Abstract
Myalgic encephalomyelitis (ME)/chronic fatigue syndrome (CFS) (ME/CFS) is a disabling and debilitating disease of unknown aetiology. It is a heterogeneous disease characterized by various inflammatory, immune, viral, neurological and endocrine symptoms. Several microbiome studies have described alterations in the bacterial component of the microbiome (dysbiosis) consistent with a possible role in disease development. However, in focusing on the bacterial components of the microbiome, these studies have neglected the viral constituent known as the virome. Viruses, particularly those infecting bacteria (bacteriophages), have the potential to alter the function and structure of the microbiome via gene transfer and host lysis. Viral-induced microbiome changes can directly and indirectly influence host health and disease. The contribution of viruses towards disease pathogenesis is therefore an important area for research in ME/CFS. Recent advancements in sequencing technology and bioinformatics now allow more comprehensive and inclusive investigations of human microbiomes. However, as the number of microbiome studies increases, the need for greater consistency in study design and analysis also increases. Comparisons between different ME/CFS microbiome studies are difficult because of differences in patient selection and diagnosis criteria, sample processing, genome sequencing and downstream bioinformatics analysis. It is therefore important that microbiome studies adopt robust, reproducible and consistent study design to enable more reliable and valid comparisons and conclusions to be made between studies. This article provides a comprehensive review of the current evidence supporting microbiome alterations in ME/CFS patients. Additionally, the pitfalls and challenges associated with microbiome studies are discussed.
Keywords: Myalgic Encephalomyelitis/Chronic Fatigue Syndrome, microbiome, Virome
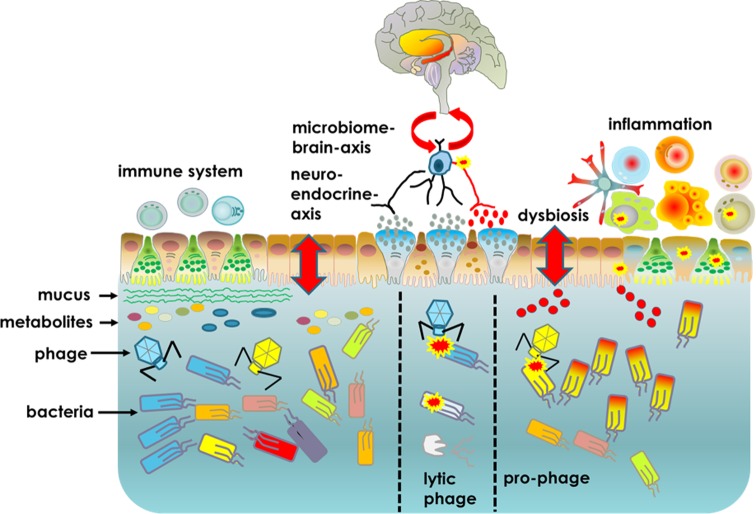
Graphical Abstract. The gut lumen contains large numbers of different bacteria which generate metabolites that together help maintain mucosal barrier function and reduce inflammation. The gut lining also contains enteroendocrine cells that secrete hormones which influence the enteric nervous system. These cells sense metabolites in the gut creating a microbiome–brain axis. Each bacterium can be infected by a bacteriophage and this can influence the microbiota. In this example, a blue bacteria is infected by a blue lytic phage and killed. The blue bacteria and its beneficial metabolites are lost from the microbiota. A second yellow bacteria is infected by a yellow prophage which integrates into the bacterial genome giving a selective advantage. Yellow bacteria and their harmful metabolites (red) predominate leading to dysbiosis, inflammation and altered enteric nervous system.
What is the microbiome?
Virtually every surface of the human body is colonized by vast populations of microbes, including prokaryotes, archaea, viruses, fungi and unicellular eukaryotes [1–3]. Bacteria of the phyla Bacteriodetes and Firmicutes dominate the diverse and complex intestinal bacteriome of most animals [4]. Microbial colonization begins rapidly at birth when the infant is first exposed to microbes in its immediate environment. The microbiome increases in diversity during the first 2–4 years of life in response to various hosts (i.e. genetics), and environmental factors including diet, lifestyle and behaviour [5–7]. It is believed that the early colonizers of the infant intestine play a key role in laying the foundations for the development of the complex and diverse adult microbiome and lifelong health [8]. In recent years, the role of microbiome in health of the host and its contribution to disease development has emerged [9–11]. It contributes to various body systems including immunity, metabolism, neurological signalling and homeostasis [12,13]. Describing the microbiome in detail is beyond the scope of this article, however several excellent review papers have been published recently [7,14–17].
The neglected virome
The vast majority of microbiome research has to date focused on its bacterial component, largely neglecting the other organisms. However, the influence of these lesser studied organisms, such as viruses are just beginning to be understood, thanks primarily to recent advancements in sequencing technology and bioinformatics capability (Table 1) [18].
Table 1. Overview of important faecal virome studies in health and disease.
| Year | Study participants | Comments | Reference |
|---|---|---|---|
| 2003 | Healthy adults | First virome metagenomics study | [168] |
| 2006 | Healthy adults | Plant RNA viruses contribute towards virome | [169] |
| 2008 | Infants | Virome establishment begins within 1 week of birth | [21] |
| 2011 | Healthy adults | Diversity and abundance of ssDNA viruses | [170] |
| 2011 | Monozygotic twins and mothers | Virome is individualized and highly stable | [22] |
| 2011 | Healthy adults | Virome is influenced by diet | [157] |
| 2012 | Healthy adults | Hypervariation driven by unique reverse transcriptase based mechanism | [171] |
| 2013 | Healthy adult | Virome is relatively stable; 80% of virome remained through 2.5-year study | [172] |
| 2013 | Pediatric CD patients | CD patients exhibited higher bacteriophage levels than controls | [49] |
| 2013 | CD patients | Similar results as above; results depend on interpretation of data | [173] |
| 2015 | Infants | Longitudinal study of virome establishment in infant twins | [174] |
| 2015 | Malnourished Malawian twins | Virome establishment affected by severe malnourishment | [178] |
| 2015 | IBD patients | Virome in IBD patients | [51] |
| 2015 | IBD patients | Increase in phage-richness abundance compared with healthy controls | [175] |
| 2015 | CD patients | Alterations in virome according to disease status and therapy | [58] |
Abbreviations: CD, Crohn’s disease; IBD; inflammatory bowel disease.
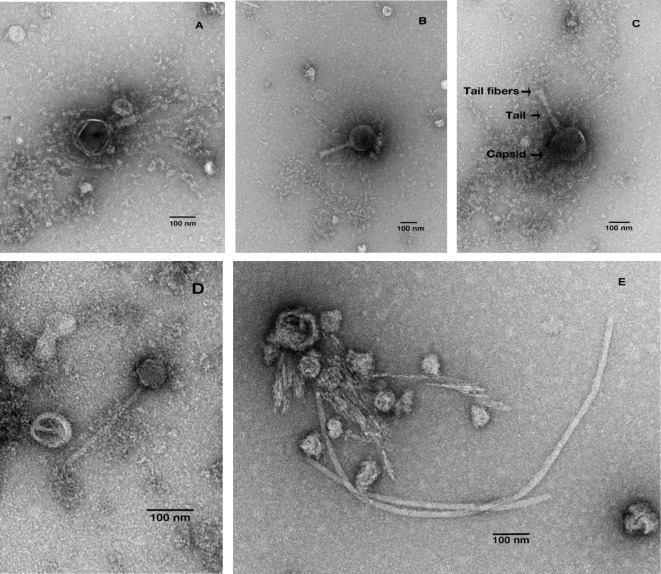
It is estimated that there are 1031 different DNA and RNA viruses on the planet; many of which remain undiscovered [19]. This collection of viruses (dsDNA, ssDNA, dsRNA and ssRNA) within an ecosystem is defined as the virome [20]. Similar to the bacteriome, the intestinal virome is established from birth and increases in diversity/complexity with age [21]. A large proportion of this complex environment consists of prokaryotic viruses (bacteriophage); with archaea-, human-, plant- and amoeba-infecting viruses found at lower frequencies [20]. The tailed, dsDNA viruses of the Order Caudovirales (Siphoviridae, Myoviridae, Podoviridae) dominate the bacteriophage portion of the virome (Figure 1) [22].
Figure 1. TEM images of Caudovirales from faecal water.
(A–C) Myoviridae and (D,E) Siphoviridae. Imaging completed by S.-Y.H. and K.C.
The microbiota in health and disease
The importance of the intestinal microbiome in maintaining health is an emerging research topic with advances in high-throughput sequencing technology allowing the identification and characterization of microbes that contribute to host health [10,11]. The microbiota has been implicated in immunomodulation, pathogen resistance, maintenance of intestine structure/function and nutrition and host metabolism [12,13]. It provides the host with a physical barrier to pathogen invasion and infection by, for example competitive exclusion and competing for nutrients, occupation of attachment sites and production of antimicrobial proteins [23–26]. Importantly, various microbiome members have been found to contribute to the intestinal metabolome, through for example vitamin synthesis, bile salt metabolism and xenobiotic degradation [27]. There is bidirectional communication between the microbiome and the local host immune system [28]. The immune system influences the composition of the microbiota and gut microbes and their products (e.g. metabolites and microbe associated molecular pattern (MAMPs) molecules) direct immune maturation and the development and possibly maintenance of immune (microbial) tolerance and homoeostasis [29,30].
There is increasing evidence that an imbalance of the intestinal microbiota (dysbiosis) may contribute to the pathogenesis of diseases affecting the gastrointestinal (GI) tract and other organ systems. Dysbiosis is characterized by a detrimental alteration of intestine microbial populations and ecology that can result in the growth and expansion of pathogenic microbes (pathobionts) and the production of factors toxic or harmful to host cells. These alterations are normally held in check by an intact microbiome but dysbiosis can result in the development and/or maintenance of chronic inflammatory infections caused by Clostridium difficile and Helicobacter pylori, metabolic syndrome and obesity, colorectal cancer, irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD) [14,31–34]. The stability of the microbiome is largely influenced by age, behaviour and lifestyle [12,35,36]. It has been hypothesized that intestinal microbial dysbiosis can lead to an imbalance in the immune system, resulting in diseases such as IBD, common variable immunodeficiency and rheumatoid arthritis [37,38]. However, to understand the significance of dysbiosis, it is necessary to establish if microbiome alterations cause, follow, precede or simply correlate with disease onset. Dysbiosis can be precipitated by drugs and medications (i.e. antibiotics), immune dysregulation, age-associated reduction in microbiota diversity, colonization by pathogenic microbes, stress and changes in diet [36,38–41]. The precise trigger or cause of dysbiosis in any disease has yet to be established but is likely to be multifactorial.
