Abstract
Narcissus tazetta L. is a bulbous ornamental plant popular for its notable fragrant flowers which make it the plant of high importance. In spite of its economic value, narcissus is found to be susceptible for a number of diseases borne by fungi, bacteria, nematodes, and viruses. A potyvirus, Cyrtanthus elatus virus-A isolate NBRI16 (CEVA-NBRI16), associated with leaf chlorotic stripe disease of N. tazetta cv. Paperwhite was reported for first time in India from our laboratory based on the partial coat protein gene sequence. In present study, the full-length genomic sequence of CEVA-NBRI16 is determined which consists of 9942 nucleotides, excluding the polyA tail, and encodes a single large polyprotein of 3102 amino acids with the genomic features typical of a potyvirus. It shares highest 93% nucleotide sequence identity and closest phylogenetic relationship with sequences of CEVA-Marijiniup7-1 and CEVA-Marijiniup7-2, both reported from Australia on Cyrtanthus elatus host. The full-length genomic sequence of CEVA from narcissus plant is being reported for the first time from India.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1189-z) contains supplementary material, which is available to authorized users.
Keywords: Cyrtanthus elatus virus A, Potyvirus, Sequence identity, Narcissus tazetta, Full-length genome
Introduction
Narcissus tazetta L. (family Amaryllidaceae) is known for attractive flowers with sweet fragrance, worldwide. It can easily be noticed anywhere by its pleasant sweet fragrance and notable cool white appearance. Due to this magnificent property, it is widely accepted as cut-flower crop and adapted by the floriculture and perfumery industry for economy. The narcissus plant is susceptible to several RNA viruses of genera Carlavirus, Maculavirus, Nepovirus, Potexvirus, etc. (Brunt 1995; Wylie and Jones 2012). Amongst all, potyviruses are the most prevalent in N. tazetta (Brunt 1995; Aminuddin and Raj 1999; Yadav and Khan 2007) and infected plants in general exhibit the symptoms of leaf chlorotic stripes, plant stunting, and flower colour breaking. The vegetative mean of propagating bulbs is a particular problem for virus disease dissemination in all bulbous crops including narcissus. These viral diseases are also reported to reduce the yield of such plants (Brunt 1977). An uncharacterized potyvirus was detected from N. tazetta cv. Paperwhite plants associated with yellow stripes symptoms (Aminuddin and Raj 1999) which was identified as Lycoris potyvirus, based on the partial 3′ sequence (Yadav and Khan 2007). Moreover, occurrence of a previously unidentified Cyrtanthus elatus virus A (CEVA) on N. tazetta cv. Paperwhite samples exhibiting chlorotic stripes were also reported on the basis of partial 3′ sequence (Kumar et al. 2015). In present study, the full-length genomic sequence of this CEVA isolate is determined. Information on complete genome sequence of CEVA from India will shed light on genetic makeup of indigenous isolate and will be a source to study the sequence diversity among potyviruses in narcissus plant.
Narcissus tazetta cv. Paperwhite plants exhibiting chlorotic stripes were observed at the experimental plot of CSIR-NBRI, Lucknow with 78% (105/137) disease incidence. Upon blooming, though the flowers in infected plants appear symptomless, the flower stalk showed light green to pale stripes (Fig. 1b, c) as compared to healthy plants having green leaves (Fig. 1a). A closeup of a single leaf clearly shown the leaf stripes symptoms (Fig. 1d). Association of such symptoms in N. tazetta was also observed earlier with potyvirus in India (Aminuddin and Raj 1999; Yadav and Khan 2007) and other countries (Wylie et al. 2014; Chen et al. 2006). For characterization of virus, infected leaf samples of narcissus were collected and stored in − 80 °C till further used. The total RNA from 100 mg infected leaf sample was isolated using TRI reagent (Sigma-Aldrich Co., MO, USA) for Reverse transcription-PCR and 5′RACE (Thermo Fisher Scientific Inc., MA, USA). For virus detection, primers MJ-I and MJ-II targeting the conserved region from MVWCIEN to QMKAAA motifs in coat protein (CP) region, were used (Marie-Jeanne et al. 2000; Grisoni et al. 2006). All the 105 infected samples produced expected size ~ 300 bp DNA band in RT-PCR. Sequencing of 18 randomly selected RT-PCR products confirmed the occurrence of potyvirus in N. tazetta.
