ABSTRACT
It is now acknowledged that extracellular vesicles (EVs) are important effectors in a vast number of biological processes through intercellular transfer of biomolecules. Increasing research efforts in the EV field have yielded an appreciation for the potential role of glycans in EV function. Indeed, recent reports show that the presence of glycoconjugates is involved in EV biogenesis, in cellular recognition and in the efficient uptake of EVs by recipient cells. It is clear that a full understanding of EV biology will require researchers to focus also on EV glycosylation through glycomics approaches. This review outlines the major glycomics techniques that have been applied to EVs in the context of the recent findings. Beyond understanding the mechanisms by which EVs mediate their physiological functions, glycosylation also provides opportunities by which to engineer EVs for therapeutic and diagnostic purposes. Studies characterising the glycan composition of EVs have highlighted glycome changes in various disease states, thus indicating potential for EV glycans as diagnostic markers. Meanwhile, glycans have been targeted as molecular handles for affinity-based isolation in both research and clinical contexts. An overview of current strategies to exploit EV glycosylation and a discussion of the implications of recent findings for the burgeoning EV industry follows the below review of glycomics and its application to EV biology.
KEYWORDS: Extracellular vesicles, exosomes, microvesicles, glycans, glycomics, glycosylation, lectins, glycoengineering
Introduction
Intercellular communication is essential to the correct functioning of multicellular organisms. Aside from chemical endocrine systems and protein growth factors, this is also achieved through the secretion and uptake of extracellular vesicles (EVs) – submicron particles consisting of a lipid bilayer with a cargo of functional proteins, RNA molecules and metabolites. The term EV comprises a taxonomy of vesicle designations, each classified mainly by their mode of biogenesis [1,2]. While apoptotic bodies are formed during programmed cell death, microvesicles and exosomes are produced by cells in normal and stressful conditions. Respectively, these latter classes originate either by directly budding from the cell membrane or through an endosomal pathway. Vesicle classification based on particle size alone is problematic as the diameters of the different vesicles types are on a continuous, overlapping scale. Although described as early as 1946 [3], EVs were often dismissed as degradation products or cellular debris and not identified as vehicles for the horizontal transfer of biomolecules until recently [4]. Early work included ultrastructural studies of subcellular compartments that defined the biogenetic pathways for these particles [5,6] and the 1996 discovery that exosomes can stimulate T-cells through antigen presentation resulted in a broad increase of interest in EV research [7]. This increase in research activity was further cemented by the 2006 finding that EVs contain RNA cargoes [8,9]. The field now enjoys an exponential growth in the number of publications [10] and EVs have now been isolated from an ever-expanding array of biofluids and cell types [1].
The capture of EVs by target cells is involved in a broad range of biological processes including: angiogenesis [11], bone development [12], interneuronal communication [13], cell migration [14] and inflammation regulation [15]. Mechanisms for these processes are diverse but generally relate to the delivery of bioactive RNA, protein, metabolite and/or lipid cargoes. Conversely, EV transmission can be co-opted to produce adverse effects and may be responsible for pathogenesis in several neurodegenerative [16], cardiovascular and metabolic diseases [17]. Specific mechanisms for these are again diverse. Intercellular transmission of β-amyloid protein by EVs has been observed in Alzheimer’s disease whilst the EV-mediated binding and concentration of coagulation factors may cause aberrant clotting and thrombosis. The study of EVs is also yielding insights to cancer biology and, in particular, metastasis [18,19]. Recent reports have illustrated how long-range transmission of exosomes leads to the preparation of pre-metastatic niches through the recruitment and perturbation of various classes of immune cells, resulting in inflammation and remodelling of the local vasculature and extracellular matrix [20,21]. Strikingly, this phenomenon of niche formation is tissue specific, as directed by integrins presented on the exosome surfaces, providing new mechanistic insights into metastatic organotropism, the propensity of some cancers to metastise to selected organs [22]. These examples of potent bioactivity and the biocompatible nature of EVs have inspired the development of vesicles as vehicles for both drug and gene therapy delivery for a large number of applications [23]. EVs may even play a direct role in evolution as studies show EV-mediated transfer of nucleic acids into sperm cells [24]. Final evidence for the importance of EVs is given by their existence across taxonomic kingdoms, from archaea through bacteria to plants and animals [1].
The strong interest in exosomes has produced hundreds of in-depth characterisation studies on EVs from a number of sources promoting the creation of dedicated databases like Vesiclepedia and Exocarta to handle the wealth of data generated [2,10,25]. The proteomic, transcriptomic and lipidomic analyses of exosomes have revealed enrichments in specific proteins including: tetraspanin, cystoskeletal and heat shock proteins, messenger and micro RNAs, and lipids such as ceramide, phosphatidylserine, cholesterol and sphingolipids. These biomolecular compositions can differ markedly between EVs from different cellular origins and also between healthy and disease states [1]. Combined with the high yield and ubiquity of EVs across all biofluids assayed, this suggests a clinical use for EVs as a source of diagnostic disease markers [26–28]. More recently however, a growing appreciation has been founded for the role of another major biomolecule in EVs – carbohydrates.
Similarly to the EV field, the study of carbohydrates in biology has historically lagged behind that of the other major biomolecules. Glycans have long been recognised as important structural components and functions in energy storage but more recently are also recognised for their function as information carriers and their role in uniquely modulated molecular recognition events. Carbohydrate structures are found conjugated to lipids and proteins as glycans or as repeating glycosaminoglycan chains in proteoglycans and these exert different functions on different size scales. These include recognition events at the cellular level and control of both intracellular trafficking and quality control of folding events at the level of individual proteins [29,30]. Proteoglycans in particular are a major constituent of the extracellular matrix and are essential to tissue architecture [31]. Aberrant glycosylation disrupts these essential functions and can aid the progression of cancers or result in lysosomal storage diseases [30]. Glycans are an essential part of the cellular make-up, underscored by the estimate that half of all human proteins are glycosylated [32]; yet the ubiquity and uses of glycan moieties in biology emerged after proteins and nucleic acids had been sequenced, structured and otherwise dissected.
The reason for this lag period is that glycan biochemistry is challenging to analyse. The constituent hexoses are stereoisomers of one another and glycosidic bonds can be formed at several points in the saccharide ring, resulting in branched structures [29] – a stark contrast to linear biopolymers composed of monomers with distinct molecular weights. These characteristics have so far complicated the development of sequencing methods, methods that were keys to unlocking the disciplines of genomics and proteomics. Moreover, there is no sequence relationship between glycans and their conjugates other than initial attachment sites and so prediction of glycan structures beyond semiconserved cores is impossible. Taking protein glycosylation as an instructive example, N-linked glycosylation occurs at the carboxyamide of asparagine residues in the well defined sequon Asn-X-Ser/Thr whilst O-linked glycosylation is less controlled. The hydroxyl group of any serine or threonine residue is a potential O-glycosylation site. N-glycosylation initiates with the co-translational addition of a 14-mer oligosaccharide in the endoplasmic reticulum. This is then remodelled though enzymatic trimming to a conserved pentasaccharide core, followed by the sequential addition of monosaccharides to create a plethora of possible structures. To date, as many as 2000 different N-glycan structures have been posited [33]. Further complicating matters, many proteins present several glycosylation sites and each site can be occupied by a variety of glycan structures leading to a heterogeneous mixture of glycoforms. Meanwhile, glycolipids possess entirely different glycan structures. The predominant class of glycolipid are glycosphingolipids, glycans conjugated to ceramide molecules [34]. Ceramide can be linked to either galactose or glucose at the endoplasmic reticulum and then sequentially extended by glycosyltransferases of the Golgi. Linear glycan chains ranging from one to six monosaccharide residues have been observed, though branching can also occur with the addition of sialic acids to form larger, branched gangliosides. All extracellular-facing ceramides have the potential to be glycosylated. Prediction of glycolipid abundance is therefore impossible and must be assayed directly, similarly to O-glycosylation.
