Abstract
Domesticated species exhibit a suite of behavioral, endocrinological, and morphological changes referred to as “domestication syndrome.” These changes may include a reduction in reactivity of the hypothalamic-pituitary-adrenal (HPA) axis and specifically reduced adrenocorticotropic hormone release from the anterior pituitary. To investigate the biological mechanisms targeted during domestication, we investigated gene expression in the pituitaries of experimentally domesticated foxes (Vulpes vulpes). RNA was sequenced from the anterior pituitary of six foxes selectively bred for tameness (“tame foxes”) and six foxes selectively bred for aggression (“aggressive foxes”). Expression, splicing, and network differences identified between the two lines indicated the importance of genes related to regulation of exocytosis, specifically mediated by cAMP, organization of pseudopodia, and cell motility. These findings provide new insights into biological mechanisms that may have been targeted when these lines of foxes were selected for behavior and suggest new directions for research into HPA axis regulation and the biological underpinnings of domestication.
Keywords: RNA-seq, pituitary, domestication, hypothalamic-pituitary-adrenal axis, Vulpes vulpes
Domesticated species exhibit greatly reduced fearfulness, increased social tolerance, and increased resistance to stress compared to their ancestral wild species (Price 2008; Campler et al. 2009; Driscoll et al. 2009). These behavioral changes are closely linked to a suite of physiological and morphological changes, including reduced reactivity of the hormonal stress response, white spotting, curling of the tail, and shortening of the cranium, which have together been referred to as the “domestication syndrome” (Hare et al. 2012; Wilkins et al. 2014). The closely correlated behavioral, endocrinological, and morphological changes across multiple species suggest that perturbations at the level of networks of genes, working together for specific biological effects, may be implicated in the domestication process. Therefore, we expect that understanding the biological basis of the endocrinological changes in one domesticated species may shed light on the mechanisms targeted during the domestication process more generally, and potentially also on the biological underpinnings of variation in resilience to stress in animals and humans (Hare et al. 2012).
The characteristic reduction in fearfulness of domesticated animals has been closely linked to reduced reactivity of the hypothalamic-pituitary-adrenal (HPA) axis—the hormonal cascade associated with the stress response in mammals (Trut et al. 2009; Hare et al. 2012; Wilkins et al. 2014). Specifically, reductions in circulating levels of adrenocorticotrophic hormone (ACTH), released by the anterior pituitary, have been demonstrated in several domesticated species (Gulevich et al. 2004; Trut et al. 2004; Kaiser et al. 2015). ACTH stimulates release of glucocorticoids by the adrenals, and reduction in glucocorticoids in domesticated species has also been shown (Albert et al. 2008; Trut et al. 2009; Kaiser et al. 2015). These differences in domesticated and wild HPA axis function have been explored in multiple animal models. Domesticated guinea pigs, in response to psychosocial stressors, have adrenal glucocorticoid responses both decreased in magnitude and returning to baseline after a shorter time (Kaiser et al. 2015). Similar findings have been demonstrated with experimentally domesticated Norway rats, which, after many generations of selection for tameness, display markedly reduced aggressiveness to humans as compared to wild rats as well as reduced glucocorticoid levels (Naumenko et al. 1989; Albert et al. 2008; Prasolova et al. 2014) and ACTH levels (Shikhevich et al. 2002). However, the mechanisms driving the difference in HPA activity between these tame animals and their wild cousins are unknown.
In perhaps the best-known domestication experiment, farmed foxes (Vulpes vulpes) have been selectively bred for >50 generations to recapitulate the dog domestication process. One population of foxes was selected for lack of fear of, and for sociability toward, humans, while a second population of foxes was selected for aggression toward humans. A comparison of these tame and aggressive fox populations demonstrates similar changes in the HPA axis as those shown between domesticated guinea pigs and their wild ancestors, as well as between tame and aggressive rats (Trut et al. 2004, 2009). Compared to the aggressive line, the tame foxes display reduced basal ACTH levels (Gulevich et al. 2004; Trut et al. 2004), decreased ACTH response to stress, and shortened time to return to baseline ACTH levels (Oskina and Plyusnina 2000), as well as markedly reduced glucocorticoid elevation and shortened time to return to baseline glucocorticoid levels after a stressor (Trut et al. 2004).
Although the hypothalamus is classically considered the “top” of the HPA axis, the pituitary, situated to amplify signals from the hypothalamus before release to the systemic circulation, is sometimes known as the “master gland” (Melmed 2010) and may contribute significantly to regulation of the HPA axis. Inhibition of the HPA hormonal cascade at the level of the pituitary may occur in at least three ways, any or all of which could have been selected for during the domestication process: reduced synthesis of ACTH, reduced secretion of ACTH, and reduction of the number of cells that synthesize ACTH (Rizzoti 2010). An examination of the details of each of these processes will provide predictions for how they might vary in wild compared to domesticated pituitaries (i.e., the pituitaries of wild vs. domesticated animals). ACTH synthesis and secretion are triggered by the binding of corticotrophin releasing hormone (CRH) to CRH receptors in the anterior pituitary (Grammatopoulos 2012). ACTH synthesis begins with the production of the proopiomelanocortin (POMC) precursor molecule, which is regulated by a suite of transcription factors (Drouin 2016) and is cleaved into ACTH by proprotein convertases (Benjannet et al. 1991), which may also be thought to regulate ACTH production. Therefore, the domesticated pituitary might contain different concentrations of CRH receptors, POMC, POMC-stimulating transcription factors, or POMC convertases compared to the aggressive pituitary.
Multiple mechanisms regulate ACTH secretion through exocytosis, but the main signal for release of hormone-filled secretory vesicles is a rise in calcium from both intra- and extracellular sources, and this rise in calcium is primarily regulated by cAMP (Burgoyne and Morgan 2003). Some pituitary hormone-releasing cells may also facilitate the release of hormones into the systemic circulation by extending pseudopodia and becoming motile to contact blood vessels (Navratil et al. 2007). Additionally, pituitary endocrine cells are known to form networks, potentially via gap junctions, to coordinate hormone release (Hodson et al. 2012a,b). Therefore, the domesticated pituitary may demonstrate different levels of motility, cellular adhesion and communication, or cAMP activity as compared to the wild pituitary.
Finally, tame and aggressive pituitaries may display different population sizes of ACTH-secreting cells (corticotrophs), with a larger population associated with greater ACTH production and/or release. Stem cells in the pituitary differentiate into specialized hormone-producing cells, including corticotrophs, throughout life at a low rate (Florio 2011). Therefore, key transcription factors involved in corticotroph cell fate determination may differ in the wild and domesticated pituitary.
