Abstract
The growth and proliferation of metazoan cells are driven by cellular nutrient status and by extracellular growth factors. Growth factor receptors on cell surfaces initiate biochemical signals that increase anabolic metabolism and macropinocytosis, an actin-dependent endocytic process in which relatively large volumes of extracellular solutes and nutrients are internalized and delivered efficiently into lysosomes. Macropinocytosis is prominent in many kinds of cancer cells, and supports the growth of cells transformed by oncogenic K-Ras. Growth factor receptor signaling and the overall metabolic status of the cell are coordinated in the cytoplasm by the mechanistic target-of-rapamycin complex-1 (mTORC1), which positively regulates protein synthesis and negatively regulates molecular salvage pathways such as autophagy. mTORC1 is activated by two distinct Ras-related small GTPases, Rag and Rheb, which associate with lysosomal membranes inside the cell. Rag recruits mTORC1 to the lysosomal surface where Rheb directly binds to and activates mTORC1. Rag is activated by both lysosomal luminal and cytosolic amino acids; Rheb activation requires phosphoinositide 3-kinase, Akt, and the tuberous sclerosis complex-1/2. Signals for activation of Rag and Rheb converge at the lysosomal membrane, and several lines of evidence support the idea that growth factor-dependent endocytosis facilitates amino acid transfer into the lysosome leading to the activation of Rag. This review summarizes evidence that growth factor-stimulated macropinocytosis is essential for amino acid-dependent activation of mTORC1, and that increased solute accumulation by macropinocytosis in transformed cells supports unchecked cell growth.
Keywords: Macropinocytosis, mTORC1, Small GTPase, Phosphoinositide, Cancer
Introduction
Macropinocytosis is an endocytic process by which cells engulf relatively large volumes of extracellular fluid solutes, including nutrients, through movements of the plasma membrane [1, 2]. Subsequent organelle fusion reactions deliver internalized solutes into endolysosomal compartments, where macromolecules may be degraded by lysosomal hydrolases into constituent subunits for anabolic metabolism. Macropinocytosis was originally called pinocytosis [3, 4], but was later renamed to distinguish it from smaller endocytic vesicles such as clathrin-coated vesicles. Growth factors, cytokines, chemokines, pathogens, and the tumor promoter phorbol myristate acetate (PMA) can induce macropinocytosis. Macrophages and dendritic cells constitutively exhibit macropinocytosis, as do cells transformed by oncogenic mutations of K-Ras and v-Src [5, 6]. Aberrant activation of macropinocytosis has been implicated in cancer progression [7, 8], neurodegenerative diseases [9], atherosclerosis [10], and renal dysfunction [11].
Extracellular nutrients and growth factors can regulate cell growth, quiescence, and survival. In response to nutrient availability and growth factor stimulation, cells grow and proliferate by increasing anabolic metabolism. Mechanistic target of rapamycin (mTOR) is an evolutionarily conserved serine/threonine kinase that plays key roles in stimulating cellular anabolic processes and inhibiting catabolic processes such as autophagy in response to growth factors and nutrient availability. TOR was originally identified in yeast as a target protein of rapamycin, a macrolide compound that is now widely used in clinical settings as an immunosuppressant, anti-restenotic, and anti-cancer agent [12–15]. mTOR forms at least two distinct multi-protein complexes termed mTOR complex 1 (mTORC1) and mTORC2 [16–20]. Both complexes contain mTOR as a core kinase and the common subunits mLST8 (also known as GβL) [20] and DEPTOR [21]. mTORC1 [15] contains the specific subunits, raptor [18, 19] and PRAS40 [22–24], while mTORC2 contains rictor [17], mSIN1 [25, 26], and PROTOR [27]. While mTORC2 plays important roles in actin cytoskeleton reorganization, cell migration, survival, and glucose metabolism, mTORC1 has been shown to be essential in cell growth and a wide array of cellular metabolic processes [28–30]. In response to a variety of stimuli, including amino acids, glucose, growth factors, cytokines, and PMA [31–33], mTORC1 stimulates cell growth and proliferation by enhancing the rate of cellular protein synthesis, and lipid and pyrimidine/purine biogenesis [34]. Aberrant activation of mTORC1 plays key pathological roles in the development of diseases such as cancer, type 2 diabetes, atherosclerosis, and neurodegeneration [28, 29, 34–37]. Thus, the mechanism of mTORC1 activation and its roles in metabolic regulation have attracted intense interest in basic and clinical sciences.
Macropinocytosis and mTORC1 activation share many common mechanisms for their induction, and recent studies have demonstrated that macropinocytosis contributes to cell growth by stimulating mTORC1 activity [2, 7, 8, 38–42]. This review compares the molecular mechanisms underlying the induction of macropinocytosis and mTORC1 activity, and discusses crucial roles of macropinocytosis in the assimilation of nutrients for cell growth.
mTORC1 activity is regulated by Rag and Rheb
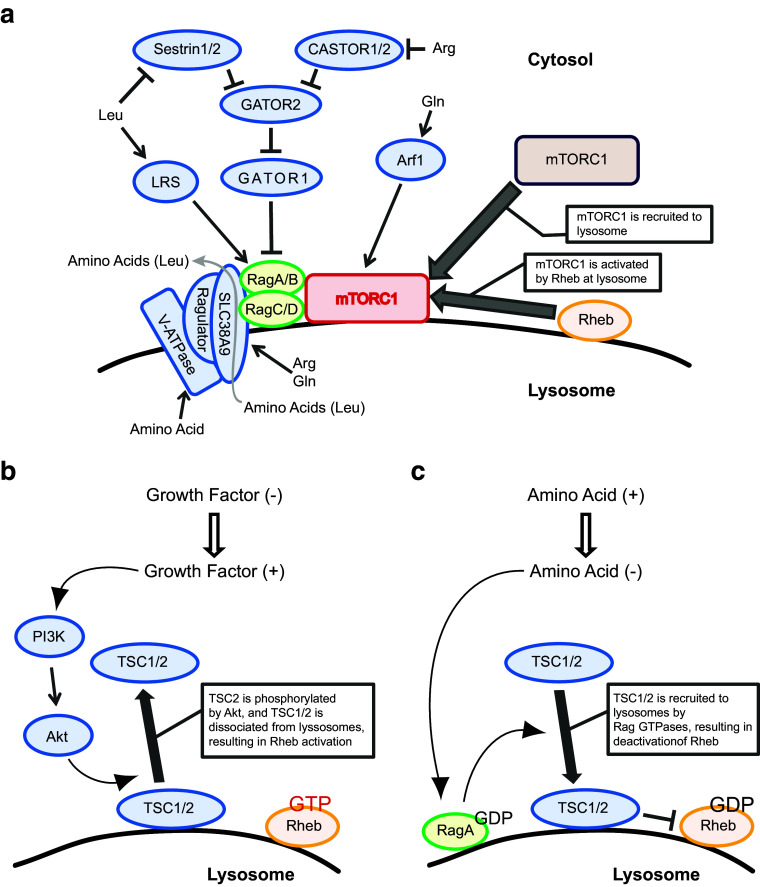
