Abstract
High blood pressure exerts its deleterious effects on health largely through acceleration of end organ diseases. Among these, progressive loss of renal function is particularly important, not only for the direct consequences of kidney damage, but also because loss of renal function is associated with amplification of other adverse cardiovascular outcomes. Genetic susceptibility to hypertension and associated end organ disease is non-Mendelian in both humans and in a rodent model, the spontaneously hypertensive rat (SHR). Here we report that hypertensive end organ disease in the inbred SHR-A3 line is attributable to genetic variation in the immunoglobulin heavy chain on chromosome 6. This variation coexists with variation in a 10 Mbase block on chromosome 17 that contains genetic variation in two genes involved in immunoglobulin Fc receptor signaling. Substitution of these genomic regions into the SHR-A3 genome from the closely-related, but injury resistant, SHR-B2 line normalizes both biomarker and histological measures of renal injury. Our findings indicate that genetic variation leads to a contribution by immune mechanisms hypertensive end organ injury and that, in this rat model, disease is influenced by differences in germ-line antibody repertoire.
Keywords: Genetics, immunoglobulin, hypertension, progressive renal disease, congenic line, proteinuria, biomarkers
Introduction
Hypertension is associated with progressive renal disease and the most effective predictor of renal disease is the presence of a close relative that has required renal dialysis 1, 2. This genetic underpinning has prompted exhaustive studies in humans 3–8, however, explanation of genetic risk remains limited. The spontaneously hypertensive rat also shows strong genetic influences on renal injury susceptibility 9, 10. The SHR-A3 line experiences progressive renal disease and has much shortened life span while other SHR lines with overlapping genetic susceptibility to hypertension, such as SHR-B2, resist renal injury and experience normal lifespans 11. The emergent pattern of injury is focal regions of combined glomerular and tubulo-interstitial damage along with albuminuria. Much normal tissue architecture remains between focal regions of injury and disease progression includes both increasing severity of injury in injury foci and increased numbers of injury foci 11. The close genetic similarity between these lines (87% identical by descent, IBD) provides an opportunity to discover and prove the genetic basis of this susceptibility 12.
In prior studies we have addressed whether the injury prone SHR-A3 line shares the same genetic architecture controlling hypertension as the SHR-B2 line 13. These lines are descended from the same founder pair 14, 15. Selective breeding fixed hypertension before the lines were separated. However, SHR-A3 is generally recognized to have higher systolic blood pressure (SBP) than injury-resistant lines 16, 17. We showed that these two lines differ in SBP and mapped a single haplotype block encompassing 10 Mbases on chromosome 17 that accounts for the SBP difference between these two lines 13. In the present study we have created a congenic line in which this chr17 block has been transferred from SHR-B2 into the SHR-A3 genetic background. We have used this to prove the effect of this segment on SBP and to investigate whether reduction of SBP to a level not different from the injury-resistant line influences the emergence of renal damage.
SHR-A3 and SHR-B2 also have extreme genetic divergence in the immunoglobulin heavy chain (IgH) locus and the haplotype block containing IgH is also associated with renal injury 18, 19. We have further tested whether this genetic difference contributes to the emergence of renal injury by creating a congenic line in which the IgH locus has been transferred from SHR-B2 into SHR-A3. We have used this congenic line to confirm that renal injury is determined, in part, by genetic variation in the IgH locus.
Finally, by crossing these two single congenic lines to create a double congenic line we have examined concurrent effects of these two loci on renal injury.
Methods
Transparency
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Animals
Studies were performed on male rats of the injury-prone spontaneously hypertensive-A3 (SHR-A3, SHRSP/Bbb) and the injury-resistant SHR-B2 inbred lines and progeny derived from crosses between these lines. The lines are maintained as closed colonies in our facility and the genetic integrity of the lines is verified using high throughput genotyping of genome-wide single nucleotide polymorphisms (SNP). The lines and their origins prior to transfer to our laboratory have been recorded at the Rat Genome Database (rgd.mcw.edu) which has applied the following identifiers: SHR-A3 – RGD ID = 8142383, Symbol = SHRSP/BbbUtx; SHR-B2 – RGD ID = 8142385, Symbol = SHR/Utx. Animals used in these studies were housed in an AAALAC-approved animal facility with sentinel monitoring to confirm the absence of transmissible pathogens in the colony. They were provided a standard rodent chow diet and drinking water ad libitum.
Genome sequencing
We have previously described the assembly and analysis of Illumina short-read genome sequences from SHR-A3 and SHR-B212. Variant call files are available on request to the corresponding author.
Congenic line construction
SHR-A3 (males) and SHR-B2 (females) parental lines were crossed to generate the F1 progeny. This progeny was backcrossed into SHR-A3 animals. Backcrossed animals were genotyped at each generation using a previously described panel of ~200 SNP markers to allow speed congenic selection of optimal animals (highest loss of SHR-B2 background alleles while retaining the introgressed chromosome 6 or 17 SHR-B2 haplotype blocks) 11. Each congenic line was then created by mating the best two male and female animals from the previous backcross and selecting their progeny that were homozygous for SHR-B2 alleles at the target loci and for SHR-A3 alleles at any locus at which heterozygosity remained in the prior backcross. Transfer of the entire chromosome 17 introgressed block was verified by genotyping the 3 SNP panel markers in the block as well as 3 additional SNP markers located at the center and extreme ends of this block. Transfer of the entire chr6 IgH haplotype block from SHR-B2 into the SHR-A3 genetic background was also verified by SNP markers located at the extreme ends of this block as well as an additional marker in the center of the block.
Serum immunoglobulin gamma sub-class measurement
We have previously reported the ELISA systems we used to measure serum immunoglobulin gamma sub-class levels 18.
Blood pressure
Blood pressure was measured by radiotelemetry devices (Data Sciences, St. Paul, MN) in adult animals implanted at 16–17 weeks of age. Catheters (Model PA-C40) were implanted under isoflurane anesthesia into the abdominal aorta above the bifurcation and below the renal arteries. Post-operative analgesic treatment with buprenorphine was provided for 2 days. Animals were allowed to recover from implantation for at least 7 days before blood pressure measurement began. Implants were calibrated under pressure (120, 160 and 200 mm Hg) at 37°C prior to implantation and again after removal and observed blood pressures were adjusted, if needed, to compensate for calibration drift. Blood pressure was measured for 24 hours once per week. During each 24 hour recording period, pressures were sampled for 30 seconds every 30 minutes.
Histological assessment of renal injury and ELISA determination of albuminuria
The divergence of histological measures of renal injury across time in SHR-A3 and SHR-B2, supported by representative photomicrographs, has been previously published 11. Quantitative histological measures of renal injury were assessed following methods we have previously described that use extensive random sampling to assess injury in both affected and unaffected renal tissue 11. Spot urines were collected because of divergent behavioral adaptation to metabolism cages observed between the two SHR lines. Urinary albumin excretion was measured by ELISA as previously described and normalized for urine creatinine levels determined by HPLC 11.
