Abstract
F-box only protein 31 (FBXO31), initially identified in 2005, is a novel subunit of the S-phase kinase associated protein 1-Cullin 1-F-box ubiquitin ligase. As with other F-box proteins, FBXO31 may interact with several proteins to promote their ubquitination and subsequent degradation in an F-box-dependent manner. It has been revealed that FBXO31 serves a crucial role in DNA damage response and tumorigenesis. However, the expression and function of FBXO31 varies in different types of human cancer. To the best of our knowledge, the present review is the first to summarize the role of FBXO31 in different types of human cancer and determine its underlying mechanisms, thereby paving the road for the design of FBXO31-targeted anticancer therapies.
Keywords: F-box protein, F-box only protein 31, tumorigenesis, DNA damage, ubiquitination, cell cycle
1. Introduction
The ubiquitin proteasome system (UPS) regulates diverse cellular processes, including cell proliferation, cell cycle progression and apoptosis (1–3). The UPS exerts its functions through the sequential action of three enzymes, namely, the ubiquitin-activating E1 enzyme, the ubiquitin-conjugating E2 enzyme and the ubiquitin-protein E3 ligase (4,5). Notably, E3 ligases determine substrate specificity for ubiquitination and subsequent degradation (4,5).
In the human genome >600 putative E3 ubiquitin ligases have been identified (6). Within this group, the cullin-RING E3 ligase (CRL) complex family is the largest. The CRL family contains eight members, including CRL1, CRL2, CRL3, CRL4A, CRL4B, CRL5, CRL7 and CRL9 (6,7). CRL1, also known as the S-phase kinase associated protein 1 (Skp1)-Cullin 1-F-box protein (SCF) E3 ligase complex, is the most well-characterized CRL family member (6,8,9).
F-box proteins (FBPs) contain one subunit of an SCF type of the E3 ubiquitin ligase complex, which serves as a substrate recognition module (10). F-box only protein (FBXO) 31 is a member of the FBP family, which may serve an important role in tumorigenesis (11,12). To the best of our knowledge, the present review is the first to summarize the role of F-box only protein 31 (FBXO31) in different types of cancer and explain its underlying mechanism of action, thereby providing the rationale to design FBXO31-targeted anticancer therapies.
2. F-box proteins
The first FBP to be identified was cyclin F, also known as FBXO1, and was first described by Bai et al (8) in 1996. Each FBP consists of at least two major functional domains, a variable carboxy-terminal domain that binds to specific substrates and the F-box motif, which is responsible for the incorporation of the FBP into the SCF complex (10). So far, a total of 69 human FBPs have been identified (10), which may be divided into three subclasses according to the presence of specific substrate recognition domains. These subclasses consist of the F-Box and WD repeat domain containing (FBXW) subclass, which contains WD40 repeat domains and is comprised of 10 proteins, including FBXW7, β-TRCP1 and β-TRCP2; the F-box and leucine rich repeat protein (FBXL) subclass, which contains leucine-rich repeat domains and is comprised of 22 proteins, including S-phase kinase-associated protein 2 (SKP2), FBXL11 and FBXL19; and the FBXO subclass, which contains various other domains and is comprised of 37 proteins, including FBXO1, FBXO11 and FBXO31 (11,13–15).
FBPs serve a crucial role in various cellular processes, including cell proliferation, the cell cycle, apoptosis, migration, invasion and metastasis, suggesting that they may be associated with tumorigenesis (11,16–18). Due to the diversity of substrates targeted by FBPs for ubiquitination and subsequent degradation, FBPs may be either oncogenic or tumor suppressive. In some instances, depending on the tissue context, FBPs may be used as prognostic markers and therapeutic targets in certain types of cancer (10–12,19).
3. FBXO31
FBXO31, a member of the FBXO subclass of FBPs, is a senescence-related gene located at chromosome 16q24.3 (20). The gene consists of 539 amino acids and has a molecular weight of 60,533 Da (20). FBXO31 is highly expressed in adipose and brain tissues, and expressed in low amounts in the bone marrow at similar levels to other human tissues, including the stomach, pancreas and breast (20).
