Abstract
Colorectal cancer is one of the most common malignancies. Aberrant expressed microRNAs (miRNAs) have been demonstrated to have strong associations with colorectal cancer by repressing their targets. Therefore, miRNAs are thought to have significant promise in the diagnosis and prognosis of colorectal cancer. Previous studies indicated that miR-155 and collagen triple helix repeat containing 1 (CTHRC1) were both involved in pathogenesis of colorectal cancer, but the underlying mechanisms of miR-155 and CTHRC1 are still unknown. The present study aimed to investigate the biological functions of miR-155 and CTHRC1 in colorectal cancer. Reverse transcription-quantitative polymerase chain reaction was used to examine miR-155 and CTHRC1 expression levels. A dual-luciferase reporter assay was applied to verify the target interaction between miR-155 and CTHRC1. Proliferation, cell cycle, apoptosis, cell migration and invasion were measured using the MTT assay, flow cytometry and Transwell assays, respectively. Results showed that miR-155 expression was decreased, but CTHRC1 expression was increased in colorectal cancer tissue and cell lines. Furthermore, it was demonstrated that miR-155 negatively regulated CTHRC1. Additionally, miR-155 overexpression suppressed cell proliferation, induced cell cycle arrest and promoted cell apoptosis, while an inhibitor of miR-155 facilitated cell proliferation and cell cycle and repressed apoptosis. Transwell experiments indicated that miR-155 inhibited the cell migratory and invasive abilities of HT-29 cells, but miR-155 inhibitor enhanced these abilities of HT-29 cells. These results suggested that miR-155 prevented colorectal cancer progression and metastasis via silencing CTHRC1 in vitro, which provides evidence for miR-155 and CTHRC1 as a novel anti-onco molecular target for the treatment of colorectal cancer in the future.
Keywords: colorectal cancer, microRNA-155, collagen triple helix repeat containing 1
Introduction
Human colorectal cancer has been ranked the third most commonly diagnosed malignancy through the world and the leading cause of cancer-related mortality in Western countries, exceeded only by lung, live and stomach cancers (1). Over the past decades, despite the mortality of colorectal cancer has been obviously improved, due to the rapid development of new drugs (i.e., irinotecan and oxaliplatin) and target therapies (i.e., bevacizumab, cetuximab, panitumab, aflibercept and regorafenib), approximately one million new cases of colorectal cancer diagnosed worldwide and half a million people died from colorectal cancer annually, which implies significant impact on public health (2). Moreover, it is now clear that excessive alcohol use, obesity, older age, chronic intestinal inflammation, family history, racial and ethnic background, environmental and genetic factors have been identified as the most important risk factors (3,4). Among these, genetic factors, mainly referred to the accumulation of both gene mutations and epigenetic modifications of the genome in colonic mucosa cells, are considered as the key components which lead to cell proliferation and metastasis and ultimately potentiate carcinogenesis of colorectal cancer (5). For example, the changes of EGF receptor (EGFR) signaling, phosphatase and tensin homolog (PTEN)/phosphatidylinositol-3-kinase (P13K) signaling and p53 signaling were implicated in pathogenesis of colorectal cancer (6). Therefore, in-depth investigations on the underlying molecular mechanisms of colorectal cancer occurrence and development and the identification of serum and tissue markers with prognostic and predictive value involved in colorectal cancer are the urgent need for the management and treatment of this disease (7). Currently, accumulating studies have highlighted that microRNA (miRNA) is a promising prognostic biomarkers and a potential therapeutic target in colorectal cancer (8,9). For instance, miR-181c was determined as a predictive molecule for recurrence of colorectal cancer patients at stage II (10).
miRNA, a discovered group of endogenous, non-coding RNA molecules approximately 22 nt in length, directly interacts with a member of the Argonaute (Ago) protein family and forms effector complexes that modulate gene expression post-transcriptionally by binding to complementary sequences in the 3′-untranslated region (UTR) of target messenger RNAs (mRNAs) (11). Given the tremendous impact of miRNA-guided gene regulation on almost all aspects of cellular processes in eukaryotic organisms, such as proliferation, cell fate determination, apoptosis, signal transduction, organ development, hematopoietic lineage differentiation and tumorigenesis, it is not surprising that miRNA deregulation is intimately related to the molecular mechanisms of various clinical diseases, including cancer (12). In the past decades, it was found that miRNA abnormalities play a pivotal role in diverse cancer subtypes, such as lung cancer, breast cancer, and glioblastoma, and different cancers have different miRNA profiles, which could reflect the developmental lineage and differentiation state of the tumors, thereby studying the specific function of these aberrant miRNAs in human carcinogenesis might be provide a powerful tool as novel clinical biomarkers for early cancer diagnosis, prognosis and targets for therapy (13,14). Numerous literatures reported that many miRNAs involved in colorectal cancer initiation and emergence were detected, for example, reduced expression of miR-143 is responsible for colorectal cancer development through derepressing Kirsten rat sarcoma viral oncogene homolog (KRAS) expression and Insulin-like growth factor 1 receptor (IGF1R) (15,16); elevated miR-92a levels promoted cell proliferation, invasion, epithelial-mesenchymal transition (EMT) via targeting phosphatase and tensin homolog (PTEN) (17).