Renal Biomarker studies
We investigated urine levels in spot urine samples of kidney injury molecule 1 (KIM-1 Havcr1), Lipocalin 2 (Lcn2, Neutrophil Gelatinase-associated Lipocalin, NGAL) and osteopontin (OPN) using the Kidney Injury Panel 1 (rat) Assay Kit manufactured by Meso Scale Diagnostics (Gaithersburg, MD). The multiplex assay plate was read on a Meso Scale Diagnostics SECTOR Imager 2400 electrochemiluminescence plate reader. Biomarkers were determined in 8 animals per group and normalized to urine creatinine levels measured by HPLC 11.
Statistical analysis
Study groups comprised of 6–26 animals, please refer to Figure and Table legends for more information on group sizes. Group means ± SEM are shown. Multiple group comparisons were performed by ANOVA followed by Scheffé test. Histological scores are arbitrary non-parametric data and were tested to verify normality using Kolmogorov-Smirnov Test. Kruskal-Wallis test was used to identify differences in histological scores across groups. Statistical significance of non-parametric data was estimated by Scheffé test for the normally distributed multiple group comparison samples.
Results
Confirmation of the physical extent of the mapped chromosome 17 blood pressure block by genome sequencing
The genomes of SHR-A3 and SHR-B2 are descended from the same outbred founder pair and shared ancestors of both lines were inbred for 8 generations before the two lines were isolated. As a result, SHR-A3 and SHR-B2 are 87% identical by descent. The 13% of the SHR-A3 and SHR-B2 genome that is descended from different ancestors has a haplotype block structure 13. A single haplotype block located from chr17:7.2Mb to chr17:15.8Mb (Rn5 assembly) was mapped and shown to explain the difference in SBP observed between these two SHR lines at 18 weeks of age. This age precedes the onset of progressive renal injury, and therefore this difference in blood pressure does not result from renal injury 13. Using next generation whole genome sequence assemblies obtained for these two SHR lines, we are able to define this block with very high resolution (chr17:6,765,076–16,742,956, Rn5 assembly), examine the genes contained within it and tabulate the genetic variation associated with those genes (Table S1). We have previously reported renal gene expression differences between SHR-A3 and SHR-B2 associated with genes in the chr17 block 13.
Creation and confirmation of congenic state of SHR-A3(chr17-SHR-B2) rat line
A congenic line was created to test the hypothesis that the chr17 block influences blood pressure in the SHR-A3 genetic background20, 21. We used a panel of ~200 dimorphic SNP markers to determine genotypes at each generation of backcrossing during creation of the SHR-A3(chr17 SHR-B2) congenic line. Congenic animals were homozygous for SHR-B2 alleles at 4 SNP markers spanning the introgressed block (marker positions chr17:7,151,820, chr17:11,561,842, chr17:13,705,857, chr17:15,630,391, Rn5 assembly) and were further verified with PCR interrogating the extreme ends of the block (SNPs located at chr17:6,815,854 and chr17:16,288,148, Fig S1). SNP genotyping across the blocks of non-identity by descent allows us to conclude that, outside the 10 Mbase introgressed chromosome 17 segment, this congenic line is greater than 99.7% genetically identical with the SHR-A3 parental line. A single region of introgressed SHR-B2 ancestry was detected outside of the chr17 target block lying between chr11:15,415,747–22,927,323 (Rn5).
Effect of congenic introgression of the mapped chromosome 17 block on blood pressure
Table 1 indicates that there is a significant difference (+16.1 mm Hg) in systolic blood pressure (SBP) between the SHR-A3 parental line and the SHR-A3(chr17 SHR-B2) congenic line, but no difference between the congenic line and SHR-B2. Table 1 also indicates levels of mean and diastolic blood pressure (DBP) and heart rate. In our earlier SHR-A3 × SHR-B2 F2 intercross mapping study this locus was detected as a highly significant SBP locus and was not a significant DBP locus. This is confirmed in the congenic. We estimated that homozygosity in F2 animals for the SHR-A3 chr17 haplotype resulted in SBP 13.6 mm Hg greater than homozygosity for SHR-B2 alleles 13. Difference in SBP between the parental SHR-A3 and SHR-B2 lines is 19 mmHg (Table 1). Thus the chr17 block appears to account for the SBP difference between these two SHR lines and the congenic line proves the role of allelic variation at the chr17 locus uncovered by mapping.
Table 1.
Blood pressures in SHR lines at 17–18 weeks of age
| SHR line | n | SBP | SEM | P | MBP | SEM | P | DBP | SEM | P | HR | SEM | p | ANOVA comparison |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SHR-A3 | 26 | 205.7 | 3.9 | * | 177.4 | 3.4 | * | 149.0 | 3.1 | † | 330.1 | 2.9 | NS | SHR-A3 vs SHR-B2 |
| SHR-A3(chr17 SHR-B2) | 25 | 190.0 | 2.6 | † | 173.8 | 2.5 | NS | 156.0 | 2.9 | NS | 321.0 | 3.4 | NS | SHR-A3 vs SHRA3(chr17 SHRB2) |
| SHR-B2 | 20 | 186.7 | 2.5 | NS | 159.1 | 2.6 | † | 135.8 | 2.1 | * | 324.9 | 2.8 | NS | SHR-B2 vs SHRA3(chr17 SHRB2) |
|
| ||||||||||||||
| LOD score in BP mapping F2 A3 × B2 | 5.23 | 3.57 | 2.97 | |||||||||||
| LOD score p value | * | ‡ | NS | |||||||||||
n = number of animals per group. SBP, MBP, DBP = systolic, mean and diastolic arterial blood pressure, HR = heart rate. BP and HR multiple group comparisons by ANOVA followed by Scheffé test when ANOVA f statistic was significant (insignificant f statistic only for HR). P values indicate
p<0.001,
p<0.01,
p<0.05,
NS = not significant. LOD scores and p values obtained in F2 intercross mapping of SBP, MBP and DBP, previously reported 13
Except for the chr17 locus no BP loci segregate between SHR-A3 and SHR-B213. This indicates that the remaining polygenic basis for hypertension is shared by SHR-A3 and SHR-B2, a finding consistent with the known genealogy and observations of hypertensive trait fixation in SHR 15. We infer that the congenic SHR-A3(chr17 SHR-B2) and SHR-B2 share an identical genetic mechanism of hypertension so that traits in which they diverge arise from the action of other genetic variation.
