Abstract
The DNA methyltransferase, Dnmt3a, is dynamically regulated throughout mammalian B cell development and upon activation by antigenic stimulation. Dnmt3a inactivation in hematopoietic stem cells has been shown to drive B cell-related malignancies, including chronic lymphocytic leukemia, and associates with specific DNA methylation patterns in transformed cells. However, while it is clear that inactivation of Dnmt3a in hematopoietic stem cells has profound functional effects, the consequences of Dnmt3a inactivation in cells of the B lineage are unclear. To assess whether loss of Dnmt3a at the earliest stages of B cell development lead to DNA methylation defects that might impair function, we selectively inactivated Dnmt3a early in mouse B cell development and then utilized whole genome bisulfite sequencing to generate base-resolution profiles of Dnmt3a+/+ and Dnmt3a−/− naïve splenic B cells. Overall, we find that global methylation patterns are largely consistent between Dnmt3a+/+ and Dnmt3a−/− naïve B cells, indicating a minimal functional effect of DNMT3A in mature B cells. However, loss of Dnmt3a induced 449 focal DNA methylation changes, dominated by loss-of-methylation events. Regions found to be hypomethylated in Dnmt3a−/− naïve splenic B cells were enriched in gene bodies of transcripts expressed in B cells, a fraction of which are implicated in B cell-related disease. Overall, the results from this study suggest that factors other than Dnmt3a are the major drivers for methylome maintenance in B cell development.
Keywords: B cell, DNA methylation, Dnmt3a, whole genome bisulfite sequencing, gene body
DNA methylation is an essential epigenetic modification in mammals (Li et al. 1992; Okano et al. 1999). Cytosine methylation, occurring predominantly in the context of CpG dinucleotides in mammalian cells, has long been hypothesized to play a critical role in the establishment and maintenance of cell type-specific gene expression (Holliday and Pugh 1975; Riggs 1975). Indeed, epigenetic modifications, including DNA methylation, are dynamically regulated throughout hematopoietic differentiation (Cabezas-Wallscheid et al. 2014; Lara-Astiaso et al. 2014). Moreover, massive perturbation of the DNA methylome occurs during B cell differentiation, maturation, and activation (Kulis et al. 2015; Lai et al. 2013; Barwick et al. 2016; Oakes et al. 2016).
The DNA methyltransferases (DNMTs), DNMT1, DNMT3A, DNMT3B, and DNMT3C establish and maintain DNA methylation patterns in mammalian cells (Jones and Liang 2009; Barau et al. 2016). Accordingly, DNMTs are dynamically regulated in B cell maturation and activation. We have previously demonstrated significant changes in expression of DNMT1, DNMT3A, and DNMT3B, concomitant with widespread alteration of the DNA methylation landscape, throughout the course of a T cell-dependent B cell immune response (Lai et al. 2013). Upon activation by antigen, DNMT1 and DNMT3B were upregulated (Lai et al. 2013), consistent with the role of DNMT1 in replication-dependent maintenance of DNA methylation (Leonhardt et al. 1992) and with the role of DNMT3B in late-stage B cell differentiation (Blanco-Betancourt et al. 2004). In contrast, DNMT3A is dramatically decreased upon activation by antigen (Lai et al. 2013). Correspondingly, extensive DNA methylation changes were observed upon activation by antigenic stimulation, between naïve and germinal center (GC) B cell populations, and the alterations were dominated by loss-of-methylation events. These site-specific DNA methylation changes were hypothesized to result from passive (rather than active) demethylation associated with a coordinated loss of DNMT3A levels, a massive burst of proliferation, and widespread alteration of nuclear architecture (Lai et al. 2013). However, the precise role of Dnmt3a in directing DNA methylation patterns in naïve B cells has not been characterized.
In addition to roles in normal B cell development, experimental and human sequencing data point to a role for DNMT3A mutation and/or loss-of-function in hematologic disease. DNMT3A is one of the most commonly mutated genes in adult hematologic malignancies (Brunetti et al. 2017; Yang et al. 2015). Loss of Dnmt3a progressively impairs hematopoietic stem cell differentiation (Challen et al. 2011) and confers a preleukemic phenotype on murine hematopoietic stem cells (Mayle et al. 2015). Further, inactivation of Dnmt3a in mouse hematopoietic stem cells induces chronic lymphocytic leukemia (CLL) and CD8-positive peripheral T cell lymphomas (Haney et al. 2016a,b; Peters et al. 2014). In transformed cells, Dnmt3a mutations and loss-of-function associate with specific DNA methylation patterns. For instance, DNMT3A mutations are associated with a specific DNA hypomethylation pattern in acute myeloid leukemia (Russler-Germain et al. 2014), and loss of DNMT3A leads to hypomethylation of hematopoietic enhancers in FLT3-ITD–associated leukemias (Yang et al. 2016). Accordingly, a cell type-specific function has been suggested for DNMT3A in cellular transformation (Haney et al. 2016a). However, while it is clear that inactivation of Dnmt3a at the hematopoietic stem cell stage has profound functional effects, the consequences of Dnmt3a inactivation in cells of the B lineage are unclear.
Here, we assess whether loss of Dnmt3a at the earliest stages of B cell development lead to DNA methylation defects that might impair function. We selectively inactivated Dnmt3a early in B cell development and then utilized whole genome bisulfite sequencing (WGBS) to characterize global DNA methylation patterns downstream of Dnmt3a in splenic naïve B cells. Overall, we find that global methylation patterns are largely consistent between Dnmt3a+/+ and Dnmt3a−/− naïve B cells, indicating a minimal functional effect of DNMT3A in mature B cells. However, loss of Dnmt3a induced a small number of focal DNA methylation changes, and the differentially methylated regions (DMRs) were found to be enriched in and around genes that are expressed in B cells and that are implicated in B cell-related disease.
Materials and Methods
Mouse lines and breeding strategy
Dnmt3aflox/flox mice containing conditional alleles for Dnmt3a (B6;129-Dnmt3atm1Jae/Mmnc, RRID:MMRRC_031042-UNC) (Nguyen et al. 2007) were obtained from the Mutant Mouse Resource Research Centers. Cd19Cre mice containing the CD19-Cre allele (B6.129P2(C)-Cd19tm1(cre)Cgn/J, RRID:IMSR_JAX:006785) (Rickert et al. 1997) were obtained from The Jackson Laboratory. Dnmt3aflox/flox animals were bred with Cd19Cre animals to generate Dnmt3aflox/+ × Cd19Cre/+ progeny. F1 Dnmt3aflox/+ × Cd19Cre/+ animals were further bred to maintain Dnmt3aflox/flox × Cd19Cre/+ (Dnmt3a−/−) and Dnmt3aflox/flox × Cd19+/+ (Dnmt3a+/+) littermate control mice. Animals were housed in a specific pathogen-free environment at the National Institute of Environmental Health Sciences (NIEHS). All animal experiments were approved by the NIEHS Institutional Animal Care and Use Committee and were performed according to National Institutes of Health guidelines for the care and use of laboratory animals. All procedures were in compliance with the Animal Welfare Act Regulations 9 CFR 1-4, with handling and treatment according to the Guide for the Care and Use of Laboratory Animals [National Research Council (US) 2011].
