Abstract
Objective:
Nucleophosmin-1 (NPM1) mutations have prognostic importance in acute myeloid leukemia (AML) patients with intermediate-risk karyotype at diagnosis. Approximately 30% of newly diagnosed cytogenetically normal AML (CN-AML) patients harbor the NPM1 mutation in India. In this study we compared the efficiency of three molecular techniques in detecting NPM1 mutation in peripheral blood and bone marrow samples.
Materials and Methods:
In a single-center cohort we analyzed 165 CN-AML bone marrow/peripheral blood samples for NPM1 mutation analysis. About 30% of the CN-AML samples revealed NPM1 mutations. For the detection, three methods were compared: Sanger sequencing, pyrosequencing, and real-time polymerase chain reaction (PCR).
Results:
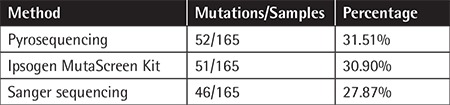
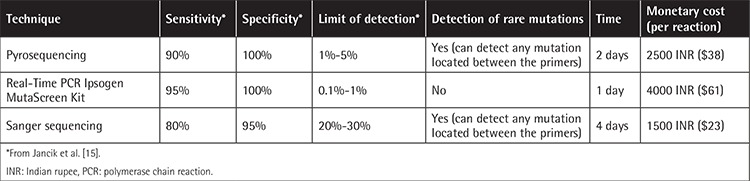
NPM1 exon 12 mutations were observed in 52 (31.51%) of all CN-AML cases. The sensitivity of Sanger sequencing, pyrosequencing, and real-time PCR was 80%, 90%, and 95%, whereas specificity was 95%, 100%, and 100%, respectively. The minimum limit of mutation detection was 20%-30% for Sanger sequencing, 1%-5% for pyrosequencing, and 0.1%-1% for real-time PCR.
Conclusion:
The sequencing method, which is the reference method, has the lowest sensitivity and is sometimes difficult to interpret. Real-time PCR is a highly sensitive method for mutation detection but is limited for specific mutation types. In our study, pyrosequencing emerged as the most suitable technique for the detection of NPM1 mutations on the basis of its easy interpretation and less time-consuming processes than Sanger sequencing.
Keywords: NPM1, Pyrosequencing, Acute myeloid leukemia, Mutation analysis
Abstract
Amaç:
Nükleofosmin-1 (NPM1) mutasyonları tanı anında orta risk akut miyeloid lösemi (AML) hastalarında prognostik öneme sahiptir. Hindistan’da, yeni teşhis normal sitogenetiğe sahip AML (CN-AML) hastalarının yaklaşık %30’u NPM1 pozitiftir. Bu çalışmada periferik kan ve kemik iliği örneklerinde NPM1 mutasyonu saptamada kullanılan üç moleküler tekniğin etkinliğini karşılaştırdık.
Gereç ve Yöntemler:
Tek merkezli bu kohortta, 165 CN-AML kemik iliği/periferik kan örneklerinde NPM1 mutasyon analizi yapıldı. CN-AML örneklerinin yaklaşık %30’unda NPM1 mutasyonu saptandı. Mutasyonun taranmasında üç yöntem karşılaştırıldı: Sanger dizileme, pirodizileme, gerçek-zamanlı polimeraz zincir reaksiyonu (PCR).
Bulgular:
Tüm CN-AML olgularının 52’sinde (%31,51) NPM1 exon12 mutasyonları gözlendi. Sanger dizileme, pirodizileme ve gerçek zamanlı PCR’nin duyarlılıkları sırasıyla %80, %90 ve %95 iken, özgünlükleri %95, %100 ve %100’dü. Mutasyonun saptanmasında minimum limit Sanger dizileme yöntemi için %20-%30, pirodizilemede %1-5, ve gerçek-zamanlı PCR için %0,1-%1 idi.
Sonuç:
Referans yöntemi olan dizileme yöntemi, en düşük duyarlılığa sahiptir ve bazen yorumlaması güçtür. Gerçek-zamanlı PCR mutasyon saptamada yüksek duyarlılığa sahip bir yöntemdir fakat özel mutasyon tipleri için sınırlıdır. Çalışmamızda, pirodizileme yönteminin kolay yorumlanması ve Sanger dizileme yönteminden daha az zaman harcanan işlem olması esasına dayanarak NPM1 mutasyonun saptanmasında en uygun teknik olduğu sonucuna varılmıştır.