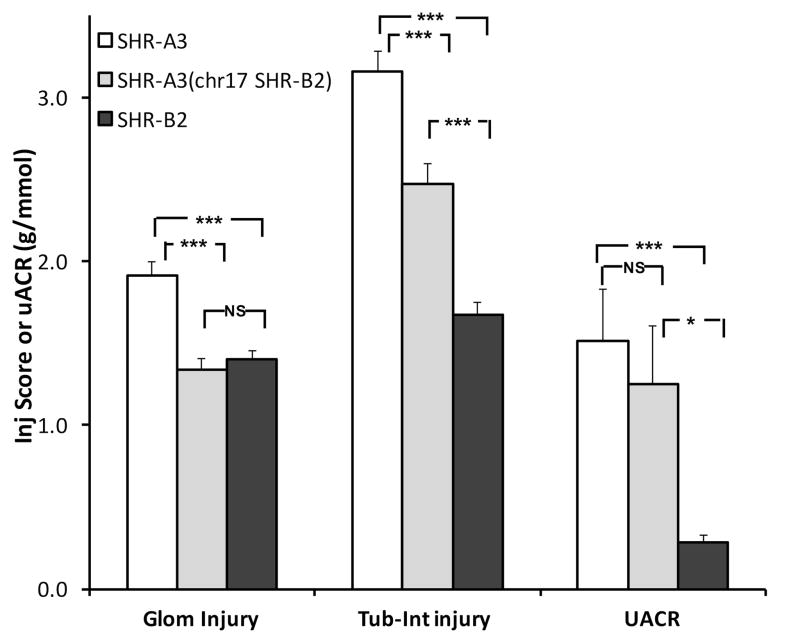
Effect of congenic introgression in chromosome 17 on renal injury
SHR-B2 strongly resists hypertensive renal injury 11. We proposed the null hypothesis that, if renal injury is driven solely by SBP differences between SHR-A3 and SHR-B2, no injury difference would be present in SHR-A3(chr17 SHR-B2) and SHR-B2. Figure 1 indicates three measures of renal injury assessed at 40 weeks of age in SHR-A3(chr17 SHR-B2) and its progenitor lines SHR-A3 and SHR-B2. Glomerular injury in the congenic line is reduced compared to SHR-A3 and is not significantly different from SHR-B2. Thus, concerning glomerular injury, our null hypothesis is supported. In contrast, tubulo-interstitial injury in the congenic line is greater than in SHR-B2, but significantly less than SHR-A3. Finally, proteinuria as reflected in urinary albumin/creatinine ratio is not different between SHR-A3 and the congenic line. This supports the conclusion that SBP has an important role in determining glomerular and tubulo-interstitial injury in SHR-A3. However, the effect is only partial regarding tubulo-interstitial injury and no effect to reduce proteinuria was observed. This may indicate other genetic influences on proteinuria arising outside of this locus.
Figure 1.
Histologically assessed glomerular and tubulo-interstitial injury scores measured in Periodic acid-Schiff-stained kidney sections and urinary albumin-creatinine ratios (UACR) and from 40 week old SHR-A3 (open), SHR-B2 (black) and congenic SHR-A3(chr17 SHR-B2) (gray) animals (SHR-A3, SHR-A3(chr17 SHR-B2), SHR-B2, n = 8, 12, 21 respectively). NS = no significant difference, * = p<0.05 and *** = p<0.001 (ANOVA, Scheffé test).
Confirmation of the physical extent of the chromosome 6 haplotype block containing the IgH locus by whole genome sequencing and haplotype differences in coding sequences
A haplotype block is located at the distal end of chromosome 6 at which genomic sequences of SHR-A3 and SHR-B2 are highly divergent. We defined the extent of this block from chr6:146,030,387–154,214,590 (Rn5 assembly of the rat genome). This block contains all components (VDJ genes, Fc isotype genes) of IgH 19. We also confirmed that the remainder of chr6 is IBD across the two SHR lines with the exception of a small block from chr6:2,028,567–4,036,845. A summary of non-IgH gene coding variation in the distal chr6 locus is provided in Table S2. Excluding IgH genes, only 18 other genes are present in this block and only 3 of these are affected by amino acid substitution. In contrast, of 230 IgH amino acid coding genes in the block, 112 IgH genes contain non-synonymous variation. There is also extensive structural variation (gene duplication and deletion) differentially affecting IgH genes in SHR-A3 compared with SHR-B219.
Creation and confirmation of congenic state of SHR-A3(IgH SHR-B2) rat line
A congenic line was created by backcrossing as above, targeting the IgH haplotype block. We genotyped animals from the resulting SHR-A3(IgH SHR-B2) congenic line to verify preservation of the entirety of the introgressed haplotype block. Our genome-wide SNP panel indicated that, outside the ~8Mbase introgressed chr6 segment, the congenic line is genetically identical with the SHR-A3 parental line.
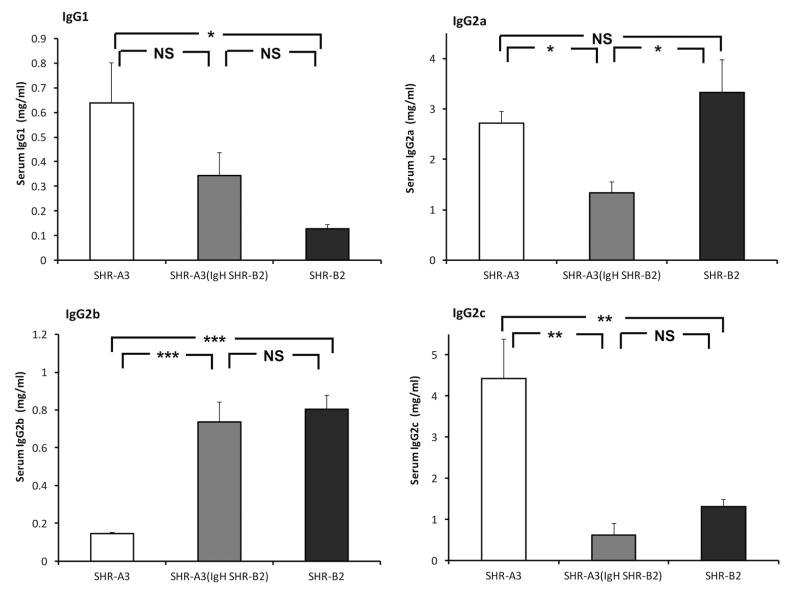
Effect of congenic introgression of the IgH locus on serum IgG levels
The rat IgG isotype exists as 4 subclasses. Serum levels of three of these subclasses (IgG1, IgG2b, IgG2c) persistently differ between SHR-A3 and SHR-B2. Our genetic mapping studies indicated a strong influence of the IgH block on serum IgG subclass levels, indicating that both coding sequence variation and regulation of immunoglobulin serum levels were determined by variation the IgH locus between SHR-A3 and SHR-B218. Congenic substitution of IgH from SHR-B2 into SHR-A3 may confer not only the coding sequences of SHR-B2 immunoglobulins on the congenic line, but may also shift the serum levels of IgG subclasses away from those observed in SHR-A3 and towards those in SHR-B2. Measurement of serum IgG subclasses confirms the effect of the IgH locus on serum IgG subclass levels (Figure 2).
Figure 2.