FBXO31 contains a 40-amino acid F-box domain and is a component of the SCF ubiquitin E3 ligases that mediate the ubiquitination of targeted proteins for proteasomal degradation (21) (Fig. 1). In SCF-FBXO31 E3 ligases, cullin 1 functions as a molecular scaffold, which interacts with the adaptor subunit Skp1 at the amino terminus and with the RING-finger protein ring-box 1 (Rbx1) at the carboxyl terminus (10). Rbx1 recruits specific E2 ubiquitin-conjugating enzymes (UBCs), including Ubc3, Ubc4 and Ubc5 (22). Conversely, Skp1 binds to the FBXO31, which specifically recognizes and binds to a number of substrates via a protein-protein interaction domain (23). By degrading various substrates, FBXO31 serves important roles in multiple pathways, including neuronal development (24,25), DNA damage response (26–29) and tumorigenesis (20,26,27,30–37). FBXO31 was initially considered to be a candidate tumor suppressor, as the decreased expression of FBXO31 was identified in breast cancer (20). However, further studies have determined that FBXO31 may be either oncogenic or tumor suppressive.
Figure 1.
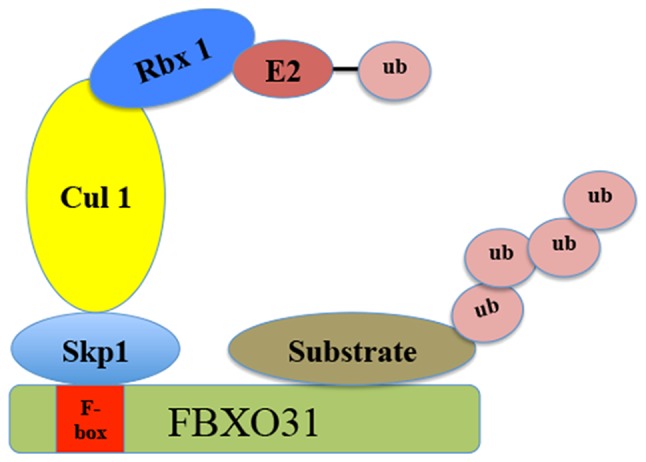
Schematic illustration of Skp1-Cullin 1-F-box E3 ligase. The Skp1-Cullin1-F box complex consists of four components: Skp1, Cul 1, Rbx1, along with the variable F-box protein family that functions as the substrate recognition component. FBXO31 is a member of the F-box protein family, which recognizes the targeted substrates. FBXO31, F-box only protein 31; Skp1, S-phase kinase associated protein 1; Rbx1, ring-box 1; Cul 1, Cullin 1; ub, ubiquitin; E2, the ubiquitin-conjugating E2 enzyme.
4. FBXO31 as a tumor suppressor
Breast cancer
Kumar et al (20) determined that FBXO31 may serve as a tumor suppressor in breast cancer. The ectopic expression of FBXO31 induced cellular senescence in the breast cancer cell line MCF-7 in an F-box domain-dependent manner. The overexpression of FBXO31 inhibited the ability of breast cancer cells to initiate colonies on plastic culture dishes and inhibited the proliferation of breast cancer cells (20). Additionally, levels of FBXO31 mRNA were increased in finite life-span human mammary epithelial cells and nonmalignant immortalized cells compared with breast cancer cell line MCF-7 (20). Furthermore, although no tumor specific mutations were identified, there was a trend associated with lower FBXO31 expression in primary tumors compared with normal breast tissue (20). Song et al (38) also indicated that FBXO31 expression was reduced in the tumor tissues of 10 patients with breast cancer, indicating that FBXO31 may serve a tumor suppressive role.
