Abstract
Hepatitis B virus (HBV) can cause chronic hepatitis B, which may lead to cirrhosis and liver cancer. Type I interferon (IFN) is an approved drug for the treatment of chronic hepatitis B. However, the fundamental mechanisms of antiviral action by type I IFN and the downstream signaling pathway are unclear. TRIM25 is an IFN-stimulated gene (ISG) that has an important role in RIG-I ubiquitination and activation. Whether TRIM25 is induced in liver cells by type I IFN to mediate anti-HBV function remains unclear. Here we report that interleukin-27 (IL-27) has a critical role in IFN-induced TRIM25 upregulation. TRIM25 induction requires both STAT1 and STAT3. In TRIM25 knockout HepG2 cells, type I IFN production was consistently attenuated and HBV replication was increased, whereas overexpression of TRIM25 in HepG2 cells resulted in elevated IFN production and reduced HBV replication. More interestingly, we found that TRIM25 expression was downregulated in HBV patients and the addition of serum samples from HBV patients could inhibit TRIM25 expression in HepG2 cells, suggesting that HBV might have involved a mechanism to inhibit antiviral ISG expression and induce IFN resistance. Collectively, our results demonstrate that type I IFN -induced TRIM25 is an important factor in inhibiting HBV replication, and the IFN-IL-27-TRIM25 axis may represent a new target for treating HBV infection.
Keywords: HBV, IL-27, STAT1, STAT3, TRIM25
HIGHLIGHTS
Induction of TRIM25 by type I IFN requires IL-27 signaling.
Both transcription factors STAT1 and STAT3 have a role in inducing TRIM25.
IL-27-dependent induction of TRIM25 inhibits HBV replication.
HBV infection attenuates TRIM25 expression.
Introduction
Despite severe side effects, type I interferon (IFN) treatment remains an antiviral option for treating chronic hepatitis B. Type I IFNs (IFN-α, IFN-β, IFN-ε and others) have been recognized as the major antiviral cytokine in vertebrates,1 constituting one of the most important innate immune responses to viral infection including hepatitis B virus (HBV) and hepatitis C virus infection. The antiviral function of type I IFN is carried out through its binding to the type I IFN receptor (IFNAR1, 2), the activation of the JAK/STAT pathway and the subsequent induction of a large number of IFN-stimulated genes (ISGs),2 which can inhibit different stages of the viral life cycle including entry, replication, assembly and budding.3, 4 However, due to co-evolution, many viruses have acquired the ability to counter the antiviral response induced by type I IFN. A thorough understanding of the interactions between host cells and HBV will improve antiviral treatment.
Classically, type I IFN stimulation leads to the formation and activation of the STAT1/2 heterodimer, and the dimer finally moves into the nucleus and binds to IFN-stimulated response elements (ISREs) together with IRF9. In addition, STAT homodimers or heterodimers formed from different combinations of STAT1, 3, 4, 5 or 6 during IFN signaling can also have roles in binding to ISREs or IFN-activated site, which could further induce ISG production.5, 6 In a sequential response, IFN induced IL-27 and then activated STAT3.7 Another report showed that in Doudi cells, which are sensitive to IFN treatment, the induction of CXCL11 by IFN depends on recruiting the transcriptional activators p65 and IRF1 through STAT3.8 These studies suggested that STAT3 may be involved in regulating the production of certain ISGs. It is not clear whether STAT3 is required to regulate ISGs production during HBV infection.
IL-27 is primarily produced by innate immune cells and can suppress T-cell immunity, including the inhibition of Th17 and Th1 differentiation in several infection models.9, 10, 11 Another function of IL-27 is to regulate transcription activity. For example, IFN-induced IL-27 secretion was responsible for both STAT1 and STAT3 activation.7, 12 In hepatic cells, IL-27 was reported to modulate CXCL9, CXCL10 and CXCL11 chemokines.13 The biological function of IL-27 in regulating ISGs induction in HBV infection is largely unknown.
The tripartite motif family (TRIM) is a protein family and is known for ring-finger E3 ubiquitin ligase activity. All family members of TRIM proteins comprise a RING domain, one or two B-box domains and an associated coiled-coil domain in the amino-terminal region.14, 15, 16 Many TRIM members are IFN -inducible genes and are important for restricting viral infection.17, 18, 19 As an important member of the TRIM family, TRIM25 was previously shown to inhibit HBV replication20; however, the mechanisms remain undefined. TRIM25 has been suggested to promote IFN-β production by activating the RIG-I signaling pathway in mouse embryonic fibroblast (MEF) cells.21 In this study, we investigated the interplays between TRIM25 and IFN signaling during HBV infection. We found that enforced expression of TRIM25 in HepG2 cells induces type I IFN expression, which could further suppress HBV replication. Interestingly, TRIM25 expression was attenuated in the peripheral blood mononuclear cells (PBMCs) of HBV patients; in addition, HBV-positive serum also downregulated TRIM25 expression in HepG2 cells. These findings may explain the resistance to IFN treatment observed in some HBV patients. IFN-TRIM25 regulatory circuitry may warrant exploration in optimizing IFN treatment.
Materials and methods
Cell culture, plasmids and reagents
Human embryonic kidney cell line HEK293T, HepG2.2.15 and HepG2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% inactivated fetal bovine serum, penicillin (100 IU/ml) and streptomycin (100 mg/ml) at 37 °C with 5% CO2. For mice bone-marrow-derived macrophage (BMM) differentiation, bone marrow cells were collected from wild-type IL27R1−/−mice and differentiated in DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin and 10 ng/ml of M-CSF for 7 days. The IL27R1−/−mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA), stock no: 018078. Antibodies against tubulin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against p-STAT3 (705), STAT3, STAT1 and TRIM25 were supplied by Cell Signaling Technology (Danvers, MA, USA). Antibody against GAPDH was obtained from Proteintech (Wuhan, Hubei, China). Human IL27 recombinant protein was supplied by eBioscience (San Diego, CA, USA) and IL-27-neutralizing antibody was from R&D SYSTEMS (Minneapolis, MN, USA). polydAdT and polyIC were purchased from Invivogen (San Diego, CA, USA). The expression construct of TRIM25 was generated by cloning the coding region sequence of human TRIM25 into VR1012 expression vector, the pHBV1.3 plasmids was kindly supplied by Dr Lishan Su in University of North Carolina.