Introduction
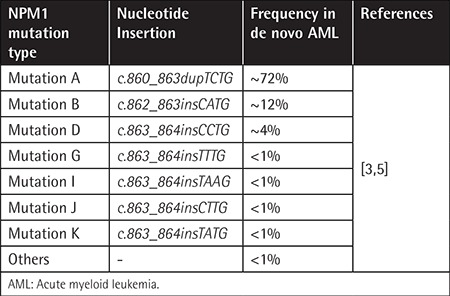
An increasing number of genetic abnormalities are revealed in acute myeloid leukemia (AML). Among these genetic alterations, potential prognostic genetic markers are the nucleophosmin 1 (NPM1) gene, FLT3 gene, and CEBPA gene [1]. Mutations in the NPM1 and FLT3 genes represent the most important diagnostic and prognostic indicators in patients with cytogenetically normal AML (CN-AML). NPM1 is a phosphoprotein that continuously shuttles between the cytoplasm and nucleus. Several functions for this protein have been described, including the binding of p53, the initiation of centrosome duplication, and ribosomal protein assembly and transport [2]. NPM1 mutations found in exon 12 code for the COOH terminal region. Frameshift mutations in the NPM1 gene result in an elongated protein that contains an additional nuclear export signal and leads to an abnormal cytoplasmic localization of the protein [3,4]. These mutations are involved in leukemogenesis and are detected in about 35%-60% of AML cases [5]. Six types of NPM1 mutation variants have been identified: NPM1 mutation A (c.860_863dupTCTG), mutation B (c.862_863insCATG), mutation D (c.863_864insCCTG), mutation I (c.863_864insTAAG), mutation J (c.863_864insCTTG), and mutation K (c.863_864insTATG). Mutation A (TCTG insertion) is the most commonly occurring variant, found in about 80% of all NPM1-mutated AML cases (Table 1) [3,5]. The effect of mutant NPM1 has been studied using gene expression profiling and studies revealed a distinctive signature of these mutations [6]. Many studies reported the prognostic significance of NPM1 mutation status in AML [7,8,9,10,11]. There are highly specific and sensitive molecular assays available for detecting NPM1 mutations, like Sanger sequencing, high-resolution melting curve analysis, real-time polymerase chain reaction (PCR), and pyrosequencing (Pyr). In this study, we evaluated the utility of Pyr in the detection of NPM1 mutation detection and also compared it with Sanger sequencing and real-time PCR in terms of assay sensitivity, specificity, limit of mutation detection, turnaround time, and assay cost [12,13].
Table 1. NPM1 mutation frequencies in de novo acute myeloid leukemia.
Materials and Methods
A total of 165 CN-AML bone marrow aspiration or peripheral blood samples taken at the time of first diagnosis were included in this study from February 2014 to September 2016. Out of these 165 patients, 79 (47.87%) were male and 86 (52.12%) were female. Twenty cases (12.12%) were pediatric cases.
DNA Extraction
Genomic DNA was extracted from the received samples using the QIAGEN DNeasy Kit (QIAGEN, Hilden, Germany) as per the manufacturer’s instructions.
NPM1 Mutation Detection by Pyr Analysis
In the Pyr method for DNA sequence analysis, inorganic phosphate released in the course of nucleotide incorporation serves as the initial substrate in a sequence of four successive enzymatic reactions. This results in the emission of light, which functions as a signal that is proportional to the number of nucleotides incorporated.
For NPM1 mutation analysis TTAACTCTCTGGTGGTAGAATG was used as a forward primer, biotin-ACATTTATCAAACACGGTAGG as a reverse primer, and TTTTCCAGGCTATTCAAGAT as the sequencing primer (Sigma-Aldrich, New Delhi, India). DNA (50 ng) was amplified using 400 nmol of forward and reverse primers in 25 µL of reaction mix with PyroMark master mix (QIAGEN). PCR conditions were as follows: initial denaturing at 95 °C for 15 min; 42 cycles of 95 °C for 20 s, 53 °C for 30 s, and 60 °C for 20 s; and final extension at 72 °C for 5 min. PCR products were electrophoresed on agarose gel to confirm successful amplification. The PCR products (10 µL) were then sequenced with the Pyr PyroMark Q24 system (QIAGEN).