The virome in health
Viruses utilize a lytic or lysogenic life cycle. In the lytic life cycle, infected host cells are destroyed during viral replication whereas in the lysogenic life cycle the virus integrates into the host chromosome as a prophage. Lytic phages can have both narrow or broad host ranges, and lysogenic phages can be converted into a lytic cycle in response to environmental stressors such as antibiotics [20]. Lytic phages can alter the microbiome by killing bacterial hosts, providing a competitive growth advantage to bacteria resistant to phages. Prophages encode mobile genetic elements which contribute to horizontal gene transfer between bacteria altering antibiotic resistance, virulence or metabolic pathways [42]. This can provide a competitive advantage by allowing bacteria to metabolize new nutrient sources or acquire antibiotic resistance [43–45]. Temperate phages, able to perform lysogenic or lytic cycle, have been shown to influence the dynamics of biofilms and dispersal by a number of important pathogens such as Pseudomonas aeruginosa (opportunistic pathogen), Streptococcus pneumoniae (e.g. pneumonia) and Bacillus anthracis (e.g. anthrax) [46–48]. For example, the presence of lysogenic phages Bcp1, Wip1, Wip4 and Frp2 in B. anthracis results in the formation of a durable, complex and viable biofilm; allowing prolonged survival of the bacterium [48]. There is a constant shift of phages between lytic and lygosenic forms that is presumed to contribute to microbiome homoeostasis and that a differential spatial distribution of phages is correlated with health.
The virome in disease
Alterations in the virome have been implicated as sources of intestinal microbial (prokaryotic) dysbiosis for several different diseases [49–52]. Prophage induction in response to various environmental stressors can induce ‘community shuffling’ which alters the ratio of symbionts to pathobionts creating an imbalance within microbial communities that can lead to occupation of symbiont niches by pathobionts [42]. These events provide an explanation for the raised number of virus-like particles (VLPs, see Figure 2) as well as microbial population shifts in patients with GI-related disorders. Of note, an experimental model of Salmonella typhimurium diarrhoea has shown that inflammation increases lysogenic conversion of prophages [53].
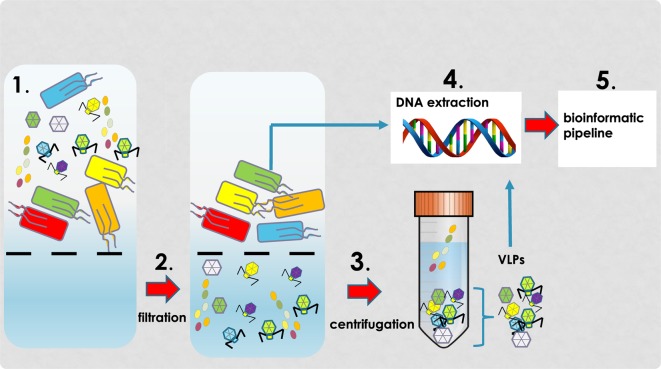
Figure 2. VLP isolation protocol.
Overview of VLP isolation protocol involving filtration (1,2) and centrifugation (3). Following isolation of concentration VLPs, DNA is extracted (4), sequencing and (5) bioinformatic tools applied to determine virome community composition.
The involvement of lytic phages in disease pathogenesis has been demonstrated through studying prophages that encode virulence factors (e.g. Shiga toxin) [54]. In the healthy intestine, toxin gene expression is silent in lysogenic phages that infect Escherichia coli. However, dysbiosis is accompanied by induction of prophages and activation of Shiga toxin genes resulting in release of the toxin into the intestine [55]. Additionally, in vitro experiments have shown phages can transmigrate across epithelial barrier cells. A recent study suggests that intestinal phages can interact directly with eukaryotic cells outside the GI tract; likely contributing to human health and immunity [56].
Although the virome is suspected to play a role in disease, relatively few studies have been undertaken with most studies focusing on IBD and HIV [49,50,52,57]. In one study, Crohn’s disease (CD) patients were shown to have a higher number of VLPs, which provide a crude estimate of phage numbers, in colonic biopsies compared with healthy controls. Patients with ulcerated mucosa had significantly fewer VLPs than non-ulcerated mucosa. The authors hypothesized that viruses had higher survival rates or a higher frequency of viruses in non-ulcerated areas. Based upon these findings, it was proposed that phages play an indirect role in the immune dysregulation evident in CD patients through microbiome alterations [57]. A later study also described differences in the virome in CD patients according to their disease status (newly diagnosed, active onset, active presurgery) and therapy. Newly diagnosed patients had a higher viral diversity in faecal and biopsy samples than those with active disease. Additionally, there were significant differences in virome diversity between patients on immunosuppressive therapy, steroids, combination therapy or no therapy. The clinical relevance of these results is unknown although more detailed studies of the alteration in virome composition in IBD patients is warranted [58]. Alterations in the enteric virome have also been reported in children susceptible to developing type 1 diabetes (T1D), prior to disease onset. Importantly, a disease-specific sequencing ‘fingerprint’ was identified in children susceptible to T1D that went on to develop the disease, compared with children that did not develop the disease. The present study reported that specific components of the virome were both directly and inversely associated with the development of T1D in these patients [59].
Additionally, a bidirectional communication network between the intestinal and central nervous system (the gut–brain–axis) is gaining research focus [60]. Its’ role is to monitor and integrate intestine functions as well as linking emotional and cognitive centres of the brain with peripheral intestinal functions and mechanisms such as immune system activation, intestinal permeability, enteric reflex, pain perception and enteroendocrine signalling [61]. Both clinical and experimental evidence suggest that the enteric microbiota has an important impact on communication pathways between the intestine and brain, also known as the gut–microbiome–brain axis. The microbiome can interact locally with intestinal cells and enteric nervous system, but also have indirect interactions with the CNS through neuroendocrine and metabolic pathways. Therefore, significant alterations in the resident microbiota or their metabolites might have a direct effect on the host nervous system and lead to neurological pathologies [62]. For example, changes in the microbiome have been associated with autism, depression, schizophrenia, Alzheimer’s disease and Parkinson’s disease [63–67].
Our own research is focused on developing a mechanistic understanding of the intestine–microbiome–brain axis and the GI tract microbiome in the pathogenesis of the neurological disorder, myalgic encephalomyelitis (ME)/chronic fatigue syndrome (CFS) (ME/CFS).
ME/CFS
Historical perspective
The causative factor(s) of ME/CFS remain elusive resulting in a lack of robust diagnostics and effective treatments [68,69]. The disease onset and progression varies from patient to patient with the onset normally associated with an acute flu-like viral infection, which is either gradual or rapid [68]. Approximately 25% of patients become house- or bed-bound with less than 10% returning to predisease levels of function [70]. The socioeconomic burden of ME/CFS is significant and estimated to be between $17 and $24 billion per annum. This considerable cost is due to direct and indirect effects of the illness, such as healthcare and loss of work for patient and/or family carers [71].
The heterogenic nature of ME/CFS suggests a multifactorial and self-sustaining disorder [72]. Several theories have been proposed including mitochondrial dysfunction, viral infection and autoimmunity [68,73,74]. Important clues for the involvement of (viral) infections in the aetiology of ME/CFS can be obtained from historical reports of epidemic or sporadic outbreaks of cases; the first of which was reported in 1934 in a suspected epidemic of poliomyelitis in Los Angeles, California [75,76]. The inconsistent disease pattern observed in patients led doctors to classify this epidemic as atypical; differing from polio cases endemic at the time by the lack of flaccid paralysis, which normally defines poliomyelitis [77]. Additionally, the affected cases were mainly older children and young adults compared with polio which affected infants and children of less than 5 years of age [78]. The disease at the onset consisted of an acute upper respiratory tract infection accompanied by muscle weakness, fever, pain, malaise and photophobia. The patients reported recurrence of fever and other symptoms during recovery, which were at a greater incidence than those in typical epidemic poliomyelitis [76].
A similar apparent epidemic of poliomyelitis appeared in Akureyri, Iceland between 1948 and 1949. There were striking similarities between this outbreak of atypical poliomyelitis and the one recorded in Los Angeles in 1934, including both overlapping symptoms and occurrence of relapse. This disease was named Iceland (or Akureyri) disease [79]. Sixty-one other outbreaks of a similar disease were reported worldwide between 1934 and 1990 [75]. The most significant outbreak was in 1955 at the Royal Free Hospital in London, where 292 hospital staff were affected by the illness. The disease when fully developed showed features of a generalized infection with involvement of the lymphoreticular system, and widespread involvement of the central nervous system. The mysterious polio-like illness (including the disease at the Royal Free Hospital) was renamed ME and later extended to CFS (ME/CFS) to include a seemingly identical disease [80,81].
What is ME/CFS?
In both historical and current cases of ME/CFS persistent fatigue is the dominant and defining symptom, which is accompanied by a range of heterogeneous symptoms that are universally present in all the patients. It is classified by the World Healthy Organization International Classification of Diseases (ICD-10) as a neurological disorder (WHO Reference 93.3). Patients often report delayed exacerbation of symptoms following mental or physical exertion and daily or weekly variations in symptom severity that have a significant impact on day-to-day living [69,82]. A standardized criterion for ME/CFS is urgently needed, with diagnosis relying heavily upon clinical observations and by exclusion of other disorders. This situation is further complicated by the use of different diagnostic criteria within the same country and between different countries. As a result, it is can take several years for sufferers to receive a diagnosis [83–85]. To date, an effective treatment for ME/CFS does not exist, with current treatments aimed at alleviating symptoms [86].
An intestinal origin for ME/CFS
The co-morbidity of ME/CFS and GI symptoms is well documented, with one study reporting 92% of patients exhibiting IBS [87]. Additional studies have reported increased mucosal and systemic levels of pro-inflammatory cytokines such as IL-6, IL-8, IL-1β and TNFα in patients with coexistent IBS [88,89]. The significant GI symptoms often experienced by ME/CFS patients has led researchers, including ourselves, to investigate the microbiome in these patients. Several studies have reported significant changes in microbiota composition of ME/CFS patients compared with controls [90–92]. However, ME/CFS microbiome studies to date have largely focused on alterations in bacterial populations. The advancement in sequencing technology and emerging influence of the virome on human health has enabled studies of the virome of ME/CFS [18].
Intestinal microbiome and ME/CFS
We completed a literature search to determine the extent of microbiome research in ME/CFS using the following search terms: ‘Myalgic encephalomyelitis’, ‘Chronic Fatigue Syndrome’, ‘CFS/ME’, ‘ME/CFS’ in combination with ‘virome’, ‘microbiome’, ‘metabolome’, ‘metagenomics’, ‘viromics’ and ‘metabolomics’. The resulting papers were screened according to abstract contents. Articles were excluded if an intervention was used (e.g. probiotics) and measurements not reported prior to the invention. This resulted in 11 papers that had examined the microbiome and/or intestinal metabolome of ME/CFS patients, dating from 1998 to 2017 (Table 2). Due to inconsistencies in study design including small sample sizes, different sequencing platforms and bioinformatics software analyses, microbial sequencing depth and a single time point‘snapshot’ of sampling and analysis; it was not possible to compare the studies statistically. However, from examining the articles individually there is sufficient evidence to support the claim of an altered intestinal microbiome in ME/CFS patients.