The underlying mechanism for the tumor suppressive function of FBX031 may be associated with the ubiquitination and degradation of cell cycle-associated substrates. Johansson et al (28) reported that FBXO31 targets chromatin licensing and DNA replication factor 1 (Cdt1), a DNA replication licensing factor, for ubiquitination and degradation in the G2 phase of the cell cycle. This process is independent of SCF-Skp2 but dependent on the CRL4-damage-specific DNA binding protein 1-associated ubiquitination of Cdt1 during the S- and G2 phases of the cell cycle. Cdt1 stabilization in FBXO31-depleted cells results in DNA re-replication, suggesting that FBXO31 may function as a tumor suppressor (21). Jeffery et al (37) demonstrated that FBXO31 is required for normal mitotic progression and genome stability as it caps forkhead box protein M1 (FXOM1) levels during the G2/M transition. Furthermore, it was suggested that FBXO31 targets mouse double minute 2 homolog (MDM2) during ubiquitination and degradation, leading to elevated p53 levels, cell cycle arrest and growth inhibition, supporting the tumor suppressive role of FBXO31 in breast cancer (27). In addition, Manne et al (39) demonstrated that FBXO31 targets the Snail family transcriptional repressor protein 2 (Slug), which is involved in the epithelial-mesenchymal transition, cell invasion, ubiquitination and degradation, and therefore represses the growth of breast cancer cells. Studies have determined that micro (mi)RNAs are dysregulated at all stages of breast cancer and serve important roles in tumorigenesis, invasion and metastasis (40,41). Liu et al (36) indicated that FBXO31 was the downstream target gene of miRNA (miR)-210 in breast cancer, miR-210 and FBXO31 are inversely expressed in breast cancer cell lines T47D and MCF-7. Subsequently, the tumor suppressive effect of miR-210 downregulation may be reversed by further depletion of FBXO31. A study by Manne et al (39) indicated that the oncogenic miRNAs miR93 and miR106a repressed FBXO31 expression, thus impairing the degradation of Slug via FBXO31.
Melanoma
A genome-wide RNA-interference screen initially identified FBXO31 as a candidate gene required for the oncogenic B-Raf serine/threonine protein (BRAF) to induce senescence in primary melanocytes (42). Subsequent results from the same group revealed that the ectopic expression of FBXO31 inhibited the growth of SK-MEL-28 melanoma cells in vitro and in mouse xenografts in vivo (43). It has been proposed that FBXO31 induces G1 arrest in melanoma cells by binding to cyclin D1, a key regulator of G1/S phase transition (44) and promotes its ubiquitination and subsequent degradation. Oncogene-induced senescence involves the activation of DNA damage response pathways (45,46). Additionally, FBXO31, a protein required for BRAF-induced senescence (42), is involved in mediating G1 arrest following DNA damage. It has been indicated that FBXO31 expression is induced following exposure to various DNA damaging agents, including ionizing radiation and is stabilized by ataxia telangiectasia mutated-mediated phosphorylation (43). Induced FBXO31 subsequently binds to and targets cyclin D1 for degradation, causing G1 arrest (43). However, FBXO31 knockdown abrogates G1 arrest following DNA damage and sensitizes melanoma cells to radiation (43). These results suggest that FBXO31 may serve as a radiosensitizing target. Therefore, a small molecule that either inhibits cyclin D1 phosphorylation at Thr286 or disrupts FBXO31-cyclin D1 binding may act as a radiosensitizer (47).
Hepatocellular carcinoma (HCC)
Loss of heterozygosity (LOH) at four microsatellite loci on chromosome 16q24.3, the region harboring FBXO31, was identified in 8.9–21.8% of HCC specimens (48). Huang et al (30) were the first to report the role of FBXO31 in HCC. The results demonstrated that FBXO31 expression was increased in the fetal liver and the fetal liver-derived cell line L02; however, low expression was observed in the majority of HCC cell lines including BEL-7402, BEL-7404, Hep3B and SMMC-7721. Additionally, FBXO31 mRNA expression was downregulated in 93.75% (30/32) of HCC specimens compared with adjacent healthy liver tissues (30). Further experiments revealed that the ectopic expression of FBXO31 inhibited cell proliferation and colony formation in the HCC cell lines HepG2 and Hep3B. These results suggest that the downregulation of cyclin D1 following FBXO31 overexpression is the proposed mechanism by which FBXO31 inhibits the development of HCC.
Gastric cancer (GC)
Mori et al (49) demonstrated that LOH occurs frequently in GC on chromosome band 16q24, including 16q24.3 where FBXO31 is encoded. Zhang et al (32) identified that levels of FBXO31 mRNA and protein were markedly decreased in GC tissue compared with adjacent non-cancerous tissue. In addition, immunohistochemical analyses revealed an association between FBXO31 levels and tumor size, tumor infiltration, clinical grade and patient prognosis. Furthermore, the results indicated that the overall survival rates of patients with negative FBXO31 expression was significantly poorer than that of patients who were FBXO31-positive. The results of previous studies using the GC cell lines BGC-823 and HGC-27 suggest that FBXO31 overexpression significantly decreases colony formation, induces G1 cell cycle arrest and inhibits the expression of cyclin D1 protein. These studies also indicate that these processes are dependent on the F-box domain of FBXO31. Furthermore, it was demonstrated that the ectopic expression of FBXO31 markedly inhibits xenograft tumor growth in nude mice exhibiting tumors with low levels of cyclin D1 expression and high levels of FBXO31 expression. This suggests that FBXO31 may function as a tumor suppressor by inhibiting cyclin D1 expression. FBXO31 knockdown induced the opposite effects (27).
