Abstract
A number of studies have attempted to elucidate the association between mircoRNAs (miRNAs/miRs) and cancer-associated processes. The aim of the present study was to determine how miR-499a-5p intervenes in human osteosarcoma cell proliferation and differentiation. The cancerous tissues and adjacent non-cancerous tissues of 62 patients with osteosarcoma (OS) were collected. miRNA microarray analysis revealed that 29 miRNAs were upregulated while 26 were downregulated, among which miR-499a-5p expression was the most decreased. Western blot analysis and reverse transcription-quantitative polymerase chain reaction demonstrated that the mRNA and protein expression of miR-499a-5p was lower, while that of protein phosphatase 1D (PPM1D) was higher in OS tissues compared with expression levels in normal tissues. Furthermore, miR-499a-5p expression was markedly decreased in the metastatic tumors and in those at stage III+IV compared with the non-metastatic tumors and those at stage I, respectively. In addition, following transfection of the human OS MG-63 cell line with an miR-499a-5p mimic, the expression of miR-499a-5p was elevated while the protein and mRNA expression of PPM1D was decreased. When combining these findings with the information obtained from the Targetscan predictive software, it was confirmed that PPM1D was targeted by miR-499a-5p. In MG-63 cells transfected with an miR-499a-5p mimic, PPM1D-associated downstream proteins phosphorylated protein kinase B (p-Akt) and phosphorylated glycogen synthase kinase 3β (p-GSK-3β) were significantly downregulated compared with the negative control (NC) group, while the expression of p-Akt and p-GSK-3β were significantly elevated in the tumor tissues compared with the adjacent non-tumor tissues. Simultaneously, the growth and proliferation activity of MG-63 cells were notably reduced when transfected with the miR-499a-5p mimic, compared with the NC group. Therefore, it may be concluded that miR-499a-5p suppresses OS cell proliferation and differentiation by targeting PPM1D through modulation of Akt/GSK-3β signaling.
Keywords: osteosarcoma, microRNA-499a-5p, protein phosphatase 1D, protein kinase B, glycogen synthase kinase 3β
Introduction
Osteosarcoma (OS), rooted in primitive bone-forming mesenchymal cells and occurring primarily in pediatric patients, is the most common malignant skeletal tumor (1,2). Generally, OS patients experience severe pain and soft-tissue swelling due to tumor cell invasion (3). In addition, the destructive effects on cancer-associated tissues and cells in the majority of cases (80–90%) are localized to the long bones, with fewer cases occurring in the axial and irregular bones (2). Although OS comprises ≤0.5% of all types of cancer (4), the risk of pathological fracture in pediatric patients is up to 15% (5). Furthermore, the 5-year survival rate of OS patients was revealed to be <60% even under the circumstances of great improvement in surgical techniques and chemotherapy and/or radiotherapy regimens (6). Therefore, on the basis of these findings, a great deal of basic studies have contributed toward understanding the mechanisms underpinning OS development. However, these mechanisms remain unclear and require further investigation in basic or clinical studies in order to be elucidated.
MicroRNAs (miRNAs/miRs) consist of 18–25 nucleotides and are a class of short non-coding RNA molecules. miRNAs, as regulators of gene expression, are able to induce the destabilization of mRNA and/or the inhibition of protein translation by directly binding to the 3′-untranslated region (UTR) of their target genes (7,8). It has been reported that miRNAs are involved in the modulation of a variety of biological processes, including cell proliferation, differentiation, apoptosis and migration, and tumorigenesis (8,9). It has also been reported that miR-499a-5p may contribute to the progression of oral squamous cell carcinoma (10). However, as of yet, there are no studies combining clinical and basic experiments regarding the function of miR-499a-5p in regulating OS tissue and cell growth, proliferation and metastasis.
In the present study, OS tissues and the adjacent normal tissues were collected from 62 patients, and the expression of protein phosphatase 1D (PPM1D), also known as wild-type p53-inducible phosphatase-1 (Wip1), which has been reported to be involved in the regulation of cellular proliferation and metastasis (11–14). The objective of the present study was to clarify the association between PPM1D and osteosarcoma.
Materials and methods
Patient tissue samples
A total of 62 primary OS and adjacent non-tumorous tissue samples were collected during surgical resection in the Department of Pathology of Xi'an Jiaotong University Affiliated Hong Hui Hospital (Xi'an, Shaanxi, China) between May 2007 and May 2016. All tissues were rapidly frozen in liquid nitrogen and maintained at −70°C. None of the patients had undergone blood transfusion, chemotherapy or radiotherapy prior to surgery. All the samples were used for experiments with the patients' written informed consent and the approval of the Ethics Committee of Hong Hui Hospital Affiliated to Xi'an Jiaotong University. In order to clarify the association between OS and miR-499a-5p expression, the 62 OS patient tissues were divided into a metastasis group (n=28) and a non-metastasis (n=34) group according to the Tumor-Node-Metastasis (TNM) staging system (15), among which the ratio of male and female was 37: 25 and the average age was 19.3±4.5 years.
miRNA microarray analysis
The total RNA of the OS tissues and the adjacent non-tumorous tissues was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. miRNA microarray analysis (v.16.0; Exiqon A/S, Vedbaek, Denmark) was applied to detect the differential expression of miRNAs in the OS tissues and the adjacent non-tumorous tissues according to the manufacturer's protocol.