NPM1 Mutation Detection by Pyr Analysis Using Ipsogen NPM1 MutaScreen Kit
The Ipsogen NPM1 MutaScreen Kit (QIAGEN) combines two techniques to screen for the presence of mutations in the target gene. The real-time quantitative PCR (qPCR) double-dye oligonucleotide hydrolysis principle uses specific primers and an internal double-dye probe with a reporter and a quencher (FAM-TAMRA) for the amplification reactions. In addition, a 3’-end modified phosphate oligonucleotide is used that perfectly matches the wild-type NPM1 gene and does not allow polymerization. The Ipsogen NPM1 MutaScreen Kit detects total NPM1 (wild-type + mutated) and mutated NPM1 and separately identifies NPM1 Mut A, Mut B, and Mut D in genomic DNA. A sample of DNA of 25 ng was used in a final reaction volume of 25 µL. The PCR profile for Rotor-Gene Q (QIAGEN) was 50 °C for 2 min, 95 °C for 10 min, and then 40 cycles of 95 °C for 15 s and 60 °C for 90 s with acquisition performed at 60 °C. Analysis was performed as per the kit’s instructions.
NPM1 Mutation Analysis by Sanger Sequencing
Analysis of NPM1 exon 12 mutations was done as described by Falini et al. [4]. A sample of DNA of 50 ng was amplified using an Applied Biosystems Veriti thermal cycler (Foster City, CA, USA) and purified PCR product was used for BigDye termination bidirectional sequencing. Results were analyzed using BioEdit sequence analysis software.
Results
NPM1 exon 12 mutation was observed in 52 (31.51%) of all CN-AML cases. As expected, the percentage of the DNA samples in which mutations were detected varied and depended upon the method of detection used. NPM1 mutation analysis by Pyr had the highest likelihood of identifying a mutation in the NPM1 gene, followed by the NPM1 MutaScreen kit and direct sequencing (Table 2). However, on the basis of our evaluation criteria (Table 1), the most sensitive tool was the Ipsogen MutaScreen kit (95%), followed by Pyr (90%) and Sanger sequencing (80%). In terms of specificity, all three methods matched equally.
Table 2. Number and percentage of mutations detected by three different methods.
Discussion
It has been found that 99% of all NPM1 mutations detected by Pyr have 4-base insertions at position 860 while the rest of the NPM1 mutations detected by Pyr were found as insertion at 862 and deletion at 863 and 861 [14]. We have examined the ability of three different methods to detect mutations in NPM1 gene exon 12 in 165 CN-AML samples. Bone marrow or peripheral blood samples with a minimum of 15% blasts were examined in this study. NPM1 mutations were found in 52 samples (31.51%), while 113 (68.48%) samples were found to be wild-type. Twenty-eight (53.84%) of the NPM1-positive patients were male while 24 (46.15%) were female. Seven (13.46%) of the NPM1-positive samples were from pediatric patients while 45 (86.53%) were from adults. Mutation type A was the most frequent mutation (~80%), followed by types B (12%) and D (6%). We also found one case of mutation type K (c.863_864insTATG) by Pyr (Figure 1). The sequencing method is considered the gold-standard technique for detection of somatic as well as generic mutations. Jancik et al. [15] compared the specificity, sensitivity, cost, and working time of five techniques including Pyr, Sanger sequencing, and real-time PCR for KRAS mutations. Ogino et al. [16] stated that the Pyr assay to detect somatic mutations from formalin-fixed paraffin embedded tissue is more sensitive than Sanger sequencing. Tsiatis et al. [17] compared Pyr, Sanger sequencing, and melting curve methods for the detection of somatic mutations like KRAS, NRAS, and BRAF and demonstrated that Sanger sequencing specificity is generally high compared with other methods, but sensitivity has been reported to differ. Real-time PCR is the most sensitive method for detecting minimal residual disease [18], but it is limited to specific detection of mutations A, B, and D. In the case of limited mutation, we can synthesize primers and probes for other mutations as well, but it will add extra cost per reaction (Table 3).