To identify mechanisms involved in the regulation of reduced reactivity of the HPA in domesticated animals, we analyzed gene expression in anterior pituitary of tame and aggressive farmed foxes. Previously, tame fox pituitaries have been shown to have comparable amounts of POMC mRNA and ACTH protein to aggressive fox pituitaries (Gulevich et al. 2004), despite decreased circulating concentrations of ACTH. Therefore, we specifically investigated whether the changes in ACTH release in tame foxes are associated with ACTH secretion (exocytosis) and/or corticotroph differentiation, rather than ACTH synthesis from POMC mRNA. Characterization of genes and gene networks with different anterior pituitary activity in tame and aggressive foxes will provide candidate mechanisms for differences in HPA axis regulation between domesticated and wild animals and may inform future studies into the biological underpinnings of domesticated animal behavior, such as reduced fearfulness, increased social tolerance, and increased resistance to stress.
Materials and Methods
Animals and sample preparation
Animals were maintained at the experimental farm of the Institute of Cytology and Genetics (ICG) in Novosibirsk, Russia. All animal procedures at the ICG complied with standards for humane care and use of laboratory animals by foreign institutions. The study was approved by the Institutional Animal Care and Use Committees (IACUC) of the University of Illinois at Urbana-Champaign. Anterior pituitary tissue was dissected from six tame and six aggressive sexually naive 1.5-yr-old males. The foxes were selected based on their relatedness and behavioral scores. Most of the selected foxes did not share parents and grandparents, with the exception of two aggressive foxes that had one common relative (a parent of one fox and a grandparent of another fox), and two pairs of tame foxes, each of which shared one grandparent. All of the selected foxes from the tame population belonged to the “elite” group of domesticated foxes, i.e., had the highest behavioral scores, while all selected foxes from aggressive population had the lowest (most aggressive) behavioral scores (Trut et al. 2004; Kukekova et al. 2014). Foxes were killed using sodium thiopental; immediately afterward, the brain case was opened with a saw and the brain was removed, leaving the pituitaries in situ. The anterior and posterior pituitaries were immediately dissected out and placed into RNAlater (Life Technologies, Grand Island, NY). The samples were stored at −70°. Total mRNA was extracted using RNeasy Mini Kit (Qiagen, Valencia, CA), according to the manufacturer’s protocol.
Sequencing and quality analysis
One microgram of high quality RNA from each sample was used for sequencing. Stranded RNA-seq libraries were prepared using TruSeq SBS Sequencing kit version 3 (Illumina, San Diego, CA). Libraries were barcoded and pooled and sequenced on two lanes on a HiSeq2500. Reads were single-end, stranded, and 100 nt in length. Sequencing results were processed by CASAVA 1.8 (Illumina, San Diego, CA).
Data quality, including base quality per position across reads, GC content, and distribution of sequence length, was initially assessed with FastQC (Andrews 2010). Reads were processed with flexbar (Dodt et al. 2012) in two passes: the first to trim adapters, remove low quality reads, and remove reads <35 bp in length, and the second to remove polyA tails. Subsequently, reads that mapped to fox mitochondrial DNA sequences from NCBI (accession numbers JN711443.1, GQ374180.1, NC_008434.1, and AM181037.1) using Bowtie2 (Langmead and Salzberg 2012) were discarded. Similarly, any remaining reads that mapped to ribosomal DNA sequences were discarded.
Evaluation and validation of differential expression
Reads were aligned to Canis familiaris 3.1 (CanFam3.1) with Ensembl 1.79 annotation using TopHat 2.0.13 (Kim et al. 2013). The Ensembl annotation used is available at ftp://ftp.ensembl.org/pub/release-79/gtf/canis_familiaris/Canis_familiaris.CanFam3.1.79.gtf.gz. The fox and the dog ancestor, the gray wolf, diverged ∼7–10 million yr ago (Wayne 1993). Therefore, to account for expected variation between our fox sequences and the CanFam3.1 reference genome, we added the following TopHat2 parameters to the command line, to allow a more robust cross-species alignment with the dog genome: -N 3 –read-edit-dist = 3. These parameters increased the number of nucleotide mismatches and maximum edit distance allowed per read. Gene expression levels were assessed using HTSeq-count 0.6.1p1 (Anders et al. 2015). Differentially expressed genes between samples from tame vs. aggressive foxes were evaluated using DESeq2 1.10.1 (Love et al. 2014), and results were corrected for multiple testing using the Benjamini-Hotchberg algorithm. Normalized gene counts (FPKMs) were assessed with CuffNorm 2.2.1 (Trapnell et al. 2013) using quartile normalization.
RNA from all 12 samples used for sequencing was also used for quantitative real-time PCR (RT-qPCR). Gene assays used were AKAP7, ANKRD6, BRCA2, CDH4, ITGA1, LAMA2, NELL2, PDE7B, and SPOCK1. Primers for qPCR (Table S1 in Supplemental Material, File S1), placed in adjacent exons and spanning exon-intron boundaries where possible, were designed with Primer3 (Koressaar and Remm 2007; Untergasser et al. 2012). cDNA was synthesized from 1 μg of RNA with random hexamers using the ThermoScript RT-PCR Kit (Invitrogen, Carlsbad, CA). RT-qPCR was performed on 96-well plates using an Applied Biosystems StepOnePlus Real Time PCR System (Applied Biosystems, Foster City, CA). Wells were loaded with 10 µl solution containing 5 µl SYBR Green reagent (Applied Biosystems), 0.1 µg cDNA, and either 2 mol forward and 2 mol reverse primers for the control gene or 0.6 mol forward and 0.6 mol reverse primers for the test genes. All samples were analyzed in triplicate. Initial denaturation was performed at 95° for 20 min, followed by 40 cycles of 95° for 3 sec and 60° for 3 sec, followed by a melting curve analysis at 60–95°. Gene expression was normalized to the expression of SDHA—a gene that did not show differential expression between tame and aggressive foxes and that had comparable expression between all foxes in the RNA sequencing results. Mean ΔΔ-Ct values for tame foxes were compared to mean ΔΔ-Ct values for aggressive foxes for each gene using the Mann-Whitney test.
Functional analysis
Functional analysis was performed for each tissue with ClueGO version 2.2.5 (Bindea et al. 2009); Gene Ontology (GO) files were downloaded on May 25, 2015. The ClueGO evaluation was performed against a background list of genes with a mean expression level in pooled tame and aggressive pituitary samples reported to be >0 by DESeq2. Resulting p-values were adjusted using the Benjamini-Hochberg correction. Enriched GO terms were reviewed for association with stimulus of hormone release, POMC regulation and processing, and corticotroph differentiation.