MG-63 cell culture and transfection
Human OS MG-63 cells, purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), were cultured in minimum essential medium/Earle's balanced salt solution supplemented with 10% fetal bovine serum, 50 U/ml penicillin and 50 mg/ml streptomycin (all Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) at 37°C in a humidified 5% CO2 atmosphere. Cells were plated onto 6-well plates (5×105 cells/well) and the medium was replaced every 3 days. Once the cells reached 70% confluence, 100 nM miR-499a-5p mimic (forward 5′-GACACGGCTCCGTCAC-3′; reverse 5′-ATCCCTGATCAACTGTCCGCC-3′) or its negative control (miR-499a-5p mimic NC) (forward 5′-AACCAGGCCTCCACCG-3′; reverse 5′-GCCACGCATGTCTTATACTGC-3′) (Sangon Biotech Co., Ltd., Shanghai, China) was transfected into the MG-63 cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After 4–6 h co-transfection, the transfection reagent was exchanged with fresh normal medium, and 24 h later, total RNA or total protein was extracted.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the OS tissues and normal cells transfected with either miR-499a-5p or its NC using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and then l mg total RNA was reverse transcribed using a PrimeScript RT Reagent Kit and a gDNA Eraser kit (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocol. Sequences of all primers were as follows: miR-499a-5p forward, 5′-ATGTAGCGTGCGACCG-3′ and reverse, 5′-CAGGCTGACGCACTCTGTGCT-3′; U6 forward, 5′-CCATCGGAAGCTCGTATACGAAATT-3′ and reverse, 5′-GGCCTCTCGAACTTGCGTGTCAG-3′; PPM1D forward, 5′-TTCAGAGCTCCATAACAAGC-3′ and reverse, 5′-GGACCTATCGCATCCGACCG-3′; and β-actin forward, 5′-GCTGTCCCTGTATGCCTCT-3′ and reverse, 5′-TGTCACGCACGATTTCC-3′. Subsequently, 1 µl miR-499a-5p or PPM1D primers (Sangon Biotech Co., Ltd.), 10 µl SYBRGreen (Takara Biotechnology Co., Ltd.), 6 µl diethylpyrocarbonate (DEPC)-treated water (Sangon Biotech Co., Ltd.) and 2 µl cDNA, with a total volume of 20 µl, was then processed in a Real-Time PCR system (Roche Diagnostics, Basel, Switzerland). The following thermocycling conditions were used: 5 sec at 95°C, followed by 40 cycles of 1 sec at 95°C and 20 sec at 65°C. The comparative 2−ΔΔCq method was used for relative quantification of gene expression (16). Each sample was analyzed ≥3 times.
Luciferase activity assay
A fragment that included the 3′-UTR of human PPM1D (NCBI Reference Sequence: NG_023265.1) was amplified from human genomic DNA using the following primer pair: 5′-TCTTGAACTCTGAGCTCAAGCGATCCACCCACCTCGGCCTCC-3′ and 5′-GTTAAGAAAGTCCCTTAATATTTCAACTGAGATCTGAGTAGC-3′. pGL3-PPM1D-wild reporter was constructed by interception of PCR products of the 3′-UTR of PPM1D, which was cut with XbaI and cloned into the corresponding sites of the pGL3 vector (Promega Corporation, Madison, WI, USA). The QuikChange II Site-Directed Mutagenesis kit (Takara Biotechnology Co., Ltd.) was used to take the mutation into the putative sites of miR-499a-5p recognition, according to the manufacturer's protocol. A total of 400 ng Luciferase reporter, 100 nM miRNA-499a-5p mimic or its NC and 30 ng pRL-TK Renilla Luciferase Reporter Vector (Promega Corporation) were added to each 24-well plate (4–5×104 cells/well). Following transfection with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h, cells were collected and luciferase activity was measured using a Dual-Luciferase® Reporter assay system to compare with renilla luciferase activity (Promega Corporation).
Bioinformatic resources
The online Targetscan software (TargetScanHuman 7.1; http://www.targetscan.org/) was used to predict the target genes of miR-499a-5p with reference to the human gene sequence. The predicted genes identified by the software program were intersected for further analysis.
