Abstract
STAT1 and STAT3 are two important members of the STAT (signal transducers and activators of transcription) protein family and play opposing roles in regulating cancer cell growth. Suppressing STAT3 and/or enhancing STAT1 signaling are considered to be attractive anticancer strategies. Cucurbitacin I (CuI) isolated from the cucurbitacin family was reported to be an inhibitor of STAT3 signaling and a disruptor of actin cytoskeleton. In this study we investigated the function and mechanisms of CuI in regulating STAT signaling in human cancer cells in vitro. CuI (0.1–10 mmol/L) dose-dependently inhibited the phosphorylation of STAT3, and enhanced the phosphorylation of STAT1 in lung adenocarcinoma A549 cells possibly through disrupting actin filaments. We further demonstrated that actin filaments physically associated with JAK2 and STAT3 in A549 cells and regulated their phosphorylation through two signaling complexes, the IL-6 receptor complex and the focal adhesion complex. Actin filaments also interacted with STAT1 in A549 cells and regulated its dephosphorylation. Taken together, this study reveals the molecular mechanisms of CuI in the regulation of STAT signaling and in a possible inhibition of human cancer cell growth. More importantly, this study uncovers a novel role of actin and actin-associated signaling complexes in regulating STAT signaling.
Keywords: cucurbitacin I, cancer cells, STAT1, STAT3, actin, phosphorylation
Introduction
The Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathways mediate the signal transduction of various cytokines and growth factors in the regulation of many physiological processes, such as immune responses, hematopoiesis, and stem cell renewal1,2,3. Dysregulation of JAK/STAT signaling often occurs in various diseases, including cancer4,5. Activating mutations in JAKs and STATs play causal roles in hematologic neoplasms6,7,8, as well as in solid tumors, and have prognostic significance9,10,11,12,13.
STAT3 promotes cell proliferation and survival, and also inhibits apoptosis, by up-regulating the expression of growth stimulatory genes, such as c-Myc, cyclin-D1, Bcl-xl, and survivin; STAT3 also functions as an oncogene in various types of cancers14,15. Furthermore, it increases the expression of VEGF, twist, and MMP, promoting tumor angiogenesis, epithelial-mesenchymal transition (EMT), and metastasis16,17,18. The major cytokine that activates STAT3 is IL-619. IL-6 binds to its receptor IL-6Rα in the plasma membrane and forms a hexamer with the signal-transducing protein GP130, which activates the GP130-associated JAKs (JAK1, JAK2 or TYK2, depending on the cell type), followed by JAK-mediated GP130 phosphorylation and STAT3 recruitment and activation20,21. Therefore, blocking IL-6/JAK/STAT3 signaling has become an attractive strategy for cancer treatment22.
In contrast to STAT3, STAT1 mediates the anti-tumor activity of IFN-γ in tumor cells to promote cell cycle arrest and apoptosis and negatively regulates angiogenesis1,23,24,25,26,27. IFN-γ binds to its receptor IFN-γR1/IFN-γR2, resulting in receptor allosteric changes and the transactivation of JAK1 and JAK2 and leading to STAT1 activation28. STAT1 mainly acts as a tumor suppressor in many tumors. For example, STAT1-deficient mice are more susceptible to chemical carcinogens29,30. High levels of STAT1 activity correlated with good prognosis in several types of cancers31,32,33,34,35. Therefore, in contrast to STAT3, enhancing STAT1 activity has also been considered to be an attractive strategy for treating cancer.
The activities of the STATs are regulated by both kinase phosphorylation and phosphatase dephosphorylation. JAK1 and JAK2 are two major tyrosine kinases involved in STAT1 and STAT3 phosphorylation, respectively. Protein tyrosine phosphatases TCPTP and SHP2 are reported to be responsible for p-STAT1 dephosphorylation, while TCPTP, SHP2, SHP1 and PTPRT are responsible for p-STAT3 dephosphorylation36.
The actin cytoskeleton plays important roles in a variety of cellular processes, including mechanical support, cytokinesis, cell migration, differentiation, and endocytosis. The actin cytoskeleton exists in different structural forms that have specific functions. Stress fibers are contractile actin bundles that are necessary for cell-cell or cell-ECM adhesion37. They often anchor to focal adhesions, through which they connect to the extracellular matrix (ECM)38. Focal adhesions are multi-protein assemblies that transduce mechanical forces and regulatory signals between the ECM and the actin cytoskeleton. Vice versa, the actin cytoskeleton directs focal adhesion assembly and disassembly and organization39. Actin is also a major component of the cell cortex, a specialized layer of cytoplasmic proteins on the inner face of the plasma membrane, and plays a central role in cell shape control and contractile ring formation during cytokinesis40. Mounting evidence indicates that the actin cytoskeleton is not only a structural protein but also a key regulator of cellular signaling41,42,43,44,45,46,47.
Cucurbitacin I (CuI) belongs to the cucurbitacin family, which are tetracyclic triterpenoids. CuI was reported to be an inhibitor of STAT3 phosphorylation48 and a disruptor of the actin cytoskeleton49,50,51. It has anti-cancer activity both in vitro and in vivo. However, the molecular mechanisms of its action and the relationships between the two functions have not yet been elucidated.
In this study, we investigated the mechanisms of Cul in the regulation of STAT signaling and actin filaments, particularly the connections between the two. We found that CuI affected not only STAT3 signaling but also STAT1 signaling, and it had opposing effects on STAT3 and STAT1, inhibiting STAT3 signaling but enhancing STAT1 signaling. Furthermore, we found a connection between the regulation of STAT signaling and actin filaments. The effects of Cul on STAT signaling were the consequence of disrupting the actin cytoskeleton. Our study not only revealed the molecular mechanisms of CuI but also uncovered novel JAK/STAT signaling complexes and a novel role of actin in their regulation.
Materials and methods
Cells and reagents
All the cell lines used were obtained from the American Type Culture Collection. A549 cells were cultured in RPMI-1640 (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% (v/v) FBS (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 100 μg/mL ampicillin and 100 μg/mL streptomycin. All cell lines were cultured at 37 °C in a humidified atmosphere of 95% air and 5% CO2.
The sources of chemicals and reagents were as follows: cucurbitacin I (# C4493; Sigma-Aldrich, Saint Louis, MO, USA), complete protease inhibitor cocktail (# P1860; Sigma-Aldrich, Saint Louis, MO, USA), biotinylated phalloidin (# P8716; Sigma-Aldrich, Saint Louis, MO, USA), jasplakinolide (# sc202191A; Santa-Cruz Biotechnology, Dallas, TX, USA), cytochalasin D (# 250255, Millipore, Billerica, MA, USA); IL-6 (# 200-02, Peprotech, Rocky Hill, NJ, USA), IFN-γ (# 300-02, Peprotech, Rocky Hill, NJ, USA), and rhodamine-phalloidin (# R415, Thermo Fisher Scientific, Waltham, MA, USA).
Western blot analysis
Western blotting was performed as previously described52. Immune complexes were detected by enhanced chemiluminescence (# WBKLS0500, Millipore, Billerica, MA, USA).
The following primary antibodies were used: rabbit anti-JAK2 (# 3230S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-pY1007/1008 JAK2 (# 3776S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-STAT3 (# 12640, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-pY705 STAT3 (# 9145S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-STAT1 (# 14994S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-pY701 STAT1 (# 7649S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-TYK2 (# 14193S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-pY1054/1055 TYK2 (# 9321S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-JAK1 (#3332S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-pY1022/1023 JAK1 (# 3331S, Cell Signaling Technology, Danvers, MA, USA), mouse anti-α-tubulin (# sc5286, Santa Cruz Biotechnology, Dallas, TX, USA), rabbit anti-IL-6Rα (# sc13947, Santa Cruz Biotechnology, Dallas, TX, USA), rabbit anti-GP130 (# sc655, Santa Cruz Biotechnology, Dallas, TX, USA), rabbit anti-SHP2 (# sc280, Santa Cruz Biotechnology, Dallas, TX, USA), rabbit anti-β-actin antibody (# P30002M, Abmart, CA, USA), rabbit anti-vinculin antibody (# ab73412, Abcam, Cambridge, UK), and mouse anti-TCPTP antibody (# PH03L, Calbiochem, Merck Millipore, Billerica, MA, USA).