Serum IgG subclass levels were assessed by ELISA in 30 week old SHR-A3, SHR-B2 and SHR-A3(IgH SHR-B2), n = 6 per group. Significant differences in IgG1, IgG2b and IgG2c in SHR-A3 versus SHR-B2 were observed. In the congenic line, transfer of the SHR-B2 IgH segment into SHR-A3 resulted in serum IgG subclass levels that closely resembled the donor (SHR-B2) levels for IgG2b and IgG2c. For IgG2a, no effect of congenic transfer was predicted based on prior mapping, though a reduction was observed in the congenic line. For IgG1, the congenic levels were intermediate between the donor and recipient levels and not significantly different from either (*** p<0.001, ** p<0.01, *, p<0.05, NS not significantly different).
Effect of congenic introgression of the IgH locus on blood pressure
Recent evidence suggests the involvement of B lymphocytes in hypertension 22. Antigen-directed immunoglobulin affinity maturation and secretion of immunoglobulin are unique functional specializations of the B cell lineage. B lymphocyte effects on blood pressure may be affected by IgH genetic variation acting through either serum immunoglobulin levels or functional effects of IgH coding sequence differences, or both. Therefore we used the IgH congenic line to test for an effect of the chromosome 6 IgH block on blood pressure. We compared blood pressure measured by telemetry in SHR-A3 and the congenic line starting prior to the age when renal injury is present (17–18 weeks) until injury becomes established at 25–26 weeks (Fig. S2). No effect on blood pressure was observed to result from substitution of the IgH locus in SHR-A3.
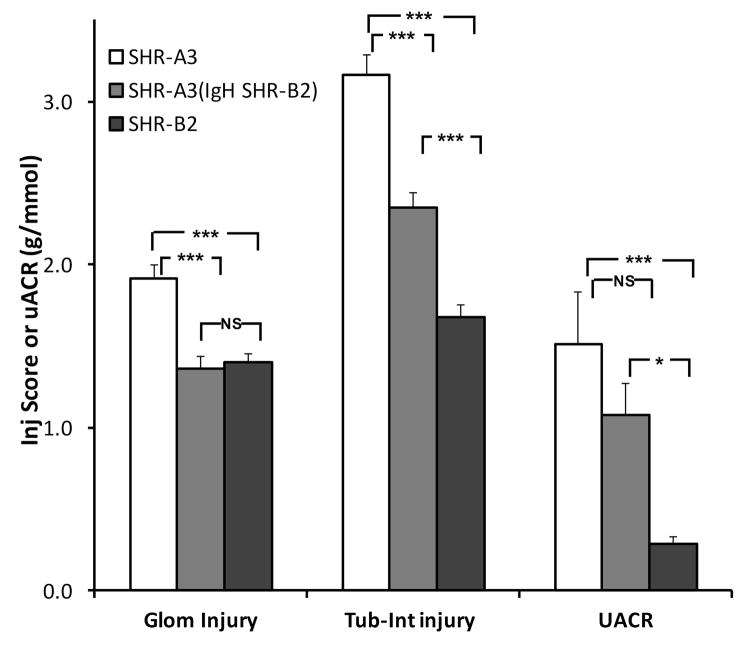
Effect of congenic introgression of the IgH locus on renal injury
Because administration of an immunosuppressive drug (mycophenolate mofetil) that impedes proliferation and maturation of T and B lymphocytes also reduces renal injury in SHR-A312, we considered it possible that IgH variation might provide a genetic influence on renal injury in SHR-A3. Figure 3 shows that glomerular injury is reduced at 40 weeks of age in the SHR-A3(IgH SHR-B2) line and approaches levels observed in SHR-B2 (SHR-A3 = 1.91 ± 0.08, SHR-A3(IgH SHR-B2) = 1.36 ± 0.08 and SHR-B2 1.20 ± 0.07). Tubulo-interstitial injury score is also significantly reduced in the congenic line (SHR-A3 = 3.16 ± 0.12, SHR-A3(IgH SHR-B2) = 2.35 ± 0.09 and SHR-B2 1.32 ± 0.12). However, no significant difference between uACR in SHR-A3 and SHR-A3(IgH SHR-B2) was observed.
Figure 3.
Histologically assessed glomerular and tubulo-interstitial injury scores and urinary albumin-creatinine ratios (UACR) and from 40 week old SHR-A3 (open), SHR-B2 (black) and the congenic SHR-A3(IgH SHR-B2) (gray) lines, n = 8, 19, 21 respectively. Glomerular and tubulo-interstitial injury were both significantly reduced in the congenic line, compared to SHR-A3, UACR was not different. NS = no significant difference, * = p<0.05 and *** = p<0.001 (ANOVA, Scheffé test)
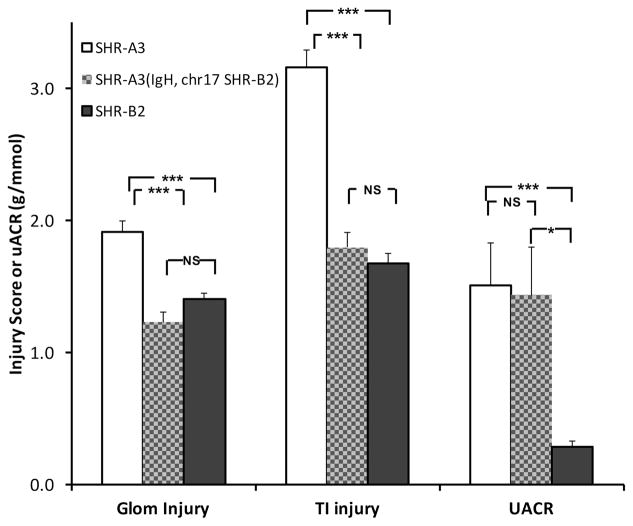
Creation and confirmation of double congenic state of SHR-A3(chr17, IgH SHR-B2) rat line
The capacity of the IgH and chr17 loci to influence renal injury independently lead us to investigate the effect of the combined actions of these two loci. We mated animals from the two single congenic lines and bred to homozygosity animals that contained the SHR-A3 genetic background, but were homozygous at both the IgH and chr17 loci for SHR-B2 alleles. We examined the development of renal injury in these double congenic animals at aged 40 weeks. Figure 4 shows that histologically assessed glomerular and tubulo-interstitial injury in the double congenic are both reduced to levels not different from SHR-B2. However, proteinuria was not reduced in the double congenic and appears to be controlled by genetic variation outside these two loci.
Figure 4.