In previous study, it was revealed that miR-155 might be implicated in formation of colorectal cancer (18), and collagen triple helix repeat containing 1 (CTHRC1) was also predicted as a potential target of miR-155 by bioinformatics analysis. Since its discovery, miR-155 was uncovered that it participated in promoting cancers of lung, liver, pancreas and gastrointestinal tract by repressing different targets (19). Furthermore, it is verified that CTHRC1 could suppress tumor growth and metastasis in colorectal cancer (20). Thus, in this study, we aimed to confirm the interaction between miR-155 and CTHRC1 and explore their roles in colorectal cancer, which might not only help elucidate the molecular mechanism of miR-155 and CTHRC1 in colorectal cancer, but also offer potential utilization as innovative therapeutics in colorectal cancer.
Materials and methods
Ethics statement and clinical samples
The procedures of this study were approved by the Clinical Ethics Management Committee of Jinshan Hospital Affiliated to Fudan University. Written informed consent was obtained from 4 participants who have hospitalized in Jinshan Hospital Affiliated to Fudan University from March, 2016 to May, 2016 and have been made a definite diagnosis by at least three pathologists (21). The detailed patients' clinical characteristics were displayed in Table I. Fresh colorectal cancer tissues and paired adjacent normal specimens were collected by surgical resection. The samples were washed with an appropriate amount of cold saline to reduce blood contamination and then immediately removed and preserved in liquid nitrogen until they were processed for miR-155 and CTHRC1 expressions detection by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assay.
Table 1.
The basic clinical characteristics of study participants.
| ID | Gender | Age (Year) | Tumor location | Pathological type | Differentiated degree | TNM classification |
|---|---|---|---|---|---|---|
| 1 | Male | 42 | Ascending colon | Adenocarcinoma | Moderately differentiated | T3aN2M1 |
| 2 | Male | 46 | Sigmoid colon | Adenocarcinoma | Moderately differentiated | T3aN0M1 |
| 3 | Female | 39 | Transverse colon | Adenocarcinoma | Moderately differentiated | T3aN0M1 |
| 4 | Female | 47 | Descending colon | Adenocarcinoma | Moderately differentiated | T3aN2M1 |
Cell line culture and transfection
Human colon cancer cells-derived cell lines HCT-8, Lovo, Colo205, HCT-116 and HT-29 were purchased from American Type Culture Collection (ATCC) and cultured in RPMI-1640 cell culture medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 2 mM glutamine, 1 mM sodium pyruvate, 100 UI/ml penicillin and 0.1 mg/ml streptomycin at 37°C in a humidified 5% CO2/95% air atmosphere until cell lines reached 90% confluence. Then, the cells washed twice with phosphate-buffered saline (PBS) and continued to passage with 0.25% trypsin. Additionally, 293T cells similarly obtained from ATCC were cultivated in RPMI-1640 cell culture medium, containing 10% FBS, 1% non-essential amino acids and 1% penicillin/streptomycin under humidified conditions at 37°C with 5% CO2 for dual luciferase assay.
For transfection with plasmids, including miR-155 mimics plasmid, miR-155 inhibitor plasmid, negative control (NC) miRNA plasmid and NC miRNA inhibitor plasmid, cells were seeded in 6-well plates with antibiotic free growth medium at a density of 1×106 cells/well. Next day, when grown to 80% confluence, cells were transfected with the above plasmid using Lipofectamine 2000 (Promega Corporation, Madison, WI, USA) following the manufacturer's guidelines. After 6 h of transfection, the cell culture medium was replaced with fresh medium and the cells were harvested at 48 h for the following experiments. The transfection efficiency was evaluated by qRT-PCR and WB. All of these plasmids used in this study were designed and purchased from Guangzhou RiboBio Co., Ltd., Guangzhou, China.