Weighted gene coexpression network analysis
Modules of genes with highly correlated expression patterns were sought using weighted gene coexpression network analysis. We expect these modules to correspond to networks of genes that interact in shared biological processes (Langfelder and Horvath 2008). The full set of genes expressed in the pituitary reads was filtered to remove genes with <10 FPKMs in each of three or more samples. We then constructed unsigned weighted gene coexpression modules using the WGCNA package in R (Langfelder and Horvath 2008). The blockwiseModules function was run with the Pearson correlation coefficient and a soft thresholding power of 12. The resulting gene groups, or modules, were named by assigning them arbitrary colors. The first principle component of each module was designated its eigengene. Eigengenes for each module were divided into samples from tame and aggressive foxes; these two groups were compared with a two-tailed Student’s t-test and their p-values were adjusted using the Benjamini-Hochberg correction. Modules selected for investigation were analyzed for gene enrichment using ClueGO as described above. Additionally, highly connected “hub” genes in these modules were identified using Cytoscape (Shannon et al. 2003) to select the genes with the largest number of connections to other genes in the same network, with the expectation that these “hub” genes would represent regulatory genes with control over many genes in that module.
Alternative splicing analysis
Differences in alternative splicing frequencies between tame and aggressive pituitary reads were explored with rMATS (Shen et al. 2014). Because rMATS requires reads of equal lengths, postfiltered reads were further filtered using SAMtools 1.1 (Li et al. 2009) to remove all reads <100 nucleotides in length. BAM files (previously aligned to CanFam3.1 for differential gene expression calling) and Ensembl annotation, version 1.79, were used with rMATS to investigate reads crossing splicing junctions. Differences in frequencies of reads including, or skipping, specific exons were used to identify exons that were skipped significantly more frequently in tame or aggressive pituitary reads. P-values were adjusted using Benjamini-Hotchberg. The list of differentially skipped exons was filtered to remove exons covered by <10 reads in either tame or aggressive samples. Differences in skipping were reported as mean percentage skipped exons, in other words, (mean number of reads skipping the exon)/(mean number of reads skipping the exon + mean number of reads including the exon). Exons identified as significantly more frequently skipped in tame or aggressive samples were further investigated by searching for their amino acid sequence using their nucleotide sequence in blastx (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify isoforms that included, or did not include, the exon in question. Conserved domains that the exon in question fell within were identified using NCBI’s Conserved Domains Database (https://www.ncbi.nlm.nih.gov/cdd/).
Data availability
The primary sequencing data have been deposited in the GenBank Sequence Read Archive (SRA) under accession number SRP107226.
Results
Gene expression in the anterior pituitary of tame and aggressive foxes
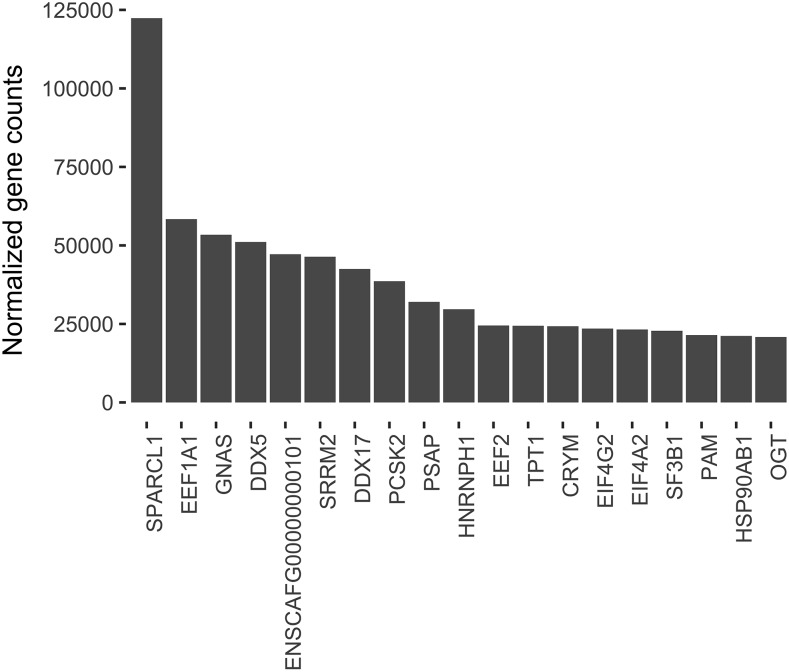
Sequencing of fox pituitaries produced 206 million reads total for six tame samples (194 million reads after filtering) and 200 million reads total for six aggressive samples (191 million reads after filtering) (Table S2 in File S1). The filtered fox reads were mapped to the Canis familiaris 3.1 reference with a mean alignment rate of 78.98% (Table S3 in File S1). The rate of multiple alignments was low, with a mean of 2.64%. Analysis of filtered pituitary reads identified 18,940 distinct genes expressed in this tissue (Table S4). Analysis of gene expression level identified 11 genes with notably higher expression (Figure 1). A GO analysis was performed on the 0.1% most highly expressed genes (180 genes). Of the most significantly enriched terms [false discovery rate (FDR) < 10.0e−9; nine terms], eight were related to RNA processing or splicing (Table S5). One of these genes, EEF1A1, was significantly more highly expressed in the tame than the aggressive fox pituitary. The most highly expressed gene, by an order of magnitude, was POMC, which codes for the precursor protein for ACTH and other peptides released by the pituitary.
Figure 1.
Most highly (0.01%) expressed genes in fox pituitary by normalized gene count, assessed by DESeq2. The most highly expressed gene, POMC, is excluded because its order of magnitude larger expression level than the next largest gene results in a scale that does not allow detailed values to be displayed for genes with smaller expression levels.