The small GTPases Rag and Rheb coordinately stimulate the activity of mTORC1 on the surface of the lysosome [43–45] (Fig. 1a). Mammalian cells contain four isoforms of Rag, Rag A, B, C, and D, which form heterodimers comprised of RagA or B with RagC or D in a functional conformation, and which are activated by amino acids such as leucine and arginine. The Rag heterodimer interacts with a pentameric protein complex called Ragulator, which consists of the proteins p18 (LAMTOR1), p14 (LAMTOR2), MP1 (LAMTOR3), C7ORF59 (LAMTOR4), and HBXIP (LAMTOR5), and associates with the lysosomal membrane [44]. Ragulator functions as a scaffold for the Rag heterodimer to localize on the lysosomal membrane and to stimulate GTP-binding by RagA or RagB through its guanine nucleotide exchange factor (GEF) activity. Amino acids in the lysosomal lumen play a key role in triggering a conformational change of the transmembrane vacuolar H+-ATPase (v-ATPase), which activates the RagA/B GEF activity of Ragulator [46, 47]. In addition, SLC38A9, a lysosomal transmembrane protein, interacts with the v-ATPase and activates Ragulator by sensing luminal arginine [48–50]. Upon binding arginine, SLC38A9 transports leucine and other amino acids from the lysosomal lumen into cytoplasm [51]. Cytosolic arginine and leucine can activate the Rag heterodimer by inhibiting the inhibitory activity of a GTPase-activating protein (GAP) for RagA/B [52] (Fig. 1a). GATOR1, a trimeric protein complex consisting of DEPDC5, Nprl2, and Nprl3, is expressed on the lysosomal membrane and functions as a GAP for RagA/B. Furthermore, GATOR1 is inhibited by another pentametric protein complex, GATOR2 [53]. Thus, GATOR2 activates the Rag heterodimer by inactivating GATOR1. Sestrin1 and/or Sestrin 2 directly interact with and inhibit GATOR2, and suppress mTORC1 function [54, 55]. Sestrin bears a leucine-binding pocket in close proximity to its GATOR2 binding site, and the binding of leucine to Sestrin relieves its inhibitory effect on GATOR2. Thus, cytosolic leucine activates mTORC1 by inhibiting GATOR1 through its binding to Sestrin1/2. Similarly, cytosolic arginine activates mTORC1 by inhibiting GATOR1 through its binding to CASTOR1. CASTOR1 forms a homodimer or a heterodimer with CASTOR2 and inhibits GATOR2. Similar to the mode of Sestrins, arginine binding to CASTOR1 blocks its interaction with GATOR2 and relieves the CASTOR1 inhibitory effect on GATOR2, thereby activating RagA/B signaling [54–57]. Glutamine also stimulates mTORC1 [58]. However, it remains unclear whether glutamine itself functions as a signaling molecule for activating mTORC1. Rather, either glutamine stimulates the influx of leucine by acting as an efflux solute through a SLC7A5–SLC3A2 heterodimeric antiporter, or the glutamine metabolite α-ketoglutarate stimulates mTORC1 by activating the Rag heterodimer [59, 60]. It has also been reported that glutamine can activate mTORC1 in a manner dependent on Arf1 but not Rag small GTPase [58]. Thus, RagA/B-dependent activation of mTORC1 occurs by amino acids detected in the cytosol but reaching mTORC1 from within lysosomes or endolysosomes.
Fig. 1.
Amino acid- and growth factor-induced mTORC1 activation. a The mechanism of amino acid-induced mTORC1 activation. mTORC1 is recruited to lysosomes by amino acid stimulation. Through V-ATPase and SLC38A9 on lysosomal membranes, amino acids such as arginine (Arg) and glutamine (Gln) modulate the function of protein complex Ragulator, leading to Rag activation. Arg and Gln are detected by SLC38A9. Once Rag is activated, mTORC1 is recruited to lysosomes via the interaction between Rag and raptor, followed by mTORC1 activation by Rheb. Upon binding arginine, SLC38A9 transports amino acids, such as leucine (Leu), from the lysosomal lumen into cytoplasm. GATOR1 and GATOR2 regulate Rag function. Rag is inhibited by GATOR1, which is inhibited by GATOR2. Sestrin1/2 and CASTOR1/2 inhibit GATOR2, and detect Leu and Arg, respectively, in cytosol. The interaction of these amino acids with their target proteins results in the reversal of inhibition by GATOR2. Leucyl-tRNA synthetase (LRS) can also activate Rag and detect Leu in the cytosol. Gln in the cytosol is detected by an Arf1-dependent mechanism, followed by Rag activation. b The mechanism of growth factor-induced Rheb activation. Growth factor stimulation induces the PI3K–Akt pathway. Akt phosphorylates TSC2, which is located at lysosomal membrane as a protein complex with TSC1. After phosphorylation, the TSC1/2 complex dissociates from the lysosome. TSC1/2 is a Rheb GAP, so loss of TSC1/2 complex from the lysosomal membrane allows Rheb to be activated (Rheb-GTP). c The mechanism of amino acid-modulated Rheb deactivation. Depletion of amino acids from culture medium induces deactivation of RagA (GDP form). Inactivated RagA triggers TSC1/2 recruitment to lysosomes, resulting in deactivation of Rheb (Rheb-GDP)
Activated Rag recruits mTORC1 to the lysosomal membrane through its interaction with Raptor [44, 61]. There, Rheb directly activates mTORC1 [15, 62, 63] (Fig. 1). Rheb itself is activated by signals from growth factor receptors [64] (Fig. 1b). Upon growth factor stimulation, active phosphoinositide 3-kinase (PI3K) synthesizes PIP3, which recruits PDK1 and Akt to the plasma membrane where Akt is phosphorylated and activated by PDK1 and mTORC2. Subsequently, active Akt on the lysosomal membrane phosphorylates and inhibits tuberous sclerosis complex 2 (TSC2), a GAP for Rheb in a larger complex comprised of TSC1, TSC2 and TBC1D7 [Tre2–Bub2–Cdc16 (TBC)1 domain family number 7] [65–67]. Alternatively, the RAS–MEK–ERK–RSK pathway phosphorylates and inactivates the TSC complex in response to growth factors, cytokines, and PMA [31, 32, 68–72]. The phosphorylation of TSC2 by Akt induces the dissociation of the TSC complex from the lysosomal membrane, consequently permitting GTP-loading of Rheb and subsequent mTORC1 activation [64, 72, 73]. The molecular mechanism by which Akt reaches the lysosome to phosphorylate TSC2, and how the phosphorylation of TSC2 leads to its dissociation from the lysosomal membrane are still unknown. Recent studies demonstrated that the dissociation of the TSC complex from lysosomes is also triggered by amino acid stimulation (Fig. 1c) [73, 74]. Under amino acid starvation conditions, the GDP-bound form of RagA (inactive) interacts with and recruits TSC2 to the lysosomal membrane. Conversely, GTP-bound RagA (active) is unable to retain the TSC complex on the lysosomal membrane. Thus, both growth factor-mediated TSC2 phosphorylation and amino acid-induced RagA activation induce the dissociation of the TSC complex and, consequently, stimulate Rheb-dependent mTORC1 activation. In addition to these mechanisms, a recent study demonstrated that arginine can directly inhibit the interaction between the TSC complex and Rheb, thereby supporting Rheb activation in response to amino acid availability [75].