RT-qpcr
Isolation of miRNAs from patient specimens and colorectal cancer cell lines was carried out using the mirVana™ miRNA Isolation kit (Thermo Fisher Scientific, Inc.) according to manufacturer's instructions. The quantity and quality of RNA were evaluated by a BioPhotometer and RNA integrity was determined by gel analysis. RNA was converted to complementary DNA (cDNA) with a miR-155-specific stem-loop primer using a PrimeScript RT Reagent Kit with cDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China) in accordance with the manufacturer's manual. Reverse transcription reaction was conducted at 16°C for 30 min and at 42°C for 40 min, followed by heat inactivation at 85°C for 5 min. Subsequently, the cDNA was in turn PCR amplified in a 96-well optical plate on an ABI PRISM® 7500 Sequence Detection System (Thermo Fisher Scientific, Inc.) with SYBR Premix Ex Taq TM II (Takara Biotechnology Co., Ltd.) at 95°C for 30 sec, followed by 40 cycles at 95°C for 15 sec and 60°C for 32 sec and dissociation at 95°C for 60 sec, 55°C for 30 sec and 95°C for 30 sec. The expression level of miR-155 and CTHRC1 was normalized to that of the housekeeping gene U6 snRNA and GAPDH, respectively. The relative fold change for miR-155 and CTHRC1 was calculated using the 2−ΔΔCt method. Sequences for miR-155, U6, CTHRC1 and GAPDH primers used in this study are following: miR-155 stem-loop primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCCCTA-3′, miR-155 forward primer, 5′-ACACTCCAGCTTAATGCTAATCGTGATAG-3′, miR-155 reverse primer, 5′-CTCAACTGGTGTCGTGGA-3′; U6 stem-loop primer, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAATATGG-3′, U6 forward primer, 5′-CTCGCTTCGGCAGCACA-3′, U6 reverse primer, 5′-AACGCTTCACGAATTTGCGT-3′; CTHRC1 forward primer, 5′-ACAATTAATATTCATCGCACT-3′, CTHRC1 reverse primer, 5′-ACAATTAATATTCATCGCACT-3′; GAPDH forward primer, 5′-CTGACTTCAACAGCGACACC-3′, GAPDH reverse primer, 5′-TCTGACTTCAACAGCGACACC-3′. The results were expressed as the mean of three individual experiments with duplicate samples.
Vector construction and dual-luciferase reporter assay
The wild-type and mutant 3′-UTR of CTHRC1 containing the seed sequence were synthesized and ordered from Sangon Biotech Co., Ltd., Shanghai, China. Then, these fragments were cloned into psi-CHECK2 Basic luciferase reporter plasmid (Promega Corporation) between the Xho I and Not I sites and the insertions were verified by sequencing. To identify the direct target relationship between miR-155 and the 3′-UTR of CTHRC1 mRNA, the 293T cells in 24-well plates were cotransfected with the above constructed plasmids concomitant with control psiCHECK-2 plasmid (that is Blank group), miR-155 mimics, miR-155 inhibitor, NC plasmid or NC inhibitor with the final dosage of 100 ng using Lipofectamine 2000 according to the manufacturer's recommendations. After 6 h, the media of the transfected cells were switched to fresh media. At last, the cells were split, and Firefly and Renilla relative luciferase activity was tested with the Dual-Luciferase Reporter Assay System (Promega Corporation) in accordance with the manufacturer's instructions at 48 h post transfection. The experiments were performed in triplicate.
MTT assay
Cell proliferation was determined 48 h after transfection treatment using a 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) kit (Promega Corporation) in a 96-well plate. The results were subsequently analyzed at 0, 1, 2 and 3 day. For the analysis, first, the cell culture medium was aspirated from the 96-well plate, and the cells were gently washed with PBS twice, followed by the addition of 20 µl of 5 mg/ml MTT reagent. After 4 h of incubation, MTT was carefully removed without disturbing the cells and 150 µl of dimethylsulfoxide (DMSO) solution was added to the wells, which was followed by continuous shaking at room temperature for 15 min to solubilise the purple formazan crystals. Finally, the optical density of each well, including the blank well without cells, was measured at a wavelength of 490 nm in a multimode plate reader (Bio-Rad, USA). Each sample was assayed in triplicate.
Flow cytometry
The flow cytometry was performed 48 h after HT-29 cells were transfected with three plasmids, namely NC plasmid, miR-155 mimics plasmid and miR-155 inhibitor plasmid, for apoptosis and cell cycle analysis. For apoptosis analysis, the harvested cells were washed by ice-cold PBS, centrifuged and labeled with FITC-Annexin V and propidium iodide (PI) in binding buffer for 15 min at room temperature in the dark according to manufacturer's guidelines. Fluorescence signals were examined within 1 h post-staining on a FACSCalibur flow cytometer. However, for cell cycle analysis, the harvested cells were washed by ice-cold PBS, centrifuged, and fixed in 70% ethanol at −20°C overnight. Then the cells were washed with PBS again and resuspended in 1 ml of PBS containing 50 mg/ml PI (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 25 mg/ml RNase A (Sigma-Aldrich; Merck KGaA) for 30 min at room temperature in the dark. A total number of 1×104 cells were subject to cell cycle analysis by flow cytometry using a FACSCalibur flow cytometer.