Analysis of this complex micro-heterogeneity requires either advanced mass spectrometry methods [35] or the use of carbohydrate-binding proteins known as lectins. Single lectins can be used much like antibodies or panels of lectins can be printed onto microarrays for wider analytical coverage [36,37]. Detailed descriptions of these methods as applied to EVs form the structure of the following review. Initial application of these techniques to EVs was pioneered in 2009 with early studies repeatedly demonstrating that EVs possess a defined repertoire of glycans [38,39] (Figure 1). This preferential enrichment signifies a role for glycans in vesiculogenesis but specific mechanisms remain uncertain. For example, the presence of N-glycans is essential for exosomal recruitment of the glycoprotein EWI-2 but the molecular recognition elements remain elusive [40]. Indeed, the L-type lectin LMAN2 was identified as a negative regulator of cargo protein recruitment in Human Embryonic Kidney (HEK) cells but whether this is due to cargo glycans or other interactions of the lectin is unclear [41, p. 2]. A recent publication presents evidence that sialic acid residues of glycans are involved in cellular recognition of exosomes [42] but perhaps the best developed story of EV glycobiology relates to the promotion of ESCRT-mediated (endosomal sorting complexes required for transport) exosome biogenesis by the syndecan family of heparan sulfate proteoglycans. Briefly, intraluminal heparanase trimming of syndecan glycosaminoglycan chains promotes clustering of these molecules and results in co-localisation of syntenin adaptor proteins and the bound ESCRT machinery [43,44].
Figure 1.
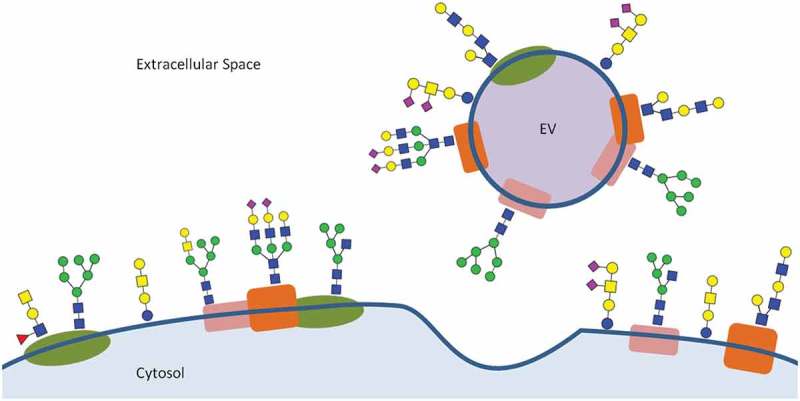
Enrichment for specific glycans to EVs.
Lectin microarrays have shown that EVs are preferentially enriched in certain glycan features compared to more diverse glycan repertoire of the producing cell membranes [38,39]. These features include polylactosamine and α2-6 linked sialic acid residues, and high mannose and complex type N-glycans. Exclusion of blood group A/B antigens was also identified. Glycans of this figure are representative of these features but are not definitive structures as these are not possible to obtain through lectin microarrays alone.
Glycomics methods are becoming increasingly well established but their application to EVs is still a novel undertaking. Hallmark features of EV biology such as cellular secretion and recognition could be mediated by glycan moieties but a concerted effort is needed to bridge broad glycome characterisation to functional studies. The field of EV glycomics is very fresh and reviews of the subject are few in number [28,45]. This current work is intended to act as a technique-led overview of the major findings to date with a focus on the protocols used therein. We hope to introduce EV researchers to the methods of lectin microarrays and MS in order to inspire research efforts in this area. Extending this practical focus, particular attention is given to works that explore the diagnostic potential of exosome glycosylation as disease biomarkers and a role for glycosylation in the burgeoning EV therapy industry is also discussed.
EV glycomics
Surface glycan profiling by lectin microarrays
Lectin microarrays are a recent development for glycomics analysis. First described in 2005 [46], these microarrays comprise a variety of carbohydrate-binding lectin proteins immobilised to a glass support. Each lectin possesses binding activity towards different glycan epitopes and incubation of the array with a fluorescently labelled sample reveals which of these epitopes are present. All surface glycoconjugates of intact vesicles can be targeted in a single analysis and such unbiased analysis is a major advantage of lectin arrays. This is in contrast to MS methods that are only compatible with individual subclasses and require laborious and destructive sample preparation steps. In a pioneering study on the EV glycome the Mahal group applied lectin microarrays to profile the surface glycans of intact T-cell-derived EVs and HIV-1 virions [38]. Comparing the microarray results of the secreted vesicles against those of the producer T-cell membranes revealed an enrichment of polylactosamine and α2-6 linked sialic acid residues, complex type N-glycans and high mannose structures. Conversely, an exclusion of O-linked A and B blood antigens from the EVs and virions was also observed. Glycan structures representative of these features are shown in Table 1. A follow-up study extended this approach and demonstrated that this EV “glycan signature” is conserved across a wider panel of cellular sources [39] (Figure 1). The panel included T-cell, carcinoma and melanoma cell lines and also a sample of human breast milk, demonstrating the validity of these findings in a physiological context. Together, these works were influential in suggesting that glycans may be involved in either EV biogenesis or cargo recruitment – hypotheses that have engendered great interest in EV glycobiology.
Table 1.
A primer of glycan features relevant to EVs.
| Glycan feature | Representative structures | EV relevance | ||
|---|---|---|---|---|
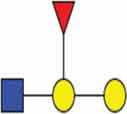
| Polylactosamine | 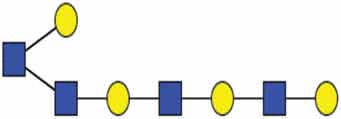 |
Enriched | ||
| α2,6 linked sialic acid |  |
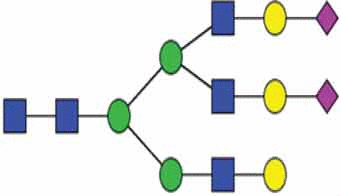 |
Enriched | |
| High mannose N-glycan | 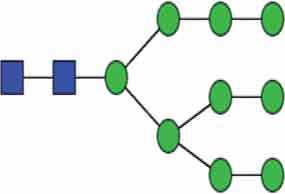 |
Enriched | ||
| Complex type N-glycan | 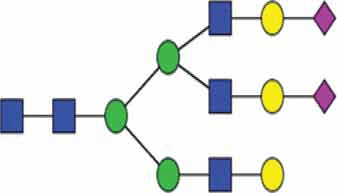 |
Enriched | ||
| O-linked blood group antigens A/B | 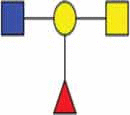 |
 |
Excluded | |
Modern publications that employ lectin microarrays should build upon these studies by not only profiling EV glycosylation but also probing the functional roles of the glycans, as exemplified by potentially impactful research from Shimoda et al. [42]. Therein, exosomes produced by human adipose derived MSCs were applied to lectin microarrays exosomes alongside purified membranes from these cells. Whilst confirming that exosomes showed strong intensities for a polylactosamine-binding lectin, STL (from Solanum tuberosum), exosomes also exhibited hugely inflated intensities for sialic-acid-binding lectins SNA, SSA and TJA-I compared to the parental membranes. The team hypothesized that these sialic acid residues were important for cell recognition and then focused on potential receptors for these – cell surface siglecs. The basal uptake of fluorescently labelled exosomes by HeLa cells was measured using confocal laser scanning microscopy and this system was then perturbed using either free sialic acid as a competitive inhibitor or by pre-incubating cells with a blocking antibody against siglec-3. In both cases cellular uptake was reduced with the effect increasing significantly with the amount of inhibitor. It is uncertain whether these sialic acid residues reside in N-glycans, O-glycans or glycolipids but this work is one of the first to describe a definite role for glycans in cellular uptake. Whether involved in cell recognition and/or active entry into cells, the precise role of these sialylated glycans is unclear. Moreover, the specificity of this effect must be probed. The presence of sialic acids is a feature of the conserved glycan signature identified for EVs and thus may be a necessary component in EV endocytosis [39]. On the other hand, different cell types express different repertoires of cell surface lectins and sialic-mediated uptake may not be universal. Further research is needed to clarify this effect and to elucidate the potential roles of other saccharide residues and receptors. It will also be important to reconcile these future findings against other classes of molecules identified as important in exosome binding and uptake. As uptake was not completely ablated, it is more likely that a synergy between different classes of molecule is needed for efficient uptake and the importance of glycans may vary between systems. Finally, extrapolation of this work reveals sialic-based glycoengineering as a potential strategy for artificial targeting or potentiating of EV therapies.