Figure 1. NPM1 mutation detection by pyrosequencing detection by pyrosequencing.
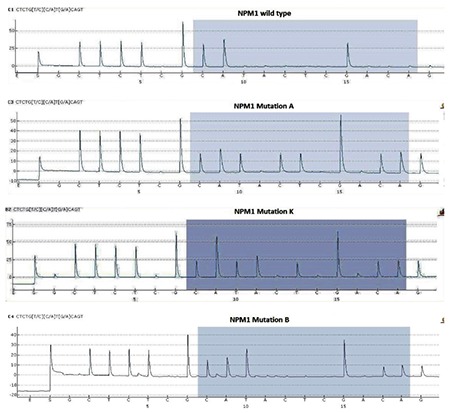
Table 3. Sensitivity, specificity, time, and monetary cost of pyrosequencing, real-time polymerase chain reaction, and Sanger sequencing.
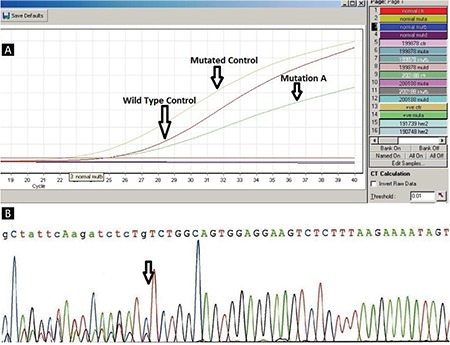
Pyr is easily capable of detecting PCR fragments that are 25-50 bp in length while longer fragments may pose a problem [15,16]. In the case of NPM1,in which 99% of mutations occur at position 956 in exon 12 [14], with Pyr we were able to detect all types of mutations (Figure 1) with lower cost than real-time PCR and less time than Sanger sequencing (Figure 2). Recently next-generation sequencing (NGS) has become popular for detection of mutations in 50 genes to 100 genes simultaneously. NGS is the method to detect mutations down to the mutational burden of 1.25%. However, even though NGS is an accurate method, it is still costly and time-consuming compared with Pyr.
Figure 2. A) NPM1 mutation detection by real-time polymerase chain reaction using Ipsogen MutaScreen Kit. B) Sanger sequencing.
Conclusion
In our study Pyr emerged as the most suitable technique for the detection of NPM1 mutations on the basis of its easy interpretation and less time-consuming processes than Sanger sequencing. However, the limit of mutation detection by real-time PCR is 0.1%-1%, the lowest of all three techniques, so real-time PCR is the best technique to determine minimal residual disease compared to Pyr, which has a limit of detection of 1%-5%. The Pyr assay can be considered as a better technique for NPM1 mutation detection.
Footnotes
Ethics
Ethics Committee Approval: This study was approved by the Gauhati University Ethical Committee with code number GUEC-12/2015.
Informed Consent: N/A.
Authorship Contributions
Concept: K.K.S., D.K.; Design: K.K.S., D.K.; Data Collection or Processing: D.K., M.K.P., S.N.; Analysis or Interpretation: D.K., K.K.S., A.M.; Literature Search: D.K., M.K.P., S.N.; Writing: D.K., M.K.P.
Conflict of Interest: The authors of this paper have no conflicts of interest, including specific financial interests, relationships, and/or affiliations relevant to the subject matter or materials included.