Splenic naïve B cell isolation
Spleen was harvested from three male Dnmt3a−/− and three male Dnmt3a+/+ mice, ranging from 2 to 4 months of age, following euthanasia by carbon dioxide asphyxiation. Single cell suspensions were prepared and cells were blocked with a cocktail of rat IgG (Jackson ImmunoResearch Laboratories) and anti-mouse CD16/CD32 (2.4G2) on ice for 10 min. Cells were then stained with allophycocyanin-B220 (clone RA3-6B2), phycoerythrin (PE)-IgD (clone 11-26c.2a), and FITC-GL7 (clone GL7) from BioLegend, eBioscience, or BD Pharmingen for 30 min in the dark. Cells were subsequently washed two times and then sorted on a BD FACSAria cell sorter (BD Biosciences). B220+IgD++GL7− B cells were sorted to a purity of 98–99%. Deletion efficiency of Dnmt3a was assessed by quantitative PCR, performed using SYBR Green on a CFX Connect Real-Time PCR Detection System (BioRad).
Quantification of lymphocyte populations in bone marrow and spleen
Bone marrow and spleen were harvested from three male Dnmt3a−/− and three male Dnmt3a+/+ mice following euthanasia by carbon dioxide asphyxiation. Single cell suspensions were prepared and cells were blocked with a cocktail of rat IgG (Jackson ImmunoResearch Laboratories) and anti-mouse CD16/CD32 (2.4G2) on ice for 10 min. Cells from bone marrow were stained with antibodies against mouse B220 (RA3-6B2), CD43 (S7), and IgM; splenocytes were stained with antibodies against mouse B220, CD19 (1D3), CD4 (GK1.5), and CD8a (53-6.7). All antibodies were purchased from BioLegend, eBioscience, Southern BioTech, or BD Pharmigen. Cells were subsequently washed two times and then analyzed on a BD LSR II Flow Cytometer (BD Biosciences) or sorted on FACSAria cell sorter (BD Biosciences). The following cell populations were analyzed from bone marrow: pro B cells (B220+CD43+IgM−), pre B cells (B220+CD43−IgM−), and immature B cells (B220+CD43−IgM+). The following cell populations were analyzed from spleen: CD4+ T cells (B220−CD19−CD4+CD8−), CD8+ T cells (B220−CD19−CD4−CD8+), B cells (B220+CD19+CD4−CD8−), and nonlymphocytes (B220−CD19−CD4−CD8−). Cell populations were sorted to a purity of 98–99%.
Nucleic acid extraction and purification
Sorted B220+IgD++GL7− B cells were homogenized in TRIzol (Thermo Fisher Scientific), and phase separation was performed according to the manufacturer’s instructions. Genomic DNA was extracted from the TRIzol organic phase by adding an equal volume of back extraction buffer, consisting of 4 M guanidine thiocyanate, 50 mM sodium citrate, and 1 M Tris base, and precipitated with isopropanol. DNA was further purified by performing phenol:chloroform:isoamyl alcohol (25:24:1) extractions (Ambion) according to the manufacturer’s instructions. DNA integrity was assessed by gel electrophoresis.
WGBS library construction and sequencing
Quantification of DNA during library production was performed using Qubit dsDNA Assay Kits (Invitrogen). For each sample, 1500 ng intact genomic DNA and 1.5 ng unmethylated λ DNA (Promega) were added to a microTUBE (Covaris) and fragmented using a Covaris S220 focused ultrasonicator according to the manufacturer’s instructions, with the following settings: target BP (peak), 300; peak incident power (W), 175; duty factor, 10%; cycles per burst, 200; treatment time (sec), 50; temperature (°C), 7; water level, 12; and sample volume (microliter), 50. Sequencing libraries were prepared using the NEXTflex Bisulfite-Seq Kit (BIOO Scientific) with a starting input of 1 μg. Steps for end repair, adenylation, and adapter ligation were performed according to the manufacturer’s instructions, following “option 1” and using NEXTflex Bisulfite-Seq Barcodes. Agarose gel size selection was implemented by electrophoresing the sample on a 2% NuSieve 3:1 agarose gel (Lonza) at 120 V for 10 min and then at 60 V for 120 min. The ladder lane was excised and visualized on a UV transilluminator, and then a gel slice was cut from each sample corresponding to 410–510 bp using a clean razor. DNA was recovered using a QIAquick Gel Extraction Kit (Qiagen) and then cleaned using an equal volume of AMPure XP (Agencourt). Bisulfite conversion was performed using an EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s instructions with the following modified thermal cycler conditions: 5 min at 95°, 25 min at 60°, 5 min at 95°, 85 min at 60°, 5 min at 95°, 175 min at 60°, 5 min at 95°, 180 min at 60°, 5 min at 95°, and 180 min at 60°, then hold at 20°. After elution, the samples were cleaned using an equal volume of AMPure XP (Agencourt). Bisulfite converted DNA product was PCR amplified for 10 cycles using KAPA HiFi Uracil+ Ready Mix (Kapa Biosystems) according to the manufacturer’s protocol. Following amplification, the libraries were cleaned using an equal volume of AMPure XP (Agencourt). The size of each amplified library was validated on Bioanalyzer High Sensitivity DNA Chips (Agilent Technologies). After library construction, all six libraries were equally multiplexed and sequenced using the Illumina NextSeq500 platform in paired-end format for 76 or 151 cycles at the NIEHS Epigenomics Core Laboratory.