Western blot analysis
Total protein was extracted from collected tissues and cells using RIPA Lysis Buffer (Beyotime Institute of Biotechnology, Haimen, China) supplemented with 100X proteinase inhibitor and phosphatase inhibitor. Protein quantity was determined using a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology) according to the manufacture's protocol. Proteins (25 µg) were separated with 10% SDS-PAGE. Subsequently, the gel was transferred onto a polyvinylidene difluoride membrane (Merck KGaA, Darmstadt, Germany) and blocked with 5% skimmed milk for 1 h at room temperature. The following rabbit monoclonal primary antibodies were used: Anti-PPM1D (cat. no. 11901; 1:1,000 dilution), anti-p-AKT (ser473; cat. no. 12694; 1:1,000 dilution), anti-p-GSK-3β (ser9; cat. no. 14026; 1:500 dilution), anti-Akt (cat. no. 2938; 1:1,000 dilution), anti-GSK-3β (cat. no. 9315; 1:500 dilution) and anti-β-actin (cat. no. 4970; 1:1,000 dilution) (all Cell Signaling Technology, Inc., Danvers, MA, USA). A secondary mouse anti-rabbit antibody was then used (1:5,000 dilution; A0208; Beyotime, Shanghai, China). The signal was visualized using the enhanced chemiluminescence kit (Invitrogen; Thermo Fisher Scientific, Inc.). The gray bands were analyzed with Image J software (version 1.46; National Institutes of Health, Bethesda, MD, USA) to compare the expression between targeted proteins and internal control of β-actin.
MG-63 cell proliferation assay
To detect the proliferation activity of MG-63 cells transfected with the miR-499a-5p mimic or its NC, the MTT assay was performed using Cell Proliferation kit I (Sigma-Aldrich; Merck KGaA) on 96-well plates at a density of 4×103 cells/well, according to the manufacturer's protocol. After 24, 48 and 72 h of incubation, MTT-formazan (Dissolved in chloroform: 10 mg/ml clear, dark violet) production was estimated by VersaMax (Molecular Devices, LLC, Sunnyvale, CA, USA) at 570 nm to evaluate the proliferation rate. The index was determined at 48 and 72 h normalized to that at 24 h. Subsequently, the images of the transfected cells were captured using a microscope (model no. DP72; Olympus Corporation, Tokyo, Japan) at ×100 magnification.
Statistical analysis
Data are presented as the mean ± standard error of the mean. All statistical analyses were performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). The differences between two groups were compared using a paired Student's t-test and the differences between three or more groups using one-way analysis of variance followed by Tukey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Low expression of miR-499a-5p and high expression of PPM1D mRNA are associated with OS
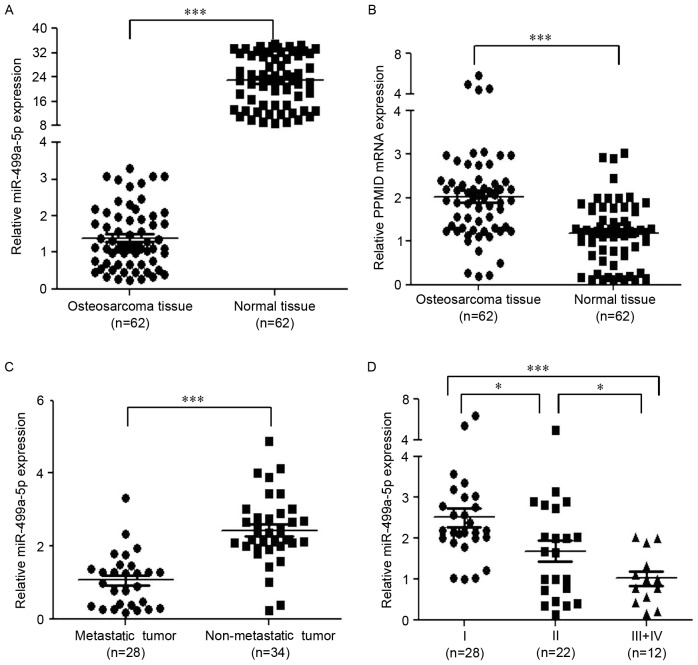
Between May 2007 and May 2016, 62 OS tissues and the adjacent non-tumorous specimens were collected. Using miRNA microarray analysis, it was determined that 29 miRNAs were significantly upregulated, while 26 miRNAs were downregulated in the OS tissues. Among the downregulated miRNAs, miR-499a-5p expression exhibited the largest decrease (Table I). Through RT-qPCR analyses, the expression of miR-499a-5p was observed to be lower (Fig. 1A), while that of PPM1D was higher (Fig. 1B) in cancerous tissues compared with that in the healthy samples. Furthermore, there were 28 metastatic and 34 non-metastatic tumor patients among all the OS patients, and the expression of miR-499a-5p was significantly downregulated in the metastatic samples compared with that in the non-metastatic samples (Fig. 1C). Additionally, with regard to TNM staging (15), tumors in stages III+IV exhibited the lowest expression of miR-499a-5p, while those in stage I, exhibited the highest expression of miR-499a-5p (Fig. 1D).