Table 2. ME/CFS microbiome articles selected following literature review.
| Number | Year | Author | Title | Area of study |
|---|---|---|---|---|
| 1 | 2017 | Armstrong [93] | The association of faecal microbiota and faecal, blood, serum and urine metabolites in ME/CFS | Microbiome and metabolites |
| 2 | 2017 | Nagy-Szakal [90] | Faecal metagenomic profiles in subgroups of patients with ME/CFS | Microbiome |
| 3 | 2016 | Giloteaux [91] | Reduced diversity and altered composition of the gut microbiota in individuals with ME/CFS | Microbiome |
| 4 | 2016 | Giloteaux [107] | A pair of identical twins discordant for ME/CFS differ in physiological parameters and gut microbiome composition | Microbiome and virome |
| 5 | 2013 | Fremont [92] | High-throughput 16S rRNA gene sequencing reveals alterations of intestinal microbiota in ME/CFS patients | Microbiome |
| 6 | 2009 | Sheedy [94] | Increased d-lactic acid intestinal bacteria in patients with CFS | Microbiome and metabolites |
| 7 | 2009 | Fremont [109] | Detection of herpes virus and parvovirus B19 in gastric and intestinal mucosa of CFS patients | Virome |
| 8 | 2008 | Chia [108] | CFS is associated with chronic enterovirus infection of the stomach | Virome |
| 9 | 2007 | Evengård [176] | Patients with CFS have higher numbers of anaerobic bacteria in the intestine compared with healthy subjects | Microbiome |
| 10 | 2001 | Butt [177] | Bacterial colonosis’ in patients with persistent fatigue | Microbiome |
| 11 | 1998 | Butt [98] | Faecal microbial growth inhibition in chronic fatigue/pain patients | Microbiome and metabolites |
Abbreviations: CFS, chronic fatigue syndrome; ME, myalgic encephalomyelitis
The literature search revealed nine articles that examined the microbiome in ME/CFS patients, with three articles also examining intestinal metabolites (Table 3); it is challenging to compare these studies because of different diagnostic criteria, patient selection, use or non-use of appropriately matched control subjects and microbial identification techniques. Of the nine articles, four used sequencing technologies, but different platforms, with five using culture-based techniques. One study (2017) performed metagenomic sequencing on 50 patient samples and was able to determine species-level differences compared with samples from control subjects [90]. A simple comparison between studies revealed eight similar results and seven conflicting results (Table 4). For example, while Giloteaux et al. [91] and Armstrong et al. [93] reported a general decrease in bacterial abundance, Sheedy et al. [94] reported an increase. These studies utilized different microbial identification techniques, which might account for the conflicting results. The lack of statistical analysis of the datasets constrains direct cross-study comparisons. From the limited cross-study analysis shown in Table 4, one finding of note was the decrease in Faecalibacterium seen in three studies. A reduction in butyrate-producing genus, which includes Faecalibacteria has been associated with dysbiosis in CD patients [95]. Butyrate has several protective properties, including improving the mucosal barrier, and immunomodulatory and anti-inflammatory effects by down-regulating pro-inflammatory cytokines [96]. However, decrease in the relative abundance of Faecalibacterium are associated with several other disorders and is not therefore specific for ME/CFS [97]. Interestingly, there was an increase in Enterobacteriaceae in two studies [91,98]. However, this may result from ME/CFS symptoms instead of a disease-specific microbial alteration. Enterobacteriaceae are dominant in the upper GI tract and are present in at low levels in the faeces of healthy individuals [99]. These taxa likely become enriched with faster stool transit time (i.e. signature of diarrhoea). The notable increase in this family would be consistent with increased transit time and reported in IBS-like symptoms in patients [100]. A depletion in the butyrate-producing family Ruminococcaceae was recorded in two studies and is also associated with diarrhoea [101]. Interestingly, Bacteroides spp. known for producing butyrate were reduced in several studies [97]. It would be beneficial to perform longitudinal studies of the microbiome in ME/CFS patients throughout the duration of the illness. This may provide more insightful clues as to the significance of any microbiome compositional changes with disease progression and severity within an individual patient. Several studies have been published on the longitudinal evaluation of ME/CFS patients; however, these focus on immune aspects, rehabilitative treatments and employment status [102–105].
Table 3. Overview of articles selected studying the microbiome in ME/CFS.
| Author | Number of patients | Number of controls | Studying | Study design |
|---|---|---|---|---|
| Armstrong [93] | 34 | 25 | Microbiome and metabolites | Culture + MS |
| Nagy-Szakal [90] | 50 | 50 | Microbiome | Metagenomics |
| Giloteaux [91] | 48 | 39 | Microbiome | 16s rRNA gene sequencing |
| Giloteaux [107] | 1 | 1 | Microbiome and virome | 16s rRNA gene sequencing |
| Fremont [92] | 43 | 36 | Microbiome | 16s rRNA gene sequencing |
| Sheedy [94] | 108 | 177 | Microbiome and metabolites | Culture |
| Evengard [176] | 10 | 10 | Microbiome | Culture |
| Butt [177] | 1390 | - | Microbiome | Culture |
| Butt [98] | 27 | 4 | Microbiome and metabolites | Culture |
Abbreviation: MS, mass spectrometry
Table 4. Basic comparison of microbiome composition alterations noted in articles selected for review.
| Article details | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Microbiome comparison | Armstrong (2017) [93] | Nagy-Szakal (2017) [90] | Giloteaux (2016) [91] | Giloteaux (2016) [107] | Fremont (2013) [92] | Sheedy (2009) [94] | Evangård (2007) [176] | Butt (2001) [177] | Butt (1998) [98] |
| Overall abundance | ↓ | ↓ | ↑ | ||||||
| Phylum Firmicutes | ↓ | ↑ | |||||||
| Phylum Proteobacteria | ↑ | ↓ | |||||||
| Family Bacteroidaceae | ↓ | ↑ | |||||||
| Family Enterobacteriacaeae | ↑ | ↑ | |||||||
| Family Prevotellaceae | ↑ | ↓ | |||||||
| Family Rickenellaceae | ↓ | ↓ | |||||||
| Family Ruminococcaceae | ↓ | ↓ | |||||||
| Genus Bacteroides | ↓ | ↓ | |||||||
| Genus Bifidobacterium | ↓ | ↓ | ↑ | ↓ | ↓ | ||||
| Genus Clostridium | ↑ | ↓ | |||||||
| Genus Coprobacillus | ↑ | ↑ | |||||||
| Genus Faecalibacterium | ↓ | ↓ | ↓ | ||||||
| Genus Haemophilus | ↓ | ↓ | |||||||
| Genus Ruminococcus | ↓ | ↓ | |||||||
| Species Enterococcus faecalis | ↑ | ↑ | |||||||
| Species E. coli | ↓ | ↓ | |||||||
Seventeen criteria were either similar or conflicting between studies (microbiome composition). The down arrows represent a decrease in patients and up arrows represent an increase in patients.
As with any microbiome study, it is difficult to determine if the alterations observed cause, precede or correlate with disease. The microbiome of a patient would exhibit disease-specific microbial signatures and general microbial changes due to an unbalanced microbiome [106]. It is important, therefore, to separate microbial alterations associated with an unbalanced microbiome from those associated with a specific disease (microbiome disease biomarkers).
Virome and ME/CFS
Of the 11 articles selected in our literature search, only 3 examined the intestinal virome of CFS/ME patients (Table 5). Of these, two articles used direct virus detection (e.g. PCR or immunostaining) and one article used a high-throughput sequencing technique (Illumina MiSeq). An increase in bacteriophage richness, particularly Siphoviridae and Myoviridae, in patients was noted in the Giloteaux et al. study [107]. However, this study is statistically underpowered due to its small sample size. Chia and Chia [108] and Frémont et al. [109] used virus detection techniques to examine the presence of eukaryotic viruses within the gastric/intestinal mucosa. These studies reported an increase in parvovirus B19, enteroviral RNA and viral capsid protein 1 in patients. Also of note, Nagy-Szakal et al. [90] used metagenomic based approach on a large (n=50) cohort of patients although they did not perform virome analysis on the dataset. The authors reported significant changes in the bacterial components of the microbiome in ME/CFS patients compared with controls [90]. Virome analysis could be performed with the available date to determine if significant changes are observed in the viral components of ME/CFS patients.
Table 5. Overview of articles selected studying the virome in ME/CFS.
Metabolomics studies
The identification of the bacterial and viral components of the microbiome is an important step forward, as is understanding how the use of nutrients by these microorganisms influences the overall metabolism within the gut. Metabolomics can be used to identify metabolites within the microbiome [110]. Only a handful of studies have attempted to characterize faecal metabolites in ME/CFS patients despite its potential for deciphering microbiome function (Table 6) [93,94,98]. There are significant challenges associated with identifying faecal metabolites due to differing metabolite properties and range of metabolite concentrations in samples [162,163]. A major challenge is not only to identify all metabolites (insufficient reference libraries available) but also to produce metadata (i.e. sample origin, tissue, experimental conditions) in a format that is easily interpreted [166]. The biological interpretation of metabolites as potential disease-associated biomarkers is often challenging as it requires data analysis and integration [167] and targeted and non-targeted metabolomics to dissect the metabolic pathway(s) and origin of metabolite(s) of interest [164]. Currently, 1H NMR is the most used analytical technique for metabolite profiling and is routinely used in clinical or pharmaceutical research and applications [165].
Table 6. Overview of articles selected for studying the metabolome in ME/CFS.
| Author | Number of patients | Number of controls | Study design |
|---|---|---|---|
| Armstrong [93] | 34 | 25 | NMR spectroscopy |
| Sheedy [94] | 108 | 177 | C13-labelled bacteria/metabolites for HPLC and NMR |
| Butt [98] | 27 | 4 | Specific metabolites |
Abbreviations: NMR, nuclear magnetic resonance; HPLC, high performance liquid chromatography
Armstrong et al. [93] quantified metabolites using high-throughput 1H NMR spectroscopy from ME patient faecal filtrates. This technique provides a non-targeted metabolic profile that measures all high concentration metabolites with non-exchangeable protons [111]. In addition to faecal metabolomics, the authors performed urine and blood serum metabolite analysis. The present study presented a robust metabolome workflow and eluded to the relationship of faecal metabolites and microbes with host blood serum and urine metabolites [93]. Two older studies used selective culture based systems to examine the metabolic output of specific bacteria [94,98]. It is difficult to draw conclusions from these older studies because of the culture-based techniques used. It is possible that the isolation of bacteria from the complex intestinal environment alters the excreted/secreted metabolites, resulting in metabolites specific to the artificial in vitro culture environment. To obtain a true picture of the faecal metabolome, samples should be prepared directly from the faecal sample.