Read processing and alignment
Raw FASTQ datasets were trimmed to remove the final low quality base (to 150 or 75 bp) and filtered to include read pairs with average base quality score of at least 20. Trimmed and filtered read pairs were aligned to the mouse (mm9/NCBI build 37) and phage λ (NC_001416.1) genomes using Bismark version 0.14.0 (Krueger and Andrews 2011) with bowtie version 0.12.8 (Langmead et al. 2009), with parameters “-X 10000--unmapped--non_bs_mm--old_flag -n 2 -l 50 -e 70--chunkmbs 1024.” A second-pass mapping step was implemented such that unmapped reads from the PE150 mapping were trimmed to a read length of 75 bp, and trimmed reads were re-submitted to Bismark as described above. For each mapped SAM file, positional bias (M-bias) was assessed using “bismark_methylation_extractor” with the following options: “--paired-end--mbias_only--include_overlap.” Positional bias trim positions were defined by first calculating the mid-read mean and SD of positional CpG methylation level for each individual read using positions 30–100 for PE150 reads and positions 20–50 for PE75 reads. Positions were retained according to the following criteria: (1) retain positions with CpG methylation level within ±3 SD of mid-read mean, (2) range must start and end with positions that meet the ±3 SD of mid-read mean criteria, and (3) single (nonconsecutive) outliers allowed as long as within ±4 SD of mid-read mean. All SAM files per biological replicate were merged then deduplicated using the Bismark “deduplicate_bismark” tool with the “--paired” option. Following deduplication, positional bias trimming was implemented, and overlapping mates were clipped. Mapped reads were split by genome (mm9/λ) and strand (plus/minus). For each library, bisulfite conversion rate was assessed using reads mapping to (1) unmethylated phage λ spike-in and (2) mitochondrial DNA (chrM). Composite bisulfite conversion rates were at least 99.84% for all libraries, and results were highly consistent between phage λ and chrM reads. Postprocessed BAM files for biological replicates (three replicates per group) were then merged using “MergeSamFiles” of Picard Tools, keeping strands separate. Sequence alignment and processing metrics are summarized in Supplemental Material, Table S1 of File S1.
Genomic context validation for CpG sites
For downstream analysis of CpG methylation, a genomic context validation strategy was implemented to identify mm9 CpG sites that are within a CpG context in all samples. Strand-specific data from all runs of all samples were combined, and each CpG dinucleotide in mm9 was assessed using reads mapped to the G on the strand opposite each cytosine. For a CpG site to be retained for downstream methylation analyses, the following criteria must be met, where N represents the cytosine position of the plus strand: (1) at least 95% of mapped bases at the N + 1 coordinate on the plus stand (opposite the expected cytosine on the minus strand) must be G, (2) at least 95% of mapped bases at the N coordinate on the minus strand (opposite the expected cytosine on the plus strand) must be G, (3) the minimum G read depth at the N + 1 position on the plus strand must be at least 5, and (4) the minimum G read depth at the N position on the minus strand must be at least 5. A total of 18,312,876 out of 21,722,957 (84.3%) CpG sites passed the genomic context validation criteria and were utilized for downstream differential methylation analyses.
Comparative methylation analyses
For comparative analysis of single-site CpG methylation levels a read depth filter was applied such that only validated CpGs with at least 10 reads were included. Differential methylation analyses were performed using MOABS v1.3.2 (Sun et al. 2014). CpG mpileup files for Dnmt3a+/+ and Dnmt3a−/− were formatted in “*.G.bed” format for input to MOABS. DMRs were identified using the “mcomp” module using default parameters except “–minNominalDif = 0.2.” Pairwise comparisons were reported using method M1. Details regarding genomic coordinates, methylation levels, and statistical significance of DMRs are listed in Tables S2 and S3 of File S1. The smoothed density scatterplot of mCG/CG was generated using the “smoothScatter” function of the “graphics” package in R, which produces a smoothed color density representation of the scatterplot, obtained through a kernel density estimate. Density color keys were generated using the “image.plot” and “tim.colors” functions of the “fields” package. Box and whisker plots of methylation levels were created using the “boxplot” function of the “stats” package using the default settings. Boxes represent the first to third quartiles of the data distribution and whiskers were drawn to the maximum data value no more than 1.5 times the interquartile distance.
Genomic feature annotation and statistical analyses
Statistical and graphical analyses were conducted using R version 3.3.2 (R Core Team 2016). Genomic feature annotation was performed using the Goldmine R package version 1.0 (Bhasin and Ting 2016). Gene model features were obtained from RefSeq annotations (February 17, 2017) (O’Leary et al. 2016). Promoters were defined as the region from 1000 bp upstream to 500 bp downstream of TSSs. Gene 3′ ends were defined as the region from 1000 bp upstream to 1000 bp downstream of TESs. CpG island coordinates for mm9 were obtained from the UCSC Table Browser (Karolchik et al. 2004). CpG island shores were defined as regions up to 2 kb flanking either CpG island edge (Doi et al. 2009; Irizarry et al. 2009). CpG island shelves were defined as regions up to 2 kb flanking CpG island shores (Bibikova et al. 2011). Genomic distance of DMRs was calculated to the nearest single TSS using the “closest” utility of Bedtools version 2.23.0 (Quinlan and Hall 2010), reporting each distance with respect to the TSS’s strand. Length-matched random control coordinates were generated using the “shuffle” utility of Bedtools using DMR coordinates with the options “-seed 1000” and “–noOverlapping” and masking mm9 gap regions, noncanonical chromosomes, and chrM. Association of genomic regions with putative target genes and the Mouse Genome Informatics (MGI) Phenotype Ontology (Blake et al. 2009) and Disease Ontology (Osborne et al. 2009) was carried out using the Genomic Regions Enrichment of Annotations Tool (GREAT version 3.0.0) (McLean et al. 2010) with the following parameters: background, whole genome background; assembly, mouse: NCBI build 37 (mm9); associated genomics regions: basal + extension, curated regulatory domains included. Depicted values for statistical enrichments for associations between genomic regions and annotations represent raw P-values using a binomial test over genomic regions with a minimum twofold region-based enrichment. For analysis of DMR overlap with bodies of genes expressed in B cells, we used a published RNA-seq dataset from Shi et al. (2015), and data were obtained from the NCBI Gene Expression Omnibus (GEO) (Edgar et al. 2002) (accession no.: GSE60927). Normalized expression values were obtained for two biological replicates (samples GSM1493786 and GSM1493787) of mouse follicular B cells (small, B220+CD21+CD23+). Genes with an average of at least one fragment per kilobase of transcript per million mapped reads were defined as expressed. EntrezID (gene ID) numbers for expressed genes were converted to RefSeq mRNA accession using the biological DataBase network (bioDBnet) database to database conversion tool (db2db) for Mus musculus (taxon ID 10090) (Mudunuri et al. 2009). Gene body coordinates for expressed genes, defined as TSS to TES, were obtained from RefSeq annotations (O’Leary et al. 2016). For comparison to Dnmt3a-mutant CLL WGBS data, gene body and promoter DMRs hypomethylated in Dnmt3aΔ/Δ CLL relative to B-1a were obtained from Haney et al. (2016a), and coordinates were converted to mm9 using liftOver. A Z-test for the difference between two proportions was utilized to test whether the proportion of hypo- or hyper-methylated DMRs overlapping a given genomic feature differs from that of a length-matched random null coordinate set.