Transwell assay
HT-29 cells with different treatments were trypsinized to generate a single-cell suspension, and 1×105 cells/well (for the migration assay) or 2×105 cells/well (for the invasion assay) were plated into 24-transwell chamber with 8 µm pore size membranes (BD Biosciences, Franklin Lakes, NJ, USA) pre-coated with or without matrigel basement membrane matrix (BD Biosciences), respectively. Meanwhile, 600 µl of culture media with 20% FBS as a chemoattractant was added to the lower chamber. Approximately 24 h later, the cells on the apical side of each transwell membranes were gently removed with the mechanical scraping, while the cells that pass through the membrane were fixed with 4% paraformaldehyde at room temperature for 30 min, stained with 0.1% crystal violet for 10 min, washed with PBS three times and ultimately counted from 4~5 randomly microscopic filed. The migration and invasion assays were repeated at least three times.
Statistical analysis
Statistical analysis of the study data was performed using SPSS v18.0 software (IBM Corp., Armonk, NY, USA). All values are expressed as the mean ± standard deviation (SD) of three independent biological experiments. Independent sample t-test and one-way analysis of variance (ANOVA) were applied to comparisons between two or more than two groups, respectively. P<0.05 was considered to indicate a statistically significant difference..
Results
Different expression levels of miR-155 and CTHRC1 in patient specimens and multiple colorectal cancer cell lines
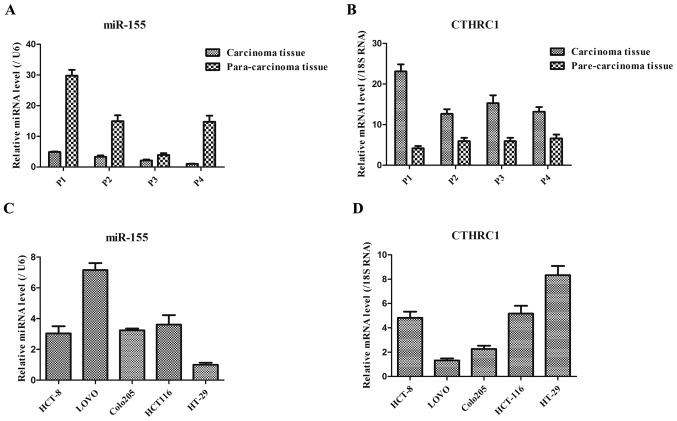
As shown in Fig. 1A, the expression level of miR-155 in carcinoma tissues of colorectal cancer patients was generally lower than that in the corresponding para-carcinoma tissues of these patients. However, the expression level of CTHRC1 in carcinoma tissues of colorectal cancer patients was generally higher than that in the corresponding para-carcinoma tissues of these patients (Fig. 1B). These results suggested that the down-regulated miR-155 and up-regulated CTHRC1 might be the key markers in colorectal cancer patients. Additionally, in five colorectal cancer cell lines, the expression levels of miR-155 in HCT8, Colo205 and HCT116 cells were basically consistent, while miR-155 expression presented lowest and highest in HT-29 and Lovo cells, respectively (Fig. 1C). Moreover, CTHRC1 expression exhibited the highest in HT-29 cells (Fig. 1D), thereby HT-29 cells were chosen as following experiment cells in this study.
Figure 1.
miR-155 expressions were assessed by RT-qPCR in (A) patient specimens and (C) multiple colorectal cancer cell lines (B). CTHRC1 expressions were assessed by RT-qPCR in (B) patient specimens and (D) multiple colorectal cancer cell lines. miR-155 expression, relative to U6 control, was determined using the 2−ΔΔCt method. P indicates patient.
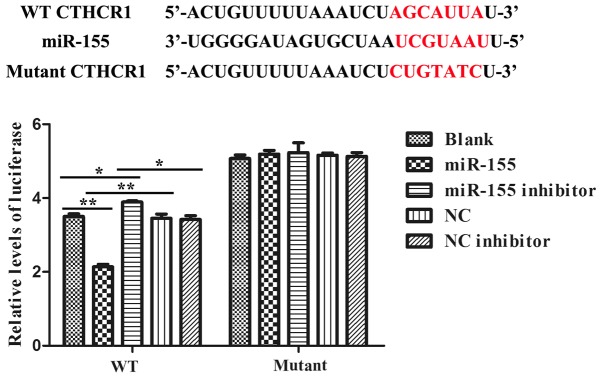
miR-155 directly targeted the 3′-UTR of CTHRC1
We constructed the dual-luciferase reporter system to determine the direct interaction between miR-155 and 3′-UTR of CTHRC1. Alignment between the predicted miR-155 target site and mutant site of the CTHRC1 3′-UTR region and miR-155 was displayed in upper part of Fig. 2. And the results of the dual-luciferase assay revealed that the activity of luciferase was remarkably decreased after co-transfection with WT constructive luciferase reporter plasmid harbouring the CTHRC1 3′UTR and miR-155 mimics plasmid. By contrast, the activity of luciferase was notably increased following co-transfection with WT constructive luciferase reporter plasmid and miR-155 inhibitor (Fig. 2). However, there were no changes in 293T cells co-transfected with the Mutant constructive luciferase reporter plasmid and other plasmid (P>0.05; Fig. 2). The results have firmly confirmed that miR-155 specifically binds to the 3′-UTR of CTHRC1.