Fig. 1.
a Healthy and b infected narcissus plant in field condition c close view of infected narcissus plant and d infected leaf showing the symptoms
The occurrence of CEVA in 18 N. tazetta samples exhibiting chlorotic leaf stripes were confirmed by nucleic acid spot hybridization (NASH) assay in a replica nitrocellulose membrane. The 2 µg RNA from a healthy N. tazetta plant (as negative control) along with 200 ng cloned DNA of CEVA (as positive controls) was also blotted. The membranes were hybridized with probes prepared by random primer labeling method (Fienberg and Vogelstein 1983); and pre-hybridization, hybridization, and washing steps were performed according to the standard methods (Sambrook et al. 1989) and exposed to X-ray films to observe hybridization signals. As a consequence, the presence of CEVA in 16 out of 18 samples showing positive signals with 88.8% disease incidence was observed (Electronic supplementary material 1).
Furthermore, a randomly chosen sample was used for full-length genome amplification taking the advantage of available degenerate primer pairs targeting different conserved motifs of potyvirus genome following the strategy as described earlier (Chen et al. 2002). The set of primer pairs: Pot-I/Pot -II (Gibbs and Mackenzie 1997); CI-F (Ha et al. 2008)/NIb-Pot-3 (Yakoubi et al. 2008); HP-F (Lucinda et al. 2010)/CI-R (Ha et al. 2008); 5′RACE and HP-R (Lucinda et al. 2010) were used. The 3′UTR to partial nuclear inclusion B (Nlb) region of ~ 1.6 kb was amplified using Pot-I/Pot-II primers which target the polyadenylated 3′ end and -GNNS- motif in Nlb region of CEVA isolate, respectively. The CI-F/NIb-Pot-3 primers were used which amplified the expected size ~ 3.0 kb band spanning the GxVGSGKST motif in cylindrical inclusion (CI) protein and targeted Nlb region. Furthermore, from partial CI to HC region of ~ 3.0 kb was amplified using HP-F/CI-R primers. For amplification of remaining 5′ end to HC region, RACE kit was employed which yielded ~ 2.0 kb DNA fragment.
All amplified DNA fragments were gel-purified and cloned in pGEM-T vector (Promega Corporation, Madison, USA). The positive transformants were screened by colony PCR and sequenced (three such positive clones for each insert). Sequences were edited and assembled using BIOEDIT tool (http://www.mbio.ncsu.edu/bioedit/bioedit.html) to eliminate sequence ambiguity. The consensus sequence for full-length genome was determined and submitted to GenBank under the accessions KX575832 (CEVA-NBRI16).
