Abstract
Background
Rheumatoid arthritis (RA) is a common worldwide public health problem, which causes a chronic, systemic inflammatory disorder of synovial joints. Paeoniflorin (PA) has achieved positive results to some extent for the treatment of RA.
Purpose
This study aimed to reveal the potential druggable targets of PA in an experimental RA model using quantitative proteomics analysis.
Study design and methods
Thirty Sprague-Dawley rats were randomly divided into a normal group, model group and PA group. PA (1 mg/kg) was used to treat collagen-induced arthritis (CIA) rats for 42 days. We used isobaric tags for relative and absolute quantitation-based quantitative proteomics to analyze the synovial tissue of rats. Ingenuity pathway analysis (IPA) software was applied to process the data. The proteins that were targeted via IPA software were verified by Western blots.
Results
We found that PA caused 86 differentially expressed proteins (≥1.2-fold or ≤0.84-fold) compared with the CIA group. Of these varied proteins, 20 significantly changed (p<0.05) proteins referred to 41 CIA-relative top pathways after IPA pathway analysis. Thirteen of the PA-regulated pathways were anchored, which intervened in 24 biological functions. Next, network analysis revealed that leukemia inhibitory factor receptor (LIFR) and asporin (ASPN), which participate in two significant networks, contributed the most to the efficacy of PA treatment. Additionally, Western blots confirmed the aforementioned druggable targets of PA for the treatment of RA.
Conclusion
The results reveal that PA may treat RA by decreasing two key proteins, LIFR and ASPN. Our research helps to identify potential agents for RA treatment.
Keywords: paeoniflorin, quantitative proteomic, ASPN, LIFR, rheumatoid arthritis
Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune disorder. It has a high prevalence with a rate of 0.24% worldwide.1 Unfortunately, current therapeutic approaches only slow the progression of RA rather than prevent it.2 To the best of our knowledge, long-term inflammation and autoimmune reaction are the primary characterizations of RA.3 Thus, therapeutic medications, such as nonsteroidal anti-inflammatory drugs and disease-modifying anti-rheumatic drugs, mostly retard the progress of this disease through anti-inflammatory and anti-immune strategies.2 However, these drugs only impact one target and are limited by a number of well-characterized clinical side-effects.4 Doctors and scientists eagerly expect new anti-RA drugs with superior efficacy and less toxicity.
With the development and exploitation of traditional Chinese medicines (TCM), some TCM with anti-RA effects have been approved gradually for use in clinical settings.5 These natural products, composed of various compounds, can have convincing curative effects6 via multiple pathways.7 Radix Paeoniae Alba, a Chinese herb, is commonly used for many diseases, especially for the treatment of RA.8 As the key compound of Radix Paeoniae Alba,9 paeoniflorin (PA) plays an important role in treating RA.10 A lot of research indicates that PA has anti-inflammatory activity, anti-spasmolytic activity, anti-oxidative activity, immune-regulatory effects, anti-apoptotic activity and neuroprotective effects.11–13 In terms of anti-inflammatory activity, our previous studies showed that PA decreased the levels of interleukin (IL)-1 and tumor necrosis factor-alpha (TNF-α) and reduced soft tissue swelling in the joints of arthritic rats.14,15 However, the multi-target pharmacological mechanism of PA therapy in RA remains unclear. Therefore, this study explores the comprehensive mechanisms of PA responsible for RA treatment.
Quantitative proteomics, one of the major components of systems biology, is used to uncover TCM mechanisms,16 discover druggable targets and find biomarkers.17 Some proteome techniques provide a valuable framework for a better understanding of systematic mapping of protein interactions.18 Isobaric tags for relative and absolute quantification (iTRAQ) is a powerful methodology for quantitative proteomics with many advantages, such as high throughput, high sensitivity and superior accuracy.19 iTRAQ is a robust method for comparative proteomics.20 Therefore, many studies use iTRAQ to profile the specific protein target(s) of a drug21,22 to gain molecular insights on the mechanism of drug action. In this study, this technique was combined with ingenuity pathway analysis (IPA) to reveal a deeper understanding of the mechanism of PA treatment in collagen-induced arthritis (CIA) rats. Thus, we believe iTRAQ-based quantitative proteomics deserves to be used in the design of our study.
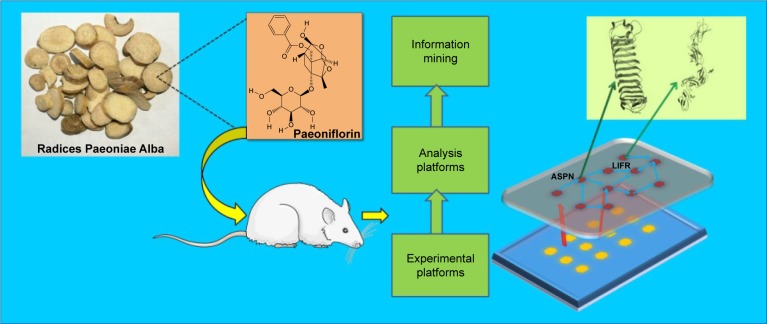
Taken together, the present study aimed to explore the druggable targets of PA in CIA rats by iTRAQ-based quantitative proteomics analysis combined with bioinformatics methods. This research will contribute to theoretical evidence for the treatment of CIA rats with PA (Figure 1).
Figure 1.
General mechanism of the collagen-induced arthritis (CIA) rat model treated by paeoniflorin through downregulation of asporin (ASPN) and leukemia inhibitory factor receptor (LIFR).
