Abstract
Background
To produce 1-propanol as a potential biofuel, metabolic engineering of microorganisms, such as E. coli, has been studied. However, 1-propanol production using metabolically engineered Saccharomyces cerevisiae, which has an amazing ability to produce ethanol and is thus alcohol-tolerant, has infrequently been reported. Therefore, in this study, we aimed to engineer S. cerevisiae strains capable of producing 1-propanol at high levels.
Results
We found that the activity of endogenous 2-keto acid decarboxylase and alcohol/aldehyde dehydrogenase is sufficient to convert 2-ketobutyrate (2 KB) to 500 mg/L 1-propanol in yeast. Production of 1-propanol could be increased by: (i) the construction of an artificial 2 KB biosynthetic pathway from pyruvate via citramalate (cimA); (ii) overexpression of threonine dehydratase (tdcB); (iii) enhancement of threonine biosynthesis from aspartate (thrA, thrB and thrC); and (iv) deletion of the GLY1 gene that regulates a competing pathway converting threonine to glycine. With high-density anaerobic fermentation of the engineered S. cerevisiae strain YG5C4231, we succeeded in producing 180 mg/L 1-propanol from glucose.
Conclusion
These results indicate that the engineering of a citramalate-mediated pathway as a production method for 1-propanol in S. cerevisiae is effective. Although optimization of the carbon flux in S. cerevisiae is necessary to harness this pathway, it is a promising candidate for the large-scale production of 1-propanol.
Electronic supplementary material
The online version of this article (10.1186/s12934-018-0883-1) contains supplementary material, which is available to authorized users.
Keywords: 1-Propanol, Yeast, S. cerevisiae, Fermentation, 2-Ketobutyrate
Background
As a means of mitigating environmental issues, such as global warming and the depletion of fossil fuels, biofuels and products from sustainable biomass resources have received significant attention in recent years. In particular, alcohols have been extensively studied, since they are already available as next-generation fuels and represent the building blocks of other chemicals. In this study, we focused on 1-propanol, which is generically used as a solvent and as a food additive, is found in paint and cosmetics, and is a chemical intermediate in the production of n-propylamine [1]. However, a natural microbial producer of 1-propanol has yet to be identified.
The budding yeast Saccharomyces cerevisiae is likely to be a good candidate for the production of 1-propanol. As S. cerevisiae has been used to produce ethanol, it is clear that the strain has tolerance to high concentrations of alcohols and other stresses during fermentation [2, 3], properties that should be useful for the industrial production of 1-propanol. Furthermore, S. cerevisiae strains that utilize not only glucose, but also xylose, as a sugar substrate have been developed to expand the range of applications of this biomass [4]. For these reasons, S. cerevisiae may be more suitable for the production of 1-propanol than other microbial hosts.
Recently, it has been reported that the production of various alcohols from α-keto acids can be achieved using 2-keto acid decarboxylase (KDC) and alcohol/aldehyde dehydrogenase (ADH) in conjunction with metabolic engineering [5]. Using this method, the authors demonstrated the production of 2.1 mM of 1-propanol from 8 g/L (78.3 mM) of 2-ketobutyrate (2 KB) in E. coli [5]. It is also possible therefore, to convert 2 KB produced from threonine into 1-propanol by reaction with KDC and ADH in S. cerevisiae (Fig. 1a). E. coli primarily produce 2 KB from threonine; however, they can learn to produce 2 KB from pyruvate via citramalate once the citramalate synthase (cimA) gene is introduced. Using this approach, the production of up to 4.5 g/L of 1-propanol has been demonstrated in E. coli [6]. A similar strategy would be expected to produce 2 KB from pyruvate via citramalate in yeast, by introducing both citramalate synthase (cimA) and methylmalate dehydratase (leuC and leuD) and overexpressing isopropyl malate dehydrogenase (LEU2) (Fig. 1b).
Fig. 1.
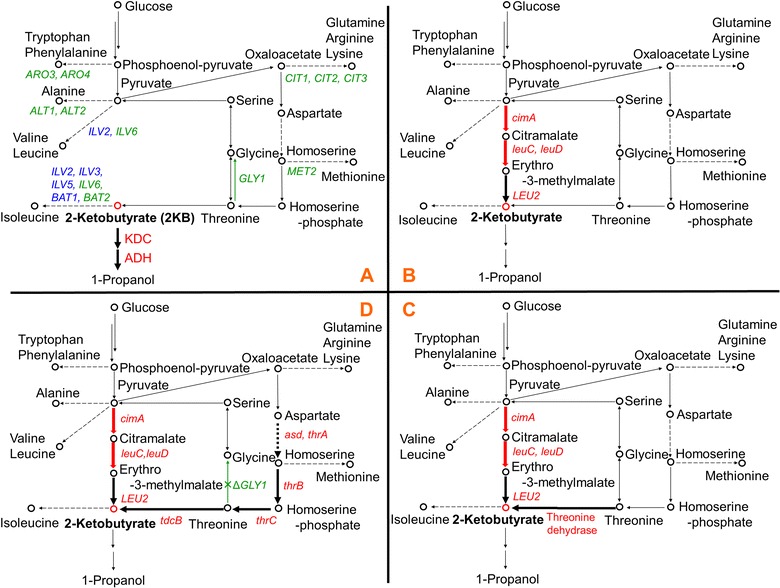
Pathways for 1-propanol production in S. cerevisiae. a–d show different theoretical methods to achieve production of this metabolite. Red letters indicate genes that are overexpressed. Blue and green letters indicate genes that are deleted. ARO3 and ARO4 encode 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase. ALT1 and ALT2 encode alanine transaminase. CIT1, CIT2 and CIT3 encode citrate synthase. MET2 encodes l-homoserine-O-acetyltransferase. GLY1 encodes threonine aldolase. ILV2 and ILV6 encode acetolactate synthase. ILV3 encodes dihydroxyacid dehydratase. ILV5 encodes acetohydroxyacid reductoisomerase. BAT1 and BAT2 encode branched-chain amino acid transaminase. cimA encodes citramalate synthase. leuC and leuD encode citramalate hydrolyase. LEU2 encodes 3-isopropylmalate dehydrogenase. asd encodes aspartate-semialdehyde dehydrogenase. thrA encodes aspartokinase and homoserine dehydrogenase I. thrB encodes homoserine kinase. thrC encodes threonine synthase. tdcB encodes threonine dehydratase
Furthermore, others have reported that deleting the acetolactate synthase gene (ILV2), which directs a competing biochemical pathway, results in the production of 60 mg/L of 1-propanol in yeast [7]. Therefore, in the present study, we aimed to develop a metabolically engineered S. cerevisiae strain suitable for the production of 1-propanol utilizing a combination of these approaches, specifically the overexpression of genes for 1-propanol biosynthesis, together with deletion of the competing metabolic pathway.