Figure 2.
Target gene, CTHRC1, is negatively regulated by miR-155. The binding site scheme of miR-155 with CTHRC1 was determined and the Dual luciferase activity of groups was indicated. Co-transfection with either the luciferase reporter plasmid of CTHRC1-3′UTR WT or CTHRC1-3′UTR Mutant and miR-155 mimic or inhibitor, or respective NC or NC inhibitor was carried out as described in materials and methods. *P<0.05 and **P<0.01 as indicated.
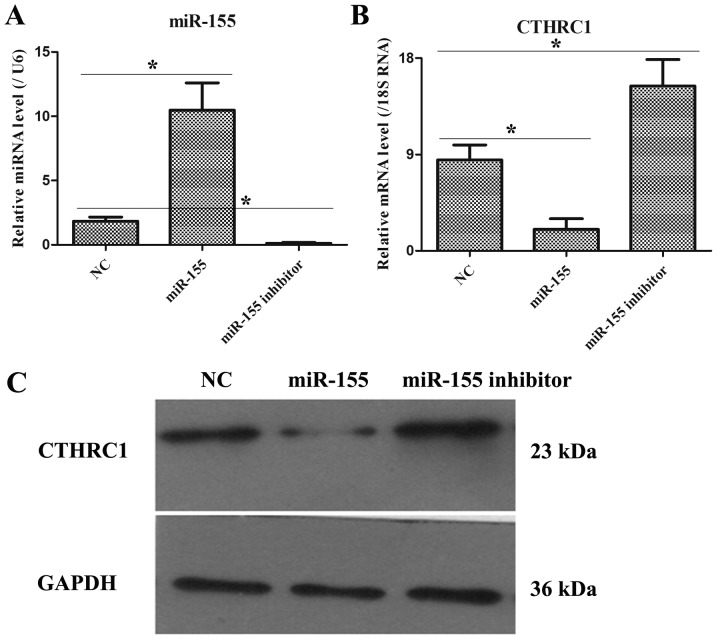
The expression levels of miR-155 and CTHRC1 in HT-29 cells after transfecting with NC, miR-155 mimic and miR-155 inhibitor plasmids
As illustrated in Fig. 3A, after transfecting 48 h, miR-155 and CTHRC1 expressions were remarkably increased and decreased, respectively, in miR-155 group as compared to NC group, but miR-155 and CTHRC1 expressions were markedly down-regulated and up-regulated, respectively, in miR-155 inhibitor group compared with NC group (Fig. 3B). Moreover, the protein level of CTHRC1 was also notably declined and elevated in miR-155 mimic and miR-155 inhibitor groups, respectively (Fig. 3C). Hence, these data pointed that the transfection efficiency of miR-155 mimic or miR-155 inhibitor was obvious.
Figure 3.
Examination of transfection efficiency. (A) The miR-155 expression was tested by qRT-PCR in HT-29 cells after transfecting with NC, miR-155 mimic and miR-155 inhibitor plasmids. (B) The CTHRC1 mRNA expression was determined by qRT-PCR in HT-29 cells after transfecting with NC, miR-155 mimic and miR-155 inhibitor plasmids. (C) The CTHRC1 protein expression was detected by qRT-PCR in HT-29 cells after transfecting with NC, miR-155 mimic and miR-155 inhibitor plasmids. *P<0.05 and **P<0.01 as indicated.
