Abstract
Lung cancer is the most commonly occurring type of cancer worldwide and also has the highest mortality rate. Although targeted therapy of non-small cell lung carcinoma (NSCLC) has become common, the majority of patients receiving first-line epithelial growth factor receptor (EGFR)-TKI treatment develop drug resistance. The EGFR T790M (NM_005228.4(EGFR):c.2369C>T (p.Thr790Met)) mutation accounts for half of all reported resistance cases; however, the molecular mechanism resulting in the drug resistance remains to be characterized. Circulating tumor DNA (ctDNA) isolated from plasma has great potential for identification of gene mutations in NSCLC. Collection of ctDNA is relatively non-invasive and can avoid the inherent disadvantages of tissue biopsy. In the present study, next-generation sequencing technology was used to detect the variation of ctDNA in the peripheral blood of patients administered with EGFR-TKI. The patients were monitored serially to establish a dynamic resistance gene detection system, with the rationale being to alter the treatment strategy as soon as the emergence of drug resistance gene mutations. A mutation spectrum of the group of patients was constructed. A driver gene mutation was identified in the ctDNA of each patient, and certain patients had clinically targetable gene mutations like EGFR, ROS proto-oncogene receptor tyrosine kinase and B-Raf proto-oncogene serine/threonine kinase. The dynamic monitoring of EGFR status indicated that the EGFR mutation rate was consistent with the tumor burden of patients. Overall, ctDNA detection is a useful method for the molecular genotyping of patients with cancer. The dynamic resistance gene detection system described in the present study is a sensitive and useful tool for the monitoring of gene status, which has potential to be used for direction of treatment strategy by detecting the emergence of drug resistance gene mutations.
Keywords: circulating tumor DNA, lung cancer, epithelial growth factor receptor T790M mutation, drug resistance
Introduction
Lung cancer is the most commonly occurring type of cancer and the leading cause of cancer-associated mortality in adults worldwide (1). Malignant lung tumors can be subdivided into small-cell lung carcinoma (SCLC) and non-small cell lung carcinoma (NSCLC). NSCLC can be further subdivided into adenocarcinoma, squamous cell carcinoma (SCC) and large cell carcinoma using conventional histopathology. Progress in the effectiveness of conventional treatment approaches to NSCLC, including chemotherapy, radiotherapy and surgery, has plateaued. The development of genetic testing technology and genotyping has provided great insight into the molecular pathology of NSCLC. Known driver mutations are identifiable in ~60% lung adenocarcinoma and ~80% of SCC (2,3). Such mutations can trigger activating cascades in signaling pathways, including the RAS-RAF-MEK-extracellular signal-regulated kinase/mitogen activated protein kinase, phosphoinositide 3-kinase (PI3K)-AKT serine/threonine kinase-mechanistic target of rapamycin, or janus kinase-signal transducer and activator of transcription 1 pathways (4–6), leading to growth, proliferation, survival, and metastasis of cancer cells. Successful targeted therapies involve the identification and inhibition of these upregulated pathways by small molecule inhibitors or receptor monoclonal antibodies (mAb). Therapeutic targeting of the interaction between epidermal growth factor receptor (EGFR) and members of its downstream pathway has been most extensively studied. The two most common EGFR-activating mutations are an exon 19 deletion and exon 21 L858R mutation, which result in the constitutive activation of the receptor (7). It has been established that EGFR activating mutations are more common in patients with specific clinicopathological characteristics: Females, non-smokers, Asiatic origin and adenocarcinoma histological subtype. Mutant EGFR can be inhibited by small molecule tyrosine kinase inhibitors (TKIs), including gefitinib, erlotinib and afatinib or mAbs, including cetuximab and panitumumab. Multiple studies have demonstrated that these drugs can significantly increase therapy response rate and improve the progression free survival (PFS), and overall survival (OS) times in patients with NSCLC (8–13). The Food and Drug Administration (FDA) have approved gefitinib, erlotinib and afatinib as the first-line drugs for patients exhibiting EGFR, and the toxicity profiles of these drugs are superior to those of standard chemotherapy drugs. However, all patients with EGFR-mutation-derived NSCLC ultimately develop resistance to TKI treatment. Almost half of the reported cases of EGFR-TKI-resistance in NSCLC are due to the EGFR T790M mutation (14–16). Other mechanisms of EGFR-TKI-resistance include amplification of MET signaling (17,18), amplification of Erb-B2 receptor tyrosine kinase 2 signaling (19), mutation of PI3K catalytic subunit a (20), mutation of B-Raf proto-oncogene serine/threonine kinase (BRAF) (21) and NSCLC transformation to small-cell lung cancer (which accounts for 3–15% of all NSCLC cases and is a rare resistance mechanism which is poorly understood (22), and epithelial-mesenchymal transition (23). TKI, AZD9291, has received FDA approval and National Comprehensive Cancer Network guideline recommendations for EGFR T790M-positive patients with non-small cell lung cancer (24). However, neoplastic tissue samples are currently required to screen patients with the EGFR T790M mutation and to identify other molecular features of patients with drug resistance. Furthermore, re-biopsies following cancer progression are problematic due to factors, including the high risk of surgery, the vulnerable health of the patients and tumor heterogeneity.