Methods
Strains, plasmids, and primers
The yeast strains used in this study are listed in Table 1. S. cerevisiae YPH499 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1, purchased from Stratagene, La Jolla, CA, USA) [8], BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the single gene deletion mutants (purchased from Thermo Scientific) [9] were used as yeast host strains. The plasmids and primers used in this study are summarized in Tables 2, 3 and Additional file 1, respectively. All plasmids were derived from the pGK and pATP vector series, in which gene expression is controlled either by the PGK1 promoter, or the ADH1, TDH1, and PGK1 promoters, respectively [10]. The cimA, leuC and leuD genes derived from Methanocaldococcus jannaschii were amplified from genomic DNA (NBRC No. 100440G, purchased from National Institute of Technology and Evaluation, Tokyo, Japan). The leuC and leuD genes derived from Clostridium beijerinckii were amplified from genomic DNA (NBRC No. 103909, purchased from National Institute of Technology and Evaluation, Tokyo, Japan). All other genes derived from S. cerevisiae and E. coli were amplified from YPH499 and BL21 (DE3) genomic DNA, respectively, using the primers shown in Table 3. The growth conditions, DNA techniques, and lithium-acetate method for transformations have been previously described [11, 12].
Table 1.
Yeast strains constructed in this study
| Strains | Genotypes |
|---|---|
| YPH499 | MATa URA3-52 LYS2-801 ADE2-101 TRP1-Δ63 HIS3-Δ200 LEU2-Δ1 |
| BY4741 | MATa HIS3Δ1 LEU2Δ0 MET15Δ0 URA3Δ0 |
| YA0K0 | YPH499/pGK426/pGK423 |
| YA0K1 | YPH499/pGK426/pGK423-kivd |
| YA0K2 | YPH499/pGK426/pGK423-ARO10 |
| YA0K3 | YPH499/pGK426/pGK423-THI3 |
| YA1K0 | YPH499/pGK426-ADH1/pGK423 |
| YA1K1 | YPH499/pGK426-ADH1/pGK423-kivd |
| YA1K2 | YPH499/pGK426-ADH1/pGK423-ARO10 |
| YA1K3 | YPH499/pGK426-ADH1/pGK423-THI3 |
| YA2K0 | YPH499/pGK426-ADH2/pGK423 |
| YA2K1 | YPH499/pGK426-ADH2/pGK423-kivd |
| YA2K2 | YPH499/pGK426-ADH2/pGK423-ARO10 |
| YA2K3 | YPH499/pGK426-ADH2/pGK423-THI3 |
| YA3K0 | YPH499/pGK426-ADH5/pGK423 |
| YA3K1 | YPH499/pGK426-ADH5/pGK423-kivd |
| YA3K2 | YPH499/pGK426-ADH5/pGK423-ARO10 |
| YA3K3 | YPH499/pGK426-ADH5/pGK423-THI3 |
| YA4K0 | YPH499/pGK426-ADH6/pGK423 |
| YA4K1 | YPH499/pGK426-ADH6/pGK423-kivd |
| YA4K2 | YPH499/pGK426-ADH6/pGK423-ARO10 |
| YA4K3 | YPH499/pGK426-ADH6/pGK423-THI3 |
| YA5K0 | YPH499/pGK426-ADH7/pGK423 |
| YA5K1 | YPH499/pGK426-ADH7/pGK423-kivd |
| YA5K2 | YPH499/pGK426-ADH7/pGK423-ARO10 |
| YA5K3 | YPH499/pGK426-ADH7/pGK423-THI3 |
| YA6K0 | YPH499/pGK426-SFA1/pGK423 |
| YA6K1 | YPH499/pGK426-SFA1/pGK423-kivd |
| YA6K2 | YPH499/pGK426-SFA1/pGK423-ARO10 |
| YA6K3 | YPH499/pGK426-SFA1/pGK423-THI3 |
| Y06C250 | YPH499/pGK406-cimA/pATP425 |
| Y06C25C | YPH499/pGK406-cimA/pATP425-leuC(Cb)-leuD(Cb) |
| Y06C25E | YPH499/pGK406-cimA/pATP425-leuC(Ec)-leuD(Ec) |
| Y06C25 M | YPH499/pGK406-cimA/pATP425-leuC(Mj)-leuD(Mj) |
| Y26C250 | YPH499/pGK426-cimA/pATP425 |
| Y26C25C | YPH499/pGK426-cimA/pATP425-leuC(Cb)-leuD(Cb) |
| Y26C25E | YPH499/pGK426-cimA/pATP425-leuC(Ec)-leuD(Ec) |
| Y26C25 M | YPH499/pGK426-cimA/pATP425-leuC(Mj)-leuD(Mj) |
| Y5040 | YPH499/pATP425/pATP424 |
| Y5041 | YPH499/pATP425/pATP424-ILV1 |
| Y5042 | YPH499/pATP425/pATP424-tdcB |
| Y5043 | YPH499/pATP425/pATP424-ilvA |
| Y5C40 | YPH499/pATP425-cimA-leuC(Cb)-leuD(Cb)/pATP424 |
| Y5C41 | YPH499/pATP425-cimA-leuC(Cb)-leuD(Cb)/pATP424-ILV1 |
| Y5C42 | YPH499/pATP425-cimA-leuC(Cb)-leuD(Cb)/pATP424-tdcB |
| Y5C43 | YPH499/pATP425-cimA-leuC(Cb)-leuD(Cb)/pATP424-ilvA |
| Y5E40 | YPH499/pATP425-cimA-leuC(Ec)-leuD(Ec)/pATP424 |
| Y5E41 | YPH499/pATP425-cimA-leuC(Ec)-leuD(Ec)/pATP424-ILV1 |
| Y5E42 | YPH499/pATP425-cimA-leuC(Ec)-leuD(Ec)/pATP424-tdcB |
| Y5E43 | YPH499/pATP425-cimA-leuC(Ec)-leuD(Ec)/pATP424-ilvA |
| Y5M40 | YPH499/pATP425-cimA-leuC(Mj)-leuD(Mj)/pATP424 |