Material and methods
Animals
An equal number of male and female Sprague-Dawley rats (n=30, 6–8 weeks old, 180–220 g), specific pathogen free, were used in this study. Rats were purchased from the animal experimental center of Hunan People’s Hospital (Hunan, People’s Republic of China). All rats were housed five per cage and received food and water ad libitum under controlled environmental conditions (room temperature 22°C±3°C, room humidity 40%–60%, background noise 4,010 dB, 12: 12 h light–dark cycles) for 1 week to adapt to the environment. We divided the 30 rats into the control group, the CIA group and the PA group randomly.
Ethics
This study was verified and approved by the Animal Ethics Committee of Hunan People’s Hospital and performed in accordance with the Hunan People’s Hospital guidelines for the care and use of laboratory animals.
Preparation of paeoniflorin
PA was purchased from The National Institute for the Control of Pharmaceutical and Biological Products (Beijing, People’s Republic of China). The purity of PA is 98% (Figure S1). It was sealed and stored at 4°C for later use.
Establishment of CIA rats
CIA was initiated according to the protocol for successful induction in rats. Bovine type II collagen (BIIC, Chondrex, Inc, Washington DC, USA) was mixed with acetic acid to form a 2 mg/mL BIIC solution. Next, 5 mL of this solution was mixed with 5 mL complete Freund’s adjuvant (CFA, Sigma-Aldrich Co., St Louis, MO, USA), to produce a 1 mg/mL BIIC solution. On day 0, the rat tail and rear foot were injected intradermally with 200 μg of the collagen/CFA emulsion for the primary immunization. On day 7, the rats were given 100 μg of collagen/incomplete Freund’s adjuvant (1 mg/mL, Sigma-Aldrich) emulsion in the same manner for the secondary immunization.
After disease onset, we recorded the clinical assessment and paw measurements of the rats. The severity of arthritis in the hind foot was scored on a scale of 0 to 3, where 0= no inflammation, 1= mild swelling of joints following redness, 2= moderate swelling and 3= severe joint swelling and the inability to be loaded. A disease score of the hind limbs was calculated for each animal (maximum score 6 per rat). The thickness of each hind paw was measured after immunization with a compass and millimeter ruler in the fixed position; body weight was monitored throughout the study. The data of the control group were measured before immunization.
For detection of serum inflammatory cytokines, blood was drawn on day 42 post-immunization (n=5), set at room temperature for 1 hour, and centrifuged at 2,000 rpm/min for 10 minutes. The serum was used in an enzyme linked immunosorbent assay (ELISA) kit (Wuhan Huamei Biotech Co., LTD) to detect the levels of IL-1β and TNF-α.23
Sample preparation
Under general anesthesia, synovial tissue was separated from the hind paws after skin, muscle, fatty tissue, bone and tendon removal in CIA, normal and PA rats on day 42 (n=10). Protein lysates were obtained using lysis buffer (8 M urea, 4% chaps, 30 mM HEPES, 1 mM phenylmethanesulfonyl fluoride (PMSF), 2 mM ethylene diamine tetraacetic acid (EDTA) and 10 mM DL-Dithiothreitol (DTT) followed by centrifugation at 20,000 rpm for 25 minutes at 4°C for 60 minutes after exposure to ultrasound (water-bath, 5 minutes). The supernatant was collected. DTT was added to a final concentration of 10 mM. We incubated the resulting solution in a water-bath at 56°C for 1 hour and added iodoacetamide quickly to a final concentration of 55 mM. Then, the solution was left in a dark room (temperature 20°C–25°C) for 1 hour. The protein samples were precipitated (−20°C, 3 hours) by the addition of precooled acetone, then centrifuged (20,000 g, 20 minutes, 4°C) and solubilized in 0.5 M triethylammonium bicarbonate plus 0.1% sodium dodecyl sulfate. The protein concentration was confirmed with a Bradford protein assay kit (Amesco).
iTRAQ method
According to the manufacturer’s protocol (Applied Biosystems), trypsin digestion and iTRAQ labeling were performed. In short, 100 μg of protein from each pooled sample was reduced, alkylated and then digested overnight at 37°C with trypsin. Ultraflex TOF/TOF (Bruker, Leipzig, Germany) was used to detect the digestion efficiency from 1 μL sample of the tryptic peptides. iTRAQ labeled the tryptic peptide solution of each sample according to the iTRAQ Reagent Multiplex Kit protocol (Applied Biosystems). The normal (N) group, CIA group and PA group-derived tryptic peptides were labeled with 113, 114 and 115, respectively. The labeling efficiency was calculated. We compared the total number of possible labeling sites (the N-termini of all peptides and lysine side chains) for iTRAQ tags with the labeled sites in the detected peptides. The iTRAQ peptide labeling efficiency was more than 97% in this study.