Pull-down of actin filaments
A549 cells grown in 100-mm dishes to 100% confluence were washed with ice-cold PBS, then scraped on ice into homogenization buffer (100 mmol/L Na2HPO4-NaH2PO4, pH 7.2; 2 mmol/L MgCl2)53 containing 0.01% NP-40, 1 mmol/L PMSF, 1 mmol/L Na3VO4, 1 mmol/L NaF, and a complete protease inhibitor cocktail, homogenized for 20 strokes with a Dounce homogenizer and centrifuged at 12000×g and 4 °C for 10 min. The supernatant was pre-cleared with monomeric avidin agarose beads (# 20267, Thermo Fisher Scientific, Waltham, MA, USA), prepared according to the manufacturer's instructions. To pull down actin filaments, biotinylated-phalloidin or biotin (negative control) was added to the pre-cleared lysates and incubated with rotation for 2 h at 4 °C, followed by incubation with monomeric avidin agarose beads for 1 h. The precipitates were washed 5 times with homogenization buffer and eluted with 10 mmol/L biotin. The elution was mixed with Laemmli 5× sample buffer and boiled for 5 min, followed by Western blot analyses.
Immunoprecipitation
A549 cells grown in 100 mm dishes to 100% confluence were harvested on ice with 50 mmol/L Tris-HCl, pH 7.4; 150 mmol/L NaCl; 1 mmol/L EDTA; 0.5% NP-40; 1 mmol/L PMSF; 1 mmol/L Na3VO4; 1 mmol/L NaF; and a complete protease inhibitor cocktail. The supernatant was pre-cleared with protein A/G-agarose beads (# sc2003, Santa Cruz Biotechnology, Dallas, TX, USA) for 1 h with agitation at 4 °C, followed by incubation with primary antibodies and general agitation at 4 °C overnight and then with protein A/G-magnetic beads (# B23201, Selleckchem, Houston, TX, USA) for another 3 h. After washing 5 times with the above lysis buffer, the pellets were mixed with Laemmli 2× sample buffer (# S3401, Sigma-Aldrich, Saint Louis, MO, USA) and boiled for 5 min, then processed for western blotting analyses.
Antibodies used for immunoprecipitation were as follows: rabbit anti-GP130 antibody (# sc656, Santa Cruz Biotechnology, Dallas, TX, USA) and mouse anti-vinculin antibody (# V9131, Sigma-Aldrich, Saint Louis, MO, USA).
In vitro kinase and phosphatase assays
A JAK2 in vitro kinase assay was performed using JAK2 immunoprecipitates. A549 cells were lysed with kinase lysis buffer (50 mmol/L HEPES, pH 7.4; 150 mmol/L NaCl; 0.15% TritonX-100; 1 mmol/L PMSF; 1 mmol/L Na3VO4; 1 mmol/L NaF; and a complete protease inhibitor cocktail) following IL-6 stimulation. Cell lysates were immunoprecipitated with goat anti-JAK2 antibody (# sc34479, Santa Cruz Biotechnology, Saint Louis, MO, USA). The precipitates were washed and resuspended in kinase reaction buffer (60 mmol/L HEPES, pH 7.5; 5 mmol/L MgCl2; 5 mmol/L MnCl2; 25 μmol/L Na3VO4; and 200 μmol/L ATP) and were then incubated with DMSO, CuI or 2-MS for 15 min at room temperature, followed by incubation with the biotinylated peptide FLT3 (Cell Signaling Technology, Danvers, MA, USA) for 30 min at 30 °C; then, the supernatants were transferred to streptavidin-coated 96-well plates, incubated for 1 h at 30 °C, rinsed with 0.1% PBST (PBS+ 0.1% Tween-20) and incubated with the anti-phosphotyrosine antibody 4G10 (# 05-321MG, Merck Millipore, Billerica, MA, USA) for 2 h at 37 °C and with HRP-conjugated secondary antibody for 30 min at 30 °C; after five washes, the TMP substrate (# 7004S, Cell Signaling Technology, Danvers, MA, USA) was added, and the reaction was stopped by adding 2 mol/L H2SO4; finally, the absorbance was measured at 450 nm.
An in vitro phosphatase assay was performed with SHP2 or TCPTP immunoprecipitates. A549 cells were lysed with PTP lysis buffer (50 mmol/L HEPES, pH 7.2; 150 mmol/L NaCl; 1% Triton X-100; 5 mmol/L EDTA; 1 mmol/L PMSF; 1 mmol/L NaF; and a complete protease inhibitor cocktail. Cell lysates were immunoprecipitated with anti-SHP2 (# sc280, Santa Cruz Biotechnology, Saint Louis, MO, USA) or anti-TCPTP (# PH03L, Merck Millipore, Billerica, MA, USA) antibodies; the precipitates were washed and re-suspended in PTP reaction buffer (100 mmol/L HEPES, pH 7.0; 150 mmol/L NaCl; and 1 mmol/L EDTA) and then incubated with DMSO, CuI or Na3VO4 for 15 min at room temperature, followed by incubation with the phosphatase substrate 4-nitrophenyl phosphate disodium (# N4645, Sigma-Aldrich, Saint Louis, MO, USA) for 3 h at 37 °C. The reaction was terminated with 1 mol/L NaOH, and the absorbance was measured at 405 nm.
STAT1 in vitro dephosphorylation assay
A549 cells receiving different treatments were lysed with hypotonic buffer (10 mmol/L HEPES, pH 7.4; 10 mmol/L KCl; and 2 mmol/L MgCl2) and nuclear extraction buffer (20 mmol/L HEPES, pH 7.4; 1.5 mmol/L MgCl2; 420 mmol/L NaCl; and 0.2 mmol/L EDTA) containing 1 mmol/L PMSF, 1 mmol/L NaF and a complete protease inhibitor cocktail, followed by centrifugation at 12000×g and 4 °C for 10 min. Cell lysates were incubated at 36 °C for different lengths of time. Laemmli 5× sample buffer was added to the mixture to facilitate Western blot analyses. For denaturation, lysates from IFN-γ-stimulated cells were boiled for 5 min and then cooled on ice.
Immunochemical staining and immunofluorescence analysis
Cells were plated on coverslips one day before use. After fixation with 4% formaldehyde for 15 min at room temperature, cells were permeabilized with 0.05% Triton X-100 in PBS for 5 min and rinsed with PBS 3 times, followed by blocking with 10% horse serum in 0.1% PBST (Tween-20) for 30 min at 37 °C and incubation with primary antibodies at 4 °C overnight and AlexaFluor 488- or AlexaFluor 594-conjugated secondary antibodies at 37 °C for 1.5 h. For actin staining, rhodamine-phalloidin was added and incubated for 1 h at room temperature. Nuclei were stained with DAPI, which was added for 5 min before washing and mounting with mounting medium (DAKO, Agilent, Santa Clara, CA, USA).
Images were captured at room temperature with a laser-scanning confocal microscope FLUOVIEW FV1000 (Olympus, PA, USA) and FV10-ASW software (Olympus, PA, USA). The microscope was equipped with a 60×(1.42 NA) PLAPON oil objective lens (Olympus, PA, USA). Tagged image file format images were processed using Photoshop software.
Antibodies used were as follows: rabbit anti-JAK2 (#3230S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-pY1007/1008 JAK2 (#3776S, Cell Signaling Technology, Danvers, MA, USA), rabbit anti-STAT3 (#12640, Cell Signaling Technology, Danvers, MA, USA), mouse anti-pY705 STAT3, rabbit anti-STAT1, rabbit anti-pY701 STAT1 (Cell Signaling Technology, Danvers, MA, USA), rabbit anti-GP130 antibody (# sc656, Santa Cruz Biotechnology, Saint Louis, MO, USA), mouse anti-Vinculin antibody (# V9131, Sigma Aldrich, Saint Louis, MO, USA), rabbit anti-vinculin antibody (# ab73412, Abcam, Cambridge, UK), secondary AlexaFluor 488-conjugated goat anti-rabbit antibody (#A11008, Thermo Fisher Scientific, Waltham, MA, USA), secondary AlexaFluor 488-conjugated goat anti-mouse antibody (#A28175, Thermo Fisher Scientific, Waltham, MA, USA), and secondary AlexaFluor 594-conjugated goat anti-mouse antibody (#GAM5942, Multi Science Biotech, Hangzhou, China).