Involvement of endocytosis and autophagy in mTORC1 activation
Given that the cytosolic face of the lysosomal membrane serves as a platform for numerous proteins and protein complexes that mediate amino acid- and growth factor signaling for mTORC1 activation, it can be hypothesized that processes important for endosomal and lysosomal trafficking play key roles in the regulation of mTORC1 activity [76–78]. In addition to Rag and Rheb, other small GTPases associated with endocytosis contribute to the activation of mTORC1. In Drosophila S2 cells [79], mTORC1 activation was decreased by knockdown of Rab5 or Arf, which are important for endocytic membrane trafficking. Similarly, knockdown of mammalian Rab5 or Arf1 decreased mTORC1 activity in HEK293 or murine embryonic fibroblast (MEF) cells. Ectopic expression of dominant-active Rab5(Q79L) in HEK293 cells specifically blocked activation of mTORC1 by amino acids but not glucose, implicating Rab5-related endocytic traffic in amino acid-dependent mTORC1 activation [79]. Ectopic expression of active Rab5 often generates unusual vesicles containing both the early endosome marker EEA1 and the late endosome/lysosome marker LAMP1, indicating that aberrant Rab5 activation causes a defect in early-to-late endosome conversion [80]. Consistent with this observation, ablation of hVps39, which plays a role in the early-to-late endosome conversion, produced hybrid endosomes and inhibited insulin-induced mTORC1 activation [80]. mTORC1 localized to these hybrid endosomes, suggesting that the maturation or integrity of the late endosome/lysosome was critical for proper activation of mTORC1. It remains unclear whether Rheb localizes to these hybrid endosomes, and whether the dissociation of the TSC complex from these organelles occurs in response to growth factor stimulation. Together, these reports suggest that the transition from early to late endosome, regulated by Rab5, is required for mTORC1 activation.
As noted above, the GTPase Ras functions as an upstream suppressor of TSC2 via the ERK pathway [31, 71]. Expression of dominant active Ras(Q61L) in HEK293T cells induced TSC2 phosphorylation [71], and stimulated mTORC1, as indicated by S6K1 phosphorylation. Thus, Ras functions upstream of Rheb to stimulate mTORC1 activity. mTORC1 activation by Ras(Q61L) was blocked by amino acid starvation in fibroblasts [65], suggesting that Ras does not act downstream of amino acid sensing machineries to activate mTORC1. However, these observations leave open the possibility that active Ras acts upstream of amino acid sensing machineries to induce mTORC1 activation. In addition, recent studies demonstrated that ablation of the GTPase Rac1 attenuated growth factor-induced mTORC1 and mTORC2 activation in MEFs and HeLa cells [40, 81]. Immunofluorescence staining showed that Rac1 co-localized with mTORC1 and mTORC2 at the plasma membrane in response to serum stimulation [81]. As both Ras and Rac regulate endocytic pathways, these reports also suggest the involvement of endosomal traffic in mTORC1 activation. Interestingly, active Ras acts upstream of Rac1 to stimulate actin cytoskeleton reorganization, membrane ruffling, and macropinocytosis [1, 82].
Another activity in which mTORC1 is responsive to lysosome function is macroautophagy, a process in which cytoplasm is sequestered into membranous autophagosomes that, like macropinosomes, fuse with lysosomes to allow macromolecule hydrolysis and nutrient recycling. Inhibition of cellular mTORC1 activity stimulates autophagy [30], and amino acids recovered by autophagy can activate mTORC1 [51, 83, 84]. Thus, both heterophagy—the assimilation of exogenous nutrients by endocytic activities—and autophagy—the degradation of cytoplasmic contents—can provide amino acids for activation or reactivation of mTORC1.
Mechanisms of macropinosome formation
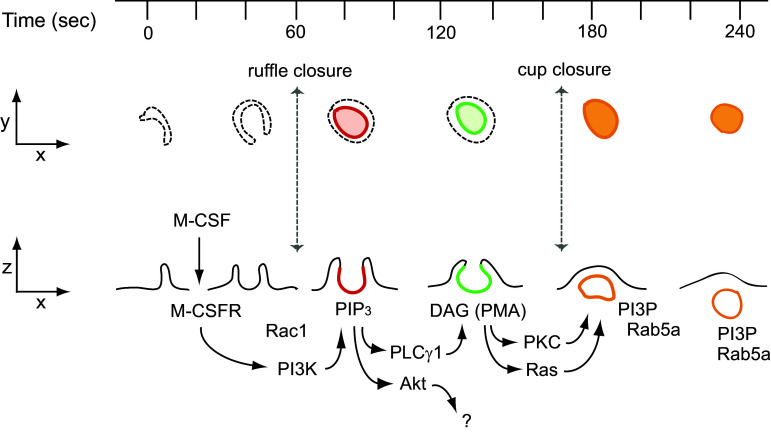
Macropinocytosis was recognized long ago as a feature of growing cells [3, 85], but its essential role in growth was only established recently [7, 8, 40]. Many of the signaling molecules necessary for mTORC1 activation also contribute to macropinocytosis. The molecular mechanism of growth factor-induced macropinocytosis has been studied with a focus on the roles of small GTPases and phosphoinositides [1, 77, 86] (Fig. 2). Treatment of macrophages with their growth factor macrophage colony-stimulating factor (M-CSF) immediately induces irregular membrane ruffles at the cell margins which transform into “C”-shaped ruffles and then “O” shaped, cup-like structures. The open area at the top of the cup later closes to form a complete macropinosome [87]. The first stage of the closing process (C- to O-shaped ruffle) is termed ruffle closure, and the second phase (cup to macropinosome) is termed cup closure [1]. Fully closed macropinosomes move toward the center of the cell via the microtubule network and fuse with the lysosome [88] or, rarely, recycle to the plasma membrane [89]. Imaging of cells expressing fluorescent protein chimeric protein probes revealed a cascade of signals corresponding to the various stages of macropinosome formation. These temporally arranged signals were all restricted to the bowl of the macropinocytic cup, likely by structural barriers to lateral diffusion in the inner leaflet of the cup membrane [90]. Förster resonance energy transfer (FRET) microscopy showed that Rac1 was active within the cup domain immediately following ruffle closure [87]. Ratiometric fluorescence microscopy showed that cyan fluorescent protein (CFP)-labeled Rab5a was recruited to the cup membrane during cup closure and persisted on the macropinosome during its movement toward the lysosome [87]. Similarly, yellow fluorescent protein (YFP)-tagged Ras-binding domain of Raf (YFP-RBD), a probe to detect activated Ras [91], was recruited to macropinocytic cups in macrophages, suggesting that Ras is active during cup closure [92]. Similar macropinocytosis signaling patterns were also reported in other cell types following stimulation with platelet-derived growth factor (PDGF) [93–97]. Thus, as for activation of mTORC1, GTPases associated with membrane traffic are required for macropinocytosis.
Fig. 2.
M-CSF-induced macropinocytosis. Interaction between M-CSF and the M-CSF receptor in macrophages activates Rac1 followed by induction of membrane ruffling. Some ruffles change into cup-like structures, in which activated PI3K then transiently generates PIP3 (red). PIP3 generation in the cup triggers the activation of PLCγ and Akt. Akt is not involved in macropinosome formation. PLCγ generates DAG in the cup (green), leading to activation of PKC and Ras. Both pathways contribute to cup closure, in which the macropinosome pinches off into the cytoplasm from the plasma membrane. Following cup closure, PI3P and Rab5a are localized at the macropinosomes (orange). Macropinosomes with these signals (orange) then move toward the center of the cells
Phosphoinositides are also essential for macropinocytosis. PI3K is required for all macropinocytosis except that stimulated by PMA [98, 99]. Fluorescence microscopy of macrophages stimulated with M-CSF showed transient recruitment of YFP-Btk-PH, which localizes PIP3, to the macropinocytic cup, indicating transient, localized PIP3 generation (PIP3 spike) [87, 92]. PI3K also regulates PDGF-induced macropinocytosis [100]. Live-cell imaging with fluorescent protein-tagged pleckstrin homology (PH)-domain chimeras demonstrated a signal transition from PI(4,5)P2 to PIP3 during epidermal growth factor (EGF)-induced macropinosome formation [86, 99]. Two well-known signal pathways are activated by PIP3: Akt and phospholipase C-γ (PLCγ). PLCγ is involved in macropinosome formation; Akt is not [101]. Imaging YFP-C1δ as a probe for the PLCγ product diacylglycerol (DAG) revealed transient generation of DAG in the cup [87, 101]. Live-cell imaging also showed that YFP-tagged protein kinase C (PKC)-α, which is activated by DAG, was recruited to cups [92]. The DAG mimetic PMA stimulates macropinocytosis in macrophages [102]. PMA-induced macropinocytosis is blocked by inhibitors of PKC and Ras but not by inhibitors of PLCγ or PI3K [101]. Additionally, the PIP3 spike was not observed in PMA-induced macropinocytic cups [40]. After cup closure, PI3P and Rab5a appeared on fully formed macropinosomes, which then moved toward the center of the cells [87]. The PKC inhibitor calphostin C blocked PDGF-induced macropinocytosis in MEFs [40]. Diacylglycerol kinase-ζ (DGKζ), which phosphorylates DAG to yield phosphatidic acid, is also necessary for macropinocytosis [103]. Knock-down of DGKζ attenuated PDGF-induced macropinocytosis. Therefore, DAG is a key signaling molecule involved in macropinocytosis. Together, these observations suggest that growth factor (GF)-induced macropinosome formation results from a signal cascade comprised of many molecules essential to growth control (Fig. 2).