Purified, intact EVs from any source are applicable to lectin microarray analysis, including those from complex matrices like biofluids. One such example is the characterisation of urinary extracellular vesicles (uEVs). Biofluid matrices are typically problematic due the presence of confounding proteins that co-purify with EV preparations. Urine in particular is highly abundant in Tamm-Horsfall protein (THP), a heavily glycosylated urinal protein that can form aggregates several million Da in size. Interestingly the study from Gerlach et al. showed that lectin microarrays can be applied to uEVs with minimal interference from THP [47]. A standard centrifugal filtration approach was used to purify the uEVs and Gerlach et al. confirmed that whilst the broad glycan signature described above is maintained in the uEVs of different individuals, a diverse variety of other glycans is present. The researchers then transposed their urine-optimised microarray protocol to the uEVs of patients with autosomal dominant polycystic kidney disease. This revealed slight but significant differences in the sample binding intensities of six lectins, providing important proof of concept for the use of lectin microarray profiles as non-invasive for the detection and monitoring of disease. More recently, uEV lectin microarray profiling has been used to develop a custom lectin bead affinity purification technology [48] (Figure 2). Ultracentrifugation is historically the major method of EV purification, requiring nonstandard, expensive instrumentation with extra purification steps depending on the complexity of the matrix. Circumventing this requirement with novel isolation methods is therefore a major objective in enabling routine, low-specialism analyses. Herein, microarray analysis was performed with a panel of 62 lectins and the polylactosamine-specific lectin STL was found to exhibit the strongest uEV binding with the least off-target THP recognition. A system of biotinylated STL with magnetic streptavidin bead capture was then devised and found to be sufficient for the purification of uEVs directly from urine. Potential issues with this approach relate to the bias of lectin specificity. EVs from different biofluids may have different glycan profiles and the lectin array analysis will need to be repeated to find the most effective binder. A corroborating study on MSC exosomes has, however, shown that similarly high binding intensities for STL [42], perhaps implying polylactosamine-containing glycans as a more universal handle for EV purification. Finally, flow cytometry characterisation of the isolated uEVs identified AQP2 positive vesicles but could not detect AQP1, another commonly detected protein in uEV. This points towards uEVs as a mixed population of vesicles with different glycan-surface patterns and that lectins can be a useful tool to isolate and characterise these subsets. Overall, coupling this facile benchtop technique with the biomarker work of Gerlach et al. does suggest an encouraging future for lectin-based EV diagnostics in the clinic.
Figure 2.
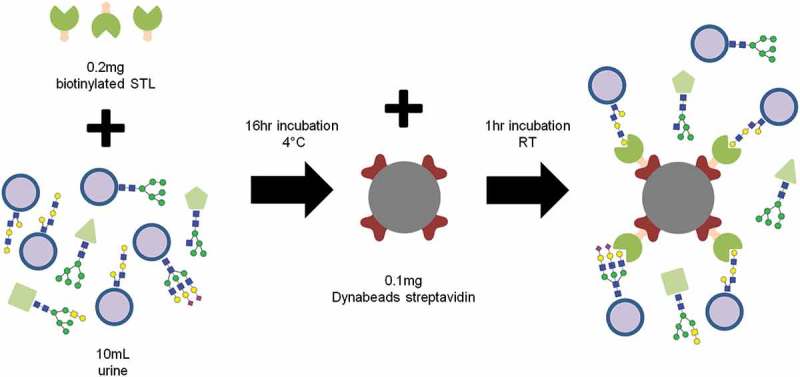
A protocol for the purification of EVs using a lectin-bead approach.
Following a lectin microarray screen, STL was found to bind to uEVs with greater specificity than urine glycoproteins [48]. Incubation of undiluted urine with biotinylated STL and streptavidin-conjugated Dynabeads allowed for magnetic recovery of uEVs.
There are some conceptual challenges with microarray glycomics. Compared with antibody recognition or nucleic acid hybridisation, the structural specifics of lectin:epitope binding is less understood. There is a strong overlap in carbohydrate binding for many lectins and the detailed characterisation of lectin specificities is still a work in progress [37]. Some lectins are multimeric and positive binding represents avidities rather than one-to-one events, limiting quantitative application. However, this can also prove a boon as lectin binding conveys not just the presence of a monosaccharide but also information as to neighbouring residues. Despite these constraints it is recommended that lectin microarrays are an excellent first line method to profile EV surface glycosylation, as full characterisation of a cell or vesicle glycome by molecular methods like mass spectrometry (MS) or nuclear magnetic resonance spectroscopy (NMR) requires the application of comprehensive workflows that target specific glycan subclasses. A major advantage of lectin arrays is the unbiased glycan analysis over all classes of carbohydrates on intact vesicle or cells which do not require any sample preparation. Compared with sequential, single lectin analyses, microarrays are of high throughput and incubation protocols are undemanding. The sensitivity of lectin arrays is extremely high, requiring sub-microgram amounts of material and modifications to microarray workflows are also possible. In particular, coupling SDS-PAGE sample separation to a lectin array has allowed more granular analysis, resolving individual glycoproteins [49] and lectins can be specifically chosen to reflect the particulars of the research – from the immune system or from prokaryotes, for example. Fabrication is a complex and expensive undertaking but array laser scanners are not uncommon instruments and commercial microarrays are now becoming available. Indeed, the most recent publication to employ lectin array profiling of EVs has sourced theirs from a commercial vendor [42] and contract analyses are also possible. Yet, use of lectin microarrays alone is insufficient to fully define complete glycan structures. Lectin binding is a functional assay and there is much information to be gained from elucidating absolute structures, particularly when considering potential glycan biomarkers.
High resolution glycomics
The structural limitations of lectin microarrays can be avoided by using high-resolution MS techniques [35,50]. Briefly, MS instruments are coupled to a previous separation technique, stratifying a sample of released glycans into fractions and facilitating structural analysis through reduced heterogeneity. Several different methods exist for both separation and MS, with sample preparation defined by the class of glycoconjugate that the researcher wishes to assay. To elaborate, different chemical linkages connect glycans to protein or lipid carriers and the glycan isolation methods that have been developed can target only one of the different glycan classes present. For example, N-glycans are readily removed from denatured glycoproteins through enzymatic digestion with PNGaseF but no efficient enzyme exists for the release of O-glycans. Sample separation is usually achieved by ultra or high-performance liquid chromatography (UPLC or HPLC) or, in some applications, capillary electrophoresis (CE). Different separation columns based on different stationary phases are used in LC, including hydrophilic-interaction chromatography (HILIC) columns or porous graphitized carbon columns, and the separation capabilities of LC allows for isomeric structures to be resolved. Moreover, released glycans are usually labelled to enhance glycan separation and mass-spectrometric detection, and several labels can be used for each analytical technique [51]. Considering those MS methods that are not coupled to a sample separation technique, Matrix-Assisted Laser Ionization Time-of-Flight Mass-Spectrometry (MALDI-TOF MS) is the most established approach due to its high-throughput and automation potential [52]. For the assignment of glycan structures tandem mass spectrometry (MS/MS) is a particularly powerful tool. Here, selected precursor ions are fragmented into product ions, usually by the collision with a gas, and the fragmentation pattern employed to gain additional insight into the analyte structure [53].