Unfortunately, it is not possible to compare these three metabolomics studies directly because different metabolites were studied. However, it is possible to make general comparisons between metabolites and microbes. For example, Sheedy et al. [94] reported an increase in lactic acid and an increase in Enterococcus faecalis, a lactic acid producing bacteria. Interestingly, Armstrong et al. [93] reported a general increase in the short chain fatty acids (SCFAs) butyrate, isovalerate and valerate. This contradicts the microbiome studies as known SCFA-producing bacteria (Faecalibacterium, Eubacteria, Roseburia and Ruminococcus) were consistently decreased across multiple studies [90–92,107]. A decrease in lactate was also reported in this study [93]. Several bacterial members of the microbiota produce lactate, which is the most common short chain hydroxyl-fatty acid in the intestinal lumen [97,112]. It can be converted into other SCFAs by a subgroup of lactate-fermenting bacterial species. Changes in these lactate-fermenting bacterial species were not noted in the current microbiome studies. Future studies will need to examine microbiome and metabolome alterations in tandem and then integrate the data to reveal a truer picture of microbiome metabolism.
Studying the microbiome: techniques and challenges
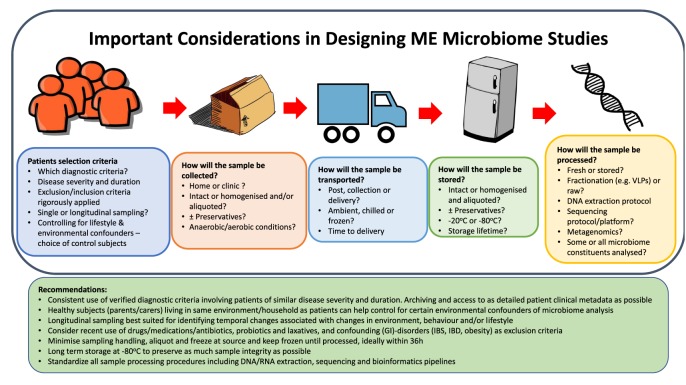
Within recent years, the increased interest in trying to understand the effect of the microbiome on health and disease has resulted in significant advancements in techniques to characterize it [9–11]. In particular, metagenomics is increasingly popular and favoured over sequencing bacterial 16s rRNA due to increased taxonomic sensitivity and potential for functional interpretation. Additionally, established techniques are being applied to microbiome research, such as metabolomics [11,113]. The research at the Quadram Institute Bioscience is focused on optimizing protocols, standardizing microbiome studies and applying this to ME/CFS. There are several pitfalls and challenges associated with microbiome studies, which need to be addressed prior to patient recruitment and sample collection [114]. The considerations that need to be made in designing microbiome studies in ME/CFS and some recommendations are outlined in Figure 3. Below we describe in some detail the particular constraints on microbiome and virome studies in ME/CFS and the approaches that can be taken to mitigate against or overcome them.
Figure 3. Important considerations in designing ME microbiome studies.
Recommendations for designing a microbiome study and important questions to consider
Patient recruitment
A standardized criterion for ME/CFS diagnosis is lacking, with diagnosis relying heavily upon clinical observations and exclusion [83,115]. Multiple diagnosis checklists have been created, with each checklist differing slightly on symptom emphasis and severity [69,83,84]. The International ME criteria and Canadian criteria place greater emphasis on the delayed exacerbation of physical and mental symptoms following exertion. However, it does not exclude psychiatric illness such as depression or anxiety [116,117]. Therefore, it is difficult to determine how many patients recruited have an accurate diagnosis of ME/CFS and how many have been misdiagnosed using inadequate criteria. Several studies have attempted to address this by using multiple diagnostic checklists, with the majority using the 1994 Fukuda diagnostic scale. The severity of ME/CFS is ranked according to impact of illness upon daily life and ranges from mild to very severe. The severity grade given to a patient is subjective and generally given by the diagnosing clinician. Several studies have used patient questionnaires to assess the level of illness [90,91,94,107]. However, four different patient questionnaires were used by four different studies (Short Form 36 Healthy Survey, Multidimensional Fatigue Inventory, Bell’s Disability Scale and McGregor 1995 questionnaire). Due to the multifactorial nature of the disease, standardization in diagnosis and disease severity are imperatives. These are a basic requirement to produce robust and reproducible microbiome studies. It is very difficult to determine if any microbiome differences are due to a true ME/CFS signature or complexities of patient recruitment. Future studies should aim to stratify patients according to disease duration and onset (sudden or gradual).
As microbiome research has increased, the need for properly matched controls has become apparent. The complexity of the GI microbiome and potential role within healthy/diseased states produce confounding factors [118]. As many of these confounding factors need to be taken into consideration in a microbiome study, and as many as possible should be accounted for or eliminated. Age, lifestyle, medications and drug use, geography and diet have all been reported to influence microbiome function and composition [5–7]. The effect of antibiotics on the microbiome is well documented [119]. However, other prescription and recreational drugs can affect microbiome analyses [120]. For example, decreasing stomach acidity with proton pump inhibitors allows upper GI microbes to move down into the intestine more readily, altering the composition of the lower GI microbiota and increasing risk of C. difficile infections [121].
Diet also influences the microbiome. Long-term dietary patterns have been linked to faecal microbiomes dominated by certain genera [118,122]. High protein/animal fat diets are associated with the prevalence of Bacteriodes, whereas diets high in carbohydrates are associated with high Prevotella [123]. To account for this, details of food consumption at least 48 h prior to sample collection should be obtained. Moreover, healthy household controls could be used to identify and exclude environmental confounders (e.g. diet, living environment); increasing the likelihood of identifying disease-specific microbiome signatures [118]. Additionally, the microbiome changes during aging and declines in diversity in the elderly [124]. Therefore, the use of age-matched controls would be beneficial to account for this important variable.
Recently, the influence of gender on the microbiome (termed as ‘microgenderome’) has become evident [125–128]. The intestine and its’ microbiome serves as a virtual endocrine organ due to the metabolites and neurotransmitters and hormones it can produce [129]. For example, early microbial exposure increases testosterone levels in male mice, leading to a protective effect against T1D [128]. Additionally, microbiome alterations are observed in pre- and post-menopausal women; highlighting hormonal cross-talk within the microbiome [130]. Certain microbes have also been discovered to be a source of hormones and neurotransmitters. Experimental models have confirmed the bidirectional relationship between the intestinal microbiota, sex hormones and the immune system and provided an explanation for sexual dimorphism in T1D [128,131]. The results of these studies revealed evidence of sex-specific microbial communities, sex-specific responses to the same microbial communities, the role of sexual maturation impacting on changes on microbial communities and that microbial communities can play a protective and therapeutic role by influencing hormonal, metabolic and immune pathways [125–128]. A 2015 study compared the microbiome of male and female patients with ME/CFS revealing significant sex-specific interactions between Firmicutes (Clostridium, Streptococcus, Lactobacillus and Enterococcus) and symptoms, regardless of compositional similarity of microbial levels across the sexes [132]. This study highlights the need for gender-matched controls to account for any gender bias from future microbiome studies.
Although it is often impractical and perhaps impossible to control for all confounding factors within a microbiome study, efforts should be made to account for as many as possible.
Sample collection, storage and processing
As the number of microbiome studies has increased, the need for consistency in sampling techniques and standard operating procedures (SOPs) has also increased. An excellent review of the critical factors for sample collection, storage, transport and ‘gold standard’ techniques for longitudinal microbiome studies in human populations was recently published [114]. The most important considerations for storing microbiome samples are to reduce changes in the original microbiota from sample collection to processing and to keep storage conditions consistent for all samples in a study [133,134]. Sample storage conditions are not always consistent due to study or research group-specific downstream applications and resource limitations. Additionally, considerations are not always taken for preserving anaerobic bacteria within an anaerobic environment. Different studies often store samples at differing temperature (e.g. 4 to −80°C), affecting the long-term preservation of certain bacteria [114]. Additionally, the length of time for which the sample is stored and frequency of freeze/thaw cycles can significantly affect the microbiome composition. For example, Bacteroides is sensitive to freezing and should be processed within 6 weeks of storage (at −80°C) to avoid bacterial degradation [135,136]. The microbiome and ME/CFS studies reviewed here used different sample collection and storage techniques; including storage at <12°C, immediate processing, −20 and −80°C. For logistical reasons, it can be difficult to standardize this across all studies. However, it is important to be aware of these limitations.
The microbe composition changes laterally and longitudinally along the GI tract, therefore it has been suggested that there is significant variation within a single faecal sample. A 2015 study reported a reduction in intrasample variation following homogenization of the whole faecal sample [137]. However, several studies use a random section of the faeces without homogenization.
Additionally, different DNA extraction techniques have been used as a prelude to sequencing (MoBio PowerSoil DNA isolation kit, QIAmp DNA Stool Mini Kit and DNeasy Blood and Tissue kit). These kits differ in protocol, bead size, reagents used and are likely to introduce unnecessary bias [138].
Identification of prokaryotes
The development of sequencing, characterization of the bacterial component of the faecal microbiome relied on culture-based techniques that allow the identification of anaerobic and aerobic bacteria using selective or non-selective culture conditions and media; albeit taxonomic resolution and sensitivity is relatively low [12]. However, this approach does inform the cultured organism’s growth requirements and substrate utilization and other physiological parameters, which cannot be obtained from sequence-based approaches [12]. Next-generation sequencing technology now makes it possible to characterize the bacterial microbiome using the 16S rRNA gene ‘fingerprint’ for identification and as an indicator of genetic diversity [4]. The 16S rRNA gene was chosen because of its relatively small size (~1.5 kb) and harbouring enough variation to distinguish between different species, yet enough similarity to assign members belonging to the same larger phylogenetic group (e.g. order, family or phylum) [5,139]. However, this approach has its limitations. It only detects and analyses a short, specific genomic region and taxonomic resolution or functional inference is therefore limited [11]. For example, this assay cannot recognize the different serovars within Salmonella enterica or detect toxin genes that could distinguish pathogenic C. difficle or distinguish pathogenic Escherichia strains from non-pathogenic strains [140]. This is particularly problematic in comparative studies of the microbiome in healthy and diseased states. It also provides no insight into functionality of the bacteriome [11].
Metagenomic sequencing is increasingly being chosen over 16S rRNA sequencing due to its higher taxonomic resolution and ability to infer functional potential [140]. It provides sequence information from the collective genomes of the microbiota, which in turn can be used to infer or predict functional contributions and biological roles of this complex community in human health and disease [11,139]. However, the absence of whole genome sequences in public databases limits the ability to identify gene function based on known sequence information. In comparison with 16S-based sequencing approaches, whole community metagenomics with an appropriate sequencing depth and coverage can be used to identify other microbes (i.e. archaea and viruses) within the microbiome [140,141]. Although it is possible to infer functional potential from metagenomic analysis through gene presence/abundance, the presence of a gene does not necessarily infer function; it is possible for the gene to be present but not transcribed [113]. Therefore, careful consideration needs to be taken when inferring functional potential from metagenomic sequences and, if possible, the predicted function should be examined using laboratory based techniques (e.g. antibiotic resistance), assuming the candidate microbe(s) can be cultured in isolation.
To date, only one study has utilized metagenomics in ME/CFS microbiome studies [90]. However, the analysis was incomplete and did not fully exploit the data produced. Whenever possible, metagenomics should be applied to microbiome studies in ME/CFS in order to achieve the required taxonomic resolution to fully examine the bacteriome and virome.