The role of Ras in macropinosome formation remains undefined. Ras-induced pinocytosis was first described as a cellular response to injection of H-Ras [85]. H-Ras(G12V) expression induced membrane ruffles and macropinocytosis in HeLa cells, which could be inhibited by the actin polymerization inhibitor cytochalasin D or by co-expression of dominant-negative Arf6(T27N) [104]. K-Ras-induced macropinocytosis in fibroblasts was blocked by cytochalasin E or by the PI3K inhibitors wortmannin and LY294002 [5]. H-Ras-induced macropinocytosis in BHK-21 cells was blocked by wortmannin or by expression of dominant negative Rab5(S34N), but not by dominant negative Rac1(S17N) [105]. The differential association of K-Ras with PI3K p110 isoforms suggests roles for Ras in ruffling and macropinosome closure. However, MEFs deficient in K-Ras, H-Ras and N-Ras are capable of generating macropinosomes in response to PDGF [106], which suggests that macropinocytosis induced by oncogenic Ras may be an aberrant cellular behavior.
Phosphoinositide signals on macropinosomes were also observed during H-Ras(G12V)-induced macropinocytosis. Live-cell imaging using YFP-AktPH and YFP-PLCδ1-PH to localize PIP3 and PI(4,5)P2, respectively, showed that H-Ras(G12V)-induced macropinosomes in COS7 cells recruited both probe proteins and indicated that, like macropinocytosis in macrophages, PI(4,5)P2 was lost from macropinosomes before the PIP3 spike appeared [104]. Live-cell imaging showed co-localization of GFP-Akt and monomeric red fluorescent protein (mRFP)-H-Ras(G12V) at macropinosomes in COS7 cells [104]. Immunofluorescence staining showed that cells co-expressing H-Ras(G12V) and Arf6(Q67L) formed macropinosomes containing phosphorylated Akt [104]. YFP-Akt-PH was recruited to M-CSF-induced macropinocytic cups in macrophages [101] and to EGF-induced macropinocytic cups in A431 cells [99]. Moreover, GFP-Akt localizes to macropinosomes in LPS-stimulated macrophages [107]. Thus, Akt is activated at the macropinocytic cup and/or macropinosomes.
Ras is also required for macropinocytosis and cell growth in axenic strains of the free-living ameba Dictyostelium discoideum which are capable of growth in nutrient broth. Those strains exhibit Ras activity localized to macropinocytic cups, which are larger than cups in wild-type amebas due to a mutation in the Ras GAP neurofibromin [108, 109]. Thus, active Ras contributes to the morphogenesis of large macropinosomes necessary for nutrient acquisition and cell growth.
Growth factor-induced macropinocytosis transfers amino acids into lysosomes to activate mTORC1
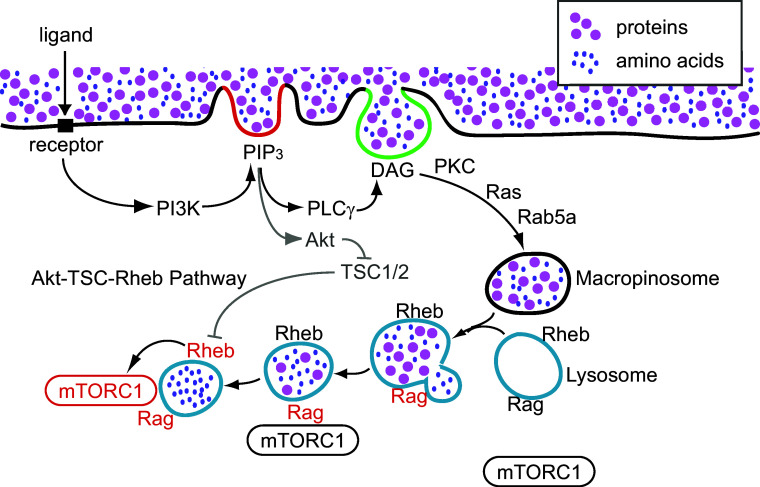
Macropinocytosis rapidly and efficiently delivers extracellular solutes into lysosomes [110]. Given that growth factors induce both mTORC1 activation and macropinocytosis, and that they share many common GTPases and signaling molecules for their induction, we proposed a model in which macropinocytosis-mediated delivery of extracellular amino acids or protein to lysosomes is essential for mTORC1 activation (Fig. 3) [40]. Biochemical studies in murine macrophages showed that M-CSF treatment induced the PI3K–Akt–TSC–Rheb–mTORC1 pathway. Live-cell imaging and quantitative fluorescence microscopy showed that M-CSF-induced macropinocytosis delivered small extracellular molecules rapidly into lysosomes, where mTORC1 was recruited and activated. Inhibition of macropinocytosis by ethyl isopropylamiloride (EIPA) [111] or with the cytoskeleton inhibitors jasplakinolide and blebbistatin (J/B) blocked M-CSF-induced mTORC1 activation without inhibiting the PI3K–Akt pathway. These results suggest that macropinocytosis provides rapid amino acid trafficking into lysosomes to activate mTORC1. Like M-CSF-induced macropinocytosis, PMA-induced macropinocytosis also increased amino acid-dependent mTORC1 activation, but without inducing Akt phosphorylation. A role for macropinocytosis in mTORC1 activation was also demonstrated in MEFs. PDGF-induced mTORC1 activation by leucine (in the absence of glucose) was blocked by EIPA, J/B, or by knock-down of Rac1, in a manner independent of the Akt–TSC pathway. PDGF treatment increased mTOR recruitment to lysosomes, as determined by the co-localization of mTOR with LAMP2, a lysosomal membrane protein.
Fig. 3.