Some of these high resolution techniques have been employed in characterising the N-glycome of exosomes secreted by the SKOV3 ovarian carcinoma line [54]. Peptide N-glycosidase F digestion was used to release glycans from the purified exosomes and 2-AB (2-aminobenzamide) used to label these for subsequent MALDI-TOF MS and HPLC-MS analyses (using HILIC and C18 columns). In this multimethodological approach, mass fragments were used to propose glycan compositions which were then confirmed using commonly available 2-AB-labelled reference standards. In broad agreement with the conserved glycan signature of exosomes, the study identified high-mannose glycans of up to nine mannose residues and complex type di-, tri- and tetraantennary structures with variable fucosylation of the proximal GlcNAc. Interestingly the complex glycans also exhibited bisecting GlcNAc residues and a high degree of terminal sialylation. The authors highlight that enrichment of these glycan types has been observed in exosomes from other cancers and more recent high resolution glycomics work from this group has confirmed their presence in exosomes of other ovarian cancer lines [55]. It is unknown whether this signifies an exosome-specific feature of cancer or simply reflects broader cancer biology. In every case these glycans are enriched compared to the glycome of the cell membranes, adding credence to the concept of glycosylation-led biogenesis mechanisms. High resolution glycan analysis has also been performed with prostatically-secreted exosomes from patients with prostate cancer. A shift from tri- and tetraantennary glycans towards bisecting GlcNAc structures was identified in a manner that could be correlated with increasing disease severity [56]. These findings further indicate potential of glycan profiles as diagnostic or prognostic biomarkers, though the question of whether glycan biomarkers are specific to different cancers remains open. Shifts in glycan content observed through high resolution uEV glycomics has also revealed new details on the pathology of galactosemia and features of other glycosylation diseases may be inferred with this non-invasive approach [57]. It should be noted that sample denaturation can be required for the efficient release of glycans for these techniques. Lysis of exosomes releases cargo glycoproteins and the data then reflects the total glycan content, not just surface glycosylation.
Turning to glycolipids, there are no published studies that directly examine the glycan structures of EV glycolipids. Glycolipids have instead been covered under lipidomic analyses. The most comprehensive of these studies is from Llorente et al. who profile the exosomes from PC-3 prostate cancer cells using quadrupole ion trap MS with C18 column separation and LC/ESI-MS/MS [58]. The study reveals a significant enrichment of glycosphingolipids in exosomes compared with the membranes of producing cells. This enrichment was observed with all glycosphingolipids assayed, including hexosylceramides, lactosylceramide and a-series gangliosides, suggesting a conserved glycolipid signature. The trend of these data is corroborated by studies with exosomes from other cell lines, averaging a 2–3 fold enrichment of glycosphingolipids [59]. A more general finding from these studies is the 8.4:1 fold ratio of lipids to proteins in these PC-3 exosomes which potentially signifies glycolipids as the major reservoir of EV glycans. The mechanisms of this glycolipid sorting are uncertain – whether passively incorporated from endo-lysosomal membranes or directly trafficked through glycan interactions. Interestingly, the core moieties of glycosphingolipids, ceramide molecules, have been proven as agents of ESCRT-independent vesiculogenesis [60]. This is achieved through induction of membrane curvature but whether ceramide-glycosphingolipid metabolism influences this process is unclear. An early investigation from Llorente´s group has shown that inhibition of this pathway by targeting glucosylceramide synthase in PC-3 cells does indeed affect the protein composition of secreted exosomes [61] and further investigation of this phenomenon is warranted.
Barriers to MS glycomics relate to the high cost equipment and also to data analysis. Branching structures, isomeric monomers and glycan anomers mean that peak assignment of MS spectra is imprecise. Until recently, this interpretation was performed manually and it was very time-consuming and it required advanced levels of skill. However, in recent years, several algorithms were developed to assist in this task and some of these are publicly available [62]. On the one hand, the freeware software suite Glycoworkbench not only provides a friendly interface for the rapid drawing of glycan pictograms but is also used for the computation and assignment of theoretical MS/MS fragments of a selected structure [63]. On the other hand, UniCarb-DB provides access to a collection of LC-MS/MS N- and O-linked glycan fragments released from glycoproteins and complemented with experimental metadata [64]. Many more such resources are available for glycan mass assignment, including the glycan database at ExPASy or the repository Toucan. Alternatively, emerging technologies will also simplify analysis. A highly promising new analytical route that can also be used is ion mobility mass spectrometry (IM-MS) [65,66]. IM-MS measures in an ms timescale the mobility of gas-phase ions under the influence of an electric field in the presence of a buffer gas. Mobility is then a function of ion size, shape and charge. As a result, it is able to separate isomeric carbohydrates thanks to their different mobilities. Furthermore, these mobilities may also be estimated computationally if the 3D structure of the molecule is known. IM-MS can also be coupled to LC, obtaining this way a multidimensional separation. However, it must be noted that none of these above described techniques can provide full unambiguous structural information. A multimethodological approach is necessary to obtain a depth understanding of complex glycomes. In this sense, the availability of well defined, pure and isotopically labelled glycan standards can solve many of the current challenges in glycan analysis, like the lack of robustness and reproducibility observed with MS, or the identification of co-eluting glycan isomers [67]. Generation of these standards may be achieved through emerging novel chemo-enzymatic glycan synthesis approaches [68]. As a final caveat, pre-established standards and databases will miss information on glycosidic linkages not cover glycans generated through atypical biosynthetic pathways. Depending on the research question and biological system, de novo glycan sequencing may be a better option but the requirement for high material amounts may preclude usage with systems that produce low quantities of EVs.
Studying EV glycoproteins
The above examples describe the parallel analysis of whole EV glycomes or the broad subclasses thereof. Characterising glycan content this way is revealing but there remains a need for single glycoprotein studies to elucidate the role of said glycans within EV biology – whether or not a specific structure is important in vesiculogenesis, for example. If the EV sample has been subjected to proteomics then glycoprotein identification can be inferred through bioinformatics approaches that search the dataset for N- and O-glycosylation sequons. Elsewise this must be achieved experimentally. A practical example of identifying EV glycoproteins for further research comes from the Mahal group [40]. Herein, DSA lectin affinity capture was used to specifically purify glycoproteins with oligolactosamine-rich complex type glycan structures from Sk-Mel-5 exosomes. Subsequent identification using gel electrophoresis and MS confirmed the enrichment of G3BP, ADAM10 and EWI-2 glycoproteins, compared with total cell membrane samples. The group then used these proteins as markers to investigate the role of these complex type N-glycans as sorting signals for exosomal proteins. They found that ablating the glycan processing capability of Sk-Mel-5 cells with the chemical inhibitor DMJ caused decreased recruitment of the triglycosylated EWI-2. This effect was further dissected through the construction of EWI-2 mutants lacking one, two or all three native N-glycosylation sequons. EWI-2 recruitment was reduced by 50%–70% depending on the number of glycosylation sites removed. This dependency could imply an avidity of multiple glycans in EWI-2 recruitment but the glycosylation profile of EWI-2 was not examined and the specific molecular interactions therefore remain unclear. A Vesiclepedia search confirms that EWI-2 is present in exosomes from a range of sources [69] but it is not known whether this mechanism is specific to a Sk-Mel-5 context or more universal. Conversely, the study also found that DMJ treatment resulted in increased G3BP recruitment. Together these findings indicate divergent mechanisms of glycan-based recruitment – a nuance that would not have emerged using glycomics alone.