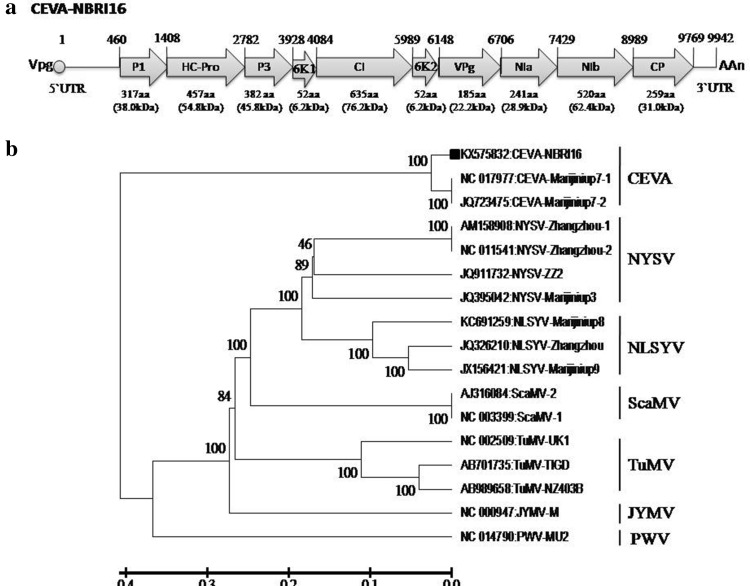
The open reading frames (ORFs) encoded by the genome were analyzed by ORF Finder (www.ncbi.nlm.nih.gov/projects/gorf/) and their putative proteins were translated by ExPasy tool (http://web.expasy.org/translate/). The sequences were compared with those of publically available sequences (Table 1) in NCBI by BLASTn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for identification. The nucleotide and amino acid identity of ORFs of selected potyvirus isolates (Table 2) was obtained by DiAlign tool (http://www.genomatix.de/cgi-bin/dialign/dialign.pl). The results of sequence analysis showed that the genome of CEVA-NBRI16 isolate is 9942 nucleotides long and contain 5`UTR, a single large predicted ORF (nucleotide positions 460-9769) encoding large polyprotein (387.12 kDa) of 3100 amino acids and 3`UTR (Fig. 2a). The polyprotein further yields the predicted ten mature proteins identified as P1, HC-Pro, P3, 6K1, CI, 6K2, VPg (viral protein genome-linked), Nla-Pro (nuclear inclusion protein a protease), Nlb, and CP having the amino acid number/molecular weight (kDa) of 317/38.0, 457/54.8, 382/45.8, 52/6.2, 635/76.2, 52/6.2, 185/22.2, 241/28.9, 520/62.4, and 259/31.0, respectively (Fig. 2a). The putative proteolytic cleavage sites in NBRI16 isolate are Y/S, G/G, Q/H, Q/S, Q/S, Q/G, E/N, Q/S, and Q/S (Electronic supplementary material 2), and concurred with Cyrtanthus elatus virus A (CEVA)-Marijiniup7-1 (NC_017977) and CEVA-Marijiniup7-2 (JQ723475) isolates (Wylie and Jones 2012). The complete nucleotide and deduced large polyprotein sequence of NBRI16 isolate when compared to previously reported potyviruses (Table 1) shared only 93 and 94% identities, respectively, with the only available full-length sequences of CEVA-Marijiniup7-1 and CEVA-Marijiniup7-2 isolates (Table 2). The pairwise sequence comparison at the level of nucleotides and deduced amino acids of all the 10 ORFs revealed 84–97% and 78–97% identities, respectively, with the Marijiniup7-1 and Marijiniup7-2 isolates.
Table 1.
Virus isolates used in present study
| aVirus | Isolate | Host | Location | Genome length (nt) | GenBank accession | Source |
|---|---|---|---|---|---|---|
| CEV-A | NBRI16 | Narcissus tazetta | India | 9942 | KX575832 | This study |
| CEV-A | Marijiniup7-1b | Cyrtanthus elatus | Australia | 9908 | NC_017977 | Wylie and Jones (2012) |
| CEV-A | Marijiniup7-2b | Cyrtanthus elatus | Australia | 9908 | JQ723475 | Wylie and Jones (2012) |
| NYSV | Zhangzhou-1 | Narcissus tazetta | China | 9650 | AM158908 | Chen et al. (2006) |
| NYSV | Zhangzhou-2 | Narcissus tazetta | China | 9650 | NC_011541 | Chen et al. (2002) |
| NYSV | ZZ2 | Narcissus tazetta | China | 9654 | JQ911732 | Unpublished |
| NYSV | Marijiniup3 | Narcissus spp. | Australia | 9647 | JQ395042 | Wylie and Jones (2012) |