Data availability
The datasets generated during the current study are available in the NCBI GEO (Edgar et al. 2002) and are accessible through GEO series accession number GSE100262 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100262). File S1 contains a summary of WGBS processing metrics (Table S1 in File S1) and summaries of hypomethylated DMRs (Table S2 in File S1) and hyper-methylated DMRs (Table S3 in File S1).
Results
Targeted knockout of Dnmt3a in mouse B cells
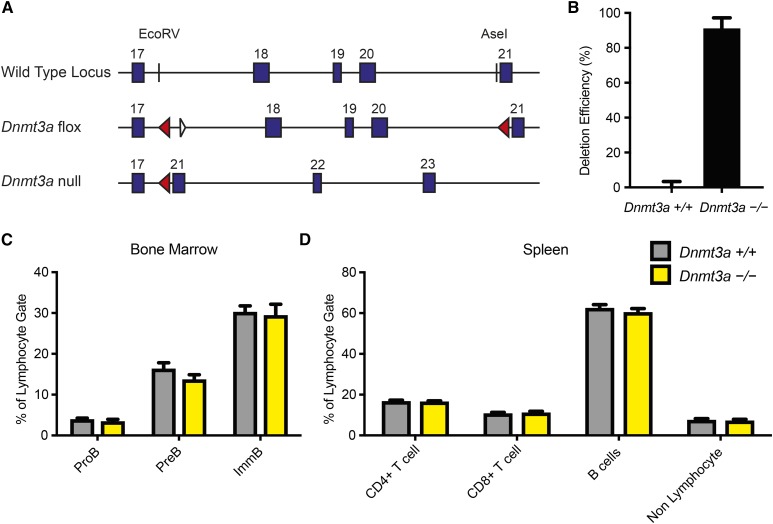
A conditional knockout allele was used to target the Dnmt3a gene, where exons encoding the catalytic domain (exons 18–20) of Dnmt3a were flanked by loxP sites (Nguyen et al. 2007), in conjunction with the Cre-loxP recombination system (Figure 1A). Dnmt3aflox/flox mice were bred to mice containing the Cd19-Cre allele (Cd19Cre/+) (Rickert et al. 1997) to selectively inactivate the Dnmt3a gene in B lymphocytes. The Cd19 promoter specifically directs Cre expression at the earliest stages and throughout B lymphocyte development and differentiation. As compared to Dnmt3aflox/flox × Cd19+/+ (Dnmt3a+/+) littermate control mice, a 91% mean deletion efficiency was observed in splenic naïve B cells (B220+IgD++GL7−) of Dnmt3aflox/flox × Cd19Cre/+ (Dnmt3a−/−) mice (Figure 1B), which is within the known range for splenic B cell deletion efficiency associated with Cd19-Cre–mediated mutagenesis (Rickert et al. 1997).
Figure 1.
Conditional knockout of Dnmt3a in mouse B cells. (A) Dnmt3a locus and gene targeting strategies. Top panel: endogenous Dnmt3a locus including relative positions of exons (blue boxes) and restriction enzyme sites (EcoRV and AseI) used to introduce the LoxP sites. Middle panel: Dnmt3a flox locus (Nguyen et al. 2007) including relative positions of the LoxP sites (red triangles) flanking exons 18 through 20 and the residual Frt site (white triangle). Bottom panel: Dnmt3a-null locus that depicts the deletion of exons 18–20. (B) Analysis of Dnmt3a deletion efficiency in naïve B cells by qPCR (N = 5 per group). (C) Quantification of the percent of pro B (ProB), pre B (PreB), and immature B (ImmB) cell populations in the bone marrow and (D) quantification of the percent of B cell, CD4+ T cell (CD4+CD8−), and CD8+ T cell (CD4− CD8+) populations in the spleen for Dnmt3a+/+ (gray bars) and Dnmt3a−/− (yellow bars) mice (N = 3 per group). Error bars indicate SEM.
To understand the phenotypic consequences of B cell-specific Dnmt3a deletion, we first characterized B cell maturation in the bone marrow by flow cytometry. The acute depletion of Dnmt3a (Dnmt3a−/−) did not disrupt the total number or frequency of developing B cells (pro B cells, pre B cells, immature B cells) as compared to littermate control mice (Dnmt3a+/+, Figure 1C). In addition, the depletion of Dnmt3a did not affect the total number or frequency of lymphocytes (B cells, T cells) in the spleen (Figure 1D). Further, characterization of lymphocyte populations in the spleen showed no change in the ratio of B to T (CD4+ or CD8+) cells in Dnmt3a-deficient mice as compared to control mice. Thus, we conclude that B cell development is not grossly affected by deletion of Dnmt3a, and since B cells develop normally, we are able to study the effects of Dnmt3a on downstream DNA methylation patterns.
Base-resolution methylomes of Dnmt3a knockout splenic naïve B cells
To assess the effect of Dnmt3a on genome-wide DNA methylation patterns, we generated base-resolution methylomes using WGBS for splenic naïve B cells (B220+IgD++GL7−) from adult Dnmt3a+/+ and Dnmt3a−/− mice. To minimize bias caused by a biological outlier, multiple animals were used, and libraries from three independent animals per genotype were sequenced to a total depth of 20–30× per genotype, as is common practice in the field (Yang et al. 2016; Kawakatsu et al. 2017). To control for variations in genetic background and to exclude polymorphic CpG sites in our mixed genetic background model (B6;129), a genomic context validation strategy was implemented to restrict downstream analyses to mm9 CpG sites that occur in CpG context in all samples (see Materials and Methods). A total of 18,312,876 out of 21,722,957 (84.3%) CpG sites passed the genomic context validation criteria and were utilized for downstream analyses. For Dnmt3a+/+ and Dnmt3a−/− cells, at least 91.2% of validated CpG sites have a read depth of at least 10 (Figure S1). A summary of sequence alignment and processing metrics is included in Table S1 of File S1.