Zhang et al (32) explored the role of miRNA on FBXO31 expression in GC and identified that miR-20a and miR-17 directly bind to the 3′-untranslated region of FBXO31 to regulate FBXO31 expression. Furthermore, a strong negative correlation between miR-20a or miR-17 and FBXO31 was observed in GC samples. These results indicate that FBXO31 may suppress GC progression via the miR-20a-miR-17-FBXO31-cyclin D1 signaling pathway.
Ovarian cancer
Launonen et al (34) assessed the frequency of LOH in 78 tumor specimens of epithelial ovarian cancer and identified that LOH at 16q24.3, a region where the FBXO31 gene is located, was associated with advanced tumor grade, serous histology, a more advanced tumor stage and metastasis. These results suggest that FBXO31 may be a potential tumor suppressor in ovarian cancer, however, further studies focusing on the role of FBXO31 in ovarian cancer are required to explore this.
Prostate cancer
Härkönen et al (35) detected the frequency of LOH in 74 tumor specimens collected from 33 patients with prostate cancer and indicated that the frequency of LOH at 16q was 68% (21/31) for primary prostate tumors, 90% (28/31) for locally recurrent tumors and 75% (9/12) for tumors of metastatic origin. Furthermore, the region 16q24.3, where FBXO31 is located, exhibited a significant development of LOH during disease recurrence, suggesting that this region harbors gene(s) associated with prostate cancer progression. However, multiple tumor suppressor genes are located at this region with FBXO31 (50–54) and no studies thus far have detected the expression and function of FBXO31 in prostate cancer; therefore, FBXO31 may only be regarded as a potential tumor suppressor.
Colon cancer
FBXO31 binds to MDM2 and targets it for ubiquitination and degradation following DNA damage (27), causing an increase in p53 expression. MDM2 and p53 expression following DNA damage did not significantly change in HCT116 colon cancer cells following FBXO31 knockdown; however, the mitotic index of these cells was markedly higher than that of control HCT116 cells at 18 and 24 h following γ-irradiation (27). In addition, the difference in mitotic index was correlated with the expression of p53 and the p53 target gene p21, which serves a critical role in p53-mediated growth arrest (55). These results suggest that FBXO31 may be a potential tumor suppressor in colon cancer.
5. FBXO31 as an oncogene
Esophageal squamous cell carcinoma (ESCC)
Kogo et al (31) indicated that the high expression of FBXO31 mRNA was associated with increased tumor invasion and a more advanced clinical stage. Furthermore, the study suggested that FBXO31 expression was an independent prognostic factor for 5-year overall in patients with ESCC following surgery, indicating that FBXO31 may act as an oncogene in ESCC. Cyclin D1 is a well-characterized oncogene in ESCC (56–58) and FBXO31 may mediate the ubiquitination and degradation of cyclin D1 (43); therefore the association between cyclin D1 and FBXO31 expression in ESCC was evaluated. The results of immunohistochemistry indicated that cyclin D1 and FBXO31 expression were increased in ESCC tissue. Additionally, a comparative genomic hybridization array indicated that cases with high cyclin D1 amplification exhibited elevated FBXO31 expression, suggesting that FBXO31 may degrade cyclin D1 as it moves from the nucleus to the cytoplasm (31). However, Liu et al (26) demonstrated that FBXO31 knockdown does not affect cyclin D1 protein expression, nor does it rescue DNA damage-induced cyclin D1 degradation in ESCC cell lines. Furthermore, the role of FBPs, including FBXO4, FBXO31, Skp2 and FBXW8, in regulating cyclin D1 stability was re-evaluated (59). The results indicated that FBXO31 knockdown did not impair cyclin D1 proteolysis in mouse embryonic fibroblasts, suggesting that FBXO31 may only regulate cyclin D1 stability in a cell type-specific manner. Therefore, the molecular mechanism involved in overexpression of FBXO31 in ESCC remains unclear. Liu et al (26) demonstrated that FBXO31 reduces stress-induced cell apoptosis by inducing lys-48-linked polyubiquitination and the degradation of mitogen-activated protein kinase kinase 6 (MKK6) in ESCC, suggesting that FBXO31 may serve an oncogenic role in ESCC by modulating cell apoptosis.