RNA extraction and quantitative real-time PCR (Q-PCR)
Total RNA was extracted from cells using an EasyPure RNA kit (TransGen, Beijing, China) and then converted to first-strand cDNA using TransScript First-Strand cDNA Synthesis SuperMix (TransGen). HBV DNA was isolated from cells according to the kit instructions (TransGen). Housekeeping gene GAPDH was used as an internal control for quantitation, and gene expression was quantified as previously described.22 The sequences of gene-specific primers used for Q-PCR are shown in Supplementary Table S1.
Immunoblotting
Immunoblotting was conducted as previously described.23 In brief, the whole cell was lysed by adding lysis buffer (20 mM HEPES, 350 mM NaCl, 20% glycerol, 1% NP40, 1 mM MgCl2, 0.5 mM EDTA, 0.1 mM EGTA and 0.5 mM dithiothreitol) together with protease inhibitors and incubated on ice for 30 min by tapping every 10 min. The protein concentration was qualified using Coomassie Plus protein assay reagent (Thermo Scientific, Rockford, IL, USA). The quantitation of immunoblotting band intensity was carried out using ChemiDoc XRS+ Molecular Imager software (Bio-Rad, Philadelphia, PA, USA).
Luciferase assay
HepG2 cells were first transfected with Dual-Luciferase construct harboring ISRE (pGL4.45) or IFNβ target sequence. After 8 h, the cells were then transfected with polyIC, polydAdT or pHBV1.3 plasmid; 16 h later, the cells were lysed with passive lysis buffer and the activity of Firefly Luciferase and Renilla Luciferase in the lysates were measured with the dual-Luciferase Assay System (Promega, Madison, WI, USA).
Eenzyme-linked immunosorbent assay
Cells were mock-transfected or transfected with TRIM25 expression plasmid. After 48 h, cells were exposed to IFNα (10 ng/ml) for 24 h. The supernatant was collected and subjected to enzyme-linked immunosorbent assay following the kit instructions (Kehua Shengwu, Shanghai, China).
CRISPR/Cas9 knockout
HepG2 or 293T cells were seeded in a 24-well plate; 16 h later, plasmids expressing Cas9 and TRIM25 or STAT1 or STAT3 single-guide RNA (sgRNA) and a plasmid with puromycin selection marker were co-transfected into cells using Viafect transfection reagent (Promega). At 36 h after transfection, cells were either selected by adding puromycin at a concentration of 2 μg/ml or subjected to immunoblotting with TRIM25-, IL27R1-,STAT1- or STAT3-specific antibodies (Cell Signaling). Two days later, living cells were diluted on a 96-well plate at ~1 cell per well. Immunoblotting was performed again to ensure that the gene knockout (KO) results after the clones were formed. DNA sequencing was performed to further confirm the gene KO results. SgRNA for CRISPR KO are shown in Supplementary Table S1.
Patient samples
Twenty-nine HBV-infected patients and 29 healthy subjects were enrolled in this study (Supplementary Table S2). Venous blood was withdrawn to prepare serum and PBMCs. These studies were approved by the institutional review board of Jilin University, The First Hospital.
Statistical analysis
The results were presented as the means±s.d. and analyzed with Student’s t-test. P<0.05 was considered statistically significant.
Results
TRIM25 was upregulated by IFN treatment in both 293T and HepG2 cells
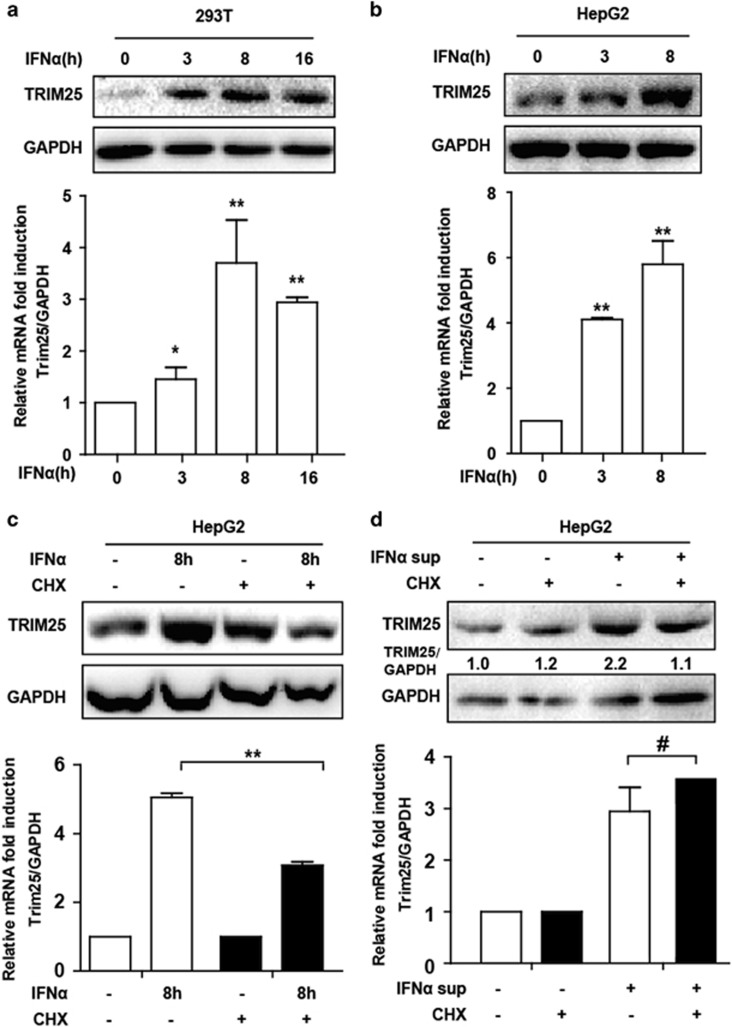
IFN antiviral function is achieved by inducing ISGs expression, which directly or indirectly inhibits different stages of the viral life cycle.3 Many TRIM proteins are also ISGs and suppress viral replication.24, 25 To investigate TRIM’s response to IFN stimulation, we examined a group of TRIM gene messenger RNAs (mRNAs), including TRIM25 with Q-PCR in IFNα-treated HepG2 cells (Supplementary Figure S1). TRIM22 expression was the highest among a panel of TRIM genes including TRIM25 in response to IFN treatment. TRIM22 was reported to inhibit HBV replication by suppressing HBV core promoter activity, whereas TRIM25 was known to mediate the ubiquitination and activation of RIG-I, which is not only an RNA sensor but also functions as a virus sensor in HBV infection.21, 26 Interestingly, TRIM25 mRNA was also significantly induced in 293T cells (Figure 1a). In addition, we performed western blot to detect the protein level. As expected, TRIM25 protein was elevated after IFNα treatment (Figures 1a and b). The reported TRIM25 antiviral response and elevated TRIM25 expression to IFN treatment prompted us to select TRIM25 for further investigation. Interestingly, the upregulation of TRIM25 by IFNα was significantly countered at the protein level when cycloheximide (CHX: a protein synthesis inhibitor that do not affect the mRNA synthesis) was added, and TRIM25 mRNA upregulation was also significantly inhibited, suggesting that IFN-induced protein expression was important in TRIM25 mRNA induction. However, CHX inhibition of TRIM25 induction appeared to wane when we treated the cells with supernatant from IFNα-stimulated HepG2 cells (Figures 1c and d), which indicated that the supernatant may contain the secreted IFNα-induced cytokines that facilitated TRIM25 expression. Collectively, these results indicated that TRIM25 expression was significantly increased in cells to respond to IFN stimulation.