Identification of viruses
Virus genomes do not encode universally conserved genes such as the 16S or 18S genes of prokaryotes and eukaryotes respectively, and they are genetically highly diverse [142]. Consequently, it is not possible to use metataxonomic approaches such as 16S rRNA gene sequencing to characterize VLPs within an ecosystem [20]. Traditionally, classical approaches of microscopy and cultivation have been used to characterize VLPs isolated from faecal samples originating in the human intestine [143,144]. The only reliable molecular method currently available for surveying the human virome is metagenomics. However, to achieve an adequate sequencing depth, lytic VLPs need to be separated from the faecal material [145,146]. An excellent review describing the human virome and its characterization was recently published [147].
The efficient isolation of VLPs is an essential step in viral metagenomics in order to obtain an accurate picture and profile of the virome [148,149]. The workflow for sequencing the nucleic acid in VLPs (Figure 2) from faecal material begins with homogenization of faeces in buffer, centrifugation to remove cell debris followed by filtration to remove bacteria [150]. Ultracentrifugation can be used to separate the sample into differing densities and a specific density containing VLPs can be selected for downstream processing. Within the intestinal microbiome community, viral genomes represent a small proportion of the total DNA compared with bacterial genomes [149,151]. It is important therefore to use a reliable, robust and efficient VLP isolation protocol with as few manipulations of the sample as possible to minimize loss of VLPs. Various VLP protocols have been published that differ in details such as filter pore size, centrifugation speed and the inclusion/omission of ultracentrifugation [148–150,152,153]. Importantly, these protocols have yet to be directly compared. It is highly desirable therefore that standardized faecal VLP isolation and DNA extraction techniques are adopted to enable direct comparisons of datasets from different virome studies.
Viral metagenomic data are analysed in a manner similar to bacterial metagenomic data [154,155]. High-quality reads are aligned to reference databases, assembly is then attempted with non-aligned reads and functional characteristics inferred [147]. However, the lack of conserved genes, high genetic variation and under-representation of virus genomes within reference databases means a minority of the reads can be taxonomically assigned. It is predicted that less than 0.001% of the predicted phage diversity is represented in global sequence databanks [156]. One virome study has reported that 98% of the generated reads did not significantly match to an identified sequence within a database [157]. Therefore, a majority of sequencing reads are unassigned to any known genomes and are considered ‘viral dark matter’. Additionally, assembly of sequencing reads is made difficult due to their short-read lengths [158,159]. Several research groups are now investigating the possibility of long-read sequencing to characterize the virome. The release of the Oxford Nanopore MinION has drastically reduced the cost of long-read sequencing (compared with PacBio sequencing). These have the potential to provide complete or near complete phage genomes without the need for alignment or representation in databases [160].
Perhaps the biggest challenge in studying the intestinal virome is the lack of bioinformatics tools for the analysis of sequence data [146,147,161]. To date, there is no easy-to-use pipeline that uses raw reads, can remove host DNA, can search for bacterial contaminants and assign taxonomy and functionality to viruses within the sample. However, efforts are being made to generate such tools. In addition to isolation and sequencing of VLPs, it is possible to identify prophages and the bacterial host(s) from metagenomic sequencing. To accurately study the virome, both techniques should be utilized to study the lytic and lysogenic phages [154,155,157]. The Norwich U.K., ME/CFS research group is currently optimizing and standardizing VLP isolation and DNA/RNA sequencing protocols in addition to developing fit-for purpose viromics pipelines to comprehensively analyse the virome in ME/CFS patients.
Concluding remarks
Several microbiome studies have been performed on ME/CFS patients in the hope of identifying disease-specific signatures. This disease should be viewed as multifactorial and that the alteration of one body system (e.g. microbiome) may not be the exclusive cause. The dysregulation of the microbiome may be variously placed in a disease progression pathway interfacing with other systems (immune, neuroendocrine and mitochondrial), tipping the body into persistent imbalance. Although studies to date often report conflicting results, microbiome dysbiosis in ME/CFS patients is evident. However, in order to discover disease-specific microbe alterations, future studies need to adopt standardized techniques and analyses. The recent advancements in sequencing technology allows the characterization of the previously neglected virome. As virome research increases, it is becoming clear that the virome can directly and indirectly affect host health, and may play a role in the pathogenesis of ME/CFS. Confirmation of such a role will be largely dependent on the adoption of robust patient selection, reproducible study design and appropriate data analyses by different research groups investigating the microbiome/virome in complex diseases such as ME/CFS.
Clinical perspectives
Several studies have reported alterations in the intestinal microbiome of ME/CFS patients, suggesting the involvement of microbial dysbiosis.
Study design needs to be consistent to allow statistical comparison between different microbiome studies.
Future microbiome studies should take account of the virome.
Acknowledgments
We thank Navena Navaneetharaja, Nadine Davies, Daniel Vipond and Lesley Hoyles for their contribution towards this research; additionally, Kathryn Cross for the TEM imaging of intestinal viruses. ME/CFS research was carried out in the laboratories of S.R.C. and T.W.
Abbreviations
- CD
Crohn’s disease
- CFS
chronic fatigue syndrome
- GI
gastrointestinal
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- IL
Interleukin
- ME
myalgic encephalomyelitis
- ME/CFS
myalgic encephalomyelitis/chronic fatigue syndrome
- SCFA
short chain fatty acid
- T1D
type 1 diabetes
- TNFα
Tumor necrosis factor alpha
- VLP
virus-like particle
- WHO
World Health Organization
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported in part by Invest in ME to S.R.C. and T.W.; and the Ph.D. funding [grant number R205102 to F.N.)].
Author contribution
F.N. wrote the manuscript with support from S.R.C. and T.W. T.W. also designed the graphical illustrations. S.-Y.H. prepared samples for TEM imaging.
References
- 1.Kunz C., Kuntz S. and Rudloff S. (2009) Intestinal flora. Adv. Exp. Med. Biol. 639, 67–79 10.1007/978-1-4020-8749-3_6 [DOI] [PubMed] [Google Scholar]
- 2.Morelli L. (2008) Postnatal development of intestinal microflora as influenced by infant nutrition. J. Nutr. 138, 1791S–1795S 10.1093/jn/138.9.1791S [DOI] [PubMed] [Google Scholar]
- 3.Neish A.S. (2009) Microbes in gastrointestinal health and disease. Gastroenterology 136, 65–80 10.1053/j.gastro.2008.10.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M. et al. (2005) Diversity of the human intestinal microbial flora. Science 308, 1635–1638, 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer C., Bik E.M., DiGiulio D.B., Relman D.A. and Brown P.O. (2007) Development of the human infant intestinal microbiota. PLoS Biol. 5, 1556–1573 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koenig J.E., Spor A., Scalfone N., Fricker A.D., Stombaugh J., Knight R. et al. (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108, 4578–4585 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamburini S., Shen N., Wu H.C. and Clemente J.C. (2016) The microbiome in early life: implications for health outcomes. Nat. Med. 22, 713–717 10.1038/nm.4142 [DOI] [PubMed] [Google Scholar]
- 8.Mackie R.I., Sghir A. and Gaskins H.R. (1999) Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69, 1035S–1045S [DOI] [PubMed] [Google Scholar]
- 9.Robinson C.J., Bohannan B.J.M. and Young V.B. (2010) From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74, 453–476, 10.1128/MMBR.00014-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassis C., Young V. and Schmidt T. (2013) Methods for characterizing microbial communities associated with the human body. In The Human Microbiota, pp. 51–74, John Wiley & Sons, Inc., Hoboken, NJ, U.S.A. [Google Scholar]
- 11.Di Bella J.M., Bao Y., Gloor G.B., Burton J.P. and Reid G. (2013) High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods 95, 401–414, 10.1016/j.mimet.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 12.Sekirov I., Russell S. and Antunes L. (2010) Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- 13.O’Hara A.M. and Shanahan F. (2006) The gut flora as a forgotten organ. EMBO Rep. 7, 688–693 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shreiner A.B., Kao J.Y. and Young V.B. (2016) The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31, 69–75 10.1097/MOG.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeiffer J.K. and Virgin H.W. (2016) Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351, aad5872 10.1126/science.aad5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim E.S., Wang D. and Holtz L.R. (2016) The bacterial microbiome and virome milestones of infant development. Trends Microbiol. 24, 801–810, 10.1016/j.tim.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 17.Columpsi P., Sacchi P., Zuccaro V., Cima S., Sarda C., Mariani M. et al. (2016) Beyond the gut bacterial microbiota: The gut virome. J. Med. Virol. 88, 1467–1472 10.1002/jmv.24508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haynes M. and Rohwer F. (2011) The human virome. In Metagenomics of the Human Body, pp. 63–77, Springer New York, New York [Google Scholar]
- 19.Breitbart M. and Rohwer F. (2005) Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13, 278–284, 10.1016/j.tim.2005.04.003 [DOI] [PubMed] [Google Scholar]
- 20.Virgin H.W. (2014) The virome in mammalian physiology and disease. Cell 157, 142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breitbart M., Haynes M., Kelley S., Angly F., Edwards R.A., Felts B. et al. (2008) Viral diversity and dynamics in an infant gut. Res. Microbiol. 159, 367–373 10.1016/j.resmic.2008.04.006 [DOI] [PubMed] [Google Scholar]
- 22.Reyes A., Haynes M., Hanson N., Angly F.E., Heath A.C., Rohwer F. et al. (2011) Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 10.1038/nature09199, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cario E., Gerken G. and Podolsky D.K. (2007) Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 132, 1359–1374, 10.1053/j.gastro.2007.02.056 [DOI] [PubMed] [Google Scholar]
- 24.Hooper L.V., Wong M.H., Thelin A., Hansson L., Falk P.G. and Gordon J.I. (2001) Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884, 10.1126/science.291.5505.881 [DOI] [PubMed] [Google Scholar]
- 25.Lutgendorff F., Akkermans L.M.A. and Söderholm J.D. (2008) The role of microbiota and probiotics in stress-induced gastro-intestinal damage. Curr. Mol. Med. 8, 282–298, 10.2174/156652408784533779 [DOI] [PubMed] [Google Scholar]