Dead cells shed DNA into the bloodstream and when this DNA is selectively isolated from the plasma, it is termed cell-free DNA (cfDNA). In patients with cancerous tumors, a fraction of the cfDNA is derived from tumor cells and is referred as circulating tumor DNA (ctDNA). The presence of circulating nucleic acids in peripheral circulation was discovered >60 years ago by Mandel and Metais (25). Leon et al (26) later described the importance of circulating tumor DNA in cancer development. The collection of ctDNA is more feasible than the collection of whole tissue via biopsy, and it is sufficient for a general screening test to characterize the genetic profile of patients with cancer. Therefore, ctDNA has the potential to promote personalized cancer therapy. In addition, particularly through real-time tumor monitoring (27), unlike localized tumor tissues, ctDNA reflects all molecular alterations of primary tumors and metastases, potentially, inclusive of all heterogeneous mutation profiles (28). ctDNA can therefore be used for the early diagnosis of cancer (29,30), real-time assessment of risk of tumor progression (31), dynamic monitoring of patient's responses to treatment (32) and drug resistance testing (33).
In the present study, a targeted sequencing approach based on the Illumina platform was employed to detect and compare NSCLC driver gene alterations in plasma samples from 32 Chinese patients with advanced NSCLC. The purpose of this project was to elucidate the association between TKI resistance and gene mutation status, and to build a dynamic mutation detection system. A particular aim was to detect the EGFR T790M mutation, which predicts EGFR-TKI drug resistance through dynamic monitoring of ctDNA. Using continuous detection methods such as that described in the present study may be useful in the clinic to anticipate required changes in treatment strategy by detecting the emergence of drug resistant gene mutations. This has the potential to improve the prognosis, quality of life and survival time of patients with NSCLC, as well as to be cost-effective in terms of healthcare.
Materials and methods
Patients and ethical statements
The present study was approved by Shenzhen People's Hospital Ethics Committee (Shenzhen, China) and the methods were performed in accordance with their guidelines. Written informed consent was obtained from all patients for the use of blood and lung tumor tissue under the approval of the Ethics Committee. All patient samples and medical data used in this study have been irreversibly anonymized.
Patients with advanced or recurrent NSCLC derived from EGFR mutations with a treatment plan to begin TKI treatment were enrolled consecutively. The exclusion criteria were as follows: i) Pregnant or lactating women; ii) patients who have received prior treatment with an MEK inhibitor; and iii) patients with a history of clinically significant interstitial lung disease or pneumonitis.
A cohort of 32 patients with advanced NSCLC treated with TKI drugs between November 2015 and December 2016 in Shenzhen People's Hospital were enrolled into the present study. The clinical characteristics of the patients are presented in Table I. Participants in the cohort, including 18 females and 14 males, had been diagnosed with stage IIIa to IV NSCLC. Patient blood samples were taken during TKI treatment. EGFR TKI was treated until disease progression, and blood samples were taken every 3 months. Treatment effectiveness was assessed every 2 months according to the RECIST 1.1 criteria (34). Testing for EGFR mutations in blood plasma and imaging studies (including chest computational tomography, brain magnetic resonance imaging, bone scans and abdominal ultrasound) were performed by two independent research teams who were blinded from each other's results until survival outcomes were analyzed. Survival status was confirmed by telephone follow-up every 3 months. The blood samples of patient H01 were taken on the 24th December 2015 and the 7th of April 2016. The blood samples of patient H02 were taken on the 31st of December 2015 and the 11th of April 2016.