Figure 1.
TRIM25 was upregulated by IFN treatment in both 293T and HepG2 cells. (a, b) 293T and HepG2 cells were seeded in a 24-well plate. Cells were treated with IFNα (10 ng/ml) as indicated. The treated cells were divided into two groups. One was subjected to immunoblotting using TRIM25 and GAPDH antibody, and the other was subjected to total RNA extraction. The mRNAs were detected using Q-PCR. Data represent the means and s.d. from three independent experiments. Student’s t-test was performed. *P<0.05,**P<0.01. (c) HepG2 cells were treated with IFNα together with CHX (10 μg/ml) or without for 8 h as indicated. Q-PCR and western blotting were performed to analyze the expression of TRIM25. (d) HepG2 cells were IFN-treated for 16 h, and supernatants were collected to add to newly seeded HepG2 cells as indicated. Cells were collected after 8 h, and Q-PCR and western blotting were performed to analyze the expression of TRIM25. Data represent the means and s.d. from three independent experiments. Student’s t-test was performed. *P<0.05, **P<0.01, #P>0.05.
IL-27 was critical for TRIM25 induction
Next, we investigated that cytokine was associated with TRIM25 induction. We previously studied the regulatory functions of IL-10 and IL-27,7, 27 and questioned if both cytokines are required. We found that both IL-10 basic expression and responsive induction were very low in HepG2 cells, whereas IL-27 was normally expressed and was elevated by IFN treatment (Figure 2a), suggesting that IL-27 was a likely candidate. To test whether IL-27 is required for TRIM25 induction, we treated the cells with IL-27-neutralizing antibody and TRIM25 induction by IFN, which was blocked at both the mRNA and protein levels (Figures 2b and c), indicating that IL-27 in the supernatant was important for TRIM25 induction by IFN. To further verify this relationship, IL27R was knocked out in 293T and HepG2 cells by the CRISPR/Cas9 system (Figure 2d). As expected, TRIM25 induction was blocked in both 293T and HepG2 IL27R KO cells (Figures 2e and f). In addition, given that BMMs are responsive to type I IFN and are the main source of the type I IFN-inducible IL-27,7, 11 we tested TRIM25 induction in wild-type and IL27R1−/− BMMs, and type I IFN-induced TRIM25 was also significantly inhibited in BMMs IL27R KO cells (Figure 2g), suggested that the IFN-IL27-TRIM25 axis was not cell-specific. We further investigated whether IL27 could directly induce TRIM25 expression, as expected, both TRIM25 mRNA and protein level were upregulated by recombinant IL27 protein stimulation, and interestingly, both TRIM25 basal and induction level was significantly reduced in STAT1 or STAT3 KO 293T cells(Figures 2h and i), which indicated that IFN-induced IL27 might elevate TRIM25 expression by activating STAT1 and STAT3. Collectively, all these data suggested that IL27 was critical for IFN-induced TRIM25 expression.
Figure 2.
IL-27 was critical for TRIM25 induction. (a) HepG2 cells were seeded in a 24-well plate and treated with IFNα (10 ng/ml) as indicated. Total RNA was extracted for IL-10 analysis; IL27 and TRIM22 expression levels were assessed using Q-PCR. (b). HepG2 cells were treated with IFNα for 8 h together with or without IL-27-neutralizing antibody as indicated. Q-PCR was performed to analyze TRIM25 expression. (c) Whole-cell lysate was immunoblotted with TRIM25 or GAPDH antibody using the same samples as in b. (d) HepG2 and 293T WT and IL27R1 KO cells were collected, and whole-cell lysate was immunoblotted with IL27R1 or GAPDH antibody as indicated. (e) WT or IL27R1 KO 293T cells were treated as indicated and immunoblotted with TRIM25 or GAPDH antibody. (f) WT and IL27R1KO HepG2 cells were treated and analyzed as in c. (g) WT or IL27R1KO BMM cells were treated with IFNα or β for 3 or 8 h or untreated, and TRIM25 expression was analyzed using Q-PCR. (h) WT, STAT1 KO and STAT3 KO 293T cells were treated with IL-27 (10 ng/ml) as indicated, RNA was extracted and Q-PCR was performed to analyze TRIM25 expression. (i) WT, STAT1 KO and STAT3 KO 293T cells were treated with IL-27 (10 ng/ml) as indicated, and whole-cell lysates were immunoblotted with the indicated antibodies. Data represent the means and s.d. from three independent experiments. Student’s t-test was performed. *P<0.05, **P<0.01.