| Y5M41 | YPH499/pATP425-cimA-leuC(Mj)-leuD(Mj)/pATP424-ILV1 |
| Y5M42 | YPH499/pATP425-cimA-leuC(Mj)-leuD(Mj)/pATP424-tdcB |
| Y5M43 | YPH499/pATP425-cimA-leuC(Mj)-leuD(Mj)/pATP424-ilvA |
| B50 | BY4741/pATP425 |
| B5C | BY4741/pATP425-cimA-leuC(Cb)-leuD(Cb) |
| B5E | BY4741/pATP425-cimA-leuC(Ec)-leuD(Ec) |
| B5M | BY4741/pATP425-cimA-leuC(Mj)-leuD(Mj) |
| BG5C | BY4741ΔGLY1/pATP425-cimA-leuC(Cb)-leuD(Cb) |
| BG5E | BY4741ΔGLY1/pATP425-cimA-leuC(Ec)-leuD(Ec) |
| BG5M | BY4741ΔGLY1/pATP425-cimA-leuC(Mj)-leuD(Mj) |
| YG5040 | YPH499ΔGLY1/pATP425/pATP424 |
| YG5C42 | YPH499ΔGLY1/pATP425-cimA-leuC(Cb)-leuD(Cb)/pATP424-tdcB |
| YG5E42 | YPH499ΔGLY1/pATP425-cimA-leuC(Ec)-leuD(Ec)/pATP424-tdcB |
| YG5M42 | YPH499ΔGLY1/pATP425-cimA-leuC(Mj)-leuD(Mj)/pATP424-tdcB |
| YG504030 | YPH499ΔGLY1/pATP425/pATP424/pATP423 |
| YG5C4231 | YPH499ΔGLY1/pATP425-cimA-leuC(Cb)-leuD(Cb)/pATP424-tdcB/pATP423-thrA-thrB-thrC |
| YG5C4232 | YPH499ΔGLY1/pATP425-cimA-leuC(Cb)-leuD(Cb)/pATP424-tdcB-asd/pATP423-thrA-thrB-thrC |
Table 2.
Plasmids used in this study
| Plasmid | Description | Source of reference |
|---|---|---|
| pGK423 | Yeast expression vector containing PGK1 promoter, 2 μ origin, HIS3 marker, no expression (control plasmid) | Ishii et al. [10] |
| pGK426 | Yeast expression vector containing PGK1 promoter, 2 μ origin, URA3 marker, no expression (control plasmid) | Ishii et al. [10] |
| pGK406 | Yeast integration vector containing PGK1 promoter, URA3 maker | Ishii et al. [10] |
| pATP425 | Yeast three gene expression vector containing ADH1, TDH3, and PGK1 promoters, 2 μ origin, LEU2 marker, no expression (control plasmid) | |
| pATP424 | Yeast three gene expression vector containing ADH1, TDH3 and PGK1 promoters, 2 μ origin, TRP1 marker, no expression (control plasmid) | |
| pATP423 | Yeast three gene expression vector containing ADH1, TDH3 and PGK1 promoters, 2 μ origin, HIS3 marker, no expression (control plasmid) | |
| pGK423-kivd | HIS3, expression of L. lactis kivd gene | Kondo et al. [14] |
| pGK423-ARO10 | HIS3, expression of S. cerevisiae ARO10 gene | Kondo et al. [14] |
| pGK423-THI3 | HIS3, expression of S. cerevisiae THI3 gene | Kondo et al. [14] |
| pGK426-ADH1 | URA3, expression of S. cerevisiae ADH1 gene | Kondo et al. [14] |
| pGK426-ADH2 | URA3, expression of S. cerevisiae ADH2 gene | Kondo et al. [14] |
| pGK426-ADH5 | URA3, expression of S. cerevisiae ADH5 gene | Kondo et al. [14] |
| pGK426-ADH6 | URA3, expression of S. cerevisiae ADH6 gene | Kondo et al. [14] |
| pGK426-ADH7 | URA3, expression of S. cerevisiae ADH7 gene | Kondo et al. [14] |
| pGK426-SFA1 | URA3, expression of S. cerevisiae SFA1 gene | Kondo et al. [14] |
| pGK426-cimA | URA3, expression of M. jannaschii cimA gene | This study |
| pGK406-cimA | URA3, genomic integration of M. jannaschii cimA gene | This study |
| pATP425-leuC(Cb)-leuD(Cb) | LEU2, co-expression of C. beijerinckii leuC and leuD genes | This study |
| pATP425-leuC(Ec)-leuD(Ec) | LEU2, co-expression of E. coli leuC and leuD genes | This study |
| pATP425-leuC(Mj)-leuD(Mj) | LEU2, co-expression of M. jannaschii leuC and leuD genes | This study |
| pATP425-cimA-leuC(Cb)-leuD(Cb) | LEU2, co-expression of M. jannaschii cimA, C. beijerinckii leuC and leuD genes | This study |
| pATP425-cimA-leuC(Ec)-leuD(Ec) | LEU2, co-expression of M. jannaschii cimA, E. coli leuC and leuD genes | This study |
| pATP425-cimA-leuC(Mj)-leuD(Mj) | LEU2, co-expression of M. jannaschii cimA, leuC and leuD genes | This study |
| pATP424-ILV1 | TRP1, expression of S. cerevisiae ILV1 gene | This study |
| pATP424-tdcB | TRP1, expression of E. coli tdcB gene | This study |
| pATP424-ilvA | TRP1, expression of E. coli ilvA gene | This study |
| pATP424-tdcB-asd | TRP1, co-expression of E. coli tdcB and asd genes | This study |
| pATP423-thrA-thrB-thrC | HIS3, co-expression of E. coli thrA, thrB and thrC genes | This study |
Table 3.
