Abstract
Dental pulp cells (DPCs) are promising candidates for use as transplantable cells in regenerative medicine. However, ex vivo expansion of these cells typically requires culture media containing fetal bovine serum, which may cause infection and immunological reaction following transplantation. In addition, the proliferation and differentiation of DPCs markedly depend upon serum batches. Therefore, the present study examined whether DPCs could be expanded under serum-free conditions. DPCs obtained from four donors were identified to proliferate actively in the serum-free medium, STK2, when compared with those cells in control medium (Dulbecco's modified Eagle's medium containing 10% serum). The high proliferative potential with STK2 was maintained through multiple successive culture passages. DNA microarray analyses demonstrated that the gene expression profile of DPCs grown in STK2 was similar to that of cells grown in the control medium; however, a number of genes related to cell proliferation, including placental growth factor and inhibin-βE, were upregulated in the STK2 cultures. Following induction of osteogenesis, DPCs grown in STK2 induced alkaline phosphatase activity and calcification at higher levels compared with the control medium cultures, indicating maintenance of differentiation potential in STK2. This serum-free culture system with DPCs may have applications in further experimental studies and as a clinical strategy in regenerative medicine.
Keywords: cell proliferation, osteogenesis, gene expression, dental pulp cells, regenerative medicine
Introduction
Dental pulp cells (DPCs), the resident mesenchymal stromal cells in dental pulp tissue, may be isolated as migrating cells from pulp tissue explants or as colony-forming cells in low-density cultures (1,2). These cells are able to differentiate into dentin, bone, periodontal ligament, fat, vascular cells and nerve-like cells in vitro and in vivo (1,3–15). DPCs and bone marrow-derived mesenchymal stem cells (BM-MSCs) have similar differentiation potentials, though the growth activity of DPCs may be greater than that of BM-MSCs (1,13,16).
DPCs, as well as BM-MSCs, are promising in cell-based therapy for various diseases including ischemia (6) and spinal cord injury (15). Fetal bovine serum (FBS) has been used for ex vivo expansion of DPCs; however, this carries a risk of contamination with prions or viruses. Furthermore, the proliferation activity and differentiation potential of DPCs depends upon the batch of serum (17). Therefore, serum-free, chemically defined media should be used for the expansion of DPCs destined for clinical application (17).
A range of serum-free media have been developed for culturing adult and embryonic stem cells (17). In the present study, a culture of DPCs under serum-free conditions was attempted using STK2, a serum-free medium for BM-MSCs. Previous studies have demonstrated that STK2 is suitable for ex vivo expansion of BM-MSCs. For instance, when compared with Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, STK2 further increased the proliferation of BM-MSCs (18), but did not promote the growth of immortalized human gingival fibroblasts or cancer cell lines (19). In addition, neural crest and endometrial carcinoma cells grown in STK2 exhibited mesenchymal-like features (20,21). Therefore, the present study examined whether STK2 could support the proliferation of human DPCs. In addition, the differentiation ability of DPCs grown in STK2 was assessed.
Materials and methods
Culture media
STK2 was purchased from DS Pharma Biomedical (Osaka Japan). DMEM (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) and 1X antibiotic-antimycotic containing 100 U/ml penicillin, 100 g/ml streptomycin and 0.25 g/ml amphotericin B (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used as control medium. α-minimal essential medium (α-MEM) supplemented with 10% FBS, 100 nM dexamethasone (Sigma-Aldrich; Merck KGaA), 50 mg/l ascorbic acid 2-phosphate (Sigma-Aldrich; Merck KGaA) and 10 mM β-glycerophosphate (Tokyo Chemical Industry, Co., Ltd., Tokyo, Japan) was used for induction of osteogenesis.
Cells
Healthy upper third molars were obtained from 4 healthy female donors (aged 23–27 years) at Hiroshima University Hospital (Hiroshima, Japan) from April 2008 to March 2009 with informed consent following a protocol approved by the Ethics Committee at Hiroshima University (approval no. D88-2). Fibroblast-like cells were grown out from tooth pulp tissue explants individually derived from the donors and were used as DPC lines (DPCs-2, DPCs-3, DPCs-4 and DPCs-5), as previously described (22). BM-MSCs (lot no. OF3853) from a 20-year-old female donor were obtained from Lonza Group, Ltd. (Basel, Switzerland).
Cell growth
The DPCs grown out from tissue explants were harvested with 0.2% trypsin and 0.02% EDTA in phosphate-buffered saline (PBS). The cells were seeded onto 10 cm plastic tissue culture dishes at a 1:5 split ratio and incubated with 10 ml DMEM supplemented with 10% FBS (DMEM/10% FBS) at 37°C in 95% air and 5% CO2. For experimentation, cells obtained from 3rd-6th passage cultures were seeded at 5×103 cells/cm2 into each well of a 12-well plate (Corning Incorporated, Corning, NY, USA) with 1.0 ml STK2 or DMEM/10% FBS. The cultures were fed with the respective media every 2–3 days. When cultures became 80–90% confluent, the cells were incubated with Accutase (Innovative Cell Technologies, Inc., San Diego, CA, USA) for 3 min at 37°C, and the number of dispersed cells was counted. These cells were then sub-cultured at the same initial density with the respective media, and cell counting was repeated as above until the experiment endpoint (37 days).