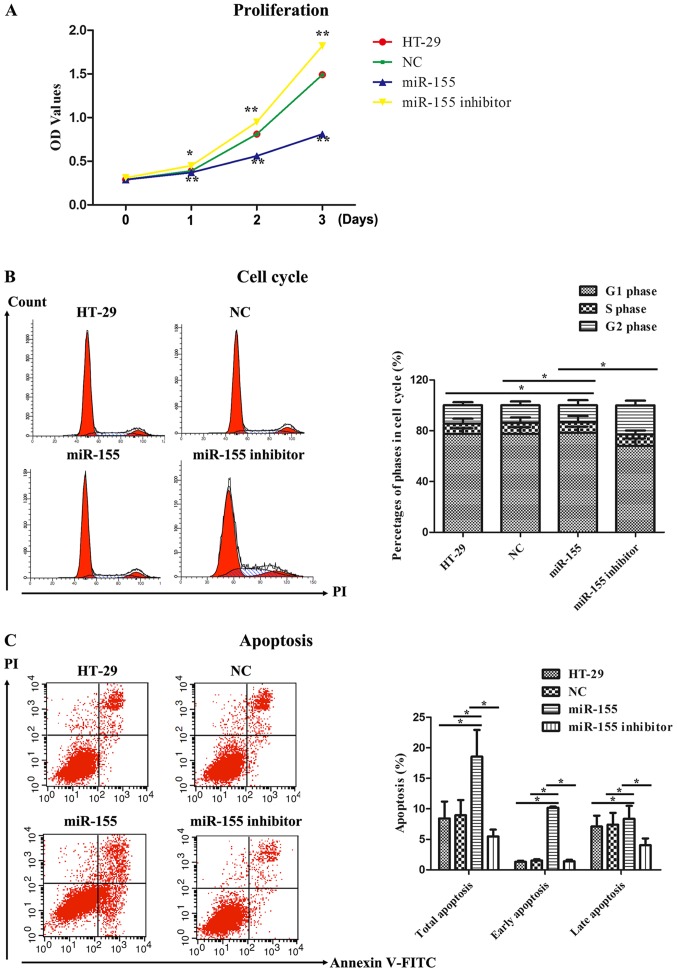
miR-155 affected cell proliferation activity, cell cycle and cell apoptosis
The proliferation activity of miR-155 on HT-29 cells was detected by MTT assay. As illustrated in Fig. 4A, the OD values representing cell proliferation activity in miR-155 group were markedly reduced compared to that in the control groups, including HT-29 cells group and NC group, at 1~3 days. Nevertheless, miR-155 inhibitor obviously reversed the proliferating effect of miR-155 on HT-29 cells. We next examined cell cycle progression in HT-29 cells treated with miR-155 mimics and miR-155 inhibitor. It was found that miR-155 induced cell cycle arrest in G0/G1 phase as comparison with HT-29 cells group and NC group, whereas miR-155 inhibitor abolished the impacts of cell cycle arrest in G0/G1 phase (Fig. 4B). Finally, cell apoptosis was further measured by flow cytometry. As displayed in Fig. 4C, Annexin V/PI dual staining was uncovered that miR-155 enhanced the early and late apoptosis rate in HT-29 cells. However, miR-155 inhibitor suppressed the early and late apoptosis rate compared to miR-155. Hence, these data indicated that miR-155 could inhibit cell proliferation, promote cell cycle arrest and facilitate cell apoptosis in HT-29 cells.
Figure 4.
miR-155 inhibited cell proliferation, promoted cell cycle arrest and facilitated cell apoptosis. (A) MTT assay was performed to analyze the effect of miR-155 on cell proliferation of HT-29 cells. Cell proliferation was expressed as the OD values. *P<0.05 vs. NC group, **P<0.01 vs. NC group. (B) Flow cytometry were performed to measure the effect of miR-155 on cell cycle of HT-29 cells. Representative flow cytometric histograms of each group showing the distribution of cell cycle were presented in left panel. *P<0.05 as indicated. (C) Flow cytometry were performed to test the effect of miR-155 on cell apoptosis of HT-29 cells. *P<0.05 as indicated.
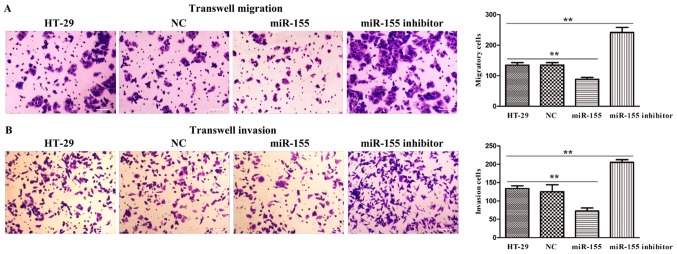
miR-155 regulated cell migration and invasion in HT-29 cells
To further investigate the biological function of miR-155 in colorectal cancer, we adopted HT-29 cells transfected with miR-155 mimics and miR-155 inhibitor plasmids to confirm cell migration and invasion activity through transwell assays. As presented in Fig. 5A, it was discovered that miR-155 was significantly declined the number of migration and invasion cells, while miR-155 inhibitor was remarkably elevated the number of migration and invasion cells (Fig. 5B). Thus, these results implied that miR-155 negatively regulated cell migratory and invasive ability of HT-29 cells.
Figure 5.
miR-155 suppressed the (A) migration and (B) invasion of HT-29 cells. Left part displayed representative images (×400) of each group in the Transwell assays, whereas right panel exhibited the statistics of the migration and invasion. Data shown are means ± SD (n=3). Scale bars=100 µm. **P<0.01.