Primers used in this study
| Target gene | Primer (5′–3′) | Restriction enzyme |
|---|---|---|
| cimA | Fw; gggGGATCCatgatggtaaggatatttgatacaa | BamHI |
| Rv; cccCCCGGGttaattcaataacatattgattcct | XmaI | |
| cimA | Fw; gggCCCGGGatgatggtaaggatatttgatacaa | XmaI |
| Rv; cccGGCGCGCCttaattcaataacatattgattcct | AscI | |
| leuC(Cb) | Fw; gggGTCGACatgggaatgacaatgactcaaaaaa | SalI |
| Rv; cccCCCGGGCGGCCGCctacactaattcaggatcagttatt | NotI | |
| leuD(Cb) | Fw; gggGTCGACCCTAGGatgagtgtaaaaggtaaagtattca | AvrII |
| Rv; cccCCCGGGCCGGCCctatctatttcttatatatccaatc | FseI | |
| leuC(Ec) | Fw; gggGTCGACatggctaagacgttatacgaaaaat | SalI |
| Rv; cccCCCGGGCGGCCGCttatttaatgttgcgaatgtcggcg | NotI | |
| leuD(Ec) | Fw; gggGTCGACCCTAGGatggcagagaaatttatcaaacaca | AvrII |
| Rv; cccCCCGGGCCGGCCttaattcataaacgcaggttgtttt | FseI | |
| leuC(Mj) | Fw; gggGTCGACatgggaatgacaattgtagagaaga | SalI |
| Rv; cccCCCGGGCGGCCGCttataaatcccttgggtcaacaagt | NotI | |
| leuD(Mj) | Fw; gggGTCGACCCTAGGatgagaagtataataaagggaagag | AvrII |
| Rv; cccCCCGGGCCGGCCttattggctttcagccatctttttc | FseI | |
| ILV1 | Fw; gggGTCGACatgtcagctactctactaaagcaac | SalI |
| Rv; cccGGATCCGCGGCCGCttaatatttcaagaatttttgataa | NotI | |
| tdcB | Fw; gggGTCGACatgcatattacatacgatctgccgg | SalI |
| Rv; cccGGATCCGCGGCCGCttaagcgtcaacgaaaccggtgatt | NotI | |
| ilvA | Fw; gggGTCGACatggctgactcgcaacccctgtccg | SalI |
| Rv; cccGGATCCGCGGCCGCctaacccgccaaaaagaacctgaac | NotI | |
| asd | Fw; CCTAGGatgaaaaatgttggttttatcggctggcgc | AvrII |
| Rv; GGCCGGCCttacgccagttgacgaagcatccgacgcag | FseI | |
| thrA | Fw; GTCGACatgcgagtgttgaagttcggcggtacatca | SalI |
| Rv; GCGGCCGCtcagactcctaacttccatgagagggtacg | NotI | |
| thrB | Fw; CCTAGGatggttaaagtttatgccccggcttccagt | AvrII |
| Rv; GGCCGGCCttagttttccagtactcgtgcgcccgccgt | FseI | |
| thrC | Fw; CCCGGGatgaaactctacaatctgaaagatcacaat | XmaI |
| Rv; GGCGCGCCttactgatgattcatcatcaatttacgcaa | AscI | |
| GLY1 | Fw; TCACTTGCCATATTCGTTCACCGGTTTTTCTTTTTATTTC | |
| Rv; caatctgctctgatgccgcatagttaagccACAAAAACCCTAACAATACACATGATGCAACTGGAACGCATGTGTTTATGTTTGCGTTTGTGTGCGGGAG | ||
| URA3 | Fw; TGTATTGTTAGGGTTTTTGTggcttaactatgcggcatcagagcagattg | |
| Rv; GAAAAAAAGGAAGAGGGTAGCAATCCTAAAACAAAAACCCTAACAATACACATGATGCAACTGGAACGCAttagttttgctggccgcatcttctcaaata |
Deletion of competing pathway
GLY1 was disrupted according to the method of Akada et al. [13]. Briefly, 300 bp of the 5′-flank of GLY1 was PCR amplified with a standard forward primer, and a reverse primer containing a 20 bp sequence of the 5′-flank followed by 40 bp sequence of the 3′-flank of GLY1. Separately, the URA3 marker cassette of pGK426 was PCR amplified with a forward primer containing a 20 bp overlap of the former PCR product and a reverse primer containing a 70 bp sequence of the 3′-flank of GLY1. Both amplified fragments were mixed and combined by PCR. The final PCR product was introduced into YPH499 using the lithium acetate method and the correctly integrated transformant was selected. The URA3 marker was then eliminated by counter selection with 5-fluoroorotic acid. Disruption of GLY1 and elimination of URA3 was confirmed by diagnostic PCR to check fragment sizes. The constructed strain, which has ΔGLY1 allele, was designated YPH499 ΔGLY1. Subsequently, double deletion strains with ΔGLY1 and other (ΔARO4, ΔALT1, ΔILV6, ΔCIT1 or ΔMET2) were constructed in common with deletion of GLY1.
Fermentation of engineered strains
The transformants were cultured for 48 h at 30 °C in 5 mL of SD minimal medium (6.7 g/L yeast nitrogen base without amino acids and 20 g/L glucose) containing the required amino acids. Following centrifugation at 3000 rpm for 5 min and removal of the supernatant, yeast cells were cultured in 5 mL of fresh SD minimal medium containing the required amino acids with/without 8 g/L 2 KB. The concentration of 1-propanol in the medium 72 h after the start of fermentation was determined using GC–MS (GCMS-QP2010 Plus; Shimadzu) following a previously described procedure [14].
For oxygen-limited fermentation, yeast transformants were anaerobically cultivated in SD minimal medium containing the required amino acids for 48 h at 30 °C. The cells were collected by centrifugation at 1000g for 5 min at 4 °C and washed twice with sterile water. The cells were then placed in 50 mL of SD minimal medium. The initial cell concentration was adjusted to OD600 = 20. All fermentations were performed at 30 °C with mild agitation in 100 mL closed bottles equipped with a bubbling CO2 outlet.