A global glycoprotein screen can instead be performed by adapting the high resolution methods discussed above. Under the title of “glycoproteomics”, glycopeptides are instead analysed following protease digestion and so information of the glycosylation site is retained, in contrast to analyses of released glycans. A representative glycoproteomic study comes from Saraswat et al., whereby a systematic analysis of uEVs identified 37 separate glycoproteins from 125 glycopeptides [70]. Here, detergent-solubilised exosomes were subjected to protease digestion to generate a pool of glycopeptides. These were then enriched using lectin affinity, and analysed by UPLC-MS/MS. The obtained fragmentation spectra were then used to calculate both glycan composition and peptide sequence. These peptides are used to identify the parental proteins, resulting in a list of glycoproteins mapped with the putative N-glycans. Data analysis here was entirely automated using the GlycopeptideID software. This tool matches the MS spectra to those of peptide and glycan databases to give the mapped glycoproteins, scored against the probability of achieving the same result through randomised sampling [71,72]. The alternative of manual data analysis is a laborious undertaking that can require months of interpretation. As such, this study well illustrates the increasing accessibility of high resolution approaches. GlycopeptideID is a freely available, open access tool, whilst contract analyses and collaborations offer data generation solutions to researchers without the requisite MS instrumentation and experience. The glycoproteins identified in the study are of diverse function and cellular localisation, including both extracellular and organelle bound proteins. A high degree of fucosylation was also observed. As the majority of fucosyltransferases are localised to the Golgi apparatus, this indicates a Golgi to endosome trafficking mechanism for cargo proteins.
Having identified glycoproteins of interest then widely used Western blot and flow cytometry techniques are appropriate for more in-depth analyses. However, whereby antibodies are the best-in-class reagents for protein recognition, their applicability to the study of glycosylation is diminished. Antibodies against N-glycans recognise families of related glycan structures as opposed to specific epitopes and monoclonal N-glycan antibodies are also exceedingly rare [73]. Indeed, in a recently compiled database of anti-glycan reagents only 25 of a total 1116 entries correspond to N-glycan antibodies [73]. In place of antibodies labelled lectins can be used in much the same manner for binding assays, though extra consideration must be given to the multimeric nature of some lectins and their weaker binding strengths. Less structurally diverse O-linked glycans and blood group antigens and more tractable towards antibody recognition and a greater repertoire of reagents is available for these. O-GlcNAc specific antibodies in particular have been used to probe the glycoproteins of EVs in colorectal cancer cell lines [74,75]. This modification is an O-linked monosaccharide addition of N-acetylglucosamine (GlcNAc) that is typically involved in nutrient sensing and cellular homeostasis but has also been implicated in cancer [76]. Through immunoblotting a heterogeneous panel of EV proteins that displayed uncharacteristically elevated O-GlcNAcylation was identified, including the significant hnRNPA2/B1 ribonucleoprotein complex responsible for loading of exosomes with miRNA [77]. Crucially, an independent study has also demonstrated that ablating O-GlcNAcylation of the cytosolic chaperone protein cryAB prevents its packaging into exosomes [78]. These findings have yet to be integrated but could reveal the presence of O-GlcNAc as another potential mechanism of glycosylation dependent cargo recruitment. Moreover, the current paradigm is that SUMOylation directs hnRNPA2/B1 recruitment [77]. There is perhaps a more complex interplay of posttranslational modifications that bears further investigation.
In summary, there are a great many tools available to study glycoproteins and there are numerous candidate molecules that warrant further study through use of these. Tetraspanins, for example, are classical exosomal markers with a defined role in exosomes biogenesis [79] whilst integrins have been shown to dictate the organotropic metastatic effect of cancer exosomes [22]. Most members of these protein families are glycosylated but whether or not these glycan are functional in the context of EVs has not been examined. In the case of integrins, glycans may affect protein conformation and therefore binding activity [80] – presenting a potential mechanism for glycan potentiated targeting of EVs to cells. Investigating the glycosylation of EV proteins is a clear source of opportunity.
Clinical perspectives on EV glycosylation and the potential for glycoengineering
As of 2015, there are seven ongoing clinical trials for EV products [23], with many more likely as the market for exosomes diagnostics and therapeutics is projected to grow from $16.1 million in 2016 to $111.8 million in 2021 [81]. If glycan interactions are indeed essential to the uptake of EVs by recipient cells or for physicochemical characteristics, there will be ramifications for this burgeoning industry as changes to EV glycosylation could therefore impact upon the efficacies of candidate therapeutics. This is especially problematic considering a switch from small scale, laboratory isolation of EVs to mass production. The established recombinant protein sector has found that a change to production processes causes a change to glycosylation of the protein product [82] and this probably applies to EV production. Moreover, whilst it is acknowledged that the protein and RNA content of EVs shifts with changing conditions and cellular stresses [83], this idea is less recognised in terms of glycosylation and may go unmonitored. In summary, pharmaceutical companies may be reducing the efficacies or half-lives of their EV products during development without understanding why. Although accounting for glycosylation is another grand challenge in an already difficult development landscape, it behooves therapy developers to address this issue as early as possible and implement some form of glycome monitoring quality control throughout the development process. It is also entirely feasible that glycome characterisation could become a regulatory requirement for EV therapies, akin to the release testing of protein biotherapeutics.
Although the role of glycoconjugates in EV biology is still being explored novel technologies that utilise EV glycosylation are already emerging. Lectin affinity chromatography has been used in the glycomics analysis of exosomes [54] and heparin-conjugated agarose beads have been used to purify vesicles from cell culture media [84], similar to the STL-streptavidin beads above [48]. Glycosylation-targeted capture of exosomes is also the basis for an upcoming plasmapheresis device, Aethlon’s hemopurifier. The hemopurifier is a hollow fibre cartridge functionalised with lectins and antibodies, to be used with dialysis systems for the extracorporeal capture of circulating exosomes from blood [85]. Tumour-derived exosomes are directly immunosuppressive in some contexts [86] and can also sequester immunotherapeutic antibodies through antigen display [87]. Depletion of tumour exosomes may therefore assist in the treatment of cancer [85]. Hemopurifier tolerance has been proven using the lectin GNA to capture hepatitis C virus particles and an ongoing trial to assess the feasibility of targeting cancer exosomes [88] means that this device may be close to entering the clinic.
Modulating the glycosylation of EVs and their cargo molecules is also an area of interest. Glycoengineering is a field in its own right and has been used to alter the stability and pharmacokinetics of protein biopharmaceuticals [89,90]. However, proof of concept for EV glycoengineering has now been shown though engineering of the exosomal membrane protein Lamp2b [91]. Targeting peptides fused to Lamp2b proteins have previously been used to target neurons [92] and breast cancer cells [93] but Hung & Leonard observed that the efficiency of peptide display varies greatly due to degradation by lysosomal proteolysis [91]. By appending an N-glycosylation sequon to the N terminus of Lamp2b constructs, peptides were protected against degradation and fusion protein expression was rescued without adversely affecting target binding capabilities [91]. Indeed, glycosylation was even shown to improve exosomes delivery twofold in the neuronally targeted system. This approach thus represents a broadly applicable strategy for improving the uptake of peptide-targeted vesicles. Regarding therapeutic glycan cargoes, exosomal delivery of glycosphingolipids has been posited as an intervention against Alzheimer’s disease [94]. Briefly, intracerebral applications of neuroblastoma-derived exosomes to Alzheimer mouse models resulted in significant reductions of β-amyloid protein levels and plaque formations. MALDI-TOF MS revealed these exosomes as highly enriched in glycosphingolipids and further investigation suggests these glycans as the specific agents of β-amyloid scavenging. With ongoing efforts to load exosomes with lipid cargoes through fusion with liposomes [95], therapeutic glycolipid cargoes possess great potential.
These few examples represent the major EV glycoengineering efforts to date and provide an encouraging platform for future work. Extrapolating the findings discussed in this review, engineered glycosylation could be used to manipulate cargo protein recruitment [40] or provide novel targeting strategies [22,42]. The physicochemical properties of EVs may also be modulated through manipulation of sialic acid residues, as excess sialylation has been shown to modify vesicle charge [96]. These ideas are currently conjecture but there is a large body of glycoengineering literature that can be applied to EVs, including the techniques of metabolic oligosaccharide engineering [97] or the transfection of producing cells with exogenous glycan-processing enzymes [98,99]. Indeed, proof of concept for the latter in an EV system has been established in prokaryotes. Glycosyltransferase gene loci from a variety of bacterial species have been introduced into non-pathogenic Escherichia coli to produce outer membrane vesicle vaccines exhibiting glycans of the donor species [100]. That these foreign glycans are readily incorporated into the vesicles is encouraging indeed for the approach and, considering the sum of these early applications, an exciting future for EV glycoengineering is certain.