For a Cell Counting Kit-8 (CCK-8) assay, DPCs obtained from 4th-6th passage cultures were seeded on 96-well plates (Corning Incorporated) at 5×103, 1×104 or 2×104 cells/cm2, and incubated in 0.1 ml STK2 or DMEM/10% FBS at 37°C in 95% air and 5% CO2. The human BM-MSCs were also used as a positive control. At 7 days after cell seeding, 0.01 ml of a WST-8 solution (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to the culture medium, and the cultures were incubated for 1 h at 37°C in 95% air and 5% CO2. The absorbance of the medium at 450 nm was measured with a microplate reader (VersaMax; Molecular Devices, LLC, Sunnyvale, CA, USA).
Alizarin Red S staining
DPCs were seeded on 24-well plates at 5×103 cells/cm2 and incubated for 7 days in 0.5 ml STK2 or DMEM/10% FBS at 37°C in 95% and 5% CO2. When the cultures became 100% confluent, cells were incubated with osteogenesis induction medium for 14, 21 and 28 days. The cell layers were washed twice with PBS, and fixed with 97% ethanol for 10 min at room temperature. They were then washed twice with PBS, and incubated for 30 min with 1% Alizarin Red S solution (Sigma-Aldrich; Merck KGaA) at room temperature. Finally, the cell layers were washed with water, and the red staining indicating calcium deposition was observed by naked eye.
Alkaline phosphatase (ALP) activity
DPCs were seeded on 48-well plates at 5×103 cells/cm2 and incubated in 0.3 ml STK2 or DMEM/10% FBS at 37°C in 95% air and 5% CO2. When the cultures became confluent, cells were incubated with osteogenesis induction medium for 0, 4, 8, 12 and 16 days. The cell layers were washed twice with saline, incubated for 15 min with 0.25 ml 1% Nonidet P-40 (NP-40) solution (Sigma-Aldrich; Merck KGaA), and then homogenized with the same solution for 1 min on ice. Using the cell extract and LabAssay ALP (Wako Pure Chemical Industries, Ltd., Osaka, Japan), ALP activity was determined according to the manufacturer's instructions. In this assay, p-nitrophenylphosphate is hydrolyzed into p-nitrophenol and phosphoric acid by ALP. The released p-nitrophenol exhibiting yellow color was optically measured at 405 nm as the enzyme activity. The quantity of DNA in the cell extract was determined using a Quant-iT PicoGreen dsDNA Assay kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Fluorescence was measured at an excitation of 480 nm and emission of 520 nm. Enzyme activity was expressed as nmol/min/µg DNA.
DNA microarray
DPCs-2, −3 and −4 from 5th passage cultures were seeded on 10 cm plastic tissue culture dishes at 5×103 cells/cm2 and incubated in STK2 or DMEM/10% FBS at 37°C in 95% air and 5% CO2. Total RNA was isolated with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,), an RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) and a TURBO DNA-Free kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) 24 h after the cultures became confluent. cDNA was synthesized using a GeneAmp RNA PCR kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). Labeled cRNA was synthesized using a Quick Amp Labeling kit (Agilent Technologies, Inc., Santa Clara, CA, USA), and hybridization was performed on a DNA microarray (SurePrint G3 Human, 8×60k; Agilent Technologies, Inc.), according to the manufacturer's protocols. Finally, the mRNA expression levels were analyzed using Agilent GeneSpring GX version 12 software (Agilent Technologies, Inc.). A correlation coefficient was calculated in GeneSpring to compare the gene expression profiles in DPCs cultured in DMEM/10% FBS with those in DPCs cultured in STK2. The resulting raw data have been deposited in the Gene Expression Omnibus database (GSE97199; http://www.ncbi.nlm.nih.gov/geo/) (23).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated as described above and cDNA was synthesized using the GeneAmp RNA PCR kit as described previously (24). Quantitative real-time PCR analysis was performed using an ABI Prism 7900HT Sequence Detection System with the included software (SDS version 2.4; Applied Biosystems; Thermo Fisher Scientific, Inc.,), based on the ΔΔCq method (25). The cDNA was amplified using a Universal PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) with appropriate forward and reverse primers (Table I) with a primer kit for eukaryotic 18S rRNA (Applied Biosystems; Thermo Fisher Scientific, Inc.) used as an endogenous control. The PCR cycling conditions comprised of incubation at 50°C for 2 min, denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. TaqMan probes (Table I) were purchased from Roche Diagnostics (Basel, Switzerland). Data were normalized against 18S ribosomal RNA levels.
Table I.
Primer and probe sequences used for reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer (5′-3′) | Roche universal probe no. |
|---|---|---|
| TXNIP | F: CTTCTGGAAGACCAGCCAAC | 85 |
| R: GAAGCTCAAAGCCGAACTTG | ||
| PGA3 | F: CCCGTCTTTGACAACATCTG | 81 |
| R: ATCACCACGCTGCCACTC | ||
| TNNT1 | F: GACTACATGGGGGAGGAACA | 17 |
| R: GCCATCAGGTCGAACTTCTC | ||
| SCRG1 | F: TGTTACTGCAACTTCAGCGAAT | 44 |
| R: TTGCAAGGAATCACGAAAGA | ||
| INHBE | F: CAGGGAGTGTGGCTCCAG | 52 |
| R: TGTAGGCTGAAGTGGAGTCTGT | ||
| GLI1 | F: CAGGGAGGAAAGCAGACTGA | 76 |
| R: ACTGCTGCAGGATGACTGG |
TXNIP, thioredoxin interacting protein; PGA3, pepsinogen 3; TNNT1, troponin T type 1 (skeletal, slow); SCRG1, stimulator of chondrogenesis 1; INHBE, inhibin-βE; GLI1, GLI family zinc finger 1; F, forward; R, reverse.