Results and discussion
Overexpression of 2-keto acid decarboxylase and alcohol/aldehyde dehydrogenase
It has been reported that various alcohols can be made from α-keto acids by two-step catalytic reactions with 2-ketoacid decarboxylase (KDC) and alcohol/aldehyde dehydrogenase (ADH) [5]. Thus, 1-propanol can be produced from 2 KB that is the intermediate metabolite of isoleucine biosynthesis (Fig. 1a). In this study, we first examined the KDC and ADH enzymes that efficiently convert 2 KB to 1-propanol in S. cerevisiae (Fig. 2). We chose three KDC enzymes (phenylpyruvate decarboxylase, ARO10I, and alpha-ketoisocaproate decarboxylase, THI3, derived from S. cerevisiae; and α-ketoisovalerate decarboxylase, Kivd, derived from Lactococcus lactis) and six ADH enzymes (ADH1, 2, 5, 6, 7, and SFA1, derived from S. cerevisiae), in reference to a previous report [14], for overexpression in S. cerevisiae.
Fig. 2.
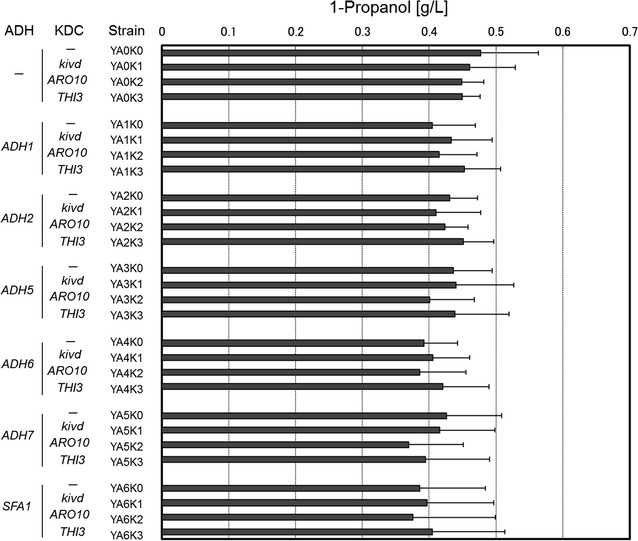
Production of 1-propanol from added 2 KB in various KDC- and ADH-overexpressing S. cerevisiae YPH499 strains
The genes encoding these KDC and ADH enzymes were co-introduced into the YPH499 yeast strain in all possible combinations, and the transformants were fermented in SD selective media containing 8 g/L of 2 KB for 72 h. We found that all transformants showed similar productivities for 1-propanol (approx. 400 mg/L) (Fig. 2). The fact that overexpression of KDC and ADH in S. cerevisiae provided no advantage for the production of 1-propanol, indicates either that the selected enzymes did not have specific activity for the conversion of 2 KB into 1-propanol, or that endogenous yeast KDC and ADH enzymes already provide sufficient activity for this purpose. Given that the negative control strain (YA0K0; exogenously overexpressing neither KDC nor ADH) also produced 1-propanol, the latter is most probable. Indeed, engineered E. coli overexpressing ARO10 (from S. cerevisiae) or Kivd (from L. lactis) with ADH2 (from S. cerevisiae) have been shown to exhibit the activity required to convert 2 KB into 1-propanol [5].
Construction of a 2-ketobutyrate biosynthesis pathway via citramalate
Since S. cerevisiae appears to have sufficient KDC and ADH activity to convert 2 KB into 1-propanol, we next tried engineering yeast metabolic pathways to increase levels of 2 KB, the precursor of 1-propanol, using glucose as a carbon source. In E. coli, 2 KB is normally produced through the enzymatic conversion of threonine by threonine dehydratase. Engineered E. coli with increased 1-propanol productivity have been developed by introducing an artificial pathway via citramalate, which can convert pyruvate into 2 KB, in addition to the original threonine-mediated pathway (Fig. 1b) [15, 16]. Since E. coli has endogenous genes encoding citramalate hydrolyase (leuC and leuD) and 3-isopropylmalate dehydrogenase (leuB), the citramalate-mediated pathway can been completed by artificially expressing the citramalate synthase (cimA) gene derived from Methanococcus jannaschii (Mj). In S. cerevisiae, 2 KB is also produced endogenously via threonine (Fig. 1a), however, it does not carry the corresponding genes for citramalate synthase (cimA) or citramalate hydrolyase (leuC and leuD). Drawing on the experience from E. coli, we therefore constructed an artificial citramalate-mediated pathway to overproduce 2 KB from pyruvate and examined the productivity of 1-propanol in S. cerevisiae (Fig. 1b).
Although S. cerevisiae has an endogenous LEU2 gene that encodes 3-isopropylmalate dehydrogenase (encoded as leuB in E. coli), the laboratory yeast strains (YPH499 and BY4741) used in this study lack the functional LEU2 gene, as they are auxotrophs for the purposes of selection after gene transfection. Therefore, we used an expression plasmid carrying the LEU2 auxotrophic marker to compensate for 3-isopropylmalate dehydrogenase activity. For citramalate hydrolyase, we selected leuC (citramalate hydrolyase, large subunit) and leuD (small subunit) genes from three different sources: thermophilic methanogenic archaea, M. jannaschii (Mj), gram-negative and facultative anaerobic bacteria, E. coli (Ec), and the gram-positive and obligate anaerobe Clostridium beijerinckii (Cb). These gene pairs were introduced into the autonomously-replicating plasmid harboring the LEU2 marker. For citramalate synthase, which catalyzes 2 KB biosynthesis via citramalate from the central metabolite pyruvate, we used the cimA gene derived from M. jannaschii, which was successfully used in E. coli in the previous study [15, 16]. Two methods of expressing cimA were tested, the first being a single-copy genomic integration into the ura3 locus to stabilize gene replication, and the second being co-integration into the LEU2 marker plasmid along with leuC and leuD, in order to increase overall expression.