Results
Cucurbitacin I inhibited STAT3 phosphorylation but enhanced STAT1 phosphorylation
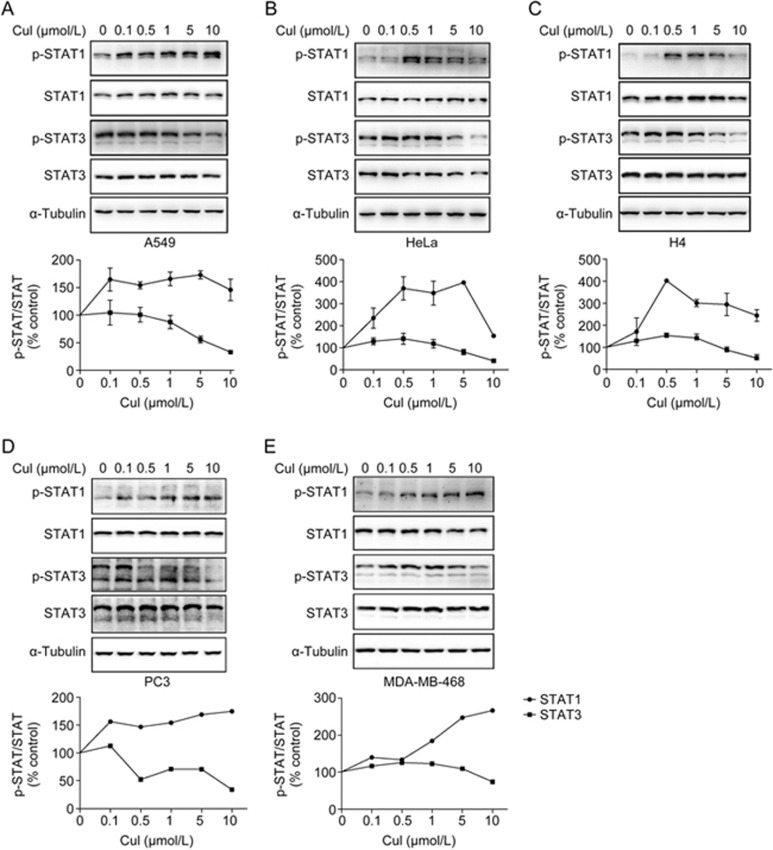
CuI was reported to inhibit STAT3 signaling and to induce apoptosis in cancer cells48. To confirm and to evaluate the specificity of CuI in STAT signaling, we analyzed the effects of CuI on the phosphorylation of STAT1, which also plays an important but opposing role in the regulation of cancer cell growth. We also found that in contrast to the inhibition of the Y705 phosphorylation of STAT3, CuI increased the Y701 phosphorylation of STAT1 in all the cancer cell lines tested (Figure 1), suggesting that CuI may have opposite effects on STAT1 and STAT3 signaling.
Figure 1.
Cucurbitacin I inhibited STAT3 phosphorylation but enhanced STAT1 phosphorylation. Lung adenocarcinoma cell line A549 (A), cervical adenocarcinoma cell line Hela (B), neuroglioma cell line H4 (C), prostate cancer cell line PC3 (D) and mammary adenocarcinoma MDA-MB-468 (E) were treated with CuI for 2 h, and whole cell lysates were processed for Western blot analyses with indicated antibodies. The levels of pY705-STAT3 and pY701-STAT1 in CuI treatments were normalized by α-tubulin and quantified to DMSO treatment, and were graphed below the corresponding blots.
Cucurbitacin I inhibited the phosphorylation of STAT3 but the dephosphorylation of p-STAT1
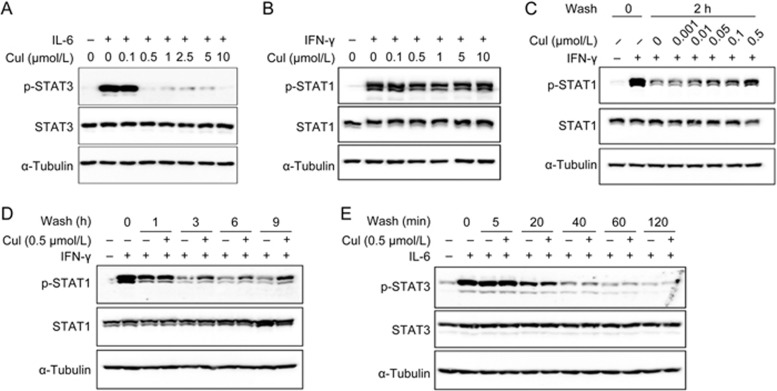
The phosphorylation status of a protein is a balanced result of kinase phosphorylation and phosphatase dephosphorylation. To clarify whether it was through the modulation of kinase phosphorylation or phosphatase dephosphorylation that CuI exerted its effects on the two STATs, we stimulated lung adenocarcinoma A549 cells with IFN-γ or IL-6 to activate STAT1 or STAT3, respectively, following CuI treatment and observed the effects of CuI on the cytokine-induced phosphorylation of the two STATs. Cul blocked IL-6-induced STAT3 phosphorylation (Figure 2A) but had little effect on IFN-γ-induced STAT1 phosphorylation (Figure 2B), suggesting that Cul affected the kinase phosphorylation process of STAT3 but not that of STAT1.
Figure 2.
Cucurbitacin I inhibited phosphorylation of STAT3 but dephosphorylation of p-STAT1. (A and B) A549 cells were treated with CuI at indicated concentrations for 2 h, followed by IL-6 (10 ng/mL, A) or IFN-γ (10 ng/mL, B) stimulation for 15 min. (C–E) Following IFN-γ (C and D) or IL-6 (E) stimulation for 30 min, A549 cells were washed with fresh medium for 3 times and then incubated with or without CuI at indicated concentrations (C) or for indicated time periods (D and E). Whole cell lysates were processed for Western blot analyses and probed with indicated antibodies.
To examine the effects of Cul on the dephosphorylation of p-STAT1 and p-STAT3, we pulse-stimulated A549 cells with IFN-γ or IL-6 and then washed off the cytokines and incubated the cells in the presence or absence of CuI for different time periods. Cul inhibited the dephosphorylation of p-STAT1 (Figure 2C and D) but had no effect on the dephosphorylation of p-STAT3 (Figure 2E). The concentration of CuI required to inhibit p-STAT1 dephosphorylation appeared to be lower than that required to inhibit STAT3 phosphorylation (Figure 2C). Thus, CuI distinctively regulated STAT3 and STAT1 phosphorylation. It inhibited the phosphorylation of STAT3 but the dephosphorylation of p-STAT1.
Cucurbitacin I did not directly inhibit STAT kinases or phosphatases
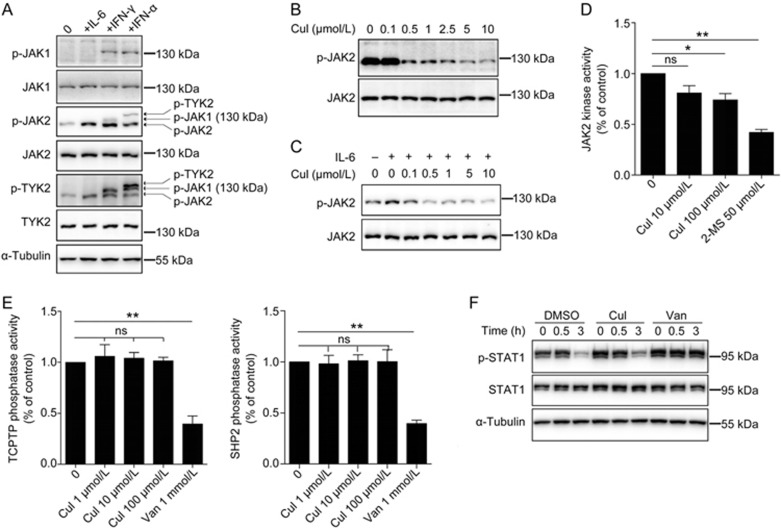
Because CuI inhibited IL-6-stimulated STAT3 phosphorylation, we examined whether it suppressed the upstream kinases of STAT3 in the IL-6 signaling pathway. JAK2 was reported to be the major kinase involved in IL-6-stimulated STAT3 signaling in the A549 cells54. However, we observed that JAK2 was constitutively phosphorylated in A549 cells and that IL-6-induced JAK2 phosphorylation could hardly be detected when the cells were cultured in adherent conditions. IL-6-induced JAK2 phosphorylation could be observed only when the cells were cultured in suspension. Therefore, we used suspended cells to examine the effects of CuI on the IL-6-induced phosphorylation of JAK2. We confirmed that IL-6 specifically activated JAK2 but not JAK1 and TYK2 in the A549 cells (Figure 3A) and found that CuI inhibited IL-6-induced and constitutive JAK2 phosphorylation in A549 cells (Figure 3B and C).