Lung cancer
Huang et al (33) analyzed the expression of FBXO31 in 50 patients with lung cancer (36 patients had adenocarcinoma and 14 patients had squamous cell cancer). The results indicated that levels of FBXO31 mRNA were increased in lung cancer tissues compared with non-cancerous lung tissues. Furthermore, the increased expression of FBXO31 was significantly associated with tumor size, tumor infiltration, clinical stage and lymph node metastasis. Additionally, the ectopic expression of FBXO31 promoted the growth, metastasis and invasion of A549 cells, whereas FBXO31 knockdown inhibited these processes. The results of a tumorigenicity assay conducted in nude mice demonstrated that FBXO31 promotes tumor growth in vivo. Therefore, these results suggest that FBXO31 may function as an oncogene in lung cancer.
6. Conclusions and perspectives
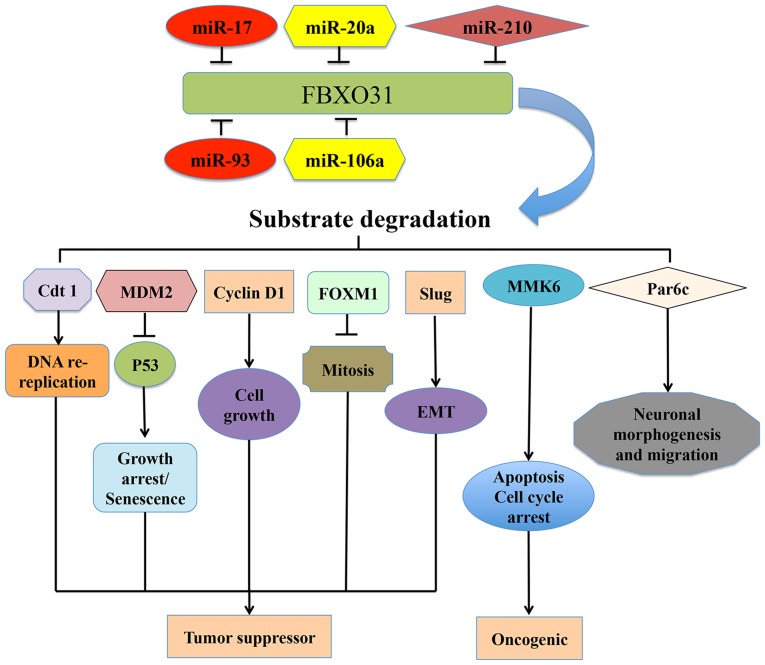
FBXO31 was initially identified in 2005 and is a poorly understood member of the FBP family (20). Furthermore, only a limited number of studies have assessed its role in various diseases, including cancer (20,24–37). With the F-box motif, FBXO31 has the ability to bind to several substrates for ubiquitination and degradation and may therefore be associated with various functions, including cell cycle progression, DNA damage and cell proliferation (25–28,37,39,43). A total of 7 FBXO31-associated substrates have been identified, including Cdt1 (28), MDM2 (27), cyclin D1 (43), MKK6 (26), FXOM1 (37), Par6c (25) and Slug (39) (Fig. 2).
Figure 2.
Upstream regulators of FBXO31 and its major downstream targets that contribute to human diseases, including cancer. FBXO31 coordinates the ubiquitin-dependent proteolysis of several proteins, including Cdt1, MDM2, cyclin D1, MKK6, FOXM1, Par6c, Slug and determines their functions in various types of cancer. Various miRNAs, including miR-17, miR-20a, miR-210, miR93 and miR106a regulate the expression of FBXO31. FBXO31, F-box only protein 31; MDM2, mouse double minute 2 homolog; MKK6, mitogen-activated protein kinase kinase 6; FXOM1, forkhead box protein M1; cdt1, chromatin licensing and DNA replication factor 1; miR, microRNA; Slug, Snail family transcriptional repressor protein 2.
