4<649::aid-mrm2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 21.Deelchand DK, Adanyeguh IM, Emir UE, Nguyen T-M, Valabregue R, Henry P-G, Mochel F, Öz G. Two-site reproducibility of cerebellar and brainstem neurochemical profiles with short-echo, single-voxel MRS at 3T. Magn Reson Med. 2015;73(5):1718–1725. doi: 10.1002/mrm.25295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reese TG, Heid O, Weisskoff RM, Wedeen VJ. Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn Reson Med. 2003;49(1):177–182. doi: 10.1002/mrm.10308. [DOI] [PubMed] [Google Scholar]
- 23.Valette J, Giraudeau C, Marchadour C, Djemai B, Geffroy F, Ghaly MA, Le Bihan D, Hantraye P, Lebon V, Lethimonnier F. A new sequence for single-shot diffusion-weighted NMR spectroscopy by the trace of the diffusion tensor. Magn Reson Med. 2012;68(6):1705–1712. doi: 10.1002/mrm.24193. [DOI] [PubMed] [Google Scholar]
- 24.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 25.Henry PG, Marjańska M, Walls JD, Valette J, Gruetter R, Ugurbil K. Proton-observed carbon-edited NMR spectroscopy in strongly coupled second-order spin systems. Magn Reson Med. 2006;55(2):250–257. doi: 10.1002/mrm.20764. [DOI] [PubMed] [Google Scholar]
- 26.Govind V, Young K, Maudsley AA. Corrigendum: Proton NMR chemical shifts and coupling constants for brain metabolites. In: Govindaraju V, Young K, Maudsley AA, editors. NMR Biomed. Vol. 13. 2000. pp. 129–153. NMR Biomed 2015; 28(7):923–924. [DOI] [PubMed] [Google Scholar]
- 27.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser LG, Marjańska M, Matson GB, Iltis I, Bush SD, Soher BJ, Mueller S, Young K. 1H MRS detection of glycine residue of reduced glutathione in vivo. J Magn Reson. 2010;202(2):259–266. doi: 10.1016/j.jmr.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maudsley AA, Govindaraju V, Young K, Aygula ZK, Pattany PM, Soher BJ, Matson GB. Numerical simulation of PRESS localized MR spectroscopy. J Magn Reson. 2005;173(1):54–63. doi: 10.1016/j.jmr.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser LG, Young K, Matson GB. Numerical simulations of localized high field 1H MR spectroscopy. J Magn Reson. 2008;195(1):67–75. doi: 10.1016/j.jmr.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn Reson Med. 1994;32(3):294–302. doi: 10.1002/mrm.1910320304. [DOI] [PubMed] [Google Scholar]
- 32.Brihuega-Moreno O, Heese FP, Hall LD. Optimization of diffusion measurements using Cramer-Rao lower bound theory and its application to articular cartilage. Magn Reson Med. 2003;50(5):1069–1076. doi: 10.1002/mrm.10628. [DOI] [PubMed] [Google Scholar]
- 33.Branzoli F, Ercan E, Webb A, Ronen I. The interaction between apparent diffusion coefficients and transverse relaxation rates of human brain metabolites and water studied by diffusion-weighted spectroscopy at 7 T. NMR Biomed. 2014;27(5):495–506. doi: 10.1002/nbm.3085. [DOI] [PubMed] [Google Scholar]
- 34.Ercan AE, Techawiboonwong A, Versluis MJ, Webb AG, Ronen I. Diffusion-weighted chemical shift imaging of human brain metabolites at 7T. Magn Reson Med. 2015;73(6):2053–2061. doi: 10.1002/mrm.25346. [DOI] [PubMed] [Google Scholar]
- 35.Ellegood J, Hanstock CC, Beaulieu C. Trace apparent diffusion coefficients of metabolites in human brain using diffusion weighted magnetic resonance spectroscopy. Magn Reson Med. 2005;53(5):1025–1032. doi: 10.1002/mrm.20427. [DOI] [PubMed] [Google Scholar]
- 36.Liu Z, Zheng D, Wang X, Zhang J, Xie S, Xiao J, Jiang X. Apparent Diffusion Coefficients of Metabolites in Patients with MELAS Using Diffusion-Weighted MR Spectroscopy. Am J Neuroradiol. 2011;32(5):898. doi: 10.3174/ajnr.A2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchadour C, Brouillet E, Hantraye P, Lebon V, Valette J. Anomalous Diffusion of Brain Metabolites Evidenced by Diffusion-Weighted Magnetic Resonance Spectroscopy in Vivo. J Cereb Blood Flow Metab. 2012;32(12):2153–2160. doi: 10.1038/jcbfm.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Upadhyay J, Hallock K, Erb K, Kim D-S, Ronen I. Diffusion properties of NAA in human corpus callosum as studied with diffusion tensor spectroscopy. Magn Reson Med. 2007;58(5):1045–1053. doi: 10.1002/mrm.21372. [DOI] [PubMed] [Google Scholar]