Statistical analysis
Experiments were performed at least in triplicate. Results are expressed as the mean ± standard deviation. Differences between groups were analyzed by two-way analysis of variance (ANOVA) with Sidak's post hoc test for multiple comparisons. In all analyses, P<0.05 indicated statistically significant differences between values.
Results
STK2 medium enhances the proliferation of DPCs
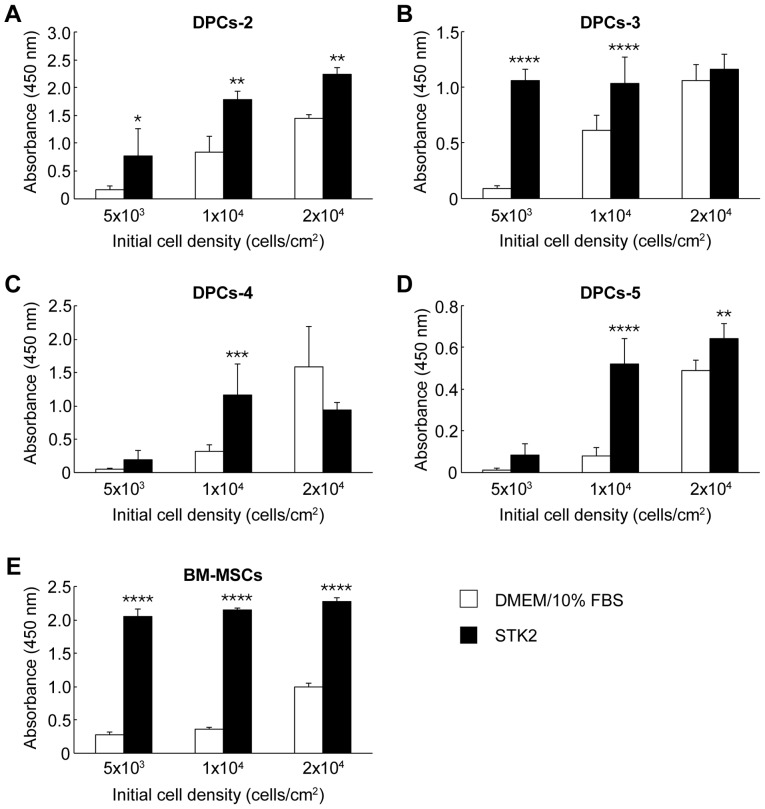
The effects of STK2 on the proliferation of four DPC lines (DPCs-2, DPCs-3, DPCs-4 and DPCs-5) were examined. These cells were seeded at low, medium or high density (5×103, 1×104 or 2×104 cells/cm2, respectively) and incubated in STK2 or DMEM/10% FBS. Cell numbers were determined by CCK-8 assay on day 7. The majority of the cell lines exhibited higher growth rates in STK2 than in DMEM/10% FBS under conditions of low and medium initial cell densities (Fig. 1A-D). However, at high initial cell density, STK2 did not enhance the growth rates of the DPCs-3 and DPCs-4 cell lines compared with DMEM/10% FBS (Fig. 1B and C). By contrast, STK2 enhanced the proliferation of human BM-MSCs at the low, medium and high initial cell densities, compared with DMEM/10% FBS (P<0.0001; Fig. 1E). In addition, at the low initial density, DPCs-5 cells obtained from 3rd-6th passage continued to proliferate actively in STK2 through successive passages (Fig. 2).
Figure 1.
Effects of STK2 on the proliferation of four DPC lines. The DPC lines (A) DPCs-2, (B) DPCs-3, (C) DPCs-4 and (D) DPCs-5, and (E) BM-MSCs were seeded at the indicated initial densities and exposed to DMEM/10% FBS or STK2. Cell number was estimated on day 7 by Cell Counting Kit-8 assay. Values are the means ± standard deviation of at least three replicate cultures. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 vs. DMEM/10% FBS at the same cell density. DPCs, dental pulp cells; BM-MSCs, bone marrow-derived mesenchymal stem cells; DMEM/10% FBS, Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Figure 2.
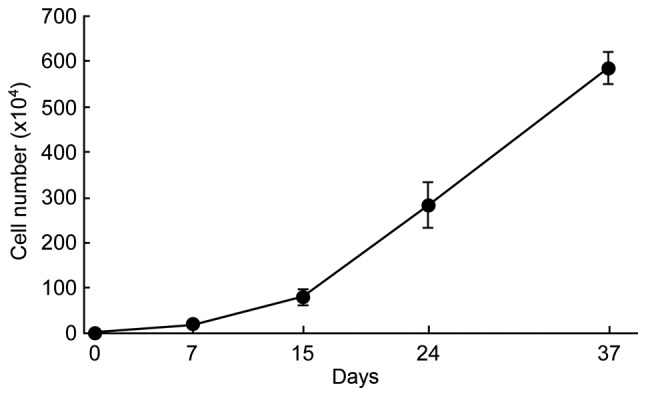
Growth curve of DPCs grown in STK2. DPCs-5 cells were seeded at low density (5×103 cells/cm2) and exposed to STK2. When the cultures approached confluence, the cells were harvested with Accutase and a population was transferred into a new dish with STK2 to allow continued cell growth. The cumulative cell numbers are shown. Values are the means ± standard deviation of four replicate cultures. DPCs, dental pulp cells.