A strong cation exchange (SCX) column (Luna SCX 100 A, Phenomenex) fractionated the mixed peptides. The mixed iTRAQ-labeled samples were diluted with 10× buffer A (25% acetone [ACN], 10 mM KH2PO4, pH 3.0), which was identical to buffer B except it did not include 2 M KCl. A linear binary gradient of 0%–100% buffer B in buffer A at a flow rate of 1 mL/min was used to perform the SCX fractionation. Based on a reverse-phase liquid chromatography column (Strata-X C18 column, 5 mm, 300 A, 100×75 mm, Phenomenex), 10 SCX fractions were collected along the gradient, dried and dissolved in buffer C (5% ACN, 0.1% FA). The high-performance liquid chromatography gradient was increased from 5% to 30% in 65 minutes at a flow rate of 400 nL/min. Mass spectrometric analysis of the iTRAQ-labeled samples was performed by Q Exactive liquid chromatography-tandem mass spectrometry (LC-MS/MS; Thermo Scientific Co.). Sequences for the peptide and reporter ions were generated to identify the protein from which the peptide originated. To diminish the effect of experimental variation, three independent MS/MS runs were performed for each sample.24
Data processing and analysis
The original MS/MS data were analyzed with the Proteome Discoverer Software (Thermo Scientific version 1.3). Mascot (version 2.3.0, Matrix Science, London, UK) and the UniProt-rat database (http://www.uniprot.org/) were used to identify proteins. The Mascot search parameters used included trypsin, peptides digested with a maximum of one missed cleavage, fixed modification (carbamidomethylation of cysteine residue), variable modifications (oxidation of methionine Gln-Pyro-Glu of N-term Q, and iTRAQ 8 plex modification of N terminal, K and Y), peptide tolerance 15 ppm and the iTRAQ fragment tolerance (0.2 Da). Pro-teinPilot Software was used to assess the false discovery rate (FDR). In this study, the resulting 1% global FDR from the fit, which corresponds to 99% correct protein identification, was used as an initial qualification criterion. The spectra were identified with 95% confidence. The quantitative protein ratio was calculated as the median of all peptide ratios, which were the ratios of the signal intensity value of the reference sample (normal sample) label to the signal strength values of the other labels. The final quantitative result was normalized to the median ratio of each label. Protein quantitative ratio was calculated as the median of all peptide ratios. The fold change of the differentially expressed proteins (DEPs) was calculated as the average value from the protein iTRAQ ratios. The DEPs were screened with the following criteria: ≥2 peptide had to be identified, proteins with an average ratio-fold change ≥1.2 or ≤0.84 between the groups. independent t-tests with p<0.05 were determined to be differentially regulated between the groups.
IPA analyses of DEPs
IPA was used to analyze the biological functions, pathways and networks of the identified DEPs. DEPs were classified based on annotations from the UniProt Knowledgebase. The data packet containing the protein IDs was uploaded into IPA. IPA analysis maps each identifier to the corresponding molecule in the Ingenuity Pathway Knowledge Base. It is available at the Ingenuity System’s web site (http://www.ingenuity. com). IPA analysis identified “immune disease” as the main disease associated with the DEPs by biological function.
Western blot validation
Western blots were performed to validate the iTRAQ-based results in N42, CIA42 and PA42 (10 samples each) mice. Protein samples (30–50 μg) were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride membranes (Bio-Rad). Following blocking with 5% skim milk, the transferred membranes were incubated overnight at 4°C with primary anti-isoform 2 of plasminogen activator inhibitor 1 RNA-binding protein (SERBP1) antibody (1:1,000; Abcam), anti-asporin (ASPN) antibody (1:200; Santa Cruz) and anti-leukemia inhibitory factor receptor (LIFR) antibody (1:500; Santa Cruz). The membranes were subsequently incubated for 1 hour at room temperature with secondary antibodies (1:3,000; Proteintech). Bands were visualized with an electrochemiluminescence detection reagent (Thermo Scientific Pierce) and quantified by densitometry using the Image-Quant image analysis system (Storm Optical Scanner, Molecular Dynamics). β-actin was detected simultaneously as a loading control. All Western blot analyses were performed in triplicate.
Statistical analyses
SPSS software (Version 18; IBM, New York, NY, USA) software and GraphPad Prism v7.0 software were used for the statistical analyses. The data are expressed as means ± SD and median and 25/75%. Group comparisons were performed by one-way analysis of variance tests and independent t-tests. A value of p<0.05 was considered statistically significant.
Results
The measurement of the degree of arthritis
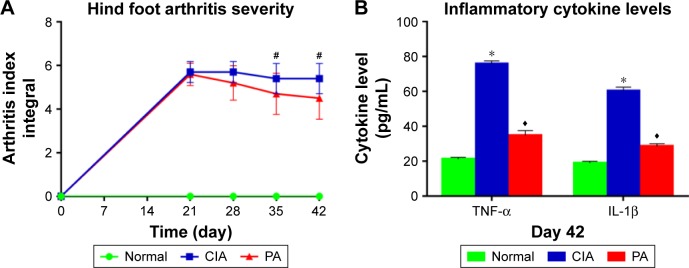
We measured the clinical index of rats, and found a significant difference between pre-immunization and post- immunization. There is no difference between the CIA group and PA group at 21 or 28 days. However, a significant change in the arthritis index was observed on days 35 and 42 (p<0.01) (Figure 2A; Table S1). The levels of TNF-α and IL-1β are shown in Figure 2B (Table S2). The expression of these two factors was significantly reduced (p<0.01) in the rats treated with PA at 42 days.
Figure 2.
The severity index in the collagen-induced arthritis (CIA) rats pre- and post-treatment. Clinical arthritis scores (A) and serum inflammatory cytokine levels (tumor necrosis factor-alpha [TNF-α], interleukin [IL-1β]) (B) were compared among the normal group, the CIA group and the paeoniflorin (PA) group. Hind foot arthritis parameters were measured following 0, 21, 28, 35 and 42 days after immunization (n=10), serum inflammatory cytokines were measured following 42 days after immunization (n=5). #p<0.01 CIA versus PA; *p<0.01 CIA versus normal; ♦p<0.01 CIA versus PA.
The DEPs are significantly changed by PA
Proteins with a ≥1.20-fold or ≤0.84-fold difference were regarded as DEPs. In all, 273 proteins were identified as DEPs between the CIA and normal control groups, and 86 proteins between the PA and CIA groups on day 42. Of these proteins, we identified proteins with a >1.50-fold or <0.80-fold difference. Twenty DEPs were significantly changed by PA (Table 1), 11 with increased expression and 9 with decreased expression. Among these DEPs, LIFR and ASPN were remarkably downregulated.