In fermentation using the engineered YPH499 strains (Table 1) in SD medium (20 g/L glucose) without 2 KB, the use of the multi-copy plasmid expressing cimA resulted in higher production of 1-propanol than direct genomic integration, in all cases tested (Fig. 3). This indicated that high expression of cimA is more successful for 1-propanol production in yeast. Comparing the biological source of leuC and leuD, Cb-derived genes showed the highest productivity of 1-propanol (Fig. 3). Yeast with genomic integration of cimA that were transfected with mock (non-LEU2 expressing) plasmid (Y06C250) produced 8.7 mg/L of 1-propanol (Fig. 3), while the engineered strain with plasmid-driven expression of cimA and Cb-derived leuC and leuD (Y26C25C) produced a much higher level of 1-propanol (20.7 mg/L). These results suggested that the exogenous expression of cimA, leuC and leuD (and the LEU2 marker) allowed 2 KB biosynthesis in S. cerevisiae via citramalate, as has previously been shown for E. coli.
Fig. 3.
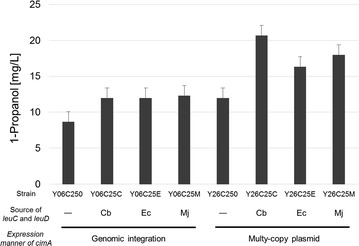
Production of 1-propanol in S. cerevisiae YPH499 strains expressing an artificially engineered pathway from pyruvate to 2 KB. (Mj: M. jannaschii, Ec: E. coli, Cb: Clostridium beijerinckii)
Overexpression of threonine dehydratase
To further increase the production of 1-propanol, we attempted to enhance the endogenous threonine-mediated pathway for 2 KB biosynthesis as shown in Fig. 1c. To do this we overexpressed threonine dehydratase, which catalyzes the conversion of threonine to 2 KB. Three types of threonine dehydratase gene were tested, namely, ILV1 from S. cerevisiae, tdcB from E. coli, and ilvA also from E. coli (Table 1 and Fig. 4). We found that strains overexpressing threonine dehydratase in the absence of cimA, leuC and leuD (Y5041 ~ 3) showed no significant increase in 1-propanol production (Fig. 4). However, all strains overexpressing threonine dehydratase in conjunction with cimA, leuC and leuD (Y5C41 ~ 3, Y5E41 ~ 3 and Y5M41 ~ 3) showed an increase in 1-propanol production, with the expression of tdcB (E. coli) having the most significant impact. The Y5C40 strain expressing cimA and Cb-derived leuC and leuD produced 23.7 mg/L of 1-propanol, while Y5C42 (representing the Y5C40 strain with co-expression of tdcB) produced 42.7 mg/L of 1-propanol. Although it is unclear why the overexpression of threonine dehydratase alone (Y5041 ~ 3) resulted in no improvement, we thought there might be no extra threonine, competing pathway from threonine to glycine might be strong or coenzyme balance might affect this result due to the reaction of LEU2 enzyme required for coenzyme. However, the findings clearly demonstrate the synergistic effect of threonine dehydratase expression and citramalate-mediated 2 KB biosynthesis for 1-propanol production in yeast.
Fig. 4.
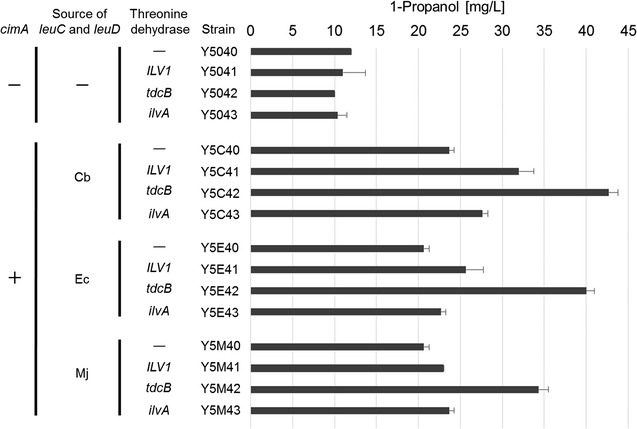
Production of 1-propanol in S. cerevisiae YPH499 strains expressing an artificially engineered pathway from pyruvate to 2 KB and overexpression of genes encoding the enzyme threonine dehydratase. (Mj: M. jannaschii, Ec: E. coli, Cb: Clostridium beijerinckii)
Deletion of competing biochemical pathways
Next, we attempted to increase the production of 1-propanol by decreasing carbon flux into competing pathways for 2 KB and amino-acid metabolism. To do this, we used yeast strains with deletion of specific biochemical pathways from a single gene deletion library of BY4741. As shown in Fig. 1a, ILV2, ILV3, ILV5 and BAT1 are candidate target genes for the knockout of the biosynthetic pathway for valine, leucine and isoleucine, however there was no strain with deletion of these genes in the library. This indicates that the deletion of each of these genes is either lethal or results in poor growth, ruling out these genes as candidates for deletion in our study.
Of the remaining candidates, 11 genes were selected for targeting (colored green in Fig. 1a). ARO3 and ARO4 encode 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from the tryptophan and phenylalanine biosynthetic pathway. ALT1 and ALT2 encode alanine transaminase from the alanine biosynthetic pathway. CIT1, CIT2 and CIT3 encode citrate synthase from the glutamine, arginine and lysine biosynthetic pathway. MET2 encodes l-homoserine-O-acetyltransferase from the methionine biosynthetic pathway. GLY1 encodes threonine aldolase, which converts threonine to glycine. ILV6 encodes the regulatory subunit of acetolactate synthase contained in the valine, leucine and isoleucine biosynthetic pathway. BAT2 encodes the branched-chain amino acid transaminase. Using BY4741 strains with individual deletions of each of these genes, we compared 1-propanol production in YPD rich medium (Fig. 5a). We found that only the strain deleting GLY1 (encoding threonine aldolase) showed an increase in 1-propanol production (Fig. 5a). Since Gly1 constitutes the main pathway to produce glycine from threonine in yeast [17], the deletion of GLY1 would decrease the loss of threonine and increase its conversion to 2 KB. GLY1 is the sole gene that encodes threonine aldolase, and the absence of isozymes no doubt enhances the efficacy of this approach. To test whether the GLY1-deleted strain could increase the production of 1-propanol via citramalate, we introduced cimA, leuC, and leuD into BY4741ΔGLY1 (Fig. 5b). Following fermentation using SD media, BY4741ΔGLY1 strains with cimA, leuC, and leuD (BG5C, BG5E, and BG5M) had higher productivity of 1-propanol than original BY4741 strains with cimA, leuC, and leuD (B5C, B5E and B5M). This result indicated that the GLY1 deletion could indeed fulfill the function of increasing 1-propanol production from the artificial citramalate pathway.