| NLSYV | Zhangzhou | Narcissus tazetta | China | 9651 | JQ326210 | Lin et al. (2012) |
| NLSYV | Marijiniup8 | Narcissus spp. | Australia | 9687 | KC691259 | Wylie et al. (2014) |
| NLSYV | Marijiniup9 | Narcissus spp. | Australia | 9577 | JX156421 | Wylie et al. (2014) |
| bScaMV | − 1 | Allium chinense | China | 9324 | NC_003399 | Chen et al. (2002) |
| bScaMV | − 2 | Allium chinense | China | 9324 | AJ316084 | Chen et al. (2002) |
| TuMV | TIGD | Tigridia spp. | Germany | 9798 | AB701735 | Nguyen et al. (2013) |
| TuMV | NZ403B | Lepidium oleraceum | New Zealand | 9796 | AB989658 | Yasaka et al. (2015) |
| TuMV | UK1 | − | UK | 9835 | NC_002509 | Jenner et al. (2000) |
| JYMV | M | Dioscorea japonica | Japan | 9760 | NC_000947 | Chen et al. (2006) |
| PWV | MU2 | Passiflora caerulea | Australia | 9682 | NC_014790 | Wylie and Jones (2012) |
aVirus Acronyms: CEV-A Cyrtanthus elatus virus A, JYMV Japanese yam mosaic virus, NLSYV narcissus late season yellows virus, NYSV narcissus yellow stripe virus, ScaMV scallion mosaic virus, TuMV turnip mosaic virus, PWV passion fruit woodiness virus
bVirus isolates abbreviated for the present study to avoid confusion
Table 2.
Percent identity of CEVA-NBRI16 isolate at the level of nucleotide and its deduced amino acid sequence of full-length genome and polyprotein, respectively, with other full length potyviruses (for which complete genome sequences are available)
| GenBank accession | Virus | Isolate | Location | Percent identity | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Complete genome (polypeptide) | Open reading frames | ||||||||||||||
| P1 | HC-Pro | P3 | PIPO | 6K1 | CI | 6K2 | VPg | NIa | NIb | CP | |||||
| NC_017977 | CEVA | Marijiniup7-1 | Australia | 93 (94) | 97 (95) | 97 (96) | 97 (94) | – | 92 (86) | 97 (96) | 87 (78) | 96 (90) | 99 (97) | 88 (93) | 84 (94) |
| JQ723475 | CEVA | Marijiniup7-2 | Australia | 93 (94) | 97 (95) | 97 (96) | 97 (94) | – | 92 (86) | 97 (96) | 87 (78) | 96 (90) | 99 (97) | 88 (93) | 84 (94) |
| AM158908 | NYSV | Zhangzhou-1 | China | 90 (97) | 99 (100) | 86 (93) | 98 (99) | 99 (100) | 99 (100) | 83 (96) | 77 (90) | 87 (97) | 95 (98) | 92 (96) | 95 (97) |
| NC_011541 | NYSV | Zhangzhou-2 | China | 90 (97) | 99 (100) | 86 (93) | 98 (99) | 99 (100) | 99 (100) | 83 (96) | 77 (90) | 87 (97) | 95 (98) | 92 (96) | 95 (97) |
| JQ911732 | NYSV | ZZ-2 | China | 59 (78) | 60 (82) | 63 (83) | 60 (66) | 56 (83) | 52 (63) | 70 (88) | 61 (71) | 68 (82) | 69 (86) | 67 (84) | 71 (82) |
| JQ395042 | NYSV | Marijiniup 3 | Australia | 58 (78) | 35 (42) | 67 (84) | 54 (59) | 53 (81) | 69 (84) | 70 (88) | 60 (83) | 65 (85) | 72 (90) | 69 (85) | 71 (81) |
| JQ326210 | NLSYV | Zhangzhou | China | 55 (74) | 33 (38) | 65 (81) | 55 (66) | 51 (77) | 71 (78) | 65 (86) | 64 (77) | 71 (87) | 71 (85) | 63 (80) | 68 (76) |
