Summary
Synaptic connections between hippocampal mossy fibers (MFs) and CA3 pyramidal neurons are essential for contextual memory encoding, but the molecular mechanisms regulating MF-CA3 synapses during memory formation and the exact nature of this regulation are poorly understood. Here we report that the activity-dependent transcription factor Npas4 selectively regulates the structure and strength of MF-CA3 synapses by restricting the number of their functional synaptic contacts, without affecting the other synaptic inputs onto CA3 pyramidal neurons. Using an activity-dependent reporter, we identified CA3 pyramidal cells that were activated by contextual learning, and found that MF inputs on these cells were selectively strengthened. Deletion of Npas4 prevented both contextual memory formation and this learning-induced synaptic modification. We further show that Npas4 regulates MF-CA3 synapses by controlling the expression of the polo-like kinase Plk2. Thus, Npas4 is a critical regulator of experience-dependent, structural and functional plasticity at MF-CA3 synapses during contextual memory formation.
Keywords: CA3, contextual memory, MF-CA3 synapses, Npas4, neuronal activity
eTOC Blurb
Weng et al. report that the transcription factor Npas4 selectively regulates the number of functional synaptic contacts between CA3 pyramidal neurons and mossy fibers, thereby allowing for learning-induced modification of MF-CA3 synapses during contextual memory formation.
Introduction
The hippocampus is critical for contextual memory formation, and specific synaptic connections within this region carry out the neuronal computations required for encoding, storing and retrieving contextual memories (Kesner and Rolls, 2015). Contextual information that flows from the entorhinal cortex (EC) into the hippocampus is processed through the trisynaptic loop formed by three interconnected but anatomically and functionally distinct sub-regions: dentate gyrus (DG), cornu ammonis 3 (CA3) and CA1. At the center of the trisynaptic loop, pyramidal neurons in CA3 receive three major excitatory synaptic inputs: mossy fiber (MF) inputs from DG granule cells, perforant path inputs directly from EC, and recurrent inputs from other CA3 pyramidal neurons. Computational models predict that the MF-CA3 connections are essential for encoding new contextual memory but are dispensable for contextual memory retrieval, which instead requires the perforant path (Treves and Rolls, 1992). Behavioral studies in which DG was lesioned or MF transmission was pharmacologically inhibited support these theoretical predictions (Jerman et al., 2006; Lassalle et al., 2000; Lee and Kesner, 2004a, b). However, the molecular mechanisms that regulate MF-CA3 synapses during memory formation and the exact nature of this regulation are currently unknown.
MF-CA3 synapses have several unique structural and functional properties: they are located on proximal dendrites of CA3 pyramidal neurons, have large presynaptic MF terminals (MFTs) with multiple release sites (Amaral and Dent, 1981) that make contact with large multi-headed postsynaptic spines (Chicurel and Harris, 1992) called complex spines or thorny excrescences (TEs), and display uniquely robust frequency facilitation (Salin et al., 1996). MF-CA3 synapses can act as “detonator” synapses, that is, under certain circumstances a single MF input can reliably cause action potential firing of CA3 pyramidal neurons (Henze et al., 2002; Urban et al., 2001; Vyleta et al., 2016). In addition, MF-CA3 synapses exhibit dramatic structural plasticity at both presynaptic MFTs and postsynaptic TEs (Wiera and Mozrzymas, 2015). Both structures are readily modified by learning, stress and aging (Galimberti et al., 2006; Gogolla et al., 2009; Magarinos et al., 1997; Maruo et al., 2016; Rekart et al., 2007; Sandi et al., 2003), and are particularly sensitive to changes in levels of activity within the hippocampus (Ben-Ari and Represa, 1990; Chierzi et al., 2012; Kim and Tsien, 2008; Lee et al., 2013; Represa and Ben-Ari, 1992a, b; Zhao et al., 2012b). Furthermore, these structural changes are correlated with functional changes, with larger morphology typically being associated with more numerous and stronger synaptic contacts between MFTs and TEs (Galimberti et al., 2006; Zhao et al., 2012a).
Long-term memory formation is believed to involve modifications in relevant neural circuits that likely occur as a result of enduring synaptic changes (Mayford et al., 2012). While it is well established that the contextual information entering CA3 through the MF pathway is critical for the encoding of contextual memory in CA3 (Lee and Kesner, 2004a, b; Rolls, 1996), and there is an abundance of evidence that MF-CA3 synapses are modified by experience (Galimberti et al., 2006; Gogolla et al., 2009; Magarinos et al., 1997; Maruo et al., 2016; Rekart et al., 2007; Sandi et al., 2003), precisely how MF-CA3 synapses are modified to contribute to contextual memory formation remains an open question. Although mechanistically diverse forms of synaptic plasticity have been identified at MF-CA3 synapses, most studies have focused on presynaptic plasticity, such as classical, NMDAR-independent long-term potentiation (LTP) (Nicoll and Schmitz, 2005). Intriguingly, MF-LTP appears to be dispensable for contextual memory formation (Ruediger et al., 2011), and it remains unclear what types of experience-dependent MF-CA3 synapse plasticity are involved in contextual learning. Moreover, molecular pathways responsible for the activity-dependent modulation of MF-CA3 synapses and the precise role of such modulation in contextual learning are poorly understood.
We have previously shown that the neuronal activity-dependent transcription factor Npas4 is required in CA3, but not in CA1, for hippocampus-dependent contextual memory formation (Ramamoorthi et al., 2011), based on the following evidence. After contextual learning, Npas4 is specifically induced in CA3, but not in CA1. Either global knockout or selective deletion of Npas4 in CA3 results in impaired contextual memory, and restoration of Npas4 in CA3 is sufficient to rescue the learning deficit in global knockout mice. In contrast, similar manipulations of Npas4 in CA1 had no effect on contextual memory. Here we report that Npas4 selectively regulates the structure and strength of MF-CA3 synapses. We also found that contextual learning selectively strengthens MF inputs on learning-activated CA3 pyramidal neurons. Deletion of Npas4 prevents this learning-induced plasticity at MF-CA3 synapses, which likely contributes to the impaired contextual memory formation. We further show that Npas4 acts through its transcriptional target polo-like kinase 2 (Plk2) to modulate MF-CA3 synapses. Expression of Plk2 in CA3 is sufficient to restore the MF-CA3 synapses and contextual memory formation in Npas4 global knockout mice. Thus, Npas4 emerges as a critical regulator of experience-dependent plasticity at MF-CA3 synapses during contextual memory formation.
Results
Excitatory Synapses on CA3 Pyramidal Neurons are Selectively Regulated by Npas4
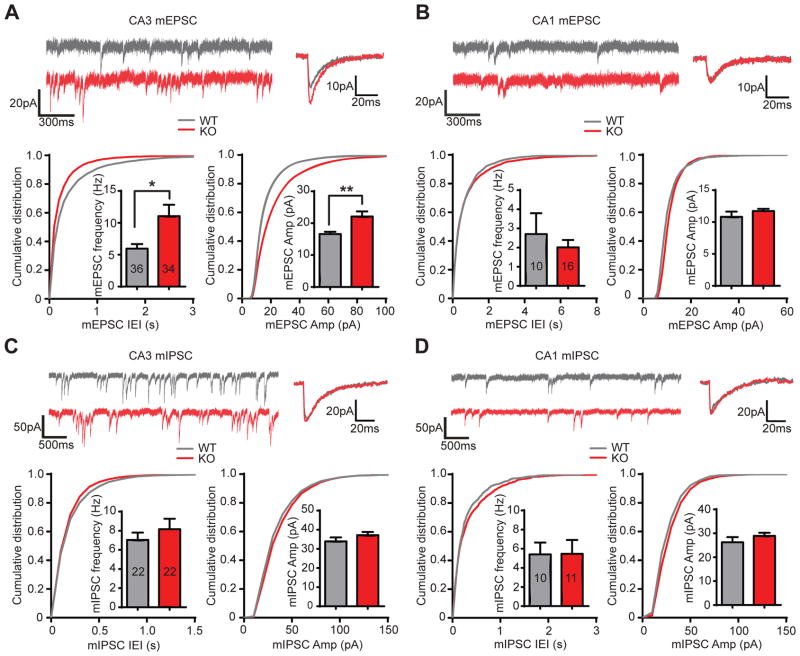
To better understand the role of Npas4 in contextual memory, we first examined miniature excitatory and inhibitory postsynaptic currents (mEPSCs and mIPSCs) by performing whole-cell patch recordings in CA3 and CA1 pyramidal neurons of acute hippocampal slices from Npas4 KO mice and their wild-type littermates. We used 8–10 week old mice, the same age at which we found that Npas4 is required in CA3 for contextual memory formation (Ramamoorthi et al., 2011). We observed a robust increase in mEPSC frequency and amplitude in CA3 pyramidal neurons of Npas4 KO mice compared to their wild-type littermates (Figure 1A). In contrast, mEPSCs were unchanged in CA1 pyramidal neurons (Figure 1B). In contrast, mIPSCs remained unchanged in Npas4 KO mice in both CA3 and CA1 pyramidal neurons (Figure 1C–D). Thus, Npas4 deletion is associated with a selective increase in excitatory miniature transmission on CA3 pyramidal neurons.
Figure 1. Excitatory miniature activity in CA3, but not CA1 pyramidal neurons is significantly increased in Npas4 KO mice, whereas inhibitory miniature activity is not affected.
(A) Sample traces, summary bar graphs, and cumulative distribution of mEPSCs recorded from hippocampal CA3 pyramidal neurons. Frequency (Hz): WT, 5.95 ± 0.71; KO, 11.05 ± 1.72; p = 0.043; Amplitude (pA): WT, 16.61 ± 0.70; KO, 22.12 ± 1.59; p = 0.002. WT: n = 36 cells, 6 animals; KO: n = 34 cells, 5 animals.
(B) Same as (A) but for CA1 pyramidal neurons. Frequency (Hz): WT, 2.70 ± 1.09; KO, 2.01 ± 0.38; p = 0.85. Amplitude (pA): WT, 10.82 ± 0.82; KO, 11.72 ± 0.35; p = 0.15. WT, n = 10 cells, 3 animals; KO, n = 16 cells, 3 animals.
(C) Sample traces, summary bar graphs, and cumulative distribution of mIPSC recorded from hippocampal CA3 neurons. Frequency (Hz): WT 7.04 ± 0.77; KO 8.16 ± 1.09; p = 0.37. Amplitude (pA): WT 33.91 ± 2.10; KO 37.14 ± 1.67; p = 0.14. WT: n = 22 cells, 3 animals; KO: n = 22 cells, 4 animals.
(D) Same as (C) but for CA1 pyramidal neurons. Frequency (Hz): WT 5.41 ± 1.23; KO 5.47 ± 1.46; p = 0.74. Amplitude (pA): WT 26.24 ± 2.13; KO 28.92 ± 1.32; p = 0.22. WT: n = 10 cells, 3 animals; KO: n = 11 cells, 3 animals.
Data are shown as mean ± SEM. *p < 0.05, **p < 0.01, Mann-Whitney U-test.
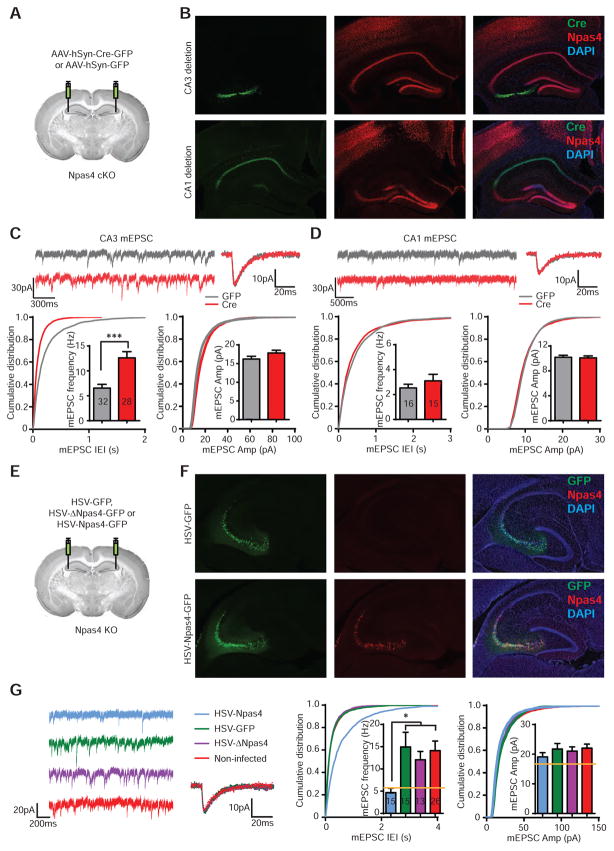
To rule out the possibility that the increase in excitatory drive on CA3 was a result of developmental compensation for global, germline loss of Npas4, we acutely removed Npas4 by stereotaxic injection of adeno-associated virus AAV-hSyn-Cre-GFP (AAV-Cre) into the CA3 or CA1 region of Npas4 conditional knockout (cKO; Npas4flx/flx) mice, with AAV-hSyn-GFP (AAV-GFP) as a control. Miniature activity was measured from CA3 and CA1 pyramidal neurons in acute hippocampal slices prepared 3 days after viral injection, when we had confirmed that the Npas4 gene had been deleted (Figure 2A–B). As in the germline knockout, mEPSC frequency in CA3 pyramidal neurons was increased more than 50% by Npas4 deletion, compared to the GFP control condition (Figure 2C). In contrast, mEPSC amplitude was not affected in cKO mice, suggesting that Npas4 does not directly regulate the mEPSC amplitude. mEPSCs in CA1 pyramidal neurons (Figure 2D) and mIPSCs in CA3 pyramidal neurons (Figure S1) were again unchanged by Npas4 deletion. Taken together, our findings indicate that Npas4 selectively modulates excitatory synaptic drive onto CA3 pyramidal neurons.