Fig. 5.
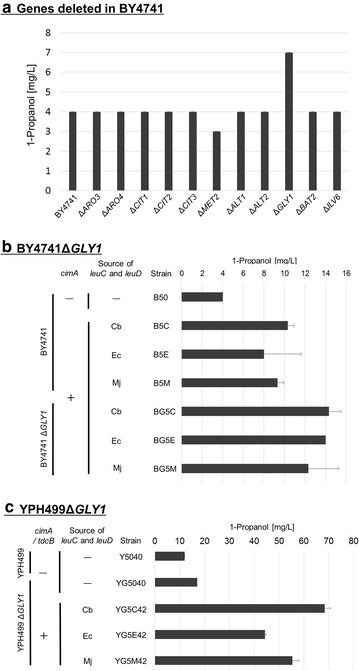
Deletion of metabolic pathways competing with 1-propanol production in yeast. a 1-Propanol production in strains from a single gene deletion library of BY4741. b Comparison of BY4741 with BY4741ΔGLY1. c Comparison of YPH499 with YPH499ΔGLY1. (Mj: M. jannaschii, Ec: E. coli, Cb: Clostridium beijerinckii)
Comparing Fig. 5b with Fig. 3, it is clear that production of 1-propanol in YPH499 was higher than that of BY4741. Therefore, we subsequently constructed a YPH499ΔGLY1 strain to enhance 2 KB biosynthesis via both the citramalate and threonine pathways. As shown in Fig. 5c, YPH499ΔGLY1 (YG5040) demonstrated higher production of 1-propanol than wildtype YPH499 (Y5040). Furthermore, YPH499ΔGLY1 with expression of cimA, leuC, leuD, and tdcB (YG5C42) produced 68.3 mg/L of 1-propanol (Fig. 5c), whereas YPH499 harboring the same genes (Y5C42) produced 42.6 mg/L (Fig. 4). Thus, just as in BY4741, the deletion of GLY1 enhanced the production of 1-propanol in YPH499 yeast strains with modifications of both the citramalate and threonine pathways.
Overexpression of threonine synthase
To further improve the production of 1-propanol, we also aimed to enhance the threonine biosynthetic pathway via aspartate as shown Fig. 1d. To increase the carbon flux from aspartate to threonine, we selected four genes (bifunctional thrA, encoding aspartokinase and homoserine dehydrogenase I; thrB, encoding homoserine kinase; thrC, encoding threonine synthase; and asd, encoding aspartate-semialdehyde dehydrogenase) derived from E. coli [15]. In addition to cimA, leuC, leuD, and tdcB, these four genes (thrA, thrB, thrC, and asd) or alternatively just three genes (thrA, thrB and thrC), were introduced into the YPH499ΔGLY1 strain to generate YG5C4232 and YG5C4231, respectively. Following fermentation in SD media, both YG5C4231 and YG5C4232 produced ~ 100 mg/L of 1-propanol, with the presence or absence of the asd gene thus appearing to make little difference (Fig. 6). Compared to YPH499ΔGLY1 expressing cimA, leuC, leuD, and tdcB (YG5C42; 68.3 mg/L in Fig. 5b), these strains therefore demonstrated an additional increase in 1-propanol production. This indicates that enhancement of aspartate-mediated threonine biosynthesis co-operates with the GLY1 deletion in regard to enhancement of 1-propanol production via the citramalate, threonine, and 2 KB pathways.
Fig. 6.
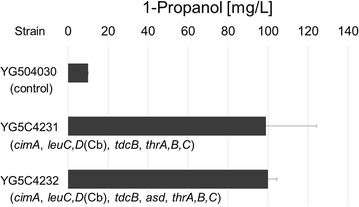
Production of 1-propanol in S. cerevisiae YPH499ΔGLY1 strains with an artificially engineered pathway from pyruvate to 2 KB and overexpression of tdcB and genes encoding for threonine synthase. (Cb: Clostridium beijerinckii)
Oxygen-limited fermentation of engineered strains
Finally, we measured the time course for 1-propanol production of engineered strains in 50 mL of SD medium (initial cell concentration, OD600 = 20) under oxygen-limited condition using fermentation bottles (Fig. 7). We found that the strains YG504030 (YPH499ΔGLY1; control), YG5C42 (cimA/leuC, leuD/tdcB) and YG5C4232 (cimA/leuC, leuD/tdcB/thrA, B, C/asd) showed a similar production of 1-propanol under limited oxygen conditions compared to their growth in test tubes. In contrast, YG5C4231 (cimA/leuC, leuD/tdcB/thrA, B, C) displayed an approximately two-fold higher productivity compared under these conditions (Fig. 7). This result suggests that the threonine biosynthetic pathway via aspartate is enhanced during oxygen-limited fermentation, resulting in yet greater 1-propanol production. We eventually obtained 179 mg/L of 1-propanol from 20 g/L of glucose using YG5C4231 (cimA/leuC, leuD/tdcB/thrA, B, C) under oxygen-limited conditions, the highest level of production observed for any of the 1-propanol-producing yeast strains. As the reason of little change from 24 to 96 h, we thought the glucose was exhausted for 24 h due to high concentration of the initial added yeast.
Fig. 7.