Comparison of gene expression levels in cells cultured in DMEM/10% FBS or STK2
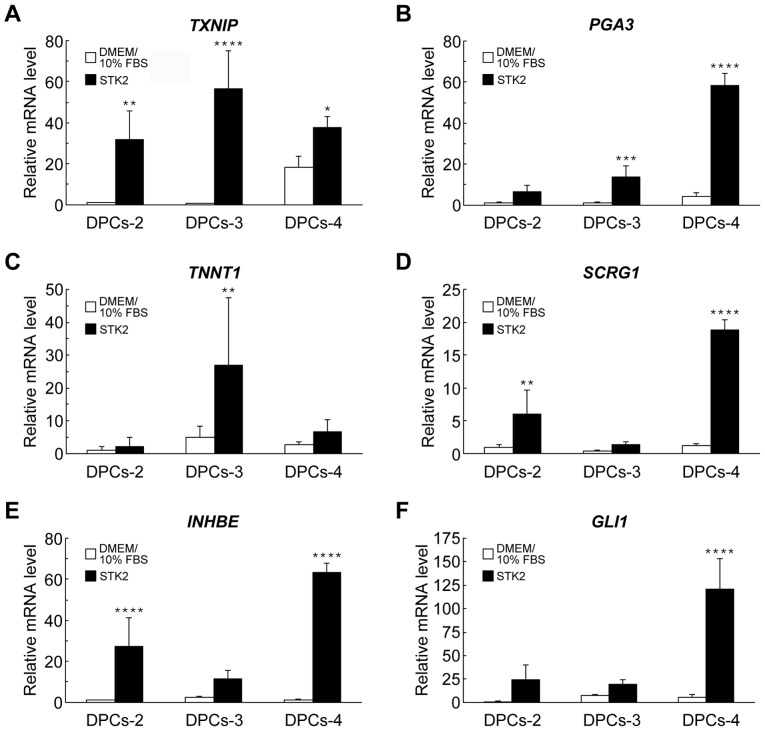
The gene expression profiles of DPCs (DPCs-2, −3 and −4) that had been grown in STK2 or DMEM/10% FBS were compared. Overall, the gene expression profile was only marginally altered by the medium (correlation coefficient between the two media = 0.97 to 0.99; data not shown). A total of 155, 38 and 8 genes were upregulated by more than 2-, 5- and 10-fold, respectively, in DPCs grown in STK2, while 254, 106 and 37 genes were downregulated by more than 2-, 5- and 10-fold, respectively. Tables II and III list the top 20 genes upregulated and downregulated by STK2, respectively. The upregulated genes included those involved in proliferation [including thioredoxin interacting protein (TXNIP), platelet factor 4 variant 1, placental growth factor and inhibin-βE (INHBE)], metabolism [including pepsinogen 3 (PGA3) and alcohol dehydrogenase 1A] and differentiation [including troponin T type 1 (TNNT1), stimulator of chondrogenesis 1 (SCRG1), hairy/enhancer-of-split related with YRPW motif 1 and GLI family zinc finger 1 (GLI1)]. Indeed, the mRNA levels of TXNIP, PGA3, TNNT1, SCRG1, INHBE and GLI1 were increased in DPCs grown in STK2, compared with their levels in DMEM/10% FBS cultures, as confirmed by RT-qPCR (Fig. 3).
Table II.
Top 20 upregulated genes in dental pulp cells grown in STK2 relative to those in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
| Gene symbol | Gene name | NCBI ID | Fold change |
|---|---|---|---|
| TXNIP | Thioredoxin interacting protein | NM_006472 | 101.0 |
| PGA3 | Pepsinogen 3 | NM_001079807 | 23.0 |
| TNNT1 | Troponin T type 1 | NM_003283 | 19.2 |
| SCRG1 | Stimulator of chondrogenesis 1 | NM_007281 | 17.9 |
| HEY1 | Hairy/enhancer-of-split related with YRPW motif 1 | NM_001040708 | 15.0 |
| ADH1A | Alcohol dehydrogenase 1A | NM_000667 | 12.7 |
| PF4V1 | Platelet factor 4 variant 1 | NM_002620 | 11.2 |
| PGF | Placental growth factor | NM_002632 | 10.8 |
| INHBE | Inhibin-βE | NM_031479 | 9.8 |
| GLI1 | GLI family zinc finger 1 | NM_005269 | 9.7 |
| PLAC1 | Placenta-specific 1 | NM_021796 | 9.2 |
| NDUFA4L2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 | NM_020142 | 8.8 |
| FER1L4 | Fer-1-like protein 4 | NR_024377 | 8.7 |
| LRRC15 | Leucine rich repeat containing 15 | NM_130830 | 8.6 |
| ZP1 | Zona pellucida glycoprotein 1 | NM_207341 | 8.4 |
| ADH1C | Alcohol dehydrogenase 1C | NM_000669 | 8.2 |
| MGC16121 | Hypothetical protein MGC16121 | NR_024607 | 8.1 |
| ANXA8L2 | Annexin A8-like 2 | NM_001630 | 8.0 |
| CA9 | Carbonic anhydrase IX | NM_001216 | 7.5 |
| BCL11A | B-cell CLL/lymphoma 11A | NM_022893 | 7.4 |
Table III.