Macropinocytosis triggers mTORC1 activation. PI3K-generated PIP3 accumulates in macropinocytic cups (red line), activating Akt and PLCγ. PLCγ generates DAG in the cup (green line), leading to Ras- and PKC-dependent pathways that close the macropinosome. Extracellular nutrients internalized by the macropinosomes are delivered rapidly into lysosomes through fusion reactions. Nutrient transfer from macropinosomes to lysosomes induces Rag activation (black to red), followed by mTORC1 recruitment to lysosomes. Meanwhile, activated Akt inhibits TSC function in a cytosolic pathway independent of macropinocytosis, resulting in Rheb activation (black to red). Rheb directly activates mTORC1 on the lysosomal membranes (black to red)
Based on these observations, it was proposed that growth factor stimulation induces macropinocytosis, leading to efficient uptake of essential amino acids via macropinosomes and subsequent delivery to the lysosome for mTORC1 activation (Fig. 3). Accordingly, growth factor- dependent mTORC1 activation is established by two distinct pathways: a PI3K–Akt–TSC–Rheb (cytosolic) pathway and a PI3K–macropinocytosis–Rag (vesicular) pathway. The cytosolic pathway is the classical Akt-dependent mTORC1 activation pathway described above: activated Akt induces TSC phosphorylation (TSC deactivation) and consequent activation of Rheb. In the vesicular pathway, PIP3 in macropinocytic cups localizes DAG synthesis and PKC activity, leading to macropinosome closure. Macropinosomes fuse with the tubular lysosomal network in macrophages or the lysosomes in MEFs, delivering ingested solutes such as proteins or amino acids. Amino acids transferred into the lysosome via macropinosome-lysosome fusion, or derived from hydrolysis of proteins in lysosomes, activate Ragulator and lead to subsequent activation of mTORC1 [40]. Therefore, growth factor receptor signaling organizes macropinosome formation, and the amino acids or proteins internalized by macropinocytosis signal to mTORC1 from inside lysosomes.
The macropinosome as a signal platform for mTORC1 signaling
Macropinocytic cups and macropinosomes may also serve as structural platforms of signaling for cell growth. In addition to small GTPases, phosphoinositides are common signaling molecules involved in mTORC1 activation and macropinocytosis [76, 112]. Phosphoinositide kinase FYVE-type zinc finger containing (PIKFYVE) catalyzes the synthesis of PI(3,5)P2 from phosphatidylinositol 3-phosphate (PI3P) [113]. PI(3,5)P2 interacts with raptor [114], indicating its involvement in mTORC1 activation [112]. In 3T3-L1 adipocytes, depletion of PIKFYVE blocked insulin-induced activation of mTORC1 (as measured by S6K phosphorylation) without affecting Akt phosphorylation [114]. Myotubularin-related phosphatase 3 (MTMR3) dephosphorylates PI3P to phosphatidylinositol [115]. Depletion of MTMR3 in HEK293T cells increased nutrient-induced mTORC1 activation, suggesting that MTMR3 suppresses mTORC1 activity by depleting PI3P [116]. Therefore, the synthesis of PI3P or PI(3,5)P2 on macropinosomes could help recruit mTORC1 to the late endosome or lysosome.
The macropinocytic cup can also localize Akt phosphorylation. Like M-CSF, the chemokine CXCL12 induces both macropinocytosis and mTORC1 activation in macrophages [38]. Unlike the response to M-CSF, however, CXCL12-induced phosphorylation of Akt and S6K (a reporter of mTORC1 activity) was dependent on actin cytoskeleton rearrangement and the formation of macropinocytic cups. Live-cell imaging showed YFP-Akt-PH recruitment to the macropinocytic cup, and western blot analysis showed that the macropinocytosis inhibitors J/B and EIPA attenuated CXCL12-induced Akt phosphorylation. Thus, Akt phosphorylation in response to CXCL12 required the formation of a macropinocytic cup. Immunofluorescence microscopy showed that Akt was phosphorylated at membrane ruffles and macropinocytic cups. The PKCα/β-specific inhibitor Gö6976 blocked macropinocytosis and S6K phosphorylation without inhibiting membrane ruffling or cup formation, suggesting that PKCα and/or PKCβ are involved in cup closure. However, Gö6976 did not inhibit CXCL12-induced Akt phosphorylation. Together these studies indicated that CXCL12-induced macropinocytic cups are signal platforms for the Akt phosphorylation required for mTORC1 activation.
To what extent does the cytosolic pathway (Akt–TSC1/2–Rheb) require macropinocytosis? The sensitivity of Akt activation by CXCL12 to cytoskeleton-inhibitors differed from Akt activation in response to M-CSF or PDGF, which was not affected by such inhibitors. The organization of the macropinocytic cup may allow localized amplification of signals from some receptors, perhaps those that require multiple inputs for signal amplification. Circular ruffles create isolated domains of plasma membrane where signal propagation can occur [92], indicating the presence of barriers to lateral diffusion in the inner leaflet of the plasma membrane of cups [90]. Maximal Akt phosphorylation observed in response to CXCL12 was less than the level of Akt phosphorylation measured in response to M-CSF. Acute stimulation of cells with M-CSF (or PDGF) may generate sufficiently high concentrations of PIP3 that a spatially organized amplification is unnecessary. However, if receptors cannot generate high PIP3 concentrations, then phosphorylation of Akt may require a mechanism based on spatial confinement of signal amplification to macropinocytic cups. Consistent with this model, a recent study identified a role for Rac-dependent macropinocytosis in the activation of the PI3K subunit p110β by G-protein coupled receptors [117].
As described above, the TSC complex inhibits Rheb function at the lysosome [64, 73, 74]. When Akt and Erk phosphorylate TSC2, the TSC complex subsequently loses its GAP activity for Rheb [31, 32, 72]. This suggests that, within a few minutes of stimulation, signal components that phosphorylate Akt and Erk reach lysosomal structures and phosphorylate TSC2. In cells co-expressing H-Ras(G12V) and Arf6(Q67L), Erk is recruited to and phosphorylated at macropinosomes [104]. Erk localizes to late endosomes and lysosomes via the protein complex p18/p14/MP1 [118]. Since macropinosomes show late endosome characteristics at this stage, growth factor/chemokine-induced macropinosomes should recruit Erk via the p18/p14/MP1 protein complex during the maturation process. Given that another important function of the p18/p14/MP1 complex is to recruit mTORC1 to the lysosome as a Ragulator, we speculate that late stage macropinosomes recruit mTORC1 directly. Together, these reports indicate that macropinosomes deliver signaling molecules to the lysosome.
How macropinocytosis could be essential to growth control
Macropinocytosis may be essential for the growth of metazoan cells [40]. Accordingly, when cells are growing in constant concentrations of growth factor, macropinosomes form stochastically as discrete units of growth factor signaling, and activation of mTORC1 follows after a bolus of extracellular protein or amino acids is delivered by macropinocytosis into the lysosomes. Moreover, Akt localization to cups and its continued association with fully formed macropinosomes could provide a route for Akt to reach its substrate tuberous sclerosis complex-1/2 (TSC1/2) on the lysosomal membrane. Thus, the magnitude of growth factor stimulation of mTORC1 may be determined in part by the volume of solute internalized by macropinocytosis, with feedback from a nutrient-sensing mechanism regulating the magnitude of Akt signaling on macropinosome membranes and the volume of nutrient delivered into the lysosome via macropinocytosis. This model predicts that macropinocytosis is necessary for cell growth and proliferation.