Both STAT1 and STAT3 were required for TRIM25 induction by IFN
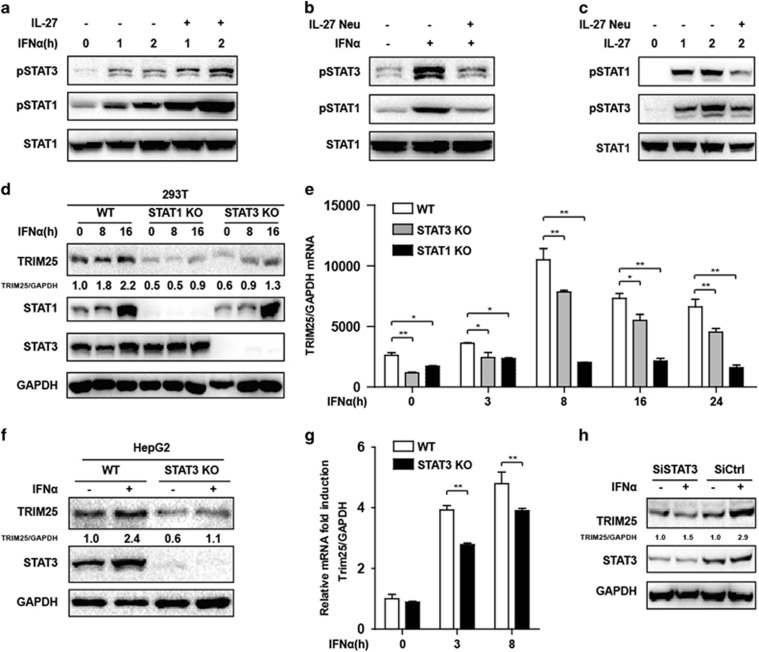
We further investigated the function of STAT1 and STAT3 in TRIM25 induction. As an ISG, we assumed that STAT1 is a major transcriptional factor in TRIM25 transcription; however, IL-27 was also reported to activate STAT3 via JAK-mediated phosphorylation in T cells28 and liver cells;29 consistently with previous reports, IL-27 protein promoted both IFN-induced STAT1 and STAT3 activation in HepG2 cells. The activation of STAT1 and STAT3 was inhibited by IL-27-neutralizing antibody upon IFN or IL-27 recombinant protein retreatment (Figures 3a–c). STAT3 was activated in IFN-treated HepG2 cells at different time points (Supplementary Figure S2a), and STAT3 activation was completely blocked by the STAT3-specific inhibitor Stattic (Supplementary Figure S2b). In addition, type I IFN-induced TRIM25 expression was significantly inhibited by Stattic (Supplementary Figures S2c and d). To confirm our result, we used the CRISPR KO technique to KO STAT1 and STAT3 in 293T cells. As expected, STAT1 KO blocked the induction of TRIM25 at the mRNA and protein levels, and STAT3 KO also inhibited TRIM25 mRNA and protein production (Figures 3d and e). A similar pattern was observed in STAT3 KO HepG2 cells (Figures 3f and g). Furthermore, small intefering RNA-mediated STAT3 knockdown in HepG2 cells also inhibited IFNα-induced TRIM25 production (Figure 3h). The basal levels of TRIM25 in both 293T and HepG2 cells were reduced after STAT3 KO or knockdown. Taken together, both STAT1 and STAT3 were indispensable for the upregulation of TRIM25 in type I IFN treatment.
Figure 3.
Both STAT1 and STAT3 were required for the IFN-induced TRIM25. (a) HepG2 cells were treated with IFNα together with or without IL-27 protein as indicated. Western blots were performed to analyze the activation of STAT1 and STAT3. (b) HepG2 cells were treated and collected as indicated, and western blots were performed to analyze the activation of STAT1 and STAT3. (c) HepG2 cells were treated by IL-27 with or without IL-27-neutralizing antibody, and analyzed as in b. (d) WT, STAT1 KO and STAT3 KO 293T cells were treated with IFNα (10 ng/ml) as indicated, and whole-cell lysates were immunoblotted with the indicated antibodies. (e) WT, STAT1 KO and STAT3 KO 293T cells were treated with IFNα (10 ng/ml) as indicated, RNA was extracted and Q-PCR was performed to analyze TRIM25 expression. (f) HepG2 WT and STAT3 knockout cells were treated with IFNα (10 ng/ml) for 3 h as indicated and analyzed with the indicated antibodies. (g) HepG2 WT and STAT3 knockout cells were treated with IFNα (10 ng/ml) or untreated. mRNA levels at 3 or 8 h after IFNα (10 ng/ml) treatment were analyzed with qRT-PCR. (h) HepG2 cells were transfected with STAT3 small interfering RNA or siCtrl. At 36 h later, the cells were treated with IFNα for 8 h and whole-cell lysates were immunoblotted with antibodies as indicated. The fold change in relative mRNA expression was expressed as the means±s.d. *P<0.05, **P<0.01.