Histologically assessed glomerular and tubulo-interstitial injury and urinary albumin-creatinine ratios (UACR) and from 40 week old SHR-A3 (open), SHR-B2 (black) and the congenic SHR-A3(IgH, chr17 SHR-B2) double congenic line (hatched) n = 8, 20, 21 respectively. Glomerular and tubulo-interstitial injury scores were not significantly different in the congenic line compared to SHR-B2, however UACR was different from SHR-B2, but not from SHR-A3. NS = no significant difference, * = p<0.05 and *** = p<0.001 (ANOVA, Scheffé test)
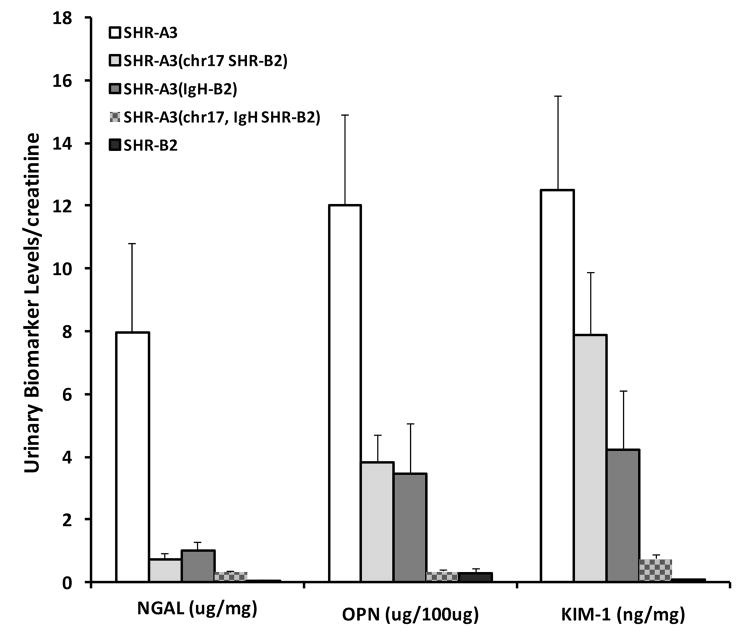
Urinary Biomarkers of Renal Injury
We assessed urinary levels of three established biomarkers of acute renal injury (Osteopontin, Neutrophil Gelatinase-associated Lipocalin and Kidney Injury Molecule-1) in SHR-A3, SHR-B2, the SHR-A3(IgH SHR-B2) congenic line, the SHR-A3(chr17 SHR-B2) congenic line and the double congenic line at 30 weeks of age. There was extreme divergence in each of the three biomarker levels between the parental lines, SHR-A3 and SHR-B2 (Figure 5). The two single congenic lines had intermediate levels of each of the biomarkers, while the double congenic line biomarker levels were all close to those observed in SHR-B2. While the precise biological significance of these markers is not fully clear, they appear to reflect predominantly injury to the proximal renal tubules and thus extend our measurement of urinary albumin excretion from what is predominantly a filtration and barrier function to a reflection of renal epithelial cellular damage 23, 24. Our observations suggest their relevance to progressive renal injury in the SHR-A3 model and support and extend the conclusion from our histological studies that the BP effect of the chr17 locus and effects arising from germ line variation in IgH are key elements of the emergence of progressive renal disease in this model of hypertension.
Figure 5.
Urinary biomarker levels normalized to urine creatinine in 30 week old SHR-A3 (open), SHR-B2 (black), the SHR-A3(chr17 SHR-B2) congenic line (light gray), the SHR-A3(IgH SHR-B2) congenic line (dark gray) and the double congenic SHR-A3(IgH, chr17 SHR-B2) line (hatched), n = 8 per group. Although presented in a single figure for clarity, the hypotheses tested by comparing each of the congenic lines with the two parental strains are independent and statistical significance testing (ANOVA, Scheffé test) reflects this. Comparing SHR-A3(IgH SHR-B2) to its parental strains: NGAL; SHR-A3 vs SHR-B2 p = 0.03,SHR-A3 vs SHR-A3(IgH SHR-B2) p = NS, SHR-A3(IgH SHR-B2) vs SHR-B2 p = NS: OPN; SHR-A3 vs SHR-B2 p = 0.001, SHR-A3 vs SHR-A3(IgH SHR-B2) p = 0.02, SHR-A3(IgH SHR-B2) vs SHR-B2 p = NS: Kim1; SHR-A3 vs SHR-B2 p = 0.001, SHR-A3 vs SHR-A3(IgH SHR-B2) p = 0.03, SHR-A3(IgH SHR-B2) vs SHR-B2 p = NS. Comparing SHR-A3(chr17 SHR-B2) to its parental strains: NGAL; SHR-A3 vs SHR-B2 p = 0.01,SHR-A3 vs SHR-A3(chr17 SHR-B2) p = 0.03, SHR-A3(chr17 SHR-B2) vs SHR-B2 p = NS: OPN; SHR-A3 vs SHR-B2 p = 0.002,SHR-A3 vs SHR-A3(chr17 SHR-B2) p = 0.04, SHR-A3(chr17 SHR-B2) vs SHR-B2 p = NS: Kim1; SHR-A3 vs SHR-B2 p = 0.002, SHR-A3 vs SHR-A3(chr17 SHR-B2) p = NS, SHR-A3(chr17 SHR-B2) vs SHR-B2 p = 0.05. Comparing SHR-A3(IgH, chr17 SHR-B2) to its parental strains: NGAL; SHR-A3 vs SHR-B2 p = 0.01, SHR-A3 vs SHR-A3(IgH, chr17 SHR-B2) p = 0.01, SHR-A3(IgH, chr17 SHR-B2) vs SHR-B2 p = NS: OPN; SHR-A3 vs SHR-B2 p = 0.0003,SHR-A3 vs SHR-A3(IgH, chr17 SHR-B2) p = 0.0003, SHR-A3(IgH, chr17 SHR-B2) vs SHR-B2 p = NS: Kim1; SHR-A3 vs SHR-B2 p = 0.0003, SHR-A3 vs SHR-A3(IgH, chr17 SHR-B2) p = 0.0005, SHR-A3(IgH, chr17 SHR-B2) vs SHR-B2 p = NS.
Discussion
SHR-A3 and SHR-B2 are descended from a single founder pair of Wistar rats whose progeny were selectively bred to fix the trait of hypertension. Separation of distinct SHR-A and SHR-B lineages occurred after fixation of hypertension so it can be expected that alleles causing hypertension in SHR are shared across SHR lines 14, 15, 25, 26. We have shown by high density single nucleotide polymorphism analysis that the genome-wide extent of shared ancestry across these two lines is ~87% and that it is comprised of blocks of shared ancestry interspersed with blocks of ancestry arising from genetically divergent progenitors 13. This structure facilitates mapping of traits at which the two lines diverge due to genetic variation. We performed mapping studies to determine whether the higher blood pressure levels in SHR-A3 than other SHR lines might be attributed to an additional genetic effect that is absent in SHR-B2. This work indicated that SHR-A3 alleles in the chr17 block segregated with blood pressure in an SHR-A3 and SHR-B2 F2 intercross when blood pressure was measured prior to the emergence of renal injury. This indicates that hypertension alleles are shared by both lines and that an additional BP effect exists only in SHR-A3 which is confined to this ~10Mb block that constitutes 0.36% of the rat genome 13. The chr17 congenic line confirms the role of this short segment of chr17 in the higher BP levels seen in SHR-A3. The present study clearly indicates that the chr17 block produces a combined effect on both blood pressure and renal injury. The injury effect may arise secondarily to the further elevation of blood pressure and it is notable that levels of blood pressure in SHR-A3 exceed the auto-regulatory range of renal blood flow and may contribute to injury initiation by disrupting renovascular function as has been suggested for other rat renal injury models 27–30.
