Abstract
Unraveling interactions among variables such as genetic, clinical, demographic and environmental factors is essential to understand the development of common and complex diseases. To increase the power to detect such variables interactions associated with clinical time-to-events outcomes, we borrowed established concepts from random survival forest (RSF) models. We introduce a novel RSF-based pairwise interaction estimator and derive a randomization method with bootstrap confidence intervals for inferring interaction significance. Using various linear and nonlinear time-to-events survival models in simulation studies, we first show the efficiency of our approach: true pairwise interaction-effects between variables are uncovered, while they may not be accompanied with their corresponding main-effects, and may not be detected by standard semi-parametric regression modeling and test statistics used in survival analysis. Moreover, using a RSF-based cross-validation scheme for generating prediction estimators, we show that informative predictors may be inferred. We applied our approach to an HIV cohort study recording key host gene polymorphisms and their association with HIV change of tropism or AIDS progression. Altogether, this shows how linear or nonlinear pairwise statistical interactions of variables may be efficiently detected with a predictive value in observational studies with time-to-event outcomes.
Keywords: epistasis, genetic variations interactions, interaction detection and modeling, random survival forest, time-to-event analysis
1 Introduction
Epistasis or gene-gene interaction, as well as gene-environment interactions, are presumed to account for a substantial fraction of missing heritability, and are essential to understand the genetic architecture of complex diseases or traits (Phillips, 2008; Cordell, 2009). Capturing the spectrum of interactions in genome-wide interaction analyses has recently been a subject of active research in statistical methodology (for review see Cordell, 2009; Cantor, Lange & Sinsheimer, 2010; Zhang et al., 2011). Modern statistical modeling techniques are required for discovering statistical interactions of multiple genetic variants in the context of potential complicating factors, such as multiple genetic markers, linkage disequilibrium (LD), and environmental influences. In addition, because of the smallness of (marginal) main-effects (Wang, Elston & Zhu, 2010), predictive variables are often associated with only a small increased risk of disease, indicating that each has only a small individual predictive value. In statistical terms, we would hypothesize that sets of weakly predictive variables can be jointly predictive. Therefore, integrating all interaction-effects of genetic variants as well as environmental factors is deemed to be necessary for a proper assessment of the risk of disease (Cordell, 2009).
Common diseases often involve a complex relationship between genotype and phenotype and have long been recognized as being potentially multifactorial. The role of epistasis in the formation and function of biological pathways and genetic networks is therefore essential (Phillips, 2008) and underscores the importance of correlation and dependence potentially existing between genetic factors that cannot be ignored for modeling purposes. In the past, however, the problem of modeling epistasis has been oversimplified or grossly overlooked. Even though investigators have realized the limitations of such approaches, prevailing analytical methods have primarily focused on univariate modeling ignoring non-additive and nonlinear effects. For instance, up until recently, GWAS or EWAS association studies have been using limited single-locus approaches, which detect single nucleotide polymorphisms (SNPs) or post-transcriptional modifications (PTM), such as methylations or phosphorylations, essentially based on their marginal associations with phenotypes. In this locus-by-locus paradigm, correlation structures and interactions between genetic variables, i.e. joint-effects, are totally ignored (Lunetta et al., 2004; Marchini, Donnelly & Cardon, 2005; Cordell, 2009). In another example, simple LD between loci alone can induce correlation between many genetic variables, and assuming independence between them seems very unrealistic. Traditional analytical methods for association and linkage studies have been therefore clearly limited and underpowered for the task (Cordell, 2009).
Methodological developments for detecting statistical interactions include recent work from R. Tibshirani’s group: in one study, a Lasso approach was used with a hierarchy restriction in that an interaction may only be included in a pairwise interaction model if one or both variables are marginally important (Bien, Taylor & Tibshirani, 2013); in another one, a permutation-based method was used for testing marginal interactions by comparing correlations between classes in a binary response (Simon & Tibshirani, 2015). Although these methods are relevant to the general problem of detecting or testing interactions, the two works are limited to continuous or binary outcomes, respectively. Another approach by the same group deals with the specific problem of estimating interaction-effect between a treatment and a large number of covariates in the context of randomized clinical trials (Tian et al., 2014), which can have potential interesting applications in personalized medicine. There are also a few Bayesian studies for modeling nonlinear, non-additive or interaction covariate effects (Chen et al., 2012; Chipman, George & McCulloch, 1998; Gustafson, 2000). Finally, there is a recent and rich literature for detecting epistasis in GWAS association studies: (Li, Horstman & Chen, 2011; Ueki & Cordell, 2012; Yung et al., 2011; Zhang et al. 2008; 2010a; 2010b; 2011). However, all these existing methods, except for the one by Chen et al. (2012), are not designed for analyzing time-to-event outcomes, possibly with censoring.
In respect to the above current limitations to detect epistasis and model non-additive and nonlinear effects, state-of-the-art machine learning methods such as decision tree-based methods and their ensemble versions are well suited because of their broader notion of statistical interaction over traditional parametric or semiparametric modeling strategies (Lunetta et al., 2004; Cordell, 2009). The capability of ensemble approaches, such as random forest (RF) (Breiman, 2001), and recent extensions to survival outcomes with random survival forest (RSF) (Ishwaran et al. 2008; 2010; 2014) to jointly integrate and prioritize genetic variants with covariates, considering both main and interaction-effects between them, was therefore especially appealing for our study. Breiman’s original RF is one of the most popular ensemble learning methods, and has very broad applications in data mining, machine learning, and biostatistics (Breiman, 2001). RSF, being derived from RF, naturally inherits many of its important properties. One of them is that, being fully non-parametric, it is model-assumption free, and, as an (ensemble) tree-based modeling approach, it is suited to adaptively discover non-monotonic, nonlinear, and non-additive or high-order interaction-effects (Ishwaran et al., 2008; Zhang et al., 2008; Li, Horstman & Chen, 2011). Therefore, these approaches provide a natural alternative to build models that bypass the need to impose parametric constraints on the underlying distributions and a way to automatically deal with high-level interactions; both of which should ultimately result in more accurate predictions and more efficient epistasis detections.
In this study, we took advantage of the aforementioned properties and capabilities of ensemble tree models, such as RF and RSF, as done by others (Zhang et al., 2008; Li, Horstman & Chen, 2011). Specifically, we used RSF to model right-censored time-to-event outcome data and introduce a novel approach that makes use of RF concepts of variable importance (Breiman, 2001) and minimal depth of maximal subtree (Ishwaran, 2007) for selecting and ranking pairwise interaction statistics. The minimal depth statistic is an order statistic related to trees, which was introduced by Ishwaran and shown to overcome several limitations hampered by variable importance [see Ishwaran et al. (2010) and Ishwaran (2007) for details]. In certain applications, it is of interest to evaluate RSF to other standard strategies for building a predictive or epistasis survival/risk model (see e.g. Mogensen, Ishwaran & Gerds, 2012). In this study, we compared prediction and epistasis (pairwise variable interactions) detection performances of RSF models versus Cox regression modeling derived from stepwise variable selection and simple testing with standard statistics non-parametric.
In observational studies, we do not really know how frequent interactions of factors under study are, whether they are observed with or without their main-effects. The consensus has often been to ignore them. This work addresses a simple practical and yet common question in observational studies, where a relatively small number of variables and a time-to-event outcome are observed. Suppose a clinician observes a collection of, e.g. genetic markers with a survival endpoint in a sample of patients, possibly along with other clinical and/or demographic covariates; and suppose that patient outcomes are affected by how these variables interact with each other, then how could he test for specific non-additive and nonlinear effects in an efficient way, without having to design new experimental studies or carry out expensive clinical trials? Can standard Cox Proportional Hazards (Cox-PH) modeling (Cox, 1972) address this problem effectively using simple stepwise variable selection with traditional test statistics? This study lays out the concepts and the approach to achieve this goal and address this question in the context of right-censored time-to-event outcomes in low-dimensional settings. Specifically, we derive estimators from RSF models for their suitability to detect statistical effects in a broader sense than traditional parametric or semi-parametric models, and for being promising alternatives in low and high-dimensional settings (Ishwaran et al. 2008; 2010).
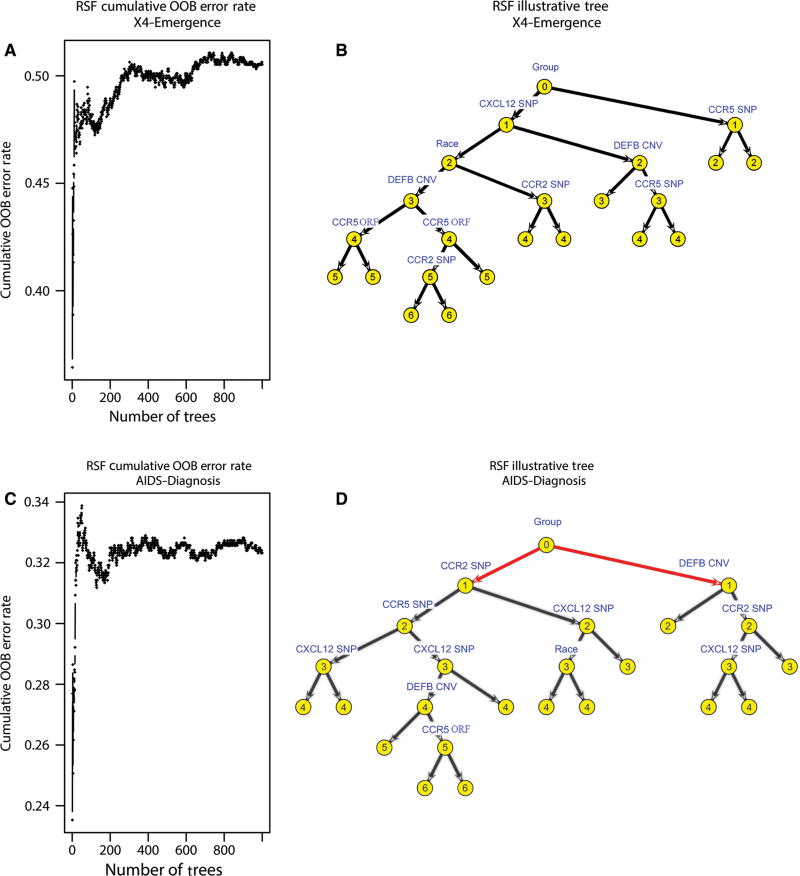
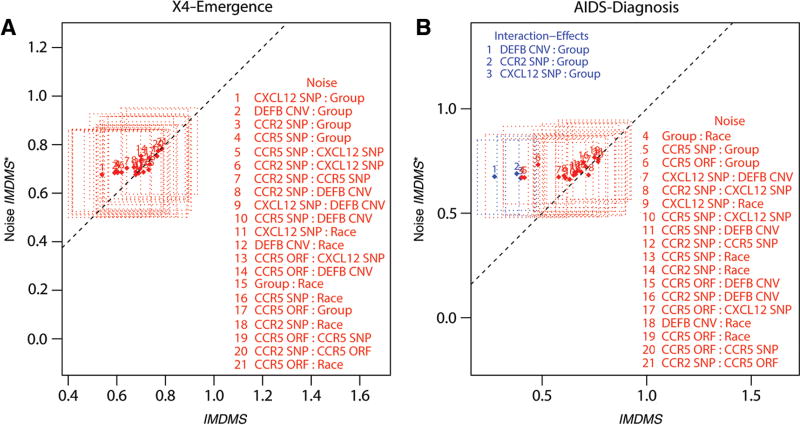
In this study, we first show how our approach works in simulation studies, then illustrate its application in a genetic interactions study from the MACS HIV cohort dataset that motivated this work. In this application study, the focus was specifically on whether copy number variation (CNV) in β-defensin genes (DEFB4/103A locus encoding hBD-2/-3) had any predictive clinical value alone or by interaction with other important known genes (polymorphisms in chemokine receptors and ligands) as well as demographics factors in the outcomes of HIV change of tropism and AIDS progression.
More generally, for datasets with right-censored time-to-event outcomes, the emphasis is not only on the ability to discover the most biologically meaningful variables and/or their interactions for association-to-event purposes, but also on finding informative variables for prediction purposes at the individual patient level. In this respect, RSF models can also be used to answer biological questions from a prediction perspective. By averaging over trees, and randomizing while growing a tree, RSF approximates complex right-censored time-to-event functions while maintaining low prediction error just like RF. In our application study of the MACS HIV cohort dataset, the goal was also to address a similar prediction problem with an RSF approach, namely, to find which key gene variants and/or their interactions are predictive of time to HIV change of tropism and time to AIDS diagnosis at the individual patient level.
Although statistical interactions of order k are possible for p (independent) variables, this study focuses on the task of finding second order (k = 2) pairwise interaction models, with or without first-order terms (main-effects), and without quadratic terms, in the context of time-to-event outcomes, possibly with right censoring. The problem of variable selection by ensemble tree methods (RSF models) is not considered here, neither is the case when the number of variables and their first-order interactions exceeds the number of samples (n < p(p + 1)/2). These problems have been previously studied by Ishwaran et al., in the same context, where he showed the power of RSF models for variable selection in high-dimensional settings, including under the n ≪ p paradigm (Ishwaran, 2007; Ishwaran et al., 2010; Chen & Ishwaran, 2012). So, the ideas we develop here can be extended to higher-dimension situations (pairwise interaction models with or without main-effects) by carrying out a RSF-based variable screening approach prior to ours.
2 Methods
2.1 Assumptions – notations – general survival framework
For all our survival analyses, we assume independent observations of a univariate right-censored risk/survival outcome, with non-competitive risks, and random (type-I or -II) non-informative censoring. We model the relationship between a right-censored survival outcome, denoted Y with a set of explanatory variables (covariates), denoted as the multivariate p-dimensional random vector X = (x1, …, xj, …,xp)T. Under these assumptions and the general random censoring survival framework, we let the true survival time (or lifetime/failure time) be the random variable T; the true censoring time be the random variable C; the observed survival time be the random variable Y = min(T,C); and the observed event indicator variable be the random variable denoted by Δ = I(T ≤ C), where I(·) denotes the indicator function throughout the paper.
Assume the training data consists of i.i.d. observations , (i.e. a SRS) drawn from an unknown joint p.d.f. Pr(Y, X) with realizations y = (y1, …, yn)T and for which each realization xi = (xi,1, …, xi,p)T is a p-dimensional vector with x = (x1, …, xn)T, for i ∈ {1, …, n}. We denote the realization of true survival time by t = (t1, …, tn)T; the realizations of true censoring time by c = (c1, …, cn)T; so that the observed data consist of realizations where yi = min(ti, ci) and δi = I(ti ≤ ci) for i ∈ {1, …, n}.
Let E denote the event of interest, whether it was observed or not, and T(E) be the time-to-event outcome that we model. Let S(t) = Pr(T(E) > t) be the probability that a subject from the population of interest will have a time-to-event T(E) exceeding t, i.e. will be free of the event until time t. The non-parametric Kaplan-Meier (Kaplan & Meier, 1958) estimator was used to estimate the “survival” probability function S(t) of time-to-event E, whether it was observed or not. The resulting “survival” probability was called event-free (EF) probability.
2.2 General regression model and goal
In the survival models under study (see also simulation design section for additional details), we consider the regression function η(X) that can take different forms depending on the type of relationship we want to simulate between the outcome Y of survival times and explanatory variables (covariates) X = (x1, …, xj, …, xp)T, whether they are continuous or discrete. Here, we limit η(·) to at most 2nd-order pairwise interactions of variables without quadratic terms (two-way interaction model), but allow it to be linear or not in its 2nd-order regression coefficients:
| (1) |
where f: ℝ → ℝ is a linear or nonlinear continuous function and where xj = (x1,j, …, xn,j)T, for j ∈ {1, …, p} and β = (β1, …, βp, …, βj,k, …)T is the p(p + 1)/2-dimensional vector of regression coefficients. The interaction summation over j < k for j, k ∈ {1, …, p} is a consequence of our notational decision to deal with the fact that the non-additive part is symmetric in its arguments. Our goal is to estimate β and specifically to correctly estimate the non-additive part of β, that is, whether the values of nonzero parameters coefficients (and only them) are different from 0 or not. We will further refer to the additive part as the main-effects terms and the non-additive part as the interaction-effects terms.