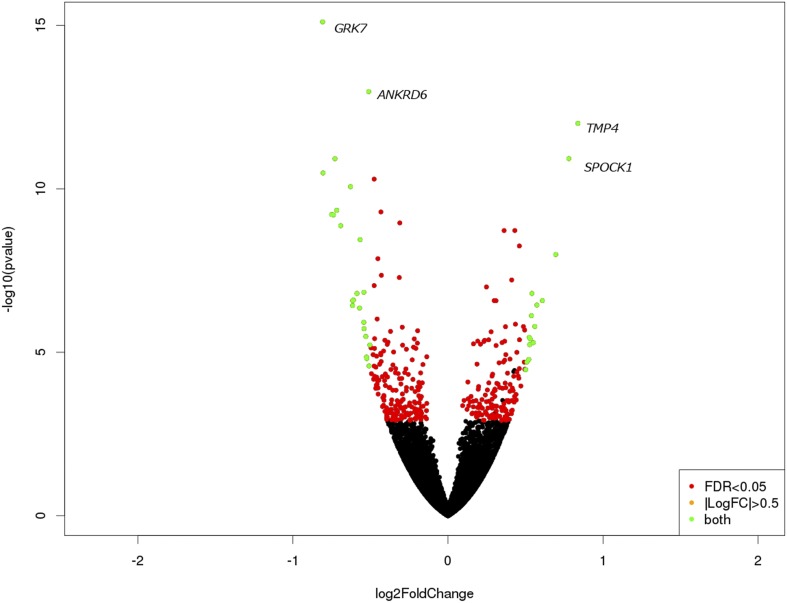
DESeq2 analysis of filtered pituitary reads identified 346 differentially expressed genes (DE genes) (FDR < 0.05); of these, 191 (55%) were upregulated in aggressive pituitary, and 155 (45%) were upregulated in tame pituitary. As predicted by Gulevich et al. (2004), POMC was not differentially expressed (expression was slightly greater in the tame samples, with log2 fold change = 0.003, FDR = 0.996). Log2 fold changes among DE genes ranged from −0.81 to 0.84 (mean −0.043, SD 0.37) (Table S6). Clustering analysis of DE genes clearly differentiated tame and aggressive samples (Figure S1 in File S1). Additional visualization using a volcano plot showed that the distribution of log2 fold changes and significance was as expected, and principal components (PC) analysis grouped tame samples separately from aggressive samples (Figure 2 and Figure S2 in File S1).
Figure 2.
Volcano plot of gene expression in fox pituitary. Red dots represent genes significantly differentially expressed in tame vs. aggressive fox lines at FDR < 0.05. Green dots represent genes that are both significantly differentially expressed and have a log2 fold change >0.5. Outlier genes are labeled with their gene symbols.
Nine DE genes (AKAP7, ANKRD6, BRCA2, CDH4, ITGA1, LAMA2, NELL2, PDE7B, and SPOCK1), with a range of FDR values and log2 fold changes, were selected for validation with quantitative PCR (qPCR). Analysis with RNA-seq data indicated that four of these genes were upregulated in aggressive foxes and five in tame foxes. Comparison of RNA-seq and qPCR results found one gene, ITGA1, which was shown to be upregulated in tame foxes according to the RNA-seq results but downregulated in tame foxes according to the qPCR results, though this result was not statistically significant. However, this gene had a very small log2 fold change in the DESeq2 output (0.21). The remaining eight of the tested genes showed expression differences in the same direction in both RNA-seq and qPCR results (see Figures S3 and S4 and Table S7 in File S1). The smallest P value (P = 0.06) in comparison of qPCR values for tame and aggressive samples was obtained for SPOCK1—the gene with the largest log2 fold change and the second smallest FDR (according to RNA-seq results) tested.
Functional analysis
ClueGO analysis:
ClueGO analysis was performed on the 328 DEG that had an associated gene symbol; a background list was used of 18,940 genes for which DESeq2 demonstrated expression in anterior pituitary. The DEG were associated with 394 GO terms, of which 74 GO terms were significantly enriched (FDR < 0.05) (Table S8).
The most significantly enriched terms in the GO analysis at FDR < 0.01 (Table 1) cluster into five groups with 11 terms total. Three of these groups are represented by a single GO term each (pseudopodium organization, regulation of dendritic cell differentiation, and granulocyte chemotaxis). The most significantly enriched GO term relates to pseudopodia—a generic term for different types of membrane projections known to function in cellular motility, as well as in release of lutenizing hormone from anterior pituitary gonadotrophs (Navratil et al. 2007). This GO term includes the gene CDC42EP2, which induces pseudopodia formation (Hirsch et al. 2001); SRGAP2 and SRGAP3, which regulate cell migration through modification of cell protrusion morphology (Guerrier et al. 2009; Endris et al. 2011); and WASH1, which promotes pseudopod extension (Zech et al. 2011).
Table 1. GO terms enriched in genes differentially expressed in tame vs. aggressive pituitary tissues at FDR < 0.01.
| Group | GO ID | Term Description | Adjusted P-Value | DEG Annotated with this Term | % Genes in this Term Differentially Expressed |
|---|---|---|---|---|---|
| 1 | GO:0031268 | Pseudopodium organization | 0.007 | CDC42EP2, SRGAP2, SRGAP3, WASH1 | 28.6 |
| 2 | GO:0007259 | JAK-STAT cascade | 0.007 | ASPN, CISH, ERBB4, FLRT1, FLRT3, IL6ST, LRRC4B, PTGER4, SOCS2, STAT3a, TNFRFSF1A | 8.3 |
| GO:0097696 | STAT cascade | 0.007 | 8.3 | ||
| GO:0046425 | Regulation of JAK-STAT cascade | 0.009 | 9.2 | ||
| GO:1904892 | Regulation of STAT cascade | 0.009 | 9.2 | ||
| 3 | GO:2001198 | Regulation of dendritic cell differentiation | 0.008 | LGALS3, LGALS9, TMEM176b | 50 |
| 4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 0.008 | RPL17, RPL23A, RPL26, RPL4, RPL7A, RPS24, RPS3A, ZFAND2Bb | 10.7 |
| GO:0006613 | Cotranslational protein targeting to membrane | 0.008 | 10.5 | ||
| GO:0045047 | Protein targeting to ER | 0.008 | 10.4 | ||
| GO:0016259 | Selenocysteine metabolic process | 0.009 | 11.7 | ||
| 5 | GO:0071621 | Granulocyte chemotaxis | 0.010 | IL17RA, IL17RC, ITGA1, LGALS3, NCKAP1L, PDE4B, TGFB2, THBS4, VAV3 | 10.1 |
STAT3 is not associated with GO:1904892.
ZFAND2B is not associated with GO:0016259.