Table I.
miRNA microarray analyses of differentially expressed miRNAs in OS tissues and adjacent normal tissues (OS tissues/normal tissues).
| miRNAs | Fold-change | P-value |
|---|---|---|
| Upregulated | ||
| hsa-miR-351-3p | 20.18 | 0.0107 |
| hsa-miR-202-3p | 19.04 | 0.0178 |
| hsa-miR-273-3p | 17.95 | 0.0117 |
| hsa-miR-121-5p | 16.77 | 0.0275 |
| hsa-miR-208-3p | 16.39 | 0.0208 |
| hsa-miR-248 | 15.28 | 0.0099 |
| hsa-miR-128-5p | 14.73 | 0.0047 |
| hsa-miR-19-3p | 14.07 | 0.0137 |
| hsa-miR-213 | 13.78 | 0.0472 |
| hsa-miR-192 | 13.07 | 0.0429 |
| hsa-miR-191 | 12.18 | 0.0378 |
| hsa-miR-576-3p | 11.45 | 0.0118 |
| hsa-miR-472-3p | 10.11 | 0.0251 |
| hsa-miR-502a-3p | 9.19 | 0.0187 |
| hsa-miR-521-3p | 8.37 | 0.0049 |
| hsa-miR-541e | 8.01 | 0.0391 |
| hsa-miR-131-5p | 6.89 | 0.0172 |
| hsa-miR-9a-5p | 6.12 | 0.0394 |
| hsa-miR-493-3p | 5.99 | 0.0187 |
| hsa-miR-702-5p | 5.08 | 0.0292 |
| hsa-miR-198 | 4.77 | 0.0031 |
| hsa-miR-130 | 4.54 | 0.0093 |
| hsa-miR-122-5p | 3.93 | 0.0172 |
| hsa-miR-122 | 3.01 | 0.0364 |
| hsa-miR-174 | 2.91 | 0.0208 |
| hsa-miR-215 | 2.66 | 0.0407 |
| hsa-miR-274 | 2.27 | 0.0099 |
| hsa-miR-176 | 2.11 | 0.0152 |
| hsa-miR-18 | 1.93 | 0.0381 |
| Downregulated | ||
| hsa-miR-197-3p | 0.99 | 0.0113 |
| hsa-miR-415-5p | 0.97 | 0.0309 |
| hsa-miR-1406 | 0.87 | 0.0293 |
| hsa-miR-13a-5p | 0.82 | 0.0183 |
| hsa-miR-311a | 0.78 | 0.0211 |
| hsa-miR-307b | 0.72 | 0.0191 |
| hsa-miR-51-5p | 0.69 | 0.0313 |
| hsa-miR-25b-5p | 0.62 | 0.0091 |
| hsa-miR-25a-5p | 0.59 | 0.0049 |
| hsa-miR-99a-3p | 0.53 | 0.0412 |
| hsa-miR-110 | 0.51 | 0.0279 |
| hsa-miR-238-3p | 0.45 | 0.0019 |
| hsa-miR-47-5p | 0.44 | 0.0373 |
| hsa-miR-29-3p | 0.42 | 0.0147 |
| hsa-miR-37-5p | 0.39 | 0.0045 |
| hsa-miR-215 | 0.37 | 0.0078 |
| hsa-miR-27-3p | 0.35 | 0.0128 |
| hsa-miR-90 | 0.29 | 0.0327 |
| hsa-miR-77-5p | 0.27 | 0.0401 |
| hsa-miR-398 | 0.16 | 0.0019 |
| hsa-miR-187 | 0.11 | 0.0108 |
| hsa-miR-150 | 0.09 | 0.0071 |
| hsa-miR-381-3p | 0.07 | 0.0131 |
| hsa-miR-294 | 0.06 | 0.0307 |
| hsa-miR-201-5p | 0.05 | 0.0287 |
| hsa-miR-499a-5p | 0.04 | 0.0175 |
miRNA, microRNA; OC, osteosarcoma.
Figure 1.
Low expression of miR-499a-5p is negatively associated with tumor metastasis and TNM classification of osteosarcoma. (A) Expression of miR-499a-5p is significantly lower in osteosarcoma tissues than in the adjacent normal tissues. (B) Expression of PPM1D mRNA is markedly higher in osteosarcoma tissues than in the adjacent normal tissues. (C) miR-499a-5p expression is lower in metastatic osteosarcoma tissues than in non-metastatic tumor tissues. (D) Relative expression of miR-499a-5p in different TNM stages. miR-499a-5p expression in stage III+IV is the lowest among the 3 stages. *P<0.05, ***P<0.001. miR, miRNA; PPM1D, protein phosphatase 1D; TNM, tumor-node-metastasis.