2.3 Random survival forest
We used RSF (Ishwaran et al., 2008) to model the survival distribution of outcomes as a function of variables and derive the concepts of interest. In each instance, a forest of B = 1000 survival trees was grown using log-rank splitting (LeBlanc & Crowley, 1993; Segal, 1988). Each survival tree was constructed from an independent bootstrap sample of the original data. The B trees were aggregated (bagged) and from this an ensemble survival function was calculated by averaging the Kaplan-Meier tree survival function, which provided individual survival estimates at the patient level. An ensemble cumulative hazard function (CHF) was estimated by averaging the Nelson-Aalen tree CHF, which provided risk estimates at the patient level. Specifically, the CHF was estimated as a function of our explanatory variables xi by:
| (2) |
where Hb(t|xi) denotes the Nelson-Aalen cumulative hazard estimate for tree b ∈ {1, …, B} and sample i ∈ {1, …, n} in a given tree terminal node. The survival forest was used for assessing variable importance (see Supplemental Material) and for identifying and ranking variable interactions (see below for details).
The rate of event E (mortality), traditionally measured by the cumulative event (death) rate, was here called “Eventuality” and measured as a vector of estimated risk for each individual obtained from the ensemble CHF, rescaled to the number of events (Ishwaran et al., 2008). Its value is interpreted here in terms of the total number of events. For example, if individual i has an eventuality value of 100, then if all individuals had the same X-values as i, we would expect an average of 100 events.
2.4 Prediction estimates
We determined RSF global prediction performance (or so-called concordance error rate) as measured by 1−C, where C is Harrell’s concordance index (Harrell, 1982). The ensemble RSF cumulative out-of-bag (OOB) error rate is calculated using the OOB data for each tree, i.e. the original data left out from the bootstrap sample used to grow the tree (100(1/e)% ≅ 36.8% of the original sample) and averaged over all trees. Essentially, the out-of-bag error estimate is an out-of-bag error estimate of the aggregated model’s error. The error rate is between 0 and 1, and measures how well the RSF ensemble correctly ranks random individuals in terms of their corresponding event-free probability (EF). A value of 0 is perfect, a value of 0.5 is no better than random guessing.
We also report the so-called RSF estimates of partial or marginal adjusted-effect of a given variable xj, for j ∈ {1, …, p}, defined here as the predicted event-free EF (survival) probability at a specific time (e.g. median or end follow-up times), after adjusting for all the other variables.
After random splitting of the data between a training and test set, estimates of cumulative OOB error rate, individualized eventuality (mortality) values, and individualized event-free EF (survival) probabilities were obtained from out-of-bag (OOB) estimates using bootstrap resampling (as described above), and by building the ensemble RSF on the training set and using the test set as follows. Traditional test set prediction estimators were first obtained by growing the forest on the training data and estimated on the test data. Additionally, so-called “overlaying” estimators were also calculated using the y-outcomes from the test data and overlaying it on the training forest. Specifically, the terminal nodes from the training forest were recalculated using the y-outcomes from the test set. This yields modified estimators in which the topology of the forest is based solely on the training data, but where the estimated values are based on the independent test data. Essentially, this method is a way to compare so-called “observed” values (“overlaying” estimators) versus predicted values (traditional estimators) for each test set-sample, i.e. how accurate predictions are on an individual level.
2.5 Cox proportional hazards modeling
To compare the performances of statistics derived from RSF modeling, we used standard semi-parametric survival modeling by Cox Proportional Hazards (Cox-PH) to produce maximum partial likelihood estimates of model parameters, as well as parametric Score (log-rank) or Wald statistics to test the significance of model regression parameters in the Cox-PH model derived from stepwise variable selection (Cox, 1972). In the Cox-PH regression, we regress the Hazard Function (HF) h(t|xi) onto our explanatory variables xi: h(t|xi) = h0(t)exp[η(xi)] for each sample i ∈ {1, …, n}. In addition to the above assumptions, we assumed proportional hazards for Cox-PH survival modeling. This assumption was checked with standard diagnostic plot (independence of time to scaled Schoenfeld residuals) (Grambsch & Therneau, 1994).
2.6 Univariate tree-based concepts of variable importance
An important advantage of RSF is that it provides rapidly computable measures of variable importance or predictiveness that can be used to rank variables. The initial measure of univariate variable importance reported is the univariate OOB error rate calculated by 1–C (Harrell, 1982), when each variable was included singly in the RSF model.
Permutation importance (Breiman-Cutler importance) is the most frequently applied importance measure (Breiman, 2001). To calculate a variable’s permutation importance, the given variable is randomly permuted in the OOB data for the tree, and the permuted OOB data are dropped down the tree. The OOB estimate of error rate is then calculated using Harrell’s C concordance index (Harrell, 1982) and random splitting (child assignment) (Cutler & Zhao, 2001; Lin & Jeon, 2006). The difference between this estimate and the OOB error without permutation, averaged over all trees, is the univariate variable importance statistic (VIMP) of variable xj, denoted VIMP(xj). The larger the permutation importance of a variable xj is, the more predictive the variable xj is. Here, variable importance was computed similar to Breiman-Cutler importance (Breiman, 2001), but rather than permutation we used a randomization scheme as described in Ishwaran (2007) and Ishwaran et al. (2008). Because variable importance is based on the concordance index, the importance of a given variable indicates how much misclassification increases, or decreases, for a new test case if the variable was not available for that case (given that the forest was grown using that variable).
We also use the tree concept referred to as the minimal depth of a maximal subtree (Ishwaran, 2007) as another univariate RSF statistic of variable importance. The minimal depth of a maximal subtree statistic (MDMS) of variable xj, MDMS(xj), further denoted by Φ(j), is the shortest distance (depth) from the root node to the parent node of the maximal subtree where the tree first splits on variable xj (0 is the smallest value possible). The smaller this depth is, the more predictive or informative xj is (Ishwaran et al., 2010). Thus, VIMP(xj) and MDMS(xj) represent two univariate measures for ranking individual variables by importance or predictiveness.
2.7 Tree-based concepts of bivariate variables interaction
From Ishwaran’s initial work on pairwise interaction statistic (Ishwaran, 2007), a paired interaction statistic between variables xj and xk, denoted VIMP(xj, xk), can be defined for j, k ∈ {1, …, p}, j < k, as follows: VIMP(xj, xk) = | PIMP(xj, xk) − AIMP(xj, xk)|, where PIMP(xj, xk) is the paired joint variable importance between variables xj and xk, defined as the amount that prediction error increases (or decreases) when xj and xk are simultaneously perturbed. The term AIMP(xj, xk) is the additive variable importance defined as the sum of each individual variable importance: AIMP(xj, xk) = VIMP(xj)+VIMP(xk). If the univariate variable importance for each variable is significantly large, a large VIMP(xj, xk) indicates a possible pairwise interaction (see Ishwaran, 2007, for more details).
However, we do not necessarily want to assume that the univariate variable importance for each variable is significantly large (i.e. both variables are marginally informative) or honor the hierarchy restriction in Bien et al.,’s sense (Bien, Taylor & Tibshirani, 2013) that an interaction may only be included in a pairwise interaction model if one or both variables are marginally important. So, we built upon the concepts of maximal subtree (Ishwaran, 2007) and minimal depth of a maximal subtree (Ishwaran et al., 2010) introduced by Ishwaran to define here an alternative bivariate interaction statistic between any two variables xj and xk, which we termed Interaction Minimal Depth Maximal Subtree (IMDMS) and denoted Ψ(j, k), for j, k ∈ {1, …, p}, j < k, as follows. Based on the original minimal depth concept, we first use the normalized minimal depth of a variable xj with respect to the maximal subtree for variable xk (normalized w.r.t. the size of xk’s maximal subtree), denoted by MDMS(xj, xk), that is, the shortest distance that xj splits under xk, where the distance is normalized with respect to the height of the tree with the xk split denoting the root node (see Ishwaran et al., 2010, for details). A small value indicates that xj is related to xk. Because MDMS(xj, xk) is not symmetric in its arguments, we then use the reciprocal MDMS(xj, xk) to define Ψ(j, k) for j, k ∈ {1, …, p}, j < k, as:
| (3) |
A small IMDMS value identifies a possible pairwise interaction.
2.8 Assessing significance of univariate and bivariate RSF statistics
To determine whether a variable’s importance measure is truly significant or not and identify only true pairwise variable interactions, it is essential to estimate statistical significance levels of our previously defined maximal subtree-based concepts of MDMS univariate and IMDMS bivariate RSF statistics, each built-from B bootstrapped trees. One must design an objective method for estimating these significance levels, because an arbitrary threshold of significance is not justified in any situation. Moreover, the estimating approach used must be properly calibrated because there is no reason to assume that a threshold of significance has to be the same overall, for all variables and outcomes. It should be estimated for each variable and each outcome from the data. To address these points, we first introduce a noise variable matched to each original variable. For each variable xj, j ∈ {1, …, p}, we introduce an additional variable , which is xj randomly permuted. So, with xj = (x1,j, …, xn,j)T,
| (4) |
where {ω(x1,j), …, ω(xn,j)} is an ordered arrangement on tuple {x1,j, …, xn,j}, and ω(·) is a bijection from {x1,j, …, xn,j} to itself. To assess significance of MDMS forest-averaged univariate RSF statistic Φ(j) of variable importance of variable xj for j ∈ {1, …, p}, we use the forest-averaged MDMS univariate RSF statistic of the corresponding noise variable , denoted Φ*(j), as a means of thresholding its significance level. Likewise, to assess significance of IMDMS forest-averaged bivariate RSF statistic Ψ(j, k) of variables interaction between variables xj and xk, for j, k ∈ {1, …, p}, j < k, we use the IMDMS forest-averaged bivariate RSF statistic of the corresponding noise variables and , denoted s, taken as a threshold of significance level. This scheme allows each resulting individual noise variable and paired noise variables to be properly calibrated with its original counterpart.
2.9 Decision rules of significance of univariate and bivariate RSF statistics
We make use of two common decision rules for making inferences about individual variable importance and variables pairwise interaction. We use the One-Standard-Error rule that is commonly used for instance in cross-validation (Hastie, Tibshirani & Friedman, 2009) as well as the more conservative rule based on confidence intervals and the hypothesis framework (Efron & Tibshirani, 1993). However, to avoid any risk of under or overfitting, we also use either the conjunction or disjunction rule based on both canonical rules as described below.
For the MDMS forest-averaged univariate RSF statistic, the One-Standard-Error (1SE) rule is that any MDMS statistic that is smaller, up to one standard-error, than its noised-up statistic is indicative of a significant predictive variable xj, for j ∈ {1, …, p}. Similarly, for the IMDMS forest-averaged bivariate RSF statistic: any IMDMS statistic that is smaller, up to one standard-error, than its noised-up statistic is indicative of a significant interaction between variables xj and xk, for j, k ∈ {1, …, p}, j < k. Then, if we denote by J = {1, …, p} the index set of variables, with obvious notations, we can formally write the 1SE index set JΦ,1SE of significant important/predictive variables, and JJΨ,1SE of significant pairs of interacting variables as:
| (5) |
| (6) |
Alternatively, to infer significance of RSF statistics, with their matched noise statistics, we can also reject the null hypothesis of no variable importance or no interaction of pairs of variables at the 2θ significance level if the corresponding 100(1 − 2θ)% confidence interval (CI) of a RSF statistic mean does not overlap with its matched noise statistic (or, equivalently, if the 100(1 − 2θ)% CI of the difference of their means does not contain zero). A distance between the CIs of these RSF statistic means can be computed as the difference between the lower bound (LB(θ)) of the CI from the original statistic and the upper bound (UB(θ)) of the CI from the noise counterpart.
| (7) |
| (8) |
We further refer to the decision rule from eqs. 5, 6 as the One Standard-Error rule, denoted 1SE, and the decision rule from eqs. 7, 8 as the Confidence Interval rule at the significance level 2θ, denoted CI(θ). We also use the disjunction or conjunction rule of both canonical rules for making RSF-based inferences about significant important/predictive variables, and significant pairs of interacting variables.
2.10 Building confidence intervals of univariate and bivariate RSF statistics
In practice, to get mean estimates of univariate RSF statistics Φ̂(j) and Φ̂*(j) for each individual variable as well as mean estimates of bivariate RSF statistics ψ̂(j, k) and ψ̂*(j, k) for each variable pair, and an approximate 100(1 − 2θ)% CI of all of these, we build percentile confidence intervals based on percentiles of their bootstrap distributions (Efron & Tibshirani, 1993). Let’s denote generically each of the above RSF statistic estimate by Ŵ(·) with its corresponding noise counterpart Ŵ*(·). To proceed, compute 100(1 − 2θ)% Bootstrap Confidence Intervals (BCI) as follows. Bag B = 1000 trees of the RSF by first generating B = 1000 bootstrap datasets of original variables {X1, …, X(b), …, X(B)} and corresponding noised variables {X*(1), …, X*(b), …, X*(B)}, further denoted and , respectively, where and for b ∈ {1, …, B} and where and , for j ∈ {1, …, p}. This yields Ŵ(b)(·), with its corresponding noise counterpart Ŵ*(b)(·). To get BCI of these statistics, repeat the sampling procedure with replacement B′ = 1000 times to generate B′ bootstrap estimates of each statistic from each RSF within each bootstrap sample with their corresponding noise counterparts . Note that each resulting bootstrap estimate is obtained from the bth bootstrap sample within the b′th bootstrap sample since a nested bootstrap is carried out with B bootstrap datasets generated from each parent bootstrap sample X(b) and X*(b). The resulting bootstrap distribution of each RSF statistic and its corresponding noise counterpart is used to derive the percentile interval. Let’s denote by Ŵ(θ)(·) and Ŵ(1−θ)(·) the θth and (1 − θ)th quantiles of the bootstrap distribution of Ŵ(·), respectively, and similarly by Ŵ*(θ)(·) and Ŵ*(1−θ)(·) for Ŵ*(·). The 100(1 − 2θ)% percentile intervals of Ŵ(·) and Ŵ*(·) are defined by the θth and (1 − θ)th quantiles of the cumulative distribution function of their respective bootstrap estimates. Since, by definition, the inverse of the cumulative distribution function of θ is the θth quantile of the bootstrap distribution, we can write the estimated percentile intervals as:
and similarly for their noise counterparts:
where LB and UB denote the lower bound and the upper bound of an interval, respectively. In practice, since we can only have a finite number B of replications, the θth empirical quantile Ŵ(θ)(·) of Ŵ(·) is taken to be the Bθth value in the ordered list of the B replications of Ŵ(·). Likewise, the (1 − θ)th empirical quantile Ŵ(1−θ)(·) of Ŵ(·) is taken to be the B(1 − θ)th value in the ordered list of the B replications of Ŵ(·) (Efron & Tibshirani, 1993). Bootstrap estimates of Ŵ(·) and Ŵ*(·) can then easily be derived by taking, for instance, the median values of the 100(1 − 2θ)% percentile confidence intervals at a certain significance level 2θ.
2.11 Design of simulated survival models
The p covariates xj = (x1,j, …, xn,j)T, for j ∈ {1, …, p}, were i.i.d. sampled either from the:
Continuous case: uniform distribution xj ~ U(a, b) on [a, b], where a < b
Discrete case: multinomial distribution xj ~ M(n, ε, π) with number of trials n, events xi,j ∈ {ε1, …, εk} and event probabilities π = (π1, …, πk)T, where 0 < πk < 1, Σ πk = 1 and Pr(xi,j = εk) = πk.
For simplification, all subsequent simulations and real data analyses were done without inter-variable correlation, with a = 1, b = 5, k = 5 and π = (1/5, …, 1/5)T. Realizations of true survival times t = (t1, …, tn)T were generated either from:
- A linear latent variable model, where T is directly proportional to a linear regression function:
- An exponential survival model, where T is i.i.d. drawn from an exponential distribution with rate parameter λ (and mean 1/λ) T ~ Exp(λ). Each individual rate λi can be directly estimated, conditionally on covariates xi = (xi,1, …, xi,p)T, from an exponential regression function:
where λi = h(t|xi) = h0(t) exp[η(xi)]
In both cases, the regression function η(xi) may include saturated models with 1st-order terms in variables, higher-order (2nd-order) pairwise interaction terms, and nonlinear transformations of terms, so that hazards or survival times will be proportional to covariates main and/or interaction-effects. To simulate a real situation problem where interactions are unknown, we model the survival with one of the simulation design above, but we fit the RSF with the 1st-order saturated regression model alone. The goal is to test whether our RSF bivariate variable interaction statistics are able (or not) to recover the true underlying pairwise interactions, and only them, from the survival outcome and 1st-order terms alone.
Under the non-informative censoring assumption, the censoring rate ρ was fixed within ρ ∈ {0.3, 0.5, 0.7} by randomly sampling from a uniformly distributed censoring time C ~ U(0, n) for ν > 0, where censoring rates are determined by υ, so that approximately 100ρ % of the simulated realizations of observed survival times yi = min(ti, ci) are censored.