The second of the three single-term GO groups at FDR < 0.01 is regulation of dendritic cell differentiation, which includes genes for two galectins related to cellular motility [LGALS3 and LGALS9 (Sano et al. 2000; Bi et al. 2011)] and a gene related to the cross-presentation of antigen [TMEM176B (Segovia et al. 2014)]. The last term of the single-term GO groups, granulocyte chemotaxis, contains two genes for receptors for interleukin 17A (IL-17), a pro-inflammatory cytokine, IL17RA and IL17RC (Gaffen 2009). This term also contains a galectin, LGALS3; an integrin, ITGA1; VAV3, a gene associated with cell migration and integrin signaling (Sachdev et al. 2002; Sindrilaru et al. 2009) and implicated in the formation of lamellipodia and membrane ruffles (Movilla and Bustelo 1999); PDE4B, a phosphodiesterase, related to cAMP regulation (Houslay and Adams 2003); and THBS4, related to cellular adhesion, migration, and attachment (Adams 2001; Mosher and Adams 2012).
Four terms among the most significant GO terms are related to the JAK-STAT pathway (JAK-STAT cascade, STAT cascade, regulation of JAK-STAT cascade, and regulation of STAT cascade). The JAK-STAT pathway is associated with the interleukin immune response (Subramaniam et al. 1999; Sansone and Bromberg 2012), cell migration (Pocha and Montell 2014), and the cAMP pathway (Chesnokova and Melmed 2002). Four other related GO terms from Table 1 (SRP-dependent cotranslational protein targeting to membrane, cotranslational protein targeting to membrane, protein targeting, and selenocysteine metabolic process) are related to protein targeting and signal recognition proteins (SRP). Protein targeting is a process by which proteins are translocated to an organelle identified by SRPs during translation.
GO analysis was also performed separately on genes significantly upregulated in the tame samples, and genes significantly upregulated in the aggressive samples. Three GO terms were significantly enriched for genes upregulated in the aggressive samples (Table S9); these terms had an overlapping set of DEG associated with them, including many of the genes in group 1 in Table 1. These three terms all related to pseudopodia. Twenty-seven GO terms were significantly enriched among the genes upregulated in tame samples (Table S10). Many of these terms had overlapping DEG, and could be sorted into four groups, containing genes associated with: white blood cell migration (containing genes overlapping with groups 3 and 5 in Table 1); protein targeting to the endoplasmic reticulum (containing genes overlapping with group 4 in Table 1); transcription and translation (also containing genes overlapping with group 4 in Table 1); and the humoral immune and complement response. This final group of terms contained genes not identified in any of the groups in Table 1, except for LGALS9, from group 3. The other genes in this group were BCL3, C1R, C1S, C7, CFI, PAXIP3, and SLC11A1.
Exploration of additional terms that were significantly enriched in DEG (Table S8) identified terms associated with cell migration (including pseudopodia and cytoskeletal organization), cell adhesion, exocytosis (including formation of exocytotic vesicles), cAMP regulation, and calcium regulation. Specific terms include: neutrophil migration, negative regulation of neuron migration, granulocyte migration, pseudopodium assembly, cell motility involved in cerebral cortex radial glia guided migration, neutrophil chemotaxis, myeloid leukocyte migration, regulation of neuron migration, leukocyte chemotaxis, and glial cell migration. Several terms are also included associated with cellular adhesion and cellular interactions with the extracellular matrix, critical components of cellular migration: extracellular structure organization, regulation of integrin activation, extracellular matrix organization, integrin activation, and positive regulation of cell adhesion mediated by integrin. Finally, several terms relate to biosynthesis of cAMP—an important signaling molecule in ACTH release by exocytosis (Reisine et al. 1985): cAMP metabolic process, cAMP biosynthetic process, positive regulation of cAMP biosynthetic process, positive regulation of cAMP metabolic process, and regulation of cAMP biosynthetic process.
As part of an investigation of the hypothesis that changes in the tame fox pituitary would primarily relate to hormone release, terms associated with CRH-dependent regulation of POMC transcription, processing of the POMC protein, and differentiation of corticotrophic cells were sought in the list of GO terms enriched for DEG. No terms related to these topics were found to be enriched at FDR < 0.05.
Analysis of genes involved in exocytosis, POMC regulation, and cell differentiation:
DEG related to exocytosis, regulation of POMC expression, and cell differentiation were sought, to investigate which of these processes were more extensively represented in the differences in gene expression between tame and aggressive fox pituitaries (Table 2). The gene with the largest log2 fold change was GRK7 (upregulated in aggressive pituitary), a member of the G-protein coupled receptor kinase family. Genes from this family are known to serve as important regulators of phosphorylation of the CRH receptor, activation of which triggers ACTH synthesis and release (Grammatopoulos 2012). AVP, which codes for arginine vasopressin—a neuropeptide with paracrine activity in the anterior pituitary known to potentiate ACTH release (Liu et al. 1994) but not synthesis (Levin et al. 1989)—was upregulated in the tame pituitary. DEG associated with membrane proteins of exocytotic vesicles included VAMP1 (Calakos et al. 1994), and two synaptotagmins [SYT10 and SYT13 (Brose et al. 1992)]. Therefore, tame and aggressive pituitaries demonstrated potential differences in regulation of ACTH release via CRH receptor regulation, vasopressin signaling, and exocytotic vesicle formation, as well as potential differences in ACTH synthesis via CRH receptor regulation.
Table 2. DEG in tame vs. aggressive fox pituitary tissue involved in exocytosis, POMC regulation, and cell differentiation.
| Gene Name | FDR | Log2 Fold Change |
|---|---|---|
| GRK7 | 1.17e−011 | −0.81 |
| NPR3 | 8.46e−005 | −0.59 |
| CALCRL | 1.02e−005 | −0.45 |
| SYT10 | 0.04 | −0.39 |
| PDE7B | 0.02 | −0.25 |
| AKAP7 | 0.002 | −0.23 |
| RAPGEF4 | 0.03 | −0.21 |
| VAMP1 | 0.04 | 0.28 |
| PDE4B | 0.02 | 0.28 |
| SYT13 | 0.01 | 0.33 |
| ADCY7 | 0.04 | 0.38 |
| AVP | 0.03 | 0.41 |
| ADCY8 | 0.01 | 0.42 |
Results are ordered by log2 fold change. Negative log2 fold change values indicate genes upregulated in aggressive samples; positive log2 fold change values indicate genes upregulated in tame samples.