TRIM25 KO in HepG2 cells facilitated HBV replication
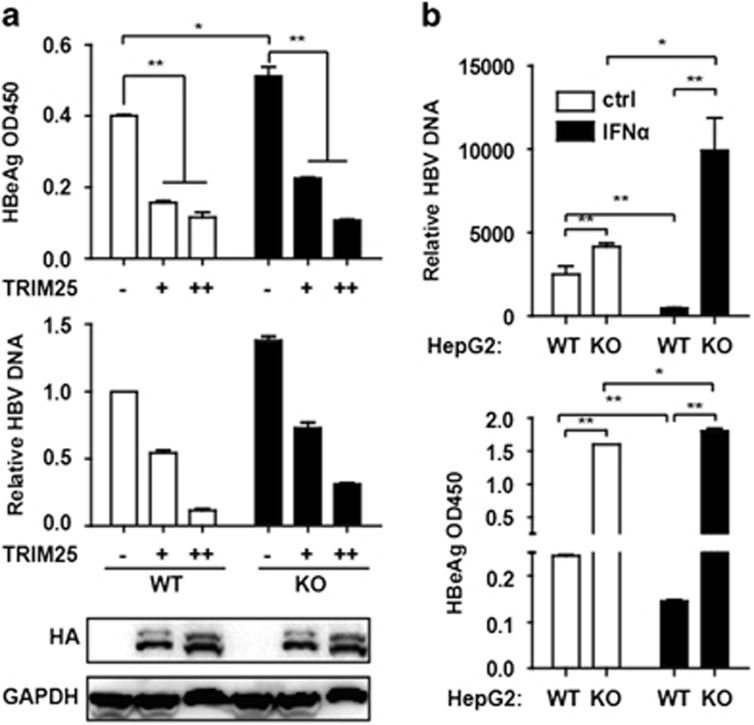
We found that the overexpression of TRIM25 with the HA tag inhibited HBeAg secretion and HBV DNA replication (Figure 4a) in HepG2 cells. To confirm the suppressive effect of TRIM25 on HBV replication, TRIM25 was completely knocked out in HepG2 cells through the CRISPR/Cas9 systems (Supplementary Figure S3a). HBeAg and HBV DNA levels were significantly increased in TRIM25 KO HepG2 cells compared with WT cells, which suggested the inhibition of HBV replication by TRIM25 (Figures 4a and b). We further investigated whether the overexpression of TRIM25 could inhibit HBV replication in HepG2.2.15 cells in which HBV keeps replicating. We found that HBeAg and HBsAg were significantly reduced in the supernatant of the overexpressed TRIM25 cells (Supplementary Figure S3b). Interestingly, HBeAg in the supernatant of pHBV1.3-transfected TRIM25 KO HepG2 cells was increased after IFNα treatment, and HBV DNA in the cells was also induced by IFNα (Figure 4b). These data indicate that the upregulation of TRIM25 was an important mechanism for mediating anti-HBV response upon IFN treatment.
Figure 4.
TRIM25 knockout in HepG2 cells promoted HBV replication. (a) HepG2 WT and TRIM25 KO cells were transfected with pHBV1.3 or co-transfected with pHBV 1.3 and different amounts of TRIM25 as indicated. At 72 h, the supernatant was collected and cells were subjected to immunoblotting with antibodies as indicated. HBV DNA and HBeAg in the supernatant were analyzed by Q-PCR or enzyme-linked immunosorbent assay (ELISA). (b) HepG2 WT and TRIM25KO cells were transfected with pHBV1.3. At 48 h, the cells were treated with IFNα or untreated, and after 24 h treatment, the supernatant was collected, and the HBV DNA and HBeAg in the supernatant were analyzed using Q-PCR or ELISA. Data represent the means and s.d. from three independent experiments. Student’s t-test was performed. *P<0.05, **P<0.01.
HBV replication increased in STAT3 KO cells
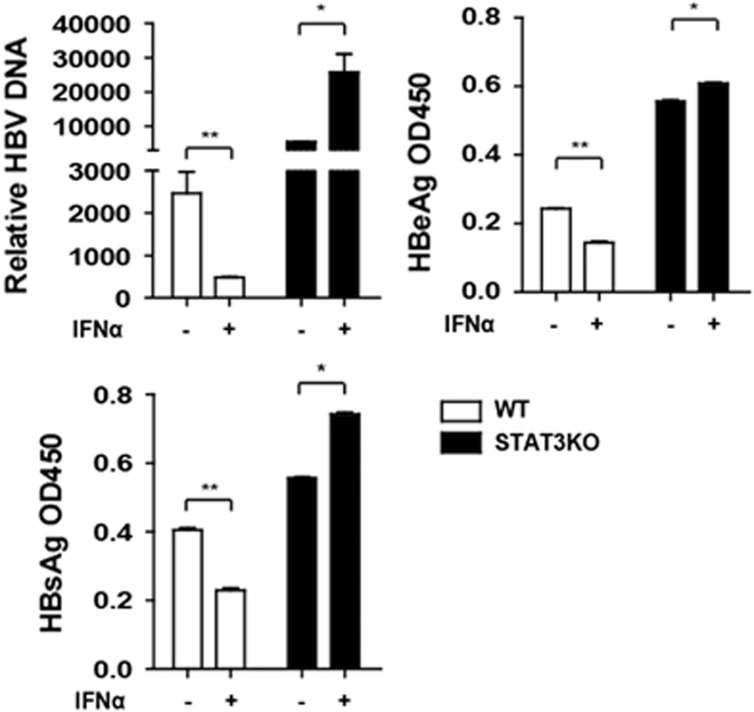
We previously showed that Apobec3G, an anti-HBV ISG, was STAT3- but not STAT1-dependent.30 We questioned the biological impact of STAT3 on HBV replication. Consistent with our previous report, we found that HBV DNA, HBeAg and HBsAg were significantly increased after STAT3 KO in HepG2 cells, and that HBV replication was enhanced by IFNα treatment (Figure 5a), indicating that STAT3 is required to inhibit HBV replication.
Figure 5.
HBV replication was increased in STAT3 knockout cells. HepG2 WT and STAT3KO cells were transfected with pHBV1.3. At 48 h, the cells were treated with IFNα or untreated, and after 24 h treatment, the supernatant was collected and HBV DNA, HBsAg and HBeAg in the supernatant were analyzed using Q-PCR or enzyme-linked immunosorbent assay. Data represent the means and s.d. from three independent experiments. Student’s t-test was performed. *P<0.05, **P<0.01.