2.12 Additional statistical methods
For the Kaplan-Meier models, we used the log-rank test to assess statistical significance of difference between distributions. In addition to the above assumptions, we assume independence of the probability of progression to the different endpoints, so they need not to be seen as competing risks (see discussion and Mehlotra et al., 2012). For ensemble prediction estimates purposes, the data was randomly split to generate unbalanced training (tr) / test (te) sets in the 4/5 ratio (ntr = 160, nte = 40) or 3/2 ratio (ntr = 30, nte = 20) for the simulated data or the MACS cohort study, respectively. To get an approximate 100(1 − θ)% CI of the medians of predicted XEF and ADF event-free probabilities, we used McGill et al.,’s approximation: The approximate 100(1 − θ)% CI of the median M extends to , where M̂ and IQ̂R are the median and interquartile range estimates, respectively, and n is the sample size (McGill, Tukey & Larsen, 1978). In a boxplot, this is given by the extent of notches. Missing values in applied dataset [DEFB4/103A copy numbers (n = 5)] were determined by simple-imputation results from the Expectation-Maximization algorithm (Dempster, Laird & Rubin, 1977), assuming missing data at random. Scatter smoothing was done by using Friedman’s “super smoother” estimator (Friedman 1984), where the span of the running lines smoother was chosen by leave-one-out cross-validation. All statistical modeling, computations, and plotting were performed in the R language and environment for statistical computing (http://www.r-project.org/). The analytical R codes for the analyses and the generation of the results are available on GitHub at https://github.com/jedazard/IRSF. Codes contain randomization, interaction modeling and prediction subroutines to be used in addition to the following R packages (http://cran.r-project.org/): “ survival” for Kaplan-Meier and Cox regression modeling, “ randomForestSRC” for RSF modeling (Ishwaran & Kogalur 2007; 2013), and “ ggRandomForests” for Random Forrest exploration/visualization (Ehrlinger, 2014). Default parameter specifications were used for all main functions.
2.13 Learning from two interacting variables
To learn how two categorical (two-level) variables interact with each other when an interaction-effect between them has previously been detected (e.g. by RSF-based estimators), we carried out additional Kaplan-Meier analyses as follows: when a difference between two levels is observed for variable #1 in a given level of variable #2, but not in any another levels of variable #2, this represents an interaction-effect between variables #1 and #2. By comparing the corresponding survival profiles, e.g. in Kaplan-Meier plots, this pattern generally allows one to learn the synergistic or antagonistic effects at play between levels of these two variables. We applied this type of analysis in addition to RSF analyses, which we referred to below as indirect interaction analyses.
3 Simulation studies
3.1 Simulation setup
In this section we analyzed the performances of our RSF-derived univariate and bivariate estimators to assess individual variables importance (1st-order main-effect) and detect pairwise interactions of variables (2nd-order interaction-effects). We also compared results with standard statistics used in Cox-PH regression modeling (CPH). In accordance with the scope of the work stated above, all simulations were carried out in low-dimensional settings (n = 200, p = 5).
To check the assumptions of our simulations, we used either the exponential survival model (EXP) or linear latent variable model (LLV), continuous or discrete variables, and various censoring rate ρ, allowed to vary within ρ ∈ {0.3, 0.5, 0.7}. In addition, we carried out these simulations in a series of regression models derived from the general one (eq. 1) to test for (i) the influence of specific main-effects as opposed to noise, corresponding to various negative and positive control situations of main-effects only, or (ii) the presence or absence of main-effects, corresponding to various negative and positive control situations of interaction-effects, as well as (iii) various transformations of the 2nd-order term, corresponding to various nonlinear models:
Regression model #1 (null model): η(X) = 0. Absence of all main-effect and interaction-effect terms: βj,k = 0 for j, k ∈ {1, …, p}, j < k and βj = 0 for j ∈ {1, …, p}.
Regression model #2: . Absence of all interaction-effect terms but presence of all main-effect terms: βj,k = 0 for j, k ∈ {1, …, p}, j < k and βj ≠ 0 for j ∈ {1, …, p} with f (u) = u.
Regression model #3: η(X) = βj + βk. Absence of all interaction-effect terms but presence of two main-effect terms: βj,k = 0 for j, k ∈ {1, …, p}, j < k and βl = 0 for l ∈ {1, …, p}\{j, k} for fixed j, k ∈ {1, …, p} with f (u) = u.
Regression model #4: η(X) = βj,kxjxk for fixed j, k ∈ {1, …, p}, j < k. Presence of one interaction-effect term between variables xj and xk with no corresponding main-effects: βj,k ≠ 0 for fixed j, k ∈ {1, …, p}, j < k and βj = 0 for j ∈ {1, …, p} with f (u) = u.
Regression model #5: η(X) = βj + βk + βj,kxjxk for fixed j, k ∈ {1, …, p}, j < k. Presence of one interaction-effect term between variables xj and xk with two corresponding main-effects: βj,k ≠ 0, βj,k ≠ 0, βk ≠ 0 and βl = 0 for l ∉ {j, k} for fixed j, k ∈ {1, …, p}, j < k with f (u) = u.
Regression model #5a: η(X) = βj + βk + exp(−βj,kxjxk), for fixed j, k ∈ {1, …, p}, j < k. Idem as model #5 with f (u) = exp(−u).
Regression model #5b: η(X) = βj + βk + Log(βj,kxjxk), for fixed j, k ∈ {1, …, p}, j < k. Idem as model #5 with f (u) = log(u).
Regression model #5c: , for fixed j, k ∈ {1, …, p}, j < k. Idem as model #5 with f (u) = √u.
Regression model #5d: η(X) = βj + βk + sin(π·βj,kxjxk), for fixed j, k ∈ {1, …, p}, j < k. Idem as model #5 with f (u) = sin(πu).
Regression model #6: for fixed j, k ∈ {1, …, p}, j < k. Presence of one interaction-effect between variables xj and xk with all main-effects: βj,k ≠ 0 for fixed j, k ∈ {1, …, p}, j < k and βj ≠ 0 for j ∈ {1, …, p} with f (u) = u.
3.2 Reporting of results and inferences
In all carried out simulations studies, bootstrap estimates of our forest-averaged MDMS univariate variable importance statistic (Φ̂(j)) and IMDMS bivariate variable interaction statistic (Ψ̂(j, k)), as well as corresponding forest-averaged noised-up statistics Φ̂*(j) and Ψ̂*(j, k), were reported with means, standard errors (SE) and confidence intervals (CI) at the θ confidence level, calculated over B = 1000 bootstrap samples, each averaged over B = 1000 bagged trees. In all corresponding figures and tables, we report these RSF statistics, either for all 1st-order terms of individual main-effects of variables xj, for j ∈ {1, …, p}, or all combinations of 2nd-order terms of pairwise interaction-effects between variables xj and xk, for j, k ∈ {1, …, p}, j < k, i.e. for p(p − 1)/2 pairs. Also reported are the corresponding Cox-PH regression p-values for testing individual main-effects of variables xj, for j ∈ {1, …, p}, or pairwise interaction-effects between variables xj and xk, for j, k ∈ {1, …, p}, j < k.
The canonical 1SE (eq. 5 or 7) and CI(θ) (eq. 6 or 8) decision rules were reported in all tables. In addition, the disjunction or conjunction rules derived from both were used to report significance of individual important/ predictive variables as well as of pairs of interacting variables, respectively (TRUE/FALSE calls in Tables and Supplemental Tables). Successful RSF inferences about individual variable importance and pairs of interacting variables are those for which only truly important/predictive variables xj are called significant, and truly interacting variables xj and xk are detected significant, respectively; that is, when either the MDMS disjunctive rule or IMDMS conjunctive rule are TRUE, respectively.
For conciseness, only results of interaction-effects by RSF bivariate estimators are shown but all results of analyses of main-effects by RSF univariate estimators are available in Supplemental Results, Figures and Tables (Supplemental Materials I). Likewise, only certain combinations of cases are shown, but all other cases are available in Supplemental Figures and Tables (Supplemental Materials I).
3.3 RSF model building and global prediction performance
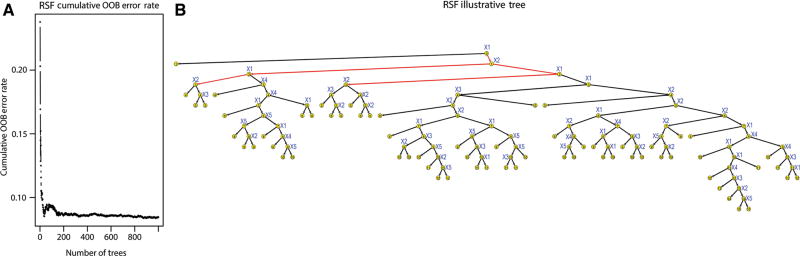
We first report RSF model building and global prediction performance for a pairwise interaction-effect regression model (#5), where a single arbitrarily-fixed 2nd-order term enter in the model, used here as positive controls of interaction-effects. The two arbitrarily-fixed variables used for simulating the pairwise interaction-effect entering into the regression model (#5) were e.g. {j = 1, k = 2}, that is, for testing a simulated interaction-effect x1x2 between variables x1 and x2. The RSF plots show that the cumulative OOB error rate stabilizes rapidly as a function of the number of trees at 8.46%, indicating that a forest of B = 1000 trees was sufficient (Figure 1A), and that the ensemble trees were well-grown for both outcomes with an average number of terminal nodes of 66.29. An exemplary tree-plot, taken at random out of the B = 1000 trees of the RSF forest, is also shown as an illustration. Although each tree of the forest represents one instance out of many, it illustrates the differences of possible ranking of variable importance as well as variables interactions. For example, the covariate x1 appears the most important (root node), and the top pairwise interactions appear those between x1 and x2, both of which were expected by simulation design (Figure 1B).
Figure 1.
RSF global prediction performance and visualization of RSF illustrative tree in simulated data. (A) Left: forest-averaged RSF cumulative OOB error rate for the ensemble as a function of number of trees. (B) Visualization of an exemplary tree (e.g. #2) out of the B = 1000 trees of the RSF forest. Result shown is for the linear latent variable survival model (LLV), with continuous variables, and fixed censoring rate (ρ = 0.5), in regression model #5 (positive control), where a single arbitrarily fixed 2nd-order term enter in the model, simulated from two arbitrarily-fixed variables (e.g. here {j = 1, k = 2}, i.e. for x1x2 – see Section 3). Note how the cumulative OOB error rate stabilizes rapidly as a function of the number of trees (at 8.46%), indicating that a forest of B = 1000 trees was sufficient with an average number of terminal nodes of 66.29. Yellow-colored nodes represent individual variables. The depth of the trees is indicated by numbers (0–15) inside of each node (0 being the root node). Each tree illustrates one possible ranking of variable importance (height vs. depth of the nodes) and variable interactions (edges between nodes in the same branch). In this example, note how top interactions involving a root node x1 with a child node x2 are detected by IMDMS bivariate RSF statistic (highlighted red edges). Also, note the proportion of true interactions x1x2 involving a parent node x1 with a child node x2 out of all top detected ones (5/7) from root to depth #3 (highlighted red branches).
3.4 Analysis of interaction-effects by RSF bivariate estimators
In this first set of simulations, we show results of our IMDMS bivariate variable interaction statistics, either in a null regression model (#1), where no terms whatsoever enter into the regression model, used here as a first negative control of interaction-effects; or in a main-effects only regression model (#2), where only 1st-order terms enter in the model, used here as second negative control of interaction-effects; or finally in a pairwise interaction-effect regression model (#4, #5, #6), where a single arbitrarily-fixed 2nd-order term enter in the model, used here as positive controls of interaction-effects. In the latter case, the two arbitrarily-fixed variables used for simulating the pairwise interaction-effect entering into regression models (#4, #5, #6) were e.g. {j = 1, k = 2}, that is, for testing a simulated interaction-effect x1x2 between variables x1 and x2. We analyzed several situations, considering the type of regression model (#1, #3, #4, #5, #6), the type of survival model (EXP vs. LLV), and the type of covariates (continuous vs. discrete) at fixed censoring rate ρ = 0.5, that is, for 20 simulations. From this first set of simulations, using the IMDMS conjunction rule (at the θ = 0.05 confidence level), results of IMDMS bivariate RSF estimator show that it was able to correctly detect true single pairs of interacting variables (and only them) in nearly all (19/20) simulations (Figure 2, Table 1 and Table 2, Supplemental Tables S18–S23).
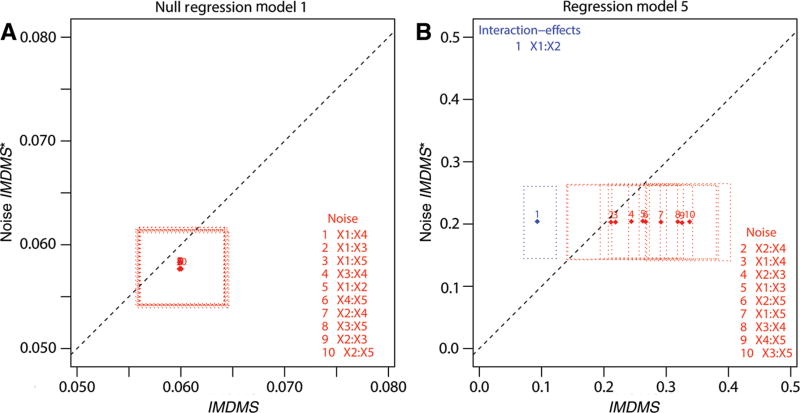
Figure 2.
Scatter plots of IMDMS bivariate RSF statistics for the detection of interaction-effects in negative and positive controls of simulated data. Results shown are for the linear latent variable survival model (LLV), with continuous variables, and fixed censoring rate (ρ = 0.5), in null regression model #1 (negative control), where no terms enter in the models, and in regression model #5 (positive control), where a single arbitrarily-fixed 2nd-order term enter in the model, simulated from two arbitrarily-fixed variables (e.g. here {j = 1, k = 2}, i.e. for x1x2 – see Section 3). (A) Left: RSF results in null regression model #1. (B) Right: RSF results in regression model #5. (A, B) For each model and each variable pair xj and xk, j, k ∈ {1, …, p}, j < k (i.e. 10 pairs, for p = 5), the IMDMS bivariate RSF statistic mean (Ψ̂(j, k) or IMDMS) is plotted against its noised-up counterpart (Ψ̂* (j, k) or Noise IMDMS*). RSF statistic means (diamonds) are numbered by order of decreasing significance. Blue or red color denotes pairs of variable with a significant or non-significant measure of IMDMS at the θ = 0.05 level, respectively. Pairs of variables with significant measures of interaction have confidence intervals on both axes (dotted boxes) farther above and left of the identity line (dashed line) without crossing it. Note the accuracy of inferences by IMDMS decision rule: IMDMS correctly does not detect any variables interaction-effects in the case of negative control regression model, and correctly detects the single true variables interaction-effect x1x2 (and only it) in the case of positive control regression model (see also Table 1 and Table 2).
Table 1.