Figure 3.
Cucurbitacin I did not inhibit the STAT kinases or phosphatases directly. (A) A549 cells in suspension were stimulated with IL-6 (10 ng/mL), IFN-γ (10 ng/mL) or IFN-α (1000 IU/mL) for 5 min. The p-JAK2 and p-TYK2 antibodies non-specifically recognized other phosphorylated JAK family members as indicated by arrows. (B and C) A549 cells in adhesion (B) or suspension (C) were treated with CuI at various concentrations for 2 h, followed by stimulation with or without 10 ng/mL IL-6 for 5 min. (D and E) JAK2, and SHP2 or TCPTP proteins immunoprecipitated from A549 cells was subjected to in vitro kinase (D) or phosphatase (E) assays respectively in the presence of indicated concentrations of CuI. 2-MS, a JAK2 inhibitor identified by our lab, and vanadate, a protein tyrosine phosphatase inhibitor, were served as positive controls. (F) Effect of Cul or vanadate on p-STAT1 dephosphorylation in A549 cells. Data are represented as mean±SD of three experiments. CuI, 2-MS and vanadate groups were compared with vehicle group by Student's t-test. ns, not significant; *P<0.05; **P<0.01. van, vanadate.
We next examined whether CuI directly inhibited JAK2 kinase activity using in vitro kinase assays. We found that CuI did not inhibit JAK2 kinase activity (Figure 3D), suggesting that CuI inhibited JAK2 through an indirect mechanism.
Two tyrosine phosphatases, TCPTP and SHP2, were reported to be responsible for p-STAT1 dephosphorylation55,56. We thus examined whether CuI inhibited these two phosphatases using an in vitro phosphatase assay. We found that CuI did not directly inhibit TCPTP or SHP2 activity (Figure 3E). We also lysed cells after IFN-γ stimulation and incubated the cell lysates in vitro in the presence or absence of CuI; we found that CuI did not inhibit p-STAT1 dephosphorylation in vitro (Figure 3F), also suggesting that CuI did not directly inhibit p-STAT1 phosphatases.
Disrupting the actin cytoskeleton inhibited STAT3 phosphorylation and p-STAT1 dephosphorylation
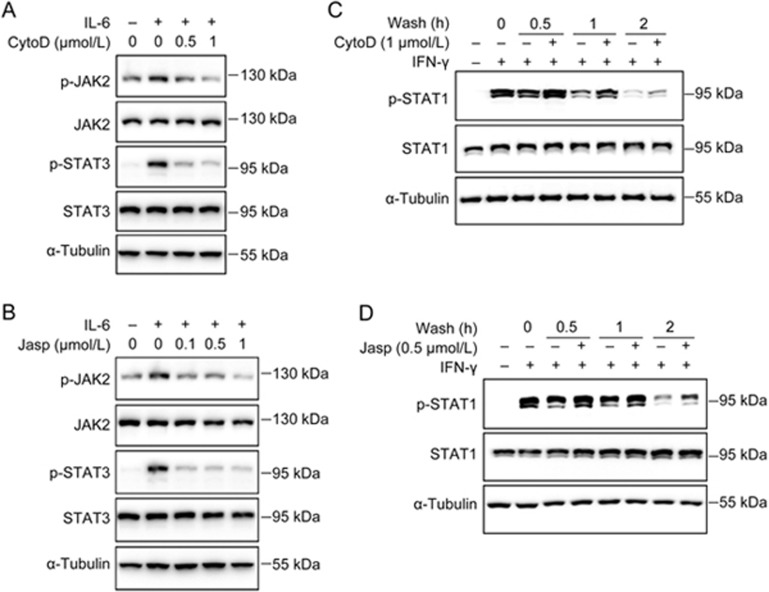
CuI was reported to covalently bind and disrupt the actin cytoskeleton50. To determine whether the disruption of the actin cytoskeleton could affect STAT signaling, we examined the effects of two known actin-disrupting agents, cytochalasin D (CytoD) and jasplakinolide (Jasp), on the phosphorylation of STAT3 and STAT1. Both CytoD and Jasp blocked the IL-6-stimulated phosphorylation of JAK2 and STAT3 (Figure 4A and B) while inhibiting the dephosphorylation of p-STAT1 (Figure 4C and D) as Cul did. These results suggested that the actin cytoskeleton regulates STAT3 and STAT1 phosphorylation and that CuI might affect JAK/STAT signaling by disrupting the actin cytoskeleton.
Figure 4.
Disrupting actin cytoskeleton inhibited STAT3 phosphorylation and p-STAT1 dephosphorylation. (A and B) A549 cells in suspension were treated with CytoD (A) or Jasp (B) at indicated concentrations for 2 h, followed by IL-6 (10 ng/mL) stimulation for 5 min. (C and D) Following IFN-γ (10 ng/mL) stimulation for 30 min, A549 cells were washed and incubated with fresh medium containing DMSO, CytoD (1 μmol/L) (C) or Jasp (0.5 μmol/L) (D) for indicated time periods.
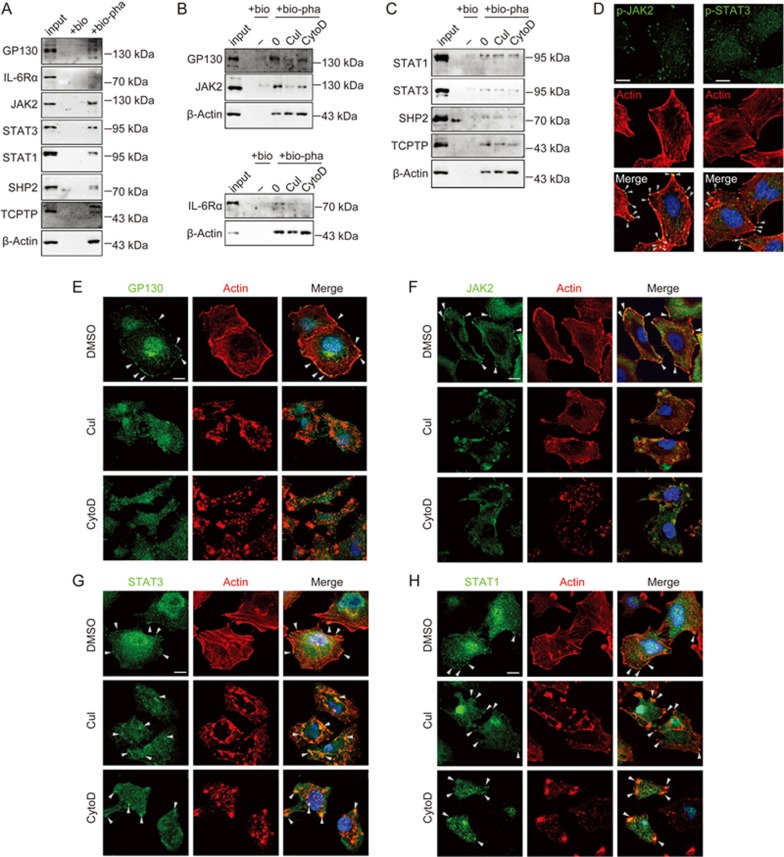
STAT3 and STAT1 signaling proteins were physically associated with actin filaments
To understand how actin filaments may regulate the STATs, we determined whether the actin cytoskeleton physically interacted with the signaling proteins that regulated STAT3 phosphorylation and STAT1 dephosphorylation. By pulling down actin filaments using biotinylated phalloidin, we found that the IL-6 receptors (IL-6Rα and GP130), JAK2, STAT3, STAT1, TCPTP, and SHP2 were all physically associated with actin filaments (Figure 5A), and that CuI or CytoD treatment disrupted the association of JAK2, GP130, and IL-6Rα with actin (Figure 5B), but not that of STAT3, STAT1, TCPTP, or SHP2 with actin (Figure 5C). Immunofluorescent staining demonstrated that GP130, JAK2/p-JAK2, and STAT3/p-STAT3 were all colocalized with actin filaments at the cell edges (Figure 5D–G arrowheads). CuI and CytoD treatment dissociated GP130 and JAK2 from actin (Figure 5E and F). However, STAT3 and STAT1 seemed to remain associated with actin after CuI or CytoD treatment (Figure 5G and H). These results suggested actin might regulate STAT signaling through physically interacting with STAT signaling proteins.