Table 1.
Differentially expressed proteins identified by iTRAQ-based quantitative proteomics
| Accession ID | Description | Score | CIA42/N42 | PA42/CIA42 |
|---|---|---|---|---|
| Q9EPH1 | Alpha-1B-glycoprotein | 292.24 | 4.132231 | 0.525 |
| G3V7K2 | Leukemia inhibitory factor receptor | 55.22 | 2.347418 | 0.671 |
| Q5XIH1 | Asporin | 781.11 | 1.915709 | 0.68 |
| Q01129 | Decorin | 2,352.43 | 1.73913 | 0.773 |
| D3ZVB7 | Osteoglycin (predicted) | 3,586.56 | 1.689189 | 0.774 |
| P31211 | Corticosteroid-binding globulin | 191.98 | 1.605136 | 0.752 |
| Q03626-2 | Isoform 2 of murinoglobulin-1 | 3,004.97 | 1.605136 | 0.679 |
| F1LV08 | Uncharacterized protein (fragment) | 44.45 | 1.55521 | 0.675 |
| P20762 | Ig gamma-2C chain C region | 318.86 | 1.510574 | 0.668 |
| Q63041 | Alpha-1-macroglobulin | 3,857.55 | 1.510574 | 0.755 |
| P02454 | Collagen alpha-1 (I) chain | 5,647.92 | 1.503759 | 0.788 |
| P08494 | Matrix Gla protein | 0 | 0.794281 | 2.504 |
| O70210 | Chondroadherin | 52.12 | 0.76864 | 2.463 |
| C0SSW8 | Eukaryotic translation initiation factor 2 subunit 3 | 57.04 | 0.76746 | 1.7 |
| F1LQI4 | Aggrecan core protein | 108.63 | 0.712251 | 1.501 |
| G3V6E7 | Fibromodulin | 815.54 | 0.665779 | 1.71 |
| P70490 | Lactadherin | 31.47 | 0.610501 | 2.278 |
| P16975 | SPARC | 166.23 | 0.57971 | 1.505 |
| F1LQ93 | Collagen alpha-1 (IX) chain | 0 | 0.493583 | 1.69 |
| F1LRM7 | Collagen alpha-1 (II) chain | 444.4 | 0.444247 | 3.623 |
Abbreviations: iTRAQ, isobaric tags for relative and absolute quantification; N, normal; CIA, collagen-induced arthritis; PA, paeoniflorin.
Bio-function analysis of the DEPs for CIA rats affected by PA
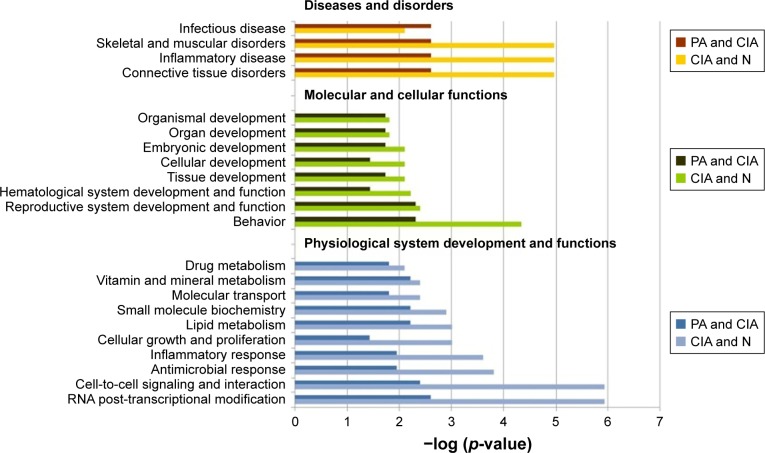
In further study, we imported the DEPs of the CIA group into the IPA software, and 56 biological functions were affected. Among them, PA significantly affected 24 biological functions. The IPA software identified biological functions from three primary categories of functions: diseases and disorders, molecular and cellular functions, and physiological system development and function.
In the diseases and disorders category, connective tissue disorders and the most relevant category of RA, were significantly improved by PA. Other categories, including inflammatory disease and skeletal and muscular disorders, were also remarkably affected after treatment with PA. For the physiological system development and function analysis, behavior, hematological system development and function, and cellular development were the top three categories after treatment with PA. For the molecular and cellular functions analysis, the DEPs for RNA post-transcriptional modification, cell-to-cell signaling and interaction, antimicrobial response, and inflammatory response were remarkably reduced after PA treatment (Figure 3).
Figure 3.
The top bio-functions of collagen-induced arthritis (CIA) day 42 versus normal (N) control day 42 versus paeoniflorin (PA) day 42 by ingenuity pathway analysis (IPA). Orange: the top function from the diseases and disorders category; green: the top function from physiological system development and functions category; blue: the top function from the molecular and cellular functions category.
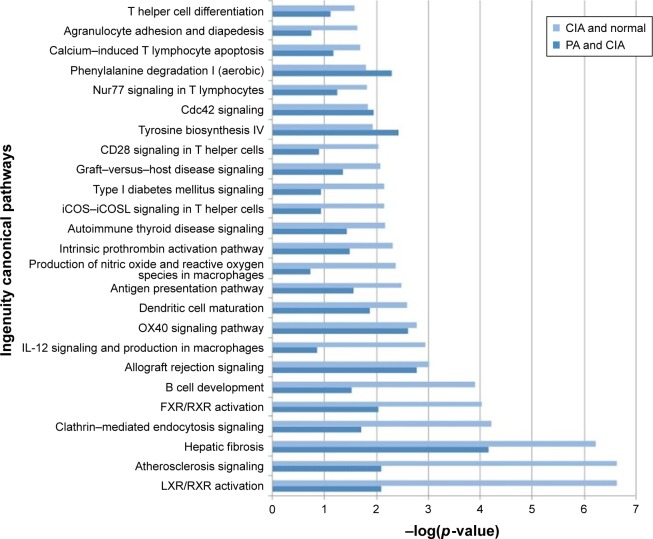
Signal pathway analysis of CIA rats treated by PA
Based on the 273 DEPs, the IPA software analysis identified 107 canonical pathways. Of these pathways, PA affected 44 signaling pathways. Forty-one canonical pathways were significantly perturbed with p<0.05, which corresponds to – log (p-value) >0.8 identified pathways (Table S3). Our results showed that liver X receptor/retinoid X receptor (LXR/RXR) activation was the pathway that was most significantly downregulated by PA (Figure 4).