In conclusion, the present review suggests that FBXO31 may act a tumor suppressor or oncogene depending on the cell type or tissue context. Furthermore, compared with other well-characterized FBPs, including Skp2, FBXW7 and β-TrCP, current understanding of FBXO31 remains limited. Therefore, further studies are warranted to fully elucidate the role of FBXO31 in various human diseases, including its involvement in the regulation of tumorigenesis and to identify novel substrates and cellular pathways regulated by FBXO31.
Acknowledgements
The present study was supported by National Science Foundation of China grants (grant no. 81502630) and partly supported by the Chinese National Key Disciplines. The authors would also like to thank Dr. Brian J. North from Beth Israel Deaconess Medical Center, Harvard Medical School (Boston, MA, USA), for assisting in the editing of the manuscript.
Glossary
Abbreviations
- FBXO31
F-box only protein 31
- SCF
Skp1-Cullin1-F box
- UPS
ubiquitin proteasome system
- CRL
Cullin-RING E3 ligase
- FBPs
F-box proteins
- MDM2
mouse double minute 2 homolog
- FXOM1
Forkhead box protein M1
- HCC
hepatocellular carcinoma
- LOH
loss of heterozygosity
- GC
gastric cancer
- ESCC
esophageal squamous cell carcinoma
- MKK6
mitogen-activated protein kinase kinase 6
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 3.Bett JS. Proteostasis regulation by the ubiquitin system. Essays Biochem. 2016;60:143–151. doi: 10.1042/EBC20160001. [DOI] [PubMed] [Google Scholar]
- 4.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: Biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 5.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 8.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/S0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 9.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong J, Lv L, Huo J. Roles of F-box proteins in human digestive system tumors (Review) Int J Oncol. 2014;45:2199–2207. doi: 10.3892/ijo.2014.2684. [DOI] [PubMed] [Google Scholar]
- 12.Zheng N, Zhou Q, Wang Z, Wei W. Recent advances in SCF ubiquitin ligase complex: Clinical implications. Biochim Biophys Acta. 2016;1866:12–22. doi: 10.1016/j.bbcan.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 14.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW. A family of mammalian F-box proteins. Curr Biol. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- 16.Diaz VM, de Herreros AG. F-box proteins: Keeping the epithelial-to-mesenchymal transition (EMT) in check. Semin Cancer Biol. 2016;36:71–79. doi: 10.1016/j.semcancer.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Heo J, Eki R, Abbas T. Deregulation of F-box proteins and its consequence on cancer development, progression and metastasis. Semin Cancer Biol. 2016;36:33–51. doi: 10.1016/j.semcancer.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randle SJ, Laman H. F-box protein interactions with the hallmark pathways in cancer. Semin Cancer Biol. 2016;36:3–17. doi: 10.1016/j.semcancer.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Zheng N, Wang Z, Wei W. Ubiquitination-mediated degradation of cell cycle-related proteins by F-box proteins. Int J Biochem Cell Biol. 2016;73:99–110. doi: 10.1016/j.biocel.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar R, Neilsen PM, Crawford J, McKirdy R, Lee J, Powell JA, Saif Z, Martin JM, Lombaerts M, Cornelisse CJ, et al. FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 2005;65:11304–11313. doi: 10.1158/0008-5472.CAN-05-0936. [DOI] [PubMed] [Google Scholar]
- 21.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/S0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 22.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardozo T, Pagano M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 24.Mir A, Sritharan K, Mittal K, Vasli N, Araujo C, Jamil T, Rafiq MA, Anwar Z, Mikhailov A, Rauf S, et al. Truncation of the E3 ubiquitin ligase component FBXO31 causes non-syndromic autosomal recessive intellectual disability in a Pakistani family. Hum Genet. 2014;133:975–984. doi: 10.1007/s00439-014-1438-0. [DOI] [PubMed] [Google Scholar]