Regulation of cAMP was specifically investigated, as cAMP serves as a major cellular signal for exocytosis. DEG associated with cAMP regulation included two members of the adenylyl cyclase family of genes that synthesize cAMP (ADCY7 and ADCY8); two members of the phosphodiesterase family of genes that break down cAMP (PDE4B and PDE7B); G-protein receptors that activate adenylyl cyclase [CALCRL/CRLR (Smith et al. 2002) and P2RY11 (Qi et al. 2001)]; a G-protein receptor which inhibits adenylyl cyclase [NPR3 (Anand-Srivastava 2005)]; a guanine nucleotide exchange factor that functions in cAMP-dependent exocytosis [RAPGEF4/EPAC2 (Sugawara et al. 2016)]; and a scaffolding protein involved in cAMP compartmentalization [AKAP7/AKAP15 (Kritzer et al. 2012)]. Additionally, the previously discussed DEG AVP is known to act through the cAMP/PKA pathway in the anterior pituitary (Liu et al. 1994) in its effects on POMC regulation. Therefore, differences in cAMP regulation are represented in multiple ways in this data set, from synthesis to metabolism to compartmentalization.
Notably, genes important in CRH-dependent regulation of POMC transcription, such as NGFIB, NURR1, and NOR1 (Bonfiglio et al. 2011), were not differentially expressed, nor were genes associated with processing of POMC into ACTH, such as PC1 and PC2 (Benjannet et al. 1991), CPE (Cool and Loh 1998), and PAM (Ciccotosto et al. 1999).
Genes associated with pituitary, and, specifically, corticotroph (ACTH-producing), cell differentiation were investigated to determine the likelihood that tame and aggressive pituitaries have different population sizes of corticotrophic cells. No genes directly associated with corticotroph differentiation were identified as significantly differentially expressed. However, TBX19/TPIT, a gene known to be essential in the establishment of the POMC-expressing lineage of corticotrophs and melanotrophs (Pulichino et al. 2003), was significantly upregulated in aggressive pituitary before adjustment at P = 0.047 (FDR = 0.32). PAX7, a gene known to suppress corticotroph differentiation in favor of melanotroph differentiation, (Budry et al. 2012), was also significantly upregulated in aggressive pituitary only before adjustment (P = 0.03, FDR = 0.28).
Weighted gene coexpression network analysis results
Weighted gene coexpression network analysis constructed 66 gene modules with activity in fox anterior pituitary. Two modules, Antique White 4 and Pale Turquoise, had significantly (FDR < 0.10) different eigengenes between tame and aggressive samples (Table 3). Both modules had a negative t-statistic, indicating that they are more active in aggressive than tame samples.
Table 3. Modules of coexpressed gene networks with significantly different activation in tame vs. aggressive fox pituitary tissue.
| Module Name | t-Statistic | FDR | Number of Genes in Module | Percent Genes in Module Differentially Expressed |
|---|---|---|---|---|
| Antique White 4 | −4.2 | 0.09 | 43 | 18.6 |
| Pale Turquoise | −4.1 | 0.09 | 80 | 20.0 |
The Antique White 4 module had eight DEG in total, including SYT10, a calcium sensor involved in exocytosis of secretory vesicles (Cao et al. 2011); RNPEP, an aminopeptidase (Piesse et al. 2002); and two genes associated with apoptosis, CPPED1/CSTP1 (Zhuo et al. 2013) and TRAF4 (Sax and El-Deiry 2003). (For DEG, see Table S6; for genes in WGCNA modules, see Table S11 and Table S12) The Antique White 4 module had no enriched GO terms, possibly due to the small number of genes (43) it contains. Hub (i.e., highly connected) genes in this module included an aminopeptidase, RNPEP (Piesse et al. 2002), which was both the most highly connected gene and upregulated in the aggressive pituitary; ZIC5, associated with neural crest cell differentiation (Nakata et al. 2000; Inoue et al. 2004); SUPT6H/SPT6, a transcription elongation factor (Diebold et al. 2010); KLHL2, involved with cell membrane process formation (Williams et al. 2005); and ATG5, involved with autophagic vesicle formation (Walczak and Martens 2013). Additional genes in this module associated with exocytosis, cell migration, and cell–cell adhesion include ANXA7 (Caohuy et al. 1996), LYN (Malik et al. 2008; Nakata et al. 2006), and RAP1B (Mandell et al. 2005); both LYN and RAP1B are also associated with integrin signaling (Malik et al. 2008; Mandell et al. 2005). Overall, this module demonstrated relationships to processes associated with exocytosis, apoptosis, vesicle formation, cell migration, and cell–cell adhesion.
The Pale Turquoise module had 16 DEG. These included a number of genes associated with exocytosis and migration, such as genes associated with the anchoring or biosynthesis of cAMP [AKAP7 (Kritzer et al. 2012) and FLNB (del Valle-Pérez et al. 2010)]; AVP, a signaling hormone for ACTH release (Denef 2008); an integrin and a laminin, two proteins known to work together in the formation of pseudopodia [ITGA6 (Mercurio et al. 2001) and LAMA2 (Gu et al. 2001)]; and a cadherin-associated cell–cell adhesion gene, PTPRT (Besco et al. 2006). There were two significantly (P < 0.01) enriched GO terms for this module, regulation of cytokinesis and intrinsic apoptotic signaling pathway (Table S13). Hub genes in this module included genes associated with puberty [WDR11 (Kim and Layman 2011)] and carnosine peptidase activity [CNDP1/CN1 (Teufel et al. 2003)]. Hub genes that were also differentially expressed included LAMA2, PTPRT, and WLS [Wnt protein regulation and secretion (Bänziger et al. 2006)]. Therefore, this module also had strong associations to genes involved in exocytosis and migration, as well as apoptosis.
Notably, the Brown module, which showed increased activation in tame samples only before correction (P = 0.03, FDR = 0.20) suggested functionality related to alternative splicing of mRNA. This module was associated with three GO terms significantly enriched for module genes that were related to this function, mRNA 3′-splice site recognition, regulation of RNA splicing, and alternative mRNA splicing, via spliceosome. The Brown module additionally contained multiple hub genes associated with mRNA alternative splicing and regulation, including TSEN2/SEN2, TNKS1BP1/TAB182, and PRCC (Trotta et al. 1997; Skalsky et al. 2001; Lau et al. 2009). The mRNA splicing functionality in this module may be associated with cell motility, as significantly enriched GO terms in the Brown module also included the terms lamellipodium organization, ruffle organization, regulation of fibroblast migration, regulation of focal adhesion assembly, wound healing/spreading of cells, and negative regulation of cell-substrate adhesion. Lamellipodia are pseudopodia formed on the leading edge of migrating cells (Krause and Gautreau 2014); membrane ruffling allows the cell to form a motile surface to network with other cells (Ridley 1994). The Brown module had a positive t-statistic of 2.7, indicating possible greater activation in tame than aggressive samples.