Figure 5.
STAT3 and STAT1 signaling proteins were physically associated with actin filaments. (A–C) A549 cells were lysed, and actin filaments were pulled-down using biotinylated-phalloidin, followed by elution with 10 mmol/L biotin. For drug treatments in B and C, A549 cells were treated with DMSO, 0.5 μmol/L CuI or 1 μmol/L CytoD for 2 h. input, 3% of the total cell lysates compared with 100% of the elution; bio, biotin; bio-pha, biotinylated-phalloidin. (D–H) Confocal micrographs of A549 cells staining with rhodamine-phalloidin (for actin, red), DAPI (for nucleus, blue), and antibodies against the indicated proteins (green). Arrowheads indicated colocalization of STATs signaling proteins with actin. For drug treatment, A549 cells were treated with DMSO, 0.5 μmol/L CuI or 1 μmol/L CytoD for 2 h prior to immunochemical staining. Scale bars, 10 μm.
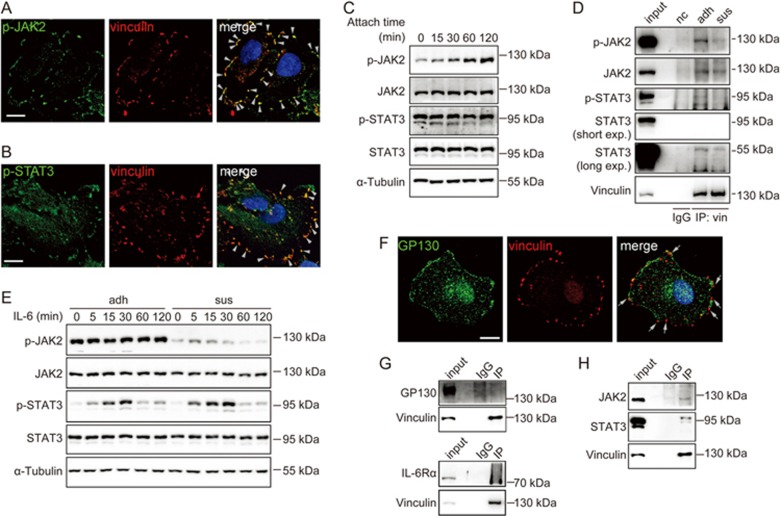
JAK2/STAT3 phosphorylation was regulated by two signaling complexes: the focal adhesion complex and the IL-6 receptor complex
The immunochemical staining data suggested that some of the STAT signaling proteins might localize at focal adhesions, in addition to their membrane localization (Figure 5D). We thus examined whether p-JAK2 and p-STAT3 colocalized with vinculin, a focal adhesion maker. Immunofluorescent staining revealed that p-JAK2 and p-STAT3 indeed colocalized with vinculin in focal adhesions (Figure 6A and B). Next, we determined whether cell adhesion could regulate the phosphorylation of JAK2 and STAT3. We examined the phosphorylation of JAK2 and STAT3 during cell attachment and found that the phosphorylation of JAK2 increased upon cell attachment, but the phosphorylation of STAT3 was unaffected (Figure 6C).
Figure 6.
JAK2/STAT3 phosphorylation were regulated by two signaling complexes: the focal adhesion complex and the IL-6 receptor complex. (A and B) Confocal micrographs of A549 cell staining with DAPI (for nucleus, blue) and antibodies against vinculin (red) and p-JAK2 (green) (A) or p-STAT3 (green) (B). Arrowheads indicated the colocalization of p-JAK2 or p-STAT3 with vinculin. Scale bars, 10 μm. (C) A549 cells were detached by 1 mmol/L EDTA and reattached to plates for various time periods. (D) A549 cells in adhesion (adh) or suspension (sus) were immunoprecipitated for vinculin, followed by immunoblotting with indicated antibodies. (E) A549 cells in adhesion (adh) or suspension (sus) were stimulated with IL-6 (10 ng/mL) for indicated time periods. (F) Confocal micrographs of A549 cell staining with DAPI (for nucleus, blue) and antibodies against vinculin (red) and GP130 (green). Arrows indicated that GP130 and vinculin were not colocalized. Scale bar, 10 μm. (G and H) A549 cell lysates were immunoprecipitated for vinculin, followed by immunoblotting with indicated antibodies. Note that JAK2 and STAT3 bonded with vinculin (H) while GP130 or IL-6Rα did not (G). input, 2.5% of the total cell lysates compared with 100% of the pellet; nc, negative control; IgG, the immunoprecipitate of IgG; IP, immunoprecipitation.
To further confirm that JAK2 and STAT3 were localized at focal adhesions and were regulated by cell adhesion, we immunoprecipitated vinculin from adherent and suspended cells, respectively, and found that JAK2 and STAT3 co-immunoprecipitated with vinculin and that the phosphorylation of vinculin-associated JAK2 and STAT3 increased in adherent cells (Figure 6D). These data suggested that JAK2 and STAT3 phosphorylation could be regulated by focal adhesions.
To determine whether focal adhesion affects IL-6 receptor-mediated JAK2/STAT3 signaling, we treated adherent and suspended cells with IL-6 and examined the phosphorylation of JAK2 and STAT3. We found that IL-6 activated JAK2/STAT3 signaling in both suspended and adherent cells (Figure 6E), indicating that IL-6R/JAK2/STAT3 signaling was independent of focal adhesions. To further strengthen this conclusion, we examined whether GP130, the IL-6 signal-transducing receptor, is present in focal adhesions. We found that GP130 was not colocalized with vinculin in focal adhesions (Figure 6F). Co-immunoprecipitation experiments also excluded the possibility of the presence of either GP130 or IL-6Rα in the focal adhesions (Figure 6G). These data suggested that two signaling complexes existed that regulated JAK2/STAT3 phosphorylation, a focal adhesion complex and a cell membrane IL-6 receptor complex, both of which had interactions with actin. JAK2 and STAT3 bonded with vinculin (Figure 6H).
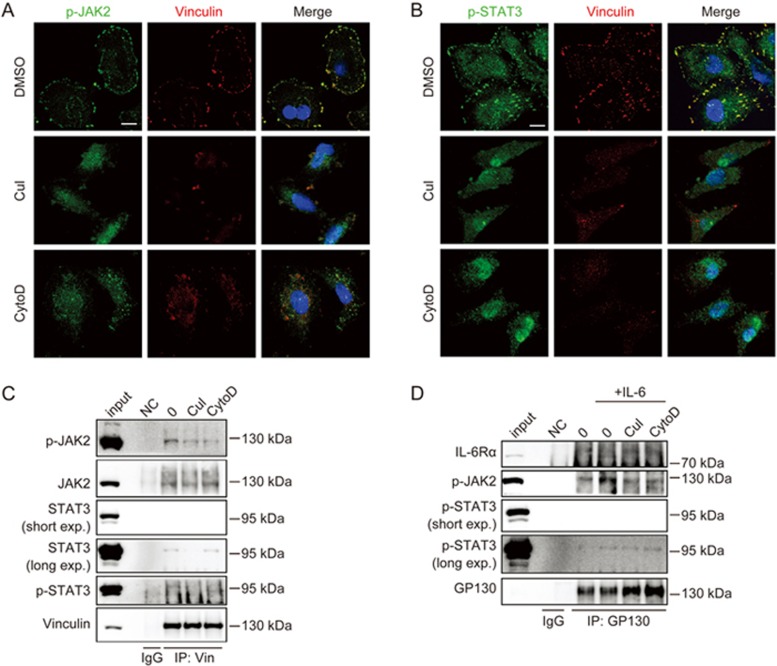
CuI and CytoD inhibited STAT3 phosphorylation in both signaling complexes by disrupting actin filaments
To determine whether STAT3 phosphorylation in both the focal adhesion complex and the IL-6 receptor complex was regulated by actin, we treated the cells with CuI and CytoD and examined the phosphorylation of JAK2/STAT3 in the two complexes. We noticed that stress fibers, the type of actin filaments that anchor to focal adhesions, were disrupted by CuI and CytoD treatment (Figure 5). No vinculin-containing focal adhesion complex existed after CuI and CytoD treatment, and focal adhesion-associated p-JAK2 and p-STAT3 decreased (Figure 7A and B). Co-immunoprecipitation experiments confirmed that CuI or CytoD decreased p-JAK2 and p-STAT3 in the vinculin/focal adhesion complex (Figure 7C). Therefore, the phosphorylation of focal adhesion-associated JAK2/STAT3 was regulated by actin stress fibers.