Figure 4.
The pathways of the significant differentially expressed proteins (DEPs) of the collagen-induced arthritis (CIA) group and the paeoniflorin (PA) group. Top 25 canonical pathways using ingenuity pathway analysis in CIA on day 42, −log (p-value) >0.8. Twenty-one pathways were involved with PA treatment.
Identification of significant networks through IPA interaction analysis
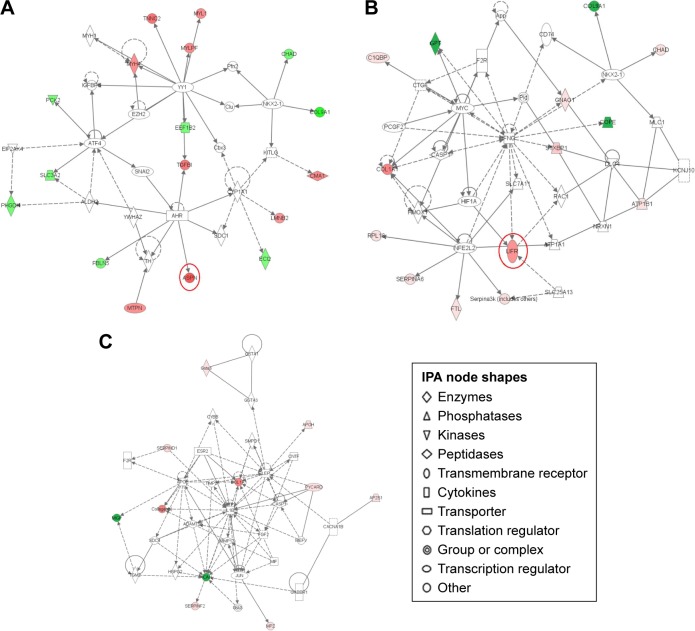
IPA interaction analysis found six networks of CIA rats after PA treatment. DEPs were mostly enriched in the three significant networks. The 35 proteins that were involved in connective tissue disorders, inflammatory disease, cystic fibrosis, acquired partial lipodystrophy and phosphoglycerate were grouped as network 1. Of these proteins, 17 proteins, including ASPN, were influenced by PA (IPA score 28, Figure 5A). The 35 proteins that were involved in kidney failure, developmental disorder, renal and urological disease, and cell death and survival were grouped as network 2. Fourteen proteins were influenced by PA (IPA score 25); LIFR was significantly downregulated (Figure 5B). In network 3, PA changed the expression level of 11 proteins, which were enriched and used in cardiovascular system development and function, organismal development and cellular development (IPA score 16, Figure 5B) (Table 2).
Figure 5.
Pathway networks were identified using ingenuity pathway analysis (IPA). Network #1 (A), network #2 (B), and network #3 (C) are shown. The node (protein) is described in the right of the figures. Colored nodes refer to proteins found in our dataset (green = upregulated, red = downregulated). Uncolored nodes were not identified as differentially expressed in our experiment and were integrated into the computationally generated IPA networks to indicate relevance to this network. Red circles indicate a significant effect by paeoniflorin (PA).
Table 2.
Networks generated using ingenuity pathways analysis
| Number | Molecules in network | Score | Focus molecules | Top diseases and functions |
|---|---|---|---|---|
| 1 | AHR, ALDH2, ASPN, ATF4, Cbx3, CHAD, Clu, CMA1, COL9A1, CYP1A1, ECI2, EEF1B2, EIF2AK4, EZH2, FBLN5, IGFBP5, KITLG, LMNB2, MTPN, MYH1, MYH4, MYL1, MYLPF, NKX2-1, PCK2, Pfn2, PHGDH, SDC1, SLC3A2, SNAI2, TGFBI, TH, TNNC2, YWHAZ, YY1 | 28 | 17 | Connective tissue disorders; inflammatory disease; cystic fibrosis; acquired partial lipodystrophy; phosphoglycerate |
| 2 | App, ATP1A1, ATP1B1, C1QBP, CASP1, CD74, CHAD, COL1A1, COL9A1, COPE, CTGF, DLG4, F2R, FTL, GNAO1, GPT, HIF1A, HMOX1, IFNG, KCNJ10, LIFR, MLC1, MYC, NFE2L2, NKX2-1, NRXN1, PCGF2, Pld, RAC1, RPL18, SERPINA6, Serpina3k (includes others), SLC25A13, SLC7A11, STXBP1 | 25 | 14 | Kidney failure; developmental disorder; renal and urological disease; cell death and survival |
| 3 | ACAN, ADAMTS5, AP2S1, APOE, APOH, CACNA1B, CASP1, CNTF, COL1A1, Collagen(s), CYBB, ESR2, F2R, FGF2, GABBR1, Gsk3, GSTA1, GSTA3, Gsta3, HSPG2, IL1B, JUN, LEP, MEFV, MGP, MIF, MMP13, MPZ, PYCARD, SDC4, SERPIND1, SERPINF2, SMPD1, TGM2, TIMP3 | 16 | 11 | Cardiovascular system development and function; organismal development; cellular development |
Verification of ASPN and LIFR
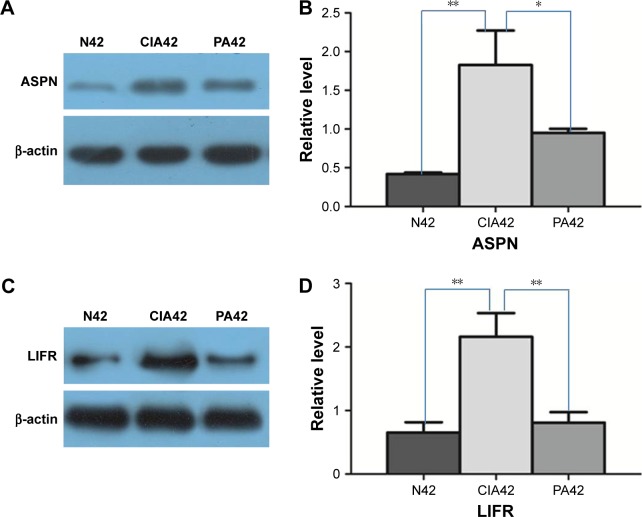
Our results provide a set of candidates for validation as biomarkers for the treatment of RA.25 We validated two detected DEPs (ASPN and LIFR) by Western blots. These two proteins were progressively upregulated at day 42 in the CIA rats versus the NC rats. On day 42 in the PA group, they were progressively downregulated (Figure 6).