Overall, gene network analysis with WGCNA demonstrated two modules, Antique White 4 and Pale Turquoise, with significantly increased activity in aggressive samples. These modules were associated with exocytosis, cell migration, and cell adhesion; both modules contained genes suggestive of the importance of integrins in these functionalities. Both modules also indicated the importance of apoptosis, suggesting that increased cellular turnover may have some significance in differing pituitary function between the lines. Finally, analysis of the Brown module suggested the importance of alternative splicing of mRNA in anterior pituitary, which may or may not show greater activity in the tame population. For this reason, an alternative splicing analysis was undertaken.
Alternative splicing analysis
rMATS was used to identify 36 genes with exons skipped at different frequencies in tame compared to aggressive fox pituitary reads at FDR < 0.05 (Table S14), and 12 genes at FDR < 0.001 (Table 4). Exon counts for these 36 genes with significantly differentially skipped exons ranged from 4 to 46 exons total (median 13, SD 9.3). Of the genes with differentially skipped exons, only one [MTHFSD, an RNA-binding protein associated with amyotrophic lateral sclerosis (MacNair et al. 2016)] is itself differentially expressed in pituitary reads. Investigation of GO terms revealed that nine terms enriched for DEG also contained genes with differentially skipped exons. Of these, four terms contained multiple genes with differentially skipped exons: establishment of protein localization to organelle (ABLIM3, SH2D4A, POT1; this term is functionally related to the highly significantly enriched group of four terms related to protein targeting); gliogenesis (ADAM22, NFIB); and extracellular matrix organization and extracellular structure organization (both containing KLKB1 and MFAP5). Nine genes with differentially skipped exons were also included in WGCNA modules associated with the state of being tame, including three genes in the Brown module (DAGLB, MYCBPAP, and STAUI). GO enrichment analysis of the genes with differentially skipped exons revealed no enriched terms.
Table 4. Genes containing exons skipped at different frequencies in tame compared to aggressive fox pituitary reads (FDR < 0.001), including conserved domains overlapping the skipped exon.
| Gene | Exon Location | Percent Reads Skipping Exon (Tame) | Percent Reads Skipping Exon (Aggr) | FDR | Skipped in Isoforms | In Conserved Domain(s) |
|---|---|---|---|---|---|---|
| MFAP5 | chr27:37,034,668-37,034,704 | 46 | 0 | 0 | X2, X3 | MAGP |
| SPICE1 | chr33:17787918-17788056 | 13 | 0 | 2.75E−10 | None | SPICE |
| MECR | chr2:71398391-71398452 | 0 | 9.8 | 1.39E−07 | None | ETR, Qor |
| SPATA20 | chr9:26,472,931-26,473,143 | 12 | 1.9 | 5.57E−06 | X4 | YyaL |
| KLKB1 | chr16:44493733-44493873 | 20 | 0 | 7.98E−00 | None | Tryp_SPc |
| THYN1 | chr5:996686-996755 | 12 | 2.3 | 1.81E−05 | None | Eve |
| FBRSL1 | chr26:542234-542303 | 13 | 2.8 | 1.29E−04 | None | None |
| TRIM65 | chr9:4716736-4716911 | 14 | 2.8 | 1.40E−04 | None | None |
| TMEM198 | chr37:26056452-26057028 | 2.6 | 0.31 | 3.22E−04 | None | DUF4203 |
| GANC | chr30:9295384-9295464 | 10 | 1.8 | 3.60E−04 | None | GH31_GANC_GANAB_alpha, Glyco_hydro_31 |
| SORBS1 | chr28:8892641-8892725 | 67 | 93 | 3.76E−04 | X7, X8, X12 | None |
| TCFL5 | chr24:46680735-46680982 | 7.1 | 1.3 | 8.40E−04 | None | None |
Of the genes listed in Table 4, MFAP5/MAGP-2 promotes cell adhesion and spreading via integrin binding (Gibson et al. 1999), and SORBS1/CAP is involved in integrin-mediated cell adhesion (Ribon et al. 1998). At FDR < 0.05, several genes related to cell migration, cell–cell adhesion, and calcium signaling contained differentially spliced exons. These included: ADAM22, a disintegrin (D’Abaco et al. 2006); CFAP97/HMW, associated with cilia and flagella (Soulavie et al. 2014); EFMA5, integrin-dependent cell-cell adhesion (Davy and Robbins 2000); ENKUR, calcium signaling and flagella (Sutton et al. 2004); ERG, adhesion and cell morphology change (McLaughlin et al. 2001); UNC5D, cell-cell adhesion/repulsion and motility (Jarjour et al. 2011). Exons alternatively spliced at FDR < 0.05 in genes related to RNA processing included ELAVL4 (Ince-Dunn et al. 2012) and FBRSL1 (Baltz et al. 2012). Additional interesting genes just above significance level were RPH3AL/NOC2 (P = 0.10), associated with exocytosis (Matsunaga et al. 2017), and POLK (P = 0.07), a DNA repair polymerase (Ogi and Lehmann 2006).
Discussion
RNA sequencing of pituitary tissue from tame and aggressive foxes identified expression differences in genes related to cAMP regulation, pseudopodia formation, and cell–cell adhesion. These findings suggest that the pituitaries of experimentally domesticated foxes may differ from the pituitaries of a lineage of foxes selected for increased aggression in regulation of exocytosis. Specifically, cAMP signaling for vesicle release may be modified in the tame foxes to reduce the release of ACTH during the stress response. Additionally, tame fox pituitary cells may differ in their motility or their ability to form networks for cell–cell communication, potentially to coordinate ACTH release, as indicated by differences in the expression of genes related to the organization of pseudopodial membrane extensions and cell adhesion. Notably, significant differences in lineage specification markers were not found between the two populations, suggesting that differentiation of pituitary cells involved in the HPA axis did not contribute to the differences in function between the tame and aggressive pituitary. However, nonsignificant differences were seen, and, due to the small relative size of the population of actively differentiating cells in the anterior pituitary (Florio 2011), significant differences may be difficult to detect in a mixed sample of cell populations such as this one. Additionally, one gene coexpression module suggested differences in alternative splicing functionality between tame and aggressive fox lines, and differentially skipped exons were identified in genes associated with cell migration, cell–cell adhesion, and calcium signaling. Therefore, differential splicing in the tame fox pituitary may affect similar functionality as do gene expression differences.