Table I.
Clinical characteristics of 32 patients with non-small cell lung cancer.
| Characteristic | No. of patients (%) |
|---|---|
| Age (years) | |
| Mean (standard deviation) | 59 (11.60) |
| Median (range) | 62 (36–85) |
| Sex | |
| Male | 18 (56.20) |
| Female | 14 (43.80) |
| Pathological diagnosis | |
| Non-small cell lung cancer | 32 (100.0) |
| Adenocarcinoma | 14 (43.80) |
| Unknown | 18 (56.20) |
| Tumor stage | |
| IIIA | 5 (15.60) |
| IIIB | 3 (9.40) |
| IV | 24 (75.0) |
| Smoking history | |
| Smoker | 10 (31.20) |
| Non-smoker | 22 (68.80) |
| Treatment history | |
| No previous treatment | 2 (6.25) |
| Previous treatment | 30 (93.75) |
| Chemotherapy history | |
| Chemotherapy undertaken | 25 (78.1) |
| No chemotherapy undertaken | 7 (21.9) |
Plasma isolation and DNA extraction
Patient blood samples were collected in tubes containing EDTA and centrifuged at 1,600 × g for 10 min at 4°C within 2 h of collection. The peripheral blood lymphocyte (PBL) debris was stored at −20°C until further use. The supernatants were further centrifuged at 10,000 × g for 10 min at 4°C, and plasma was harvested and stored at −80°C until further use. DNA from PBLs was extracted using the RelaxGene Blood DNA system (Tiangen Biotech Co., Ltd., Beijing, China) and ctDNA was extracted from at least 2 ml plasma using the QIAamp Circulating Nucleic Acid kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturers' instructions. DNA was quantified with the Qubit 2.0 Fluorometer and the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to manufacturer's instructions.
Next-generation sequencing (NGS) library construction
DNA collected from PBLs was sheared using enzyme dsDNA Fragmentase (New England BioLabs, Inc., Ipswich, MA, USA) according to the manufacturer's instructions. Size selection for 150–250 bp fragments was performed with AMPure XP beads (Beckman Coulter, Inc., Brea, CA, USA), which has a high recovery efficiency (the amount of DNA harvested from the size selection procedure is relatively large). DNA fragments and ctDNA were used to construct a library using the KAPA Library Preparation kit (Kapa Biosystems, Inc., Wilmington, MA, USA) according to the manufacturer's instructions. Agencourt AMPure XP beads were used for the purification of the DNA/ctDNA. Generally, following fragmentation, the End Repair and 3′-end dA-tailing ensued. T-tailed adapters were used and a 3′dA overhang was added enzymatically onto the fragmented DNA sample, using the KAPA Library Preparation kit (Kapa Biosystems, Inc.). Ligation was performed for 15 min at 20°C. Single-step size selection was performed by adding 50 µl (1X) PEG/NaCl SPRI solution buffer (Beckman Coulter, Inc.) to enrich for ligated DNA fragments. The ligated fragments were then amplified using 1X KAPA HiFi Hot Start Ready mix (Kapa Biosystems, Inc.) and Pre-LM-PCR Oligos (Kapa Biosystems, Inc.) in 50µl reactions and 7–8 PCR cycles, depending on the input DNA mass. The thermocycling conditions were as follows: Initial Denaturation, 98°C for 30 sec; 7–8 cycles at 98°C for 10 sec; 60°C for 10 sec; 68°C for 30 sec, and the final extension at 68°C for 60 sec. Library purity and concentration was assessed by Qubit 2.0 Fluorometer using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Fragment length was determined on a 4200 Bioanalyzer using the DNA 1000 kit (both from Agilent Technologies, Inc., Santa Clara, CA, USA).
Library design for hybrid selection
Hybrid selection was performed with custom biotinylated DNA probes for 104 genes associated with lung cancer (Roche Diagnostics, Basel, Switzerland). The library of amplified samples was hybridized with the SeqCap EZ Library (Roche Diagnostics) according to the manufacturer's instructions for 16–20 h at 47°C. Following hybrid selection, the captured DNA fragments were amplified with 12–14 cycles of PCR using 1X KAPA HiFi Hot Start Ready mix and Post-LM-PCR Oligos in two separate 50 µl reactions. The thermocycling conditions were as follows: Initial denaturation, 98°C for 30 sec; 7–8 cycles at 98°C for 10 sec; 60°C for 10 sec; 68°C for 30 sec, and the final extension at 68°C for 60 sec. The reactions were then pooled and purified using Agencourt AMPure XP beads.