Figure 6.
Result of Western blot verification: A and B show the expressional changes of asporin (ASPN) on day 42 of collagen-induced arthritis (CIA), normal (N) control and paeoniflorin (PA); C and D show the expressional changes of leukemia inhibitory factor receptor (LIFR) on day 42 of CIA, the normal control and PA.
Notes: *p<0.05; **p<0.01.
Discussion
The present study demonstrated that ASPN and LIFR, which were involved in two significant networks, contributed mostly to the efficacy of PA to treat CIA rats. These two proteins, related to inflammation, immunoreactions and metabolism, were significantly downregulated by PA. Further, Western blots confirmed the aforementioned druggable targets of PA treatment. This is the first study to elucidate the molecular mechanism for PA treatment of CIA rats.
The results of the bio-functional analysis indicated that more than one-third of RA-related functions are affected by PA, including connective tissue disorders, inflammatory disease, skeletal and muscular disorders and lipid metabolism. Furthermore, these functions are mostly connected with inflammation, immunoreactions and metabolism. During PA challenge with RA, proteins, including LIFR and ASPN, enriched in these functions were significantly downregulated in synovial tissue. With the help of pathway analysis, we uncovered 41 significantly enriched canonical pathways after PA treatment. Interestingly, the majority of the pathways contribute to inflammation, metabolism and immune response. Based on previous research, LXR/RXR activation is a key pathway related to inflammation and metabolism.25
LXR/RXR activation primarily increases the expression of genes involved in cholesterol and lipid metabolism26 and the inflammatory response.27 This pathway induces the expression of sterol regulatory element-binding SREBF1 and SREBF2,28 leading to a decrease in fatty acid synthesis. Our previous study demonstrated that LXR/RXR activation is a considerable pathogenesis mechanism in CIA rats.25 Some studies indicate that activation of the LXR/RXR heterodimer through either LXR or RXR prevents pathogen-induced macrophage apoptosis, a microbial strategy for eluding innate immune responses. The LXR/RXR heterodimer promotes macrophage survival by enhancing the expression of anti-apoptotic regulators (eg, AIM/CT2, Bcl-XL, Birc1a and SPa) and inhibiting the expression of proapoptotic regulators (eg, caspases 1, 4, 11, 7, and 12 and Fas ligand) and thereby might have utility in the therapy of systemic lupus erythematosus or allergic asthma.29,30 Thus, the inhibition of this pathway in the synovial tissue may reduce the level of lipid synthesis and/or inflammation. This study clearly indicates that PA downregulated the expression of proteins in LXR/RXR activation. Therefore, PA may have a role in anti-inflammation or lipogenic mechanisms in synovial tissue.
The aforementioned results showed that PA could improve the RA-related pathways implicated in inflammation, metabolism and immune response. Similarly, the IPA interaction analysis indicated that PA substantially down-regulated ASPN and LIFR, which were involved in two essential networks after PA treatment. Evidence indicates that these two proteins mostly contribute to inflammation, metabolism and immune response.31,32 ASPN, a class I small leucine rich proteoglycan (SLRP), is expressed in articular cartilage.31 Many SLRPs influence cellular functions through binding and modulating members of the transforming growth factor β (TGF-β) superfamily.33 Activated TGF-β (aTGF-β) interacts with and signals through TGF-β type I and type II serine/threonine kinase receptors (TGF-β RI and RII).33 ASPN directly links with TGF-β type I, which subsequently leads to a loss of aTGF-β and subsequently influences cellular functions.33 High levels of ASPN in the synovial tissue reduces the expression of TGF-β1 and promotes the dysregu-lation of collagen fibril metabolism of the joint.34 Therefore, the expression of ASPN is positively correlated with the severity of RA. LIFR is a subunit of a receptor for LIF. LIF, produced by muscle cells, is a soluble IL-6 family cytokine that regulates a number of physiologic functions.35 In addition, LIF affects skeletal muscle regeneration by stimulating the production of pro-inflammatory factors in the joint.36 Therefore, overexpression of LIFR could activate the ligand and common signaling mediators to aggravate inflammation of the chondral cells in synovial tissue.37 Interestingly, the present study highlights PA inhibition of ASPN and LIFR in CIA rats. Furthermore, Western blots confirmed the downregulation of LIFR and ASPN after intervention with PA. Taken together, this study provides the evidence that PA could alleviate pathological changes in the synovial tissue due to RA through the inhibition of the proteins involved in inflammation, metabolism and immune response.