Conclusions
The role of glycans in EV biology is less understood than that of proteins, lipids and nucleic acids. Fortunately, there is an expanding body of literature that details the glycan composition of vesicles from a range of sources. This knowledge is already producing technological advances and appears to be a novel source of EV biomarkers [28], although validation of these is ongoing. Those EV researchers who are interested in probing EV glycosylation should first define the scope of their research question – whether complete characterisation is warranted or it is sufficient to analyse a given subclass of glycoconjugate. In the former case, lectin microarrays can assay glycosylation on proteins and lipids in parallel with a sensitivity that is adequate for small sample volumes and with sufficient reproducibility for comparative analyses in disease marker studies. For more in-depth structural analyses then high resolution methods are suggested, however it must again be highlighted that specific glycan subclasses must be targeted and experimental workflows differ for these. Both approaches should be viewed as complementary and can be useful in validating the findings of each other. Characterisation alone is not sufficient to elucidate the elements of EV biology that are encoded in glycosylation. More functional studies are needed to bridge these.
The current review describes a general toolbox for EV researchers interested in probing glycosylation. This is not an exhaustive survey and mostly focuses on approaches for studying bulk glycosylation profiles and individual glycoproteins. In a reflection of the current literature for EV glycomics, less prominence is given to the glycoconjugate classes of proteoglycans and glycolipids. Proteoglycans in particular have been implicated in EV biogenesis and cellular attachment [43,44,101,102] but with current technologies it is difficult to characterise the glycosaminoglycan chains of these molecules due to high size heterogeneity and physicochemical characteristics that are not favourable towards MS [103]. Glycolipids, meanwhile, are more amenable towards high-resolution analyses but beyond characterisation of general glycolipids classes, there are few studies examining their function in EVs or presenting structural glycolipidomics. However, the findings of these publications are potentially influential and more attention should be given to EV glycolipids. A notable UHPLC-MS/MS study has given proof of concept for EV glycolipids as biomarkers as they found that levels of short-chain sulfatides were significantly elevated in the exosome fraction of plasma from MS patients when compared with control samples [104]. Wider validation studies are required to determine the robustness of this marker but extending this approach to other diseases could be fruitful. Finally, complimentary and emerging techniques for studying glycosylation have not been discussed, with NMR and Raman spectroscopy showing promise here [105–107].
An open question for the field of EV glycomics is whether a given sample will be uniformly glycosylated or whether intervesicle differences are important. For example, total EV population analyses are the norm for the studies described herein whereas stratification of vesicle samples into subpopulations may reveal a more nuanced glycosylation landscape. Single vesicle glycan analyses are an attractive concept in this regard but glycomics methods have not yet reached sufficient sensitivity to achieve this. The hypothesis of glycan subpopulations is supported by a recent publication from the Griffin group showing different glycan profiles for uEVs isolated through either tetraspanin-targeted antibodies or a broad mixture of lectins [108]. This research links to a broader question on the impact of isolation methods on EV samples. To elaborate, many comparison studies have been performed to systematically examine the impact of methods such as ultracentrifugation, chromatography, ultrafiltration and commercial isolation kits on the protein and RNA cargoes of EVs [109–111] but these have not addressed exosome glycosylation. Lectin-based purifications could confer a bias in this regard but for other methods the effect of glycosylation on exosome isolation is less certain. Earlier work from the Griffin group found that uEVs isolated by ultracentrifugation, ultrafiltration and density gradient purification all exhibit similar glycosylation profiles, as assayed by lectin microarrays [47]. Further studies that employ other glycomics techniques are needed to confirm that isolation methods do not significantly impact glycan content.
EV glycobiology holds yet more open questions. In terms of quantity, the relative contribution of the different glycoconjugates to the extravesicular surface is uncertain. However, one estimate states that there are more glycosphingolipids molecules than proteins on the surface of PC-3 exosomes [58]. The fate of EV glycans is also a potential matter of interest – what happens to the glycans following uptake by recipient cells. They may be functioning as metabolic regulators, particularly in the case of O-GlcNAcylated cargo proteins. This is a nontrivial possibility as intracellular glycans have even been implicated in cell apoptosis pathways [112]. As glycomics gains greater prominence in the EV field, it is expected that these questions and more will be answered.
Responsible Editor Emanuele Cocucci, Ohio State University, USA
Funding Statement
This work has been funded by the Ramón Areces Foundation to JMF and is co-supported by CIC bioGUNE and CIC biomaGUNE.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1]. Yáñez-Mó M, Siljander PR-M, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLOS Biol. 2012;10:e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Chargaff E, West R.. The biological significance of the thromboplastic protein of blood. J Biol Chem. 1946;166:189–14. [PubMed] [Google Scholar]
- [4]. Cocucci E, Racchetti G, Meldolesi J.. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. [DOI] [PubMed] [Google Scholar]
- [5]. Harding C, Stahl P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem Biophys Res Commun. 1983;113:650–658. [DOI] [PubMed] [Google Scholar]
- [6]. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. [DOI] [PubMed] [Google Scholar]
- [7]. Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. [DOI] [PubMed] [Google Scholar]
- [9]. Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- [10]. Kim D-K, Kang B, Kim OY, et al. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013;2:20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Ribeiro MF, Zhu H, Millard RW, et al. Exosomes function in pro- and anti-angiogenesis. Curr Angiogenesis. 2013;2:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Cui Y, Luan J, Li H, et al. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett. 2016;590:185–192. [DOI] [PubMed] [Google Scholar]
- [13]. Schiera G, Di Liegro CM, Di Liegro I. Extracellular membrane vesicles as vehicles for brain cell-to-cell interactions in physiological as well as pathological conditions. BioMed Res Int. 2015;2015:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Sung BH, Ketova T, Hoshino D, et al. Directional cell movement through tissues is controlled by exosome secretion. Nat Commun. 2015;6:7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Alexander M, Hu R, Runtsch MC, et al. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun. 2015;6:7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Thompson AG, Gray E, Heman-Ackah SM, et al. Extracellular vesicles in neurodegenerative disease - pathogenesis to biomarkers. Nat Rev Neurol. 2016;12:346–357. [DOI] [PubMed] [Google Scholar]
- [17]. Lawson C, Vicencio JM, Yellon DM, et al. Microvesicles and exosomes: new players in metabolic and cardiovascular disease. J Endocrinol. 2016;228:R57–R71. [DOI] [PubMed] [Google Scholar]
- [18]. Becker A, Thakur BK, Weiss JM, et al. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Dos Anjos Pultz B, Andrés Cordero da Luz F, Socorro Faria S, et al. The multifaceted role of extracellular vesicles in metastasis: priming the soil for seeding. Int J Cancer. 2017;140:2397–2407. [DOI] [PubMed] [Google Scholar]
- [20]. Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Liu Y, Gu Y, Han Y, et al. Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell. 2016;30:243–256. [DOI] [PubMed] [Google Scholar]