Our prior work indicates that difference in injury susceptibility appears to result, at least in part, from genetic variation in SHR-A3 that influences renal inflammation 12. This raises the question of whether substitution of the chr17 block affecting SBP and renal injury alters an immune component of renal injury in SHR-A3. Although the chr17 block is relatively rich in genes, two immune signaling genes (Syk and Dok3) lie in this block and exist as dimorphic alleles across SHR-A3 and SHR-B2. Furthermore, these genes both participate in inflammatory signaling networks arising from Fc receptors for which immunoglobulins are the activating ligand 31–33. Further refinement of this locus by sub-congenic line creation may clarify the possible involvement of one or both of these genes in contributing to blood pressure elevation and renal injury.
There is extensive genetic variation between the germ-line IgH sequences across SHR-B2 and SHR-A319. Immunosuppression reduces hypertensive renal injury in SHR-A3 line 12 and this suggested the possible involvement of B cell mediated immunity in hypertensive renal injury. The SHR-A3(IgH SHR-B2) congenic line provides clear evidence that germ-line genetic variation in IgH influences hypertensive renal injury and that such influence does not arise from a resulting difference in SBP between SHR-A3 and SHR-B2.
Congenic substitution approaches are subject to confounding effects arising from undetected gene variants transferred during backcrossing from outside the targeted congenic segment. Such a possibility appears to be small in this case because of the high levels of genetic identity between the SHR-A3 and SHR-B2 lines and because we have extensively genotyped across the haplotype blocks from which the SHR-A3 and SHR-B2 lines arise from divergent ancestors. IgH sequence variation seems likely to be the cause of trait variation in the comparisons made, because IgH comprises essentially all of the protein coding variation in the transferred block. Analysis of whole genome sequences for SHR-A3 and SHR-B2 has allowed us to determine the extent of non-synonymous protein coding variation between these two closely-related rat lines. Across the entire genome, excluding the IgH locus, we have identified 652 genes containing non-synonymous sequence variation comparing these lines with each other (~3% of all genes). In contrast, there are 112 immunoglobulin gene segments with non-synonymous variation in the IgH locus (49% of IgH genes) and potentially even more variation arising from the frequent structural genetic variation and gene duplication known to occur in the IgH locus and that is poorly resolved by next-generation, short-read sequencing. Thus, at the whole genome level, at least 17% of non-synonymous genetic variation between SHR-A3 and SHR-B2 occurs in the 0.3% of the genome that constitutes the IgH locus. An assembled, structurally complete genome sequence of the highly variable IgH locus in humans has only recently been achieved and none is available for the rat 34. Germ line immunoglobulin sequence diversity provides the starting point for antigen recognition 35. Germ line variation in IgH in SHR-A3 may favor the development of antibodies to antigens exposed in the hypertensive kidney.
The double congenic line we have created indicates that renal injury in SHR-A3 can be reduced to the same low levels observed in SHR-B2 by substitution of just two genomic loci comprising 18 Mbases of the 2,870 Mbase rat genome. SBP levels in SHR-A3 that are nearly 20 mmHg greater than SHR-B2 may play an important role in the initiation of injury. This additional degree of SBP elevation may overwhelm renal blood flow auto-regulatory processes which appear to retain function until ~180 mmHg 36. The resulting damage might initiate injury in both glomerular and tubulo-interstitial compartments that is amplified when the germ line SHR-A3 IgH sequences are present compared with those in SHR-B2. The persistence of high levels of proteinuria in the double congenic is unexpected and indicates a potential genetic influence from the SHR-A3 genome outside of the loci we have isolated, though other explanations may be possible.
In conclusion, these studies have identified two loci that determine susceptibility to renal injury in this rat model of hypertension. The pathway appears to involve both increased injury due to genetic variation driving higher blood pressure and to the involvement of genetic variation in immunoglobulin biology arising from within the IgH locus. There is possible interaction between gene variation affecting renal injury because two diallelic immunoglobulin signaling genes are present in the BP locus. These studies pose questions regarding the potential interaction between immunoglobulin genetic variation and hypertensive renal injury in humans. The difficulty of addressing this possibility is highlighted by the fact that the extreme genetic diversity of the immunoglobulin loci reduces the capacity of contemporary genome-wide association or genome sequencing studies to detect this involvement 37, 38. This may contribute to the “missing heritability” observed in GWAS studies of renal function 39. It is well recognized that there is common genetic variation in humans that influences the likelihood of formation of self-reactive antibodies 40–44. Such variation might interact with germ-line immunoglobulin variation to contribute to risk of disease in hypertensive patients. This rat model proposes novel, discrete disease targets and mechanisms for investigation in humans.
Supplementary Material
Perspectives.
SHR lines exist that differ in susceptibility to hypertensive end organ injury and provide an opportunity to advance understanding of genetic mechanisms that create injury susceptibility. We have identified two loci that control the emergence of renal injury in the SHR strain. One is associated with a greater rise in blood pressure (chr17:7.2–15.8 Mb). The other (chr6:146.0–154.2 Mb) includes all immunoglobulin VDJ and constant genes of the IgH locus which are highly divergent between SHR lines. The presence of genetic variation in the chr17 block that affects genes involved in B cell immune signaling also suggest a role for immunoglobulin structural and/or functional divergence in the mechanism of disease. These observations tie the suspected role of immune-mediated damage in end organ disease to specific aspects of immune function. Interestingly, the structural complexity of the immunoglobulin locus has prevented it from being investigated in human population genetic studies of end organ disease and it may be an important source of unexplained heritability of disease susceptibility.
Novelty and Significance.
1. What Is New?
Hypertensive kidney injury has a strong heritable basis that remains largely undefined. In a hypertensive rat model, we show that injury in the susceptible SHR-A3 line is strongly attenuated by replacement of two loci representing less than 0.7% of the genome from the SHR-B2 injury-resistant line. One of these loci (chr17) contributes to additional elevation of blood pressure. The other locus contains genes encoding the immunoglobulin heavy chain (chr6, IgH).
2. What Is Relevant?
These studies indicate that prior observations implicating immunological processes in hypertensive renal disease can arise genetically. The IgH has a complex structure that cannot be systematically investigated in human GWAS.
3. Summary
These studies show interaction between increased blood pressure and renal injury and indicate that genetic variation in the antibody repertoire can contribute to hypertensive renal disease.
Acknowledgments
Sources of funding: This research was supported by grants from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (NIH-NIDDK DK081866 and DK114235) to PAD.