Denaturing and diluting libraries for sequencing
Multiplexed libraries were denatured by 0.2M NaOH and diluted by Tris-HCl according to the manufacturer's instructions (Illumina, Inc., San Diego, CA, USA). The libraries were sequenced using 150-bp paired-end runs using a NextSeq 500 system (Illumina, Inc.). All genes captured by NGS are listed in Table II.
Table II.
The gene list for sequencing in our NGS method.
Variant calling and analysis
Sequencing data were demultiplexed and aligned to the hg19 genome (GRch37) using Burrows-Wheeler Aligner (35) version 0.7.15-r1140. Pileup files for properly paired reads with mapping quality >=60 were generated using Samtools (http://www.htslib.org/) (36). Somatic variants were created using VarScan2 (http://varscan.sourceforge.net/) (37) (-min-coverage=100; -min-var-freq=0.05; -somatic-P-value=0.01; -strand-filter 1; otherwise, default parameters were used). Allele Frequency (AFs) were calculated for all Q30 bases. Using a custom Python script, previously identified tumor DNA mutation were intersected with a SAMtools mpileup file generated for each plasma DNA sample, and the number and frequency of supporting reads were calculated for each mutation. For screening plasma DNA without knowledge of tumor DNA, a mutation was identified if >=4 mutant reads were revealed in plasma with >=1 read on each strand, and no mutant reads were observed in germline DNA or in a prior plasma sample with >=10-fold coverage.
Results
Patient characteristics
A total of 14/32 patients (43.8%) presented with adenocarcinoma, the remaining 18 patients (56.2%) were of unknown histopathology. The majority of patients were non-smokers (22/32, 68.8%), and the majority of patients were treated with EGFR-TKI (30/32, 93.75%).
The peripheral blood samples of these patients were collected during the outpatient evaluation follow-up. Secondary peripheral blood samples were collected from 4 patients, resulting in a total of 36 plasma samples from 32 patients for EGFR mutation testing.
Mutation spectrums of the 32 patients with non-small cell lung cancer
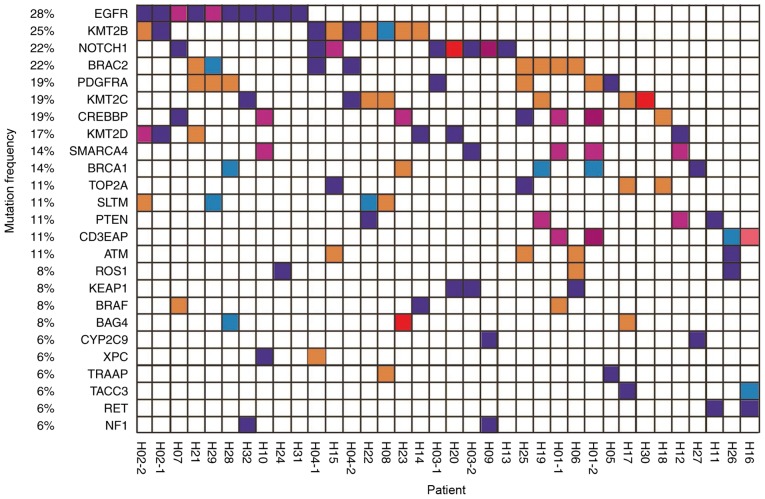
To elucidate the association between the effectiveness of EGFR-TKI treatment and gene status, NGS technology was used to detect variations in ctDNA in the peripheral blood of a cohort of 32 patients. Mutation spectrums were drawn for 36 samples from these patients (Fig. 1).
Figure 1.
Mutation spectrum of 36 plasma samples from 32 patients with non-small cell lung cancer. The genes with a high mutation frequency among all patients are listed on the left and individual patients are represented by the columns. First and second blood draws are indicated by −1 and −2, respectively. Mutation types of nonsynonymous single nucleotide variant, stop-gain, frameshift insertion, non-frameshift deletion, and non-frameshift insertion are represented by purple, blue, dark red, red, orange and pink respectively.