PPM1D is targeted by miR-499a-5p in MG-63 cells
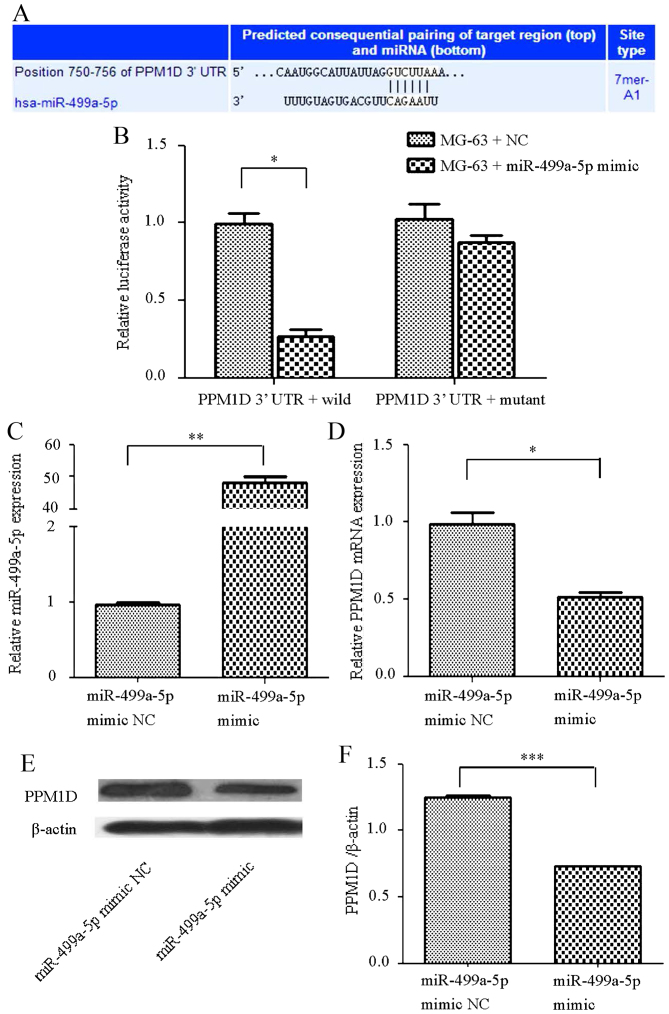
In order to confirm that PPM1D is targeted by miR-499a-5p, the potential targets of miR-499a-5p were examined using Targetscan software and the conserved predicted consequential pairing of the PPM1D 3′-UTR of miR-499a-5p was observed (Fig. 2A). In addition, human OS MG-63 cells were transfected with an miR-499a-5p mimic or an NC, and the luciferase activity was notably suppressed in the PPM1D 3′-UTR wild-type group. However, no statistically significant differences were observed in the mutant group (Fig. 2B). When cells were transfected with the mimic, miR-499a-5p expression was elevated (Fig. 2C), while PPM1D mRNA and protein expression was significantly decreased compared with that of the NC (Fig. 2D-F).
Figure 2.
PPM1D is inversely targeted by miR-499a-5p in the human MG-63 osteosarcoma cell line. (A) The predicted consequential pairing of miR-499a-5p and 3′-UTR of PPM1D mRNA. (B) Following transfection of the MG-63 cell line with an miR-499a-5p mimic in the wild-type 3′-UTR of PPM1D, the relative luciferase activity was markedly suppressed in comparison with its NC. No statistical significance was observed when the MG-63 cell line was transfected with miR-499a-5p in the mutant 3′UTR of PPM1D. Relative expression of (C) miR-499a-5p was notably upregulated and relative expression of (D) PPM1D mRNA was downregulated when MG-63 cells were transfected with an miR-499a-5p mimic. (E) Western blot analysis and (F) statistical analysis demonstrating that the relative expression of PPM1D protein was significantly downregulated by transfection of MG-63 cells with an miR-499a-5p mimic compared with the NC group. *P<0.05, **P<0.01, ***P<0.001. PPM1D, protein phosphatase 1D; UTR, untranslated region, NC, negative control; miR, microRNA.
miR-499a-5p inversely regulates MG-63 cell growth and proliferation by targeting PPM1D through Akt/GSK-3β signaling
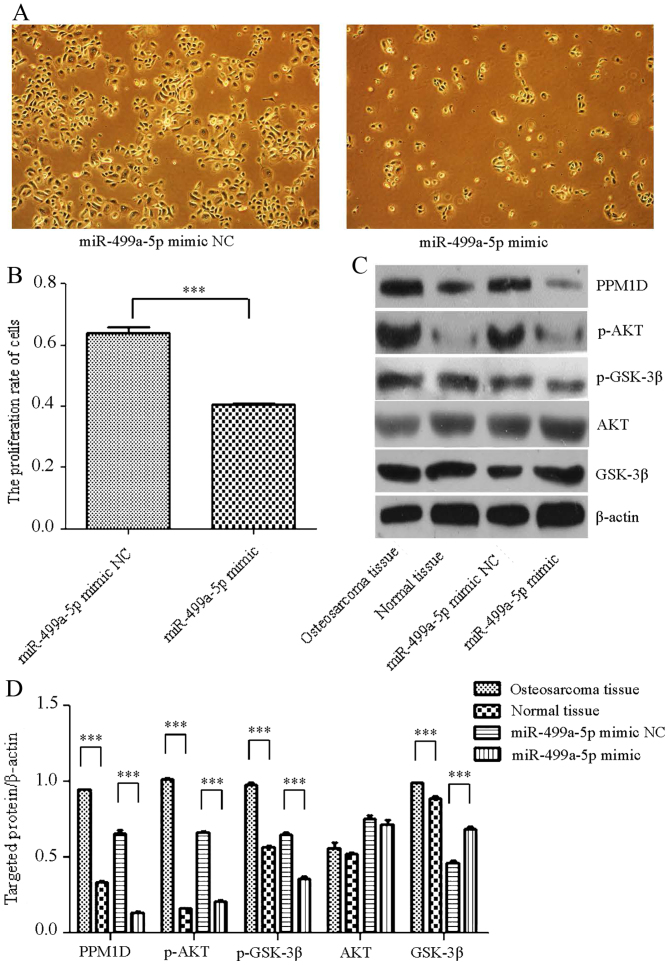
Following transfection of MG-63 cells with an miR-499a-5p mimic, fewer cells were observed in the transfection group compared with the number in the NC group (Fig. 3A). Using the Cell Proliferation kit, it was observed that the proliferation rate of the cells transfected with the miR-499a-5p mimic was significantly decreased (Fig. 3B). In addition, total protein was extracted from the tumorous tissues, the non-tumorous tissues and the transfected cells. Western blot analysis indicated that the expression of PPM1D, as well as that of the downstream proteins of p-AKT and p-GSK-3β, was upregulated in OS tissues and downregulated in cells transfected with the miR-499a-5p mimic compared with expression in the normal tissues and the miR-499a-5p mimic NC group, respectively (Fig. 3C). Image J software analyses indicated that the expression of PPM1D, p-AKT and p-GSK-3β was significantly altered (Fig. 3D).
Figure 3.
miR-499a-5p targeting PPM1D suppresses the growth of osteosarcoma tissues and cells through Akt/GSK-3β signaling. (A) Microscopy and (B) statistical analysis demonstrating that the proliferation rate of MG-63 cells transfected with an miR-499a-5p mimic was significantly lower than that in the NC group (×100 magnification). (C) Western blot analysis and (D) statistical analysis demonstrating that the expression of PPM1D and downstream proteins p-Akt and p-GSK-3β was markedly higher in osteosarcoma tissues than in the adjacent NC tissues. However, expression was markedly reduced by transfection of the human osteosarcoma cell line with an miR-499a-5p mimic compared with that in the NC group. ***P<0.001. PPM1D, protein phosphatase 1D; Akt, protein kinase B; GSK-3β, glycogen synthase kinase 3β; NC, negative control; miR, microRNA; p, phosphorylated.