B cell-specific deletion of Dnmt3a does not globally alter DNA methylation distribution in splenic naïve B cells
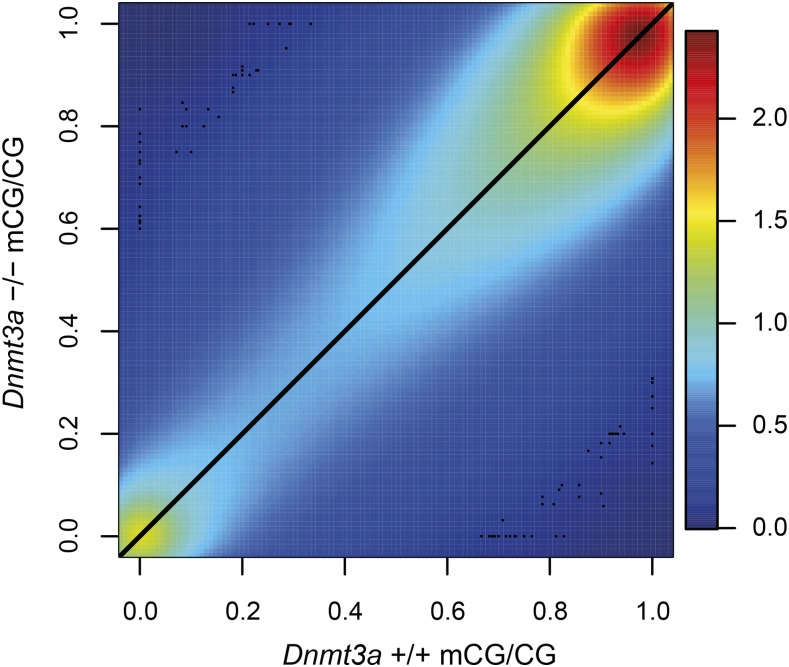
Using the base-resolution methylomes, we first asked if B cell-specific deletion of Dnmt3a effects DNA methylation levels at a global scale in splenic naïve B cells. The baseline distribution of CpG methylation in wild-type mouse naïve B cells was largely bimodal, and the vast majority of CpG sites were found to be methylated (Figure S2A), consistent with reported methylomes of sorted human naïve B cells (Kulis et al. 2015). We found global methylation distributions to be extremely consistent between Dnmt3a+/+ and Dnmt3a−/− B cells (Figure S2, A and B). In both Dnmt3a+/+ and Dnmt3a−/− B cells, 82% of validated CpG sites with read depth of at least 10 were largely methylated (mCG/CG > 0.75) whereas 7% were largely unmethylated (mCG/CG < 0.25). CpGs with intermediate methylation level (mCG/CG between 0.25 and 0.75) accounted for 11% of all validated sites assayed in each sample. Comparison of single-CpG methylation levels across the genome in naïve B cells with and without Dnmt3a confirmed that knockout of Dnmt3a does not induce a gross global shift of DNA methylation, as site-specific mCG/CG values exhibit a strong positive linear relationship between Dnmt3a+/+ and Dnmt3a−/− B cells (Figure 2).
Figure 2.
Comparison of single-site CpG methylation levels in splenic naïve B cells with and without Dnmt3a. Smoothed kernel density scatterplot of Dnmt3a+/+ mCG/CG (x-axis) vs. Dnmt3a−/− mCG/CG (y-axis) for all validated CpG sites with read depth of at least 10 in both groups (N = 16,233,681). Points to the upper left of the diagonal reflect CpGs with greater methylation in Dnmt3a−/− and points to the lower right of the diagonal reflect CpGs with greater methylation in Dnmt3a+/+.
B cell-specific deletion of Dnmt3a induces a limited number of site-specific DNA methylation alterations in splenic naïve B cells
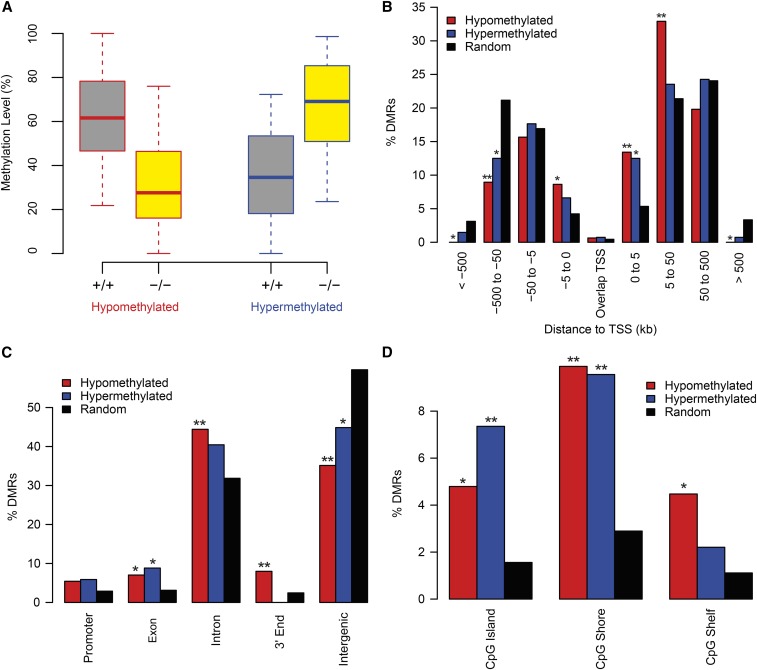
We next asked if B cell-specific deletion of Dnmt3a induces focal, site-specific DNA methylation alterations in splenic naïve B cells. To define regions that are differentially methylated between Dnmt3a+/+ and Dnmt3a−/− B cells, we utilized the differential methylation analysis algorithm, MOABS (Model-based Analysis of Bisulfite Sequencing data) (Sun et al. 2014). In total, 449 DMRs were identified, and both hypomethylated DMRs (Table S2 in File S1) and hypermethylated DMRs (Table S3 in File S1) were identified. However, regions hypomethylated in Dnmt3a−/− cells (N = 313) were over twofold more frequently observed than hypermethylated regions (N = 136), consistent with loss of methyltransferase activity. Loci that became hypo- or hypermethylated in Dnmt3a−/− cells were found to occur at regions spanning a broad range of methylation levels in Dnmt3a+/+ cells, specifically 22–100% for hypomethylated loci and 0–76% for hypermethylated loci (Figure 3A). At the majority of DMRs, intermediate regional methylation levels are observed in both Dnmt3a+/+ and Dnmt3a−/− cells. Together, the relatively small number of DMRs occurring in both directions (hypo- and hypermethylation) across a broad range of methylation levels are consistent with the lack of an observed systematic effect on global methylation distribution.
Figure 3.