- 25.Vadhvani M, Schwedhelm-Domeyer N, Mukherjee C, Stegmüller J. The centrosomal E3 ubiquitin ligase FBXO31-SCF regulates neuronal morphogenesis and migration. PLoS One. 2013;8:e57530. doi: 10.1371/journal.pone.0057530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Han L, Li B, Yang J, Huen MS, Pan X, Tsao SW, Cheung AL. F-box only protein 31 (FBXO31) negatively regulates p38 mitogen-activated protein kinase (MAPK) signaling by mediating lysine 48-linked ubiquitination and degradation of mitogen-activated protein kinase kinase 6 (MKK6) J Biol Chem. 2014;289:21508–21518. doi: 10.1074/jbc.M114.560342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malonia SK, Dutta P, Santra MK, Green MR. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress; Proc Natl Acad Sci USA; 2015; pp. 8632–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson P, Jeffery J, Al-Ejeh F, Schulz RB, Callen DF, Kumar R, Khanna KK. SCF-FBXO31 E3 ligase targets DNA replication factor Cdt1 for proteolysis in the G2 phase of cell cycle to prevent re-replication. J Biol Chem. 2014;289:18514–18525. doi: 10.1074/jbc.M114.559930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiloh Y. FBXO31: A new player in the ever-expanding DNA damage response orchestra. Sci Signal. 2009;2:pe73. doi: 10.1126/scisignal.296pe73. [DOI] [PubMed] [Google Scholar]
- 30.Huang HL, Zheng WL, Zhao R, Zhang B, Ma WL. FBXO31 is down-regulated and may function as a tumor suppressor in hepatocellular carcinoma. Oncol Rep. 2010;24:715–720. doi: 10.3892/or_00000912. [DOI] [PubMed] [Google Scholar]
- 31.Kogo R, Mimori K, Tanaka F, Komune S, Mori M. FBXO31 determines poor prognosis in esophageal squamous cell carcinoma. Int J Oncol. 2011;39:155–159. doi: 10.3892/ijo.2011.1018. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Kong Y, Xu X, Xing H, Zhang Y, Han F, Li W, Yang Q, Zeng J, Jia J, Liu Z. F-box protein FBXO31 is down-regulated in gastric cancer and negatively regulated by miR-17 and miR-20a. Oncotarget. 2014;5:6178–6190. doi: 10.18632/oncotarget.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang HL, Jiang Y, Wang YH, Chen T, He HJ, Liu T, Yang T, Yang LW, Chen J, Song ZQ, et al. FBXO31 promotes cell proliferation, metastasis and invasion in lung cancer. Am J Cancer Res. 2015;5:1814–1822. [PMC free article] [PubMed] [Google Scholar]
- 34.Launonen V, Mannermaa A, Stenbäck F, Kosma VM, Puistola U, Huusko P, Anttila M, Bloigu R, Saarikoski S, Kauppila A, Winqvist R. Loss of heterozygosity at chromosomes 3, 6, 8, 11, 16, and 17 in ovarian cancer: Correlation to clinicopathological variables. Cancer Genet Cytogenet. 2000;122:49–54. doi: 10.1016/S0165-4608(00)00279-X. [DOI] [PubMed] [Google Scholar]
- 35.Härkönen P, Kyllönen AP, Nordling S, Vihko P. Loss of heterozygosity in chromosomal region 16q24.3 associated with progression of prostate cancer. Prostate. 2005;62:267–274. doi: 10.1002/pros.20147. [DOI] [PubMed] [Google Scholar]
- 36.Liu D, Xia H, Wang F, Chen C, Long J. MicroRNA-210 interacts with FBXO31 to regulate cancer proliferation cell cycle and migration in human breast cancer. Onco Targets Ther. 2016;9:5245–5255. doi: 10.2147/OTT.S110969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeffery JM, Kalimutho M, Johansson P, Cardenas DG, Kumar R, Khanna KK. FBXO31 protects against genomic instability by capping FOXM1 levels at the G2/M transition. Oncogene. 2017;36:1012–1022. doi: 10.1038/onc.2016.268. [DOI] [PubMed] [Google Scholar]
- 38.Song Q, Jing H, Wu H, Zhou G, Kajiyama T, Kambara H. Gene expression analysis on a photodiode array-based bioluminescence analyzer by using sensitivity-improved SRPP. Analyst. 2010;135:1315–1319. doi: 10.1039/c0an00012d. [DOI] [PubMed] [Google Scholar]