Detection and ranking of pairwise interaction-effects by IMDMS bivariate RSF statistic vs standard Cox-PH regression in negative controls of simulated data.
| Reg. model type |
Var.
pair xjxk |
Ψ̂(j,
k) (SE) |
Ψ̂(j,
k) [BCI] |
Ψ̂*
(j, k) (SE) |
Ψ̂*
(j, k) [BCI] |
1SE rule |
CI(θ) rule |
IMDMS 1SE CI(θ) rule |
CPH. p-value |
|---|---|---|---|---|---|---|---|---|---|
| #1 | |||||||||
| x1x4 | 0.0598 (0.00249) | [0.0560, 0.0642] | 0.0577 (0.00220) | [0.0542, 0.0617] | FALSE | FALSE | FALSE | 1.00E+00 | |
| x1x3 | 0.0599 (0.00249) | [0.0559, 0.0642] | 0.0576 (0.00222) | [0.0542, 0.0613] | FALSE | FALSE | FALSE | 1.00E+00 | |
| x1x5 | 0.0599 (0.00251) | [0.0561, 0.0641] | 0.0576 (0.00220) | [0.0542, 0.0614] | FALSE | FALSE | FALSE | 1.00E+00 | |
| x3x4 | 0.0599 (0.00260) | [0.0557, 0.0642] | 0.0577 (0.00220) | [0.0542, 0.0615] | FALSE | FALSE | FALSE | 1.00E+00 | |
| x1x2 | 0.0599 (0.00251) | [0.0560, 0.0642] | 0.0577 (0.00220) | [0.0542, 0.0613] | FALSE | FALSE | FALSE | 1.00E+00 | |
| x4x5 | 0.0599 (0.00257) | [0.0558, 0.0643] | 0.0577 (0.00228) | [0.0541, 0.0614] | FALSE | FALSE | FALSE | 1.00E+00 | |
| x2x4 | 0.0599 (0.00250) | [0.0559, 0.0642] | 0.0577 (0.00219) | [0.0543, 0.0614] | FALSE | FALSE | FALSE | 1.00E+00 | |
| x3x5 | 0.0600 (0.00257) | [0.0560, 0.0644] | 0.0577 (0.00230) | [0.0543, 0.0617] | FALSE | FALSE | FALSE | 1.00E+00 | |
| x2x3 | 0.0600 (0.00259) | [0.0559, 0.0645] | 0.0577 (0.00223) | [0.0539, 0.0614] | FALSE | FALSE | FALSE | 1.00E+00 | |
| x2x5 | 0.0600 (0.00252) | [0.0560, 0.0646] | 0.0577 (0.00218) | [0.0542, 0.0612] | FALSE | FALSE | FALSE | 1.00E+00 | |
| #2 | |||||||||
| x3x5 | 0.139 (0.0278) | [0.0927, 0.186] | 0.202 (0.0333) | [0.146, 0.257] | TRUE | FALSE | FALSE | 3.21E-01 | |
| x3x4 | 0.143 (0.0288) | [0.0997, 0.192] | 0.200 (0.0344) | [0.142, 0.255] | TRUE | FALSE | FALSE | 1.34E-01 | |
| x1x3 | 0.163 (0.0332) | [0.1136, 0.220] | 0.200 (0.0345) | [0.140, 0.254] | TRUE | FALSE | FALSE | 3.21E-01 | |
| x2x3 | 0.165 (0.0291) | [0.1148, 0.212] | 0.200 (0.0340) | [0.146, 0.257] | TRUE | FALSE | FALSE | 1.20E-01 | |
| x4x5 | 0.170 (0.0262) | [0.1320, 0.216] | 0.203 (0.0353) | [0.144, 0.260] | TRUE | FALSE | FALSE | 5.09E-01 | |
| x2x5 | 0.171 (0.0311) | [0.1255, 0.225] | 0.203 (0.0347) | [0.145, 0.259] | TRUE | FALSE | FALSE | 9.42E-01 | |
| x2x4 | 0.175 (0.0308) | [0.1258, 0.227] | 0.202 (0.0344) | [0.145, 0.259] | FALSE | FALSE | FALSE | 1.84E-02 | |
| x1x5 | 0.184 (0.0348) | [0.1265, 0.241] | 0.203 (0.0335) | [0.149, 0.258] | FALSE | FALSE | FALSE | 5.09E-02 | |
| x1x4 | 0.198 (0.0304) | [0.1581, 0.258] | 0.202 (0.0352) | [0.143, 0.260] | FALSE | FALSE | FALSE | 6.44E-02 | |
| x1x2 | 0.205 (0.0341) | [0.1560, 0.268] | 0.202 (0.0344) | [0.145, 0.257] | FALSE | FALSE | FALSE | 6.54E-01 |
Results shown are for the linear latent variable survival model (LLV), with continuous variables, and fixed censoring rate (ρ = 0.5), in null regression models #1, #2 (negative controls), where either no terms or no 2nd-order terms enter in the models, respectively (see Section 3). Other cases are shown in Supplemental Tables S18, S19, S20. IMDMS bivariate RSF statistics means (Ψ̂(j, k)) are reported with standard errors (SE) and bootstrap confidence intervals (BCI) for all possible pairs of variable xj and xk, for j, k ∈ {1, …, p}, j < k (i.e. 10 pairs, for p = 5) by increasing order of IMDMS from top to bottom. The corresponding noised-up statistic (Ψ̂* (j, k)) as well as IMDMS decision rule (1SE ∧ CI(θ)) at the θ level are shown in the adjacent columns. Also reported are the corresponding Cox-PH regression p-values testing 2nd-order pairwise interaction-effect terms at the θ level. NA value stands for situations when the fitting algorithm did not converge. In bold are the significant decision rules or p-values at the θ = 0.05 level. Note the accuracy of inferences by IMDMS decision rule as compared to Cox-PH regression inference: IMDMS correctly does not detect any variables interaction-effects in both cases of negative control regression models (see also Figure 2). Bold values stands for statistically significant or TRUE values.
Table 2.
Detection and ranking of pairwise interaction-effects by IMDMS bivariate RSF statistic vs standard Cox-PH regression in positive controls of simulated data.
| Reg.model type |
Var.
pair xjxk |
Ψ̂(j,
k) (SE) |
Ψ̂(j,
k) [BCI] |
Ψ̂*
(j, k) (SE) |
Ψ̂*
(j, k) [BCI] |
1SE rule |
CI(θ) rule |
IMDMS 1SE CI(θ) rule |
CPH. p-value |
|---|---|---|---|---|---|---|---|---|---|
| #4 | |||||||||
| x1x2 | 0.094 (0.0182) | [0.0702, 0.130] | 0.205 (0.0356) | [0.147, 0.263] | TRUE | TRUE | TRUE | NA | |
| x2x4 | 0.221 (0.0354) | [0.1644, 0.278] | 0.204 (0.0366) | [0.145, 0.265] | FALSE | FALSE | FALSE | 8.10E-01 | |
| x1x4 | 0.243 (0.0393) | [0.1793, 0.304] | 0.205 (0.0367) | [0.147, 0.266] | FALSE | FALSE | FALSE | 8.08E-01 | |
| x2x3 | 0.250 (0.0356) | [0.1931, 0.308] | 0.206 (0.0371) | [0.147, 0.268] | FALSE | FALSE | FALSE | 9.06E-01 | |
| x2x5 | 0.261 (0.0396) | [0.1937, 0.325] | 0.204 (0.0363) | [0.145, 0.263] | FALSE | FALSE | FALSE | 2.09E-01 | |
| x1x3 | 0.267 (0.0384) | [0.2029, 0.326] | 0.206 (0.0383) | [0.142, 0.268] | FALSE | FALSE | FALSE | 8.05E-01 | |
| x1x5 | 0.288 (0.0340) | [0.2287, 0.344] | 0.204 (0.0372) | [0.147, 0.269] | FALSE | FALSE | FALSE | 1.16E-01 | |
| x4x5 | 0.331 (0.0380) | [0.2690, 0.392] | 0.203 (0.0367) | [0.142, 0.264] | FALSE | FALSE | FALSE | 7.63E-01 | |
| x3x4 | 0.333 (0.0401) | [0.2668, 0.397] | 0.205 (0.0372) | [0.144, 0.264] | FALSE | FALSE | FALSE | 2.42E-01 | |
| x3x5 | 0.372 (0.0454) | [0.2929, 0.443] | 0.204 (0.0360) | [0.144, 0.266] | FALSE | FALSE | FALSE | 6.77E-03 | |
| #5 | |||||||||
| x1x2 | 0.093 (0.0168) | [0.0715, 0.123] | 0.204 (0.0354) | [0.145, 0.260] | TRUE | TRUE | TRUE | NA | |
| x2x4 | 0.211 (0.0384) | [0.1422, 0.267] | 0.203 (0.0362) | [0.143, 0.264] | FALSE | FALSE | FALSE | 8.38E-01 | |
| x1x4 | 0.219 (0.0455) | [0.1403, 0.290] | 0.203 (0.0354) | [0.143, 0.262] | FALSE | FALSE | FALSE | 9.49E-01 | |
| x2x3 | 0.244 (0.0329) | [0.1940, 0.300] | 0.204 (0.0357) | [0.145, 0.263] | FALSE | FALSE | FALSE | 1.44E-01 | |
| x1x3 | 0.263 (0.0345) | [0.2071, 0.318] | 0.205 (0.0361) | [0.145, 0.264] | FALSE | FALSE | FALSE | 9.97E-01 | |
| x2x5 | 0.267 (0.0350) | [0.2128, 0.328] | 0.204 (0.0357) | [0.146, 0.262] | FALSE | FALSE | FALSE | 2.90E-01 | |
| x1x5 | 0.292 (0.0315) | [0.2396, 0.342] | 0.203 (0.0363) | [0.143, 0.263] | FALSE | FALSE | FALSE | 1.74E-01 | |
| x3x4 | 0.319 (0.0390) | [0.2565, 0.383] | 0.204 (0.0355) | [0.144, 0.262] | FALSE | FALSE | FALSE | 4.70E-02 | |
| x4x5 | 0.325 (0.0337) | [0.2732, 0.380] | 0.202 (0.0363) | [0.144, 0.263] | FALSE | FALSE | FALSE | 4.83E-01 | |
| x3x5 | 0.338 (0.0393) | [0.2685, 0.403] | 0.204 (0.0374) | [0.141, 0.265] | FALSE | FALSE | FALSE | 7.15E-02 | |
| #6 | |||||||||
| x1x2 | 0.0927 (0.0118) | [0.0777, 0.114] | 0.202 (0.0353) | [0.145, 0.259] | TRUE | TRUE | TRUE | 1.31E-05 | |
| x2x3 | 0.1689 (0.0239) | [0.1326, 0.211] | 0.201 (0.0357) | [0.141, 0.262] | TRUE | FALSE | FALSE | 1.87E-02 | |
| x1x3 | 0.1727 (0.0350) | [0.1251, 0.236] | 0.202 (0.0346) | [0.145, 0.258] | FALSE | FALSE | FALSE | 2.40E-01 | |
| x1x4 | 0.2050 (0.0379) | [0.1398, 0.264] | 0.202 (0.0349) | [0.143, 0.259] | FALSE | FALSE | FALSE | 9.83E-01 | |
| x2x4 | 0.2075 (0.0281) | [0.1583, 0.249] | 0.201 (0.0364) | [0.139, 0.258] | FALSE | FALSE | FALSE | 3.66E-01 | |
| x2x5 | 0.2104 (0.0223) | [0.1763, 0.247] | 0.202 (0.0360) | [0.143, 0.258] | FALSE | FALSE | FALSE | 8.18E-02 | |
| x1x5 | 0.2376 (0.0350) | [0.1760, 0.289] | 0.202 (0.0374) | [0.142, 0.265] | FALSE | FALSE | FALSE | 2.37E-01 | |
| x3x4 | 0.2533 (0.0296) | [0.2063, 0.304] | 0.202 (0.0340) | [0.144, 0.258] | FALSE | FALSE | FALSE | 2.80E-02 | |
| x3x5 | 0.2547 (0.0295) | [0.2072, 0.305] | 0.201 (0.0342) | [0.147, 0.258] | FALSE | FALSE | FALSE | 4.79E-02 | |
| x4x5 | 0.2900 (0.0357) | [0.2312, 0.350] | 0.201 (0.0351) | [0.140, 0.258] | FALSE | FALSE | FALSE | 6.76E-01 |
Results shown are for the linear latent variable survival model (LLV), with continuous variables, and fixed censoring rate (ρ = 0.5) in regression models #4, #5, #6 (positive controls), where a single arbitrarily-fixed 2nd-order term enter in the model, simulated from two arbitrarily-fixed variables (e.g. here {j = 1, k = 2}, i.e. for x1x2 – see Section 3). Other cases are shown in Supplemental Tables S21, S22, S23. IMDMS bivariate RSF statistics means (Ψ̂(j, k)) are reported with standard errors (SE) and bootstrap confidence intervals (BCI) for all possible pairs of variable xj and xk, for j, k ∈ {1, …, p}, j < k (i.e. 10 pairs, for p = 5) by increasing order of IMDMS from top to bottom. The corresponding noised-up statistic (Ψ̂* (j, k)) as well as IMDMS decision rule (1SE ∧ CI(θ)) at the θ level are shown in the adjacent columns. Also reported are the corresponding Cox-PH regression p-values testing 2nd-order pairwise interaction-effect terms at the θ level. NA value stands for situations when the fitting algorithm did not converge. In bold are the significant decision rules or p-values at the θ = 0.05 level. Note the accuracy of inferences by IMDMS decision rule as compared to Cox-PH regression inference: IMDMS correctly detects the true single variables interaction-effect x1x2 (and only it) in all cases of positive control regression models (see also Figure 2). Bold values stands for statistically significant or TRUE values.
From now on, since bivariate RSF inferences of interaction-effects are fairly robust to the presence or absence of any associated main-effects, we focused on a specific regression model with, for instance, one interaction-effect term between variables xj and xk accompanied by two corresponding main-effects (i.e. model #5).
In the second set of simulations, we tested the influence of discrete or continuous types of covariates random variables as well as the type of survival model used for modeling censored survival times. We analyzed several situations, using continuous or discrete covariates, for both types of survival models (EXP vs. LLV), at fixed censoring rate ρ = 0.5, in regression model #5, for 4 simulations. Using the IMDMS conjunction rule (at the θ = 0.05 confidence level), IMDMS bivariate RSF statistics inferences were found correct in all (4/4) simulations (Table 3 and Table 4, Supplemental Tables S24–S25).
Table 3.
Detection and ranking of pairwise interaction-effects by IMDMS bivariate RSF statistic vs standard Cox-PH regression in simulated data testing for influence of covariates types.
| Covariate type |
Var.
pair xjxk |
Ψ̂(j,
k) (SE) |
Ψ̂(j,
k) [BCI] |
Ψ̂*
(j, k) (SE) |
Ψ̂*
(j, k) [BCI] |
1SE rule |
CI(θ) rule |
IMDMS 1SE CI(θ) rule |
CPH. p-value |
|---|---|---|---|---|---|---|---|---|---|
| Cont. | |||||||||
| x1x2 | 0.093 (0.0168) | [0.0715, 0.123] | 0.204 (0.0354) | [0.145, 0.260] | TRUE | TRUE | TRUE | NA | |
| x2x4 | 0.211 (0.0384) | [0.1422, 0.267] | 0.203 (0.0362) | [0.143, 0.264] | FALSE | FALSE | FALSE | 8.38E-01 | |
| x1x4 | 0.219 (0.0455) | [0.1403, 0.290] | 0.203 (0.0354) | [0.143, 0.262] | FALSE | FALSE | FALSE | 9.49E-01 | |
| x2x3 | 0.244 (0.0329) | [0.1940, 0.300] | 0.204 (0.0357) | [0.145, 0.263] | FALSE | FALSE | FALSE | 1.44E-01 | |
| x1x3 | 0.263 (0.0345) | [0.2071, 0.318] | 0.205 (0.0361) | [0.145, 0.264] | FALSE | FALSE | FALSE | 9.97E-01 | |
| x2x5 | 0.267 (0.0350) | [0.2128, 0.328] | 0.204 (0.0357) | [0.146, 0.262] | FALSE | FALSE | FALSE | 2.90E-01 | |
| x1x5 | 0.292 (0.0315) | [0.2396, 0.342] | 0.203 (0.0363) | [0.143, 0.263] | FALSE | FALSE | FALSE | 1.74E-01 | |
| x3x4 | 0.319 (0.0390) | [0.2565, 0.383] | 0.204 (0.0355) | [0.144, 0.262] | FALSE | FALSE | FALSE | 4.70E-02 | |
| x4x5 | 0.325 (0.0337) | [0.2732, 0.380] | 0.202 (0.0363) | [0.144, 0.263] | FALSE | FALSE | FALSE | 4.83E-01 | |
| x3x5 | 0.338 (0.0393) | [0.2685, 0.403] | 0.204 (0.0374) | [0.141, 0.265] | FALSE | FALSE | FALSE | 7.15E-02 | |
| Disc. | |||||||||
| x1x2 | 0.103 (0.00707) | [0.0923, 0.115] | 0.228 (0.0333) | [0.177, 0.284] | TRUE | TRUE | TRUE | NA | |
| x1x4 | 0.304 (0.03089) | [0.2533, 0.353] | 0.229 (0.0347) | [0.180, 0.292] | FALSE | FALSE | FALSE | 8.32E-01 | |
| x1x5 | 0.307 (0.02900) | [0.2554, 0.354] | 0.228 (0.0341) | [0.176, 0.287] | FALSE | FALSE | FALSE | 6.91E-01 | |
| x1x3 | 0.319 (0.02827) | [0.2729, 0.366] | 0.229 (0.0339) | [0.174, 0.287] | FALSE | FALSE | FALSE | 7.69E-01 | |
| x2x4 | 0.322 (0.03014) | [0.2718, 0.370] | 0.228 (0.0325) | [0.178, 0.283] | FALSE | FALSE | FALSE | 9.75E-01 | |
| x2x5 | 0.325 (0.02859) | [0.2777, 0.372] | 0.228 (0.0327) | [0.178, 0.285] | FALSE | FALSE | FALSE | 8.71E-01 | |
| x2x3 | 0.337 (0.02903) | [0.2888, 0.381] | 0.230 (0.0350) | [0.179, 0.291] | FALSE | FALSE | FALSE | 6.23E-01 | |
| x3x5 | 0.390 (0.04163) | [0.3256, 0.462] | 0.231 (0.0341) | [0.180, 0.288] | FALSE | FALSE | FALSE | 7.91E-01 | |
| x4x5 | 0.393 (0.03906) | [0.3335, 0.462] | 0.230 (0.0340) | [0.180, 0.289] | FALSE | FALSE | FALSE | 4.25E-02 | |
| x3x4 | 0.417 (0.03707) | [0.3601, 0.484] | 0.229 (0.0324) | [0.180, 0.287] | FALSE | FALSE | FALSE | 8.37E-01 |
Results shown are for the linear latent variable survival model (LLV), with continuous (Cont.) or discrete (Disc.) covariates, and fixed censoring rate (ρ = 0.5) in regression model #5 (positive control), where a single arbitrarily-fixed 2nd-order term enter in the model, simulated from two arbitrarily-fixed variables (e.g. here {j = 1, k = 2}, i.e. for x1x2 – see Section 3). Other cases are shown in Supplemental Table S24. Estimated forest-averaged IMDMS bivariate RSF statistics means (ψ̂(j, k)) are reported with standard errors (SE) and bootstrap confidence intervals (BCI) for all possible pairs of variable xj and xk, for j, k ∈ {1, …, p}, j < k (i.e. 10 pairs, for p = 5) by increasing order of IMDMS from top to bottom. The corresponding noised-up statistic (ψ̂* (j, k)) as well as IMDMS decision rule (1SE ∧ CI(θ)) at the θ level are shown in the adjacent columns. Also reported are the corresponding Cox-PH regression p-values testing 2nd-order pairwise interaction-effect terms at the θ level. NA value stands for situations when the fitting algorithm did not converge. In bold are the significant decision rules or p-values at the θ = 0.05 level. Note the accuracy of inferences by IMDMS decision rule as compared to Cox-PH regression inference: IMDMS correctly detects the true single variables interaction-effect x1x2 (and only it) for both tested types of covariates. Bold values stands for statistically significant or TRUE values.