Figure 2. mEPSC frequency in CA3 pyramidal neurons is significantly altered by acute manipulations of Npas4 expression.
(A) AAV1-hSyn-Cre-eGFP (Cre) or AAV1-hSyn-eGFP (GFP) was stereotaxically injected into the CA3 or CA1 region of Npas4flx/flx mice at 8–10 weeks of age. Whole cell voltage clamp recordings were performed 3 days after viral injection.
(B) Representative images showing complete Npas4 gene deletion from the CA3 (top) or CA1 (bottom) region in Npas4flx/flx mice 3 days after injection of Cre. Npas4 expression (red) following kainic acid-induced seizure was completely absent in regions expressing Cre (green). Scale bar corresponds to 300 μm.
(C) Sample traces, summary bar graphs, and cumulative distribution of mEPSCs recorded from hippocampal CA3 pyramidal neurons from Npas4flx/flx mice injected with Cre or GFP. Frequency (Hz): GFP 6.55 ± 0.75; Cre 12.61 ± 1.26; p = 0.0002. Amplitude (pA): GFP 16.22 ±0.73; Cre 17.83 ± 0.74; p = 0.087. GFP: n = 32 cells, 6 animals; Cre: n = 28 cells, 6 animals.
(D) Same as (C) but for CA1 pyramidal neurons. Frequency (Hz): GFP 2.53 ± 0.30; Cre 3.10 ± 0.52; p = 0.68. Amplitude (pA): GFP 10.19 ± 0.32; Cre 10.09 ± 0.32; p = 0.82. GFP: n = 16 cells, 4 animals; Cre: n = 15 cells, 3 animals.
(E) HSV-eGFP, HSV-Npas4-eGFP or HSV-ΔNpas4-eGFP were stereotaxically injected into the CA3 region of Npas4 KO mice at 8–10 weeks of age. Whole cell voltage clamp recordings were performed 3 days after viral injection.
(F) Representative images showing acute re-expression of Npas4 in CA3 of Npas4 KO mice using HSV-Npas4-eGFP (bottom panel). HSV-eGFP was used as a control (top panel). Scale bar corresponds to 300 μm.
(G) Sample traces, cumulative distribution and summary bar graphs of mEPSCs recorded from hippocampal CA3 pyramidal cells of Npas4 KO mice after expression of Npas4, GFP or ΔNpas4, or from uninfected cells. Horizontal lines indicate the levels in wild-type animals (from Figure 1A). Frequency (Hz): Npas4 4.68 ± 0.98; GFP 14.94 ± 3.30; ΔNpas4 12.05 ±1.88; uninfected 14.10 ± 2.16; F (3, 65) = 3.739; p = 0.015. Amplitude (pA): Npas4 19.09 ± 1.43; GFP 21.75 ± 1.84; ΔNpas4 21.04 ± 1.43; uninfected 22.04 ± 1.36; F (3, 65) = 0.7318; p = 0.54. Npas4: n = 15 cells, 3 animals; GFP: n =15 cells, 3 animals; ΔNpas4: n = 13 cells, 3 animals; uninfected: n = 26 cells, 9 animals.
Data are shown as mean ± SEM. (C) & (D): ***p < 0.001, Mann-Whitney U-test. (G): *p < 0.05, one-way ANOVA, Dunnett’s post-hoc test. See also Figure S1.
Abnormally elevated excitatory drive onto CA3 could be at least partly responsible for the impaired contextual memory we observed in Npas4-deficient animals (Ramamoorthi et al., 2011). If so, re-expression of Npas4 in CA3 of Npas4 KO mice, a manipulation that rescued the deficit in contextual memory formation (Ramamoorthi et al., 2011), should also restore CA3 excitatory synaptic transmission to a normal level. Indeed, 3 days after a herpes simplex virus (HSV) encoding Npas4 was stereotaxically injected into CA3 of Npas4 KO mice, mEPSC frequency in infected CA3 pyramidal neurons was reduced almost to the level seen in wild-type mice (Figure 2E–G). Expression of GFP or a truncated, transcriptionally inactive form of Npas4 (ΔNpas4), neither of which rescued the learning and memory deficit (Ramamoorthi et al., 2011), failed to restore mEPSC activity (Figure 2G). mEPSC amplitudes were similar in all experiments. These results suggest that the increase of excitatory inputs onto CA3 pyramidal neurons by Npas4 deletion interferes contextual memory formation.
Npas4 Selectively Regulates the Mossy Fiber Inputs on CA3 Pyramidal Neurons
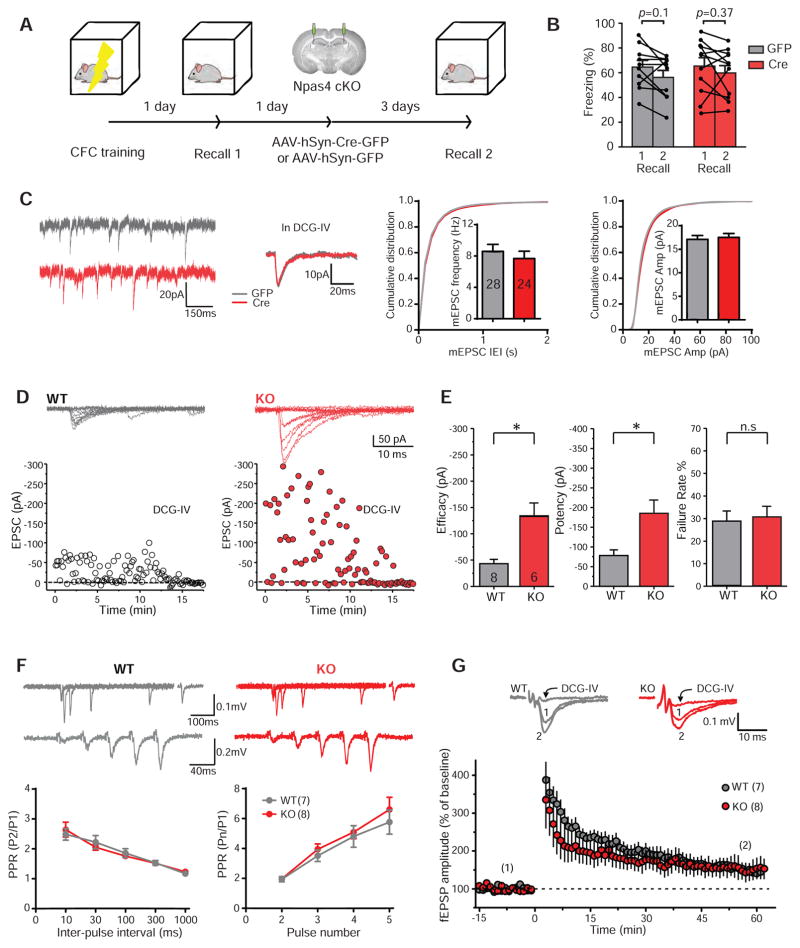
We considered the possibility that Npas4 might selectively regulate a specific subset of the three types of synaptic inputs received by CA3 pyramidal neurons, i.e. MF, recurrent or perforant path inputs. We have previously shown that deletion of Npas4 in CA3 does not affect short-term memory formation (Ramamoorthi et al., 2011), a process likely depending on the recurrent inputs from other CA3 neurons (Kesner, 2007). Contextual memory encoding and retrieval depend on the MF and perforant path inputs, respectively (Jerman et al., 2006; Lassalle et al., 2000; Lee and Kesner, 2004a). Since deletion of Npas4 in CA3 before contextual fear conditioning (CFC) training impairs formation of long-term contextual memory (Ramamoorthi et al., 2011), it suggests that Npas4 could be involved in either encoding or retrieval of contextual memory, or both. We next investigated whether Npas4 could be selectively involved in memory encoding or retrieval. Npas4 cKO mice were subjected to a CFC training and their contextual memory examined the following day by re-exposing them to the same context (Recall 1, Figure 3A). One day later, the animals were injected in CA3 with AAV-Cre to delete Npas4, or with AAV-GFP as control. Three days after injection, their contextual memory was examined again (Recall 2). We found that levels of freezing behavior were similar at Recall 1 and Recall 2 for all mice, regardless of whether Npas4 was deleted or not (Figure 3A–B). This result, combined with our previous findings (Ramamoorthi et al., 2011), indicates that Npas4 is required in CA3 for memory encoding but not retrieval, consistent with the idea that Npas4 modulates MF-CA3 synapses and not the perforant path.
Figure 3. Deletion of Npas4 in CA3 selectively alters the excitatory inputs received by CA3 pyramidal neurons from mossy fibers, while short-term and long-term plasticity remains unchanged.
(A) – (B) Npas4 is not required in CA3 for retrieval of contextual memory.
(A) Npas4 cKO mice underwent CFC training and their contextual memories were recalled the following day (Recall 1). Npas4 was deleted 1 day after Recall 1 and contextual memories were re-examined 3 days after surgery (Recall 2).
(B) All animals, regardless of the presence or absence of Npas4, displayed similar freezing levels during both recalls. GFP freezing level (%): Recall 1, 64.58 ± 5.22%, Recall 2, 56.25 ± 5.35%; p = 0.1. Cre freezing level (%): Recall 1, 65.34 ± 6.85, Recall 2, 59.85 ± 5.94; p = 0.27. Wilcoxon matched-pairs signed rank tests were used to compare Recall 1 and Recall 2 for each treatment.
(C) AAV1-hSyn-eGFP (GFP) or AAV1-hSyn-Cre-eGFP (Cre) was stereotaxically injected into the CA3 region of Npas4flx/flx mice at 8–10 weeks of age. Whole cell voltage clamp recordings were performed 3 days after viral injection. In the presence of 1 μM DCG-IV (to block MF transmission), Npas4 deletion had no effect on the frequency or amplitude of mEPSCs recorded from CA3 pyramidal neurons. Sample traces, summary bar graphs and cumulative distributions are shown. Frequency (Hz): GFP 8.60 ± 0.89; Cre 7.68 ± 0.93; p = 0.46. Amplitude (pA): GFP 17.10 ± 0.82; Cre 17.53 ± 0.83; p = 0.65. GFP: n = 28 cells, 7 animals; Cre: n = 24 cells, 7 animals.
(D) Representative experiments of MF-CA3 EPSCs elicited by minimal stimulation in WT and Npas4 KO mice. Superimposed individual traces (top) and time-course plots (bottom). Adding DCG-IV (1μM) at the end of the experiments blocked synaptic transmission (traces not shown).
(E) Summary data showing efficacy (pA): WT: 43.3 ± 8.9; KO: 132.8 ± 25.7, p = 0.0027; potency (pA): WT: 78 ± 13.4, KO: 184 ± 34.1; p = 0.003, and failure rate (%): WT: 28.7 ± 4.6, KO: 30.5 ± 4.8, p = 0.68. WT: 8 slices, 6 mice; KO: 6 slices, 4 mice.
(F) Short-term plasticity was not altered in Npas4 KO mice. Extracellular MF synaptic responses (fEPSPs) were recorded in acute hippocampal slices from Npas4 KO mice and their WT littermates at 8–10 weeks of age. Sample traces and summary plots are shown. Left, paired-pulse ratio was assessed by delivering paired-pulses at 10, 30, 100, 300 and 1000 ms inter-stimulus intervals, and P2/P1 are plotted. Right, frequency facilitation was probed by 5 consecutive pulses at 40 ms intervals, and Pn/P1 are plotted. WT: n = 7 slices, 3 animals; KO: n = 8 slices, 3 animals.
(G) LTP at MF-CA3 synapses was normal in Npas4 KO mice. LTP was induced with a train of 125 stimuli at 25 Hz. DCG-IV (1 μM) was added at the end of each experiment. Representative traces are shown on the top and summary plots on the bottom. LTP (% of the baseline for the last 10 minutes): WT: 152.16 ± 2.52; n = 7 slices, 3 animals; KO: 152.74 ± 2.01; n = 8 slices, 3 animals. p = 0.55.
All summary data are shown as mean ± SEM. (C) to (G): **p < 0.01, Mann-Whitney U-test. n.s: non-significant. See also Figure S2 and S3.