Discussion
A large volume of evidence has demonstrated that miRNAs may regulate human tumor development and may progress as oncogenes or cancer suppressors through mediating the expression of their targets (17,18). Therefore, in order to fully understand the precise mechanisms associated with pathological and physiological processes and associated diseases, miRNAs may provide a novel solution for the diagnosis and therapy of various types of disease, particularly cancer (19). Numerous studies have revealed that miRNAs are involved in the development and progression of OS. Certain aspects of these studies have focused on the role of miRNAs in promoting the development of OS. For example, increased expression of miRNA-148a in OS promotes cancer cell growth by targeting phosphatase and tensin homolog (20), miRNA-181a promotes proliferation and inhibits apoptosis by suppressing component of the cleavage factor Im 25 in OS (21), and miR-26a induces OS cell growth and metastasis via the Wnt/β-catenin pathway (22). However, the majority of studies have illustrated that high levels of miRNAs exert an inhibitory effect on OS. For instance, miRNA-101 inhibited OS cell migration and invasion through regulation of the enhancer of zeste homolog 2 (23), elevated expression of miRNA-646 suppressed OS cell proliferation, migration and invasion by inversely targeting fibroblast growth factor-2 (24), and miRNA-145 inhibited OS cell growth by suppressing the expression of ρ-associated, coiled-coil-containing protein kinase 1 (25). In addition, transfecting OS cells with a plasmid with upregulated miRNA-410 suppressed cell proliferation or promoted cell apoptosis by targeting vascular endothelial growth factor (26). Furthermore, miR-497 has been revealed to be strongly associated with a human OS cell line and its downregulation is believed to contribute toward tumor cell growth and proliferation (19,27). However, no studies have demonstrated an association between the expression of miR-499a-5p and OS. In the present study, 62 patient OS tissues and the adjacent non-tumorous tissues were collected between May 2007 and May 2016. miRNA microarray analysis demonstrated that 29 miRNAs were significantly upregulated, while 26 were downregulated in the OS tissues. Among the upregulated miRNAs, miR-351-3p was elevated ~20-fold and was the most promoted. In addition, among the downregulated miRNAs, miR-499a-5p was decreased 25-fold and was the most declined. Therefore, miR-499a-5p was selected for the subsequent experiments. It was identified that in the metastasis group, miR-499a-5p expression was markedly lower than that in the non-metastasis group. Furthermore, as the patients were further divided into stage I, II or III+IV groups according to the TNM stage classification system (15), miR-499a-5p expression was notably declined in the stage III+IV tumors compared with that in stage I. These data indicate that miR-499a-5p was inversely associated with tumor tissue differentiation and metastasis.