References
- 1.Naoe T, Suzuki T, Kiyoi H, Urano T. Nucleophosmin: a versatile molecule associated with hematological malignancies. Cancer Sci. 2006;97:963–969. doi: 10.1111/j.1349-7006.2006.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao T, Zhu HH, Wang J, Jia JS, Yang SM, Jiang H, Lu J, Chen H, Xu LP, Zhang XH, Jiang B, Ruan GR, Wang DB, Huang XJ, Jiang Q. Prognostic significance of early assessment of minimal residual disease in acute myeloid leukemia with mutated NPM1 patients. Zhonghua Xue Ye Xue Za Zhi. 2013;38:10–16. doi: 10.3760/cma.j.issn.0253-2727.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, Uitterlinden AG, Erpelinck CA, Delwel R, Löwenberg B, Valk PJ. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106:3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 4.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, Bigerna B, Pacini R, Pucciarini A, Liso A, Vignetti M, Fazi P, Meani N, Pettirossi V, Saglio G, Mandelli F, Lo-Coco F, Pelicci PG, Martelli MF GIMEMA Acute Leukemia Working Party. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 5.Thiede C, Creutzig E, Reinhardt D, Ehninger G, Creutzig U. Different types of NPM1 mutations in children and adults: evidence for an effect of patient age on the prevalence of the TCTG-tandem duplication in NPM1-exon 12. Leukemia. 2007;21:366–367. doi: 10.1038/sj.leu.2404519. [DOI] [PubMed] [Google Scholar]
- 6.Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, Meani N, Diverio D, Bernard L, Tizzoni L, Volorio S, Luzi L, Colombo E, Lo Coco F, Mecucci C, Falini B, Pelicci PG. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem cell maintenance. Blood. 2005;106:899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, He P, Liu F, Shi L, Zhu H, Zhao J, Wang Y, Cheng X, Zhang M. Prognostic significance of NPM1 mutations in acute myeloid leukemia: a meta-analysis. Mol Clin Oncol. 2014;2:275–281. doi: 10.3892/mco.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki T, Kiyoi H, Ozeki K, Tomita A, Yamaji S, Suzuki R, Kodera Y, Miyawaki S, Asou N, Kuriyama K, Yagasaki F, Shimazaki C, Akiyama H, Nishimura M, Motoji T, Shinagawa K, Takeshita A, Ueda R, Kinoshita T, Emi N, Naoe T. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106:2854–2861. doi: 10.1182/blood-2005-04-1733. [DOI] [PubMed] [Google Scholar]
- 9.Gale RE, Green C, Allen C, Mead AJ, Burnett AK, Hills RK, Linch DC Medical Research Council Adult Leukaemia Working Party. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 10.Becker H, Marcucci G, Maharry K, Radmacher MD, Margeson KM, Whitman SP, Wu YZ, Schwind S, Paschka P, Powell BL, Carter TH, Kolitz ZE, Wetzler M, Carrol AJ, Baer MR, Caligiuri MA, Larson RA, Bloomfield CD. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boonthimat C, Thongnoppakhun W, Auewarakul CU. Nucleophosmin mutation in Southeast Asian acute myeloid leukemia: eight novel variants, FLT3 coexistence and prognostic impact of NPM1/FLT3 mutations. Haematologica. 2008;93:1565–1569. doi: 10.3324/haematol.12937. [DOI] [PubMed] [Google Scholar]
- 12.Falini B, Martelli MP, Pileri SA, Mecucci C. Molecular and alternative methods for diagnosis of acute myeloid leukemia with mutated NPM1: flexibility may help. Haematologica. 2010;95:529–534. doi: 10.3324/haematol.2009.017822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorello P, Cazzaniga G, Alberti F, Dell’Oro MG, Gottardi E, Specchia G, Roti G, Rosati R, Martelli MF, Diverio D, Lo Coco F, Biondi A, Saglio G, Mecucci C, Falini B. Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia. 2006;20:1103–1108. doi: 10.1038/sj.leu.2404149. [DOI] [PubMed] [Google Scholar]
- 14.Schnittger S, Kern W, Tschulik C, Weiss T, Dicker F, Falini B, Haferlach C, Haferlach T. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 2009;114:2220–2231. doi: 10.1182/blood-2009-03-213389. [DOI] [PubMed] [Google Scholar]
- 15.Jancik S, Drabek J, Berkovcova J, Xu YZ, Stankova M, Klein J, Kolek V, Skarda J, Tichy T, Grygarkova I, Radzioch D, Hajduch M. A comparison of direct sequencing, pyrosequencing, high resolution melting analysis, TheraScreen DxS, and the K-ras StripAssay for detecting KRAS mutations in non small cell lung carcinomas. J Exp Clin Cancer Res. 2012;31:79. doi: 10.1186/1756-9966-31-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010;12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falini B, Martelli MP, Bolli N, Sportoletti P, Liso A, Tiacci E, Haferlach T. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011;117:1109–1120. doi: 10.1182/blood-2010-08-299990. [DOI] [PubMed] [Google Scholar]