| KC691259 | NLSYV | Marijiniup8 | Australia | 54 (75) | 32 (36) | 64 (81) | 56 (66) | 49 (69) | 63 (78) | 67 (85) | 68 (81) | 67 (86) | 66 (84) | 64 (79) | 68 (75) |
| JX156421 | NLSYV | Marijiniup9 | Australia | 54 (75) | 35 (39) | 67 (81) | 56 (66) | 49 (69) | 73 (78) | 65 (86) | 66 (79) | 69 (87) | 68 (85) | 65 (79) | 68 (75) |
| AJ316084 | ScaMV | – | China | 43 (63) | 25 (22) | 56 (65) | 41 (50) | 44 (64) | 60 (67) | 53 (72) | 45 (54) | 61 (71) | 57 (71) | 58 (71) | 58 (69) |
| NC_003399 | ScaMV | – | China | 43 (63) | 25 (22) | 56 (65) | 41 (50) | 44 (64) | 60 (67) | 53 (72) | 45 (54) | 61 (71) | 57 (71) | 58 (71) | 58 (69) |
| AB701735 | TuMV | TIGD | – | 42 (59) | 15 (15) | 58 (66) | 25 (36) | 41 (60) | 56 (76) | 55 (71) | 48 (50) | 53 (67) | 60 (73) | 62 (73) | 64 (67) |
| AB989658 | TuMV | NZ403B | New Zealand | 39 (60) | 21 (23) | 55 (66) | 30 (36) | 40 (58) | 64 (78) | 55 (71) | 54 (50) | 54 (67) | 59 (73) | 59 (74) | 60 (66) |
| NC_002509 | TuMV | – | – | 41 (59) | 20 (19) | 53 (66) | 31 (34) | 40 (58) | 64 (76) | 55 (70) | 50 (58) | 56 (69) | 57 (71) | 55 (73) | 65 (67) |
| NC_000947 | JYMV | – | – | 40 (57) | 19 (19) | 53 (63) | 40 (38) | 39 (62) | 58 (67) | 55 (68) | 52 (43) | 55 (65) | 50 (60) | 55 (69) | 56 (64) |
| NC_014790 | PWV | PWV-MU2 | Australia | 25 (42) | 13 (13) | 35 (45) | 19 (20) | 10 (40) | 23 (32) | 44 (54) | 40 (35) | 37 (47) | 37 (51) | 44 (57) | 50 (59) |
Fig. 2.
a Schematic representation of full length genome of narcissus isolate CEVA-NBRI16. Arrow represents the orientation of ORFs present in virus genome. The numbers above the ORF indicate their starting site, while the length (amino acids) and predicted molecular weight (in kDa) of ORF are shown below. b Phylogenetic tree showing closest relationship of CEVA-NBRI16 isolates with CEVA-Marijiniup7-1 and Marijiniup7-2. Tree was constructed employing MEGA v6.1 tool using Maximum-likelihood method based on the Tamura–Nei model with 1000 bootstrap replicates. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site and the percentage of trees in which the associated taxa clustered together is shown next to the branches. Bar at the bottom represents the nucleotide substitutions per site
Phylogenetic relationship of NBRI16 isolate under study was inferred with other narcissus infecting full-length potyvirus sequences (Table 1) using maximum-likelihood method and the phylogram was obtained using Tamura-Nei model in MEGA v6.1 program (Tamura et al. 2013). Phylogeny revealed close relationship of NBRI16 isolate with CEVA and grouped it with CEVA-Marijiniup7-1 and Marijiniup7-2 (Fig. 2b) forming a discrete cluster. Phylogeny revealed its distant relationship with narcissus infecting Narcissus yellow stripe virus (NYSV) and Narcissus late season yellows virus (NLSYV) isolates reported from Australia (Wylie et al. 2010).