- 26.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S. and Medzhitov R. (2004) Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241, 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 27.Hooper L.V., Bry L., Falk P.G. and Gordon J.I. (1998) Host-microbial symbiosis in the mammalian intestine: exploring an internal ecosystem. Bioessays 20, 336–343, 10.1002/(SICI)1521-1878(199804)20:4%3c336::AID-BIES10%3e3.0.CO;2-3 [DOI] [PubMed] [Google Scholar]
- 28.Rooks M.G. and Garrett W.S. (2016) Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352, 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooper L.V. and Macpherson A.J. (2010) Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169, 10.1038/nri2710 [DOI] [PubMed] [Google Scholar]
- 30.Kabat A.M., Srinivasan N. and Maloy K.J. (2014) Modulation of immune development and function by intestinal microbiota. Trends Immunol. 35, 507–517, 10.1016/j.it.2014.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M. et al. (2013) Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215, 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kostic A.D., Xavier R.J. and Gevers D. (2014) The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499, 10.1053/j.gastro.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G. et al. (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 34.Distrutti E., Monaldi L., Ricci P. and Fiorucci S. (2016) Gut microbiota role in irritable bowel syndrome: New therapeutic strategies. World J. Gastroenterol. 22, 2219–2241, 10.3748/wjg.v22.i7.2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajilić-Stojanović M., Heilig H.G.H.J., Tims S., Zoetendal E.G. and de Vos W.M. (2013) Long-term monitoring of the human intestinal microbiota composition. Environ. Microbiol. 15, 1146–1159, 10.1111/1462-2920.12023 [DOI] [PubMed] [Google Scholar]
- 36.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E. et al. (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563, 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scher J.U., Littman D.R. and Abramson S.B. (2016) Microbiome in inflammatory arthritis and human rheumatic diseases. Arthritis Rheumatol. 68, 35–45, 10.1002/art.39259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berbers R.M., Nierkens S., van Laar J.M., Bogaert D. and Leavis H.L. (2017) Microbial dysbiosis in common variable immune deficiencies: evidence, causes, and consequences. Trends Immunol. 38, 206–216, 10.1016/j.it.2016.11.008 [DOI] [PubMed] [Google Scholar]
- 39.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M. et al. (2012) Human gut microbiome viewed across age and geography. Nature 486, 222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster J.A., Rinaman L. and Cryan J.F. (2017) Stress & the gut-brain axis: regulation by the microbiome. Neurobiol. Stress 7, 124–136, 10.1016/j.ynstr.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dethlefsen L., Huse S., Sogin M.L. and Relman D.A. (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16s rRNA sequencing. PLoS Biol. 6, e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Paepe M., Leclerc M., Tinsley C.R. and Petit M.-A. (2014) Bacteriophages: an underestimated role in human and animal health? Front. Cell Infect. Microbiol. 4, 39 10.3389/fcimb.2014.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Modi S.R., Lee H.H., Spina C.S. and Collins J.J. (2013) Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222, 10.1038/nature12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meessen-Pinard M., Sekulovic O. and Fortier L.C. (2012) Evidence of in vivo prophage induction during Clostridium difficile infection. Appl. Environ. Microbiol. 78, 7662–7670, 10.1128/AEM.02275-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matos R.C., Lapaque N., Rigottier-Gois L., Debarbieux L., Meylheuc T., Gonzalez-Zorn B. et al. (2013) Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 9, e1003539, 10.1371/journal.pgen.1003539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb J.S., Lau M. and Kjelleberg S. (2004) Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186, 8066–8073, 10.1128/JB.186.23.8066-8073.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrolo M., Frias M.J., Pinto F.R., Melo-Cristino J. and Ramirez M. (2010) Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS ONE 5, e15678, 10.1371/journal.pone.0015678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuch R. and Fischetti V.A. (2009) The secret life of the anthrax agent Bacillus anthracis: Bacteriophage-mediated ecological adaptations. PLoS ONE 4, e6532, 10.1371/journal.pone.0006532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner J., Maksimovic J., Farries G., Sim W.H., Bishop R.F., Cameron D.J. et al. (2013) Bacteriophages in gut samples from pediatric Crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm. Bowel Dis. 19, 1598–1608, 10.1097/MIB.0b013e318292477c [DOI] [PubMed] [Google Scholar]
- 50.Pérez-Brocal V., García-López R., Vázquez-Castellanos J.F., Nos P., Beltrán B., Latorre A. et al. (2013) Study of the viral and microbial communities associated with Crohn’s disease: a metagenomic approach. Clin. Transl. Gastroenterol. 4, e36 10.1038/ctg.2013.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W., Jovel J., Halloran B., Wine E., Patterson J., Ford G. et al. (2015) Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria. Inflamm. Bowel Dis. 21, 1419–1427, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monaco C.L., Gootenberg D.B., Zhao G., Handley S.A., Ghebremichael M.S., Lim E.S. et al. (2016) Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 19, 311–322, 10.1016/j.chom.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diard M., Bakkeren E., Cornuault J.K., Moor K., Hausmann A., Sellin M.E. et al. (2017) Inflammation boosts bacteriophage transfer between Salmonella spp. Science 355, 1211–1215, 10.1126/science.aaf8451 [DOI] [PubMed] [Google Scholar]
- 54.Boyd E.F. (2012) Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions. Adv. Virus Res. 82, 91–118, 10.1016/B978-0-12-394621-8.00014-5 [DOI] [PubMed] [Google Scholar]
- 55.Gamage S.D., Patton A.K., Hanson J.F. and Weiss A.A. (2004) Diversity and host range of Shiga toxin-encoding phage. Infect. Immun. 72, 7131–7139, 10.1128/IAI.72.12.7131-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen S., Baker K., Padman B.S., Patwa R., Dunstan R.A., Weston T.A. et al. (2017) Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. MBio 8, e01874–17, 10.1128/mBio.01874-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lepage P., Colombet J., Marteau P., Sime-Ngando T., Doré J. and Leclerc M. (2008) Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut. 57, 424–425, 10.1136/gut.2007.134668 [DOI] [PubMed] [Google Scholar]
- 58.Pérez-Brocal V., García-López R., Nos P., Beltrán B., Moret I. and Moya A. (2015) Metagenomic analysis of Crohn’s disease patients identifies changes in the virome and microbiome related to disease status and therapy, and detects potential interactions and biomarkers. Inflamm. Bowel Dis. 21, 2515–2532, 10.1097/MIB.0000000000000549 [DOI] [PubMed] [Google Scholar]
- 59.Kramná L., Kolářová K., Oikarinen S., Pursiheimo J.P., Ilonen J., Simell O. et al. (2015) Gut virome sequencing in children with early islet autoimmunity. Diabetes Care 38, 930–933 10.2337/dc14-2490 [DOI] [PubMed] [Google Scholar]
- 60.Rhee S.H., Pothoulakis C. and Mayer E.A. (2009) Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat. Rev. Gastroenterol. Hepatology 6, 306–314, 10.1038/nrgastro.2009.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carabotti M., Scirocco A., Maselli M.A. and Severi C. (2015) The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 28, 203–209, [PMC free article] [PubMed] [Google Scholar]
- 62.Grenham S., Clarke G., Cryan J.F. and Dinan T.G. (2011) Brain-gut-microbe communication in health and disease. Front. Physiol. 2, 94, 10.3389/fphys.2011.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulle J.G., Sharp W.G. and Cubells J.F., and (2013) The gut microbiome: a new frontier in autism research. Curr. Psychiatry Rep. 15, 337, 10.1007/s11920-012-0337-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evrensel A. and Ceylan M.E. (2015) The gut-brain axis: the missing link in depression. Clin. Psychopharmacol. Neurosci. 13, 239–244, 10.9758/cpn.2015.13.3.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dickerson F., Severance E. and Yolken R. (2017) The microbiome, immunity, and schizophrenia and bipolar disorder. Brain Behav. Immun. 62, 46–52, 10.1016/j.bbi.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogt N.M., Kerby R.L., Dill-McFarland K.A., Harding S.J., Merluzzi A.P., Johnson S.C. et al. (2017) Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 7, 13537, 10.1038/s41598-017-13601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sampson T.R., Debelius J.W., Thron T., Janssen S., Shastri G.G., Ilhan Z.E. et al. (2016) Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469.e12–1480.e12, 10.1016/j.cell.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bansal A.S., Bradley A.S., Bishop K.N., Kiani-Alikhan S. and Ford B. (2012) Chronic fatigue syndrome, the immune system and viral infection. Brain Behav. Immun. 26, 24–31 10.1016/j.bbi.2011.06.016 [DOI] [PubMed] [Google Scholar]
- 69.Bansal A.S. (2016) Investigating unexplained fatigue in general practice with a particular focus on CFS/ME. BMC Fam. Pract. 17, 81 10.1186/s12875-016-0493-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kerr J.R., Cunniffe V.S., Kelleher P., Bernstein R.M. and Bruce I.N. (2003) Successful intravenous immunoglobulin therapy in 3 cases of parvovirus B19-associated chronic fatigue syndrome. Clin. Infect. Dis. 36, e100–6, 10.1086/374666 [DOI] [PubMed] [Google Scholar]
- 71.Jason L.A., Benton M.C., Valentine L., Johnson A. and Torres-Harding S. (2008) The economic impact of ME/CFS: individual and societal costs. Dyn. Med. 7, 6 10.1186/1476-5918-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hickie I., Lloyd A., Hadzi-Pavlovic D., Parker G., Bird K. and Wakefield D. (1995) Can the chronic fatigue syndrome be defined by distinct clinical features? Psychol. Med. 25, 925–935, 10.1017/S0033291700037417 [DOI] [PubMed] [Google Scholar]
- 73.Sanders P. and Korf J. (2008) Neuroaetiology of chronic fatigue syndrome: an overview. World J. Biol. Psychiatry 9, 165–171, 10.1080/15622970701310971 [DOI] [PubMed] [Google Scholar]
- 74.Morris G., Berk M., Galecki P. and Maes M. (2014) The emerging role of autoimmunity in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). Mol. Neurobiol. 49, 741–756, 10.1007/s12035-013-8553-0 [DOI] [PubMed] [Google Scholar]
- 75.Underhill R.A. (2015) Myalgic encephalomyelitis, chronic fatigue syndrome: an infectious disease. Med. Hypotheses. 85, 765–773, 10.1016/j.mehy.2015.10.011 [DOI] [PubMed] [Google Scholar]
- 76.Meals R.W., Hauser V.F. and Bower A.G. (1935) Poliomyelitis-The Los Angeles Epidemic of 1934 : part I. Cal. West. Med. 43, 123–125, [PMC free article] [PubMed] [Google Scholar]
- 77.Mateen F.J. and Black R.E. (2013) Expansion of acute flaccid paralysis surveillance: beyond poliomyelitis. Trop. Med. Int. Health 18, 1421–1422, 10.1111/tmi.12181 [DOI] [PubMed] [Google Scholar]
- 78.Hall W.J., Nathanson N. and Langmuir A.D. (1957) The age distribution of poliomyelitis in the United States in 1955. Am. J. Hyg. 66, 214–234, [DOI] [PubMed] [Google Scholar]
- 79.Sigurdsson B., Sigurjónsson J., Sigurdsson J.H.J., Thorkelsson J. and Gudmundsson K.R. (1950) A disease epidemic in Iceland simulating poliomyelitis. Am. J. Epidemiol. 52, 222–238, 10.1093/oxfordjournals.aje.a119421 [DOI] [PubMed] [Google Scholar]