TRIM25 promoted type I IFN production in HepG2 cells
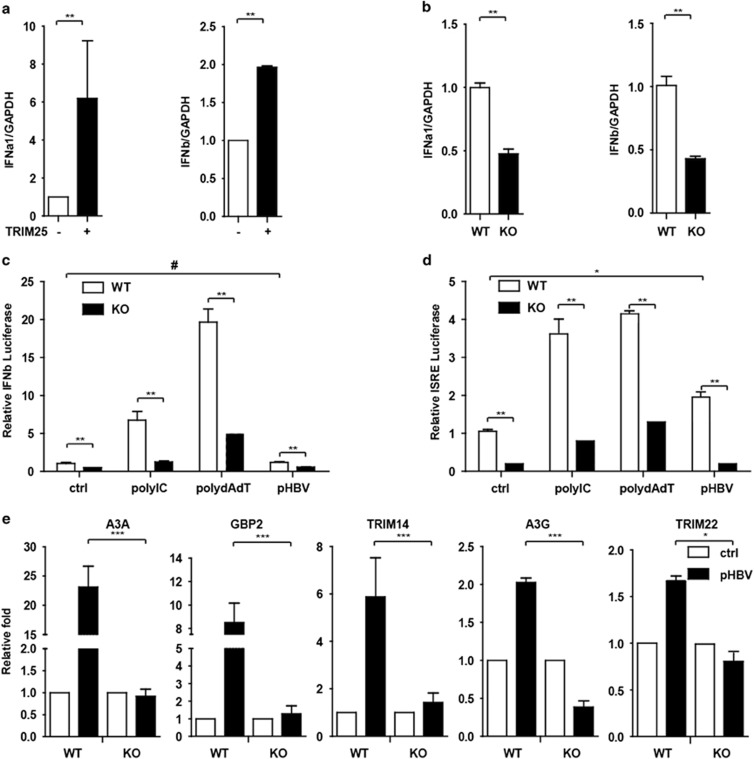
A previous report showed that IFNβ production was blocked in TRIM25−/− MEFs.21 To understand how TRIM25 inhibits HBV replication, we sought to determine whether TRIM25 increases IFN production. Notably, type I IFN was increased in TRIM25-overexpressed cells, and TRIM25 KO reduced the production of type I IFN in HepG2 cells (Figures 6a and b). In addition, we questioned whether TRIM25 KO could reduce downstream ISG expression in liver cells. As expected, after transfection with polydAdT, polyIC or pHBV1.3 plasmids, an IFNβ or ISRE-Luc reporter assay indicated that luciferase activity induction was significantly inhibited in TRIM25 KO HepG2 cells (Figures 6c and d), and the induction of ISGs by pHBV1.3 transfection was also significantly inhibited (Figure 6e).
Figure 6.
TRIM25 promoted type I interferon production in HepG2 cells. (a) HepG2 cells were transfected with TRIM25 or no transfection. IFNα, β and GAPDH mRNA levels at 36 h after transfection were analyzed with qRT-PCR, and the fold change in relative mRNA expression is shown as the means±s.d. (b) IFNα, β mRNA levels in HepG2 WT and TRIM25 KO cells were analyzed with qRT-PCR. (c, d) HepG2 WT and TRIM25 KO cells were transfected with IFNb-Luc or ISRE-Luc together with pGL4.7 TK-Luc reporter. After 8 h, cells were transfected with polyIC, polydAdT or pHBV1.3 plasmids and luciferase activity was quantified at 16 h after transfection. (e) HepG2 WT and TRIM25 KO cells were treated for 16 h and subjected to Q-PCR to analyze related gene expression as indicated. Data represent the means and s.d. from three independent experiments. Student’s t-test was performed. *P<0.05, **P<0.01, ***P<0.001.
HBV infection inhibited TRIM25 expression
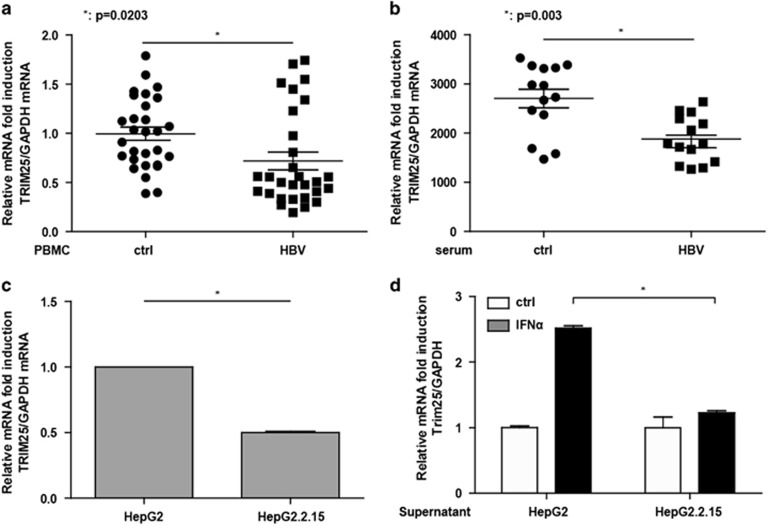
We next investigated whether HBV could inhibit TRIM25 expression in HBV patients. We found that TRIM25 expression in PBMCs isolated from HBV patients was significantly decreased compared with healthy controls (Figure 7a). To confirm this finding in liver cells, we found that the addition of HBV serum could remarkably inhibit TRIM25 expression in HepG2 cells (Figure 7b). In addition, the expression of TRIM25 in HepG2.2.15 cells in which HBV keeps replicating was significantly inhibited compared with HepG2 cells (Figure 7c). The induction of TRIM25 by IFNα stimulation was inhibited after the HepG2 cells were co-cultured with HepG2.2.15 supernatants (Figure 7d). HBV appears to resist IFN treatment by inhibiting TRIM25 expression.
Figure 7.
HBV infection inhibited TRIM25 expression. (a) Total RNA was extracted from PBMCs isolated from 28 healthy controls and 29 HBV patients. The TRIM25 expression level was analyzed using Q-PCR. (b) HepG2 cells were treated with serum from healthy controls or HBV patients for 24 h, and Q-PCR was performed to analyze the TRIM25 mRNA level. (c) Total RNA was extracted from HepG2 and HepG2.2.15 cells, and the TRIM25 expression level was analyzed using Q-PCR. (d) HepG2 cells were either treated with IFNa or untreated and either cultured with HepG2.2.15 supernatant or normal culture medium. After 8 h, the TRIM25 mRNA level was analyzed using Q-PCR. Data from three independent experiments were pooled and are shown as the means±s.d. *P<0.05, **P<0.01.