Top 20 downregulated genes in dental pulp cells grown in STK2 relative to those in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
| Gene symbol | Gene name | NCBI ID | Fold change |
|---|---|---|---|
| HTR2B | 5-Ηydroxytryptamine receptor 2B | NM_000867 | 51.0 |
| GPNMB | Glycoprotein nmb | NM_001005340 | 45.4 |
| NPTX1 | Neuronal pentraxin I | NM_002522 | 38.8 |
| NTN1 | Netrin 1 | NM_004822 | 36.3 |
| SLC16A6 | Solute carrier family 16 member 6 | NM_004694 | 35.5 |
| PTGDS | Prostaglandin D2 synthase | NM_000954 | 32.8 |
| RRAGD | Ras-related GTP binding D | NM_021244 | 30.4 |
| A2M | α2-macroglobulin | NM_000014 | 30.1 |
| CLDN1 | Claudin 1 | NM_021101 | 24.0 |
| SPON2 | Spondin 2 | NM_012445 | 23.2 |
| NFIB | Nuclear factor I/B | NM_005596 | 23.2 |
| BIRC3 | Baculoviral IAP repeat-containing 3 | NM_001165 | 22.2 |
| KCNJ2 | Potassium voltage-gated channel subfamily J member 2 | NM_000891 | 21.2 |
| MALL | Mal, T-cell differentiation protein-like | NM_005434 | 19.5 |
| GRIA4 | Glutamate ionotropic receptor AMPA type subunit 4 | NM_001077244 | 18.5 |
| SLC7A14 | Solute carrier family 7 member 14 | NM_020949 | 18.1 |
| SCG2 | Secretogranin II | NM_003469 | 17.9 |
| AGT | Angiotensinogen | NM_000029 | 17.9 |
| GPX3 | Glutathione peroxidase 3 | NM_002084 | 17.3 |
| ANO1 | Anoctamin 1 | NM_018043 | 15.4 |
Figure 3.
RT-qPCR analyses of gene expression in DPCs grown in DMEM/10% FBS or STK2. DPCs (DPCs-2, DPCs-3 and DPCs-4) were cultured in DMEM/10% FBS or STK2 for 7 days. The mRNA levels of (A) TXNIP, (B) PGA3, (C) TNNT1, (D) SCRG1, (E) INHBF and (F) GLI1 were quantified by RT-qPCR analysis. Values are the means ± standard deviation of four replicate cultures. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 vs. DMEM/10% FBS in the same cell line. DPCs, dental pulp cells; DMEM/10% FBS, Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum; TXNIP, thioredoxin interacting protein; PGA3, pepsinogen 3; TNNT1, troponin T type 1; SCRG1, stimulator of chondrogenesis 1; INHBE, inhibin-βE; GLI1, GLI family zinc finger 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
STK2 medium enhances osteogenic differentiation in DPCs
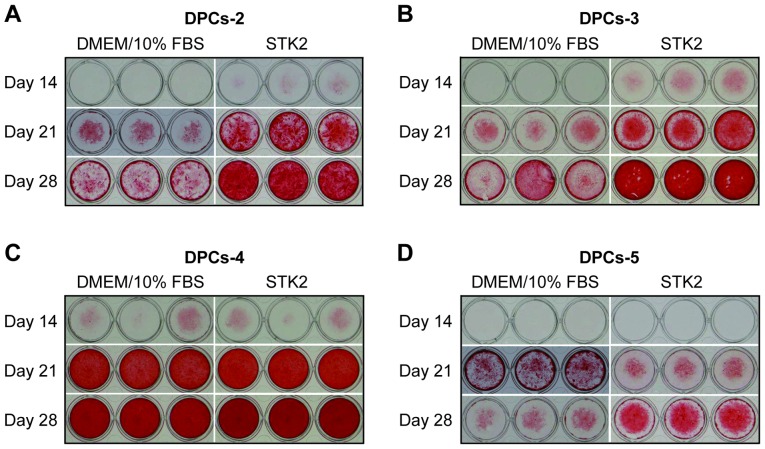
Subsequently, it was determined whether DPCs incubated in STK2 maintained their differentiation potential via Alizarin Red S staining (Fig. 4). DPCs (DPCs-2, −3, −4 and −5) were seeded at a low density and expanded for 7 days in STK2 or DMEM/10% FBS. When these cultures became confluent, they were exposed to osteogenesis induction medium for up to 28 days. On days 21 and 28, DPCs-2 and DPCs-3 expanded in STK2 exhibited more extensive calcification than those expanded in DMEM/10% FBS (Fig. 4A and B), while DPCs-4 exhibited extensive calcification on days 21 and 28 irrespective of the medium (Fig. 4C), probably due to the higher differentiation potential of this cell line. The calcification level of DPCs-5 grown in STK2 was higher than that of cells grown in DMEM/10 FBS on day 28 but lower on day 21 (Fig. 4D).
Figure 4.