Figure 7.
CuI and CytoD inhibited STAT3 phosphorylation in both signaling complexes through disrupting actin filaments. (A and B) A549 cells were treated with DMSO, CuI (0.5 μmol/L) or CytoD (1 μmol/L) for 2 h, fixed, permeabilized, and stained with DAPI (nucleus, blue) and antibodies against vinculin and p-JAK2 (green) (A) or p-STAT3 (green) (B). Scale bars, 10 μm. (C) Following treatment with DMSO, CuI (0.5 μmol/L) or CytoD (1 μmol/L) for 2 h, A549 cells were lysed and immunoprecipitated for vinculin, and the indicated proteins were detected by immunoblotting using indicated antibodies. (D) A549 cells were treated with DMSO, CuI (0.5 μmol/L) or CytoD (1 μmol/L) for 2 h, followed by stimulation with or without IL-6 (10 ng/mL) for 5 min, and were lysed and immunoprecipitated for GP130. The immunoprecipitates were processed for Western blot analysis. input, 2.5% of the total cell lysates compared with 100% of the pellet; nc, negative control; IgG, the immunoprecipitate of IgG; IP, immunoprecipitation.
We next examined whether the phosphorylation of JAK2/STAT3 in the IL-6 receptor complex was also regulated by actin using co-immunoprecipitation experiments; we found that CuI and CytoD inhibited the IL-6-induced phosphorylation of JAK2 in the GP130 complex (Figure 7D), suggesting that IL-6 receptor-mediated JAK2/STAT3 phosphorylation was also regulated by actin.
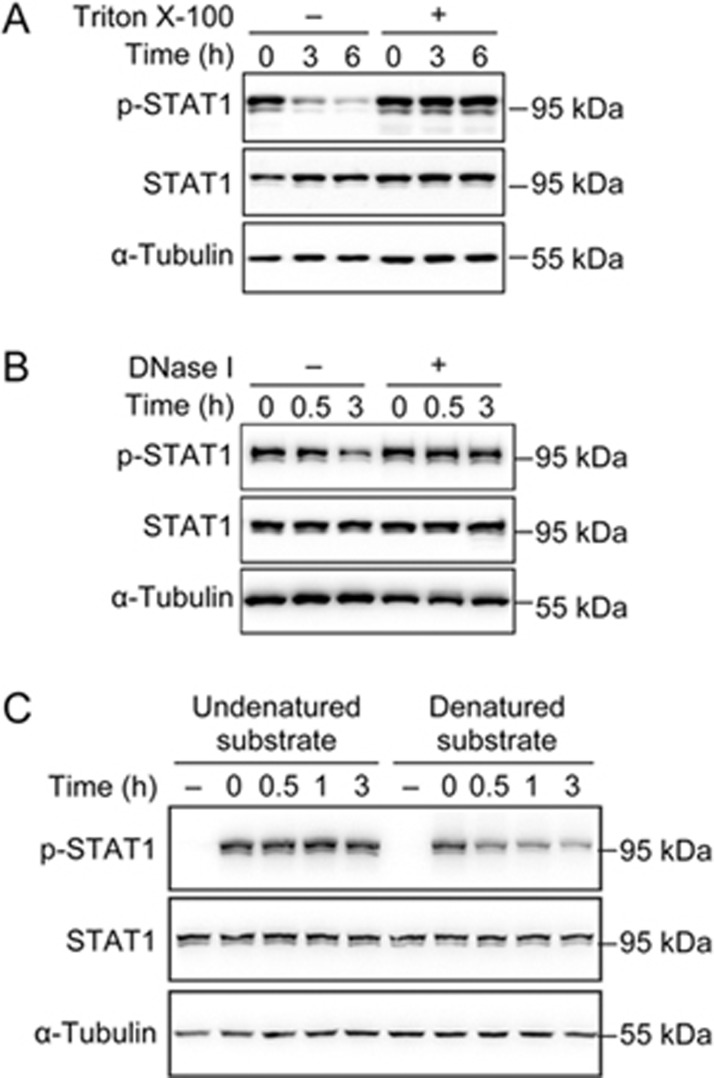
Actin filaments regulated p-STAT1 dephosphorylation by modulating p-STAT1 proteins, rather than by modulating the activity of protein tyrosine phosphatases (PTPs)
To understand the role and mechanisms of the actin cytoskeleton in the regulation of p-STAT1 dephosphorylation, we conducted an in vitro p-STAT1 dephosphorylation experiment in the presence or absence of actin filaments. We stimulated the cells with IFN-γ, then lysed the cells and incubated the lysates in the absence or presence of 1% Triton X-100, which was used to precipitate actin filaments57. We found that p-STAT1 underwent dephosphorylation in vitro, which was inhibited by Triton X-100 (Figure 8A). To further confirm that it was the actin filaments that regulated the p-STAT1 dephosphorylation, the cell lysates were incubated with DNase I, which can bind to actin monomers and depolymerize actin filaments58. We found that p-STAT1 could not be dephosphorylated in the presence of DNase I (Figure 8B). These data demonstrated that filamentous actin was involved in p-STAT1 dephosphorylation.
Figure 8.
Actin filaments regulated p-STAT1 dephosphorylation through modulating p-STAT1 proteins, rather than the activity of protein tyrosine phosphatases (PTPs). (A and B) A549 cells were treated with IFN-γ (10 ng/mL) for 30 min, and then were lysed and incubated in vitro at 36 °C in the presence or absence of 1% Triton X-100 (A) or DNase I (B) for indicated time periods. Following incubation, the lysates were mixed with laemmli 2×sample buffer and processed for Western blot analyses with indicated antibodies. (C) Two dishes of A549 cells were stimulated with or without IFN-γ (10 ng/mL) respectively for 30 min and lysed. The IFN-γ-stimulated lysate was divided into two parts, one of which was heat-denatured while the other undenatured, and then were mixed with unstimulated cell lysates respectively and incubated at 36 °C for indicated time periods.
To understand the mechanism of actin in regulating the dephosphorylation of p-STAT1, we heat-denatured the IFN-γ-stimulated cell lysate and mixed it with an unstimulated cell lysate in the presence of Triton X-100. We observed that although p-STAT1 in the undenatured cell lysate could not be dephosphorylated, in the presence of Triton X-100, p-STAT1 in the heat-denatured cell lysate was dephosphorylated (Figure 8C), demonstrating that actin was not required for p-STAT1 dephosphorylation when it was denatured. The actin filaments might regulate the conformation of p-STAT1 to expose the phosphorylated tyrosine to be dephosphorylated by phosphatases, which could also be achieved by denaturation59.
Discussion
Cucurbitacin I was initially identified as a potent inhibitor of cancer cell growth48. Investigations into its molecular mechanisms of action have found that actin is one of its direct targets and that the JAK2/STAT3 signaling pathway is the primary signaling pathway that is influenced by cucurbitacin I48,50. However, the connections between the two activities and the mechanisms of regulation of JAK2/STAT3 signaling have not yet been elucidated. Here, we present evidence demonstrating that cucurbitacin I inhibits JAK2/STAT3 phosphorylation, as well as p-STAT1 dephosphorylation, by disrupting actin filaments. The actin filaments were found to interact with distinct JAK/STAT signaling complexes located in different regions of a cell, such as the membrane cytokine receptor complexes and the focal adhesion complex, to regulate their activities. We propose that actin filaments function not only in the structure support, mobility, and contraction of cells but also as a signaling organizer and mediator to coordinate cellular signaling. By disrupting actin, cucurbitacin I, as well as other actin disruptors, altered JAK/STAT signaling and possibly caused cell cycle arrest and cell death60,61.
STAT1 and STAT3 are known to play opposite roles in the regulation of cell growth; STAT3 transduces growth stimulatory signals and STAT1 generates growth inhibitory signals62. In this regard, it is interesting to note that disrupting actin filaments affected STAT1 and STAT3 phosphorylation in opposite ways, inhibiting the phosphorylation of STAT3 but the dephosphorylation of p-STAT1, and leading to decreased STAT3 activity but sustained STAT1 activity, which synergized with each other to inhibit cell growth and to induce cell death. These data again suggest that actin filaments function to coordinate different signals to control cell growth.