Discussion
In this study, we found: (1) IFN elevates TRIM25 expression via an IL-27-dependent regulatory loop, (2) both transcription factors STAT1 and STAT3 is required for the induction of TRIM25, (3) STAT3 is important for inhibiting HBV replication and (4) HBV infection/replication may inhibit TRIM25 expression. Our results provide a possible explanation for the poor response to IFN treatment in a portion of HBV-infected patients.
STAT3 is an important transcriptional factor in regulating ISGs expression and generally acts as a positive regulator.31 For instance, the upregulation of CXCL11 (an IFN-inducible gene) by IFN was blocked in STAT3 KO cells.8 However, STAT3 can also negatively regulate type I IFN -mediated antiviral response in MEFs.32 For example, the expression of PKR, OAS and IRF1, which are well-characterized ISGs, was increased in STAT3 KO MEFs. Thus, the impact of STAT3 on ISGs can be bi-directional. As shown by Ho and Ivashkiv,33 on the one hand, STAT3 downregulated the IFNα-mediated induction of inflammatory mediators such as the chemokines CXCL9 and CXCL10; on the other hand, IFNα-activated STAT3 supported the ISGF-3-dependent induction of antiviral genes.
Using CRISPR/Cas9 systems, we knocked out STAT1 and STAT3 in HepG2 cells. As expected, STAT1 KO blocked TRIM25 induction. Notably, TRIM25 upregulation was inhibited in STAT3-deficient HepG2 cells. Similarly, a treatment with a STAT3-specific inhibitor blocked the induction of TRIM25 by IFN stimulation. Collectively, both STAT1 and STAT3 are important for type I IFN -induced TRIM25 upregulation. In addition, our data show that HBV replication was not inhibited by IFN after STAT3 KO despite the continued expression of STAT1 in HepG2 cells. This suggested that STAT1 and STAT3 may function in tandem to deliver an antiviral response.30
In response to RNA virus, TRIM25 E3 ubiquitin ligase induces the Lys 63-linked ubiquitination of RIG-I, which is a key component utilized by the cytosolic RIG-I signaling pathway in mounting host antiviral innate immunity.21 RIG-I, an RNA sensor, could also act as an innate sensor and antiviral factor for HBV by counteracting HBV polymerase (P protein).26 In HIV infection, TRIM5α, another member of the TRIM family, can interact with incoming HIV capsid and send it for hydrolysis through the ubiquitin pathway. It would be interesting to investigate whether TRIM25 can exert direct anti-HBV activity. In this study, we showed that type I IFN was significantly reduced in TRIM25 KO HepG2 cells and that the overexpression of TRIM25 in HepG2 cells promoted IFN production. These data indicate that TRIM25 may also inhibit HBV replication by boosting type I IFN signaling and thereby further amplifying downstream anti-HBV ISGs production.
Unexpectedly, TRIM25 expression was significantly inhibited in PBMCs isolated from HBV-infected patients, and the addition of HBV patient serum inhibited TRIM25 expression in HepG2 cells. Our results suggest that HBV life cycle might utilize a mechanism inhibiting ISG expression. This finding was consistent with a previous report showing that HBV can suppress the TLR-induced antiviral activity in liver cells.34 Major vault protein (MVP) is a novel virus-induced host factor that upregulates type I IFN production. A recent report showed that HBV inhibited the MVP- and MyD88-induced activation of NF-κB and IFN-β by disrupting MVP/MyD88 interactions.35 In this study, IFNα was not reduced in the PBMCs of HBV patients; the TRIM25 reduction in HBV patients may show that HBV products inhibit the activation of some transcriptional factors including STAT1 and STAT3. Notably, both STAT1 and STAT3 were required for IFN -induced TRIM25 upregulation; thus, it is conceivable that HBV may inhibit TRIM25 by suppressing STAT1 and STAT3 activation. This understanding is consistent with our earlier study showing that HBVs could inhibit STAT3-S727 phosphorylation and further reduce APOBEC3G induction by IFN in HepG2 cells.30
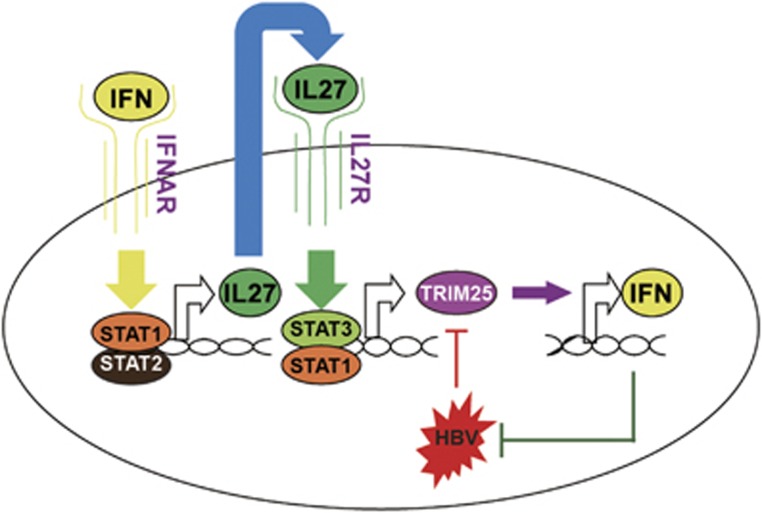
Based on these findings, we propose a working model for TRIM25 in mediating the crosstalk between type I IFN and HBV infection (Figure 8). In conclusion, we found that the type I IFNs induced IL-27, which activated STAT1 and STAT3 to promote TRIM25 gene expression. The elevated TRIM25 inhibits HBV replication by further inducing IFNα/β production. HBV inhibits IFN -induced TRIM25 expression both in vitro and in vivo. An increase in TRIM25 expression may benefit HBV treatment.
Figure 8.
Model depicting the crosstalk between type I Interferon and HBV through TRIM25.