Effects of pre-culture in STK2 on calcification in DPCs. The DPC lines (A) DPCs-2, (B) DPCs-3, (C) DPCs-4 and (D) DPCs-5 were seeded at low density (5×103 cells/cm2) and pre-cultured for 7 days in DMEM/10% FBS or STK2. When cultures reached confluence, the cells were cultured in osteogenesis induction medium for up to 28 days. The calcified matrix was stained with Alizarin Red S on the indicated days following the exposure to osteogenesis induction medium. DPCs, dental pulp cells; DMEM/10% FBS, Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
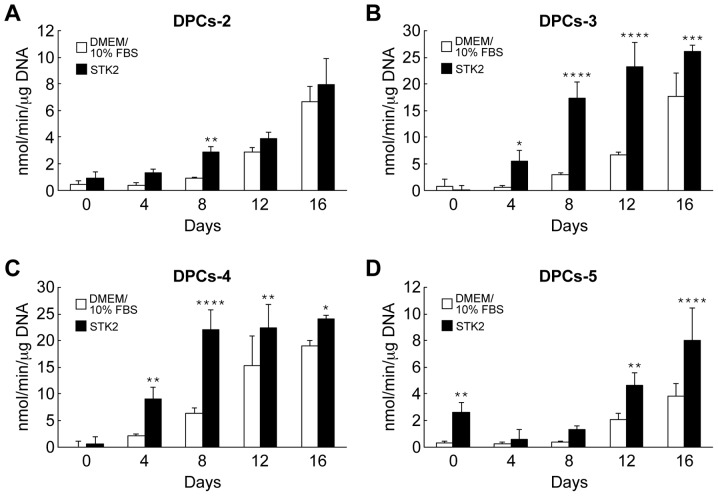
The effects of STK2 pre-culture on ALP activity were also assessed. Following exposure of the cell cultures to osteogenesis induction medium, ALP activity began to increase on day 4 or 8, depending on the cell line and medium (Fig. 5). All cell lines grown in STK2 exhibited greater ALP activity than those grown in DMEM/10% FBS (Fig. 5).
Figure 5.
Effects of pre-culture in STK2 on ALP activity in DPCs. The dental pulp cell lines (A) DPCs-2, (B) DPCs-3, (C) DPCs-4 and (D) DPCs-5 were seeded at low density (5×103 cells/cm2) and pre-cultured for 7 days in DMEM/10% FBS or STK2. When cultures reached confluence, the cells were cultured in osteogenesis induction medium for up to 16 days. ALP activity was determined on the indicated days following the exposure to osteogenesis induction medium. Values are the means ± standard deviation of four replicate cultures. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 vs. DMEM/10% FBS on the same day. ALP, alkaline phosphatase; DPCs, dental pulp cells; DMEM/10% FBS, Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Discussion
DPCs have been reported to have stem cell-like properties (6,7,10,15,26), and are thereby expected to be a source of transplantable cells in regenerative medicine for dental (7,9,27), bone (10,11), vascular (6,12) and nerve diseases (26,28). However, it is necessary for safe and stable cell therapy to establish a serum-free culture system for DPCs, since the use of serum has the potential risk of viral contamination and the components in serum vary depending on batch. In the present study, human DPCs were successfully cultured in a serum-free condition. DPCs were efficiently grown in STK2 with maintained osteogenic potential. This finding may promote clinical applications of DPCs in regenerative medicine.
Various serum-free media have a lower proliferation-promoting activity than serum-containing media. However, in the current study, DPCs generally proliferated at a higher rate in STK2 than in DMEM/10% FBS. The increased proliferation in STK2 was consistently observed with the four DPC lines examined, even though an optimum batch of FBS was used, selected from more than 10 batches. It may be speculated that FBS contains not only growth factors but also growth inhibitors and proteases, which decrease the growth rate of DPCs.
DNA microarray analysis demonstrated that the gene expression profiles of DPCs grown in STK2 and DMEM/10% FBS were similar. Thus, STK2 did not induce gross changes in the phenotypic expression of DPCs even in the absence of serum, suggesting that STK2 may be used for routine cultures of DPCs. It was identified that the number of downregulated genes was greater than that of upregulated genes in STK2 cultures, relative to DMEM/10% FBS cultures, which may be due to the absence of certain serum compounds, including proteins and other biologically active molecules, in the serum-free medium. While most genes exhibited no marked changes in gene expression levels, 38 genes were upregulated more than 5-fold in STK2-cultured cells. Some of these genes, including placental growth factor and INHBE, may account for the growth-promoting effect of STK2. Further experiments, including small interfering RNA knockdowns, are required to reveal which upregulated genes serve a role in the STK2-mediated simulation of proliferation.
Another important finding was the maintenance of the differentiation potential of DPCs under serum-free conditions. DPCs expanded in STK2 generally exhibited greater increases in ALP activity and calcified matrix following exposure to osteogenesis induction medium than cells expanded in DMEM/10% FBS. This may be due to the presence of certain proteins in serum, but not in STK2, that decrease the osteogenic potential of DPCs. DPCs are able to differentiate into adipocytes, chondrocytes and neural cells as well as osteoblasts (1,11). In future studies, our group plans to assess whether DPCs expanded in STK2 maintain their potency for differentiation into these cell types.
STK2 was originally developed using human BM-MSCs in order to create a serum-free culture medium for MSCs (17). Indeed, STK2 has been reported to exert a significantly greater proliferation effect on BM-MSCs compared with DMEM/10% FBS or Lonza MSCGMTM MSC growth medium (18). In addition, Sawada et al (29) demonstrated that a number of genes related to cell proliferation, including insulin-like growth factor binding protein 6, NRAS proto-oncogene, phosphoinositide-3-kinase regulatory subunit 3 and Jun proto-oncogene, were upregulated in BM-MSCs following culture in STK2 through DNA microarray analysis. However, these upregulated genes were not similar to those of the present study, probably due to differences in experimental conditions. The present study assessed DPCs in STK2 culture for 7 days, whereas Sawada's group used BM-MSCs cultured in STK2 for 50 days. Although it is difficult to directly compare the effect of STK2 on human DPCs and BM-MSCs due to inter-individual variations, STK2 appeared to be more effective for BM-MSCs than for DPCs in the current assays, since STK2 enhanced the proliferation of human BM-MSCs under all conditions of initial cell density. Modifications of the composition of STK2 may be required to optimize the expansion and differentiation of DPCs, despite STK2 being sufficient in supporting proliferation and maintaining the differentiation potency of DPCs.