The primary sites for the cytokine-induced phosphorylation of STATs are in the cytokine receptor complexes at the plasma membrane. However, both STAT1 and STAT3 have also been found in focal adhesions63,64. Disrupting actin inhibited STAT3 phosphorylation in both the receptor signaling complexes and the focal adhesions, suggesting that actin is involved in regulating JAK2/STAT3 at both sites. The signals and mechanisms through which JAK2 and STAT3 are phosphorylated in the focal adhesions and the mechanism through which actin regulates the phosphorylation of focal adhesion-associated JAK2 and STAT3 remain unclear. However, there is some evidence suggesting that β1-integrin plays a role in activating focal adhesion-associated JAK265 as both JAK2 and STAT3 are physically associated with β1-integrin (unpublished data), and actin may participate by regulating the affinity of integrin66. Further studies are needed to clarify these hypotheses.
Disrupting the actin filaments affected only the dephosphorylation, and not the phosphorylation, of STAT1. Phosphorylated STAT1 is translocated into the nuclei, and the dephosphorylation of p-STAT1 is believed to occur in the nuclei67,68; the major phosphatases involved in p-STAT1 dephosphorylation are TCPTP and SHP255,56. Our data demonstrated that actin filaments are required for the dephosphorylation of p-STAT1 both in vivo and in vitro. However, actin filaments did not affect the enzymatic activity of TCPTP or SHP2. Instead, they appeared to affect the conformation of p-STAT1 to make the tyrosine phosphate accessible to the phosphatase as the heat-denatured p-STAT1 could be dephosphorylated in the absence of actin filaments. Therefore, actin in nuclei may regulate the dephosphorylation of p-STAT1 by changing its conformation. Disrupting actin may send a signal through STAT1 to stop cell cycle progression to avoid inappropriate cell division.
Cucurbitacin I is a potent inhibitor of cancer cell growth and has been considered as an anti-cancer drug candidate48. Our data and others suggest that actin filaments may be the major target of cucurbitacin I. It is not clear what other proteins cucurbitacin I may interact with in addition to actin, but disrupting actin filaments alone may cause severe adverse effects in cancer patients. Therefore, caution should be taken when considering cucurbitacin I as a disease-treating drug.
In summary, our study has revealed the molecular mechanisms of cucurbitacin I and has uncovered a novel role of actin in the regulation of STAT1 and STAT3 signaling. Actin is essential for the phosphorylation of STAT3 and the dephosphorylation of p-STAT1 and may regulate the two signaling pathways synergistically to control cell survival and growth. Our study also suggests the existence of different JAK/STAT signaling complexes in different subcellular regions of a cell. Actin filaments may play a key role in coordinating the different signaling complexes to direct various cellular activities. These findings shed new light on understanding JAK/STAT signaling and regulation, as well as the function and mechanisms of actin.
Author contribution
Hui GUO conducted all experiments, analyzed the results, and wrote most of the paper. Shan KUANG screened cucurbitacin I. Qiao-ling SONG, Man LIU and Xiao-xiao SUN provided advice regarding the experiments. Qiang YU conceived the idea for the project and wrote the paper with Hui GUO.
Acknowledgments
This work was supported by the China Ministry of Science and Technology Key New Drug Creation and Manufacturing Program (No 2014ZX9102001002 to Qiang YU), the China National Key Basic Research Program (No 2013CB910900 to Qiang YU), and the National Natural Science Foundation of China (No 81373447 and 81673465 to Qiang YU).
References
- Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, et al. Chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell 2010; 18: 556–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 2012; 36: 542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AC, Touw I, Yoshimura A. The Jak-Stat pathway in normal and perturbed hematopoiesis. Blood 2000; 95: 19–29. [PubMed] [Google Scholar]
- Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene 2013; 32: 2601–13. [DOI] [PubMed] [Google Scholar]
- Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer 2015; 113: 365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005; 434: 1144–8. [DOI] [PubMed] [Google Scholar]
- Lacronique V, Boureux A, Monni R, Dumon S, Mauchauffe M, Mayeux P, et al. Transforming properties of chimeric TEL-JAK proteins in Ba/F3 cells. Blood 2000; 95: 2076–83. [PubMed] [Google Scholar]
- Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med 2012; 366: 1905–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck AR, Witkiewicz AK, Liu C, Stringer GA, Klimowicz AC, Pequignot E, et al. Loss of nuclear localized and tyrosine phosphorylated Stat5 in breast cancer predicts poor clinical outcome and increased risk of antiestrogen therapy failure. J Clin Oncol 2011; 29: 2448–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macha MA, Matta A, Kaur J, Chauhan SS, Thakar A, Shukla NK, et al. Prognostic significance of nuclear pSTAT3 in oral cancer. Head Neck 2011; 33: 482–9. [DOI] [PubMed] [Google Scholar]
- Sonnenblick A, Uziely B, Nechushtan H, Kadouri L, Galun E, Axelrod JH, et al. Tumor STAT3 tyrosine phosphorylation status, as a predictor of benefit from adjuvant chemotherapy for breast cancer. Breast Cancer Res Treat 2013; 138: 407–13. [DOI] [PubMed] [Google Scholar]
- Mirtti T, Leiby BE, Abdulghani J, Aaltonen E, Pavela M, Mamtani A, et al. Nuclear Stat5a/b predicts early recurrence and prostate cancer-specific death in patients treated by radical prostatectomy. Hum Pathol 2013; 44: 310–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Lu S. A meta-analysis of STAT3 and phospho-STAT3 expression and survival of patients with non-small-cell lung cancer. Eur J Surg Oncol 2014; 40: 311–7. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell 1999; 98: 295–303. [DOI] [PubMed] [Google Scholar]
- Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer 2004; 4: 97–105. [DOI] [PubMed] [Google Scholar]
- Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene 2002; 21: 2000–8. [DOI] [PubMed] [Google Scholar]
- Cheng GZ, Zhang WZ, Sun M, Wang Q, Coppola D, Mansour M, et al. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J Biol Chem 2008; 283: 14665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene 2004; 23: 3550–60. [DOI] [PubMed] [Google Scholar]
- Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 2014; 14: 736–46. [DOI] [PubMed] [Google Scholar]
- Guschin D, Rogers N, Briscoe J, Witthuhn B, Watling D, Horn F, et al. A major role for the protein tyrosine kinase JAK1 in the JAK/STAT signal transduction pathway in response to interleukin-6. EMBO J 1995; 14: 1421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narazaki M, Witthuhn BA, Yoshida K, Silvennoinen O, Yasukawa K, Ihle JN, et al. Activation of JAK2 kinase mediated by the interleukin 6 signal transducer gp130. Proc Natl Acad Sci U S A 1994; 91: 2285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorff TB, Goldman B, Pinski JK, Mack PC, Lara PN Jr, Van Veldhuizen PJ Jr, et al. Clinical and correlative results of SWOG S0354: a phase II trial of CNTO328 (siltuximab), a monoclonal antibody against interleukin-6, in chemotherapy-pretreated patients with castration-resistant prostate cancer. Clin Cancer Res 2010; 16: 3028–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JF, Williams V, Wang S, Tremblay ML, Muller WJ, Durbin JE, et al. Stat1 is a suppressor of ErbB2/Neu-mediated cellular transformation and mouse mammary gland tumor formation. Cell Cycle 2011; 10: 794–804. [DOI] [PubMed] [Google Scholar]
- Dimco G, Knight RA, Latchman DS, Stephanou A. STAT1 interacts directly with cyclin D1/Cdk4 and mediates cell cycle arrest. Cell Cycle 2010; 9: 4638–49. [DOI] [PubMed] [Google Scholar]
- Penafuerte C, Bautista-Lopez N, Bouchentouf M, Birman E, Forner K, Galipeau J. Novel TGF-beta antagonist inhibits tumor growth and angiogenesis by inducing IL-2 receptor-driven STAT1 activation. J Immunol 2011; 186: 6933–44. [DOI] [PubMed] [Google Scholar]