Table 4.
Detection and ranking of pairwise interaction-effects by IMDMS bivariate RSF statistic vs standard Cox-PH regression in simulated data testing for influence of survival model type.
| Surv. model type |
Var.
pair xjxk |
Ψ̂(j,k) (SE) |
Ψ̂(j,k) [BCI] |
Ψ̂*
(j,k) (SE) |
Ψ̂*
(j,k) [BCI] |
1SE rule |
CI(θ) rule |
IMDMS 1SECI(θ) rule |
CPH. p-value |
|---|---|---|---|---|---|---|---|---|---|
| LLV | |||||||||
| x1x2 | 0.093 (0.0168) | [0.0715, 0.123] | 0.204 (0.0354) | [0.145, 0.260] | TRUE | TRUE | TRUE | NA | |
| x2x4 | 0.211 (0.0384) | [0.1422, 0.267] | 0.203 (0.0362) | [0.143, 0.264] | FALSE | FALSE | FALSE | 8.38E-01 | |
| x1x4 | 0.219 (0.0455) | [0.1403, 0.290] | 0.203 (0.0354) | [0.143, 0.262] | FALSE | FALSE | FALSE | 9.49E-01 | |
| x2x3 | 0.244 (0.0329) | [0.1940, 0.300] | 0.204 (0.0357) | [0.145, 0.263] | FALSE | FALSE | FALSE | 1.44E-01 | |
| x1x3 | 0.263 (0.0345) | [0.2071, 0.318] | 0.205 (0.0361) | [0.145, 0.264] | FALSE | FALSE | FALSE | 9.97E-01 | |
| x2x5 | 0.267 (0.0350) | [0.2128, 0.328] | 0.204 (0.0357) | [0.146, 0.262] | FALSE | FALSE | FALSE | 2.90E-01 | |
| x1x5 | 0.292 (0.0315) | [0.2396, 0.342] | 0.203 (0.0363) | [0.143, 0.263] | FALSE | FALSE | FALSE | 1.74E-01 | |
| x3x4 | 0.319 (0.0390) | [0.2565, 0.383] | 0.204 (0.0355) | [0.144, 0.262] | FALSE | FALSE | FALSE | 4.70E-02 | |
| x4x5 | 0.325 (0.0337) | [0.2732, 0.380] | 0.202 (0.0363) | [0.144, 0.263] | FALSE | FALSE | FALSE | 4.83E-01 | |
| x3x5 | 0.338 (0.0393) | [0.2685, 0.403] | 0.204 (0.0374) | [0.141, 0.265] | FALSE | FALSE | FALSE | 7.15E-02 | |
| EXP | |||||||||
| x1x2 | 0.128 (0.0176) | [0.104, 0.160] | 0.251 (0.0543) | [0.167, 0.344] | TRUE | TRUE | TRUE | NA | |
| x2x3 | 0.312 (0.0585) | [0.213, 0.403] | 0.250 (0.0544) | [0.166, 0.346] | FALSE | FALSE | FALSE | 2.40E-11 | |
| x1x3 | 0.316 (0.0497) | [0.237, 0.397] | 0.252 (0.0556) | [0.169, 0.349] | FALSE | FALSE | FALSE | 4.42E-01 | |
| x1x4 | 0.344 (0.0390) | [0.285, 0.410] | 0.253 (0.0559) | [0.168, 0.351] | FALSE | FALSE | FALSE | 7.80E-01 | |
| x2x4 | 0.347 (0.0572) | [0.251, 0.437] | 0.251 (0.0556) | [0.163, 0.343] | FALSE | FALSE | FALSE | 5.03E-01 | |
| x1x5 | 0.355 (0.0417) | [0.286, 0.420] | 0.251 (0.0559) | [0.167, 0.350] | FALSE | FALSE | FALSE | 1.56E-01 | |
| x2x5 | 0.384 (0.0438) | [0.315, 0.452] | 0.250 (0.0567) | [0.162, 0.349] | FALSE | FALSE | FALSE | 7.27E-01 | |
| x3x4 | 0.534 (0.0802) | [0.398, 0.656] | 0.254 (0.0554) | [0.168, 0.346] | FALSE | FALSE | FALSE | 3.19E-01 | |
| x3x5 | 0.544 (0.0746) | [0.425, 0.667] | 0.252 (0.0536) | [0.169, 0.350] | FALSE | FALSE | FALSE | 1.62E-01 | |
| x4x5 | 0.582 (0.0689) | [0.472, 0.696] | 0.253 (0.0547) | [0.172, 0.353] | FALSE | FALSE | FALSE | 2.57E-02 |
Results shown are for the linear latent variable (LLV) and exponential (EXP) survival models, with continuous variables, and fixed censoring rate (θ = 0.5) in regression model #5 (positive control), where a single arbitrarily-fixed 2nd-order term enter in the model, simulated from two arbitrarily-fixed variables (e.g. here {j = 1, k = 2}, i.e. for x1x2 – see Section 3). Other cases are shown in Supplemental Table S25. IMDMS bivariate RSF statistics means (Ψ̂(j, k)) are reported with standard errors (SE) and bootstrap confidence intervals (BCI) for all possible pairs of variable xj and xk, for j, k ∈ {1, …, p}, j < k (i.e. 10 pairs, for p = 5) by increasing order of IMDMS from top to bottom. The corresponding noised-up statistic (Ψ̂* (j, k)) as well as IMDMS decision rule (1SE ∧ CI(θ)) at the θ level are shown in the adjacent columns. Also reported are the corresponding Cox-PH regression p-values testing 2nd-order pairwise interaction-effect terms at the θ level. NA value stands for situations when the fitting algorithm did not converge. In bold are the significant decision rules or p-values at the θ = 0.05 level. Note the accuracy of inferences by IMDMS decision rule as compared to Cox-PH regression inference: IMDMS correctly detects the true single variables interaction-effect x1x2 (and only it) for both tested types of survival models. Bold values stands for statistically significant or TRUE values.
In a third set of simulation, we tested the influence of the censoring rate ρ that we allowed to vary within ρ ∈ {0.3, 0.5, 0.7}. We carried out similar analyses as above using both types of survival models (EXP vs. LLV), as well as both types of continuous or discrete covariates, in regression model #5, for 12 simulations. Using the IMDMS conjunction rule (at the θ = 0.05 confidence level), we show that IMDMS bivariate RSF statistics inferences were correct in nearly all (11/12) simulations (Table 5, Supplemental Tables S26–S28). Since bivariate RSF inferences of interaction-effects are robust to the effect of increasing censoring rate ρ, we validate the above results at fixed censoring rate ρ = 0.5.
Table 5.
Detection and ranking of pairwise interaction-effects by IMDMS bivariate RSF statistic vs standard Cox-PH regression in simulated data testing for influence of censoring rate.
| Cens. rate |
Var.
pair xjxk |
Ψ̂(j,
k) (SE) |
Ψ̂(j,
k) [BCI] |
Ψ̂*
(j, k) (SE) |
Ψ̂*
(j, k) [BCI] |
1SE rule |
CI(θ) rule |
IMDMS 1SE CI(θ) rule |
CPH. p-value |
|---|---|---|---|---|---|---|---|---|---|
| 30% | |||||||||
| x1x2 | 0.110 (0.0161) | [0.0871, 0.138] | 0.230 (0.0491) | [0.152, 0.315] | TRUE | TRUE | TRUE | NA | |
| x1x4 | 0.217 (0.0519) | [0.1279, 0.300] | 0.231 (0.0486) | [0.154, 0.318] | FALSE | FALSE | FALSE | 9.15E-01 | |
| x2x3 | 0.219 (0.0368) | [0.1603, 0.280] | 0.228 (0.0489) | [0.152, 0.311] | FALSE | FALSE | FALSE | 5.63E-01 | |
| x2x4 | 0.231 (0.0384) | [0.1697, 0.292] | 0.232 (0.0488) | [0.153, 0.315] | FALSE | FALSE | FALSE | 7.67E-01 | |
| x2x5 | 0.241 (0.0320) | [0.1882, 0.292] | 0.231 (0.0495) | [0.155, 0.312] | FALSE | FALSE | FALSE | 1.02E-01 | |
| x1x3 | 0.244 (0.0437) | [0.1723, 0.312] | 0.229 (0.0486) | [0.154, 0.313] | FALSE | FALSE | FALSE | 5.03E-01 | |
| x1x5 | 0.292 (0.0382) | [0.2238, 0.353] | 0.230 (0.0498) | [0.153, 0.316] | FALSE | FALSE | FALSE | 7.12E-01 | |
| x3x4 | 0.343 (0.0710) | [0.2337, 0.466] | 0.229 (0.0471) | [0.154, 0.312] | FALSE | FALSE | FALSE | 4.92E-01 | |
| x4x5 | 0.357 (0.0642) | [0.2650, 0.473] | 0.232 (0.0491) | [0.157, 0.320] | FALSE | FALSE | FALSE | 6.80E-01 | |
| x3x5 | 0.362 (0.0598) | [0.2664, 0.465] | 0.229 (0.0481) | [0.157, 0.313] | FALSE | FALSE | FALSE | 2.62E-01 | |
| 50% | |||||||||
| x1x2 | 0.0916 (0.0161) | [0.0716, 0.121] | 0.203 (0.0353) | [0.145, 0.264] | TRUE | TRUE | TRUE | NA | |
| x2x4 | 0.2129 (0.0381) | [0.1485, 0.275] | 0.204 (0.0361) | [0.145, 0.263] | FALSE | FALSE | FALSE | 8.38E-01 | |
| x1x4 | 0.2208 (0.0463) | [0.1423, 0.295] | 0.205 (0.0369) | [0.143, 0.263] | FALSE | FALSE | FALSE | 9.49E-01 | |
| x2x3 | 0.2413 (0.0341) | [0.1839, 0.298] | 0.202 (0.0368) | [0.141, 0.262] | FALSE | FALSE | FALSE | 1.44E-01 | |
| x1x3 | 0.2593 (0.0363) | [0.2009, 0.319] | 0.203 (0.0368) | [0.143, 0.264] | FALSE | FALSE | FALSE | 9.97E-01 | |
| x2x5 | 0.2682 (0.0341) | [0.2151, 0.327] | 0.203 (0.0363) | [0.141, 0.265] | FALSE | FALSE | FALSE | 2.90E-01 | |
| x1x5 | 0.2919 (0.0324) | [0.2415, 0.347] | 0.204 (0.0367) | [0.145, 0.264] | FALSE | FALSE | FALSE | 1.74E-01 | |
| x3x4 | 0.3186 (0.0398) | [0.2547, 0.382] | 0.204 (0.0368) | [0.143, 0.264] | FALSE | FALSE | FALSE | 4.70E-02 | |
| x4x5 | 0.3249 (0.0329) | [0.2715, 0.377] | 0.206 (0.0346) | [0.149, 0.263] | FALSE | FALSE | FALSE | 4.83E-01 | |
| x3x5 | 0.3372 (0.0411) | [0.2687, 0.409] | 0.203 (0.0357) | [0.146, 0.262] | FALSE | FALSE | FALSE | 7.15E-02 | |
| 70% | |||||||||
| x1x2 | 0.0955 (0.0139) | [0.0773, 0.122] | 0.197 (0.0346) | [0.139, 0.255] | TRUE | TRUE | TRUE | NA | |
| x2x4 | 0.2541 (0.0365) | [0.1894, 0.307] | 0.198 (0.0344) | [0.143, 0.254] | FALSE | FALSE | FALSE | 6.14E-01 | |
| x1x4 | 0.2614 (0.0446) | [0.1801, 0.326] | 0.196 (0.0349) | [0.137, 0.253] | FALSE | FALSE | FALSE | 8.48E-01 | |
| x2x3 | 0.2688 (0.0344) | [0.2129, 0.324] | 0.197 (0.0354) | [0.137, 0.253] | FALSE | FALSE | FALSE | 4.88E-01 | |
| x1x3 | 0.2835 (0.0370) | [0.2230, 0.342] | 0.197 (0.0337) | [0.142, 0.250] | FALSE | FALSE | FALSE | 3.99E-01 | |
| x2x5 | 0.2841 (0.0362) | [0.2285, 0.347] | 0.197 (0.0356) | [0.136, 0.254] | FALSE | FALSE | FALSE | 5.76E-01 | |
| x1x5 | 0.3063 (0.0347) | [0.2495, 0.368] | 0.197 (0.0338) | [0.143, 0.254] | FALSE | FALSE | FALSE | 2.77E-01 | |
| x4x5 | 0.3908 (0.0431) | [0.3225, 0.464] | 0.196 (0.0334) | [0.141, 0.251] | FALSE | FALSE | FALSE | 7.36E-01 | |
| x3x4 | 0.3915 (0.0466) | [0.3167, 0.467] | 0.196 (0.0336) | [0.141, 0.251] | FALSE | FALSE | FALSE | 8.98E-02 | |
| x3x5 | 0.3947 (0.0484) | [0.3159, 0.474] | 0.196 (0.0344) | [0.140, 0.251] | FALSE | FALSE | FALSE | 4.50E-03 |
Results shown are for the linear latent variable survival model (LLV), with continuous variables, and variable censoring rate (ρ ∈ {0.3, 0.5, 0.7}) in regression model #5 (positive control), where a single arbitrarily-fixed 2nd-order term enter in the model, simulated from two arbitrarily-fixed variables (e.g. here {j = 1, k = 2}, i.e. for x1x2 – see Section 3). Other cases are shown in Supplemental Tables S26, S27, S28. IMDMS bivariate RSF statistics means (Ψ̂(j, k)) are reported with standard errors (SE) and bootstrap confidence intervals (BCI) for all possible pairs of variable xj and xk, for j, k ∈ {1, …, p}, j < k (i.e. 10 pairs, for p = 5) by increasing order of IMDMS from top to bottom. The corresponding noised-up statistic (Ψ̂* (j, k)) as well as IMDMS decision rule (1SE ∧ CI(θ)) at the θ level are shown in the adjacent columns. Also reported are the corresponding Cox-PH regression p-values testing 2nd-order pairwise interaction-effect terms at the θ level. NA value stands for situations when the fitting algorithm did not converge. In bold are the significant decision rules or p-values at the θ = 0.05 level. Note the accuracy of inferences by IMDMS decision rule as compared to Cox-PH regression inference: IMDMS correctly detects the true single variables interaction-effect x1x2 (and only it) for all tested rates of censoring. Bold values stands for statistically significant or TRUE values.
In a fourth set of simulation, we tested the influence of nonlinear regression models with transformations of 2nd-order term by functions such as exponential, logarithm, power and trigonometric. We carried out similar analyses as above using both types of survival models (EXP vs. LLV), as well as both types of continuous or discrete covariates, in regression model #5, that is, for 16 simulations. Using the IMDMS conjunction rule (at the θ = 0.05 confidence level), we show that IMDMS bivariate RSF statistics inferences were correct in all (16/16) simulations (Table 6, Supplemental Tables S29–S31).
Table 6.