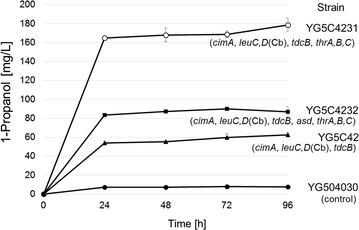
Oxygen-limited fermentation of engineered YPH499ΔGLY1 strains. (Cb: Clostridium beijerinckii)
Conclusions
In the present study, we modified metabolic pathways of S. cerevisiae to engineer yeast strains producing 1-propanol. Firstly, we observed that the activity of endogenous yeast KDC and ADH is sufficient to convert 2 KB to 1-propanol. Secondly, we found that 1-propanol production could be increased by constructing an artificial 2 KB biosynthetic pathway from pyruvate via citramalate, with the introduction of cimA and leuC/leuD genes from M. jannaschii and C. beijerinckii, respectively. Furthermore, in addition to the overexpression of threonine dehydratase (with the introduction of tdcB), and enhancement of threonine biosynthesis from aspartate (with the introduction of thrA, thrB and thrC), 1-propanol production was greatly increased by deletion of the GLY1 gene that regulates a competing pathway converting threonine to glycine. While the control YPH499 strain (Y5040) produced only 12 mg/L of 1-propanol in test tubes, the engineered strain YG5C4231 produced 99 mg/L. Moreover, in the context of high-density anaerobic fermentation, we succeeded in producing 179 mg/L of 1-propanol using this strain. These results demonstrate that construction of a citramalate-mediated pathway as the production method of 1-propanol in S. cerevisiae is effective. For yet further improvement of 1-propanol production in S. cerevisiae, it may be necessary to engineer the carbon flux from ethanol to 2 KB and oxidoreduction balance due to coenzyme. For example, as shown in Additional file 2 using yeast strains of Additional file 3, double deletion of competing pathway have the potential to increase 1-propanol production.
Additional files
Additional file 1. Primers used for the construction of double deletion strains in Additional file 2.
Additional file 2. Double deletion of metabolic pathways competing with 1-propanol production in YPH499.
Additional file 3. Yeast strains used in Additional file 2.
Authors’ contributions
Conceived and designed the experiments: YN, JI, AK. Performed the experiments: YN and TM. Analyzed the data: YN. Wrote the paper: YN and JI. Supervised the whole work: AK. All authors read and approved the final manuscript.
Acknowledgements
This work was supported in part by a Special Coordination Fund for Promoting Science and Technology, Creation of Innovative Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe) from the Ministry of Education, Culture, Sports and Technology (MEXT; to AK), and by Science Research Grants from the Ministry of Health, Labor and Welfare, Japan (to AK).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data supporting the conclusions of this article are included with the article. Strains examined are available from the corresponding author.
Consent for publication
The authors provide consent for publication.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12934-018-0883-1) contains supplementary material, which is available to authorized users.
Contributor Information
Yuya Nishimura, Email: nyuya@landscape.kobe-u.ac.jp.
Jun Ishii, Email: junjun@port.kobe-u.ac.jp.
Akihiko Kondo, Phone: +81 78 803 6196, Email: akondo@kobe-u.ac.jp.
References
- 1.Ohno S, Furusawa C, Shimizu H. In silico screening of triple reaction knockout Escherichia coli strains for overproduction of useful metabolites. J Biosci Bioeng. 2013;115:2. doi: 10.1016/j.jbiosc.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Hong ME, Lee KS, Yu BJ, et al. Identification of gene targets eliciting improved alcohol tolerance in Saccharomyces cerevisiae through inverse metabolic engineering. J Biotechnol. 2010;149(1):52–59. doi: 10.1016/j.jbiotec.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Fischer CR, Klein-Marcuschamer D, Stephanopoulos G. Selection and optimization of microbial hosts for biofuels production. Metab Eng. 2008;10:6. doi: 10.1016/j.ymben.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Jo SE, Seong YJ, Lee HS, et al. Microaerobic conversion of xylose to ethanol in recombinant Saccharomyces cerevisiae SX6MUT expressing cofactor-balanced xylose metabolic enzymes and deficient in ALD6. J Biotechnol. 2016;227:72–78. doi: 10.1016/j.jbiotec.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:7174. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 6.Shen CR, Liao JC. Synergy as design principle for metabolic engineering of 1-propanol production in Escherichia coli. Metab Eng. 2013;17:1. doi: 10.1016/j.ymben.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Eden A, Van Nedervelde L, Drukker M, Benvenisty N, Debourg A. Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl Microbiol Biotechnol. 2001;55:3. doi: 10.1007/s002530000506. [DOI] [PubMed] [Google Scholar]
- 8.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:1. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brachmann CB, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:2. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Ishii J, Izawa K, Matsumura S, et al. A simple and immediate method for simultaneously evaluating expression level and plasmid maintenance in yeast. J Biochem. 2009;145:6. doi: 10.1093/jb/mvp028. [DOI] [PubMed] [Google Scholar]
- 11.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:1. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katahira S, Mizuike A, Fukuda H, Kondo A. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl Microbiol Biotechnol. 2006;72:6. doi: 10.1007/s00253-006-0402-x. [DOI] [PubMed] [Google Scholar]
- 13.Akada R, Kitagawa T, Kaneko S, et al. PCR-mediated seamless gene deletion and marker recycling in Saccharomyces cerevisiae. Yeast. 2006;23:5. doi: 10.1002/yea.1365. [DOI] [PubMed] [Google Scholar]
- 14.Kondo T, Tezuka H, Ishii J, Matsuda F, Ogino C, Kondo A. Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J Biotechnol. 2012;159:1–2. doi: 10.1016/j.jbiotec.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 15.Jun Choi Y, Hwan Park J, Yong Kim T, Yup Lee S. Metabolic engineering of Escherichia coli for the production of 1-propanol. Metab Eng. 2012;14:5. doi: 10.1016/j.ymben.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Atsumi S, Liao JC. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl Environ Microbiol. 2008;74:24. doi: 10.1128/AEM.02046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monschau N, Stahmann KP, Sahm H, McNeil JB, Bognar AL. Identification of Saccharomyces cerevisiae GLY1 as a threonine aldolase: a key enzyme in glycine biosynthesis. FEMS Microbiol Lett. 1997;150:1. doi: 10.1016/S0378-1097(97)00096-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Primers used for the construction of double deletion strains in Additional file 2.
Additional file 2. Double deletion of metabolic pathways competing with 1-propanol production in YPH499.
Additional file 3. Yeast strains used in Additional file 2.
Data Availability Statement
The data supporting the conclusions of this article are included with the article. Strains examined are available from the corresponding author.