Consistent with the idea that Npas4 selectively modulates MF-CA3 synapses, we found that a significantly large fraction of mEPSC events with short rise times (< 2.5ms) in the Npas4 KO CA3 neurons (Figure S2), as expected for events originated close to the cell soma, like those generated by MF-CA3 synapses (Claiborne et al., 1993; Jonas et al., 1993). Moreover, the elevated mEPSC activity observed in Npas4-deficient neurons (Figure 2C) was abolished in the presence of the metabotropic glutamate receptor subtypes 2/3 (mGluR2/3)-specific agonist DCG-IV (1μM) (Figure 3C), a manipulation known to specifically suppress glutamate release from MFs but not recurrent or perforant path inputs (Kamiya and Ozawa, 1999; Kamiya et al., 1996). Furthermore, using minimal stimulation of MF inputs (Figure S3) (Hofmann et al., 2008; Jonas et al., 1993), we found that MF-CA3 synaptic efficacy was increased in Npas4 KO mice compared to their wild-type littermates, and that this enhancement was associated with increased potency but no change in failure rate (Figure 3D–E). In addition, paired-pulse ratio (PPR) and burst-induced facilitation of evoked MF-CA3 synaptic responses, two forms of short-term synaptic plasticity that are inversely correlated with the probability of neurotransmitter release (Pr), were undistinguishable in Npas4 KO and wild-type littermates (Figure 3F). Classical presynaptic LTP at MF-CA3 synapses was also normal in Npas4 KO mice (Figure 3G). Taken together, our results therefore suggest that the selective increase in MF-CA3 transmission observed in Npas4-deficient neurons likely results from an increase in the number of functional synaptic contacts established by each MF. In contrast, Npas4 deletion does not affect MF-CA3 basal Pr or classical forms of presynaptic plasticity.
Npas4 Selectively Restricts the Number of MF-CA3 Synaptic Contacts
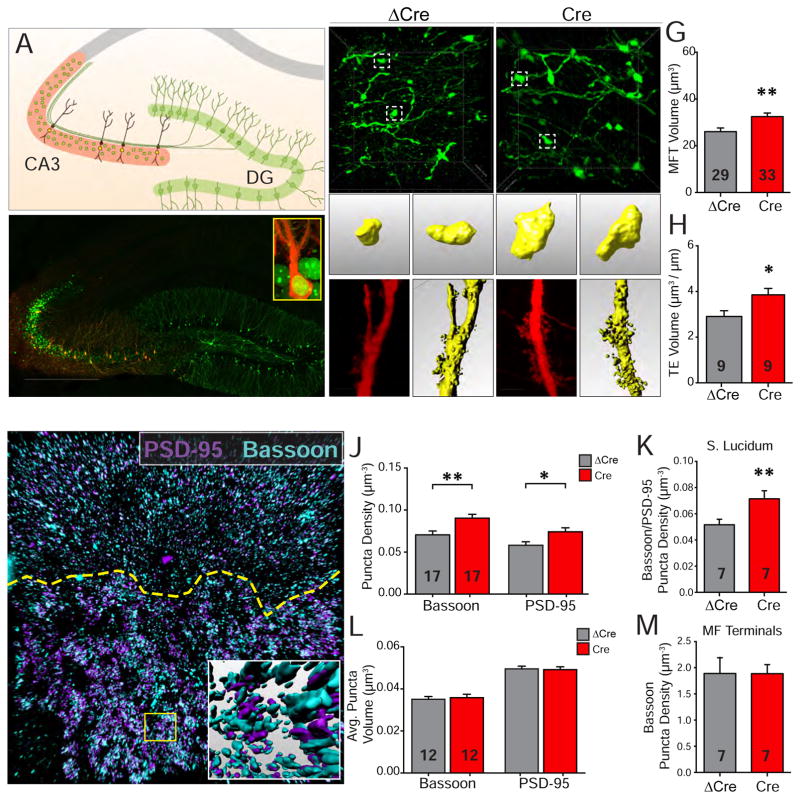
The number of conventional glutamatergic synapses on pyramidal neurons is correlated with the density of dendritic spines (Alvarez and Sabatini, 2007; Yuste and Bonhoeffer, 2001). To test whether changes in the number of MF-CA3 synaptic contacts is correlated with morphological changes, we designed a strategy to sparsely and brightly label MF terminals and TEs in different colors, using DNA recombinase Flp∘-dependent reporters expressing both cytosolic and membrane-bound farnesylated GFP (AAV-hSyn-fDIO-GFP-P2A-farnGFP) or tdTomato (AAV-hSyn-fDIO-tdTomato) (Figure 4A; see Methods for details). We performed bilateral double injections on Npas4 cKO mice. One injection, targeting dorsal CA3, delivered AAV-hSyn-Cre-GFP (to delete Npas4) or AAV-hSyn-ΔCre-GFP (containing a truncated and inactive version of Cre as the control), low-titer AAV-EF1α-Flp∘ (for sparse labeling) and AAV-hSyn-fDIO-tdTomato. This resulted in nuclear GFP labeling of the majority of dorsal CA3 neurons, by either Cre-GFP or ΔCre-GFP, and a sparse population of CA3 neurons labeled red by tdTomato (Figure 4A–B). A second injection was targeted to dorsal DG, delivering low-titer AAV-EF1α-Flp∘ and AAV-hSyn-fDIO-GFP-P2A-farnGFP, to label MFTs in green (Figure 4A–B).
Figure 4. Npas4 modifies the number and structure of MF-CA3 synapses.
(A) Schematic of labeling strategy with CA3 pyramidal cells expressing nuclear-localized Cre-GFP or ΔCre-GFP and cytosolic tdTomato to label morphology. Mossy fiber projections from DG are labeled with both cytosolic and membrane-bound farnesylated GFP.
(B) Representative low-resolution image of two color labeling of CA3 pyramidal cells (red, tdtomato) and DG cells and their MF projections (green, GFP). Inset, zoomed-in representation of a CA3 pyramidal cell with nuclear expression of either Cre or ΔCre (green). Scale bar = 500 μm.
(C–D) Top, representative images of MF terminals expressing GFP in stratum lucidum of CA3 in ΔCre (C) or Cre (D) conditions. Bottom, examples of 3D surface reconstruction of large single MF terminals. Scale bar = 10 μm and 1 μm.
(E–F) Left, representative images of TEs on a proximal CA3 dendrite segment and (right) partial 3D surface reconstruction of a dendritic segment for ΔCre (E) and Cre (F) conditions. Scale bar = 5 μm.
(G) MF terminals that synapse onto Npas4 deleted cells have significantly increased volumes. Volume: ΔCre 26.06 ± 1.56, n = 29 MF terminals, 3 animals; Cre 32.48 ± 1.53, n = 33 MF terminals, 3 animals; p = 0.0065.
(H) Volume of TE structures is significantly increased in Npas4 deleted cells. Volume: ΔCre 2.91 ± 0.25, n = 9, 3 animals; Cre 3.85 ± 0.28, n = 9, 3 animals; p = 0.04.
(I) Representative image of MAP-processed sections with labeling of synaptic markers PSD-95 (magenta) and Bassoon (cyan). Inset shows 3D surface reconstruction of synaptic marker puncta in the region marked with a yellow box. Dashed line indicates the boundary between stratum lucidum and radiatum in CA3. Scale bars = 10 μm and 1 μm.
(J) Density of pre- and post-synaptic marker puncta is significantly increased in stratum lucidum of Npas4 deleted cells. Synaptic density: Bassoon, ΔCre 0.07 ± 0.004, n = 17 regions, 6 animals; Bassoon, Cre 0.09 ± 0.004, n = 17 regions, 6 animals; p = 0.008; PSD-95, ΔCre 0.06 ± 0.004, n = 17 regions, 6 animals; PSD-95, Cre 0.07 ± 0.005, n = 17 regions, 6 animals; p = 0.02.
(K) Density of co-localized Bassoon and PSD-95 puncta is significantly increased in lucidum of Npas4-deleted mice. Synaptic density: Bassoon, ΔCre 0.05 ± 0.004, n = 17 regions, 6 animals; Cre 0.07 ± 0.006, n = 17 regions, 6 animals; p = 0.007.
(L) Average synaptic puncta volume does not change when Npas4 is deleted. Avg. Volume: Bassoon, ΔCre 0.05 ± 0.004, n = 12 regions, 6 animals; Cre 0.05 ±0.001, n = 12 regions, 6 animals; p = 0.57; PSD-95, ΔCre 0.04 ± 0.003, n = 12 regions, 6 animals; PSD-95, Cre 0.04 ± 0.002, n = 12 regions, 6 animals; p = 0.42.
(M) Density of Bassoon puncta localized within MF terminals does not change in Npas4-deleted cells. Synaptic density: Bassoon, ΔCre 1.89 ± 0.31, n = 12 regions, 6 animals; Cre 1.89 ± 0.17, n = 12 regions, 6 animals; p = 0.98.
Summary data are shown as mean ± SEM. *p < 0.05, **p < 0.01, Mann-Whitney U-test.
To analyze the 3D structures of MFTs and TEs we adopted the SeeDB2 procedure to immuno-stain and clarify brain slices for confocal imaging (Ke et al., 2016). SeeDB2 appears to be superior to other tissue clearing procedures in preserving synaptic ultrastructure such as dendritic spines (K.Z. & W.X., unpublished observation). The 3D structures of MF terminals and TEs were reconstructed and their total volumes quantified (Figure 4C–F). We found that deletion of Npas4 in CA3 results in significantly larger MFT and TE structures (Figure 4G–H), supporting our hypothesis that loss of Npas4 increases the number of synaptic contacts between MFTs and TEs.
To unequivocally determine that Npas4 deletion in CA3 increases the total number of MF-CA3 synaptic contacts, as opposed to increasing the proportion of functional synapses, we examined the density of synapses within the stratum lucidum, where MF-CA3 synapses reside. We adopted a recently developed tissue processing technique called Magnified Analysis of Proteome (MAP) (Ku et al., 2016). By combining tissue expansion, clarification and immunostaining, MAP allows 3D analysis of the subcellular localization of individual synaptic components using a conventional confocal microscope (Figure 4I). We first measured densities within randomly selected cubic regions of the stratum lucidum. Npas4 deletion increased the densities of the pre-synaptic active zone marker Bassoon, the post-synaptic marker PSD-95, and the density of synapses defined by overlapped Bassoon and PSD-95 puncta, whereas the average sizes of the Bassoon and PSD-95 puncta remained unchanged (Figure 4J–L). We also found that the density of synapses within individual MF terminals remained unchanged regardless of the presence of Npas4 (Figure 4M), suggesting that the enlargement of synaptic structure in Npas4-deficient CA3 neurons is associated with an increase in synaptic contacts per MF terminal/TE, and this structural change likely underlies the increase in MF-CA3 synaptic transmission observed in the absence of Npas4.
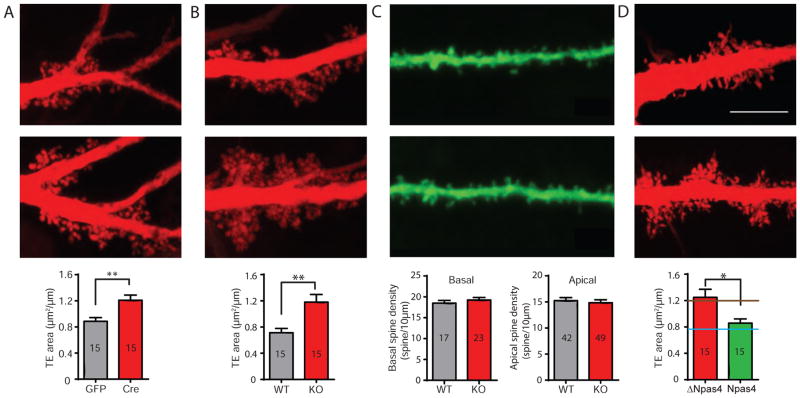
A more conventional and higher throughput 2D morphological analysis of TE size using the “stick-and-stain” method (Wallace and Bear, 2004) also showed a significant increase in TE size in both Npas4 KOs and cKOs (Figure 5A–B). In contrast, dendritic spine densities in apical and basal distal dendrites (i.e. more than 30μm from the soma), where recurrent and perforant path inputs are found, were indistinguishable in Npas4 KO and wild-type (Figure 5C), consistent with our electrophysiological findings indicating unchanged inputs from these two pathways (Figure 3E). Remarkably, expressing Npas4 in CA3 of Npas4 KO mice, which rescues the contextual learning deficit (Ramamoorthi et al., 2011) and restores changes in CA3 mEPSCs (Figure 2E–G), reduced TE sizes to wild-type levels, while expressing the truncated, transcriptionally inactive form ΔNpas4 had no such effect (Figure 5D). Altogether, our data indicate that Npas4 selectively restricts the size of MFTs and TEs and the number of functional contacts between them.
Figure 5. Npas4 bi-directionally regulates thorny excrescence (TE) size.
(A) Representative images and summary bar graphs showing that total area of CA3 TE is significantly increased when Npas4 is deleted by AAV-Cre in Npas4flx/flx mice, compared to littermate controls injected with AAV-GFP. TE area (μm2/μm): GFP 0.89 ± 0.06, n =15, 3 animals; Cre 1.21 ± 0.08, n = 15, 3 animals; p = 0.0014.