Detection and ranking of pairwise interaction-effects by IMDMS bivariate RSF statistic vs standard Cox-PH regression in simulated data testing for influence of nonlinear transformation.
| Reg. model type |
Var.
pair xjxk |
Ψ̂(j,
k) (SE) |
Ψ̂(j,
k) [BCI] |
Ψ̂*
(j, k) (SE) |
Ψ̂*
(j, k) [BCI] |
1SE rule |
CI(θ) rule |
IMDMS 1SE CI(θ) rule |
CPH. p-value |
|---|---|---|---|---|---|---|---|---|---|
| #5a | |||||||||
| x1x2 | 0.0958 (0.0164) | [0.0738, 0.126] | 0.201 (0.0354) | [0.143, 0.261] | TRUE | TRUE | TRUE | NA | |
| x1x5 | 0.1670 (0.0177) | [0.1372, 0.198] | 0.201 (0.0343) | [0.147, 0.258] | TRUE | FALSE | FALSE | 8.51E-01 | |
| x1x4 | 0.1751 (0.0186) | [0.1423, 0.204] | 0.200 (0.0345) | [0.144, 0.256] | TRUE | FALSE | FALSE | 7.28E-01 | |
| x1x3 | 0.1779 (0.0158) | [0.1534, 0.203] | 0.201 (0.0332) | [0.145, 0.252] | TRUE | FALSE | FALSE | 8.60E-01 | |
| x2x5 | 0.1868 (0.0288) | [0.1385, 0.232] | 0.201 (0.0350) | [0.143, 0.259] | FALSE | FALSE | FALSE | 9.13E-01 | |
| x2x3 | 0.1948 (0.0276) | [0.1474, 0.238] | 0.200 (0.0341) | [0.144, 0.254] | FALSE | FALSE | FALSE | 8.27E-01 | |
| x2x4 | 0.1995 (0.0274) | [0.1543, 0.243] | 0.201 (0.0345) | [0.145, 0.259] | FALSE | FALSE | FALSE | 8.46E-01 | |
| x3x5 | 0.2873 (0.0333) | [0.2339, 0.344] | 0.201 (0.0337) | [0.144, 0.255] | FALSE | FALSE | FALSE | 1.68E-03 | |
| x3x4 | 0.2927 (0.0335) | [0.2418, 0.350] | 0.200 (0.0344) | [0.143, 0.256] | FALSE | FALSE | FALSE | 1.48E-01 | |
| x4x5 | 0.3019 (0.0333) | [0.2478, 0.356] | 0.202 (0.0350) | [0.145, 0.259] | FALSE | FALSE | FALSE | 5.17E-01 | |
| #5b | |||||||||
| x1x2 | 0.0905 (0.0159) | [0.0677, 0.121] | 0.202 (0.0340) | [0.146, 0.258] | TRUE | TRUE | TRUE | 2.06E-05 | |
| x1x5 | 0.1638 (0.0193) | [0.1318, 0.196] | 0.201 (0.0345) | [0.147, 0.257] | TRUE | FALSE | FALSE | 9.85E-01 | |
| x1x4 | 0.1660 (0.0170) | [0.1398, 0.194] | 0.199 (0.0333) | [0.145, 0.251] | TRUE | FALSE | FALSE | 9.98E-01 | |
| x1x3 | 0.1678 (0.0201) | [0.1344, 0.201] | 0.202 (0.0341) | [0.146, 0.256] | TRUE | FALSE | FALSE | 6.71E-01 | |
| x2x5 | 0.1803 (0.0283) | [0.1293, 0.224] | 0.202 (0.0344) | [0.146, 0.257] | FALSE | FALSE | FALSE | 7.50E-01 | |
| x2x3 | 0.1849 (0.0270) | [0.1403, 0.228] | 0.200 (0.0340) | [0.142, 0.254] | FALSE | FALSE | FALSE | 8.19E-01 | |
| x2x4 | 0.2004 (0.0264) | [0.1559, 0.241] | 0.203 (0.0352) | [0.145, 0.262] | FALSE | FALSE | FALSE | 6.40E-01 | |
| x3x5 | 0.2877 (0.0333) | [0.2342, 0.343] | 0.200 (0.0331) | [0.143, 0.255] | FALSE | FALSE | FALSE | 1.18E-01 | |
| x3x4 | 0.2878 (0.0316) | [0.2396, 0.342] | 0.200 (0.0342) | [0.145, 0.255] | FALSE | FALSE | FALSE | 2.18E-03 | |
| x4x5 | 0.3000 (0.0362) | [0.2425, 0.360] | 0.203 (0.0342) | [0.147, 0.261] | FALSE | FALSE | FALSE | 7.60E-01 | |
| #5c | |||||||||
| x1x2 | 0.102 (0.0183) | [0.0744, 0.134] | 0.202 (0.0344) | [0.146, 0.257] | TRUE | TRUE | TRUE | NA | |
| x1x5 | 0.170 (0.0170) | [0.1424, 0.199] | 0.202 (0.0365) | [0.140, 0.261] | TRUE | FALSE | FALSE | 9.16E-01 | |
| x1x3 | 0.172 (0.0192) | [0.1388, 0.202] | 0.203 (0.0348) | [0.147, 0.260] | TRUE | FALSE | FALSE | 7.85E-01 | |
| x1x4 | 0.177 (0.0157) | [0.1497, 0.202] | 0.201 (0.0350) | [0.141, 0.258] | TRUE | FALSE | FALSE | 8.97E-01 | |
| x2x5 | 0.186 (0.0288) | [0.1362, 0.232] | 0.200 (0.0344) | [0.138, 0.254] | FALSE | FALSE | FALSE | 8.80E-01 | |
| x2x3 | 0.189 (0.0283) | [0.1408, 0.232] | 0.200 (0.0338) | [0.145, 0.255] | FALSE | FALSE | FALSE | 6.23E-01 | |
| x2x4 | 0.205 (0.0254) | [0.1607, 0.244] | 0.202 (0.0345) | [0.143, 0.255] | FALSE | FALSE | FALSE | 6.53E-01 | |
| x3x4 | 0.297 (0.0325) | [0.2443, 0.351] | 0.199 (0.0345) | [0.140, 0.256] | FALSE | FALSE | FALSE | 1.79E-03 | |
| x3x5 | 0.298 (0.0348) | [0.2387, 0.356] | 0.201 (0.0353) | [0.142, 0.259] | FALSE | FALSE | FALSE | 1.62E-01 | |
| x4x5 | 0.311 (0.0377) | [0.2480, 0.372] | 0.201 (0.0349) | [0.143, 0.257] | FALSE | FALSE | FALSE | 5.35E-01 | |
| #5d | |||||||||
| x1x2 | 0.0865 (0.0147) | [0.0642, 0.113] | 0.200 (0.0352) | [0.144, 0.258] | TRUE | TRUE | TRUE | 2.99E-01 | |
| x2x4 | 0.1520 (0.0274) | [0.1094, 0.198] | 0.200 (0.0349) | [0.143, 0.258] | TRUE | FALSE | FALSE | 8.14E-01 | |
| x1x4 | 0.1553 (0.0227) | [0.1178, 0.192] | 0.202 (0.0344) | [0.143, 0.260] | TRUE | FALSE | FALSE | 7.08E-01 | |
| x1x3 | 0.1614 (0.0262) | [0.1193, 0.205] | 0.203 (0.0342) | [0.146, 0.261] | TRUE | FALSE | FALSE | 9.04E-01 | |
| x2x3 | 0.1774 (0.0293) | [0.1299, 0.226] | 0.200 (0.0337) | [0.143, 0.254] | FALSE | FALSE | FALSE | 4.42E-01 | |
| x1x5 | 0.1815 (0.0255) | [0.1351, 0.220] | 0.202 (0.0345) | [0.146, 0.258] | FALSE | FALSE | FALSE | 8.09E-02 | |
| x2x5 | 0.1919 (0.0301) | [0.1371, 0.237] | 0.200 (0.0344) | [0.144, 0.257] | FALSE | FALSE | FALSE | 2.28E-01 | |
| x3x4 | 0.2292 (0.0330) | [0.1760, 0.284] | 0.203 (0.0340) | [0.148, 0.260] | FALSE | FALSE | FALSE | 1.21E-01 | |
| x4x5 | 0.2519 (0.0300) | [0.2058, 0.304] | 0.200 (0.0347) | [0.146, 0.256] | FALSE | FALSE | FALSE | 8.98E-01 | |
| x3x5 | 0.2586 (0.0323) | [0.2066, 0.312] | 0.201 (0.0355) | [0.143, 0.256] | FALSE | FALSE | FALSE | 1.01E-02 |
Results shown are for the linear latent variable survival model (LLV), with continuous variables, and fixed censoring rate (ρ = 0.5) in nonlinear regression models #5a, #5b, #5c, #5d (positive controls), where a single arbitrarily-fixed 2nd-order term enter in the model, simulated from two arbitrarily-fixed variables (e.g. here {j = 1, k = 2}, i.e. for x1x2 – see Section 3). Other cases are shown in Supplemental Tables S29, S30, S31. IMDMS bivariate RSF statistics means (Ψ̂ (j, k)) are reported with standard errors (SE) and bootstrap confidence intervals (BCI) for all possible pairs of variable xj and xk, for j, k ∈ {1, …, p}, j < k (i.e. 10 pairs, for p = 5) by increasing order of IMDMS from top to bottom. The corresponding noised-up statistic (Ψ̂* (j, k)) as well as IMDMS decision rule (1SE ∧ CI(θ)) at the θ level are shown in the adjacent columns. Also reported are the corresponding Cox-PH regression p-values testing 2nd-order pairwise interaction-effect terms at the θ level. NA value stands for situations when the fitting algorithm did not converge. In bold are the significant decision rules or p-values at the θ = 0.05 level. Note the accuracy of inferences by IMDMS decision rule as compared to Cox-PH regression inference: IMDMS correctly detects the true single variables interaction-effect x1x2 (and only it) in all tested types of nonlinear regression models. Bold values stands for statistically significant or TRUE values.
Finally, in this last set of simulations, we tested the potential selection bias that could be induced by arbitrarily fixing two non-null variables {j, k} with their corresponding interaction-effect xjxk entering into regression model #5. We carried out similar analyses as above using both types of survival models (EXP vs. LLV), as well as both types of continuous or discrete covariates, at fixed censoring rate (ρ = 0.5). We tested for all 2nd-order pairwise interaction terms out of all combinations of variables pairs {j, k} in type-#5 regression model, that is, for 40 simulations. Here, successful RSF inferences are those for which only one variable pair is found significant and matches the tested 2nd-order term of true pairwise interaction-effect. Using the IMDMS conjunction rule (at the θ = 0.05 confidence level), except for two (2) situations where RSF inference failed, result show that the arbitrary choice of {j, k} does not matter. This is consistent with the fact that variables xj for j ∈ {1, …, p} were i.i.d., that is, without inter-variable correlation (Table 7, Supplemental Tables S32–S34).
Table 7.
Detections of pairwise interaction-effects by IMDMS bivariate RSF statistic vs standard Cox-PH regression in simulated data testing for all possible pairwise interaction terms.
| Tested type-#5 reg. model βj,k ≠ 0 |
1SE rule | CI(θ) rule |
IMDMS 1SE
CI(θ) rule |
Signif. var. pair found by CPH |
CPH. p-value |
|---|---|---|---|---|---|
| β1,2 ≠ 0 | {x1x2} | {x1x2} | {x1x2} | {∅} | NA |
| {x3x4} | 4.70E-02 | ||||
| β1,3 ≠ 0 | {x1x3} | {x1x3} | {x1x3} | {∅} | NA |
| β1,4 ≠ 0 | {x1x4} | {x1x4} | {x1x4} | {∅} | NA |
| {x3x4} | 3.68E-02 | ||||
| β1,5 ≠ 0 | {x1x5} | {x1x5} | {x1x5} | {∅} | NA |
| β2,3 ≠ 0 | {x2x3} | {x2x3} | {x2x3} | {∅} | NA |
| β2,4 ≠ 0 | {x2x4} | {x2x4} | {x2x4} | {∅} | NA |
| {x1x3} | 1.43E-02 | ||||
| β2,5 ≠ 0 | {x2x5} | {x2x5} | {x2x5} | {∅} | NA |
| β3,4 ≠ 0 | {x3x4} | {x3x4} | {x3x4} | {∅} | NA |
| {x1x5} | 3.15E-02 | ||||
| β3,5 ≠ 0 | {x3x5} | {x3x5} | {x3x5} | {∅} | NA |
| β4,5 ≠ 0 | {x4x5} | {x4x5} | {x4x5} | {∅} | NA |
Results shown are for the linear latent variable survival model (LLV), with continuous variables, and fixed censoring rate (ρ = 0.5) in regression model #5 (positive control), where a single arbitrarily-fixed 2nd-order term enter in the model, simulated from two arbitrarily-fixed variables chosen between all possible pairs of variable xj and xk, for j, k ∈ {1, …, p}, j < k (i.e. for testing xjxk interaction-effect with corresponding xj and xk main-effects – see Section 3). Other cases are shown in Supplemental Tables S32, S33, S34. For each tested regression model #5 (row βj,k ≠ 0), the IMDMS decision rule (1SE ∧ CI(θ)) corresponding to all possible pairs of variables was determined, and those pairs, whose IMDMS bivariate RSF statistics were found significant by IMDMS decision rule at the θ = 0.05 level, are reported. If multiple pairs of variables were found significant for a given test, a list is reported by increasing order of IMDMS from top to bottom. Also reported are the corresponding significant Cox-PH regression p-values testing 2nd-order pairwise interaction-effect terms at the θ = 0.05 level. NA value stands for situations when the fitting algorithm did not converge. Note the difference of accuracy of inferences by IMDMS decision rule as compared to Cox-PH regression inference: in contrast to Cox-PH regression, IMDMS correctly detects all true variables interaction-effects (and only them) in all tested type-#5 regression models.
3.5 Summary of pairwise interaction-effects inferences and comparison to Cox-PH modeling
Overall, empirical evidences in simulation studies show that the IMDMS bivariate RSF statistic with its conjunction rule (at a given significance level θ) was successful in detecting nearly all true pairwise interaction-effects between variables with the exception of four (4) ones that were missed out of all eight-eight (88) possible cases (4.5%). This points to the power of the RSF approach to detect pairwise interaction-effects associated with a time-to-event outcome, regardless of the accompaniment of corresponding main-effects, the type of linear or nonlinear regression model used, the type of survival model used, the type of continuous or discrete covariates, and the rate of censoring (Table 3–Table 7 and Supplemental Tables S18–S34).
Further, results show that the randomization method used, along with the generation of CI of IMDMS bivariate RSF statistics, and the use of a simple decision rule, were altogether essential parts of our RSF approach for making accurate inferences of pairwise interaction-effects between variables by since many spurious would have been falsely detected otherwise (by simply comparing one forest-averaged RSF statistic to its noised-up counterpart).
Finally, successful RSF inferences about pairwise interaction-effects also point to the superiority of our RSF approach to detect more subtle or complex pairwise interaction-effects as compared to standard Cox-PH regression inferences. In contrast to RSF modeling, empirical evidences in our simulation studies show that Cox-PH regression often failed to detect true pairwise interactions, especially in the case of nonlinear regression models/ transformations of 2nd order term (Table 6, Supplemental Tables S29, S30, S31). In addition, Cox-PH regression inferences also appear generally less robust to noise since several spurious pairwise interaction-effects were also wrongly called significant at the same significance level θ = 0.05 (Table 3–Table 7 and Supplemental Tables S18–S34).
3.6 Individualized predictions
In our simulated data analyses as well as in real data application (see next), we first calculated the accuracy of our ensemble prediction approach to make individualized predictions in terms of eventuality (mortality) values and event-free (survival) probabilities, using an independent test dataset of individuals (see Section 2). This was done for all situations of types of survival models, variables, and fixed censoring rate (ρ = 0.5), in regression model #5, as well as in real data analyses for both outcomes (see next section). For instance, RSF prediction error estimates for the LLV survival model, with continuous variables, and fixed censoring rate (ρ = 0.5) were found to be 30.8%, as well as 32.9% and 29.1% for time-to-X4-Emergence and time-to-AIDS-Diagnosis respectively.
Second, we show how our ensemble approach can be used to calculate and plot predicted individual eventuality (mortality) values and individual event-free (survival) probabilities for each of the test-set individuals as a function of follow-up time. In both simulated and real data analyses, each event time point and curve of the plots corresponds to a test-set individual’s ensemble prediction estimate (i.e. values obtained by overlaying the test set outcomes on the training forest), evaluated at each observed time point (see Section 2). Again, this was done for all situations of types of survival models, variables, and fixed censoring rate (ρ = 0.5), in regression model #5, and we illustrate the approach in one case (Supplemental Figure S7) as well as in real data analyses for both outcomes (Supplemental Figure S12).