Conclusion
Our results confirm the feasibility of using iTRAQ-based quantitative proteomics to identify DEPs. First, 20 identified proteins showed progressive changes with PA treatment. Next, in the 107 biological functions associated with CIA rats, 44 were found to be affected by PA. In addition, PA significantly downregulated the expression of proteins in LXR/RXR activation. Moreover, the network analysis indicated that PA may be an efficacious herb to treat RA. Of these candidate biomarkers of CIA rats in networks, PA anticipated the expression mechanism of ASPN and LIFR. Our study is the first to confirm the druggable targets of CIA rats with PA treatment through iTRAQ-based quantitative proteomics. These findings may serve as potential therapeutic agents against RA.
Acknowledgments
This work was funded by grants from the National Natural Science Foundation of China (81673719, 81303074, and 81102564), the Doctoral Fund of the State Ministry of Education (20110162120004), and the Provincial Science and Technology Project (2014sk3035). We especially thank Rong Fan, Wei Huang, and Chunhu Zhang for contributing to this research.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cross M, Smith E, Hoy D, et al. The global burden of rheumatoid arthritis: estimates from the global burden of disease 2010 study. Ann Rheum Dis. 2014;73(7):1316–1322. doi: 10.1136/annrheumdis-2013-204627. [DOI] [PubMed] [Google Scholar]
- 2.Smolen JS, Landewe R, Breedveld FC, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2013 update. Ann Rheum Dis. 2014;73(3):492–509. doi: 10.1136/annrheumdis-2013-204573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423(6937):356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 4.Yue R, Zhao L, Hu Y, et al. Rapid-resolution liquid chromatography TOF-MS for urine metabolomic analysis of collagen-induced arthritis in rats and its applications. J Ethnopharmacol. 2013;145(2):465–475. doi: 10.1016/j.jep.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Gu Y, Lu C, Zha Q, et al. Plasma metabonomics study of rheumatoid arthritis and its Chinese medicine subtypes by using liquid chromatography and gas chromatography coupled with mass spectrometry. Mol Biosyst. 2012;8(5):1535–1543. doi: 10.1039/c2mb25022e. [DOI] [PubMed] [Google Scholar]
- 6.Lu SW, Wang QS, Li GY, Sun S, Guo YY, Kuang HX. The treatment of rheumatoid arthritis using Chinese medicinal plants: from pharmacology to potential molecular mechanisms. J Ethnopharmacol. 2015;176:177–206. doi: 10.1016/j.jep.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Huang X, Zhong MZ, et al. Pretreatment of Shaoyao Gancao Decoction () alters pharmacokinetics of intravenous paclitaxel in rats. Chin J Integr Med. 2017;23(1):70–75. doi: 10.1007/s11655-016-2619-6. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Chen CX, Shen YH. Effects of total glucosides from paeony (Paeonia lactiflora Pall) roots on experimental atherosclerosis in rats. J Ethnopharmacol. 2011;135(2):469–475. doi: 10.1016/j.jep.2011.03.045. [DOI] [PubMed] [Google Scholar]
- 9.Jiang D, Chen Y, Hou X, Xu J, Mu X, Chen W. Influence of Paeonia lactiflora roots extract on cAMP-phosphodiesterase activity and related anti-inflammatory action. J Ethnopharmacol. 2011;137(1):914–920. doi: 10.1016/j.jep.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 10.He DY, Dai SM. Anti-inflammatory and immunomodulatory effects of Paeonia lactiflora Pall., a traditional Chinese herbal medicine. Front Pharmacol. 2011;2:10. doi: 10.3389/fphar.2011.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu P, Zhu L, Liu Y, Zhang L, Liu J, Shen H. Protective effects of paeoniflorin on TNBS-induced ulcerative colitis through inhibiting NF-kappaB pathway and apoptosis in mice. Int Immunopharmacol. 2017;50:152–160. doi: 10.1016/j.intimp.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Dou W, Zhang E, et al. Paeoniflorin abrogates DSS-induced colitis via a TLR4-dependent pathway. Am J Physiol Gastrointest Liver Physiol. 2014;306(1):G27–G36. doi: 10.1152/ajpgi.00465.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi EM, Suh KS, Rhee SY, Kim YS. Inhibitory effect of paeoniflorin on methylglyoxal-mediated oxidative stress in osteoblastic MC3T3-E1 cells. Phytomedicine. 2014;21(10):1170–1177. doi: 10.1016/j.phymed.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Dai SM. Mechanisms involved in the therapeutic effects of Paeonia lactiflora Pallas in rheumatoid arthritis. Int Immunopharmacol. 2012;14(1):27–31. doi: 10.1016/j.intimp.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Wu D, Chen J, Zhu H, et al. UPLC-PDA determination of paeoniflorin in rat plasma following the oral administration of Radix Paeoniae Alba and its effects on rats with collagen-induced arthritis. Exp Ther Med. 2014;7(1):209–217. doi: 10.3892/etm.2013.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suo T, Wang H, Li Z. Application of proteomics in research on traditional Chinese medicine. Expert Rev Proteomics. 2016;13(9):873–881. doi: 10.1080/14789450.2016.1220837. [DOI] [PubMed] [Google Scholar]