Summary characteristics of Dnmt3a-dependent DMRs. (A) Methylation levels at Dnmt3a-dependent DMRs by direction of methylation change. Box and whisker plots represent methylation levels for Dnmt3a+/+ (gray fill) and Dnmt3a−/− (yellow fill) B cells at hypomethylated DMRs (red outline) and hyper-methylated DMRs (blue outline). (B) Percent of DMRs that fall in distance-dependent bins relative to the single nearest TSS. (C) Percent of DMRs that fall in gene model contexts for promoter, exon, intron, 3′ end, and intergenic regions. (D) Percent of DMRs that overlap with CpG-islands (UCSC genome browser), CpG-shores (±2 kb from islands), and CpG shelves (±2 kb from shores). Hypomethylated (red) and hypermethylated (blue) DMRs are compared with a length-matched random genomic null region set (black). Significant differences from the random set are noted; * P < 0.05, ** P < 0.001, two-sample Z-test for the difference in proportions.
However, the hypo- and hypermethylated DMRs exhibit differential distributions relative to genomic context. The percentage of hyper-methylated DMRs that fall in distance-dependent bins relative to the nearest TSS closely resemble the percentage of length-matched random genomic null regions (Figure 3B). However, hypermethylated DMRs exhibited significant enrichment within 5 kb downstream of TSSs (Figure 3B). Alternatively, hypomethylated DMRs were highly enriched between 0 and 50 kb downstream of TSSs. Further, hypomethylated DMRs were notably depleted at TSS-distal regions and were completely absent in gene-poor regions, i.e., >500 kb away from the nearest TSS (Figure 3B). Likewise, hypomethylated DMRs were enriched in gene body elements, including exons, introns, and 3′-ends while being highly depleted in intergenic regions (Figure 3C). Consistent with enrichment of both hypo- and hypermethylated DMRs within 5 kb of TSSs, both were highly enriched near CpG islands, particularly at CpG shores (Figure 3D). Together, these results support a model in which B cell-specific methylation by DNMT3A is targeted to gene-centric regions.
Dnmt3a−/−-associated hypomethylation targets genes involved in B cell-related phenotypes and disease
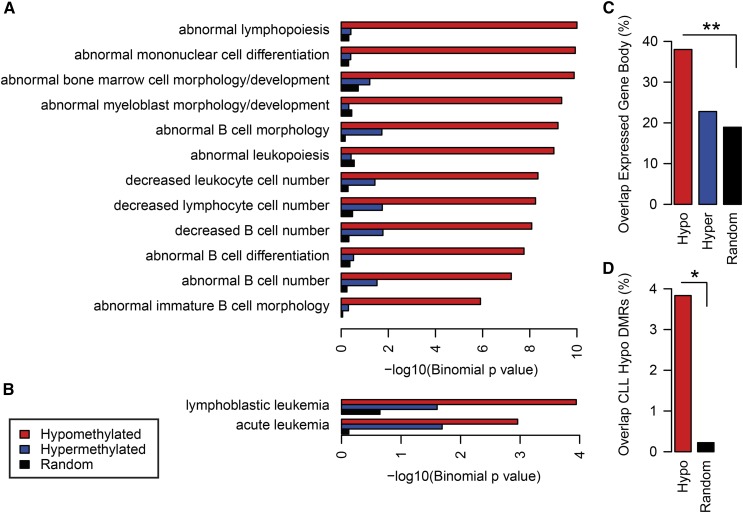
To gain insight on the functional significance of genes associated with Dnmt3a-dependent hypo- and hypermethylation events, we performed gene ontology analyses using GREAT (McLean et al. 2010), which enables prediction of function for cis-regulatory regions. Interestingly, statistical enrichments for association between putative target genes of Dnmt3a-dependent hypomethylated DMRs and annotations of the MGI Phenotype Ontology (Blake et al. 2009) indicated significant links to B cell-related mouse phenotypes such as abnormal B cell morphology, cell number, and differentiation (Figure 4A). Conversely, significant enrichments of B cell-related mouse phenotypes were not observed for genes associated with hypermethylated DMRs or random null control regions. Furthermore, upon examination of associations with terms of the Disease Ontology, a standardized ontology for human disease, putative target genes of Dnmt3a-dependent hypomethylated DMRs were found to be enriched for genes involved in lymphoblastic leukemia, a term including the B cell malignancy, CLL (Figure 4B). Again, functional enrichment of B cell-related phenotypes and disease were solely driven by hypomethylated DMRs, rather than hypermethylated DMRs.
Figure 4.
Hypomethylated Dnmt3a-dependent DMRs are located in and around genes with functional roles in B cell development and disease. Statistical enrichments for association between putative target genes of Dnmt3a-dependent DMRs and annotations of the (A) Mouse Genome Informatics (MGI) Phenotype Ontology and (B) the Disease Ontology. The x-axis corresponds to raw P values (binomial test, in logarithmic scale). (C) Percent overlap of hypomethylated (red) and hypermethylated (blue) DMRs with gene bodies of transcripts expressed in B cells, as compared with length-matched random genomic null regions (black). (D) Percent overlap of hypomethylated DMRs (red) with hypomethylated promoter and gene body DMRs in Dnmt3aΔ/Δ CLL relative to B-1a cells (Haney et al. 2016a), as compared with length-matched random genomic null regions (black). Significant differences from the random set are noted; * P = 1.5 × 10−4, ** P = 4.8 × 10−9, two-sample Z-test for the difference in proportions.
Based on the functional enrichment of Dnmt3a-dependent DMRs in B cell-related phenotypes and on previous work supporting the role of DNMT3A in catalyzing DNA methylation in gene bodies of transcriptionally permissive genes (Wu et al. 2010), we hypothesized that in B cell development, DNMT3A is likely catalyzing DNA methylation in gene bodies of transcripts expressed in B cells. To test this hypothesis, we first utilized existing RNA-seq data for follicular B cells (small B220+CD21+CD23+) (Shi et al. 2015), which represent the majority of naive splenic B cells, to define the set of genes expressed in B cells. Next, we asked whether Dnmt3a-dependent DMRs overlap gene bodies, defined as the region from the TSS to the TES, of B cell-expressed genes. Indeed, nearly 40% of hypomethylated DMRs were found to overlap gene bodies of transcripts expressed in B cells. Hypomethylated DMRs were found to overlap B cell-expressed gene bodies approximately twofold more frequently than hypermethylated regions or a length-matched random genomic null region set (Figure 4C).