Pathogenic functions of macropinocytosis in K-Ras-induced cancer
Dysregulation of Ras and mTORC1 are involved in cancer development [15, 29]. Pathologic functions of macropinocytosis in oncogenic K-Ras-expressing cancer cells have been described. Human carcinoma cells expressing K-Ras(G12C) or H-Ras(G12V) showed increased macropinocytosis, similar to NIH 3T3 cells expressing K-Ras(G12V). Extracellular proteins ingested by macropinocytosis in cells expressing oncogenic K-Ras were degraded and their constituent amino acids were used for anabolic metabolism [7]. The macropinocytosis inhibitor EIPA blocked albumin-dependent cell proliferation [7], indicating that ingestion of albumin by K-Ras(G12D)-induced macropinocytosis and subsequent hydrolysis of proteins in lysosomes were sufficient to provide the essential amino acids (EAA) necessary for cell proliferation [39]. Moreover, the growth of cells in nutrient-poor regions of pancreatic tumors was supported by scavenging of extracellular proteins [119]. Other groups have reported that H-Ras(G12V)-induced macropinocytosis is necessary for albumin-dependent cell growth of MEFs and that inhibition of mTORC1 activation increases the rate of macropinocytosis in carcinoma cells (MIA PaCa-2 K Ras mutant) [41, 42]. Additionally, inhibition of DOCK1, a Rac-activating protein required for macropinocytosis, reduces survival of Ras-driven cell growth [120]. Thus, macropinocytosis-mediated ingestion of extracellular protein is now considered a hallmark of cancer metabolism [121].
However, unlike the responses observed in macrophages and MEFs, mTORC1 activation by EAA in K-Ras transformed cells was not inhibited by EIPA [8]. This indicates that macropinocytosis in Ras-transformed cells is not the primary route by which free amino acids reach the cytosolic SESTRIN1/2 and CASTOR detection systems.
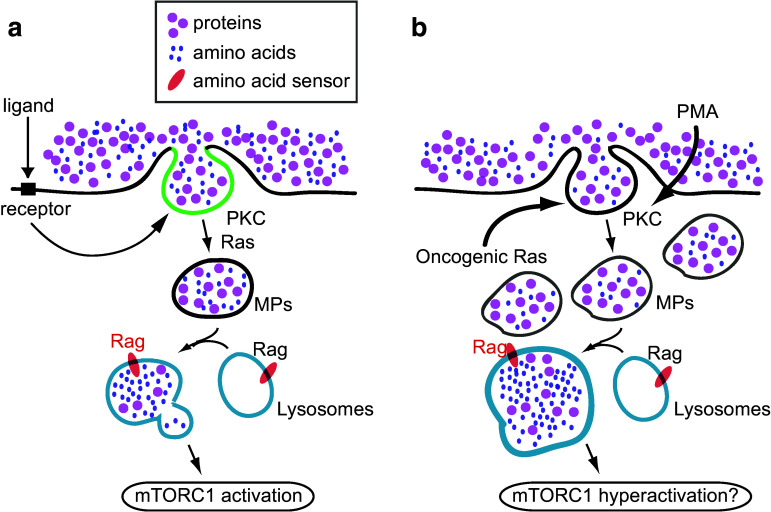
In sum, these studies suggest that macropinosomes serve as organizational units of a signal transduction pathway that is induced by extracellular stimuli such as growth factors and chemokines (Fig. 4a). If this is the case, constitutive macropinocytosis induced by oncogenic K-Ras or cSrc may hyperactivate mTORC1, resulting in unrestrained growth (Fig. 4b). Similarly, the tumor promoting activity of PMA may be partly attributable to its activation of mTORC1 via macropinocytosis.
Fig. 4.
Two models of macropinocytosis-regulated mTORC1 activation. a Role of macropinocytosis in ligand-induced mTORC1 activation. Signals derived from DAG (green) modulate macropinosome (MP) formation via the activation of PKC and Ras. Formed macropinosomes convey extracellular nutrients into lysosomes, where Rag is activated. b Proposed hypothesis of the function of oncogenic protein-induced macropinocytosis and mTORC1 activation. Over-expression of oncogenic Ras continuously induces macropinosomes, resulting in an overload of nutrients in the lysosomes. Because of this, following Rag activation, mTORC1 is hyperactivated. PMA treatment directly induces PKC activation, which would also lead to increased nutrient uptake via macropinocytosis
Future directions
Significant questions remain to be answered about the relationship between macropinocytosis and mTORC1. To what extent does macropinocytosis support growth of non-neoplastic cells? Why is mTORC1 activation by EAA in K-Ras-transformed cells independent of macropinocytosis? Does membrane traffic unrelated to macropinocytosis regulate mTORC1 activity? Does the activity of mTORC1 or the nutrient status of the cell regulate macropinosome formation or fusion with the lysosomes? The studies of Palm et al. [8, 106] indicated that active mTORC1 inhibits protein delivery into lysosomes via macropinocytosis, whereas Nofal et al. [122], showed that mTORC1 activation does not affect degradation of extracellular protein. These studies suggest that mTORC1 or the cytosolic concentrations of amino acids regulate the uptake and degradation of extracellular solutes by macropinocytosis (i.e., heterophagy) in a manner analogous to its role in protein recycling and degradation by autophagy.
Alternative macropinocytosis-specific inhibitors are needed, both for better understanding of macropinocytosis biology and for the potential therapeutic manipulation of the macropinocytosis signaling pathway. Although EIPA does not block other types of endocytosis, such as phagocytosis and clathrin-dependent endocytosis, it is reasonable to expect it to affect other signal pathways related to cell growth and differentiation. Drugs targeting macropinocytosis could attenuate growth of neoplastic cells or related mosaic disorders resulting from mutations in the signals leading to mTORC1 [123].
Acknowledgements
The authors are grateful for the editorial suggestions of Dr. David Friedman. This work was supported by NIH Grants R01 GM110215 (J.S), GM110019 (K.I), DK083491 (K.I), and US Department of Defense Grant TS140055 (K.I).
References
- 1.Swanson JA. Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol. 2008;9(8):639–649. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloomfield G, Kay RR. Uses and abuses of macropinocytosis. J Cell Sci. 2016;129(14):2697–2705. doi: 10.1242/jcs.176149. [DOI] [PubMed] [Google Scholar]
- 3.Lewis WH. Pinocytosis. B Johns Hopkins Hosp. 1931;49:17–27. [Google Scholar]
- 4.Cohn ZA, Parks E. The regulation of pinocytosis in mouse macrophages. IV. The immunological induction of pinocytic vesicles, secondary lysosomes, and hydrolytic enzymes. J Exp Med. 1967;125(6):1091–1104. doi: 10.1084/jem.125.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amyere M, Payrastre B, Krause U, Van Der Smissen P, Veithen A, Courtoy PJ. Constitutive macropinocytosis in oncogene-transformed fibroblasts depends on sequential permanent activation of phosphoinositide 3-kinase and phospholipase C. Mol Biol Cell. 2000;11(10):3453–3467. doi: 10.1091/mbc.11.10.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veithen A, Cupers P, Baudhuin P, Courtoy PJ. v-Src induces constitutive macropinocytosis in rat fibroblasts. J Cell Sci. 1996;109(Pt 8):2005–2012. doi: 10.1242/jcs.109.8.2005. [DOI] [PubMed] [Google Scholar]
- 7.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, Bar-Sagi D. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB. The utilization of extracellular proteins as nutrients is suppressed by mTORC1. Cell. 2015;162(2):259–270. doi: 10.1016/j.cell.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeineddine R, Yerbury JJ. The role of macropinocytosis in the propagation of protein aggregation associated with neurodegenerative diseases. Front Physiol. 2015;6:277. doi: 10.3389/fphys.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruth HS, Jones NL, Huang W, Zhao B, Ishii I, Chang J, Combs CA, Malide D, Zhang WY. Macropinocytosis is the endocytic pathway that mediates macrophage foam cell formation with native low density lipoprotein. J Biol Chem. 2005;280(3):2352–2360. doi: 10.1074/jbc.M407167200. [DOI] [PubMed] [Google Scholar]
- 11.Chung JJ, Huber TB, Godel M, Jarad G, Hartleben B, Kwoh C, Keil A, Karpitskiy A, Hu J, Huh CJ, Cella M, Gross RW, Miner JH, Shaw AS. Albumin-associated free fatty acids induce macropinocytosis in podocytes. J Clin Investig. 2015;125(6):2307–2316. doi: 10.1172/JCI79641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 13.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78(1):35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 14.Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270(2):815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 15.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6(11):1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 17.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 18.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 19.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 20.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10(3):457–468. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 21.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137(5):873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9(3):316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 23.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25(6):903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, Kikkawa U, Yonezawa K. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282(28):20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20(20):2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16(18):1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405(3):513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 2012;18(9):524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev. 2013;23(1):53–62. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 2013;38(5):233–242. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendoza MC, Er EE, Blenis J. The Ras–ERK and PI3K–mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36(6):320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15(6):555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24(7):400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao Y, Inoki K. The role of mechanistic target of rapamycin in maintenance of glomerular epithelial cells. Curr Opin Nephrol Hypertens. 2016;25(1):28–34. doi: 10.1097/MNH.0000000000000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurdi A, De Meyer GR, Martinet W. Potential therapeutic effects of mTOR inhibition in atherosclerosis. Br J Clin Pharmacol. 2016;82(5):1267–1279. doi: 10.1111/bcp.12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perluigi M, Di Domenico F, Butterfield DA. mTOR signaling in aging and neurodegeneration: at the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiol Dis. 2015;84:39–49. doi: 10.1016/j.nbd.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Pacitto R, Di Gaeta I, Swanson JA, Yoshida S. CXCL12-induced macropinocytosis modulates two distinct pathways to activate mTORC1 in macrophages. J Leukoc Biol. 2017;101:683–692. doi: 10.1189/jlb.2A0316-141RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zwartkruis FJ, Burgering BM. Ras and macropinocytosis: trick and treat. Cell Res. 2013;23(8):982–983. doi: 10.1038/cr.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida S, Pacitto R, Yao Y, Inoki K, Swanson JA. Growth factor signaling to mTORC1 by amino acid-laden macropinosomes. J Cell Biol. 2015;211(1):159–172. doi: 10.1083/jcb.201504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sung S, Choi J, Cheong H. Catabolic pathways regulated by mTORC1 are pivotal for survival and growth of cancer cells expressing mutant Ras. Oncotarget. 2015;6(38):40405–40417. doi: 10.18632/oncotarget.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheong H. mTORC1 regulates nutrient access in Ras-mediated tumors. Aging. 2016;8(6):1165–1166. doi: 10.18632/aging.100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem. 2005;137(3):423–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- 44.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol. 2013;203(4):563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150(6):1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, Wang T, Bar-Peled L, Zoncu R, Straub C, Kim C, Park J, Sabatini BL, Sabatini DM. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347(6218):188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, Scorzoni S, Filipek PA, Huber KV, Bigenzahn JW, Heinz LX, Kraft C, Bennett KL, Indiveri C, Huber LA, Superti-Furga G. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519(7544):477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung J, Genau HM, Behrends C. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol Cell Biol. 2015;35(14):2479–2494. doi: 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, Vander Heiden MG, Sabatini DM. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. 2017;171(3):642–654 e612. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hallett JE, Manning BD. CASTORing new light on amino acid sensing. Cell. 2016;165(1):15–17. doi: 10.1016/j.cell.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. The sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014;9(1):1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng M, Yin N, Li MO. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell. 2014;159(1):122–133. doi: 10.1016/j.cell.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351(6268):43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, Sabatini DM. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell. 2016;165(1):153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jewell JL, Kim YC, Russell RC, Yu FX, Park HW, Plouffe SW, Tagliabracci VS, Guan KL. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015;347(6218):194–198. doi: 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136(3):521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell. 2012;47(3):349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 61.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5(6):566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 63.Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5(6):559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 64.Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156(4):771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17(15):1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11(6):1457–1466. doi: 10.1016/S1097-2765(03)00220-X. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5(6):578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 68.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4(9):648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 69.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4(9):658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 70.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10(1):151–162. doi: 10.1016/S1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 71.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci USA. 2004;101(37):13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25(9):545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benjamin D, Hall MN. mTORC1: turning off is just as important as turning on. Cell. 2014;156(4):627–628. doi: 10.1016/j.cell.2014.01.057. [DOI] [PubMed] [Google Scholar]
- 74.Demetriades C, Doumpas N, Teleman AA. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell. 2014;156(4):786–799. doi: 10.1016/j.cell.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carroll B, Maetzel D, Maddocks OD, Otten G, Ratcliff M, Smith GR, Dunlop EA, Passos JF, Davies OR, Jaenisch R, Tee AR, Sarkar S, Korolchuk VI. Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. eLife. 2016 doi: 10.7554/eLife.11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swanson JA. Phosphoinositides and engulfment. Cell Microbiol. 2014;16(10):1473–1483. doi: 10.1111/cmi.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mayor S, Parton RG, Donaldson JG. Clathrin-independent pathways of endocytosis. Cold Spring Harbor Perspect Biol. 2014 doi: 10.1101/cshperspect.a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shibutani S, Okazaki H, Iwata H. Dynamin-dependent amino acid endocytosis activates mechanistic target of rapamycin complex 1 (mTORC1) J Biol Chem. 2017 doi: 10.1074/jbc.M117.776443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, Guan KL. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285(26):19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flinn RJ, Yan Y, Goswami S, Parker PJ, Backer JM. The late endosome is essential for mTORC1 signaling. Mol Biol Cell. 2010;21(5):833–841. doi: 10.1091/mbc.E09-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saci A, Cantley LC, Carpenter CL. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell. 2011;42(1):50–61. doi: 10.1016/j.molcel.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buckley CM, King JS. Drinking problems: mechanisms of macropinosome formation and maturation. Febs J. 2017 doi: 10.1111/febs.14115. [DOI] [PubMed] [Google Scholar]