- [22]. Hoshino A, Costa-Silva B, Shen T-L, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Lener T, Gimona M, Aigner L, et al. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J Extracell Vesicles. 2015;4:30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Johnson GD, Mackie P, Jodar M, et al. Chromatin and extracellular vesicle associated sperm RNAs. Nucleic Acids Res. 2015;43:6847–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. [DOI] [PubMed] [Google Scholar]
- [26]. González E, Falcón-Pérez JM. Cell-derived extracellular vesicles as a platform to identify low-invasive disease biomarkers. Expert Rev Mol Diagn. 2015;15:907–923. [DOI] [PubMed] [Google Scholar]
- [27]. Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med. 2013;7:769–778. [DOI] [PubMed] [Google Scholar]
- [28]. Costa J. Glycoconjugates from extracellular vesicles: structures, functions and emerging potential as cancer biomarkers. Biochim Biophys Acta. 2017;1868:157–166. [DOI] [PubMed] [Google Scholar]
- [29]. Ghazarian H, Idoni B, Oppenheimer SB. A glycobiology review: carbohydrates, lectins, and implications in cancer therapeutics. Acta Histochem. 2011;113:236–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. [DOI] [PubMed] [Google Scholar]
- [31]. Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. [DOI] [PubMed] [Google Scholar]
- [33]. Aoki KF, Yamaguchi A, Okuno Y, et al. Efficient tree-matching methods for accurate carbohydrate database queries. Genome Inform Int Conf Genome Inform. 2003;14:134–143. [PubMed] [Google Scholar]
- [34]. Schnaar RL, Suzuki A, Stanley P. Glycosphingolipids In: Varki A, Cummings RD, Esko JD, et al editors. Essentials of glycobiology. [Internet]. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. [cited 2017 August3]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1909/ [Google Scholar]
- [35]. Zaia J. Mass spectrometry and glycomics. OMICS J Integr Biol. 2010;14:401–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Hirabayashi J, Yamada M, Kuno A, et al. Lectin microarrays: concept, principle and applications. Chem Soc Rev. 2013;42:4443–4458. [DOI] [PubMed] [Google Scholar]
- [37]. Cummings RD, Etzler ME. Antibodies and lectins in glycan analysis In: Varki A, Cummings RD, Esko JD, et al editors. Essentials of glycobiology. [Internet]. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. [cited 2017 August3]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1919/ [Google Scholar]
- [38]. Krishnamoorthy L, Bess JW, Preston AB, et al. HIV-1 and microvesicles from T-cells share a common glycome, arguing for a common origin. Nat Chem Biol. 2009;5:244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Batista BS, Eng WS, Pilobello KT, et al. Identification of a conserved glycan signature for microvesicles. J Proteome Res. 2011;10:4624–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Liang Y, Eng WS, Colquhoun DR, et al. Complex N-linked glycans serve as a determinant for exosome/microvesicle cargo recruitment. J Biol Chem. 2014;289:32526–32537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Kwon S-H, Oh S, Nacke M, et al. Adaptor protein CD2AP and L-type lectin LMAN2 regulate exosome cargo protein trafficking through the golgi complex. J Biol Chem. 2016;291:25462–25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Shimoda A, Tahara Y, Sawada S-I, et al. Glycan profiling analysis using evanescent-field fluorescence-assisted lectin array: importance of sugar recognition for cellular uptake of exosomes from mesenchymal stem cells. Biochem Biophys Res Commun. 2017;491:701–707. [DOI] [PubMed] [Google Scholar]
- [43]. Baietti MF, Zhang Z, Mortier E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. [DOI] [PubMed] [Google Scholar]
- [44]. Roucourt B, Meeussen S, Bao J, et al. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015;25:412–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Gerlach JQ, Griffin MD. Getting to know the extracellular vesicle glycome. Mol Biosyst. 2016;12:1071–1081. [DOI] [PubMed] [Google Scholar]
- [46]. Pilobello KT, Krishnamoorthy L, Slawek D, et al. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem Eur J Chem Biol. 2005;6:985–989. [DOI] [PubMed] [Google Scholar]
- [47]. Gerlach JQ, Krüger A, Gallogly S, et al. Surface glycosylation profiles of urine extracellular vesicles. PLoS One. 2013;8:e74801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Echevarria J, Royo F, Pazos R, et al. Microarray-based identification of lectins for the purification of human urinary extracellular vesicles directly from urine samples. Chembiochem Eur J Chem Biol. 2014;15:1621–1626. [DOI] [PubMed] [Google Scholar]
- [49]. Etxebarria J, Calvo J, Martin-Lomas M, et al. Lectin-array blotting: profiling protein glycosylation in complex mixtures. ACS Chem Biol. 2012;7:1729–1737. [DOI] [PubMed] [Google Scholar]
- [50]. Krishnamoorthy L, Mahal LK. Glycomic analysis: an array of technologies. ACS Chem Biol. 2009;4:715–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Ruhaak LR, Zauner G, Huhn C, et al. Glycan labeling strategies and their use in identification and quantification. Anal Bioanal Chem. 2010;397:3457–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Reiding KR, Ruhaak LR, Uh H-W, et al. Human plasma N-glycosylation as analyzed by matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance-MS associates with markers of inflammation and metabolic health. Mol Cell Proteomics MCP. 2017;16:228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Alley WR, Novotny MV. Structural glycomic analyses at high sensitivity: a decade of progress. Annu Rev Anal Chem Palo Alto Calif. 2013;6:237–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Escrevente C, Grammel N, Kandzia S, et al. Sialoglycoproteins and N-glycans from secreted exosomes of ovarian carcinoma cells. PloS One. 2013;8:e78631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Gomes J, Gomes-Alves P, Carvalho SB, et al. Extracellular vesicles from ovarian carcinoma cells display specific glycosignatures. Biomolecules. 2015;5:1741–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56]. Nyalwidhe JO, Betesh LR, Powers TW, et al. Increased bisecting N-acetylglucosamine and decreased branched chain glycans of N-linked glycoproteins in expressed prostatic secretions associated with prostate cancer progression. Proteomics Clin Appl. 2013;7:677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Staubach S, Schadewaldt P, Wendel U, et al. Differential glycomics of epithelial membrane glycoproteins from urinary exovesicles reveals shifts toward complex-type N-glycosylation in classical galactosemia. J Proteome Res. 2012;11:906–916. [DOI] [PubMed] [Google Scholar]
- [58]. Llorente A, Skotland T, Sylvänne T, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 2013;1831:1302–1309. [DOI] [PubMed] [Google Scholar]
- [59]. Skotland T, Sandvig K, Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog Lipid Res. 2017;66:30–41. [DOI] [PubMed] [Google Scholar]
- [60]. Trajkovic K, Hsu C, Chiantia S, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. [DOI] [PubMed] [Google Scholar]
- [61]. Phuyal S, Hessvik NP, Skotland T, et al. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J. 2014;281:2214–2227. [DOI] [PubMed] [Google Scholar]
- [62]. Lisacek F, Mariethoz J, Alocci D, et al. Databases and associated tools for glycomics and glycoproteomics. Methods Mol Biol Clifton NJ. 2017;1503:235–264. [DOI] [PubMed] [Google Scholar]
- [63]. Ceroni A, Maass K, Geyer H, et al. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 2008;7:1650–1659. [DOI] [PubMed] [Google Scholar]
- [64]. Campbell MP, Nguyen-Khuong T, Hayes CA, et al. Validation of the curation pipeline of UniCarb-DB: building a global glycan reference MS/MS repository. Biochim Biophys Acta. 2014;1844:108–116. [DOI] [PubMed] [Google Scholar]
- [65]. Pagel K, Harvey DJ. Ion mobility-mass spectrometry of complex carbohydrates: collision cross sections of sodiated N-linked glycans. Anal Chem. 2013;85:5138–5145. [DOI] [PubMed] [Google Scholar]
- [66]. Hofmann J, Hahm HS, Seeberger PH, et al. Identification of carbohydrate anomers using ion mobility-mass spectrometry. Nature. 2015;526:241–244. [DOI] [PubMed] [Google Scholar]
- [67]. Etxebarria J, Reichardt N-C. Methods for the absolute quantification of N-glycan biomarkers. Biochim Biophys Acta. 2016;1860:1676–1687. [DOI] [PubMed] [Google Scholar]