- 80.Staff M. (1957) An outbreak of encephalomyelitis in the royal free hospital group, London, in 1955. Br. Med. J. 2, 895–904, 10.1136/bmj.2.5050.895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Committee on the Diagnostic Criteria for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome; Board on the Health of Select Populations and Institute of Medicine (2015) In Beyond Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Redefining an Illness, National Academies Press, Washington (DC) [PubMed] [Google Scholar]
- 82.Meeus M., Van Eupen I., Van Baarle E., De Boeck V., Luyckx A., Kos D. et al. (2011) Symptom fluctuations and daily physical activity in patients with chronic fatigue syndrome: a case-control study. Arch. Phys. Med. Rehabil. 92, 1820–1826 10.1016/j.apmr.2011.06.023 [DOI] [PubMed] [Google Scholar]
- 83.Twisk F.N. (2015) Accurate diagnosis of myalgic encephalomyelitis and chronic fatigue syndrome based upon objective test methods for characteristic symptoms. World J. Methodol. 5, 68–87 10.5662/wjm.v5.i2.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fukuda K., Straus S.E., Hickie I., Sharpe M.C., Dobbins J.G. and Komaroff A. (1994) The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann. Intern. Med. 121, 953–959 [DOI] [PubMed] [Google Scholar]
- 85.Capelli E., Zola R., Lorusso L., Venturini L., Sardi F. and Ricevuti G. (2010) Chronic fatigue syndrome/myalgic encephalomyelitis: an update. Int. J. Immunopathol. Pharmacol. 23, 981–989 10.1177/039463201002300402 [DOI] [PubMed] [Google Scholar]
- 86.Whiting P., Bagnall A.-M., Sowden A.J., Cornell J.E., Mulrow C.D. and Ramírez G. (2001) Interventions for the treatment and management of chronic fatigue syndrome. JAMA 286, 1360, 10.1001/jama.286.11.1360 [DOI] [PubMed] [Google Scholar]
- 87.Aaron L.A., Burke M.M. and Buchwald D. (2000) Overlapping conditions among patients with chronic fatigue syndrome, fibromyalgia, and temporomandibular disorder. Arch. Intern. Med. 160, 221, 10.1001/archinte.160.2.221 [DOI] [PubMed] [Google Scholar]
- 88.Quigley E.M.M. (2011) The enteric microbiota in the pathogenesis and management of constipation. Best Pract. Res. Clin. Gastroenterol. 25, 119–126, 10.1016/j.bpg.2011.01.003 [DOI] [PubMed] [Google Scholar]
- 89.Quigley E.M. (2011) Gut microbiota and the role of probiotics in therapy. Curr. Opin. Pharmacol. 11, 593–603, 10.1016/j.coph.2011.09.010 [DOI] [PubMed] [Google Scholar]
- 90.Nagy-Szakal D., Williams B.L., Mishra N., Che X., Lee B., Bateman L. et al. (2017) Fecal metagenomic profiles in subgroups of patients with myalgic encephalomyelitis/ chronic fatigue syndrome. Microbiome 5, 44, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Giloteaux L., Goodrich J.K., Walters W.A., Levine S.M., Ley R.E., Hanson M.R. et al. (2016) Reduced diversity and altered composition of the gut microbiome in individuals with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome 4, 953–959 10.1186/s40168-016-0171-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fremont M., Coomans D., Massart S. and De Meirleir K. (2013) High-throughput 16S rRNA gene sequencing reveals alterations of intestinal microbiota in myalgic encephalomyelitis/chronic fatigue syndrome patients. Anaerobe 22, 50–56 10.1016/j.anaerobe.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 93.Armstrong C.W., McGregor N.R., Lewis D.P., Butt H.L. and Gooley P.R. (2017) The association of fecal microbiota and fecal, blood serum and urine metabolites in myalgic encephalomyelitis/chronic fatigue syndrome. Metabolomics 13, 8, 10.1007/s11306-016-1145-z [DOI] [Google Scholar]
- 94.Sheedy J.R., Wettenhall R.E.H., Scanlon D., Gooley P.R., Lewis D.P., Mcgregor N. et al. (2009) Increased d-lactic acid intestinal bacteria in patients with chronic fatigue syndrome. In Vivo 23, 621–628 [PubMed] [Google Scholar]
- 95.Wright E.K., Kamm M.A., Teo S.M., Inouye M., Wagner J. and Kirkwood C.D. (2015) Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: a systematic review. Inflamm. Bowel Dis. 21, 1219–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V. et al. (2014) A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283, 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- 97.Ríos-Covián D., Ruas-Madiedo P., Margolles A., Gueimonde M., De los Reyes-Gavilán C.G. and Salazar N. (2016) Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Butt H., Dunstan R., McGregor N. and Roberts T. (1998) Faecal microbial growth inhibition in chronic fatigue/pain patients. Proceedings of the AHMF International Clinical and Scientific Conference, Newcastle, Australia [Google Scholar]
- 99.Gall A., Fero J., McCoy C., Claywell B.C., Sanchez C.A., Blount P.L. et al. (2015) Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett’s esophagus cohort. PLoS ONE 10, e0129055, 10.1371/journal.pone.0129055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carroll I.M., Ringel-Kulka T., Siddle J.P. and Ringel Y. (2012) Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 24, 521–530, e248, 10.1111/j.1365-2982.2012.01891.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vandeputte D., Falony G., Vieira-Silva S., Tito R.Y., Joossens M. and Raes J. (2016) Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65, 57–62, 10.1136/gutjnl-2015-309618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goldsmith K., Chalder T., White P., Sharpe M. and Pickles A. (2015) Longitudinal mediation in the PACE randomised clinical trial of rehabilitative treatments for chronic fatigue syndrome: modelling and design considerations. Trials 16 (Suppl. 2), O43, 10.1186/1745-6215-16-S2-O43 [DOI] [Google Scholar]
- 103.Hardcastle S.L., Brenu E.W., Johnston S., Nguyen T., Huth T., Ramos S. et al. (2015) Longitudinal analysis of immune abnormalities in varying severities of Chronic Fatigue Syndrome/Myalgic Encephalomyelitis patients. J. Transl. Med. 13, 299, 10.1186/s12967-015-0653-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nyland M., Naess H., Birkeland J.S. and Nyland H. (2014) Longitudinal follow-up of employment status in patients with chronic fatigue syndrome after mononucleosis. BMJ Open 4, e005798, 10.1136/bmjopen-2014-005798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brenu E.W., van Driel M.L., Staines D.R., Ashton K.J., Hardcastle S.L., Keane J. et al. (2012) Longitudinal investigation of natural killer cells and cytokines in chronic fatigue syndrome/myalgic encephalomyelitis. J. Transl. Med. 10, 88, 10.1186/1479-5876-10-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A. and Alm E.J. (2017) Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 8, 1784, 10.1038/s41467-017-01973-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Giloteaux L., Hanson M.R. and Keller B.A. (2016) A pair of identical twins discordant for myalgic encephalomyelitis/chronic fatigue syndrome differ in physiological parameters and gut microbiome composition. Am. J. Case Rep. 17, 720–729 10.12659/AJCR.900314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chia J.K.S. and Chia A.Y. (2007) Chronic fatigue syndrome is associated with chronic enterovirus infection of the stomach. J. Clin. Pathol. 61, 43–48, 10.1136/jcp.2007.050054 [DOI] [PubMed] [Google Scholar]
- 109.Frémont M., Metzger K., Rady H., Hulstaert J. and De Meirleir K. (2009) Detection of herpesviruses and parvovirus B19 in gastric and intestinal mucosa of chronic fatigue syndrome patients. In Vivo 23, 209–213 [PubMed] [Google Scholar]
- 110.Aguiar-Pulido V., Huang W., Suarez-Ulloa V., Cickovski T., Mathee K. and Narasimhan G. (2016) Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol. Bioinform. Online 12 (Suppl. 1), 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matysik S., Le Roy C.I., Liebisch G. and Claus S.P. (2016) Metabolomics of fecal samples: a practical consideration. Trends Food Sci. Technol. 57, 244–255, 10.1016/j.tifs.2016.05.011 [DOI] [Google Scholar]
- 112.Duncan S.H., Louis P. and Flint H.J. (2004) Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70, 5810–5817, 10.1128/AEM.70.10.5810-5817.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morgan X.C. and Huttenhower C. (2014) Meta’omic analytic techniques for studying the intestinal microbiome. Gastroenterology 146, 1437.e1–1448.e1 10.1053/j.gastro.2014.01.049 [DOI] [PubMed] [Google Scholar]
- 114.Vandeputte D., Tito R.Y., Vanleeuwen R., Falony G. and Raes J. (2017) Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 41 (Supp_1), S154–S167, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Attree E.A., Arroll M.A., Dancey C.P., Griffith C. and Bansal A.S. (2014) Psychosocial factors involved in memory and cognitive failures in people with myalgic encephalomyelitis/chronic fatigue syndrome. Psychol. Res. Behav. Manag. 7, 67–76, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carruthers B.M. (2007) Definitions and aetiology of myalgic encephalomyelitis: how the Canadian consensus clinical definition of myalgic encephalomyelitis works. J. Clin. Pathol. 60, 117–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carruthers B.M., Van de Sande M.I., De Meirleir K.L., Klimas N.G., Broderick G., Mitchell T. et al. (2011) Myalgic encephalomyelitis: International Consensus Criteria. J. Intern. Med. 270, 327–338, 10.1111/j.1365-2796.2011.02428.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim D., Hofstaedter C.E., Zhao C., Mattei L., Tanes C., Clarke E. et al. (2017) Optimizing methods and dodging pitfalls in microbiome research. Microbiome 5, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Langdon A., Crook N. and Dantas G. (2016) The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 8, 39, 10.1186/s13073-016-0294-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wilson I.D. and Nicholson J.K. (2017) Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 179, 204–222, 10.1016/j.trsl.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Minalyan A., Gabrielyan L., Scott D., Jacobs J. and Pisegna J.R. (2017) The gastric and intestinal microbiome: role of proton pump inhibitors. Curr. Gastroenterol. Rep. 19, 42, 10.1007/s11894-017-0577-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Singh R.K., Chang H.-W., Yan D., Lee K.M., Ucmak D., Wong K. et al. (2017) Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 15, 73, 10.1186/s12967-017-1175-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A. et al. (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108, 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Claesson M.J., Cusack S., O’Sullivan O., Greene-Diniz R., de Weerd H., Flannery E. et al. (2011) Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U.S.A. 108, 4586–4591, 10.1073/pnas.1000097107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mueller S., Saunier K., Hanisch C., Norin E., Alm L., Midtvedt T. et al. (2006) Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl. Environ. Microbiol. 72, 1027–1033, 10.1128/AEM.72.2.1027-1033.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haro C., Rangel-Zúñiga O.A., Alcalá-Díaz J.F., Gómez-Delgado F., Pérez-Martínez P., Delgado-Lista J. et al. (2016) Intestinal microbiota is influenced by gender and body mass index. PLoS ONE 11, e0154090, 10.1371/journal.pone.0154090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yurkovetskiy L., Burrows M., Khan A.A., Graham L., Volchkov P., Becker L. et al. (2013) Gender bias in autoimmunity is influenced by microbiota. Immunity 39, 400–412, 10.1016/j.immuni.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Markle J.G.M., Frank D.N., Mortin-Toth S., Robertson C.E., Feazel L.M., Rolle-Kampczyk U. et al. (2013) Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088, 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- 129.Sudo N. (2014) Microbiome, HPA axis and production of endocrine hormones in the gut. Adv. Exp. Med. Biol. 817, 177–194, 10.1007/978-1-4939-0897-4_8 [DOI] [PubMed] [Google Scholar]