A recent study demonstrated that STK2 could induce differentiation into mesenchymal-like cells; by culturing in STK2, human pluripotent stem cells-derived neural crest cells were induced into MSCs, which expressed specific cell surface markers and were able to differentiate into osteogenic, chondrogenic and adipogenic cells (20). In addition, STK2 induced the epithelial-mesenchymal transition when endometrial carcinoma cell lines transformed into mesenchymal-like cells (21). In the current study, it was demonstrated that DPCs proliferated more actively in STK2 than in DMEM/10% FBS while maintaining their osteogenic potential. Therefore, the serum-free culture system with STK2 may be useful for basic experimental research and cell therapy applications with DPCs as well as BM-MSCs.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- DPCs
dental pulp cells
- BM-MSCs
bone marrow-derived mesenchymal stem cells
- FBS
fetal bovine serum
- DMEM
Dulbecco's modified Eagle's medium
- α-MEM
α-minimal essential medium
- PBS
phosphate-buffered saline
- DMEM/10% FBS
DMEM supplemented with 10% FBS
- CCK-8
Cell Counting Kit-8
- ALP
alkaline phosphatase
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- TXNIP
thioredoxin interacting protein
- PGA3
pepsinogen 3
- TNNT1
troponin T type 1
- SCRG1
stimulator of chondrogenesis 1
- INHBE
inhibin-βE
- GLI1
GLI family zinc finger 1
Funding
The present study was supported by the Grant-in-Aid for Challenging Exploratory Research (grant no. 24659876) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, with accession number GSE97199 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97199).
Authors' contributions
SF, KF, FN and YK conceived and designed the experiments. SF, NG, AI, KK, MK, AS and KS performed the experiments. SF, KF, YA, AI and MK analyzed the data, and SF, KF, NG, JS, MK and YK interpreted the data. JS and KK contributed reagents/materials/analysis tools. SF, KF and YK wrote the paper.
Ethics approval and consent to participate
Ethical approval for the study protocols was provided by Hiroshima University according to the guidelines of its Ethics Committee (approval no. D88-2). The healthy samples from donors at Hiroshima University Hospital were obtained with informed consent. The donor sample from Lonza Group, Ltd. (Basel, Switzerland) was initially donated after obtaining permission for research use by informed consent and legal authorization.
Consent for publication
Not applicable.
Competing interests
YK is a director of Two Cells Co., Ltd., and JS is an employee of Two Cells Co., Ltd. All other authors declare no competing interests.
References
- 1.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo; Proc Natl Acad Sci USA; 2000; pp. 13625–13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanislawski L, Carreau JP, Pouchelet M, Chen ZH, Goldberg M. In vitro culture of human dental pulp cells: Some aspects of cells emerging early from the explant. Clin Oral Investig. 1997;1:131–140. doi: 10.1007/s007840050024. [DOI] [PubMed] [Google Scholar]
- 3.Couble ML, Farges JC, Bleicher F, Perrat-Mabillon B, Boudeulle M, Magloire H. Odontoblast differentiation of human dental pulp cells in explant cultures. Calcif Tissue Int. 2000;66:129–138. doi: 10.1007/PL00005833. [DOI] [PubMed] [Google Scholar]
- 4.He H, Yu J, Liu Y, Lu S, Liu H, Shi J, Jin Y. Effects of FGF2 and TGFbeta1 on the differentiation of human dental pulp stem cells in vitro. Cell Biol Int. 2008;32:827–834. doi: 10.1016/j.cellbi.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Iohara K, Zheng L, Ito M, Tomokiyo A, Matsushita K, Nakashima M. Side population cells isolated from porcine dental pulp tissue with self-renewal and multipotency for dentinogenesis, chondrogenesis, adipogenesis, and neurogenesis. Stem Cells. 2006;24:2493–2503. doi: 10.1634/stemcells.2006-0161. [DOI] [PubMed] [Google Scholar]
- 6.Iohara K, Zheng L, Wake H, Ito M, Nabekura J, Wakita H, Nakamura H, Into T, Matsushita K, Nakashima M. A novel stem cell source for vasculogenesis in ischemia: Subfraction of side population cells from dental pulp. Stem Cells. 2008;26:2408–2418. doi: 10.1634/stemcells.2008-0393. [DOI] [PubMed] [Google Scholar]
- 7.Iohara K, Imabayashi K, Ishizaka R, Watanabe A, Nabekura J, Ito M, Matsushita K, Nakamura H, Nakashima M. Complete pulp regeneration after pulpectomy by transplantation of CD105+ stem cells with stromal cell-derived factor-1. Tissue Eng Part A. 2011;17:1911–1920. doi: 10.1089/ten.tea.2010.0615. [DOI] [PubMed] [Google Scholar]
- 8.Min JH, Ko SY, Cho YB, Ryu CJ, Jang YJ. Dentinogenic potential of human adult dental pulp cells during the extended primary culture. Hum Cell. 2011;24:43–50. doi: 10.1007/s13577-011-0010-7. [DOI] [PubMed] [Google Scholar]
- 9.Shi S, Bartold PM, Miura M, Seo BM, Robey PG, Gronthos S. The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod Craniofac Res. 2005;8:191–199. doi: 10.1111/j.1601-6343.2005.00331.x. [DOI] [PubMed] [Google Scholar]
- 10.Sloan AJ, Waddington RJ. Dental pulp stem cells: What, where, how? Int J Paediatr Dent. 2009;19:61–70. doi: 10.1111/j.1365-263X.2008.00964.x. [DOI] [PubMed] [Google Scholar]