- 39.Manne RK, Agrawal Y, Bargale A, Patel A, Paul D, Gupta NA, Rapole S, Seshadri V, Subramanyam D, Shetty P, Santra MK. A microRNA/Ubiquitin ligase feedback loop regulates slug-mediated invasion in breast cancer. Neoplasia. 2017;19:483–495. doi: 10.1016/j.neo.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bahrami A, Aledavood A, Anvari K, Hassanian SM, Maftouh M, Yaghobzade A, Salarzaee O, ShahidSales S, Avan A. The prognostic and therapeutic application of microRNAs in breast cancer: Tissue and circulating microRNAs. J Cell Physiol. 2017 Jan 21; doi: 10.1002/jcp.25813. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 41.Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34–49. doi: 10.1016/j.pharmthera.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 42.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alao JP. The regulation of cyclin D1 degradation: Roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 46.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre' M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 47.Jia L, Sun Y. F-box proteins FBXO31 and FBX4 in regulation of cyclin D1 degradation upon DNA damage. Pigment Cell Melanoma Res. 2009;22:518–519. doi: 10.1111/j.1755-148X.2009.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin YW, Lee IN, Chen CH, Huang GT, Lee HS, Lee PH, Lu FJ, Sheu JC. Deletion mapping of chromosome 16q24 in hepatocellular carcinoma in Taiwan and mutational analysis of the 17-beta-HSD gene localized to the region. Int J Cancer. 2001;93:74–79. doi: 10.1002/ijc.1287. [DOI] [PubMed] [Google Scholar]
- 49.Mori Y, Matsunaga M, Abe T, Fukushige S, Miura K, Sunamura M, Shiiba K, Sato M, Nukiwa T, Horii A. Chromosome band 16q24 is frequently deleted in human gastric cancer. Br J Cancer. 1999;80:556–562. doi: 10.1038/sj.bjc.6690391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kremmidiotis G, Baker E, Crawford J, Eyre HJ, Nahmias J, Callen DF. Localization of human cadherin genes to chromosome regions exhibiting cancer-related loss of heterozygosity. Genomics. 1998;49:467–471. doi: 10.1006/geno.1998.5281. [DOI] [PubMed] [Google Scholar]
- 51.Whitmore SA, Settasatian C, Crawford J, Lower KM, McCallum B, Seshadri R, Cornelisse CJ, Moerland EW, Cleton-Jansen AM, Tipping AJ, et al. Characterization and screening for mutations of the growth arrest-specific 11 (GAS11) and C16orf3 genes at 16q24.3 in breast cancer. Genomics. 1998;52:325–331. doi: 10.1006/geno.1998.5457. [DOI] [PubMed] [Google Scholar]
- 52.Pullman WE, Bodmer WF. Cloning and characterization of a gene that regulates cell adhesion. Nature. 1992;356:529–532. doi: 10.1038/356529a0. [DOI] [PubMed] [Google Scholar]
- 53.Graña X, Claudio PP, De Luca A, Sang N, Giordano A. PISSLRE, a human novel CDC2-related protein kinase. Oncogene. 1994;9:2097–2103. [PubMed] [Google Scholar]
- 54.Powell JA, Gardner AE, Bais AJ, Hinze SJ, Baker E, Whitmore S, Crawford J, Kochetkova M, Spendlove HE, Doggett NA, et al. Sequencing, transcript identification, and quantitative gene expression profiling in the breast cancer loss of heterozygosity region 16q24.3 reveal three potential tumor-suppressor genes. Genomics. 2002;80:303–310. doi: 10.1006/geno.2002.6828. [DOI] [PubMed] [Google Scholar]
- 55.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 56.Nakagawa H, Zukerberg L, Togawa K, Meltzer SJ, Nishihara T, Rustgi AK. Human cyclin D1 oncogene and esophageal squamous cell carcinoma. Cancer. 1995;76:541–549. doi: 10.1002/1097-0142(19950815)76:4<541::AID-CNCR2820760402>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 57.Shinozaki H, Ozawa S, Ando N, Tsuruta H, Terada M, Ueda M, Kitajima M. Cyclin D1 amplification as a new predictive classification for squamous cell carcinoma of the esophagus, adding gene information. Clin Cancer Res. 1996;2:1155–1161. [PubMed] [Google Scholar]
- 58.Hou X, Liang RB, Wei JC, Xu Y, Fu JH, Luo RZ, He JH, Zhang LJ, Lin P, Yang HX. Cyclin D1 expression predicts postoperative distant metastasis and survival in resectable esophageal squamous cell carcinoma. Oncotarget. 2016;7:31088–31096. doi: 10.18632/oncotarget.9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanie T, Onoyama I, Matsumoto A, Yamada M, Nakatsumi H, Tateishi Y, Yamamura S, Tsunematsu R, Matsumoto M, Nakayama KI. Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation. Mol Cell Biol. 2012;32:590–605. doi: 10.1128/MCB.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]