All patients exhibited ‘driver gene’ mutations in the ctDNA, and a number of patients exhibited actionable mutations in genes including EGFR, ROS proto-oncogene receptor tyrosine kinase and BRAF. Thus, it was demonstrated that the ctDNA tool is useful for the molecular genotyping of patients with cancer.
Dynamic detection of EGFR gene mutations in blood plasma
Patients were monitored for 24 months via consultation, with blood collected every 3 months, in order to create a dynamic resistance gene detection system. The somatic mutation profile of all 32 patients is presented in Fig. 1. Their blood samples were collected during TKI treatment and 4 underwent a second blood draw. The results demonstrate that EGFR mutation rate was associated with disease progression.
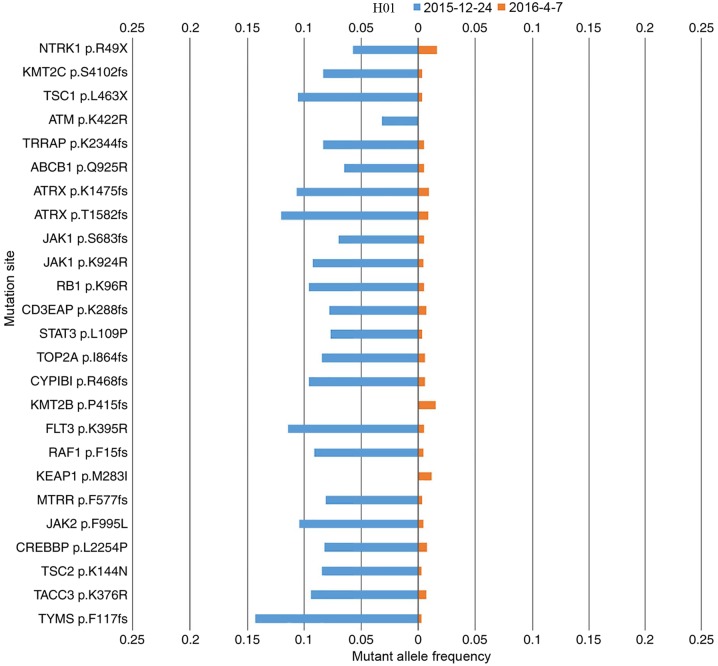
Patient H01 presented with adenocarcinoma and an EGFR exon 19 deletion was also detected by ARMS-PCR prior to the study. After 7 months of gefitinib treatment, the ctDNA of this patient was measured, followed by a second measurement 4 months later. The ctDNA mutation landscape for each measurement is compared in Fig. 2. Upon the second measurement, the EGFR exon 19 deletion was not identified, but the mutation rate had declined, which was consistent with the partial response (PR) clinical status of the patient following gefitinib treatment. The mutation variation between first and second measurements for patients H03, and H04 was similar to that of H01.
Figure 2.
The cell-free tumor DNA mutation landscape of patient H01 in two different measurements. The gene mutation rate of the two cell-free DNA samples from patient 1 extracted on 2015-12-24 and 2016-04-07 are compared in blue and orange, respectively.