Discussion
Colorectal cancer is known as one of main types of gastrointestinal cancers which remain an important public health problem in different populations (22). Although recent advances in medicine have significantly improved the survival of patients with early-stage colorectal cancer, patients with advanced colorectal cancer have poor prognosis mainly due to frequent tumor metastasis and tumor recurrence after surgical resection (8,23). Emerging studies show that chromosomal abnormalities, genetic alterations, epigenetic modifications and unhealthy lifestyle are decisive factors that initiate and drive the occurrence and development of colorectal cancer (4,5). Moreover, miRNAs increasingly reported to be differentially expressed either as oncogenes (e.g., miR-21, miR-191) or tumor suppressors (e.g., let-7, miR-34) play a pivotal role in the pathogenesis and tumorigenicity of colorectal cancer (24,25). Otherwise, aberrant miR-155 expression was previously observed in colorectal cancer (18,26), but the biological function and regulatory mechanisms of miR-155 in colorectal cancer is still largely unknown. In this study, we found that expressions of miR-155 and CTHRC1 in colorectal cancer specimens were remarkably lower and higher than those in corresponding adjacent tissues, respectively, suggesting miR-155 and CTHRC1 might exert a crucial role in colorectal cancer. However, higher expression levels of miR-155 have been detected in other human malignancies, including lung cancer, cervical cancer, hematological malignancies and thyroid carcinoma (18,19). This distinction may be caused by tissue specificity, ethnic diversity or different tumor stages. Subsequently, we examined miR-155 and CTHRC1 expressions in five different colorectal cancer cell lines by qRT-PCR. The result displayed the lowest expression of miR-155 and the highest expression of CTHRC1 in HT-29 cells, which was chosen as the following experiment cell line. Moreover, the opposite expression trend between miR-155 and CTHRC1 also implied that miR-155 might target CTHRC1. Functional assays revealed that restoration of miR-155 notably inhibited cell proliferation, migration, and invasion, and promoted cell arrested at G0/G1 stage and cell apoptosis in HT-29 cells. The precise regulation of cell proliferation, cell cycle and apoptosis is basic premise for cellular normal growth. Once dysregulation in cell proliferation, cell cycle and apoptosis, it might lead to cellular abnormal changes, which ultimately might trigger cell canceration (27). Thus, the alterations of cell proliferation, cell cycle and apoptosis indicated that miR-155 suppressed the growth and progression of colorectal cancer in vitro. Additionally, migratory and invasive abilities are closely associated with tumor metastasis which has been widely recognized as the important reason for the unsatisfactory prognosis of colorectal cancer patients (28,29). Hence, our data implied that miR-155 decelerated the tumor metastasis of HT-29 cells by inhibiting cell migratory and invasion behaviors.
CTHRC1, initially screened from differentially expressed genes in balloon-injured vs. normal rat arteries, is a well-known regulator of the growth and metastasis of human cancers, such as lung cancer, pancreas cancer, breast cancer, cervix cancer, and liver cancer (30). In the present study, it was confirmed that miR-155 could directly interact with the 3′-UTR of CTHRC1 using luciferase reporter assay, suggesting that CTHRC1 is a direct target of miR-155. Furthermore, inhibition of miR-155 have an opposite effects on these features related to tumor growth and metastasis compared with miR-155 treatment in HT-29 cells. Based on the targeted interaction between miR-155 and CTHRC1, as well as the CTHRC1 expression levels after transfecting with miR-155 mimic and miR-155 inhibitor as shown in Fig. 3, the function of miR-155 inhibitor should be similar to that of CTHRC1 over-expression, thereby these results indirectly reflected that CTHRC1 might be an oncogenic gene in colorectal cancer.
In conclusion, in the current study, we validated the significance of down-regulated miR-155 expression in colorectal cancer patients and its tumor suppressive role in HT-29 cells by attenuating cell growth and metastasis. Additionally, CTHRC1 was also verified to be downstream target of miR-155 and we adopted an indirectly method (i.e., utilizing miR-155 inhibitor to replace CTHCR1 over-expression) to investigate the oncogenic role of CTHRC1 in colorectal cancer in vitro. Taken together, these findings implicated that miR-155 and its target CTHCR1 might be used as a prognostic indicator and therapeutic target in colorectal cancer patients.
Acknowledgements
The present study is supported by Shanghai Emerging Frontier Technology Joint Research Program (SHDC12016104).