- Kachroo P, Lee MH, Zhang L, Baratelli F, Lee G, Srivastava MK, et al. IL-27 inhibits epithelial-mesenchymal transition and angiogenic factor production in a STAT1-dominant pathway in human non-small cell lung cancer. J Exp Clin Cancer Res 2013; 32: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, Yun H, Lai R, Su M. Correlation of STAT1 with apoptosis and cell-cycle markers in esophageal squamous cell carcinoma. PLoS One 2014; 9: e113928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin CM, Hamon Y, Gonnord P, Boularan C, Kagan J, Viaris de Lesegno C, et al. Glycosylation-dependent IFN-gamma partitioning in lipid and actin nanodomains is critical for JAK activation. Cell 2016; 166: 920–34. [DOI] [PubMed] [Google Scholar]
- Koromilas AE, Sexl V. The tumor suppressor function of STAT1 in breast cancer. JAKSTAT 2013; 2: e23353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A 1998; 95: 7556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JA, Al-Attar A, Watson NF, Scholefield JH, Ilyas M, Durrant LG. Intratumoral T cell infiltration, MHC class I and STAT1 as biomarkers of good prognosis in colorectal cancer. Gut 2010; 59: 926–33. [DOI] [PubMed] [Google Scholar]
- Chen G, Wang H, Xie S, Ma J, Wang G. STAT1 negatively regulates hepatocellular carcinoma cell proliferation. Oncol Rep 2013; 29: 2303–10. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Molavi O, Su M, Lai R. The clinical and biological significance of STAT1 in esophageal squamous cell carcinoma. BMC Cancer 2014; 14: 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yang S, Sun N, Chen J. Differential expression of STAT1 and p21 proteins predicts pancreatic cancer progression and prognosis. Pancreas 2014; 43: 619–23. [DOI] [PubMed] [Google Scholar]
- Osborn JL, Greer SF. Metastatic melanoma cells evade immune detection by silencing STAT1. Int J Mol Sci 2015; 16: 4343–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmer FD, Friedrich K. Protein tyrosine phosphatases as wardens of STAT signaling. JAKSTAT 2014; 3: e28087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Lappalainen P. Actin stress fibers--assembly, dynamics and biological roles. J Cell Sci 2012; 125: 1855–64. [DOI] [PubMed] [Google Scholar]
- Naumanen P, Lappalainen P, Hotulainen P. Mechanisms of actin stress fibre assembly. J Microsc 2008; 231: 446–54. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Choi CK, Horwitz AR. Integrins in cell migration–the actin connection. J Cell Sci 2009; 122: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salbreux G, Charras G, Paluch E. Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol 2012; 22: 536–45. [DOI] [PubMed] [Google Scholar]
- Tang J, Gross DJ. Regulated EGF receptor binding to F-actin modulates receptor phosphorylation. Biochem Biophys Res Commun 2003; 312: 930–6. [DOI] [PubMed] [Google Scholar]
- Samarakoon R, Higgins PJ. MEK/ERK pathway mediates cell-shape-dependent plasminogen activator inhibitor type 1 gene expression upon drug-induced disruption of the microfilament and microtubule networks. J Cell Sci 2002; 115: 3093–103. [DOI] [PubMed] [Google Scholar]
- Are AF, Galkin VE, Pospelova TV, Pinaev GP. The p65/RelA subunit of NF-kappaB interacts with actin-containing structures. Exp Cell Res 2000; 256: 533–44. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Batista FD, Treanor B. Dynamics of the actin cytoskeleton mediates receptor cross talk: an emerging concept in tuning receptor signaling. J Cell Biol 2016; 212: 267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudnakova T, Spraggon L, Slight J, Hastie N. Actin: a novel interaction partner of WT1 influencing its cell dynamic properties. Oncogene 2010; 29: 1085–92. [DOI] [PubMed] [Google Scholar]
- Treanor B, Depoil D, Gonzalez-Granja A, Barral P, Weber M, Dushek O, et al. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity 2010; 32: 187–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillet AH, Lavergne V, Pasquier V, Gesbert F, Theze J, Rose T. IL-2 induces conformational changes in its preassembled receptor core, which then migrates in lipid raft and binds to the cytoskeleton meshwork. J Mol Biol 2010; 403: 671–92. [DOI] [PubMed] [Google Scholar]
- Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res 2003; 63: 1270–9. [PubMed] [Google Scholar]
- Zhang Y, Ouyang D, Xu L, Ji Y, Zha Q, Cai J, et al. Cucurbitacin B induces rapid depletion of the G-actin pool through reactive oxygen species-dependent actin aggregation in melanoma cells. Acta Biochim Biophys Sin (Shanghai) 2011; 43: 556–67. [DOI] [PubMed] [Google Scholar]
- Sorensen PM, Iacob RE, Fritzsche M, Engen JR, Brieher WM, Charras G, et al. The natural product cucurbitacin E inhibits depolymerization of actin filaments. ACS Chem Biol 2012; 7: 1502–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima S, Matsuda H, Kurume A, Oda Y, Nakamura S, Yamashita M, et al. Cucurbitacin E as a new inhibitor of cofilin phosphorylation in human leukemia U937 cells. Bioorg Med Chem Lett 2010; 20: 2994–7. [DOI] [PubMed] [Google Scholar]
- Sun X, Ai M, Wang Y, Shen S, Gu Y, Jin Y, et al. Selective induction of tumor cell apoptosis by a novel P450-mediated reactive oxygen species (ROS) inducer methyl 3-(4-nitrophenyl) propiolate. J Biol Chem 2013; 288: 8826–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Elson-Schwab I, Khurana V, Steinhilb ML, Spires TL, Hyman BT, et al. Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat Cell Biol 2007; 9: 139–48. [DOI] [PubMed] [Google Scholar]
- Mitzel DN, Jaramillo RJ, Stout-Delgado H, Senft AP, Harrod KS. Human metapneumovirus inhibits the IL-6-induced JAK/STAT3 signalling cascade in airway epithelium. J Gen Virol 2014; 95: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TR, Hong YK, Wang XD, Ling MY, Dragoi AM, Chung AS, et al. SHP-2 is a dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J Biol Chem 2002; 277: 47572–80. [DOI] [PubMed] [Google Scholar]
- ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, Zhu W, Tremblay M, David M, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol 2002; 22: 5662–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujfalusi-Pozsonyi K, Hild G, Grof P, Gutay-Toth Z, Bacso Z, Nyitrai M. The effects of detergents on the polymerization properties of actin. Cytometry A 2010; 77: 447–56. [DOI] [PubMed] [Google Scholar]
- Hitchcock SE, Carisson L, Lindberg U. Depolymerization of F-actin by deoxyribonuclease I. Cell 1976; 7: 531–42. [DOI] [PubMed] [Google Scholar]
- Mertens C, Zhong M, Krishnaraj R, Zou W, Chen X, Darnell JE Jr. Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev 2006; 20: 3372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar DR, Jane EP, Pollack IF. Cucurbitacin-I inhibits Aurora kinase A, Aurora kinase B and survivin, induces defects in cell cycle progression and promotes ABT-737-induced cell death in a caspase-independent manner in malignant human glioma cells. Cancer Biol Ther 2015; 16: 233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZG, Liu N, Song HS, Li JQ, Jiang J, Zhu JY, et al. Cytochalasin B inhibits the proliferation of human glioma U251 cells through cell cycle arrest and apoptosis. Genet Mol Res 2014; 13: 10811–22. [DOI] [PubMed] [Google Scholar]
- Avalle L, Pensa S, Regis G, Novelli F, Poli V. STAT1 and STAT3 in tumorigenesis: A matter of balance. JAKSTAT 2012; 1: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res 2004; 64: 3550–8. [DOI] [PubMed] [Google Scholar]
- Xie B, Zhao J, Kitagawa M, Durbin J, Madri JA, Guan JL, et al. Focal adhesion kinase activates Stat1 in integrin-mediated cell migration and adhesion. J Biol Chem 2001; 276: 19512–23. [DOI] [PubMed] [Google Scholar]
- Balanis N, Wendt MK, Schiemann BJ, Wang Z, Schiemann WP, Carlin CR. Epithelial to mesenchymal transition promotes breast cancer progression via a fibronectin-dependent STAT3 signaling pathway. J Biol Chem 2013; 288: 17954–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JS, Zigmond S, Vilaire G, Cunningham ME, Bednar B. The platelet cytoskeleton regulates the affinity of the integrin alpha(IIb)beta(3) for fibrinogen. J Biol Chem 1999; 274: 25301–7. [DOI] [PubMed] [Google Scholar]
- Meyer T, Marg A, Lemke P, Wiesner B, Vinkemeier U. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev 2003; 17: 1992–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol 2006; 6: 602–12. [DOI] [PubMed] [Google Scholar]