Footnotes
Conflicts of Interest
None
References
- 1.Freedman BI, Soucie JM, McClellan WM. Family history of end-stage renal disease among incident dialysis patients. Journal of the American Society of Nephrology: JASN. 1997;8:1942–1945. doi: 10.1681/ASN.V8121942. [DOI] [PubMed] [Google Scholar]
- 2.Freedman BI, Volkova NV, Satko SG, Krisher J, Jurkovitz C, Soucie JM, McClellan WM. Population-based screening for family history of end-stage renal disease among incident dialysis patients. Am J Nephrol. 2005;25:529–535. doi: 10.1159/000088491. [DOI] [PubMed] [Google Scholar]
- 3.Kottgen A, Kao WH, Hwang SJ, Boerwinkle E, Yang Q, Levy D, Benjamin EJ, Larson MG, Astor BC, Coresh J, Fox CS. Genome-wide association study for renal traits in the Framingham Heart and Atherosclerosis Risk in Communities Studies. BMC Med Genet. 2008;9:49. doi: 10.1186/1471-2350-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kottgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, Smith AV, Arking DE, Astor BC, Boerwinkle E, Ehret GB, Ruczinski I, Scharpf RB, Chen YD, de Boer IH, Haritunians T, Lumley T, Sarnak M, Siscovick D, Benjamin EJ, Levy D, Upadhyay A, Aulchenko YS, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, Chasman DI, Pare G, Ridker PM, Kao WH, Witteman JC, Coresh J, Shlipak MG, Fox CS. Multiple loci associated with indices of renal function and chronic kidney disease. Nature Genetics. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bostrom MA, Lu L, Chou J, Hicks PJ, Xu J, Langefeld CD, Bowden DW, Freedman BI. Candidate genes for non-diabetic ESRD in African Americans: a genome-wide association study using pooled DNA. Hum Genet. 2010;128:195–204. doi: 10.1007/s00439-010-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kottgen A. Genome-wide association studies in nephrology research. American journal of kidney diseases: the official journal of the National Kidney Foundation. 2010;56:743–758. doi: 10.1053/j.ajkd.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Kottgen A, Hwang SJ, Larson MG, Van Eyk JE, Fu Q, Benjamin EJ, Dehghan A, Glazer NL, Kao WH, Harris TB, Gudnason V, Shlipak MG, Yang Q, Coresh J, Levy D, Fox CS. Uromodulin levels associate with a common UMOD variant and risk for incident CKD. Journal of the American Society of Nephrology. 2010;21:337–344. doi: 10.1681/ASN.2009070725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wuttke M, Kottgen A. Insights into kidney diseases from genome-wide association studies. Nat Rev Nephrol. 2016;12:549–562. doi: 10.1038/nrneph.2016.107. [DOI] [PubMed] [Google Scholar]
- 9.Gigante B, Rubattu S, Stanzione R, Lombardi A, Baldi A, Baldi F, Volpe M. Contribution of genetic factors to renal lesions in the stroke-prone spontaneously hypertensive rat. Hypertension. 2003;42:702–706. doi: 10.1161/01.HYP.0000084635.01667.8A. [DOI] [PubMed] [Google Scholar]
- 10.Churchill PC, Churchill MC, Griffin KA, Picken M, Webb RC, Kurtz TW, Bidani AK. Increased genetic susceptibility to renal damage in the stroke-prone spontaneously hypertensive rat. Kidney Int. 2002;61:1794–1800. doi: 10.1046/j.1523-1755.2002.00321.x. [DOI] [PubMed] [Google Scholar]
- 11.Braun MC, Herring SM, Gokul N, Monita M, Bell R, Hicks MJ, Wenderfer SE, Doris PA. Hypertensive Renal Disease: Susceptibility And Resistance In Inbred Hypertensive Rat Lines. J Hypertension. 2013;31:2050–2059. doi: 10.1097/HJH.0b013e328362f9a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun MC, Herring SM, Gokul N, Monita M, Bell R, Zhu Y, Gonzalez-Garay ML, Wenderfer SE, Doris PA. Hypertensive renal injury is associated with gene variation affecting immune signaling. Circ Cardiovasc Genet. 2014;7:903–910. doi: 10.1161/CIRCGENETICS.114.000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell R, Herring SM, Gokul N, Monita M, Grove ML, Boerwinkle E, Doris PA. High-resolution identity by descent mapping uncovers the genetic basis for blood pressure differences between spontaneously hypertensive rat lines. Circ Cardiovasc Genet. 2011;4:223–231. doi: 10.1161/CIRCGENETICS.110.958934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamoto K, Aoki K. Development of a strain of spontaneously hypertensive rats. Jpn Circ J. 1963;27:282–293. doi: 10.1253/jcj.27.282. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto K, Yamori Y, Nagaoka A. Establishment of the Stroke-prone Spontaneously Hypertensive Rat (SHR) Circ Res. 1974;14:I143–I153. [Google Scholar]
- 16.Inomata H, Watanabe T, Iizuka Y, Liang YQ, Mashimo T, Nabika T, Ikeda K, Yanai K, Gotoda T, Yamori Y, Isobe M, Kato N. Identification of quantitative trait loci for cardiac hypertrophy in two different strains of the spontaneously hypertensive rat. Hypertens Res. 2005;28:273–281. doi: 10.1291/hypres.28.273. [DOI] [PubMed] [Google Scholar]
- 17.Nagaoka A, Iwatsuka H, Suzuoki Z, Okamoto K. Genetic predisposition to stroke in spontaneously hypertensive rats. Am J Physiol. 1976;230:1354–1359. doi: 10.1152/ajplegacy.1976.230.5.1354. [DOI] [PubMed] [Google Scholar]
- 18.Herring SM, Gokul N, Monita M, Bell R, Boerwinkle E, Wenderfer SE, Braun MC, Doris PA. Immunoglobulin locus associates with serum IgG levels and albuminuria. J Am Soc Nephrol. 2011;22:881–889. doi: 10.1681/ASN.2010111148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Garay ML, Cranford SM, Braun MC, Doris PA. Diversity in the preimmune immunoglobulin repertoire of SHR lines susceptible and resistant to end-organ injury. Genes Immun. 2014;15:528–533. doi: 10.1038/gene.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagrange D, Fournie GJ. Generation of congenic and consomic rat strains. Methods Mol Biol. 2010;597:243–266. doi: 10.1007/978-1-60327-389-3_17. [DOI] [PubMed] [Google Scholar]
- 21.Markel P, Shu P, Ebeling C, Carlson GA, Nagle DL, Smutko JS, Moore KJ. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat Genet. 1997;17:280–284. doi: 10.1038/ng1197-280. [DOI] [PubMed] [Google Scholar]
- 22.Chan CT, Sobey CG, Lieu M, Ferens D, Kett MM, Diep H, Kim HA, Krishnan SM, Lewis CV, Salimova E, Tipping P, Vinh A, Samuel CS, Peter K, Guzik TJ, Kyaw TS, Toh BH, Bobik A, Drummond GR. Obligatory Role for B Cells in the Development of Angiotensin II-Dependent Hypertension. Hypertension. 2015;66:1023–1033. doi: 10.1161/HYPERTENSIONAHA.115.05779. [DOI] [PubMed] [Google Scholar]