- 130.Baker J.M., Al-Nakkash L. and Herbst-Kralovetz M.M. (2017) Estrogen-gut microbiome axis: Physiological and clinical implications. Maturitas 103, 45–53, 10.1016/j.maturitas.2017.06.025 [DOI] [PubMed] [Google Scholar]
- 131.Fransen F., van Beek A.A., Borghuis T., Meijer B., Hugenholtz F., van der Gaast-de Jongh C. et al. (2017) The impact of gut microbiota on gender-specific differences in immunity. Front. Immunol. 8, 754, 10.3389/fimmu.2017.00754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wallis A., Butt H., Ball M., Lewis D.P. and Bruck D. (2016) Support for the microgenderome: associations in a human clinical population. Sci. Rep. 6, 19171 10.1038/srep19171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Choo J.M., Leong L.E. and Rogers G.B. (2015) Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 5, 16350, 10.1038/srep16350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tedjo D.I., Jonkers D.M.A.E., Savelkoul P.H., Masclee A.A., Van Best N., Pierik M.J. et al. (2015) The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS ONE 10, e0126685, 10.1371/journal.pone.0126685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rochet V., Rigottier-Gois L., Rabot S. and Doré J. (2004) Validation of fluorescent in situ hybridization combined with flow cytometry for assessing interindividual variation in the composition of human fecal microflora during long-term storage of samples. J. Microbiol. Methods 59, 263–270, 10.1016/j.mimet.2004.07.012 [DOI] [PubMed] [Google Scholar]
- 136.Jia J., Frantz N., Khoo C., Gibson G.R., Rastall R.A. and McCartney A.L. (2010) Investigation of the faecal microbiota associated with canine chronic diarrhea. FEMS Microbiol. Ecol. 71, 304–312, 10.1111/j.1574-6941.2009.00812.x [DOI] [PubMed] [Google Scholar]
- 137.Gorzelak M.A., Gill S.K., Tasnim N., Ahmadi-Vand Z., Jay M. and Gibson D.L. (2015) Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS ONE 10, e0134802, 10.1371/journal.pone.0134802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bag S., Saha B., Mehta O., Anbumani D., Kumar N., Dayal M. et al. (2016) An improved method for high quality metagenomics DNA extraction from human and environmental samples. Sci. Rep. 6, 26775, 10.1038/srep26775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Peterson D.A., Frank D.N., Pace N.R. and Gordon J.I. (2008) Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3, 417–427, 10.1016/j.chom.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ranjan R., Rani A., Metwally A., McGee H.S. and Perkins D.L. (2016) Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 469, 967–977, 10.1016/j.bbrc.2015.12.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jovel J., Patterson J., Wang W., Hotte N., O’Keefe S., Mitchel T. et al. (2016) Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 7, 459, 10.3389/fmicb.2016.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Woolhouse ME., Howey R., Gaunt E., Reilly L., Chase-Topping M. and Savill N. (2008) Temporal trends in the discovery of human viruses. Proc. Biol. Sci. 275, 2111–2115, 10.1098/rspb.2008.0294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Flewett T.H., Bryden A.S. and Davies H. (1974) Diagnostic electron microscopy of faeces: I The viral flora of the faeces as seen by electron microscopy. J. Clin. Pathol. 27, 603–608, 10.1136/jcp.27.8.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mitgutsch K., Schirra S. and Verrilli S. (2013) Movers and shakers. CHI’13 Ext. Abstr. Hum. Factors Comput. Syst. CHI EA’13 976, 715 [Google Scholar]
- 145.Lecuit M. and Eloit M. (2013) The human virome: new tools and concepts. Trends Microbiol. 21, 510–515 10.1016/j.tim.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wylie K.M., Weinstock G.M. and Storch G.A. (2013) Virome genomics: a tool for defining the human virome. Curr. Opin. Microbiol. 16, 479–484 10.1016/j.mib.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Carding S.R., Davis N. and Hoyles L. (2017) Review article: the human intestinal virome in health and disease. Aliment. Pharmacol. Ther. 46, 800–815, 10.1111/apt.14280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Castro-Mejía J.L., Muhammed M.K., Kot W., Neve H., Franz C.M.A.P., Hansen L.H. et al. (2015) Optimizing protocols for extraction of bacteriophages prior to metagenomic analyses of phage communities in the human gut. Microbiome 3, 64 10.1186/s40168-015-0131-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kleiner M., Hooper L.V. and Duerkop B.A. (2015) Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genomics 16, 7 10.1186/s12864-014-1207-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hoyles L., McCartney A.L., Neve H., Gibson G.R., Sanderson J.D., Heller K.J. et al. (2014) Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res. Microbiol. 165, 803–812 10.1016/j.resmic.2014.10.006 [DOI] [PubMed] [Google Scholar]
- 151.Reyes A., Semenkovich N.P., Whiteson K., Rohwer F. and Gordon J.I. (2012) Going viral: next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 10, 607–617 10.1038/nrmicro2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Conceição-Neto N., Zeller M., Lefrère H., De Bruyn P., Beller L., Deboutte W. et al. (2015) Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci. Rep. 5, 16532 10.1038/srep16532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hall R.J., Wang J., Todd A.K., Bissielo A.B., Yen S., Strydom H. et al. (2014) Evaluation of rapid and simple techniques for the enrichment of viruses prior to metagenomic virus discovery. J. Virol. Methods 195, 194–204 10.1016/j.jviromet.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bikel S., Valdez-Lara A., Cornejo-Granados F., Rico K., Canizales-Quinteros S., Soberón X. et al. (2015) Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: Towards a systems-level understanding of human microbiome. Comput. Struct. Biotechnol. J. 13, 390–401 10.1016/j.csbj.2015.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hurwitz B.L., U’Ren J.M. and Youens-Clark K. (2016) Computational prospecting the great viral unknown. FEMS Microbiol. Lett. 363, 10.1093/femsle/fnw077 [DOI] [PubMed] [Google Scholar]
- 156.Ogilvie L.A. and Jones B.V. (2015) The human gut virome: a multifaceted majority. Front. Microbiol. 6, 10.3389/fmicb.2015.00918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Minot S., Sinha R., Chen J., Li H., Keilbaugh S.A., Wu G.D. et al. (2011) The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 21 1616–1625 10.1101/gr.122705.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Brum J.R. and Sullivan M.B. (2015) Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 13, 147–159 10.1038/nrmicro3404 [DOI] [PubMed] [Google Scholar]
- 159.Mizuno C.M., Rodriguez-Valera F., Kimes N.E. and Ghai R. (2013) Expanding the marine virosphere using metagenomics. PLoS Genet. 9, e1003987, 10.1371/journal.pgen.1003987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lu H., Giordano F. and Ning Z. (2016) Oxford Nanopore MinION sequencing and genome assembly. Genomics Proteomics Bioinformatics 14, 265–279, 10.1016/j.gpb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wylie K.M., Weinstock G.M. and Storch G.A. (2012) Emerging view of the human virome. Transl. Res. 160, 283–290 10.1016/j.trsl.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.De Preter V. and Verbeke K. (2013) Metabolomics as a diagnostic tool in gastroenterology. World J. Gastrointest. Pharmacol. Ther. 4, 97, 10.4292/wjgpt.v4.i4.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Smirnov K.S., Maier T.V., Walker A., Heinzmann S.S., Forcisi S., Martinez I. et al. (2016) Challenges of metabolomics in human gut microbiota research. Int. J. Med. Microbiol. 306, 266–279, 10.1016/j.ijmm.2016.03.006 [DOI] [PubMed] [Google Scholar]
- 164.Vernocchi P., Del Chierico F. and Putignani L. (2016) Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front. Microbiol. 7, 10.3389/fmicb.2016.01144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang C., Yin A., Li H., Wang R., Wu G., Shen J. et al. (2015) Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBio Med. 2, 968–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Weckwerth W. and Morgenthal K. (2005) Metabolomics: from pattern recognition to biological interpretation. Drug Discov. Today 10, 1551–1558, 10.1016/S1359-6446(05)03609-3 [DOI] [PubMed] [Google Scholar]
- 167.Nassar A.-E.F. and Talaat R.E. (2004) Strategies for dealing with metabolite elucidation in drug discovery and development. Drug Discov. Today 9, 317–327, 10.1016/S1359-6446(03)03018-6 [DOI] [PubMed] [Google Scholar]
- 168.Breitbart M., Hewson I., Felts B., Mahaffy J.M., Nulton J., Salamon P. et al. (2003) Metagenomic analyses of an uncultured viral community from human feces metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 10.1128/JB.185.20.6220-6223.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang T., Breitbart M., Lee W.H., Run J.Q., Wei C.L., Soh S.W.L. et al. (2006) RNA viral community in human feces: Prevalence of plant pathogenic viruses. PLoS Biol. 4, 0108–0118, 10.1371/journal.pbio.0040003, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kim M.-S., Park E.-J., Roh S.W. and Bae J.-W. (2011) Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 77, 8062–8070 10.1128/AEM.06331-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Minot S., Grunberg S., Wu G.D., Lewis J.D. and Bushman F.D. (2012) Hypervariable loci in the human gut virome. Proc. Natl. Acad. Sci. U.S.A. 109, 3962–3966, 10.1073/pnas.1119061109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Minot S. and Bryson A. (2013) Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U.S.A. 110, 12450–12455, 10.1073/pnas.1300833110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pérez-Brocal V., García-López R., Vázquez-Castellanos J.F., Nos P., Beltrán B., Latorre A. et al. (2013) Study of the viral and microbial communities associated with Crohn’s disease: a metagenomic approach. Clin. Transl. Gastroenterol. 4, e36, 10.1038/ctg.2013.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lim E.S., Zhou Y., Zhao G., Bauer I.K., Droit L., Ndao I.M. et al. (2015) Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234 10.1038/nm.3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Norman J.M., Handley S.A., Baldridge M.T., Droit L., Liu C.Y., Keller B.C. et al. (2015) Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 10.1016/j.cell.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Evengård B., Nord C.E. and Sullivan  (2007) P1239 Patients with chronic fatigue syndrome have higher numbers of anaerobic bacteria in the intestine compared to healthy subjects. Int. J. Antimicrob. Agents 29, S340, 10.1016/S0924-8579(07)71079-8 [DOI] [Google Scholar]
- 177.Butt H., Dunstan R., McGregor N. and Roberts T. (2001) Bacterial colonosis in patients with persistent fatigue. Proceedings of the AHMF International Clinical and Scientific Conference, Sydney, Australia [Google Scholar]
- 178.Reyes A., Blanton L., Cao S., Zhao G., Manary M., Trehan I. et al. (2015) Gut viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl. Acad. Sci. U.S.A. 112, 11941–11946 10.1073/pnas.1514285112 [DOI] [PMC free article] [PubMed] [Google Scholar]