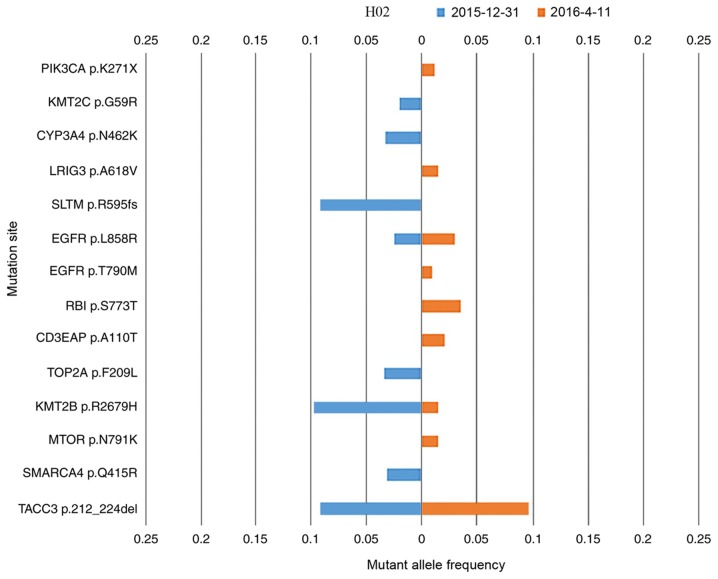
Patient H02 presented with adenocarcinoma; however, no EGFR mutation was detected by ARMS-PCR. This patient was treated with erlotinib blindly, which was initially effective. After 7 months of erlotinib treatment, the ctDNA of this patient was measured, followed by a second measurement 3 months later. The mutation landscapes for each measurement are compared in Fig. 3. The EGFR L858R (NM_005228.4(EGFR):c.2573T>G (p.Leu858Arg)) mutation was detected upon both measurements, and may explain the PR clinical status following initial treatment with erlotinib. Furthermore, the EGFR T790M [NM_005228.4(EGFR):c.2369C>T (p.Thr790Met)] mutation was detected in the second measurement. EGFR T790M is detected as a ‘second-site mutation’ in >50% EGFR-mutation-derived types of lung cancer that have developed resistance to erlotinib or gefitinib (38). Patient H02 demonstrated a case of EGFR p.T790M-induced drug resistance. Other novel driver gene mutations were detected in the second measurement, which may explain the progressive disease clinical status assigned at this stage. ARMS-PCR for two loci is sufficient to detect the presence or absence of specific mutations; however, NGS is able to identify high numbers of simultaneous mutations (including common actionable mutations, EGFR p.L858R and EGFR p.T790M (Fig. 3) which ARMS-PCR cannot achieve (39).
Figure 3.
The cell-free tumor DNA mutation landscape of patient H02 in two different measurements. The gene mutation rate of the two cell-free DNA samples from patient H02 extracted at 2015-12-31 and 2016-04-11 are compared in blue and orange, respectively.
Discussion
In the present study, a targeted sequencing approach based on the Illumina platform was used to test variations in NSCLC driver genes, including point mutations, insertions, deletions and gene rearrangements simultaneously in plasma samples from 32 Chinese patients with advanced NSCLC. To elucidate the association between drug effect and mutation status, mutation profiles were constructed for 32 patients with NSCLC. Furthermore, the EGFR status was dynamically monitored in order to construct a dynamic resistance gene detection system.
As the results demonstrated, for patients with NSCLC, a large number of gene mutations may occur. Besides these common actionable mutations, including NM_005228.4(EGFR):c.2573T>G (p.Leu858Arg), EGFR Exon 19 deletions and NM_005228.4(EGFR):c.2369C>T (p.Thr790Met), others may also be used as biomarkers for prognosis. Single or small numbers of target mutations can be detected using conventional single locus testing methods such as ARMS-PCR; however, such techniques do not indicate other, potentially unexpected, mutations (40). On the other hand, NGS is able to detect numerous gene mutations simultaneously (40,41). The present study investigated >25 genes, constructed overall mutation landscapes for individual patients and performed blood collection rather than tissue biopsy. Another advantage of the present study is the use of continuous samples from the same individuals instead of isolated samples.
In recent years, ctDNA has been demonstrated to be an effective material for tumor DNA monitoring. In the present study, it was demonstrated that ctDNA may be used to measure tumor burden (number of cancer cells, the size of a tumor or the amount of cancer in the body). The concordance between ctDNA and tissue immunohistochemistry testing of mutation variation is ~50%; considering that the occurrence of novel mutations and the disappearance of existing mutations is constant due to evolving tumors, this is a relatively high similarity rate (42). This may be due to release of genomic DNA from necrotic white blood cells into blood, and diluted ctDNA in the plasma (43). The amount of ctDNA in patients is also likely to be associated with metastasis, vascularity, cellular turnover and response to therapy (44,45). In addition, ctDNA may be influenced by clearance, degradation and other physiological filtering events involving blood, and lymphatic circulation (43).
While the sample size of 36 is not large, all samples represented advanced NSCLC and were associated with complete clinical information. Therefore, the samples used in the present study were of high quality and consistency. The clinical consequence of EGFR-mutation-derived NSCLC demonstrated inter-patient variations following the initially rapid response to EGFR-TKIs. For example, certain patients experience a rapid disease flare just subsequent to the cessation of EGFR-TKI treatment, while others do not (46). Repeated treatment with an EGFR-TKI is effective in certain patients, while ineffective in others (47). These results indicate that the contribution of driver mutations vary between patients and change during the clinical courses. Therefore, the monitoring of EGFR gene mutation status is critical. Currently, re-biopsy is recommended to achieve such monitoring by tissue sample sequencing. However, tumor biopsy is usually invasive and difficult to repeat in solid tumors. Since genetic changes appear to accumulate over time in patients with NSCLC and different portions of a tumor carry different genetic profiles, single biopsy of a tumor is spatially and temporally limited, and may fail to monitor dynamics of cancer progression (48,49). The advantages of using ctDNA for dynamic genetic monitoring studies are demonstrated in this study, which suggests that this method should be applied more widely in clinical practice.