PPM1D is overexpressed in a number of human cancer types, including breast carcinomas, neuroblastomas, pancreatic adenocarcinomas, gastric carcinomas and medulloblastomas (28–32). However, in certain types of cancer, it has been reported that PPM1D negatively regulates neutrophil migration and inflammation (33) and suppresses ovarian cancer metastasis (12). The function of PPM1D in cancer metastasis appears to be type-specific. In the present study, not only was an inverse association between miR-499a-5p expression and OS observed, but protein and mRNA levels of PPM1D were revealed to be upregulated in OS tissues. Additionally, in MG-43 cells transfected with miR-499a-5p, the mRNA and protein expression of PPM1D was downregulated. The results of the Targetscan predictive software and transfection of MG-63 cells with an miR-499a-5p mimic demonstrated that PPM1D was targeted by miR-499a-5p. In the human OS SAOS-2 cell line, the downregulation of miR-497 contributes toward cell growth and cisplatin resistance through the Akt pathway (18), and in ovarian cancer lines, Wip1 suppresses ovarian cancer metastasis through the ataxia telangiectasia-mutated protein/Akt/Snail-mediated signaling pathway (12). In the present study, the proliferation rate of MG-63 cells was significantly decreased following transfection with an miR-499a-5p mimic which promoted the expression of miR-499a-5p compared with its NC group. In the OS tissues and the MG-63 cells, expression of PPM1D and downstream proteins p-Akt and p-GSK-3β were all inversely regulated by miR-499a-5p.
In conclusion, the present study provides novel and significant basic evidence demonstrating that miR-499a-5p may target-regulate OS tissue and cell proliferation and differentiation via the PPM1D/Akt/GSK-3β signaling pathways. However, the exact mechanism underpinning this may require further investigation in the future.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Zheng H, Min J. Role of long noncoding RNA HOTAIR in the growth and apoptosis of osteosarcoma cell MG-63. Biomed Res Int. 2016;2016:5757641. doi: 10.1155/2016/5757641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angiero F, Moltrasio F, Cattoretti G, Valente MG. Clinical and histopathological profile of primary or secondary osteosarcoma of the jaws. Anticancer Res. 2011;31:4485–4489. [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 5.Picci P. Osteosarcoma (osteogenic sarcoma) Orphanet J Rare Dis. 2007;2:6. doi: 10.1186/1750-1172-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desandes E. Survival from adolescent cancer. Cancer Treat Rev. 2007;33:609–615. doi: 10.1016/j.ctrv.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Maeda Y, Farina NH, Matzelle MM, Fanning PJ, Lian JB, Gravallese EM. Synovium-derived MicroRNAs regulate bone pathways in rheumatoid arthritis. J Bone Miner Res. 2017;32:461–472. doi: 10.1002/jbmr.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou W, Zou B, Liu L, Cui K, Gao J, Yuan S, Cong N. MicroRNA-98 acts as a tumor suppressor in hepatocellular carcinoma via targeting SALL4. Oncotarget. 2016;7:74059–74073. doi: 10.18632/oncotarget.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi E, Choi E, Hwang KC. MicroRNAs as novel regulators of stem cell fate. World J Stem Cells. 2013;5:172–187. doi: 10.4252/wjsc.v5.i4.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou YY, Lee JH, Chen HC, Yang CM, Huang SJ, Liou HH, Chi CC, Tsai KW, Ger LP. The association between miR-499a polymorphism and oral squamous cell carcinoma progression. Oral Dis. 2015;21:195–206. doi: 10.1111/odi.12241. [DOI] [PubMed] [Google Scholar]
- 11.Zhao M, Zhang H, Zhu G, Liang J, Chen N, Yang Y, Liang X, Cai H, Liu W. Association between overexpression of Wip1 and prognosis of patients with non-small cell lung cancer. Oncol Lett. 2016;11:2365–2370. doi: 10.3892/ol.2016.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin S, Wang P, Yang L, Liu Y, Wang Y, Liu M, Qi Z, Meng J, Shi TY, Yang G, Zang R. Wip1 suppresses ovarian cancer metastasis through the ATM/AKT/Snail mediated signaling. Oncotarget. 2016;7:29359–29370. doi: 10.18632/oncotarget.8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70:7176–7186. doi: 10.1158/0008-5472.CAN-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, Qi L, Han W, Wan X, Jiang S, Li Y, Xie Y, Liu L, Zeng F, Liu Z, Zu X. Overexpression of wip1 is associated with biologic behavior in human clear cell renal cell carcinoma. PLoS One. 2014;9:e110218. doi: 10.1371/journal.pone.0110218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng J, Zhang R, Pan Y, Wang B, Wu L, Jiao X, Bao T, Hao X, Liang H. Comparison of the staging of regional lymph nodes using the sixth and seventh editions of the tumor-node-metastasis (TNM) classification system for the evaluation of overall survival in gastric cancer patients: Findings of a case-control analysis involving a single institution in China. Surgery. 