- 83.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465(7300):942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan HWS, Sim AYL, Long YC. Glutamine metabolism regulates autophagy-dependent mTORC1 reactivation during amino acid starvation. Nat Commun. 2017;8(1):338. doi: 10.1038/s41467-017-00369-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science. 1986;233(4768):1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 86.Egami Y, Taguchi T, Maekawa M, Arai H, Araki N. Small GTPases and phosphoinositides in the regulatory mechanisms of macropinosome formation and maturation. Front Physiol. 2014;5:374. doi: 10.3389/fphys.2014.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoshida S, Hoppe AD, Araki N, Swanson JA. Sequential signaling in plasma-membrane domains during macropinosome formation in macrophages. J Cell Sci. 2009;122(Pt 18):3250–3261. doi: 10.1242/jcs.053207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Racoosin EL, Swanson JA. Macropinosome maturation and fusion with tubular lysosomes in macrophages. J Cell Biol. 1993;121(5):1011–1020. doi: 10.1083/jcb.121.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hewlett LJ, Prescott AR, Watts C. The coated pit and macropinocytic pathways serve distinct endosome populations. J Cell Biol. 1994;124(5):689–703. doi: 10.1083/jcb.124.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Welliver TP, Chang SL, Linderman JJ, Swanson JA. Ruffles limit diffusion in the plasma membrane during macropinosome formation. J Cell Sci. 2011;124(Pt 23):4106–4114. doi: 10.1242/jcs.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Rooij J, Bos JL. Minimal Ras-binding domain of Raf1 can be used as an activation-specific probe for Ras. Oncogene. 1997;14(5):623–625. doi: 10.1038/sj.onc.1201005. [DOI] [PubMed] [Google Scholar]
- 92.Welliver TP, Swanson JA. A growth factor signaling cascade confined to circular ruffles in macrophages. Biol Open. 2012;1(8):754–760. doi: 10.1242/bio.20121784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dubielecka PM, Cui P, Xiong X, Hossain S, Heck S, Angelov L, Kotula L. Differential regulation of macropinocytosis by Abi1/Hssh3bp1 isoforms. PLoS One. 2010;5(5):e10430. doi: 10.1371/journal.pone.0010430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schlunck G, Damke H, Kiosses WB, Rusk N, Symons MH, Waterman-Storer CM, Schmid SL, Schwartz MA. Modulation of Rac localization and function by dynamin. Mol Biol Cell. 2004;15(1):256–267. doi: 10.1091/mbc.E03-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lanzetti L, Palamidessi A, Areces L, Scita G, Di Fiore PP. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 2004;429(6989):309–314. doi: 10.1038/nature02542. [DOI] [PubMed] [Google Scholar]
- 96.Hoon JL, Wong WK, Koh CG. Functions and regulation of circular dorsal ruffles. Mol Cell Biol. 2012;32(21):4246–4257. doi: 10.1128/MCB.00551-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Itoh T, Hasegawa J. Mechanistic insights into the regulation of circular dorsal ruffle formation. J Biochem. 2013;153(1):21–29. doi: 10.1093/jb/mvs138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Araki N, Johnson MT, Swanson JA. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J Cell Biol. 1996;135(5):1249–1260. doi: 10.1083/jcb.135.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Araki N, Egami Y, Watanabe Y, Hatae T. Phosphoinositide metabolism during membrane ruffling and macropinosome formation in EGF-stimulated A431 cells. Exp Cell Res. 2007;313(7):1496–1507. doi: 10.1016/j.yexcr.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 100.Dubielecka PM, Machida K, Xiong X, Hossain S, Ogiue-Ikeda M, Carrera AC, Mayer BJ, Kotula L. Abi1/Hssh3bp1 pY213 links Abl kinase signaling to p85 regulatory subunit of PI-3 kinase in regulation of macropinocytosis in LNCaP cells. FEBS Lett. 2010;584(15):3279–3286. doi: 10.1016/j.febslet.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoshida S, Gaeta I, Pacitto R, Krienke L, Alge O, Gregorka B, Swanson JA. Differential signaling during macropinocytosis in response to M-CSF and PMA in macrophages. Front Physiol. 2015;6:8. doi: 10.3389/fphys.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu WS, Heckman CA. The sevenfold way of PKC regulation. Cell Signal. 1998;10(8):529–542. doi: 10.1016/S0898-6568(98)00012-6. [DOI] [PubMed] [Google Scholar]
- 103.Ard R, Mulatz K, Pomoransky JL, Parks RJ, Trinkle-Mulcahy L, Bell JC, Gee SH. Regulation of macropinocytosis by diacylglycerol kinase zeta. PLoS One. 2015;10(12):e0144942. doi: 10.1371/journal.pone.0144942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Porat-Shliom N, Kloog Y, Donaldson JG. A unique platform for H-Ras signaling involving clathrin-independent endocytosis. Mol Biol Cell. 2008;19(3):765–775. doi: 10.1091/mbc.E07-08-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li G, D’Souza-Schorey C, Barbieri MA, Cooper JA, Stahl PD. Uncoupling of membrane ruffling and pinocytosis during Ras signal transduction. J Biol Chem. 1997;272(16):10337–10340. doi: 10.1074/jbc.272.16.10337. [DOI] [PubMed] [Google Scholar]
- 106.Palm W, Araki J, King B, DeMatteo RG, Thompson CB. Critical role for PI3-kinase in regulating the use of proteins as an amino acid source. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1712726114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wall AA, Luo L, Hung Y, Tong SJ, Condon ND, Blumenthal A, Sweet MJ, Stow JL. Small GTPase Rab8a-recruited phosphatidylinositol 3-kinase gamma regulates signaling and cytokine outputs from endosomal toll-like receptors. J Biol Chem. 2017;292(11):4411–4422. doi: 10.1074/jbc.M116.766337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bloomfield G, Traynor D, Sander SP, Veltman DM, Pachebat JA, Kay RR. Neurofibromin controls macropinocytosis and phagocytosis in Dictyostelium. eLife. 2015 doi: 10.7554/eLife.04940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Veltman DM, Williams TD, Bloomfield G, Chen BC, Betzig E, Insall RH, Kay RR. A plasma membrane template for macropinocytic cups. eLife. 2016 doi: 10.7554/eLife.20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Swanson JA. Phorbol esters stimulate macropinocytosis and solute flow through macrophages. J Cell Sci. 1989;94(Pt 1):135–142. doi: 10.1242/jcs.94.1.135. [DOI] [PubMed] [Google Scholar]
- 111.Koivusalo M, Welch C, Hayashi H, Scott CC, Kim M, Alexander T, Touret N, Hahn KM, Grinstein S. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J Cell Biol. 2010;188(4):547–563. doi: 10.1083/jcb.200908086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marat AL, Haucke V. Phosphatidylinositol 3-phosphates-at the interface between cell signalling and membrane traffic. EMBO J. 2016;35(6):561–579. doi: 10.15252/embj.201593564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shisheva A. PIKfyve: partners, significance, debates and paradoxes. Cell Biol Int. 2008;32(6):591–604. doi: 10.1016/j.cellbi.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bridges D, Ma JT, Park S, Inoki K, Weisman LS, Saltiel AR. Phosphatidylinositol 3,5-bisphosphate plays a role in the activation and subcellular localization of mechanistic target of rapamycin 1. Mol Biol Cell. 2012;23(15):2955–2962. doi: 10.1091/mbc.E11-12-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Robinson FL, Dixon JE. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 2006;16(8):403–412. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 116.Hao F, Itoh T, Morita E, Shirahama-Noda K, Yoshimori T, Noda T. The PtdIns3-phosphatase MTMR3 interacts with mTORC1 and suppresses its activity. FEBS Lett. 2016;590(1):161–173. doi: 10.1002/1873-3468.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Erami Z, Khalil BD, Salloum G, Yao Y, LoPiccolo J, Shymanets A, Nurnberg B, Bresnick AR, Backer JM. Rac1-stimulated macropinocytosis enhances Gβγ activation of PI3Kβ. Biochem J. 2017 doi: 10.1042/BCJ20170279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK–ERK pathway to late endosomes. EMBO J. 2009;28(5):477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, Thompson CB, Rabinowitz JD. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Can Res. 2015;75(3):544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tajiri H, Uruno T, Shirai T, Takaya D, Matsunaga S, Setoyama D, Watanabe M, Kukimoto-Niino M, Oisaki K, Ushijima M, Sanematsu F, Honma T, Terada T, Oki E, Shirasawa S, Maehara Y, Kang D, Cote JF, Yokoyama S, Kanai M, Fukui Y. Targeting Ras-driven cancer cell survival and invasion through selective inhibition of DOCK1. Cell Rep. 2017;19(5):969–980. doi: 10.1016/j.celrep.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 121.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nofal M, Zhang K, Han S, Rabinowitz JD. mTOR inhibition restores amino acid balance in cells dependent on catabolism of extracellular protein. Mol Cell. 2017;67(6):936–946 e935. doi: 10.1016/j.molcel.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nathan N, Keppler-Noreuil KM, Biesecker LG, Moss J, Darling TN. Mosaic disorders of the PI3K/PTEN/AKT/TSC/mTORC1 signaling pathway. Dermatol Clin. 2017;35(1):51–60. doi: 10.1016/j.det.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]