(B) Representative images and summary bar graphs showing significantly larger total area of CA3 TE in Npas4 KO mice than their WT littermates. TE area (μm2/μm): WT 0.71 ± 0.07, n =15 neurons, 3 animals; KO 1.18 ± 0.12, n = 15 neurons, 3 animals; p = 0.0032.
(C) Representative images and summary bar graphs showing comparable spine density on distal dendrites of CA3 pyramidal neurons in Npas4 KO mice and their WT littermates. # basal spines/10 μm: WT 18.49 ± 0.64; KO 19.25 ± 0.59; p = 0.43. # apical spines/10 μm: WT 15.25 ± 0.58; KO 14.84 ± 0.57; p = 0.64. WT basal: n = 17 dendrites, 2 animals; KO basal: n = 23 dendrites, 3 animals; WT apical: n = 42 dendrites, 7 animals; KO apical: n = 49 dendrites, 8 animals.
(D) Representative images and summary bar graphs showing Npas4 expression in Npas4 KO mice reduced CA3 TE size to the wild type level. TE area (μm2/μm): ΔNpas4 1.25 ± 0.12; Npas4 0.86 ± 0.06; n = 15 neurons, 3 animals for each condition; p = 0.013. The blue and brown lines indicate the TE area values of Npas4 WT and KO mice, respectively.
Scale bars correspond to 5 μm. Summary data are shown as mean ± SEM. *p < 0.05, **p < 0.01, Mann-Whitney U-test.
Contextual Learning Selectively Strengthens MF Inputs on Learning Activated CA3 Pyramidal Neurons
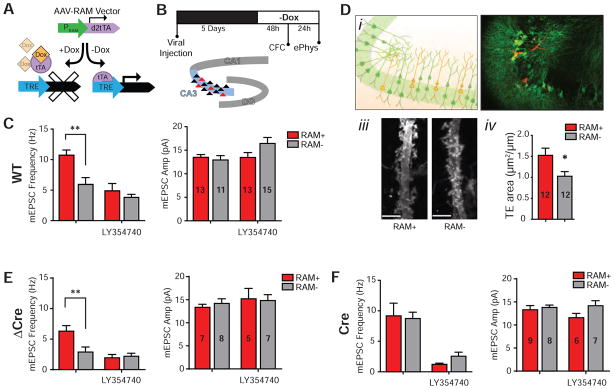
Given that Npas4 is required in CA3 for contextual memory formation (Ramamoorthi et al., 2011), and our new findings indicating that Npas4 selectively regulates MF-CA3 synapses, we hypothesized that these synapses are modulated during contextual memory formation. To identify synaptic changes directly associated with a contextual learning experience, we took advantage of our recently developed Robust Activity Marking (RAM) system (Sorensen et al., 2016), which allows us to tag neurons selectively activated by CFC. The RAM system is composed of a synthetic promoter that is strongly activated by neuronal activity and a downstream reporter gene to allow subsequent investigation and manipulation (Figure 6A). A modified doxycycline (Dox)-dependent Tet-Off system provides the temporal control to label neurons that are activated by a specific experience, which occurs in the absence of Dox. We have recently shown that the RAM system selectively labels neuron ensembles activated by contextual learning in various brain regions, including CA3 (Sorensen et al., 2016).
Figure 6. Contextual learning selectively strengthens MF inputs and Npas4 deletion prevents this experience-dependent modulation.
(A) The design of the RAM system. The RAM promoter PRAM drives the expression of a destabilized version of the tetracycline transactivator (d2tTA), which subsequently drives the expression of the reporter gene under the control of a tTA-responsie element (TRE) in the promoter. The binding of d2tTA to TRE, and as a result the expression of the reporter gene, can be blocked by the antibiotic doxycycline (Dox).
(B) Experimental timeline. Animals were taken off Dox diet 5 days after viral injection and 48h before CFC training and sacrificed 24h later for electrophysiological recordings.
(C) Wild type C57BL/6 mice were co-injected with AAV-RAM-mKate2 and AAV-EF1α-GFP (as infection control). RAM+ CA3 pyramidal neurons have significant higher mEPSC frequency than their neighboring RAM− neurons, while LY354740 (300 nM) abolishes this difference. Frequency (Hz): control RAM+ 10.72 ± 0.86; control RAM− 5.93 ± 1.13; LY RAM+ 4.88 ± 1.21; LY RAM− 3.81 ± 0.5; F (1, 48) = 3.975, p = 0.05; Amplitude (pA): control RAM+ 13.47 ± 0.6; control RAM− 12.92 ± 0.92; LY RAM+ 13.45 ± 1.05; LY RAM− 16.42 ± 1.3; F (1, 48) = 2.829, p = 0.1. Control RAM+: n = 13, control RAM−: n = 11, LY RAM+: n = 13, LY RAM−: n = 15; 6 animals.
(D) 2D morphological analysis of RAM+ and RAM− neurons. (i) Schematic showing sparse labeling strategy in CA3 pyramidal cells with RAM+ cells labeled in orange (GFP and tdTomato) and RAM− cells labeled in green (GFP only). (ii) Representative image of CA3 following labeling scheme in (i). (iii) Representative images of dendritic segments with TEs of RAM+ and RAM− cells. (iv) 2D quantification of TE areas. TE Area: RAM+ 1.52 ± 0.17, n = 12 dendrites, 5 animals; RAM− 1.03 ± 0.11, n = 12 dendrites, 5 animals; p = 0.0296.
(E) Npas4 cKO mice were co-injected with AAV-RAM-mKate2 and AAV-hSyn-ΔCre-GFP. RAM+ CA3 pyramidal neurons have significant higher mEPSC frequency than their neighboring RAM− neurons, while LY354740 (300 nM) abolishes this difference. Frequency (Hz): control RAM+ 6.31 ± 0.91; control RAM− 2.88 ± 0.82; LY RAM+ 1.95 ± 0.54; LY RAM− 2.19 ± 0.51; F (1, 23) = 5.653, p = 0.026; Amplitude (pA): control RAM+ 13.36 ± 0.67; control RAM− 14.2 ± 1.01; LY RAM+ 14.82 ± 1.51; LY RAM− 15.21 ± 1.9; F (1, 23) = 0.2331, p = 0.63. Control RAM+: n = 7, control RAM−: n = 8, LY RAM+: n = 5, LY RAM−: n = 7; 5 animals.
(F) Npas4 cKO mice were co-injected with AAV-RAM-mKate2 and AAV-hSyn-Cre-GFP. mEPSC frequency for both RAM+ and RAM− was elevated by Npas4 deletion but the difference between them was abolished. Frequency (Hz): control RAM+ 9.19 ± 2.08; control RAM− 8.73 ± 1.03; LY RAM+ 1.24 ± 0.19; LY RAM− 2.57 ± 0.65; F (1, 26) = 0.3899, p = 0.54; Amplitude (pA): control RAM+ 13.31 ± 0.9; control RAM− 13.81 ± 0.52; LY RAM+ 11.58 ± 0.94; LY RAM− 14.17 ± 1.1; F (1, 26) = 1.39, p = 0.25. Control RAM+: n = 9, control RAM−: n = 8, LY RAM+: n = 6, LY RAM−: n = 7; 3 animals.
Summary data are shown as mean ± SEM. (C) (E) (F) **p < 0.01, Two-way ANOVA, Bonferroni’s post-hoc test. (D) *p < 0.05, Mann-Whitney U-test. See also Figure S4.
In the process of developing the RAM system, we had previously found that CA3 neurons activated by exposure to an enriched environment (EE), and hence labeled by RAM (Figure S4A), have significantly higher mEPSC frequencies (Figure S4B) and a modest decrease in their mIPSC amplitude (Figure S4C). This result suggests that contextual learning may selectively strengthen MF-CA3 connections on the activated CA3 neurons, and leads to the hypothesis that Npas4 deletion abolishes this learning-induced synaptic modification, leading to the observed impairment in contextual memory formation.
To identify neurons that are activated by CFC, we stereotaxically injected AAV-RAM-mKate2, which expresses the reporter gene mKate2 in activated neurons, into CA3 of wild-type C57BL/6 mice. Mice were then placed on a Dox diet for 5 days, switched to Dox-free food for 48 hours, then subjected to CFC; mEPSC measurements was carried out 24 hours later (Figure 6B). CA3 neurons activated during CFC and hence labeled by mKate2 (RAM+) had a substantially higher mEPSC frequency than neighboring unlabeled (RAM−) pyramidal neurons (Figure 6C). This difference was abolished when mEPSC measurements were carried out in the presence of the selective mGluR2/3 agonist LY354740, which, like DCG-IV, blocks MF-CA3 transmission (Figure 6C). This result suggests that contextual learning selectively strengthens MF inputs onto the ensemble of CA3 neurons that are activated during CFC. In addition, TEs were significantly larger on RAM+ compared to RAM− CA3 pyramidal neurons (Figure 6D), suggesting an increase in the number of MF-CA3 synaptic contacts. Taken together, our results provide the first evidence that contextual learning selectively increases MF-CA3 transmission on learning-activated CA3 pyramidal neurons, consistent with the postulated role of MF inputs in encoding contextual memory (Kesner and Rolls, 2015).
Npas4 Deletion Prevents Learning-induced Modification of MF-CA3 Synapses
We next examined whether the contextual learning-induced changes in MF-CA3 synapses are abolished when Npas4 is acutely deleted in CA3 pyramidal neurons. AAV–Cre-GFP or AAV–ΔCre-GFP was stereotaxically co-injected with AAV-RAM-mKate2 into CA3. Neurons activated by CFC were labeled using a similar procedure to that described above (Figure 6B). In Npas4 cKO mice injected with the control virus AAV-ΔCre-GFP, which does not cause deletion of Npas4, RAM+ neurons showed significantly higher mEPSC frequency compared to neighboring RAM− neurons, which was abolished by LY354740 treatment, and no difference in mEPSC amplitude (Figure 6E), equivalent to our results in wild-type mice (Figure 6C). However, after deletion of Npas4, there was no difference in either mEPSC frequency or amplitude between RAM+ and RAM− neurons, presumably due to the robust elevation of mEPSC frequency in both RAM+ and RAM− neurons that results from Npas4 deletion (Figure 6F). These data suggest that Npas4 deletion prevents the learning-induced modification of MF-CA3 synapses, which may be responsible for the impairment in contextual memory formation that we observed upon Npas4 deletion.
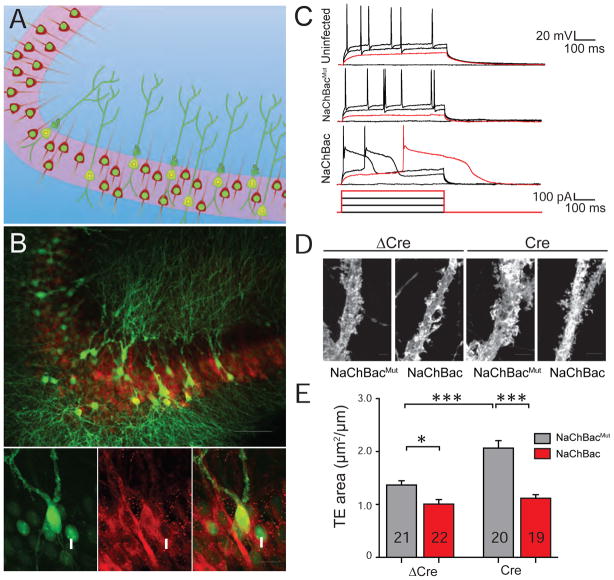
Thus far, our results suggest that Npas4 selectively restricts the number of functional synaptic contact established by a MF terminal onto a CA3 pyramidal neuron. Because Npas4 expression is selectively regulated by neuronal activity (Lin et al., 2008; Ramamoorthi et al., 2011), and MF-CA3 synapses are very sensitive to changes in the levels of neuronal activity (Kim and Tsien, 2008; Lee et al., 2013), we wondered whether Npas4 is involved in homeostatic shrinkage of TEs in response to neuronal activity within CA3. To address this question, we artificially increased the activity level in CA3 pyramidal neurons by expressing the bacterial sodium channel NaChBac, which renders neurons hyperexcitable (Ren et al., 2001) and has been used to induce homeostatic changes in neurons (Sim et al., 2013; Xue et al., 2014). AAV-CaMKII-NaChBac-mCherry or control AAV-CaMKII-NaChBacmut-mCherry expressing the non-conducting mutant form NaChBacmut (Yue et al., 2002), was stereotaxically injected into Npas4 cKO mice, together with either AAV-Cre-GFP or AAV-ΔCre-GFP (to express GFP in the nucleus). To visualize TE morphology in a sparse population of neurons, we co-injected diluted AAV-EF1α-Flp∘ and AAV-hSyn-fDIO-GFP-P2A-farnGFP (Figure 7A–B). Ten days after viral injection, neurons expressing NaChBac displayed prolonged action potentials and hyperexcitability, as reported previously (Ren et al., 2001; Sim et al., 2013; Xue et al., 2014), compared to those expressing NaChBacmut, which have similar properties to uninfected neurons (Figure 7C). As expected, deletion of Npas4 significantly increased TE size (Figure 7E, compare Cre and ΔCre conditions with expression of NaChBacMut). Interestingly, NaChBac expression caused a significant homeostatic reduction in TE size both with and without Npas4 (Figure 7E). This observation suggests Npas4 is not generally required for activity-dependent homeostatic reduction of MF-CA3 synapses, but also the impaired contextual memory formation in the absence of Npas4 is likely due to loss of the specific learning-induced modulation of MF-CA3 synapses, rather than a general disruption of homeostatic balance in CA3.