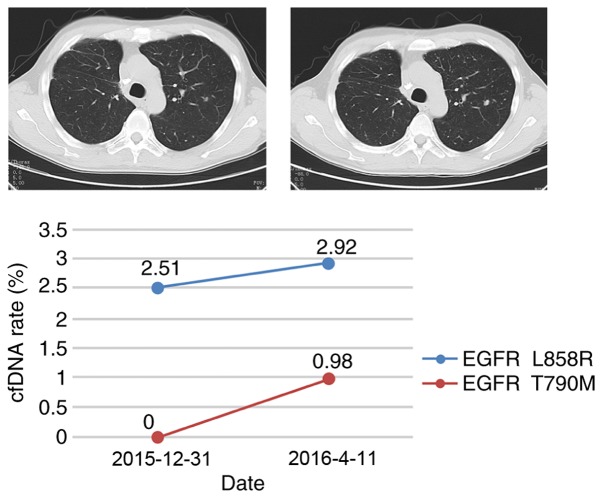
Figure 4.
The driver mutations, EGFR L858R and EGFR T790M, vary in patient H02 between two measurements taken on 2015-12-31 and 2016-04-11. The computed tomography scan images were taken on these dates. The scans demonstrate disease progression. The graph illustrates the circulating tumor DNA mutation rate of EGFR L858R and EGFR T790M, indicated by the blue and red lines, respectively. EGFR, endothelial growth factor receptor; cfDNA, cell-free DNA.
Acknowledgements
The authors would like to thank Liwei Deng and Cheng Jin for their input to diagram presentation. The present study was supported by the National Science Foundation of China (grant no. 61472411) for high performance servers and publication cost. The Technology Development and Creative Design Program of Nanshan Shenzhen (grant no. KC2015JSJS0028A) funded data collection, and the Special Funds for Future Industries of Shenzhen (grant no. JSGG20160229123927512) funded the sequencer and reagents.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Alamgeer M, Ganju V, Watkins DN. Novel therapeutic targets in non-small cell lung cancer. Curr Opin Pharmacol. 2013;13:394–401. doi: 10.1016/j.coph.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Savas P, Hughes B, Solomon B. Targeted therapy in lung cancer: IPASS and beyond, keeping abreast of the explosion of targeted therapies for lung cancer. J Thorac Dis. 2013;5(Suppl 5):S579–S592. doi: 10.3978/j.issn.2072-1439.2013.08.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Dearden S, Stevens J, Wu YL, Blowers D. Mutation incidence and coincidence in non small-cell lung cancer: Meta-analyses by ethnicity and histology (mutMap) Ann Oncol. 2013;24:2371–2376. doi: 10.1093/annonc/mdt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arteaga CL. The epidermal growth factor receptor: From mutant oncogene in nonhuman cancers to therapeutic target in human neoplasia. J Clin Oncol. 2001;19(Suppl 1):32S–40S. [PubMed] [Google Scholar]
- 7.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 8.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 9.Yang JC, Hirsh V, Schuler M, Yamamoto N, O'Byrne KJ, Mok TS, Zazulina V, Shahidi M, Lungershausen J, Massey D, et al. Symptom control and quality of life in LUX-Lung 3: A phase III study of afatinib or cisplatin/pemetrexed in patients with advanced lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3342–3350. doi: 10.1200/JCO.2012.46.1764. [DOI] [PubMed] [Google Scholar]
- 10.Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, Li W, Hou M, Shi JH, Lee KY, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213–222. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 11.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 12.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 13.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 15.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, Chiang A, Yang G, Ouerfelli O, Kris MG, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 17.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib; Proc Natl Acad Sci USA; 2007; pp. 20932–20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 19.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: Learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 20.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, Pan Y, Wang L, de Stanchina E, Shien K, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1; Proc Natl Acad Sci USA; 2012; pp. E2127–E2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacher AG, Jänne PA, Oxnard GR. Management of acquired resistance to epidermal growth factor receptor kinase inhibitors in patients with advanced non-small cell lung cancer. Cancer. 2014;120:2289–2298. doi: 10.1002/cncr.28723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zakowski MF, Ladanyi M, Kris MG, Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome Group EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355:213–215. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 24.Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel P, Metais P. Les acides nucléiques du plasma sanguine chez l'homme. CR Seances Soc Biol Fil. 1948;142:241–243. (In Undetermined Language) [PubMed] [Google Scholar]