4 Application to the MACS HIV cohort study
4.1 Background
For our objectives, we utilized samples from the previously published MACS cohort study, which provides longitudinal account of viral tropism in relation to the HIV full spectrum of rates of HIV-1 disease progression (Shepherd et al., 2008). To our knowledge, this cohort provides a unique dataset with well characterized clinical information for analyzing associations between host genetic variation and viral tropism as well as disease progression. Here, we determined whether CNV in β-defensin and its interactions with certain polymorphisms in chemokine receptors and ligand genes are associated, either alone or jointly, with clinical events in HIV-seropositive patients, such as time to HIV change of tropism or time to AIDS diagnosis. Additional descriptions of the dataset and materials used are provided in the Supplemental Material
4.2 Variables and outcomes
The variables included in the MACS cohort study were five genetic variants (DEFB4/103A CNV [2–5], CCR2 SNP [190G>A], CCR5 [SNP −2459G>A, ORF], CXCL12 SNP [801G>A]) and two non-genetic variables, taken as two additional covariates. All variables were categorical with no more than three levels (experimental groups) each. We used genetic variables with original and aggregated categories as follows: DEFB CNV [CNV = 2 or CNV > 2]; CCR2 SNP [GG or GA], CCR5 SNP [GG or GA]; CCR5 ORF [WT or Δ32], CXCL12 SNP [GG or GA]. For justification see Supplemental Materials and Methods with references therein (Supplemental Materials II). The first covariate was the two-level disease progression Group variable [Fast, Slow], and the second was the three-level Race/Ethnicity variable [White, Hispanic, Black]. For each observation i ∈ {1, …, n}, we denote the jth variable by the n-dimensional vector xj = (x1,j, …, xn,j)T, where j ∈ {1, …, p}. Here, p denotes the number of variables. Hereafter, we denoted the p = 7 included variables as follows: x1 = DEFB CNV, x2 = CCR2 SNP, x3 = CCR5 SNP, x4 = CCR5 ORF, x5 = CXCL12 SNP, x6 = Group, x7 = Race. Whenever we needed to fix a certain level of a variable, we used the expression “conditioning on”. The time-to-event outcomes included in the MACS cohort study, generically denoted E, were the time-to-X4-Emergence (denoted XE) and the time-to-AIDS-Diagnosis (denoted AD), whether each was observed or not during each patient’s follow-up time (Supplemental Figure S8). The corresponding event-free (EF) (“survival”) probability function S(t) of time-to-event E≔XE (X4-Emergence) or E≔AD (AIDS-Diagnosis),were called X4-Emergence-Free (E≔XEF) or AIDS-Diagnosis-Free (E≔ADF) probability.
4.3 RSF model building and global prediction performance
Here, we report RSF model building and global prediction performance for the X4-Emergence (Figure 3A, B) and AIDS-Diagnosis (Figure 3C, D) outcomes of the MACS cohort study. The RSF plots show that the cumulative OOB error rate stabilizes rapidly as a function of the number of trees, indicating that both forests of B = 1000 trees were sufficient, and that the ensemble trees were well-grown for both outcomes with an average number of terminal nodes of 14.2 and 14.5 for the X4-Emergence and AIDS-Diagnosis outcomes, respectively. However, the error rates convergence limits of 50.53% and 32.32% for the X4-Emergence (Figure 3A) and AIDS-Diagnosis (Figure 3C) outcomes, respectively, indicate that modeling of RSF-based pairwise interactions will be reliable for the later outcome only. Exemplary tree-plots are also shown for the X4-Emergence (Figure 3B) and AIDS-Diagnosis (Figure 3D) outcomes, each taken at random out of the B = 1000 trees of the RSF forest. Although both trees of the forests represent one instance out of many, they illustrate the differences of possible ranking of variable importance as well as variables interactions for each outcome. For example, in both outcomes, the covariate Group appears the most important (root nodes), and the top interactions appear to be those between Group with genetic variants CXCL12 801, DEFB4/103A CNV and CCR2 190 SNP (Figure 3B, D).
Figure 3.
RSF Global Prediction Performance and Visualization of RSF Illustrative Tree in Both Outcomes of the MACS Cohort Study (A, B) Left: forest-averaged RSF cumulative OOB error rates for the ensemble as a function of number of trees. (C, D) Visualization of an exemplary tree (e.g. #38, C; e.g. #11, D) out of the B = 1000 trees of the RSF forests for the time-to-X4-Emergence (C) and time-to-AIDS-Diagnosis (D) outcomes, respectively. Note how cumulative OOB error rates stabilize rapidly as a function of the number of trees (at 50.53% and 32.32%), indicating that a forest of B = 1000 trees was sufficient for both outcomes with an average number of terminal nodes of 14.2 and 14.5 for the X4-Emergence and AIDS-Diagnosis outcomes, respectively. Yellow-colored nodes represent individual variables. The depth of the trees is indicated by numbers (0–6) inside of each node (0 being the root node). Each tree illustrates one possible ranking of variable importance (height vs. depth of the nodes) and variable interactions (edges between nodes in the same branch). In this example, note how top interactions involving root node Group with a child node of another genetic variant are detected by IMDMS bivariate RSF statistic (highlighted red edges).
4.4 Reporting of results and inferences
Estimates of our IMDMS bivariate variable interaction statistics were calculated as described in Methods for all combinations of 2nd-order term (pairwise) interactions between variables xj and xk, for j, k ∈ {1, …, p}, j < k (and noised-up counterparts), i.e. for 21 pairs, with p = 7. Also reported are the corresponding Cox-PH regression p-values testing the 2nd-order pairwise interaction-effect terms between all possible pairs of variables xj and xk (Table 8 and Table 9).
Table 8.
Detection and ranking of pairwise interaction-effects by IMDMS bivariate RSF statistic vs standard Cox-PH regression in X4-emergence outcome of the MACS cohort study.
| Var.
pair xjxk |
Ψ̂(j,
k) (SE) |
Ψ̂(j,
k) [BCI] |
Ψ̂*
(j, k) (SE) |
Ψ̂*
(j, k) [BCI] |
1SE rule | CI(θ) rule |
IMDMS 1SE CI(θ) rule |
CPH. p-value |
|---|---|---|---|---|---|---|---|---|
| CXCL12 SNP: Group | 0.539 (0.1172) | [0.401, 0.698] | 0.676 (0.136) | [0.499, 0.859] | TRUE | FALSE | FALSE | 3.90E-01 |
| DEFB CNV: Group | 0.591 (0.1355) | [0.419, 0.782] | 0.684 (0.132) | [0.510, 0.863] | FALSE | FALSE | FALSE | 9.10E-01 |
| CCR2 SNP: Group | 0.597 (0.1480) | [0.416, 0.808] | 0.692 (0.136) | [0.514, 0.858] | FALSE | FALSE | FALSE | 1.27E-01 |
| CCR5 SNP: Group | 0.602 (0.1504) | [0.416, 0.819] | 0.689 (0.129) | [0.524, 0.863] | FALSE | FALSE | FALSE | 6.48E-01 |
| CCR5 SNP: CXCL12 SNP | 0.603 (0.1082) | [0.466, 0.742] | 0.685 (0.129) | [0.514, 0.854] | FALSE | FALSE | FALSE | 3.44E-01 |
| CCR2 SNP: CXCL12 SNP | 0.620 (0.0886) | [0.515, 0.736] | 0.686 (0.140) | [0.502, 0.872] | FALSE | FALSE | FALSE | 4.01E-01 |
| CCR2 SNP: CCR5 SNP | 0.641 (0.1155) | [0.505, 0.790] | 0.703 (0.132) | [0.526, 0.868] | FALSE | FALSE | FALSE | 1.98E-01 |
| CCR2 SNP: DEFB CNV | 0.670 (0.0973) | [0.542, 0.792] | 0.706 (0.132) | [0.534, 0.879] | FALSE | FALSE | FALSE | 1.79E-01 |
| CXCL12 SNP: DEFB CNV | 0.678 (0.0894) | [0.566, 0.794] | 0.676 (0.137) | [0.502, 0.861] | FALSE | FALSE | FALSE | 4.80E-01 |
| CCR5 SNP: DEFB CNV | 0.678 (0.1045) | [0.544, 0.813] | 0.687 (0.134) | [0.518, 0.862] | FALSE | FALSE | FALSE | 4.01E-01 |
| CXCL12 SNP: Race | 0.691 (0.1255) | [0.522, 0.848] | 0.682 (0.143) | [0.503, 0.880] | FALSE | FALSE | FALSE | NA |
| DEFB CNV: Race | 0.694 (0.1205) | [0.531, 0.844] | 0.699 (0.143) | [0.513, 0.894] | FALSE | FALSE | FALSE | 5.70E-01 |
| CCR5 ORF: CXCL12 SNP | 0.699 (0.1031) | [0.563, 0.833] | 0.736 (0.139) | [0.553, 0.917] | FALSE | FALSE | FALSE | 8.85E-01 |
| CCR5 ORF: DEFB CNV | 0.699 (0.1452) | [0.488, 0.881] | 0.753 (0.134) | [0.576, 0.926] | FALSE | FALSE | FALSE | NA |
| Group: Race | 0.710 (0.1446) | [0.516, 0.889] | 0.687 (0.143) | [0.500, 0.882] | FALSE | FALSE | FALSE | 1.31E-01 |
| CCR5 SNP: Race | 0.731 (0.1287) | [0.549, 0.893] | 0.696 (0.139) | [0.514, 0.880] | FALSE | FALSE | FALSE | 7.97E-01 |
| CCR5 ORF: Group | 0.734 (0.1372) | [0.542, 0.903] | 0.745 (0.137) | [0.563, 0.915] | FALSE | FALSE | FALSE | 3.92E-02 |
| CCR2 SNP: Race | 0.736 (0.1409) | [0.532, 0.907] | 0.719 (0.143) | [0.529, 0.915] | FALSE | FALSE | FALSE | 1.42E-01 |
| CCR5 ORF: CCR5 SNP | 0.766 (0.1077) | [0.624, 0.896] | 0.753 (0.133) | [0.572, 0.921] | FALSE | FALSE | FALSE | NA |
| CCR2 SNP: CCR5 ORF | 0.770 (0.0985) | [0.641, 0.893] | 0.774 (0.135) | [0.584, 0.946] | FALSE | FALSE | FALSE | 6.48E-01 |
| CCR5 ORF: Race | 0.786 (0.1243) | [0.618, 0.932] | 0.782 (0.135) | [0.595, 0.951] | FALSE | FALSE | FALSE | NA |
IMDMS bivariate RSF statistics means (Ψ̂(j, k)) are reported with standard errors (SE) and bootstrap confidence intervals (BCI) for all possible pairs of variable xj and xk, for j, k ∈ {1, …, p}, j < k (i.e. 21 pairs, for p = 7) by increasing order from top to bottom. The corresponding noised-up statistic (Ψ̂* (j, k)) as well as IMDMS decision rule (1SE ∧ CI(θ)) at the θ level are shown in the adjacent columns. Also reported are the corresponding Cox-PH regression p-values testing 2nd-order pairwise interaction-effect terms at the θ level. NA value stands for situations when the fitting algorithm did not converge. In bold are the significant decision rules or p-values at the θ = 0.10 level. Note the differences of inferences between RSF and Cox-PH regression modeling (see also Figure 4). Bold values stands for statistically significant or TRUE values.
Table 9.
Detection and ranking of pairwise interaction-effects by IMDMS bivariate RSF statistic vs standard Cox-PH regression in AIDS-diagnosis outcome of the MACS cohort study.
| Var.
pair xjxk |
Ψ̂(j,
k) (SE) |
Ψ̂(j,
k) [BCI] |
Ψ̂*
(j, k) (SE) |
Ψ̂*
(j, k) [BCI] |
1SE rule | CI(θ) rule |
IMDMS 1SE CI(θ) rule |
CPH. p-value |
|---|---|---|---|---|---|---|---|---|
| DEFB CNV: Group | 0.272 (0.0812) | [0.190, 0.389] | 0.676 (0.138) | [0.495, 0.851] | TRUE | TRUE | TRUE | NA |
| CCR2 SNP: Group | 0.378 (0.0756) | [0.284, 0.473] | 0.690 (0.134) | [0.513, 0.870] | TRUE | TRUE | TRUE | NA |
| CXCL12 SNP: Group | 0.397 (0.0547) | [0.325, 0.465] | 0.669 (0.137) | [0.487, 0.851] | TRUE | TRUE | TRUE | NA |
| Group: Race | 0.403 (0.1523) | [0.228, 0.616] | 0.672 (0.150) | [0.479, 0.878] | TRUE | FALSE | FALSE | NA |
| CCR5 SNP: Group | 0.416 (0.0765) | [0.314, 0.513] | 0.671 (0.139) | [0.492, 0.857] | TRUE | FALSE | FALSE | NA |
| CCR5 ORF: Group | 0.480 (0.1269) | [0.314, 0.647] | 0.732 (0.144) | [0.544, 0.916] | TRUE | FALSE | FALSE | NA |
| CXCL12 SNP: DEFB CNV | 0.579 (0.1009) | [0.457, 0.709] | 0.673 (0.137) | [0.495, 0.853] | FALSE | FALSE | FALSE | 4.84E-01 |
| CCR2 SNP: CXCL12 SNP | 0.605 (0.1070) | [0.482, 0.743] | 0.679 (0.139) | [0.493, 0.857] | FALSE | FALSE | FALSE | 5.20E-01 |
| CXCL12 SNP: Race | 0.611 (0.1418) | [0.434, 0.814] | 0.669 (0.145) | [0.483, 0.870] | FALSE | FALSE | FALSE | 5.94E-01 |
| CCR5 SNP: CXCL12 SNP | 0.631 (0.1125) | [0.489, 0.771] | 0.663 (0.137) | [0.483, 0.839] | FALSE | FALSE | FALSE | 1.25E-01 |
| CCR5 SNP: DEFB CNV | 0.650 (0.1118) | [0.507, 0.803] | 0.682 (0.139) | [0.504, 0.864] | FALSE | FALSE | FALSE | 7.78E-02 |
| CCR2 SNP: CCR5 SNP | 0.656 (0.1424) | [0.467, 0.847] | 0.695 (0.142) | [0.509, 0.874] | FALSE | FALSE | FALSE | 6.85E-02 |
| CCR5 SNP: Race | 0.660 (0.1578) | [0.461, 0.881] | 0.687 (0.149) | [0.489, 0.882] | FALSE | FALSE | FALSE | 7.30E-02 |
| CCR2 SNP: Race | 0.670 (0.1646) | [0.459, 0.897] | 0.703 (0.147) | [0.510, 0.899] | FALSE | FALSE | FALSE | 1.74E-01 |
| CCR5 ORF: DEFB CNV | 0.686 (0.1244) | [0.522, 0.852] | 0.699 (0.137) | [0.527, 0.871] | FALSE | FALSE | FALSE | 7.98E-01 |
| CCR2 SNP: DEFB CNV | 0.698 (0.1368) | [0.501, 0.867] | 0.743 (0.141) | [0.549, 0.927] | FALSE | FALSE | FALSE | 4.05E-01 |
| CCR5 ORF: CXCL12 SNP | 0.708 (0.1093) | [0.564, 0.847] | 0.725 (0.144) | [0.527, 0.912] | FALSE | FALSE | FALSE | NA |
| DEFB CNV: Race | 0.722 (0.1256) | [0.565, 0.899] | 0.683 (0.149) | [0.495, 0.890] | FALSE | FALSE | FALSE | 4.33E-03 |
| CCR5 ORF: Race | 0.750 (0.1314) | [0.578, 0.925] | 0.767 (0.142) | [0.567, 0.946] | FALSE | FALSE | FALSE | NA |
| CCR5 ORF: CCR5 SNP | 0.767 (0.1083) | [0.622, 0.906] | 0.748 (0.143) | [0.557, 0.932] | FALSE | FALSE | FALSE | NA |
| CCR2 SNP: CCR5 ORF | 0.769 (0.1194) | [0.596, 0.920] | 0.759 (0.139) | [0.566, 0.936] | FALSE | FALSE | FALSE | 1.96E-01 |
IMDMS bivariate RSF statistics means (Ψ̂(j, k)) are reported with standard errors (SE) and bootstrap confidence intervals (BCI) for all possible pairs of variable xj and xk, for j, k ∈ {1, …, p}, j < k (i.e. 21 pairs, for p = 7) by increasing order from top to bottom. The corresponding noised-up statistic (Ψ̂* (j, k)) as well as IMDMS decision rule (1SE ∧ CI(θ)) at the θ level are shown in the adjacent columns. Also reported are the corresponding Cox-PH regression p-values testing 2nd-order pairwise interaction-effect terms at the θ level. NA value stands for situations when the fitting algorithm did not converge. In bold are the significant decision rules or p-values at the θ = 0.10 level. Note the differences of inferences between RSF and Cox-PH regression modeling (see also Figure 4). Bold values stands for statistically significant or TRUE values.