Figure 7. Npas4 is not required for activity-dependent homeostatic shrinkage of TE structures.
(A) Schematic showing sparse labeling strategy in CA3 pyramidal cells with morphology in green (GFP) and NaChBac or NaChBacmut cells labeled in red (tdTomato). CA3 pyramidal cells also expressed nuclear-localized Cre-GFP, or ΔCre-GFP as a control.
(B) Representative image of CA3 cells following labeling scheme in A. Bottom: high-magnification image of a cell expressing cytosolic GFP to label morphology (arrowhead) and nuclear Cre-GFP (*); neighboring cells expressing only nuclear Cre-GFP (arrow). Cells also expressed mCherry in the presence of NaChBac or NaChBachMut. Scale bars = 100 and 10 μm respectively.
(C) Representative traces from electrophysiological recordings of uninfected cells and cells expressing NaChBac or NaChBacMut, 10 days after viral injection.
(D) Representative images of dendritic segments with TE structures. Scale bar = 5 μm.
(E) 2D quantification of TE areas. TE area (μm2/μm): ΔCre + NaChBac 1.01 ± 0.09, n = 21 cells; ΔCre + NaChBacMut 1.37 ± 0.08, n = 22 cells; Cre + NaChBac 1.12 ± 0.07, n = 20 cells; Cre + NaChBacMut 2.07 ± 0.14, n = 19 cells. 3 mice per condition. F(1,78)=9.343. p = 0.0031.p* < 0.05, **p < 0.01, ***p < 0.001, Two-way ANOVA with Tukey’s post-hoc multiple comparisons test.
Npas4 Regulates MF-CA3 Synapses Via Its Transcriptional Target Plk2
Npas4 is a transcription factor and exerts its effects on neural circuits by regulating the expression of downstream transcriptional targets. One such target, brain-derived neurotrophic factor (BDNF), has been shown to mediate the effect of Npas4 on GABAergic inhibitory synapses in CA1 (Bloodgood et al., 2013; Lin et al., 2008; Ramamoorthi et al., 2011). To determine whether BDNF is also involved in the regulation of MF-CA3 synapses by Npas4, we removed the BDNF gene from CA3 pyramidal neurons of BDNF cKO (BDNFflx/flx) mice (Rios et al., 2001) by stereotaxic injection of AAV-Cre. This manipulation caused a significant reduction in mIPSC amplitude in CA3 pyramidal neurons, indicating effective deletion of BDNF. However, deletion of BDNF in CA3 had no effect on mEPSCs, suggesting that a different transcriptional target of Npas4 mediates its effect on MF-CA3 synapses (Figure S5).
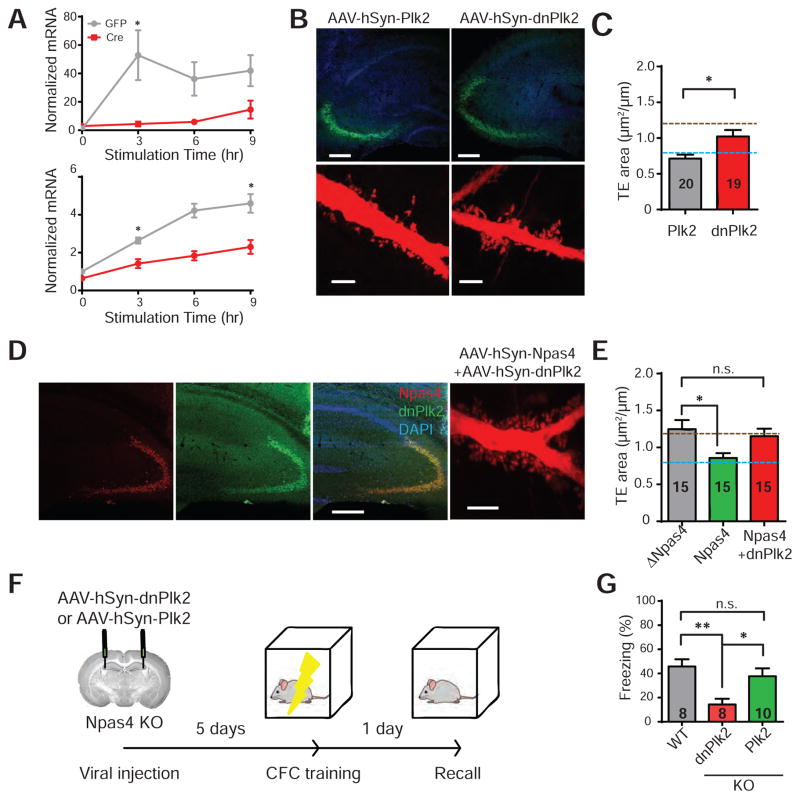
The activity-regulated gene polo-like kinase-2 (Plk2) has been implicated in activity-dependent shrinkage of dendritic spines (Evers et al., 2010; Lee et al., 2011; Pak and Sheng, 2003; Seeburg et al., 2008), and a dominant-negative form of Plk2 (dnPlk2) has been reported to enlarge TEs (Lee et al., 2013). Npas4 ChIP-Seq data (Kim et al., 2010) indicates that Npas4 binds to the promoter and enhancers of Plk2, suggesting that Plk2 is a direct transcriptional target of Npas4 (Figure S6). Consistent with this idea, Npas4 deletion from hippocampal neurons prepared from Npas4 cKO mice significantly reduced the activity-dependent expression of Plk2 (Figure 8A).
Figure 8. Npas4 mediates activity-dependent modulation of thorny excrescences (TEs) through its transcriptional target Plk2.
(A) Npas4 controls the activity-dependent expression of Plk2. Cultured hippocampal neurons from Npas4flx/flx mice were infected with AAV1-hSyn-GFP-Cre (Cre) or AAV1-hSyn-GFP (GFP) virus on DIV9, then stimulated with 10 μM kainic acid on DIV15 for 3, 6 and 9 hours before being lysed for RT-PCR analysis. 0hr represents the unstimulated conditions. mRNA levels were normalized to Tuj1 expression levels. Data were derived from 4 independent experiments. *p < 0.05, Mann-Whitney U-test.
(B) – (C) Selective expression of Plk2 in CA3 reduces TE size in Npas4 KO mice, while expression of the dominant-negative form of Plk2 (dnPlk2) fails to increase TE size further. AAV-hSyn-Plk2-flag or AAV-hSyn-dnPlk2-flag were stereotaxically injected into the hippocampus of Npas4 KO mice.
(B) Representative images showing Plk2 and dnPlk2 overexpression and the corresponding TEs in CA3 of Npas4 KO mice. Scale bars correspond to 200 μm on the top and 5 μm on the bottom.
(C) Summary bar graphs of TE size analysis in Npas4 KO mice injected with AAV-hSyn-Plk2-flag or AAV-hSyn-dnPlk2-flag. TE area (μm2/μm): Plk2 0.71 ± 0.05; dnPlk2 0.99 ± 0.09; p = 0.026; n = 20 and 19 neurons, respectively, 4 animals for each condition. *p < 0.05, Mann-Whitney U-test.
(D) – (E) The effect of Npas4 expression in CA3 of Npas4 KO, which previously reduced TE size to wild-type level, was blocked by dnPlk2. Npas4 KO mice were stereotaxically injected with AAV-hSyn-ΔNpas4-HA, AAV-hSyn-Npas4-HA or AAV-hSyn-Npas4-HA and AAV-hSyn-dnPlk2-flag.
(D) Representative images showing Npas4 (red) and Plk2 (green) expression in CA3 of Npas4 KO mice co-injected with AAV-hSyn-Npas4-HA and AAV-hSyn-dnPlk2-flag. Scale bar corresponds to 300 μm. A representative image of TEs is shown on the right and the scale bar here corresponds to 5 μm.
(E) Summary bar graph of TE size in Npas4 KO mice infected with AAV-hSyn-ΔNpas4-HA, AAV-hSyn-Npas4-HA or co-infected with AAV-hSyn-Npas4-HA and AAV-hSyn-dnPlk2-flag. TE area (μm2/μm): ΔNpas4 1.25 ± 0.12; Npas4 0.86 ± 0.06; Npas4 + dnPlk2 1.15 ± 0.10; F (2, 42) = 4.396; p = 0.019; n = 15 neurons, 3 animals per condition. *p < 0.05, one way ANOVA, Dunnett’s post-hoc test. Data for Npas4 and ΔNpas4 conditions are re-plotted from Figure 5D.
(F) – (G) Resetting TE size in Npas4 KO mice rescues the contextual memory deficit.
(F) Npas4 KO mice were stereotaxically injected with AAV1-hSyn-dnPlk2 or AAV1-hSyn-Plk2 in CA3. Five days after surgery, mice underwent CFC and their contextual memories were tested 24 hours later.
(G) Summary bar graph showing that Npas4 KO mice injected with Plk2 displayed similar freezing levels to their wild-type littermates, while those injected with dnPlk2 continued to display impaired contextual memory. Freezing level (%): WT 45.83 ± 6.85%; KO + dnPlk 2 14.32 ± 5.44%; KO + Plk2 37.71 ± 7.24%; F (2, 23) = 7.135; p = 0.0039. *p < 0.05, **p < 0.01, one way ANOVA, Tukey’s post-hoc test.
The blue and brown horizontal lines in (C) and (E) indicate the TE areas seen in Npas4 wild-type and KO mice, respectively (Figure 5C). All summary data are shown as mean ± SEM. See also Figure S5–7.
We then designed epistasis experiments to test the hypothesis that Plk2 acts downstream of Npas4 to modulate TE size in MF-CA3 synapses. If this hypothesis is true, abolishing activity of both Npas4 and Plk2 should not have an additive effect, therefore blocking Plk2 function in Npas4 KO mice by expressing dnPlk2 should not cause any further increase in TE size. To test this, AAVs expressing dnPlk2 or wild-type Plk2 were stereotaxically delivered to CA3 of Npas4 KO mice and TE areas were measured 3 days later (Figure 8B). Over-expression of dnPlk2 did not cause any additional enlargement of TEs, consistent with Plk2 functioning downstream of Npas4 (Figure 8C). Over-expression of wild-type Plk2, which bypasses the requirement for Npas4, significantly reduced TE area (Figure 8C). If Plk2 mediates the effect of Npas4 on TE size, dnPlk2 should block this effect. Re-expressing Npas4 in CA3 of Npas4 KO mice not only rescues the contextual learning deficit (Ramamoorthi et al., 2011), but also restores changes in mEPSCs (Figure 2E–G), and reduces the TE size to wild-type levels (Figure 5D). However, co-expression of dnPlk2 completely prevented Npas4 from restoring TE sizes to normal (Figure 8E). We conclude that Npas4 likely exerts its effect on MF-CA3 TEs by activating Plk2 expression.
Our findings suggest that contextual memory formation depends on Npas4 restricting the number of MF-CA3 synaptic contacts. If this is true, resetting MF-CA3 synapses to wild-type level by expressing the Npas4 downstream target Plk2 should restore contextual memory formation. Indeed, we found that Plk2 overexpression in CA3 of Npas4 KO mice (Figure 8F) restored TEs to a size comparable to wild-type littermates (Figure 8C). Some mice in each behavioral cohort were injected with dnPlk2 as controls. Remarkably, while over-expression of either Plk2 or dnPlk2 did not affect the behavior of wild-type mice (Figure S7), Npas4 KO mice over-expressing wild-type Plk2 were able to form contextual memories, exhibiting similar freezing behavior to their wild-type littermates (Figure 8G). Npas4 KO mice over-expressing dnPlk2 remained defective in forming contextual memories (Figure 8G). These results indicate that restriction of the number of MF-CA3 synaptic contacts by Npas4 is critical for the encoding of new contextual information.