Conclusion
This study reports the first full-length genomic sequence of CEVA isolate of N. tazetta from India. The information on complete genome will allow the characterization and identification of other CEVA isolates and assist to study the occurring genetic diversity. Since N. tazetta is prone to infection with other virus also (Chen et al. 2003; Pearson et al. 2009), CEVA may not be the sole agent causing the chlorotic stripe disease and required further advanced unbiased studies like Illumina sequencing (Wylie and Jones 2011) to fish out the other potyvirus species. The vegetative propagation of CEVA-infected bulbs along with untrained culture practices may be the reason for viral disease dissemination and may exacerbate the cultivation problem in narcissus and other vegetatively propagating plant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (DOCX 207 kb)
Acknowledgements
This study was funded by the Council of Scientific and Industrial Research, New Delhi, India (Grant Number BSC-0117). RR is thankful to University Grant Commission for Rajiv Ghandi National fellowship.
Compliance with ethical standards
Conflict of interest
There is no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1189-z) contains supplementary material, which is available to authorized users.
References
- Aminuddin Khan JA, Raj SK. Association of an unknown potyvirus isolate with severe mosaic of Narcissus tazetta L. Indian J Exp Biol. 1999;37:1034–1036. [Google Scholar]
- Brunt AA. Some hosts and properties of narcissus latent virus, a carlavirus commonly infecting narcissus and bulbous iris. Ann Appl Biol. 1977 [Google Scholar]
- Brunt AA. Narcissus. In: Loebenstein G, Lawson RH, Brunt AA, editors. Virus and virus-like diseases of bulb and flower crops. Chichester: Wiley; 1995. pp. 322–334. [Google Scholar]
- Chen J, Zheng HY, Chen JP, Adams MJ. Characterisation of a potyvirus and a potexvirus from Chinese scallion. Arch Virol. 2002;147:683–693. doi: 10.1007/s007050200018. [DOI] [PubMed] [Google Scholar]
- Chen J, Chen JP, Langeveld SA, Derks AFLM, Adams MJ. Molecular characterization of carla- and potyviruses from narcissus in China. J Phytopathol. 2003;151:1–4. doi: 10.1046/j.1439-0434.2003.00674.x. [DOI] [Google Scholar]
- Chen J, Lu YW, Shi YH, Adams MJ, Chen JP. Complete nucleotide sequence of the genomic RNA of Narcissus yellow stripe virus from Chinese narcissus in Zhangzhou city, China. Arch Virol. 2006;151:1673–1677. doi: 10.1007/s00705-006-0788-x. [DOI] [PubMed] [Google Scholar]
- Fienberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Ann Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gibbs A, Mackenzie A. A primer pair for amplifying part of the genome of all potyvirids by RT-PCR. J Virol Methods. 1997;63:9–16. doi: 10.1016/S0166-0934(96)02103-9. [DOI] [PubMed] [Google Scholar]
- Grisoni M, Moles M, Farreyrol K, Rassaby L, Davis R, Pearson M. Identification of Potyviruses infecting vanilla by direct sequencing of a short RT-PCR amplicon. Plant Pathol. 2006;55:523–529. doi: 10.1111/j.1365-3059.2006.01397.x. [DOI] [Google Scholar]
- Ha C, Coombs S, Revill PA, Harding RM, Vu M, Dale JL. Design and application of two novel degenerate primer pairs for the detection and complete genomic characterization of potyviruses. Arch Virol. 2008;153:25–36. doi: 10.1007/s00705-007-1053-7. [DOI] [PubMed] [Google Scholar]
- Jenner CE, SanchezF Nettleship SB, Foster GD, Ponz F, Walsh JA. The cylindrical inclusion gene of Turnip mosaic virus encodes a pathogenic determinant to the Brassica resistance gene TuRB01. Mol Plant Microbe Interact. 2000;13(10):1102–1108. doi: 10.1094/MPMI.2000.13.10.1102. [DOI] [PubMed] [Google Scholar]