Next, we asked to what extent do the Dnmt3a-dependent DMRs observed here overlap with DNA methylation alterations observed in a mouse model of Dnmt3a-null CLL. To do this, we assessed DMRs recently reported by Haney et al., contrasting splenic B-1a cells from a Dnmt3a+/+ control mouse and a EμSRα-tTA;Teto-Cre;Dnmt3afl/fl;Rosa26LOXPEGFP/EGFP (Dnmt3aΔ/Δ) mouse with CLL (Haney et al. 2016a). Hypo- and hypermethylated Dnmt3aΔ/Δ CLL DMRs located in promoter and gene body regions were compared with Dnmt3a−/− naïve B cell hypo- and hypermethylated DMRs, respectively. A total of ∼4% of hypomethylated Dnmt3a−/− naïve B cell DMRs directly overlapped with hypomethylated Dnmt3aΔ/Δ CLL DMRs, which was significantly greater than the overlap observed with a random genomic null region set (Figure 4D). Conversely, no overlap was observed between the hypermethylated DMR sets. Together, these data indicate that the limited number of Dnmt3a−/−-associated hypomethylation events target genes involved in B cell-related phenotypes, primarily in gene bodies of transcripts expressed in B cells, a small fraction of which are associated with B cell malignancies.
Discussion
Here, we report high-quality base-resolution CpG methylation profiles of Dnmt3a+/+ and Dnmt3a−/− mouse naïve splenic B cells. Globally, the observed bimodal CpG methylation distribution in naïve B cells is largely consistent with features of the mouse ES cell methylome (Stadler et al. 2011) and with previously reported genome-wide profiles in human naïve B cells (Kulis et al. 2015). Similar to human uncommitted hematopoietic progenitor cells, pre-BII cells, and naïve B cells (Kulis et al. 2015), examination of global DNA methylation distributions revealed that the vast majority of CpG sites (82%) in mouse naïve splenic B cells are methylated, while only a small proportion (7%) are unmethylated. In contrast, DNA methylation is known to be remodeled in GC B cells, memory B cells, and plasma cells during a B cell immune response, and is characterized by hypomethylation and increased methylome diversity in conjunction with dynamic regulation of three methyltransferases: Dnmt1, Dnmt3b, and Dnmt3a (Barwick et al. 2016; Dominguez et al. 2015; Kulis et al. 2015; Lai et al. 2013; Shaknovich et al. 2011).
Examining the effects of Dnmt3a on DNA methylation patterns in B cell development, we generally observed modest changes in naïve B cells lacking Dnmt3a. Methylation changes downstream of B cell-specific Dnmt3a knockout occur at a focal scale rather than by affecting DNA methylation at a global scale. The observed DMRs represent a small fraction of the genome (<1%), which is consistent with our observation that B cells lacking Dnmt3a develop normally. However, while B cell-specific deletion of Dnmt3a affected only a small fraction of the genome, the regions which are differentially methylated reflect a connection to B cell biology.
Based on the data presented here, DNA methylation changes associated with B cell-specific Dnmt3a knockout most frequently correspond to hypomethylation, which is consistent with loss of DNMT activity. Further, the higher proportion of hypomethylation in naïve B cells is consistent with observations of hypomethylation in DNMT3A-mutant and Dnmt3a-null leukemias (Haney et al. 2016a,b; Russler-Germain et al. 2014; Yang et al. 2016). Contrary to observations of promoter hypomethylation in Dnmt3a+/− and Dnmt3a−/− CLL (Haney et al. 2016b), promoters were minimally affected in Dnmt3a−/− naïve B cells. This difference in outcomes may be influenced by DMRs resulting from the pathogenesis of CLL itself rather than loss of Dnmt3a.
The hypomethylation events observed in Dnmt3a-null B cells were enriched in gene bodies, particularly in bodies of genes that are poised for transcription in B cells. Correspondingly, gene-poor regions (gene deserts) are devoid of hypomethylated DMRs in Dnmt3a−/− cells. Together, this evidence is consistent with existing models that one function of DNMT3A is to methylate gene bodies (Wu et al. 2010). Hypermethylation events observed in this work were enriched in promoter proximal regions within gene bodies (Figure 4B). While the mechanistic basis of these events is currently unclear, it is conceivable that these loci are preferential targets for methylation by other DNMTs (including DNMT3B) that are revealed upon loss of DNMT3A activity.
Further, the results presented here allow us to speculate on comparisons of Dnmt3a loss-of-function in B cell-derived cells and early hematopoietic cell types. It is clear that DNMT3A is crucial to embryonic stem (ES) cell and hematopoietic stem cell (HSC) differentiation and regulates the function of stem cells. DNMT3A mutations occur in human HSCs, in which they act as a preleukemic lesion (Yang et al. 2015). In the mouse, loss of Dnmt3a in ES cells progressively impairs HSC differentiation over serial transplantation, causes HSC expansion (Challen et al. 2011), and predisposes HSCs to malignant transformation (Mayle et al. 2015). Further, conditional inactivation of Dnmt3a in HSCs and progenitors results in the development of CLL (Peters et al. 2014). The functional and epigenomic data here are consistent with the hypothesis that the cells of origin for these tumors are early hematopoietic cell types rather than B cell-derived descendants.
Dnmt3a, in conjunction with Dnmt1 and Dnmt3b, is known to be dynamically regulated throughout normal B cell development and during a B cell immune response, and its inactivation in hematopoietic stem cells has been shown to drive B cell-related malignancies. Here, we tested whether B cell-specific loss of Dnmt3a leads to DNA methylation defects in B cells that might impair function. While we identified a subset of CpGs that require Dnmt3a to maintain DNA methylation patterns in naïve B cells, we do not observe DNA methylation changes that would indicate large functional changes and predict that phenotypic consequences would be minimal. The results from this study suggest that factors other than Dnmt3a are the major drivers for methylome maintenance in B cell development.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300446/-/DC1.
Acknowledgments
Sequencing was performed at the National Institute of Environmental Health Sciences (NIEHS) Epigenomics Core Laboratory, and we thank Gregory G. Solomon, Jason A. Malphurs, Nicole Reeves, and John T. Otstot for support related to next-generation sequencing efforts. The authors express gratitude to Adam B. Burkholder for initial sequence quality control and processing. We gratefully acknowledge David C. Fargo and Frank Day for outstanding scientific computing support. The authors express gratitude to members of the Wade laboratory for useful comments throughout the course of this work. This work was supported by the Division of Intramural Research of the NIEHS, and the Division of the National Toxicology Program, NIEHS, National Institutes of Health, Research Triangle Park, North Carolina (ES101965 and ES103146 to P.A.W.). The statements, opinions, or conclusions contained therein do not necessarily represent the statements, opinions, or conclusions of NIEHS, National Institutes of Health, or the US Government.