Discussion
Here we provide evidence for a molecular pathway that selectively regulates the critical excitatory MF input into CA3 for encoding contextual memory. To the best of our knowledge, Npas4, which we previously showed to be required in CA3 for contextual memory formation (Ramamoorthi et al., 2011), emerges as the first gene with such a specific role in CA3-dependent contextual memory formation. In addition, our study also uncovers a form of learning-induced plasticity that occurs specifically at the MF-CA3 synapses. Furthermore, this experience-dependent synaptic plasticity at MF-CA3 synapses is under the control of Npas4. Lastly, we have identified Plk2 as the downstream transcriptional target of Npas4 that mediates its effect on MF-CA3 synapses. Altogether, our findings reveal a novel molecular and synaptic mechanism underlying long-term contextual memory formation in CA3. Our results suggest a molecular cascade in which Npas4 is activated by neuronal activity and induces the expression of Plk2, which in turn restricts the number of functional contacts between MF terminals and CA3 pyramidal neurons. Notably, Npas4 selectively regulates MF inputs but not other excitatory (or inhibitory) synaptic inputs converging on CA3 pyramidal neurons. Considering Npas4 is selectively induced by neuronal activity (Lin et al., 2008; Ramamoorthi et al., 2011), its selective regulation of MF-CA3 synapses might be determined by the unique features of MF transmission, including robust frequency facilitation and strong postsynaptic drive, which are distinct from those received from the perforant and recurrent paths (Henze et al., 2000; Henze et al., 2002; Urban et al., 2001). In other words, Npas4 may be preferentially induced by activity input from MF pathways. This could explain why, although it appears to homeostatically regulate MF-CA3 synapses, Npas4 is not required for homeostatic change at those synapses in response to a global, non-MF-specific, activity increase caused by NaChBac expression (Figure 7).
Npas4 has previously been shown to engage different downstream transcriptional targets in different cell types and brain regions to modify synaptic connections (Bloodgood et al., 2013; Lin et al., 2008; Sim et al., 2013; Spiegel et al., 2014; Yoshihara et al., 2014). Most notably, BDNF mediates the effects of Npas4 in recruiting GABAergic inhibitory synapses onto CA1 pyramidal neurons in response to activity (Bloodgood et al., 2013; Lin et al., 2008). Intriguingly, Npas4 does not appear to modulate GABAergic synapses on CA3 pyramidal neurons, at least under the conditions we have tested. We currently do not know what additional cellular mechanisms determine the varying effects of Npas4 in different cell types and brain regions.
Our study also uncovered a form of learning-induced, structural and functional plasticity that occurs specifically at the MF-CA3 synapses during contextual memory formation. We have shown that contextual learning selectively strengthens MF inputs onto the group of CA3 pyramidal neurons that are activated during learning. These so-called ensemble neurons have been shown to be involved in memory encoding (Denny et al., 2014; Liu et al., 2012), but synaptic plasticity on these neurons specifically induced by learning has not been widely explored. While previous studies have provided an abundance of evidence that MF-CA3 synapses are modified by experience (Galimberti et al., 2006; Gogolla et al., 2009; Magarinos et al., 1997; Maruo et al., 2016; Rekart et al., 2007; Sandi et al., 2003), they have typically relied on chronic behavioral manipulations to induce changes in MF-CA3 synapses. Consequently, it is difficult to determine whether these changes are the result of specific learning experience or a consequence of network-wide homeostatic adaptation, especially because MF-CA3 synapses are highly sensitive to homeostatic changes (Chater and Goda, 2013; Kim and Tsien, 2008; Lee et al., 2013). To the best of our knowledge, we are the first to demonstrate a synaptic change on CA3 ensemble neurons that is specifically induced by contextual learning.
MF inputs to CA3 pyramidal neurons are required for contextual memory encoding (Jerman et al., 2006; Lassalle et al., 2000; Lee and Kesner, 2004a) and the learning-induced functional and structural change we describe here appears to selectively occur at MF-CA3 synapses. The fact that Npas4 deletion prevents MF-CA3 synaptic modulation and also impairs contextual memory formation suggests that the learning-induced synaptic modulation we found plays an important role in contextual memory formation. Npas4 deletion in CA3 strongly increases the number of synaptic contacts established by a single MF terminal onto a CA3 pyramidal neuron. In the absence of Npas4, strengthened MF inputs could reach the ceiling of their dynamic range and no longer be modified by learning, thereby disrupting the formation of a sparse engram. Thus, Npas4 could be required for maintaining MF inputs within their normal dynamic range. While we identify Npas4 as a critical regulator of the experience-dependent plasticity at MF-CA3 synapses, the molecular pathways responsible for the selective strengthening of the MF input caused by learning are currently unknown.
Enhanced connectivity between DG granule cells to CA3 pyramidal neurons has long been postulated a key feature of the computational mechanism by which CA3 encodes new contextual representations transmitted from DG (Cerasti and Treves, 2010; Treves and Rolls, 1992). The robust increase in the number of MF-CA3 synaptic contacts induced by Npas4 deletion could also include enhanced DG-CA3 connectivity, a possibility we cannot completely rule out. It is intriguing that Npas4 regulates the number of MF-CA3 functional contacts, but not their dynamic properties (i.e. short-term and long-term plasticity). Contextual learning itself also increases the number of functional MF-CA3 synaptic contacts on CA3 pyramidal neurons activated by learning. The computational benefit of modulating MF-CA3 synapse number selectively is currently unknown and warrants future investigation.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the lead contact, Dr. Yingxi Lin (yingxi@mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Npas4−/− (KO) and Npas4flx/flx mice were generated previously (Lin et al., 2008) and BDNFflx/flx mice (Rios et al., 2001) were generously provided by Dr. Qiang Chang at the University of Wisconsin, Madison. C57BL/6 mice were purchased from Charles River Laboratory. Heterozygous mice were bred to produce Npas4−/− and Npas4+/+ littermates, and homozygous mice were bred to produce Npas4flx/flx (cKO) and BDNFflx/flx (BDNF cKO) animals. Mice were weaned at postnatal day 21 and housed by sex in groups of 3–4 until being used for experiments. For CFC, mice were single-housed for 3–5 days prior to conditioning. All mice were housed with a 12 hour light-dark cycle. As standard practice, behavior was done with males, electrophysiology was done with both male and female. Genders of the experimental animals were recorded, but no difference were detected from the data obtained from male and female animals, therefore data were pooled together. No further analysis of gender influence was performed. Rat or mouse pups of either gender were used to produce primary glia or neuron cultures, respectively. Animal protocols were performed in accordance with NIH guidelines and approved by the Massachusetts Institute of Technology and the Albert Einstein College of Medicine Committees on Animal Care.
METHOD DETAILS
Acute slice preparation
Npas4−/− mice of either sex, male Npas4flx/flx, BDNFflx/flx, and C57BL/6 mice were used at 8–10 weeks of age. Mice were anesthetized and decapitated, their brains were removed and immediately immersed in carbogenated (95% O2, 5% CO2) ice-cold cutting solution containing (in mM): 105 NMDG, 2.5 KCl, 1.24 NaH2PO4, 10 MgCl2, 0.5 CaCl2, 26 NaHCO3, 15 Glucose and 1 Na-ascorbate, 310 mOsm osmolarity, pH adjusted to 7.3 with HCl. Brains were then rapidly blocked and 350μm transverse slices were cut in the same solution using a vibratome (VT1200, Leica). Slices containing dorsal hippocampus were transferred to an incubation chamber filled with warm (32°C) carbogenated cutting solution and moved to room temperature (~23°C) for recovery. Slices were then transferred to a holding chamber filled with carbogenated room temperature ACSF containing (in mM): 119 NaCl, 2.5 KCl, 1.24 NaH2PO4, 1.3 MgCl2, 2.5 CaCl2, 26 NaHCO3, and 10 Glucose, 300 mOsm osmolarity, pH 7.3, for storage. Slices were kept in ACSF for at least 1 hour before recording.
Electrophysiology
After recovery, slices were transferred to a recording chamber perfused with carbogenated ACSF at room temperature at a flow rate of 2ml/min. A borosilicate glass pipette (3–6MΩ tip resistance) was filled with one of the following internal solutions (in mM). Cs based internal solution for mEPSCs: 130 CsMeSO3, 10 phosphocreatine, 1 MgCl2, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 0.5 Na-GFP, pH adjusted to 7.25 with CsOH, 290 mOsm osmolarity; high Cl internal solution for mIPSCs: 103 CsCl, 12 CsMeSO3, 5 TEA-Cl, 10 HEPES, 4 Mg-ATP, 0.5 Na-GTP, 0.5 EGTA, 1 MgCl2, pH adjusted to 7.25 with CsOH, 290 mOsm osmolarity. Data were collected using a Multiclamp 700B (Molecular Devices), filtered at 3kHz and digitized at 10kHz using a Digidata 1440A and Clampex 10.2 software (Molecular Devices). mEPSCs were recorded in voltage clamp mode and pharmacologically isolated with 0.5μM tetrodotoxin (TTX, Tocris) and 50μM picrotoxin (Tocris) in the ACSF. mIPSCs were recorded in voltage clamp mode with 0.5μM TTX, 50μM APV (Tocris) and 20μM DNQX (Tocris) in the ACSF. To record mEPSCs and mIPSCs, CA3 and CA1 cells were held at −70mV with a −5mV pulse delivered every 30–60s to monitor access resistance. Recordings with access resistance greater than 25MΩ or changes exceeding 15% were discarded.
In the mossy fibers blockade experiments mEPSCs and mIPSCs were recorded with 1 μM DCG-IV (Tocris) or 300 nM LY354740 (Tocris) in the ACSF. Evoked fEPSPs were recorded in current clamp mode with no drugs added to the ACSF. Recording and stimulating pipettes (< 2MΩ tip resistance) were filled with ACSF. Recording pipettes were placed in the stratum lucidum while stimulating pipettes were placed in the DG granule cell layer. Stimulation intensity was adjusted to produce 0.2 – 0.4mV fEPSP amplitudes. Data were filtered at 1kHz, digitized at 10kHz and analyzed with MiniAnalysis (Synaptosoft), Clampfit 10.2 (Molecular Devices) or custom software written in IgorPro (Wavemetrics).
Minimal stimulation of mossy fibers was performed as described (Hofmann et al., 2008; Jonas et al., 1993). Briefly, a theta glass-type stimulating pipette was placed in stratum lucidum 50–100 μm away from the recorded CA3 pyramidal cell. Stimulation intensity was set in order to elicit a synaptic response and then intensity was reduced until failures were detected. All responses had fast 10–90% rise time (<2 ms) and were blocked by bath application of 1 μM DCG-IV. To minimize contamination from recurrent associational/commissural (a/c) inputs, AMPAR-EPSC were recorded in presence of the CB1 receptor agonist WIN55,212-2 (5 μM), which selectively reduces the strength of a/c inputs onto CA3 pyramidal cells (Hofmann et al., 2008). The GABAA receptor antagonist picrotoxin (30 μM) was present in the bath to block fast inhibition. Potency was calculated as the average EPSC amplitude of successful trials while efficacy included both successes and failures. Failure rate was determined as the ratio of failures over total number of trials expressed as percentage.
Experimenters were blind to genotypes or injected viruses. Experimental group sizes were based on generally accepted criteria in the field. Different experimental groups were statistically compared using Mann-Whitney U-tests or ANOVA for more than two groups or conditions.
In the case of acute deletion in cKO mice, we inspect every transverse slices, and only animals with viral infection in overall more than 80–85% of dorsal CA3 and less than 10–15% dorsal DG were used. In addition, Npas4 cKO animals were excluded if any of their sections indicated Npas4 was deleted in less than 75% of dorsal CA3 and more than 20% of dorsal DG.
Viral vectors
AAV-hSyn-eGFP and AAV-hSyn-Cre-eGFP were obtained from the University of Pennsylvania Vector Core. HSV-Npas4-eGFP and HSV-ΔNpas4-eGFP were prepared as previously described (Ramamoorthi et al.). HSV-eGFP was obtained from the Viral Gene Transfer Core at MIT. All other AAVs were produced in our lab.
Stereotaxic injection
Npas4 KO, Npas4flx/flx, BDNFflx/flx, and C57BL/6 mice were gas-anesthetized with 1.5% isoflurane in O2 and received bilateral injections of 150–500nl of AAV or 1000nl of HSV into dorsal hippocampal CA3, CA1 or DG. Coordinates for target sites, using bregma as a reference point, were as follows. CA3: AP −1.8, ML ±2.25, DV −2.35; CA1: AP −2, ML ±1.5, DV −1.2; DG: AP −1.9, ML ± 1.0, DV −2.1. All injected mice, except in the RAM experiments, were used for recording on post-injection day 3 or 5. GFP and mKate2 expression were verified in acute slices and brains. Mice or tissues with off-target expression were excluded from further analysis.
Only animals with viral infection and effective deletion or overexpression in more than 80–85% of dorsal CA3 and less than 10–15% of DG, averaged over all sections, were included in the analysis. Animals were also excluded if any of their sections indicated deletion or overexpression in less than 75% of dorsal CA3 or more than 20% of DG. All experiments and analysis of data were performed in a “blind” manner by investigators that were unaware of the genotype or manipulation.