As in above simulation studies, the canonical 1SE (eqs. 5 or 7) and CI(θ) (eqs. 6 or 8) decision rules were reported in all Tables. In addition, the disjunction or conjunction rules derived from both were used to report significance of individual important/predictive variables as well as of pairs of interacting variables, respectively (TRUE/FALSE calls in Tables and Supplemental Tables). Successful RSF inferences about individual variable importance and pairs of interacting variables are as stated in above section.
For conciseness, only results of interaction-effects by RSF bivariate estimators are shown, but all results of analyses of main-effects by RSF univariate estimators are available in Supplemental Results, Discussion, Figures and Tables (Supplemental Materials II).
4.5 Analysis of interaction-effects by RSF bivariate estimators
Bivariate RSF statistic IMDMS revealed no significant interaction-effects between all variables for the time-to-X4-Emergence outcome (Table 8, Figure 4A), and three significant interaction-effects between Group and variables DEFB4/103A CNV, CCR2 190 SNP, CXCL12 801 SNP (by order of significance) for the time-to-AIDS-Diagnosis outcome (Table 9, Figure 4B). Of note, significant RSF interaction-effects are not always accompanied by corresponding significant RSF main-effects (Table 8 and Table 9, Figure 4, Supplemental Figures S9, S10). This underscores the limitations of focusing on only univariate measures of variable importance, and of fitting interactions to variables with significant main-effects only. Even though more interaction-effects are detected using the more liberal 1SE rule for both outcomes, results from this rule are to be taken with caution due to the fact that they are not confirmed by the CI(θ) rule and because empirical evinces in our simulations studies showed that the more conservative CI(θ) rule is in general more accurate. In fact, the additional interaction-effect between Group and Race in time-to-AIDS-Diagnosis (but not in time-to-X4-Emergence) is to be carefully interpreted due to uneven racial/ethnic distributions in covariate Race, which reflects the fact that the MACS cohort is predominantly White.
Figure 4.
Scatter plots of IMDMS bivariate RSF statistics for the detection of interaction-effects in both outcomes of the MACS cohort study. (A) Left: RSF results for time-to-X4-Emergence outcome. (B) Right: RSF results for time-to-AIDS-Diagnosis outcome. (A, B) For each outcome and each variable pair xj and xk, for j, k ∈ {1, …, p}, j < k, the IMDMS bivariate RSF statistic mean (Ψ̂(j, k) or IMDMS) is plotted against its noised-up counterpart (Ψ̂* (j, k) or Noise IMDMS*). RSF statistic means (diamonds) are numbered by order of decreasing significance. Blue or red color denotes a significant or non-significant measure of IMDMS at the θ = 0.10 level, respectively. Pairs of variables with significant measures of interaction have confidence intervals on both axes (dotted boxes) farther above and left of the identity line (dashed line) without crossing it (see also Table 8, Table 9).
4.6 Indirect interaction analyses of DEFB4/103A CNV with AIDS progression groups
Using indirect interaction analyses of DEFB4/103A CNV with Group, we replicated the interaction-effect result in our previously published study (Mehlotra et al., 2012) in that the difference of time-to-AIDS-Diagnosis between CNV copy number of DEFB4/103A depends on whether patients are in the Slow (log-rank p = 2.09E-4, CPH p = 3.43E-3) vs. Fast (log-rank p = 0.536, Cox p = 0.537) AIDS progression groups for the time-to-AIDS-Diagnosis outcome (Supplemental Figure S11CD). We carried out similar analyses of DEFB4/103A CNV with Group regarding the time-to-X4-Emergence. Interestingly, both RSF modeling and indirect interaction analyses failed to detect any difference of DEFB4/103A CNV (CNV = 2 vs. CNV>2) in the Slow (log-rank p = 0.227, CPH p = 0.238) vs. Fast (log-rank p = 0.374, CPH p = 0.378) AIDS progression groups for the time-to-X4-Emergence outcome (Supplemental Figure S11AB). We interpret that higher copy number of DEFB4/103A (CNV>2) has a protective effect in the slow progressors only, and that this was specific to the time-to-AIDS-Diagnosis outcome.
4.7 Summary of pairwise interaction-effects inferences and comparison to Cox-PH modeling
In contrast to RSF modeling, Cox-PH regression did not detect the same interaction-effects between genetic and environment factors at all (Figure 3 and Figure 4, Table 8 and Table 9). Several of these involves covariate Race: e.g. between Race and CCR5 −2459 SNP or DEFB4/103A CNV for the time-to-AIDS-Diagnosis outcome (Table 9), which are both questionable for the same reliability reason about covariate Race as mentioned above.
Collectively, these results prompt two conclusions. First, this confirms the power of RSF bivariate estimators to identify potentially complex and subtle pairwise interaction-effects, and possibly meaningful, over Cox-PH regression and Kaplan-Meier estimators. Second, indirect interaction analyses can help explain/interpret, in some instances, how two variables interact with each other when an interaction-effect between them has previously been detected by RSF modeling.
5 Conclusion
In this study, we developed a novel RSF modeling approach and use simple decision rules to determine statistical significance of univariate and bivariate RSF-based estimators of main and interaction-effects. Several reasons explain why RSF models are especially suited to handle interactions arising from gene networks and biological pathways. As an ensemble tree-based model, RSF is naturally able to identify non-monotonic, nonlinear, and high-order interactions (Breiman, 2001; Cordell, 2009). RSF models are also known to inherit the grouping property of trees (Ishwaran et al., 2010), a property similar to the Elastic Net (Zou & Hastie, 2005) that makes it possible to analyze data, which may be highly correlated with high interactivity. This grouping property is highly desirable because a group of correlated genes often reflects an underlying biological pathway or process. Another advantage of RSF over other survival model is that it is highly data-adaptive and fully non-parametric (Ishwaran et al., 2008). Thus, RSF is model-assumption free and, unlike traditional Cox regression, does not assume proportional hazards or other assumptions about the data. At the same time, there is always the concern of misspecification in parametric models, i.e. whether associations between variables and hazards have been modeled appropriately, and whether or not nonlinear effects or high-order interactions for variables should be included. This task is even further complicated by the masking problem in that a true interaction-effect between variables can be masked by one or several main-effect(s), whether they involve the interacting variables or not. Altogether, these properties of RSF were especially appealing for our study and resulted in successful identification of gene-covariate interactions.
Using our IMDMS bivariate RSF statistic for pairwise interaction-effects and a simple decision rule derived from the RSF model, we were able to efficiently detect significant statistical pairwise interactions in association with a time-to-event outcome of interest. Importantly, these interactions are not always accompanied by their corresponding main-effects and may be difficult to detect when using standard statistical methods by which we examine the effects of factors one or two at a time, due to a lack of sensitivity or specificity, violation of modeling assumptions, and misspecifications.
Although RSF inferences about individual variable importance/predictiveness as well as variable pairwise interactions can be made in each context using either canonical decision rule (1SE or CI(θ)), empirical evidences show differences between the two. We found the CI(θ) decision rule to be more conservative when making inferences by MDMS univariate RSF statistic, and the 1SE decision rule to be more liberal when making inferences by IMDMS bivariate RSF statistic. Evidences also showed that the disjunction decision rule (1SE ∨ CI(θ)) and conjunction decision rule (1SE ∧ CI(θ)) derived from both canonical rules were appropriate for making RSF-based inferences about significant important/predictive variables (MDMS decision rule), and significant pairs of interacting variables (IMDMS decision rule), respectively. Therefore, in practice, we recommend using these decision rules in these contexts.
Further, using a novel cross-validation scheme for generating modified prediction estimators from a RSF model, we show how our model may be of predictive clinical value, we show how RSF and the developments that we made from these models can be successfully exploited not only to uncover potential complex genes and covariates interactions and detect epistasis phenomena in association to the outcome of interest, but also useful to make predictions at an individual level on new patients (Supplemental Figures S7, S12).
In our application study, our results further our understanding of how genetic predisposition influences AIDS progression with possible underlying biological mechanisms. Specifically, we postulate a disease model whereby higher DEFB4/103A copy number is an additional genetic factor among others that is not associated with time-to-X4-Emergence, but with a delayed onset of AIDS-Diagnosis. (Supplemental Figure S13). Finally, based on our ensemble prediction approach at an individual level, we envision useful clinical applications for AIDS patients. Once new incoming HIV-seropositive patients are genotyped for these genetic variants, it is possible to accurately predict patient’s time-to-X4-Emergence and time-to-AIDS-Diagnosis, and offer prognosis on how they may fare under various precise treatment strategies in the HAART era. Eventually, this approach could be translated and potentially validated in a clinical setting for HIV/AIDS studies or other complex diseases.
Supplementary Material
Acknowledgments
This work made use of the High Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University. We are thankful to Ms. Janet Schollenberger, Senior Project Coordinator, CAMACS, as well as Dr. Jeremy J. Martinson, Sudhir Penugonda, Shehnaz K. Hussain, Jay H. Bream, and Priya Duggal, for providing us the data related to the samples analyzed in the present study. Data in this manuscript were collected by the Multicenter AIDS Cohort Study (MACS at http://www.statepi.jhsph.edu/macs/macs.html) with centers at Baltimore, Chicago, Los Angeles, Pittsburgh, and the Data Coordinating Center: The Johns Hopkins University Bloomberg School of Public Health. The MACS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional co-funding from the National Cancer Institute (NCI), the National Heart, Lung, and Blood Institute (NHLBI), and the National Institute on Deafness and Communication Disorders (NIDCD). MACS data collection is also supported by Johns Hopkins University CTSA. This study was supported by two grants from the National Institute of Health: NIDCR P01DE019759 (Aaron Weinberg, Peter Zimmerman, Richard J. Jurevic, Mark Chance) and NCI R01CA163739 (Hemant Ishwaran). The work was also partly supported by the National Science Foundation grant DMS 1148991 (Hemant Ishwaran) and the Center for AIDS Research grant P30AI036219 (Mark Chance). The contents of this publication are solely the responsibility of the authors and do not represent the official views of the granting agencies and institutions. The funders had also no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest: The authors do not have a commercial or other association that might pose a conflict of interest.
Supplemental Material: The online version of this article offers supplementary material (DOI: https://doi.org/10.1515/sagmb-2017-0038).
References
- Bien J, Taylor J, Tibshirani R. A lasso for hierarchical interactions. Ann. Stat. 2013;41:1111–1141. doi: 10.1214/13-AOS1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am. J. Hum. Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Ghosh D, Raghunathan TE, Norkin M, Sargent DJ, Bepler G. On Bayesian methods of exploring qualitative interactions for targeted treatment. Stat. Med. 2012;31:3693–3707. doi: 10.1002/sim.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99:323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman HA, George EI, McCulloch RE. Bayesian cart model search. J. Am. Stat. Assoc. 1998;93:935–948. [Google Scholar]
- Cordell HJ. Detecting gene–gene interactions that underlie human diseases. Nat. Rev. Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR. Regression models and life-tables. J. R. Stat. Soc. Ser. B. 1972;34:187–220. [Google Scholar]
- Cutler A, Zhao G. Pert-perfect random tree ensembles. Comput. Sci. Stat. 2001;33:490–497. [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B Stat. Methodol. 1977;39:1–38. [Google Scholar]
- Efron B, Tibshirani R. In: An introduction to the bootstrap. Hall Ca., editor. London: CRC Press; 1993. [Google Scholar]
- Ehrlinger J. Contributed R package: ggRandomForests for visually exploring random forests. The Comprehensive R Archive Network. 2014 DOI: https://cran.r-project.org/web/packages/ggRandomForests/index.html.
- Friedman JH. SLAC PUB-3477 STAN-LCS 005. Technical Report, Stanford University; 1984. [Accessed on October 1984]. A variable span scatterplot smoother. [Google Scholar]
- Grambsch P, Therneau T. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–526. [Google Scholar]
- Gustafson P. Bayesian regression modeling with interactions and smooth effects. J. Am. Stat. Assoc. 2000;95:795–806. [Google Scholar]
- Harrell FE. Evaluating the yield of medical tests. J. Am. Med. Assoc. 1982;247:2543–2546. [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. In: The elements of statistical learning: data mining, inference, and prediction. 2. Statistics, S. S. i, editor. New York: Springer Science; 2009. [Google Scholar]
- Ishwaran H. Variable importance in binary regression trees and forests. Electron. J. Stat. 2007;1:519–537. [Google Scholar]
- Ishwaran H, Kogalur UB. Random survival forests for R. RNews. 2007;7:25–31. [Google Scholar]
- Ishwaran H, Kogalur UB. Contributed R package randomForestSRC: random forests for survival, regression and classification (RFSRC. The Comprehensive R Archive Network. 2013 DOI: https://CRAN.R-project.org/package=randomForestSRC.
- Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann. Appl. Stat. 2008;2:841–860. [Google Scholar]
- Ishwaran H, Kogalur UB, Gorodeski EZ, Minn AJ, Lauer MS. High-dimensional variable selection for survival data. J. Am. Stat. Assoc. 2010;105:205–217. [Google Scholar]
- Ishwaran H, Gerds TA, Kogalur UB, Moore RD, Gange SJ, Lau BM. Random survival forests for competing risks. Biostatistics. 2014;15:757–773. doi: 10.1093/biostatistics/kxu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- LeBlanc M, Crowley J. Survival trees by goodness of split. J. Am. Stat. Assoc. 1993;88:457–467. [Google Scholar]
- Li J, Horstman B, Chen Y. Detecting epistatic effects in association studies at a genomic level based on an ensemble approach. Bioinformatics. 2011;27:i222–i229. doi: 10.1093/bioinformatics/btr227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Jeon Y. Random forests and adaptive nearest neighbors. J. Am. Stat. Assoc. 2006;101:578–590. [Google Scholar]
- Lunetta KL, Hayward LB, Segal J, Van Eerdewegh P. Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- McGill R, Tukey JW, Larsen WA. Variations of box plots. Am. Stat. 1978;32:12–16. [Google Scholar]
- Mehlotra RK, Dazard J-E, John B, Zimmerman PA, Weinberg A, Jurevic RJ. Copy number variation within human β-Defensin gene cluster influences progression to AIDS in the multicenter AIDS cohort study. AIDS Clin. ResJ. AIDS Clin. Res. 2012;3:10. doi: 10.4172/2155-6113.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen UB, Ishwaran H, Gerds TA. Evaluating random forests for survival analysis using prediction error curves. J. Stat. Softw. 2012;50:1–23. doi: 10.18637/jss.v050.i11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal MR. Regression trees for censored data. Biometrics. 1988;44:35–47. [Google Scholar]
- Shepherd JC, Jacobson LP, Qiao W, Jamieson BD, Phair JP, Piazza P, Quinn TC, Margolick JB. Emergence and persistence of CXCR4-Tropic Hiv-1 in a population of men from the multicenter AIDS cohort study. J. Infect. Dis. 2008;198:1104–1112. doi: 10.1086/591623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon N, Tibshirani R. A permutation approach to testing interactions for binary response by comparing correlations between classes. J. Am. Stat. Assoc. 2015;110:1707–1716. [Google Scholar]
- Tian L, Alizadeh AA, Gentles AJ, Tibshirani R. A simple method for estimating interactions between a treatment and a large number of covariates. J. Am. Stat. Assoc. 2014;109:1517–1532. doi: 10.1080/01621459.2014.951443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki M, Cordell HJ. Improved statistics for genome-wide interaction analysis. PLoS Genet. 2012;8:e1002625. doi: 10.1371/journal.pgen.1002625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elston RC, Zhu X. The meaning of interaction. Hum. Hered. 2010;70:269–277. doi: 10.1159/000321967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung LS, Yang C, Wan X, Yu W. GBOOST: a GPU-based tool for detecting gene–gene interactions in genome-wide case control studies. Bioinformatics. 2011;27:1309–1310. doi: 10.1093/bioinformatics/btr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang S, Wong MY, Wareham NJ, Sha Q. An ensemble learning approach jointly modeling main and interaction effects in genetic association studies. Genet. Epidemiol. 2008;32:285–300. doi: 10.1002/gepi.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Pan F, Xie Y, Zou F, Wang W. COE: a general approach for efficient genome-wide two-locus epistasis test in disease association study. J. Comput. Biol. 2010a;17:401–415. doi: 10.1089/cmb.2009.0155. [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang S, Zou F, Wang W. Team: efficient two-locus epistasis tests in human genome-wide association study. Bioinformatics. 2010b;26:i217–i227. doi: 10.1093/bioinformatics/btq186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Huang S, Zou F, Wang W. Tools for efficient epistasis detection in genome-wide association study. Source Code Biol. Med. 2011;6:1. doi: 10.1186/1751-0473-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. 2005;67:301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.