- 17.Ji Q, Zhu FS, Liu X, Li Q, Su SB. Recent advance in applications of proteomics technologies on traditional Chinese medicine research. Evid Based Complement Alternat Med. 2015;2015:983139. doi: 10.1155/2015/983139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stelzl U, Worm U, Lalowski M, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122(6):957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Xia F, Yao X, Tang W, Xiao C, Yang M, Zhou B. Isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis of Hugan Qingzhi and its protective properties against free fatty acid-induced L02 hepatocyte injury. Front Pharmacol. 2017;8:99. doi: 10.3389/fphar.2017.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruppen I, Grau L, Orenes-Pinero E, et al. Differential protein expression profiling by iTRAQ-two-dimensional LC-MS/MS of human bladder cancer EJ138 cells transfected with the metastasis suppressor KiSS-1 gene. Mol Cell Proteomics. 2010;9(10):2276–2291. doi: 10.1074/mcp.M900255-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Gao L, Lee YM, et al. Target identification of natural and traditional medicines with quantitative chemical proteomics approaches. Pharmacol Ther. 2016;162:10–22. doi: 10.1016/j.pharmthera.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Xiong XG, Liang Q, Zhang C, et al. Serum proteome alterations in patients with cognitive impairment after traumatic brain injury revealed by iTRAQ-based quantitative proteomics. Biomed Res Int. 2017;2017:8572509. doi: 10.1155/2017/8572509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo YJ, Chen J, Xiong XG, Wu D, Zhu H, Liang QH. Effect of Bizhongxiao decoction and its dismantled formulae on IL-1 and TNF levels in collagen-induced arthritis in rat synovial joints. Theor Biol Med Model. 2012;9:47. doi: 10.1186/1742-4682-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serada S, Fujimoto M, Ogata A, et al. iTRAQ-based proteomic identification of leucine-rich alpha-2 glycoprotein as a novel inflammatory bio-marker in autoimmune diseases. Ann Rheum Dis. 2010;69(4):770–774. doi: 10.1136/ard.2009.118919. [DOI] [PubMed] [Google Scholar]
- 25.Huang W, Liang QH, Chen J, et al. Quantitative proteomic analysis of synovial tissue from rats with collagen-induced arthritis. RSC Adv. 2015;5(105):86088–86101. [Google Scholar]
- 26.Shen Q, Bai Y, Chang KC, et al. Liver X receptor-retinoid X receptor (LXR-RXR) heterodimer cistrome reveals coordination of LXR and AP1 signaling in keratinocytes. J Biol Chem. 2011;286(16):14554–14563. doi: 10.1074/jbc.M110.165704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghaddab-Zroud R, Seugnet I, Steffensen KR, Demeneix BA, Clerget-Froidevaux MS. Liver X receptor regulation of thyrotropin-releasing hormone transcription in mouse hypothalamus is dependent on thyroid status. PLoS One. 2014;9(9):e106983. doi: 10.1371/journal.pone.0106983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moyes KM, Drackley JK, Morin DE, et al. Gene network and pathway analysis of bovine mammary tissue challenged with Streptococcus uberis reveals induction of cell proliferation and inhibition of PPAR gamma signaling as potential mechanism for the negative relationships between immune response and lipid metabolism. BMC Genomics. 2009;10:542. doi: 10.1186/1471-2164-10-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A-Gonzalez N, Bensinger SJ, Hong C, et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity. 2009;31(2):245–258. doi: 10.1016/j.immuni.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiss M, Czimmerer Z, Nagy L. The role of lipid-activated nuclear receptors in shaping macrophage and dendritic cell function: from physiology to pathology. J Allergy Clin Immunol. 2013;132(2):264–286. doi: 10.1016/j.jaci.2013.05.044. [DOI] [PubMed] [Google Scholar]
- 31.Ueda M, Goto T, Kuroishi KN, et al. Asporin in compressed periodontal ligament cells inhibits bone formation. Arch Oral Biol. 2016;62:86–92. doi: 10.1016/j.archoralbio.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Janssens K, Van den Haute C, Baekelandt V, et al. Leukemia inhibitory factor tips the immune balance towards regulatory T cells in multiple sclerosis. Brain Behav Immun. 2015;45:180–188. doi: 10.1016/j.bbi.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Maris P, Blomme A, Palacios AP, et al. Asporin is a fibroblast-derived TGF-beta1 inhibitor and a tumor suppressor associated with good prognosis in breast cancer. PLoS Med. 2015;12(9):e1001871. doi: 10.1371/journal.pmed.1001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu L, Li Z, Liu SY, Xu SY, Ni GX. Asporin and osteoarthritis. Osteoarthr Cartil. 2015;23(6):933–939. doi: 10.1016/j.joca.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Sims NA, Johnson RW. Leukemia inhibitory factor: a paracrine mediator of bone metabolism. Growth Factors. 2012;30(2):76–87. doi: 10.3109/08977194.2012.656760. [DOI] [PubMed] [Google Scholar]
- 36.Hunt LC, Upadhyay A, Jazayeri JA, Tudor EM, White JD. An anti-inflammatory role for leukemia inhibitory factor receptor signaling in regenerating skeletal muscle. Histochem Cell Biol. 2013;139(1):13–34. doi: 10.1007/s00418-012-1018-0. [DOI] [PubMed] [Google Scholar]
- 37.Jazayeri JA, De Weerd N, Raye W, Kivivuori S, Zabihi E, Carroll GJ. In vitro evaluation of leukemia inhibitory factor receptor antagonists as candidate therapeutics for inflammatory arthritis. J Interferon Cytokine Res. 2007;27(4):281–289. doi: 10.1089/jir.2006.0138. [DOI] [PubMed] [Google Scholar]