- 23.Alderson HV, Ritchie JP, Pagano S, Middleton RJ, Pruijm M, Vuilleumier N, Kalra PA. The Associations of Blood Kidney Injury Molecule-1 and Neutrophil Gelatinase-Associated Lipocalin with Progression from CKD to ESRD. Clin J Am Soc Nephrol. 2016;11:2141–2149. doi: 10.2215/CJN.02670316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waikar SS, Sabbisetti V, Arnlov J, Carlsson AC, Coresh J, Feldman HI, Foster MC, Fufaa GD, Helmersson-Karlqvist J, Hsu CY, Kimmel PL, Larsson A, Liu Y, Lind L, Liu KD, Mifflin TE, Nelson RG, Riserus U, Vasan RS, Xie D, Zhang X, Bonventre JV Chronic Kidney Disease Biomarkers Consortium I. Relationship of proximal tubular injury to chronic kidney disease as assessed by urinary kidney injury molecule-1 in five cohort studies. Nephrol Dial Transplant. 2016;31:1460–1470. doi: 10.1093/ndt/gfw203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto K, Yamori Y, Nosaka S, Ooshima A, Hazama F. Studies on hypertension in spontaneously hypertensive rats. Clin Sci Mol Med Suppl. 1973;45(Suppl 1):11s–14. doi: 10.1042/cs045011s. [DOI] [PubMed] [Google Scholar]
- 26.Doris PA. The genetics of hypertension: an assessment of progress in the spontaneously hypertensive rat. Physiol Genomics. 2017 doi: 10.1152/physiolgenomics.00065.2017. physiolgenomics 00065 02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge Y, Fan F, Didion SP, Roman RJ. Impaired myogenic response of the afferent arteriole contributes to the increased susceptibility to renal disease in Milan normotensive rats. Physiol Rep. 2017:5. doi: 10.14814/phy2.13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller B, Palygin O, Rufanova VA, Chong A, Lazar J, Jacob HJ, Mattson D, Roman RJ, Williams JM, Cowley AW, Jr, Geurts AM, Staruschenko A, Imig JD, Sorokin A. p66Shc regulates renal vascular tone in hypertension-induced nephropathy. J Clin Invest. 2016;126:2533–2546. doi: 10.1172/JCI75079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vavrinec P, Henning RH, Goris M, Landheer SW, Buikema H, van Dokkum RP. Renal myogenic constriction protects the kidney from age-related hypertensive renal damage in the Fawn-Hooded rat. J Hypertens. 2013;31:1637–1645. doi: 10.1097/HJH.0b013e328361d506. [DOI] [PubMed] [Google Scholar]
- 30.Wright KD, Staruschenko A, Sorokin A. IR- Role of adaptor protein p66Shc in renal pathologies. Am J Physiol Renal Physiol. 2017 doi: 10.1152/ajprenal.00414.2017. ajprenal 00414 02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daeron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 32.Getahun A, Cambier JC. Of ITIMs, ITAMs, and ITAMis: revisiting immunoglobulin Fc receptor signaling. Immunol Rev. 2015;268:66–73. doi: 10.1111/imr.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemay S, Davidson D, Latour S, Veillette A. Dok-3, a novel adapter molecule involved in the negative regulation of immunoreceptor signaling. Mol Cell Biol. 2000;20:2743–2754. doi: 10.1128/mcb.20.8.2743-2754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J, Willsey AJ, Joy JB, Scott JK, Graves TA, Wilson RK, Holt RA, Eichler EE, Breden F. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet. 2013;92:530–546. doi: 10.1016/j.ajhg.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herzog S, Jumaa H. Self-recognition and clonal selection: autoreactivity drives the generation of B cells. Curr Opin Immunol. 2012;24:166–172. doi: 10.1016/j.coi.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Izzard AS, Graham D, Burnham MP, Heerkens EH, Dominiczak AF, Heagerty AM. Myogenic and structural properties of cerebral arteries from the stroke-prone spontaneously hypertensive rat. Am J Physiol Heart Circ Physiol. 2003;285:H1489–1494. doi: 10.1152/ajpheart.00352.2003. [DOI] [PubMed] [Google Scholar]
- 37.Watson CT, Glanville J, Marasco WA. The Individual and Population Genetics of Antibody Immunity. Trends Immunol. 2017;38:459–470. doi: 10.1016/j.it.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson CT, Matsen FAt, Jackson KJL, Bashir A, Smith ML, Glanville J, Breden F, Kleinstein SH, Collins AM, Busse CE. Comment on “A Database of Human Immune Receptor Alleles Recovered from Population Sequencing Data”. J Immunol. 2017;198:3371–3373. doi: 10.4049/jimmunol.1700306. [DOI] [PubMed] [Google Scholar]
- 39.O’Seaghdha CM, Fox CS. Genome-wide association studies of chronic kidney disease: what have we learned? Nat Rev Nephrol. 2011;8:89–99. doi: 10.1038/nrneph.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cantaert T, Schickel JN, Bannock JM, Ng YS, Massad C, Delmotte FR, Yamakawa N, Glauzy S, Chamberlain N, Kinnunen T, Menard L, Lavoie A, Walter JE, Notarangelo LD, Bruneau J, Al-Herz W, Kilic SS, Ochs HD, Cunningham-Rundles C, van der Burg M, Kuijpers TW, Kracker S, Kaneko H, Sekinaka Y, Nonoyama S, Durandy A, Meffre E. Decreased somatic hypermutation induces an impaired peripheral B cell tolerance checkpoint. J Clin Invest. 2016;126:4289–4302. doi: 10.1172/JCI84645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho JH, Feldman M. Heterogeneity of autoimmune diseases: pathophysiologic insights from genetics and implications for new therapies. Nat Med. 2015;21:730–738. doi: 10.1038/nm.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Martin A, Adams BD, Lai M, Shepherd J, Salvador-Bernaldez M, Salvador JM, Lu J, Nemazee D, Xiao C. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nature immunology. 2016;17:433–440. doi: 10.1038/ni.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massaad MJ, Zhou J, Tsuchimoto D, Chou J, Jabara H, Janssen E, Glauzy S, Olson BG, Morbach H, Ohsumi TK, Schmitz K, Kyriacos M, Kane J, Torisu K, Nakabeppu Y, Notarangelo LD, Chouery E, Megarbane A, Kang PB, Al-Idrissi E, Aldhekri H, Meffre E, Mizui M, Tsokos GC, Manis JP, Al-Herz W, Wallace SS, Geha RS. Deficiency of base excision repair enzyme NEIL3 drives increased predisposition to autoimmunity. J Clin Invest. 2016;126:4219–4236. doi: 10.1172/JCI85647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schickel JN, Kuhny M, Baldo A, Bannock JM, Massad C, Wang H, Katz N, Oe T, Menard L, Soulas-Sprauel P, Strowig T, Flavell R, Meffre E. PTPN22 inhibition resets defective human central B cell tolerance. Sci Immunol. 2016;1:aaf7153. doi: 10.1126/sciimmunol.aaf7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.