- [68]. Wang Z, Chinoy ZS, Ambre SG, et al. A general strategy for the chemoenzymatic synthesis of asymmetrically branched N-glycans. Science. 2013;341:379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69]. Vesiclepedia EWI2 gene summary [Internet]. [cited 2017 March15]. Available from: http://microvesicles.org/gene_summary?gene_id=93185
- [70]. Saraswat M, Joenväära S, Musante L, et al. N-linked (N-) glycoproteomics of urinary exosomes. Mol Cell Proteomics MCP. 2015;14:2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71]. Joenväärä S, Ritamo I, Peltoniemi H, et al. N-glycoproteomics - an automated workflow approach. Glycobiology. 2008;18:339–349. [DOI] [PubMed] [Google Scholar]
- [72]. Peltoniemi H, Natunen S, Ritamo I, et al. Novel data analysis tool for semiquantitative LC-MS-MS2 profiling of N-glycans. Glycoconj J. 2013;30:159–170. [DOI] [PubMed] [Google Scholar]
- [73]. Sterner E, Flanagan N, Gildersleeve JC. Perspectives on anti-glycan antibodies gleaned from development of a community resource database. ACS Chem Biol. 2016;11:1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74]. Phueaouan T, Chaiyawat P, Netsirisawan P, et al. Aberrant O-GlcNAc-modified proteins expressed in primary colorectal cancer. Oncol Rep. 2013;30:2929–2936. [DOI] [PubMed] [Google Scholar]
- [75]. Chaiyawat P, Weeraphan C, Netsirisawan P, et al. Elevated O-GlcNAcylation of extracellular vesicle proteins derived from metastatic colorectal cancer cells. Cancer Genomics Proteomics. 2016;13:387–398. [PMC free article] [PubMed] [Google Scholar]
- [76]. Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011;11:678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77]. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78]. Kore RA, Abraham EC. Phosphorylation negatively regulates exosome mediated secretion of cryAB in glioma cells. Biochim Biophys Acta. 2016;1863:368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Janik ME, Lityńska A, Vereecken P. Cell migration-the role of integrin glycosylation. Biochim Biophys Acta. 2010;1800:545–555. [DOI] [PubMed] [Google Scholar]
- [81]. Research and Markets Exosome diagnostics and therapeutics: global markets [Internet] [cited 2017 March15]. Available from: http://www.researchandmarkets.com/reports/3633268/exosome-diagnostics-and-therapeutics-global
- [82]. Costa AR, Rodrigues ME, Henriques M, et al. Glycosylation: impact, control and improvement during therapeutic protein production. Crit Rev Biotechnol. 2014;34:281–299. [DOI] [PubMed] [Google Scholar]
- [83]. De Jong OG, Verhaar MC, Chen Y, et al. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012;1:18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84]. Balaj L, Atai NA, Chen W, et al. Heparin affinity purification of extracellular vesicles. Sci Rep. 2015;5:10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85]. Marleau AM, Chen C-S, Joyce JA, et al. Exosome removal as a therapeutic adjuvant in cancer. J Transl Med. 2012;10:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86]. Kunigelis KE, Graner MW. The dichotomy of tumor exosomes (TEX) in cancer immunity: is it all in the ConTEXt? Vaccines. 2015;3:1019–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87]. Ciravolo V, Huber V, Ghedini GC, et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol. 2012;227:658–667. [DOI] [PubMed] [Google Scholar]
- [88]. UC Irvine Health Plasma exosome concentration in cancer patients undergoing treatment [Internet]. [cited 2017 March15]. Available from: http://www.ucirvinehealth.org/clinical-trials/cancer-clinical-trial/breast-female-breast-male-colon-lip-oral-cavity-and-pharynx-liver-lung-melanoma-skin-other-digestiv/plasma-exosome-concentration-in-cancer-patients-undergoing-treatment/
- [89]. Solá RJ, Griebenow K. Glycosylation of therapeutic proteins: an effective strategy to optimize efficacy. BioDrugs Clin Immunother Biopharm Gene Ther. 2010;24:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90]. Solá RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci. 2009;98:1223–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91]. Hung ME, Leonard JN. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem. 2015;290:8166–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92]. Alvarez-Erviti L, Seow Y, Yin H, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. [DOI] [PubMed] [Google Scholar]
- [93]. Tian Y, Li S, Song J, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35:2383–2390. [DOI] [PubMed] [Google Scholar]
- [94]. Yuyama K, Sun H, Sakai S, et al. Decreased amyloid-β pathologies by intracerebral loading of glycosphingolipid-enriched exosomes in Alzheimer model mice. J Biol Chem. 2014;289:24488–24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95]. Sato YT, Umezaki K, Sawada S, et al. Engineering hybrid exosomes by membrane fusion with liposomes. Sci Rep. 2016;6:21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96]. Akagi T, Kato K, Kobayashi M, et al. On-chip immunoelectrophoresis of extracellular vesicles released from human breast cancer cells. PloS One. 2015;10:e0123603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97]. Sminia TJ, Zuilhof H, Wennekes T. Getting a grip on glycans: a current overview of the metabolic oligosaccharide engineering toolbox. Carbohydr Res. 2016;435:121–141. [DOI] [PubMed] [Google Scholar]
- [98]. Baker JL, Çelik E, DeLisa MP. Expanding the glycoengineering toolbox: the rise of bacterial N-linked protein glycosylation. Trends Biotechnol. 2013;31:313–323. [DOI] [PubMed] [Google Scholar]
- [99]. Wang L-X, Lomino JV. Emerging technologies for making glycan-defined glycoproteins. ACS Chem Biol. 2012;7:110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100]. Price NL, Goyette-Desjardins G, Nothaft H, et al. Glycoengineered outer membrane vesicles: a novel platform for bacterial vaccines. Sci Rep. 2016;6:24931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101]. Christianson HC, Svensson KJ, van Kuppevelt TH, et al. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci U S A. 2013;110:17380–17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102]. Chen L, Brigstock DR. Integrins and heparan sulfate proteoglycans on hepatic stellate cells (HSC) are novel receptors for HSC-derived exosomes. FEBS Lett. 2016;590:4263–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103]. Ly M, Laremore TN, Linhardt RJ. Proteoglycomics: recent progress and future challenges. OMICS J Integr Biol. 2010;14:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104]. Moyano AL, Li G, Boullerne AI, et al. Sulfatides in extracellular vesicles isolated from plasma of multiple sclerosis patients. J Neurosci Res. 2016;94:1579–1587. [DOI] [PubMed] [Google Scholar]
- [105]. Brisson J-R, Vinogradov E, McNally DJ, et al. The application of NMR spectroscopy to functional glycomics. Methods Mol Biol Clifton NJ. 2010;600:155–173. [DOI] [PubMed] [Google Scholar]
- [106]. Irague R, Massou S, Moulis C, et al. NMR-based structural glycomics for high-throughput screening of carbohydrate-active enzyme specificity. Anal Chem. 2011;83:1202–1206. [DOI] [PubMed] [Google Scholar]
- [107]. Vangala K, Yanney M, Hsiao C-T, et al. Sensitive carbohydrate detection using surface enhanced Raman tagging. Anal Chem. 2010;82:10164–10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108]. Gerlach JQ, Maguire CM, Krüger A, et al. Urinary nanovesicles captured by lectins or antibodies demonstrate variations in size and surface glycosylation profile. Nanomed. 2017;12:1217–1229. [DOI] [PubMed] [Google Scholar]
- [109]. Royo F, Zuñiga-Garcia P, Sanchez-Mosquera P, et al. Different EV enrichment methods suitable for clinical settings yield different subpopulations of urinary extracellular vesicles from human samples. J Extracell Vesicles. 2016;5:29497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110]. Andreu Z, Rivas E, Sanguino-Pascual A, et al. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J Extracell Vesicles [Internet]. 2016;5 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4916259/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111]. Szatanek R, Baran J, Siedlar M, et al. Isolation of extracellular vesicles: determining the correct approach. Int J Mol Med. 2015;36:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112]. Lichtenstein RG, Rabinovich GA. Glycobiology of cell death: when glycans and lectins govern cell fate. Cell Death Differ. 2013;20:976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]