Acknowledgments
This work was supported in part by the National Natural Science Foundation, China, Jilin Provincial Science and Technology Department of the Youth Fund Project, and Jilin University Bethune training program (grant no. 81401290, 20160520161JH, 470110000456 to GT). CAMS Initiative for Innovative Medicine (no. CAMS-I2M) to FXQ and GC.
Footnotes
Supplementary Information for this article can be found on the Cellular & Molecular Immunology website (http://www.nature.com/cmi)
The authors declare no conflict of interest.
Supplementary Material
References
- Mutimer DJ, Lok A. Management of HBV- and HCV-induced end stage liver disease. Gut 2012; 61 (Suppl 1): i59–i67. [DOI] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol 2014; 14: 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci USA 2012; 109: 4239–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 2013; 38: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 2005; 5: 375–386. [DOI] [PubMed] [Google Scholar]
- Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 2013; 41 (Database issue): D1040–D1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SS, Ghaffari AA, Cheng G. Lipopolysaccharide-mediated IL-10 transcriptional regulation requires sequential induction of type I IFNs and IL-27 in macrophages. J Immunol 2010; 185: 6599–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Wei L, Pfeffer SR, Du Z, Murti A, Valentine WJ et al. Identification of CXCL11 as a STAT3-dependent gene induced by IFN. J Immunol 2007; 178: 986–992. [DOI] [PubMed] [Google Scholar]
- Stumhofer JS, Laurence A, Wilson EH, Huang E, Tato CM, Johnson LM et al. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat Immunol 2006; 7: 937–945. [DOI] [PubMed] [Google Scholar]
- Hunter CA. New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat Rev Immunol 2005; 5: 521–531. [DOI] [PubMed] [Google Scholar]
- Guo B, Chang EY, Cheng G. The type I IFN induction pathway constrains Th17-mediated autoimmune inflammation in mice. J Clin Invest 2008; 118: 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA et al. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat Immunol 2007; 8: 1363–1371. [DOI] [PubMed] [Google Scholar]
- Basset L, Chevalier S, Danger Y, Arshad MI, Piquet-Pellorce C, Gascan H et al. Interleukin-27 and IFNgamma regulate the expression of CXCL9, CXCL10, and CXCL11 in hepatitis. J Mol Med 2015; 93: 1355–1367. [DOI] [PubMed] [Google Scholar]
- Torok M, Etkin LD. Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation 2001; 67: 63–71. [DOI] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 2008; 8: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Garcia-Sastre A, Versteeg GA. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol 2014; 426: 1265–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M et al. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 2013; 38: 384–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, Luban J et al. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol 2013; 87: 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Zhao X, Sun D, Yang L, Chong C, Pan Y et al. Interferon alpha (IFNalpha)-induced TRIM22 interrupts HCV replication by ubiquitinating NS5A. Cell Mol Immunol 2016; 13: 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Guo JT, Wu JZ, Yang G. Identification and characterization of multiple TRIM proteins that inhibit hepatitis B virus transcription. PLoS One 2013; 8: e70001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007; 446: 916–920. [DOI] [PubMed] [Google Scholar]
- Tan G, Niu J, Shi Y, Ouyang H, Wu ZH. NF-kappaB-dependent microRNA-125b up-regulation promotes cell survival by targeting p38alpha upon ultraviolet radiation. J Biol Chem 2012; 287: 33036–33047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G, Shi Y, Wu ZH. MicroRNA-22 promotes cell survival upon UV radiation by repressing PTEN. Biochem Biophys Res Commun 2012; 417: 546–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004; 427: 848–853. [DOI] [PubMed] [Google Scholar]
- Gao B, Duan Z, Xu W, Xiong S. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 2009; 50: 424–433. [DOI] [PubMed] [Google Scholar]
- Sato S, Li K, Kameyama T, Hayashi T, Ishida Y, Murakami S et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 2015; 42: 123–132. [DOI] [PubMed] [Google Scholar]
- Teles RM, Kelly-Scumpia KM, Sarno EN, Rea TH, Ochoa MT, Cheng G et al. IL-27 Suppresses antimicrobial activity in human leprosy. J Invest Dermatol 2015; 135: 2410–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Takeda A, Hamano S, Miyazaki Y, Kinjyo I, Ishibashi T et al. Two-sided roles of IL-27: induction of Th1 differentiation on naive CD4+ T cells versus suppression of proinflammatory cytokine production including IL-23-induced IL-17 on activated CD4+ T cells partially through STAT3-dependent mechanism. J Immunol 2006; 177: 5377–5385. [DOI] [PubMed] [Google Scholar]
- Bender H, Wiesinger MY, Nordhoff C, Schoenherr C, Haan C, Ludwig S et al. Interleukin-27 displays interferon-gamma-like functions in human hepatoma cells and hepatocytes. Hepatology 2009; 50: 585–591. [DOI] [PubMed] [Google Scholar]
- Xu F, Song H, Li N, Tan G. HBsAg blocks TYPE I IFN induced up-regulation of A3G through inhibition of STAT3. Biochem Biophys Res Commun 2016; 473: 219–223. [DOI] [PubMed] [Google Scholar]
- Levy DE, Lee CK. What does Stat3 do? J Clin Invest 2002; 109: 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WB, Levy DE, Lee CK. STAT3 negatively regulates type I IFN-mediated antiviral response. J Immunol 2011; 187: 2578–2585. [DOI] [PubMed] [Google Scholar]
- Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem 2006; 281: 14111–14118. [DOI] [PubMed] [Google Scholar]
- Wu J, Meng Z, Jiang M, Pei R, Trippler M, Broering R et al. Hepatitis B virus suppresses toll-like receptor-mediated innate immune responses in murine parenchymal and nonparenchymal liver cells. Hepatology 2009; 49: 1132–1140. [DOI] [PubMed] [Google Scholar]
- Liu S, Peng N, Xie J, Hao Q, Zhang M, Zhang Y et al. Human hepatitis B virus surface and e antigens inhibit major vault protein signaling in interferon induction pathways. J Hepatol 2015; 62: 1015–1023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.