Footnotes
Communicating editor: C. Pikaard
Literature Cited
- Barau J., Teissandier A., Zamudio N., Roy S., Nalesso V., et al. , 2016. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354: 909–912. [DOI] [PubMed] [Google Scholar]
- Barwick B. G., Scharer C. D., Bally A. P., Boss J. M., 2016. Plasma cell differentiation is coupled to division-dependent DNA hypomethylation and gene regulation. Nat. Immunol. 17: 1216–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin J. M., Ting A. H., 2016. Goldmine integrates information placing genomic ranges into meaningful biological contexts. Nucleic Acids Res. 44: 5550–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., et al. , 2011. High density DNA methylation array with single CpG site resolution. Genomics 98: 288–295. [DOI] [PubMed] [Google Scholar]
- Blake J. A., Bult C. J., Eppig J. T., Kadin J. A., Richardson J. E., et al. , 2009. The mouse genome database genotypes:phenotypes. Nucleic Acids Res. 37: D712–D719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Betancourt C. E., Moncla A., Milili M., Jiang Y. L., Viegas-Pequignot E. M., et al. , 2004. Defective B-cell-negative selection and terminal differentiation in the ICF syndrome. Blood 103: 2683–2690. [DOI] [PubMed] [Google Scholar]
- Brunetti L., Gundry M. C., Goodell M. A., 2017. DNMT3A in leukemia. Cold Spring Harb. Perspect. Med. 7: a030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N., Klimmeck D., Hansson J., Lipka D. B., Reyes A., et al. , 2014. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15: 507–522. [DOI] [PubMed] [Google Scholar]
- Challen G. A., Sun D., Jeong M., Luo M., Jelinek J., et al. , 2011. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44: 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A., Park I. H., Wen B., Murakami P., Aryee M. J., et al. , 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41: 1350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez P. M., Teater M., Chambwe N., Kormaksson M., Redmond D., et al. , 2015. DNA methylation dynamics of germinal center B cells are mediated by AID. Cell Rep. 12: 2086–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E., 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney S. L., Upchurch G. M., Opavska J., Klinkebiel D., Appiah A. K., et al. , 2016a Loss of Dnmt3a induces CLL and PTCL with distinct methylomes and transcriptomes in mice. Sci. Rep. 6: 34222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney S. L., Upchurch G. M., Opavska J., Klinkebiel D., Hlady R. A., et al. , 2016b Promoter hypomethylation and expression is conserved in mouse chronic lymphocytic leukemia induced by decreased or inactivated Dnmt3a. Cell Rep. 15: 1190–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E., 1975. DNA modification mechanisms and gene activity during development. Science 187: 226–232. [PubMed] [Google Scholar]
- Irizarry R. A., Ladd-Acosta C., Wen B., Wu Z., Montano C., et al. , 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Liang G., 2009. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 10: 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A. S., Furey T. S., Roskin K. M., Sugnet C. W., et al. , 2004. The UCSC table browser data retrieval tool. Nucleic Acids Res. 32: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., Nery J. R., Castanon R., Ecker J. R., 2017. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 18: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F., Andrews S. R., 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M., Merkel A., Heath S., Queiros A. C., Schuyler R. P., et al. , 2015. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat. Genet. 47: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A. Y., Mav D., Shah R., Grimm S. A., Phadke D., et al. , 2013. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B-cell activation. Genome Res. 23: 2030–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D. A., et al. , 2014. Immunogenetics. Chromatin state dynamics during blood formation. Science 345: 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H., Page A. W., Weier H. U., Bestor T. H., 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71: 865–873. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R., 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926. [DOI] [PubMed] [Google Scholar]
- Mayle A., Yang L., Rodriguez B., Zhou T., Chang E., et al. , 2015. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 125: 629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., et al. , 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudunuri U., Che A., Yi M., Stephens R. M., 2009. bioDBnet: the biological database network. Bioinformatics 25: 555–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (US) , 2011. Guide for the Care and Use of Laboratory Animals. National Academies Press, Washington, DC. [Google Scholar]
- Nguyen S., Meletis K., Fu D., Jhaveri S., Jaenisch R., 2007. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev. Dyn. 236: 1663–1676. [DOI] [PubMed] [Google Scholar]
- Oakes C. C., Seifert M., Assenov Y., Gu L., Przekopowitz M., et al. , 2016. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat. Genet. 48: 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Bell D. W., Haber D. A., Li E., 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. [DOI] [PubMed] [Google Scholar]
- O’Leary N. A., Wright M. W., Brister J. R., Ciufo S., Haddad D., et al. , 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44: D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne J. D., Flatow J., Holko M., Lin S. M., Kibbe W. A., et al. , 2009. Annotating the human genome with disease ontology. BMC Genomics 10(Suppl. 1): S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S. L., Hlady R. A., Opavska J., Klinkebiel D., Pirruccello S. J., et al. , 2014. Tumor suppressor functions of Dnmt3a and Dnmt3b in the prevention of malignant mouse lymphopoiesis. Leukemia 28: 1138–1142. [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2016. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Rickert R. C., Roes J., Rajewsky K., 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25: 1317–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., 1975. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 14: 9–25. [DOI] [PubMed] [Google Scholar]
- Russler-Germain D. A., Spencer D. H., Young M. A., Lamprecht T. L., Miller C. A., et al. , 2014. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25: 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaknovich R., Cerchietti L., Tsikitas L., Kormaksson M., De S., et al. , 2011. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 118: 3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Liao Y., Willis S. N., Taubenheim N., Inouye M., et al. , 2015. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 16: 663–673. [DOI] [PubMed] [Google Scholar]
- Stadler M. B., Murr R., Burger L., Ivanek R., Lienert F., et al. , 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. [DOI] [PubMed] [Google Scholar]
- Sun D., Xi Y., Rodriguez B., Park H. J., Tong P., et al. , 2014. MOABS: model based analysis of bisulfite sequencing data. Genome Biol. 15: R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Coskun V., Tao J., Xie W., Ge W., et al. , 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Rau R., Goodell M. A., 2015. DNMT3A in haematological malignancies. Nat. Rev. Cancer 15: 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Rodriguez B., Mayle A., Park H. J., Lin X., et al. , 2016. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell 29: 922–934 (erratum: Cancer Cell 30: 363–365). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the NCBI GEO (Edgar et al. 2002) and are accessible through GEO series accession number GSE100262 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100262). File S1 contains a summary of WGBS processing metrics (Table S1 in File S1) and summaries of hypomethylated DMRs (Table S2 in File S1) and hyper-methylated DMRs (Table S3 in File S1).