Sparse cell labeling strategy
To sparsely label cell for morphological analysis, AAV-hSyn-fDIO-TdTomato or AAV-hSyn-fDIO-GFP-P2A-farnGFP was mixed with 1000–2000 diluted AAV-ef1a-Flp∘ (final titer ~1010 gc/ml) and co-injected with other AAVs. For Npas4 acute deletion experiments, AAV-hSyn-fDIO-GFP-P2A-farnGFP+AAV-ef1a-Flp∘ mixture was directly injected in DG to sparsely label granule cells and their axon. AAV-hSyn-fDIO-TdTomato + AAV-ef1a-Flpo mixture was co-injected with AAV-hSyn-Cre-GFP or AAV-hSyn-ΔCre-GFP in CA3 to knockdown Npas4 in whole CA3 population but only sparsely label few CA3 cells and their dendritic spines. For RAM morphology experiments, C57BL/6 mice were injected with AAV-RAM-d2-tTA-TRE-mKate2 + low-titer AAV-EF1α-Flp∘ + AAV-hSyn-fDIO-GFP-P2A-farnGFP in CA3. For NaChBac experiments, AAV-hSyn-fDIO-GFP-p2a-farnGFP + AAV-ef1a-Flp∘ mixture was coinjected with AAV-hSyn-Cre-GFP or AAV-hSyn-ΔCre-GFP + AAV-CaMkII-NaChBac-mKate2 or AAV-CaMkII-NaChBacmut-mKate2 to knockdown and manipulate neuronal activity in whole CA3 population but sparsely label few CA3 cells. In both case Cre-GFP and ΔCre-GFP expression is restricted to nuclei of CA3 cells, it does not interfere with the dendritic or axonal GFP expression in the lucidum.
Seizure induction and in vivo activity manipulation
For Npas4 deletion verification, Npas4 cKO mice and Npas4 KO mice or their littermates were intraperitoneally (i.p.) injected with kainic acid (Sigma, 12–15 mg/kg dissolved in sterile saline, pH~7.2) to induce visible seizure and processed 2 hours after injection. Mice were used for experiments at 8–10 weeks of age.
Immunohistochemistry
Animals were transcardial perfused with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) and brains were post-fixed in 4% PFA at 4°C overnight. Brains were then sectioned at 50μm or 100 μm thickness using a vibratome. Coronal brain sections containing hippocampi were blocked in solution containing 1% Triton X-100 and 10% goat serum in PBS for 1 hour at room temperature, then incubated with primary antibody in solution containing 0.1% Triton X-100 and 5% goat serum in PBS overnight at 4°C. Sections were washed in PBS for 10 minutes 3 times then incubated with secondary antibody in the solution containing 0.1% Triton X-100 and 5% goat serum in PBS at room temperature for 1 hour. Sections were then washed in PBS for 10 minutes 3 times and mounted on microscope slides with DAPI Fluoromount-G (Southern Biotech) and allowed to dry for at least 2 hours.
Antibodies used: rabbit anti-Npas4 (1:10,000, homemade) (Lin et al., 2008), rat anti-Flag (1:200, Thermo Fisher), mouse anti-HA (1:1,000, Covance), mouse anti-PSD-95 (1:100, NeuroMab), guinea pig anti-Bassoon (1:100, SySy), chicken anti-GFP (1:1,000, Thermos Fisher), rabbit anti-RFP (1:500, Rockland) and Alexa 488, 555, 647 (1:500, Invitrogen).
Stick and stain
For spine density and TE morphology, we used the “stick-and-stain” protocol as previously described (Wallace and Bear, 2004 Wallace and Bear, 2004). Mice were perfused with warm (37°C) 4% PFA in phosphate buffer (PB) and brains were vibratome sectioned into 150μm slices. Brain slices containing dorsal hippocampi were microinjected in the CA3 region with Alexa Fluor 568 Hydrazide (Invitrogen) using glass pipettes (resistance ~95MΩ). Slices were submerged in ice-cold PBS and dye propagation was aided by 5μA current injection through the pipettes. Once cells were filled with dye, slices were post-fixed with 4% PFA overnight. The post-fixed slices were washed 3 times in PBS and mounted or subjected immunohistochemistry before mounting. After immunohistochemistry or stick-and-stain brain slices containing hippocampi were mounted for image acquisition. Confocal Z-series images were acquired using a Fluoview 1000 confocal microscope (Olympus).
SeeDB2
We used the SeeDB2 protocol to quantify structural changes to MF-CA3 synapse morphology Briefly, brain slices (100 μm) were prepared and then fixed overnight at 4°C with 4% PFA followed by permeabilization with 2% Triton X-100 at room temperature for 12–16 hours. The slices were clarified by soaking them in Omnipaque 350 solution (Sigma, catalog #1344600-200MG) at room temperature for 12 hours as described previously (Ke et al., 2016). Images of cleared slices were recorded by laser Airyscan confocal microscopy (ZEISS LSM 880 with Airyscan). Fifty to seventy optical sections were taken with 63x immersion objective (NA 1.40; WD 0.19 mm, Zeiss).
MAP
In order to quantify changes in synaptic marker densities, we used the MAP protocol as previously described (Ku et al., 2016). In order to quantify changes in synaptic marker densities, we used the MAP protocol as previously harvested, embedded in hydrogel, and sliced into 100 μm thick (original thickness) transverse sections, which are then cleared, denatured, and immunostained. MAP-processed hippocampal slices were incubated with primary antibodies in PBS with 0.1% Triton X-100 (PBST) at 37 °C overnight, followed by washing at 37 °C for 2 hr in PBST three times. The tissue was then incubated with secondary antibodies in PBST at 37 °C overnight, followed by washing at 37 °C for 2 hr in PBST three times. Antibodies against pre- and post-synaptic markers (Bassoon and PSD-95) were used to label excitatory synapses. Finally, at least 1 hr before imaging, sections were linearly expanded 4 fold, preserving fine sub-cellular structures and protein localizations. With this expansion, high-resolution confocal z-stack imaging of synaptic marker localization was carried out using a Leica TCS SP8 microscope system with 63x, 1.30 NA glycerol-immersion objective (WD 0.3 mm).
2D Spine density and TE morphology
For distal spine density measurements, dendritic segments of dye-filled CA3 cells were chosen. Segments were 20μm in length and at least 30μm away from the CA3 cell layer. The spine density was calculated as the total spine number per 10μm. For TE morphology measurements, a proximal dendritic segment was chosen within 30μm of the soma (lucidum layer). The area of TEs was calculated using Metamorph software (Molecular Devices) by measuring the total area (TEs + dendrites) within the 30μm length of a Z-stack image then subtracting the dendritic area from the total. For each cell, TE density was calculated as the total TE area per total length measured (approximately 30μm). Experimenters were blind to genotypes or injected viruses. Analyzers were blind to the experimental conditions. Experimental group sizes were based on generally accepted criteria in the field. Different experimental groups were statistically compared using unpaired-Student’s t-tests or ANOVA for more than two groups or conditions.
3D Reconstruction of MF, TE and synaptic puncta
Reconstruction and analysis of synaptic structures were carried out in Imaris (Bitplane, UK). For morphological analysis of the TE spines and MF terminals, Z-stacks were imported and the surface tool was used to generate 3-dimensional dendritic and synaptic structures, which uses an automatic smoothing of the image by application of a Gaussian filter. The volume of individual TE structures was calculated by subtracting the volume of the dendritic shaft, while individual MF terminal volumes can be directly measured from the reconstructions. To quantify synaptic protein density, surface reconstructions of synaptic marker puncta (PSD-95 and Bassoon) was performed using the same creation parameters and the total volume, average puncta volume, and total number of reconstructed puncta was measured from a volume within the stratum lucidum.
Contextual fear conditioning
After viral injection, mice were handled to habituate daily for 3 days prior to CFC training. On CFC day 1, mice were placed in a novel chamber, allowed to explore for 58s and then given a 2s 0.55mA footshock. The explore-shock cycle was repeated 3 times and the 4th cycle ended with no shock. After the 4 minute training, mice were returned to their home cages. On CFC day 2, mice were returned to the conditioning chamber for 4 minutes to test memory recall. In experiments with a second recall (Fig 3A–B), mice were returned to the chamber for another 4 minutes 4 days after the first recall (3 days after surgery). Contextual memory was measured by scoring freezing behavior over the 4 minute recall period, with freezing defined as the total absence of movement apart from respiration. Scorers were blind to the experimental conditions and experimenters were blind to genotypes or injected viruses. Experimental group sizes were based on generally accepted criteria in the field. Different experimental groups were statistically compared using unpaired-Student’s t-tests or ANOVA for more than two groups or conditions.
Neuronal cultures and qRT-PCR
Astrocytes derived from P1-P2 rat cortices were plated at low density in DMEM + 10% FBS in poly-D-lysine-coated 6 well plates and stored and maintained at 37°C in a humidified incubator with 10% CO2. Once the glial cells had formed a confluent monolayer, approximately 7 days after plating, dissociated hippocampal neurons from mouse Npas4 cKO P0-P1 pups of either sex were plated at a density of 0.5 million cells per well, and the media was changed to NBA supplemented with B27 and Glutamax. Ara-C (5μM, Sigma) was added to the culture the next day, and conditional media was supplemented with fresh media on DIV7.
For qRT-PCR experiments, hippocampal Npas4-cKO neurons were infected with either AAV-hSyn-GFP-Cre or AAV-hSyn-GFP control virus on DIV9. Cultures were then stimulated with 10μM kainic acid on DIV15 for up to 9 hours before being lysed in 600μl Buffer RLT (RNeasy Mini Kit, Qiagen). Lysates were homogenized using QIAShredder (Qiagen) and RNA was purified from these lysates using the RNeasy Mini Kit (Qiagen). Reverse transcription was performed using the SuperScript® III First-Strand Synthesis System (Invitrogen), and all samples were then diluted to equalize the amount of RNA used. For qRT-PCR we used 10μl SsoFast EvaGreen Supermix (Bio-Rad) per reaction, 1μl of each primer, and 0.7μl cDNA. Reactions were performed in a Bio-Rad CFX96 real-time PCR detection system using the following protocol: 95°C for 30s, 45 cycles of 95°C for 5s and 60°C for 30s.
RT-PCR analysis
Analysis was performed using the relative standard curve method, using Tuj1 as the calibration gene for normalization. Primer sequences (forward, reverse) were as follows:
Plk2 (5′-AGCAGCGAATGCCTTGAAG-3′, 5′-TCCTCGAAGGACTCTTGCCA-3′)
Npas4 (5′-CTGCATCTACACTCGCAAGG-3′, 5′-GCCACAATGTCTTCAAGCTCT-3′)
Tuj1 (5′-CAGTGCGGCAACCAGATAG-3′, 5′-GCAGGTCTGAGTCCCCTACA-3′).
To compare gene induction across samples, we then normalized the data for each set of samples to the unstimulated, GFP-infected samples.
QANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± SEM unless otherwise stated in text and figure legends. All statistical tests are described in the corresponding figure legends and were performed using Prism (Graphpad). All experiments and analysis of data were performed in a “blind” manner by investigators that were unaware of the genotype or manipulation. In all experiments, “n” indicates the number of cells used. The total number of animals used per experiment is also explicitly reported in the figure legends. Comparisons were done using the Mann-Whitney U test unless otherwise stated. All comparisons are two-sided.
Supplementary Material
Highlights.
Npas4 selectively restricts the number of MF-CA3 synaptic contacts
Contextual learning selectively strengthens MF inputs on CA3 pyramidal neurons
Npas4 deletion prevents learning-induced strengthening of MF inputs
PlK2 functions downstream of Npas4 to modulate MF-CA3 synapses
Acknowledgments
We thank C. Mark Fletcher and Charles Jennings for critical reading of the manuscript, Matthew Wilson, Howard Eichenbaum and members of the Lin lab for discussions and comments on the study, Qiang Chang for kindly providing the BDNFflx/flx mice, Mollie Meffert for kindly providing the Plk2 and dnPlk2 constructs, and Rachel Schecter for help with the stick-and-stain technique. We are grateful to Li-Huei Tsai and the Picower Institute for Learning and Memory for sharing their microscopy resources. This work was funded by a Swedish Brain Foundation Research Fellowship (A.T.S.), Swiss national science foundation SNF171978 (K.Z.), JPB foundation (W.X.), NIH grants DA017392 and MH081935 (P.E.C.), and the James H. Ferry Fund and NIH grants MH091220 and NS088421 (Y.L.). The authors declare no competing financial interests.
Footnotes
Author Contributions
F.W. designed and conducted most of the electrophysiology, morphology and behavior experiments, R.I.G carried out all the 3D morphological reconstruction and high resolution imaging of MAP processed samples, as well as some other imaging analysis. Y.Z. and D.R. contributed to the morphology and behavior experiments, M.D. conducted RT-PCR experiments, D.G.D. and M.H. contributed to the spine density measurement, T.D.Y. and A.T.S. assisted the electrophysiology and behavior experiments, T.K. processed samples using MAP for high resolution synaptic analysis, K.Z. carried out sample processing and imaging using the SeeDB2 procedure, S.L. K.A. and P.E.C assessed the function of evoked mossy fiber transmission, Y.L. designed and supervised the study, Y.L. and F.W. wrote the paper with input from all authors.