- Kumar S, Raj R, Kaur C, Raj SK, Roy RK. First report of Cyrtanthus elatus virus A in Narcissus tazetta in India. Plant Dis. 2015;99:1658. doi: 10.1094/PDIS-04-15-0492-PDN. [DOI] [Google Scholar]
- Lin SQ, Shen JG, Gao FL, Cai W, Huang Z, Xie LY, Wu ZJ. Complete genome sequence of narcissus late season yellows virus infecting Chinese narcissus in China. Arch Virol. 2012;157(9):1821–1824. doi: 10.1007/s00705-012-1328-5. [DOI] [PubMed] [Google Scholar]
- Lucinda N, Inoue-Nagata AK, Kitajima EW, Nagata T. Complete genome sequence of Brugmansia suaveolens mottle virus, a potyvirus from an ornamental shrub. Arch Virol. 2010;155:1729–1732. doi: 10.1007/s00705-010-0798-6. [DOI] [PubMed] [Google Scholar]
- Marie-Jeanne V, Loos R, Peyre J, Alliot B, Signoret P. Differentiation of Poaceae potyviruses by reverse transcription polymerase chain reaction and restriction analysis. J Phytopathol. 2000;148:141–151. doi: 10.1046/j.1439-0434.2000.00473.x. [DOI] [Google Scholar]
- Nguyen HD, Tomitaka Y, Ho SYW, Duchêne S, Vetten H-J, Lesemann D, Walsh JA, Gibbs AJ, Ohshima K. Turnip mosaic potyvirus probably first spread to Eurasian brassica crops from wild orchids about 1000 years ago. PLoS One. 2013;8(2):e55336. doi: 10.1371/journal.pone.0055336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson MN, Cohen D, Cowell SJ, Jones D, Blouin A, Lebas BSM, Shiller JB, Clover GRC. A survey of viruses of flower bulbs in New Zealand. Aust Plant Pathol. 2009;38:305–309. doi: 10.1071/AP09006. [DOI] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 60. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie SJ, Jones MGK. The complete genome sequence of a Passion fruit woodiness virus isolate from Australia determined using deep sequencing, and its relationship to other potyviruses. Arch Virol. 2011;156:479–482. doi: 10.1007/s00705-010-0845-3. [DOI] [PubMed] [Google Scholar]
- Wylie SJ, Jones MGK. Complete genome sequences of seven carlavirus and potyvirus isolates from Narcissus and Hippeastrum plants in Australia, and proposals to clarify their naming. Arch Virol. 2012;157:1471–1480. doi: 10.1007/s00705-012-1319-6. [DOI] [PubMed] [Google Scholar]
- Wylie SJ, Nouri S, Coutts BA, Jones MGK. Narcissus late season yellows virus and Vallota speciosa virus found infecting domestic and wild populations of Narcissus species in Australia. Arch Virol. 2010;155:1171–1174. doi: 10.1007/s00705-010-0682-4. [DOI] [PubMed] [Google Scholar]
- Wylie SJ, Li H, Sivasithamparam K, Jones MGK. Complete genome analysis of three isolates of Narcissus late season yellows virus and two of Narcissus yellow stripe virus: three species or one? Arch Virol. 2014;159:1521–1525. doi: 10.1007/s00705-013-1969-z. [DOI] [PubMed] [Google Scholar]
- Yadav N, Khan JA. Identification of a potyvirus associated with mosaic disease of Narcissus sp. in India. New Dis Rep. 2007;14:56. [Google Scholar]
- Yakoubi S, Lecoq H, Desbiez C. Algerian watermelon mosaic virus (AWMV): a new potyvirus species in the PRSV cluster. Virus Genes. 2008;37:103–109. doi: 10.1007/s11262-008-0237-x. [DOI] [PubMed] [Google Scholar]
- Yasaka R, Ohba K, Schwinghamer M, Fletcher J, Ochoa-Corona F, Thomas J, Ho S, Gibbs A, Ohshima KJ. Phylodynamic evidence of the migration of turnip mosaic potyvirus from Europe to Australia and New Zealand. J Gen Virol. 2015;96(3):701–713. doi: 10.1099/jgv.0.000007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (DOCX 207 kb)