2014;156:64–74. doi: 10.1016/j.surg.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Dang X, Ma A, Yang L, Hu H, Zhu B, Shang D, Chen T, Luo Y. MicroRNA-26a regulates tumorigenic properties of EZH2 in human lung carcinoma cells. Cancer Genet. 2012;205:113–123. doi: 10.1016/j.cancergen.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. doi: 10.1371/journal.pone.0028486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao XJ, Miao MH, Xue J, Xue J, Ji XQ, Zhu H. The down-regulation of MicroRNA-497 contributes to cell growth and cisplatin resistance through PI3K/Akt pathway in osteosarcoma. Cell Physiol Biochem. 2015;36:2051–2062. doi: 10.1159/000430172. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Wang Y, Xu T, Li C, Wu J, He Q, Wang G, Ding C, Liu K, Tang H, Ji F. Increased expression of microRNA-148a in osteosarcoma promotes cancer cell growth by targeting PTEN. Oncol Lett. 2016;12:3208–3214. doi: 10.3892/ol.2016.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng S, Gu L, Ju F, Zhang H, Wang Y, Tang H, Bi Z, Yang C. MicroRNA-224 promotes the sensitivity of osteosarcoma cells to cisplatin by targeting Rac1. J Cell Mol Med. 2016;20:1611–1619. doi: 10.1111/jcmm.12852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Qu F, Li CB, Yuan BT, Qi W, Li HL, Shen XZ, Zhao G, Wang JT, Liu YJ. MicroRNA-26a induces osteosarcoma cell growth and metastasis via the Wnt/β-catenin pathway. Oncol Lett. 2016;11:1592–1596. doi: 10.3892/ol.2015.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang K, Zhang Y, Ren K, Zhao G, Yan K, Ma B. MicroRNA-101 inhibits the metastasis of osteosarcoma cells by downregulation of EZH2 expression. Oncol Rep. 2014;32:2143–2149. doi: 10.3892/or.2014.3459. [DOI] [PubMed] [Google Scholar]
- 24.Sun XH, Geng XL, Zhang J, Zhang C. miRNA-646 suppresses osteosarcoma cell metastasis by downregulating fibroblast growth factor 2 (FGF2) Tumour Biol. 2015;36:2127–2134. doi: 10.1007/s13277-014-2822-z. [DOI] [PubMed] [Google Scholar]
- 25.Li E, Zhang J, Yuan T, Ma B. MiR-145 inhibits osteosarcoma cells proliferation and invasion by targeting ROCK1. Tumour Biol. 2014;35:7645–7650. doi: 10.1007/s13277-014-2031-9. [DOI] [PubMed] [Google Scholar]
- 26.Zhao D, Jia P, Wang W, Zhang G. VEGF-mediated suppression of cell proliferation and invasion by miR-410 in osteosarcoma. Mol Cell Biochem. 2015;400:87–95. doi: 10.1007/s11010-014-2265-2. [DOI] [PubMed] [Google Scholar]
- 27.Ruan WD, Wang P, Feng S, Xue Y, Zhang B. MicroRNA-497 inhibits cell proliferation, migration, and invasion by targeting AMOT in human osteosarcoma cells. Onco Targets Ther. 2016;9:303–313. doi: 10.2147/OTT.S95204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, Gabriele T, McCurrach ME, Marks JR, Hoey T, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 29.Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, Yokoyama T, Nozawa S, Inazawa J, Imoto I. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res. 2003;9:1995–2004. [PubMed] [Google Scholar]
- 30.Loukopoulos P, Shibata T, Katoh H, Kokubu A, Sakamoto M, Yamazaki K, Kosuge T, Kanai Y, Hosoda F, Imoto I, et al. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: Identification of genetic indicators that predict patient outcome. Cancer Sci. 2007;98:392–400. doi: 10.1111/j.1349-7006.2007.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrbrecht A, Müller U, Wolter M, Hoischen A, Koch A, Radlwimmer B, Actor B, Mincheva A, Pietsch T, Lichter P, et al. Comprehensive genomic analysis of desmoplastic medulloblastomas: Identification of novel amplified genes and separate evaluation of the different histological components. J Pathol. 2006;208:554–563. doi: 10.1002/path.1925. [DOI] [PubMed] [Google Scholar]
- 32.Fuku T, Semba S, Yutori H, Yokozaki H. Increased wild-type p53-induced phosphatase 1 (Wip1 or PPM1D) expression correlated with downregulation of checkpoint kinase 2 in human gastric carcinoma. Pathol Int. 2007;57:566–571. doi: 10.1111/j.1440-1827.2007.02140.x. [DOI] [PubMed] [Google Scholar]
- 33.Sun B, Hu X, Liu G, Ma B, Xu Y, Yang T, Shi J, Yang F, Li H, Zhang L, Zhao Y. Phosphatase Wip1 negatively regulates neutrophil migration and inflammation. J Immunol. 2014;192:1184–1195. doi: 10.4049/jimmunol.1300656. [DOI] [PubMed] [Google Scholar]