- 26.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 27.Qiu M, Wang J, Xu Y, Ding X, Li M, Jiang F, Xu L, Yin R. Circulating tumor DNA Is effective for the detection of EGFR mutation in non-small cell lung cancer: A meta-analysis. Cancer Epidemiol Biomarkers Prev. 2015;24:206–212. doi: 10.1158/1055-9965.EPI-14-0895. [DOI] [PubMed] [Google Scholar]
- 28.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 29.Chen KZ, Lou F, Yang F, Zhang JB, Ye H, Chen W, Guan T, Zhao MY, Su XX, Shi R, et al. Circulating tumor DNA detection in early-stage non-small cell lung cancer patients by targeted sequencing. Sci Rep. 2016;6:31985. doi: 10.1038/srep31985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beaver JA, Jelovac D, Balukrishna S, Cochran R, Croessmann S, Zabransky DJ, Wong HY, Toro PV, Cidado J, Blair BG, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014;20:2643–2650. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng D, Ye X, Zhang MZ, Sun Y, Wang JY, Ni J, Zhang HP, Zhang L, Luo J, Zhang J, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913. doi: 10.1038/srep20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorensen BS, Wu L, Wei W, Tsai J, Weber B, Nexo E, Meldgaard P. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer. 2014;120:3896–3901. doi: 10.1002/cncr.28964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, Lai Z, Markovets A, Vivancos A, Kuang Y, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koboldt DC, Larson DE, Wilson RK. Using VarScan 2 for germline variant calling and somatic mutation detection. Curr Protoc Bioinformatics. 2013;44:15.4.1–17. doi: 10.1002/0471250953.bi1504s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, Hammett T, Cantarini M, Barrett JC. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of technologies to support the clinical development of AZD9291. J Clin Oncol. 2015;90:509–515. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Xu T, Kang X, You X, Dai L, Tian D, Yan W, Yang Y, Xiong H, Liang Z, Zhao GQ, et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor dna from non-small cell lung carcinoma patient plasma. Theranostics. 2017;7:1437–1446. doi: 10.7150/thno.16558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O'Connell A, Feeney N, Feeney N, Mach SL, Jänne PA, Oxnard GR. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2:1014–1022. doi: 10.1001/jamaoncol.2016.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jänne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, Ahn MJ, Kim SW, Su WC, Horn L, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 42.Fenizia F, De Luca A, Pasquale R, Sacco A, Forgione L, Lambiase M, Iannaccone A, Chicchinelli N, Franco R, Rossi A, et al. EGFR mutations in lung cancer: From tissue testing to liquid biopsy. Future Oncol. 2015;11:1611–1623. doi: 10.2217/fon.15.23. [DOI] [PubMed] [Google Scholar]
- 43.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 44.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohler C, Barekati Z, Radpour R, Zhong XY. Cell-free DNA in the circulation as a potential cancer biomarker. Anticancer Res. 2011;31:2623–2628. [PubMed] [Google Scholar]
- 46.Chaft JE, Oxnard GR, Sima CS, Kris MG, Miller VA, Riely GJ. Disease flare after tyrosine kinase inhibitor discontinuation in patients with EGFR-mutant lung cancer and acquired resistance to erlotinib or gefitinib: Implications for clinical trial design. Clin Cancer Res. 2011;17:6298–6303. doi: 10.1158/1078-0432.CCR-11-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishino K, Imamura F, Morita S, Mori M, Komuta K, Kijima T, Namba Y, Kumagai T, Yamamoto S, Tachibana I, et al. A retrospective analysis of 335 Japanese lung cancer patients who responded to initial gefitinib treatment. Lung Cancer. 2013;82:299–304. doi: 10.1016/j.lungcan.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 48.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]