- 11.Spath L, Rotilio V, Alessandrini M, Gambara G, De Angelis L, Mancini M, Mitsiadis TA, Vivarelli E, Naro F, Filippini A, Papaccio G. Explant-derived human dental pulp stem cells enhance differentiation and proliferation potentials. J Cell Mol Med. 2010;14(6b):1–1644. doi: 10.1111/j.1582-4934.2009.00848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran-Hung L, Mathieu S, About I. Role of human pulp fibroblasts in angiogenesis. J Dent Res. 2006;85:819–823. doi: 10.1177/154405910608500908. [DOI] [PubMed] [Google Scholar]
- 13.Yamada Y, Fujimoto A, Ito A, Yoshimi R, Ueda M. Cluster analysis and gene expression profiles: A cDNA microarray system-based comparison between human dental pulp stem cells (hDPSCs) and human mesenchymal stem cells (hMSCs) for tissue engineering cell therapy. Biomaterials. 2006;27:3766–3781. doi: 10.1016/j.biomaterials.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Gronthos S, Shi S. Dental pulp stem cells. Methods Enzymol. 2006;419:99–113. doi: 10.1016/S0076-6879(06)19005-9. [DOI] [PubMed] [Google Scholar]
- 15.Sakai K, Yamamoto A, Matsubara K, Nakamura S, Naruse M, Yamagata M, Sakamoto K, Tauchi R, Wakao N, Imagama S, et al. Human dental pulp-derived stem cells promote locomotor recovery after complete transection of the rat spinal cord by multiple neuro-regenerative mechanisms. J Clin Invest. 2012;122:80–90. doi: 10.1172/JCI59251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura S, Yamada Y, Katagiri W, Sugito T, Ito K, Ueda M. Stem cell proliferation pathways comparison between human exfoliated deciduous teeth and dental pulp stem cells by gene expression profile from promising dental pulp. J Endod. 2009;35:1536–1542. doi: 10.1016/j.joen.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Gottipamula S, Muttigi MS, Kolkundkar U, Seetharam RN. Serum-free media for the production of human mesenchymal stromal cells: A review. Cell Prolif. 2013;46:608–627. doi: 10.1111/cpr.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa I, Sawada R, Kato Y, Tsuji K, Shao J, Yamada T, Kato R, Tsuchiya T. Effectivity of the novel serum-free medium STK2 for proliferating human mesenchymal stem cells. Yakugaku Zasshi. 2009;129:381–384. doi: 10.1248/yakushi.129.381. [DOI] [PubMed] [Google Scholar]
- 19.Tsugeno Y, Sato F, Muragaki Y, Kato Y. Cell culture of human gingival fibroblasts, oral cancer cells and mesothelioma cells with serum-free media, STK1 and STK2. Biomed Rep. 2014;2:644–648. doi: 10.3892/br.2014.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuta M, Nakai Y, Kirino K, Nakagawa M, Sekiguchi K, Nagata S, Matsumoto Y, Yamamoto T, Umeda K, Heike T, et al. Derivation of mesenchymal stromal cells from pluripotent stem cells through a neural crest lineage using small molecule compounds with defined media. PLoS One. 2014;9:e112291. doi: 10.1371/journal.pone.0112291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue H, Takahashi H, Hashimura M, Eshima K, Akiya M, Matsumoto T, Saegusa M. Cooperation of Sox4 with β-catenin/p300 complex in transcriptional regulation of the Slug gene during divergent sarcomatous differentiation in uterine carcinosarcoma. BMC Cancer. 2016;16:53. doi: 10.1186/s12885-016-2090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii S, Fujimoto K, Goto N, Kanawa M, Kawamoto T, Pan H, Srivatanakul P, Rakdang W, Pornprasitwech J, Saskianti T, et al. Characteristic expression of MSX1, MSX2, TBX2 and ENTPD1 in dental pulp cells. Biomed Rep. 2015;3:566–572. doi: 10.3892/br.2015.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasamoto T, Fujimoto K, Kanawa M, Kimura J, Takeuchi J, Harada N, Goto N, Kawamoto T, Noshiro M, Suardita K, et al. DEC2 is a negative regulator for the proliferation and differentiation of chondrocyte lineage-committed mesenchymal stem cells. Int J Mol Med. 2016;38:876–884. doi: 10.3892/ijmm.2016.2660. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Nakashima M, Iohara K, Sugiyama M. Human dental pulp stem cells with highly angiogenic and neurogenic potential for possible use in pulp regeneration. Cytokine Growth Factor Rev. 2009;20:435–440. doi: 10.1016/j.cytogfr.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Nakashima M, Iohara K, Murakami M, Nakamura H, Sato Y, Ariji Y, Matsushita K. Pulp regeneration by transplantation of dental pulp stem cells in pulpitis: A pilot clinical study. Stem Cell Res Ther. 2017;8:61. doi: 10.1186/s13287-017-0506-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song M, Lee JH, Bae J, Bu Y, Kim EC. Human dental pulp stem cells are more effective than human bone marrow-derived mesenchymal stem cells in cerebral ischemic injury. Cell Transplant. 2017;26:1001–1016. doi: 10.3727/096368916X694391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawada R, Yamada T, Tsuchiya T, Matsuoka A. Microarray analysis of the effects of serum-free medium on gene expression changes in human mesenchymal stem cells during the in vitro culture. Yakugaku Zasshi. 2010;130:1387–1393. doi: 10.1248/yakushi.130.1387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, with accession number GSE97199 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97199).