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annual Review of Neuroscience. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Dent JA. Development of the mossy fibers of the dentate gyrus: I. A light and electron microscopic study of the mossy fibers and their expansions. J Comp Neurol. 1981;195:51–86. doi: 10.1002/cne.901950106. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y, Represa A. Brief seizure episodes induce long-term potentiation and mossy fibre sprouting in the hippocampus. Trends Neurosci. 1990;13:312–318. doi: 10.1016/0166-2236(90)90135-w. [DOI] [PubMed] [Google Scholar]
- Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–125. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerasti E, Treves A. How informative are spatial CA3 representations established by the dentate gyrus? PLoS Comput Biol. 2010;6:e1000759. doi: 10.1371/journal.pcbi.1000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater TE, Goda Y. CA3 mossy fiber connections: giant synapses that gain control. Neuron. 2013;77:4–6. doi: 10.1016/j.neuron.2012.12.024. [DOI] [PubMed] [Google Scholar]
- Chicurel ME, Harris KM. Three-dimensional analysis of the structure and composition of CA3 branched dendritic spines and their synaptic relationships with mossy fiber boutons in the rat hippocampus. J Comp Neurol. 1992;325:169–182. doi: 10.1002/cne.903250204. [DOI] [PubMed] [Google Scholar]
- Chierzi S, Stachniak TJ, Trudel E, Bourque CW, Murai KK. Activity maintains structural plasticity of mossy fiber terminals in the hippocampus. Molecular and Cellular Neuroscience. 2012;50:260–271. doi: 10.1016/j.mcn.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Claiborne BJ, Xiang Z, Brown TH. Hippocampal circuitry complicates analysis of long-term potentiation in mossy fiber synapses. Hippocampus. 1993;3:115–121. doi: 10.1002/hipo.450030202. [DOI] [PubMed] [Google Scholar]
- Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, Tomm NK, Turi GF, Losonczy A, Hen R. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83:189–201. doi: 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers DM, Matta JA, Hoe HS, Zarkowsky D, Lee SH, Isaac JT, Pak DT. Plk2 attachment to NSF induces homeostatic removal of GluA2 during chronic overexcitation. Nat Neurosci. 2010;13:1199–1207. doi: 10.1038/nn.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimberti I, Gogolla N, Alberi S, Santos AF, Muller D, Caroni P. Long-term rearrangements of hippocampal mossy fiber terminal connectivity in the adult regulated by experience. Neuron. 2006;50:749–763. doi: 10.1016/j.neuron.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Gogolla N, Galimberti I, Deguchi Y, Caroni P. Wnt signaling mediates experience-related regulation of synapse numbers and mossy fiber connectivities in the adult hippocampus. Neuron. 2009;62:510–525. doi: 10.1016/j.neuron.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Henze DA, Urban NN, Barrionuevo G. The multifarious hippocampal mossy fiber pathway: a review. Neuroscience. 2000;98:407–427. doi: 10.1016/s0306-4522(00)00146-9. [DOI] [PubMed] [Google Scholar]
- Henze DA, Wittner L, Buzsaki G. Single granule cells reliably discharge targets in the hippocampal CA3 network in vivo. Nat Neurosci. 2002;5:790–795. doi: 10.1038/nn887. [DOI] [PubMed] [Google Scholar]
- Hofmann ME, Nahir B, Frazier CJ. Excitatory afferents to CA3 pyramidal cells display differential sensitivity to CB1 dependent inhibition of synaptic transmission. Neuropharmacology. 2008;55:1140–1146. doi: 10.1016/j.neuropharm.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerman T, Kesner RP, Hunsaker MR. Disconnection analysis of CA3 and DG in mediating encoding but not retrieval in a spatial maze learning task. Learn Mem. 2006;13:458–464. doi: 10.1101/lm.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas P, Major G, Sakmann B. Quantal components of unitary EPSCs at the mossy fibre synapse on CA3 pyramidal cells of rat hippocampus. J Physiol. 1993;472:615–663. doi: 10.1113/jphysiol.1993.sp019965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya H, Ozawa S. Dual mechanism for presynaptic modulation by axonal metabotropic glutamate receptor at the mouse mossy fibre-CA3 synapse. J Physiol. 1999;518(Pt 2):497–506. doi: 10.1111/j.1469-7793.1999.0497p.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya H, Shinozaki H, Yamamoto C. Activation of metabotropic glutamate receptor type 2/3 suppresses transmission at rat hippocampal mossy fibre synapses. J Physiol. 1996;493(Pt 2):447–455. doi: 10.1113/jphysiol.1996.sp021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke MT, Nakai Y, Fujimoto S, Takayama R, Yoshida S, Kitajima TS, Sato M, Imai T. Super-Resolution Mapping of Neuronal Circuitry With an Index-Optimized Clearing Agent. Cell Rep. 2016;14:2718–2732. doi: 10.1016/j.celrep.2016.02.057. [DOI] [PubMed] [Google Scholar]
- Kesner RP. Behavioral functions of the CA3 subregion of the hippocampus. Learn Mem. 2007;14:771–781. doi: 10.1101/lm.688207. [DOI] [PubMed] [Google Scholar]
- Kesner RP, Rolls ET. A computational theory of hippocampal function, and tests of the theory: new developments. Neurosci Biobehav Rev. 2015;48:92–147. doi: 10.1016/j.neubiorev.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Kim J, Tsien RW. Synapse-specific adaptations to inactivity in hippocampal circuits achieve homeostatic gain control while dampening network reverberation. Neuron. 2008;58:925–937. doi: 10.1016/j.neuron.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku T, Swaney J, Park JY, Albanese A, Murray E, Cho JH, Park YG, Mangena V, Chen J, Chung K. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol. 2016;34:973–981. doi: 10.1038/nbt.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle JM, Bataille T, Halley H. Reversible inactivation of the hippocampal mossy fiber synapses in mice impairs spatial learning, but neither consolidation nor memory retrieval, in the Morris navigation task. Neurobiol Learn Mem. 2000;73:243–257. doi: 10.1006/nlme.1999.3931. [DOI] [PubMed] [Google Scholar]
- Lee I, Kesner RP. Differential contributions of dorsal hippocampal subregions to memory acquisition and retrieval in contextual fear-conditioning. Hippocampus. 2004a;14:301–310. doi: 10.1002/hipo.10177. [DOI] [PubMed] [Google Scholar]
- Lee I, Kesner RP. Encoding versus retrieval of spatial memory: double dissociation between the dentate gyrus and the perforant path inputs into CA3 in the dorsal hippocampus. Hippocampus. 2004b;14:66–76. doi: 10.1002/hipo.10167. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Lee Y, Rozeboom A, Lee JY, Udagawa N, Hoe HS, Pak DT. Requirement for Plk2 in orchestrated ras and rap signaling, homeostatic structural plasticity, and memory. Neuron. 2011;69:957–973. doi: 10.1016/j.neuron.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJ, Queenan BN, Rozeboom AM, Bellmore R, Lim ST, Vicini S, Pak DT. Mossy fiber-CA3 synapses mediate homeostatic plasticity in mature hippocampal neurons. Neuron. 2013;77:99–114. doi: 10.1016/j.neuron.2012.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magarinos AM, Verdugo JM, McEwen BS. Chronic stress alters synaptic terminal structure in hippocampus. Proc Natl Acad Sci U S A. 1997;94:14002–14008. doi: 10.1073/pnas.94.25.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo T, Mandai K, Takai Y, Mori M. Activity-dependent alteration of the morphology of a hippocampal giant synapse. Mol Cell Neurosci. 2016;71:25–33. doi: 10.1016/j.mcn.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Mayford M, Siegelbaum SA, Kandel ER. Synapses and memory storage. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll RA, Schmitz D. Synaptic plasticity at hippocampal mossy fibre synapses. Nat Rev Neurosci. 2005;6:863–876. doi: 10.1038/nrn1786. [DOI] [PubMed] [Google Scholar]
- Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, Otto T, Lin Y. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334:1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekart JL, Sandoval CJ, Bermudez-Rattoni F, Routtenberg A. Remodeling of hippocampal mossy fibers is selectivity induced seven days after the acquisition of a spatial but not a cued reference memory task. Learn Memory. 2007;14:416–421. doi: 10.1101/lm.516507. [DOI] [PubMed] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- Represa A, Ben-Ari Y. Kindling is associated with the formation of novel mossy fibre synapses in the CA3 region. Exp Brain Res. 1992a;92:69–78. doi: 10.1007/BF00230384. [DOI] [PubMed] [Google Scholar]
- Represa A, Ben-Ari Y. Long-term potentiation and sprouting of mossy fibers produced by brief episodes of hyperactivity. Epilepsy Res Suppl. 1992b;7:261–269. [PubMed] [Google Scholar]
- Rios M, Fan G, Fekete C, Kelly J, Bates B, Kuehn R, Lechan RM, Jaenisch R. Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol Endocrinol. 2001;15:1748–1757. doi: 10.1210/mend.15.10.0706. [DOI] [PubMed] [Google Scholar]
- Rolls ET. A theory of hippocampal function in memory. Hippocampus. 1996;6:601–620. doi: 10.1002/(SICI)1098-1063(1996)6:6<601::AID-HIPO5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ruediger S, Vittori C, Bednarek E, Genoud C, Strata P, Sacchetti B, Caroni P. Learning-related feedforward inhibitory connectivity growth required for memory precision. Nature. 2011;473:514–518. doi: 10.1038/nature09946. [DOI] [PubMed] [Google Scholar]
- Salin PA, Scanziani M, Malenka RC, Nicoll RA. Distinct short-term plasticity at two excitatory synapses in the hippocampus. Proc Natl Acad Sci U S A. 1996;93:13304–13309. doi: 10.1073/pnas.93.23.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandi C, Davies HA, Cordero MI, Rodriguez JJ, Popov VI, Stewart MG. Rapid reversal of stress induced loss of synapses in CA3 of rat hippocampus following water maze training. Eur J Neurosci. 2003;17:2447–2456. doi: 10.1046/j.1460-9568.2003.02675.x. [DOI] [PubMed] [Google Scholar]
- Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DT, Sheng M. Critical role of CDK5 and Polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–583. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Antolin S, Lin CW, Lin Y, Lois C. Increased cell-intrinsic excitability induces synaptic changes in new neurons in the adult dentate gyrus that require Npas4. J Neurosci. 2013;33:7928–7940. doi: 10.1523/JNEUROSCI.1571-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AT, Cooper YA, Baratta MV, Weng FJ, Zhang Y, Ramamoorthi K, Fropf R, LaVerriere E, Xue J, Young A, et al. A robust activity marking system for exploring active neuronal ensembles. Elife. 2016;5 doi: 10.7554/eLife.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves A, Rolls ET. Computational constraints suggest the need for two distinct input systems to the hippocampal CA3 network. Hippocampus. 1992;2:189–199. doi: 10.1002/hipo.450020209. [DOI] [PubMed] [Google Scholar]
- Urban NN, Henze DA, Barrionuevo G. Revisiting the role of the hippocampal mossy fiber synapse. Hippocampus. 2001;11:408–417. doi: 10.1002/hipo.1055. [DOI] [PubMed] [Google Scholar]
- Vyleta NP, Borges-Merjane C, Jonas P. Plasticity-dependent, full detonation at hippocampal mossy fiber-CA3 pyramidal neuron synapses. Elife. 2016;5 doi: 10.7554/eLife.17977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace W, Bear MF. A morphological correlate of synaptic scaling in visual cortex. J Neurosci. 2004;24:6928–6938. doi: 10.1523/JNEUROSCI.1110-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiera G, Mozrzymas JW. Extracellular proteolysis in structural and functional plasticity of mossy fiber synapses in hippocampus. Front Cell Neurosci. 2015;9:427. doi: 10.3389/fncel.2015.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Atallah BV, Scanziani M. Equalizing excitation-inhibition ratios across visual cortical neurons. Nature. 2014;511:596–600. doi: 10.1038/nature13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara S, Takahashi H, Nishimura N, Kinoshita M, Asahina R, Kitsuki M, Tatsumi K, Furukawa-Hibi Y, Hirai H, Nagai T, et al. Npas4 regulates Mdm2 and thus Dcx in experience-dependent dendritic spine development of newborn olfactory bulb interneurons. Cell Rep. 2014;8:843–857. doi: 10.1016/j.celrep.2014.06.056. [DOI] [PubMed] [Google Scholar]
- Yue L, Navarro B, Ren D, Ramos A, Clapham DE. The cation selectivity filter of the bacterial sodium channel, NaChBac. J Gen Physiol. 2002;120:845–853. doi: 10.1085/jgp.20028699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- Zhao S, Studer D, Chai X, Graber W, Brose N, Nestel S, Young C, Rodriguez EP, Saetzler K, Frotscher M. Structural plasticity of hippocampal mossy fiber synapses as revealed by high-pressure freezing. J Comp Neurol. 2012a;520:2340–2351. doi: 10.1002/cne.23040. [DOI] [PubMed] [Google Scholar]
- Zhao S, Studer D, Chai X, Graber W, Brose N, Nestel S, Young C, Rodriguez EP, Saetzler K, Frotscher M. Structural plasticity of spines at giant mossy fiber synapses. Front Neural Circuits. 2012b;6:103. doi: 10.3389/fncir.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.