References
- 1.Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18(pii):E197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolocan A, Ion D, Ciocan DN, Paduraru DN. Prognostic and predictive factors in colorectal cancer. Chirurgia (Bucur) 2012;107:555–563. [PubMed] [Google Scholar]
- 3.De Rosa M, Rega D, Costabile V, Duraturo F, Niglio A, Izzo P, Pace U, Delrio P. The biological complexity of colorectal cancer: Insights into biomarkers for early detection and personalized care. Therap Adv Gastroenterol. 2016;9:861–886. doi: 10.1177/1756283X16659790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fumery M, Dulai PS, Gupta S, Prokop LJ, Ramamoorthy S, Sandborn WJ, Singh S. Incidence, risk factors, and outcomes of colorectal cancer in patients with ulcerative colitis with low-grade dysplasia: A systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2017;15:665–674.e5. doi: 10.1016/j.cgh.2016.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coppedè F, Lopomo A, Spisni R, Migliore L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J Gastroenterol. 2014;20:943–956. doi: 10.3748/wjg.v20.i4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Du Y, Liu X, Cho WC, Yang Y. MicroRNAs as regulator of signaling networks in metastatic colon cancer. Biomed Res Int. 2015;2015:823620. doi: 10.1155/2015/823620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berretta M, Alessandrini L, De Divitiis C, Nasti G, Lleshi A, Di Francia R, Facchini G, Cavaliere C, Buonerba C, Canzonieri V. Serum and tissue markers in colorectal cancer: State of art. Crit Rev Oncol Hematol. 2017;111:103–116. doi: 10.1016/j.critrevonc.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol. 2016;22:1745–1755. doi: 10.3748/wjg.v22.i5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lech G, Slotwinski R, Krasnodebski IW. The role of tumor markers and biomarkers in colorectal cancer. Neoplasma. 2014;61:1–8. doi: 10.4149/neo_2014_003. [DOI] [PubMed] [Google Scholar]
- 10.Yamazaki N, Koga Y, Taniguchi H, Kojima M, Kanemitsu Y, Saito N, Matsumura Y. High expression of miR-181c as a predictive marker of recurrence in stage II colorectal cancer. Oncotarget. 2017;8:6970–6983. doi: 10.18632/oncotarget.14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 12.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–R44. [PubMed] [Google Scholar]
- 13.Ruan K, Fang X, Ouyang G. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 14.Giannakakis A, Coukos G, Hatzigeorgiou A, Sandaltzopoulos R, Zhang L. miRNA genetic alterations in human cancers. Expert Opin Biol Ther. 2007;7:1375–1386. doi: 10.1517/14712598.7.9.1375. [DOI] [PubMed] [Google Scholar]
- 15.Pagliuca A, Valvo C, Fabrizi E, di Martino S, Biffoni M, Runci D, Forte S, De Maria R, Ricci-Vitiani L. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene. 2013;32:4806–4813. doi: 10.1038/onc.2012.495. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 17.Zhang G, Zhou H, Xiao H, Liu Z, Tian H, Zhou T. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig Dis Sci. 2014;59:98–107. doi: 10.1007/s10620-013-2858-8. [DOI] [PubMed] [Google Scholar]
- 18.Wan J, Xia L, Xu W, Lu N. Expression and function of miR-155 in diseases of the gastrointestinal tract. Int J Mol Sci. 2016;17(pii):E709. doi: 10.3390/ijms17050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou Y, Wang J, Wang X, Shi S, Wang W, Chen Z. Appraising MicroRNA-155 as a noninvasive diagnostic biomarker for cancer detection: A meta-analysis. Medicine (Baltimore) 2016;95:e2450. doi: 10.1097/MD.0000000000002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan L, Yu J, Tan F, Ye GT, Shen ZY, Liu H, Zhang Y, Wang JF, Zhu XJ, Li GX. SP1-mediated microRNA-520d-5p suppresses tumor growth and metastasis in colorectal cancer by targeting CTHRC1. Am J Cancer Res. 2015;5:1447–1459. [PMC free article] [PubMed] [Google Scholar]
- 21.Wang JP. Chinese standard for the diagnosis and treatment of colorectal cancer (2010) Zhonghua Wei Chang Wai Ke Za Zhi. 2011;14:1–4. (In Chinese) [PubMed] [Google Scholar]
- 22.Binefa G, Rodriguez-Moranta F, Teule A, Medina-Hayas M. Colorectal cancer: From prevention to personalized medicine. World J Gastroenterol. 2014;20:6786–6808. doi: 10.3748/wjg.v20.i22.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sideris M, Papagrigoriadis S. Molecular biomarkers and classification models in the evaluation of the prognosis of colorectal cancer. Anticancer Res. 2014;34:2061–2068. [PubMed] [Google Scholar]
- 24.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 25.Moridikia A, Mirzaei H, Sahebkar A, Salimian J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol. 2018;233:901–913. doi: 10.1002/jcp.25801. [DOI] [PubMed] [Google Scholar]
- 26.He B, Gao SQ, Huang LD, Huang YH, Zhang QY, Zhou MT, Shi HQ, Song QT, Shan YF. MicroRNA-155 promotes the proliferation and invasion abilities of colon cancer cells by targeting quaking. Mol Med Rep. 2015;11:2355–2359. doi: 10.3892/mmr.2014.2994. [DOI] [PubMed] [Google Scholar]
- 27.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang X, Jiang L, Sun Z, Miao Z, Xu H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Z, Yu L, Luo S, Li M, Li J, Li Q, Sun Y, Wang C. miR-296 inhibits the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100A4. BMC Cancer. 2017;17:140. doi: 10.1186/s12885-017-3121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang L, Dai DL, Su M, Martinka M, Li G, Zhou Y. Aberrant expression of collagen triple helix repeat containing 1 in human solid cancers. Clin Cancer Res. 2006;12:3716–3722. doi: 10.1158/1078-0432.CCR-06-0030. [DOI] [PubMed] [Google Scholar]