Alignment and pituitary gene expression
POMC was by far the most highly expressed gene in the pituitary, with an expression level an order of magnitude higher than the next most highly expressed gene. The robust expression of POMC was expected in anterior pituitary tissue, as it codes for a prohormone that is spliced into the primary hormonal product of both melanotrophs and corticotrophs (Drouin 2016). PCSK2, a convertase for processing POMC (Chrétien and Mbikay 2016), was also highly expressed. The third and fourth most highly expressed genes, EEF1A1, a translation elongation factor that serves to target actin mRNA to pseudopodial cell projections for translation (Liu et al. 2002), and GNAS, an adenylyl cyclase regulator (Weinstein et al. 2004), represented interesting findings in light of the importance of pseudopodium formation and cAMP regulation in differences between tame and aggressive fox pituitary function described by other analyses in this study. Of particular interest was the significant upregulation of EEF1A1 in the tame fox pituitary. Other highly expressed genes were related to transcription and mRNA splicing [DDX5/P68, SRRM2, DDX17, and HNRNPH1 (Jurica et al. 2002; Lin et al. 2005; Fuller-Pace and Ali 2008; Russo et al. 2010; Wang et al. 2012)]. Along with the highly enriched GO terms associated with RNA processing and splicing for the top 0.1% most highly expressed genes, these findings suggest that the fox pituitary is highly active in RNA production. This enrichment is noteworthy due to the association of the Brown module with mRNA splicing functionality and the differentially spliced exons identified in the two populations of foxes.
Pseudopodia, cell motility, and networks
GO analysis identified pseudopodium organization as the most significant term associated with DEG in pituitary tissue, and an analysis separating upregulated genes in tame from aggressive samples identified enriched terms related to pseudopodium organization specifically in aggressive samples. In addition to the DEG included in this GO term, several genes known to be important for pseudopodium formation and functioning are not included in this term but are differentially expressed between pituitary samples of tame and aggressive foxes. These genes code for laminins (LAMA2 and LAMA5), a major component of the extracellular protein matrix, and integrins (ITGA1 and ITGA6), transmembrane receptors known to frequently interact with laminins to promote the formation of pseudopodia (Gu et al. 2001), and to function in epithelial cell migration through pseudopod formation (Mercurio et al. 2001). Two cadherin-family genes, CDH4 (R-cadherin or retinal cadherin) and CDH13 (T-cadherin, H-cadherin, or heart cadherin), are also differentially expressed; cadherins are implicated in cell motility, adhesion, and morphological changes, and these two cadherins particularly have been implicated in vascular remodeling (Dorrell et al. 2002; Ivanov et al. 2004). Weighted gene coexpression network analysis similarly described functionality related to cell migration and adhesion, particularly in the Antique White 4, Pale Turquoise, and Brown modules, with statistically significant increases in activity in the first two modules associated with aggressive samples. Significantly alternatively spliced genes in the tame vs. aggressive fox pituitary also demonstrated associations to integrins [MFAP5 (Gibson et al. 1999)] and cell adhesion [SORBS1 (Ribon et al. 1998); EFNA5 (Davy and Robbins 2000); UNC5D (Jarjour et al. 2011)]. Together, these results demonstrate a recurrent theme of cell adhesion and pseudopodia formation throughout analyses of differential gene expression, GO term enrichment, weighted gene coexpression network analysis, and alternative splicing analysis, suggesting that this finding is a robust one.
Regulation of dendritic cell differentiation—another GO term highly significantly enriched for differentially expressed genes in the tame vs. aggressive pituitary—also contains genes associated with pseudopodia. Dendritic cells are antigen-presenting white blood cells that pass through the final stages of differentiation in tissues after being activated by exposure to antigen (Ziegler-Heitbrock et al. 2010). Enrichment in genes related to regulation of their differentiation in the pituitary is a surprising finding; examination of the specific genes reveals two galectins related to cellular motility [LGALS3, LGALS9 (Sano et al. 2000; Bi et al. 2011)] and an intracellular protein associated with cross-presentation of antigen [TMEM176B (Segovia et al. 2014)]. These findings may result from the migration of dendritic cells through the bloodstream to the pituitary gland and subsequent differentiation there. However, galectins and TMEM176B are not solely found in dendritic cells but are represented in numerous other tissues (Wada et al. 1997; Stillman et al. 2006; Cuajungco et al. 2012). Their functionalities are not limited to dendritic cell differentiation; for example, galectin 3 has been associated with vesicle formation (Mehul and Hughes 1997) and localizes in dendritic cell lamellipodia (a type of pseudopodium) during migration (Hsu et al. 2009); galectin 9 associates with integrins to regulate cell adhesion (Kasamatsu et al. 2005; Nobumoto et al. 2008). Therefore, while the enrichment of a GO term related to dendritic cell differentiation is difficult to explain in relationship to pituitary functionality, examination of specific DEG reveals functionality related to vesicle formation (associated with exocytosis) and pseudopodia formation and cell adhesion (associated with cellular networking of hormone-releasing cells).
Another highly enriched GO term, granulocyte chemotaxis, again describes a cell type (granulocytic white blood cell) that is not expected to be present in large quantities in the pituitary. This term contains two genes for receptors for interleukin 17A (IL-17), a pro-inflammatory cytokine, IL17RA and IL17RC (Gaffen 2009). While IL-17 is most commonly expressed in T cells, it is also widely expressed in other tissues, including epithelial tissue (Chesné et al. 2014), which is the tissue type of the anterior pituitary (Conklin 1968). Interleukins signal through the JAK-STAT pathway (Subramaniam et al. 1999; Sansone and Bromberg 2012), which is also represented by enriched GO terms (described below). IL-17 has different functions in different tissues, so its function in pituitary is difficult to ascertain, but it is known to enhance the production of tight junctions in epithelial tissue (Cua and Tato 2010). Tight junctions can control cell permeability and are associated with cell signaling and cell–cell adhesion (Steed et al. 2010). In addition to receptors for the pro-inflammatory cytokine IL-17, this term also includes the pro- and anti-inflammatory cytokine TGFB2. Inflammatory cytokines, released during the acute phase of inflammation, have effects on the brain leading to behavioral changes. Specifically, they are known to cause “sickness behavior,” including social withdrawal (Parnet et al. 2002, Dantzer et al. 2008). They also interact with the hypothalamic-pituitary-adrenal axis, both regulating, and being regulated by, glucocorticoid release (Parnet et al. 2002). This term may therefore relate more to cell migration, adhesion, cytokine effects on behavior, and hormone release than to granulocyte chemotaxis in this tissue.
These findings are suggestive of differential function of pseudopodia, mediated by differences in integrin, laminin, and galectin expression, in tame and aggressive fox pituitaries. Pituitary lactotrophs and gonadotrophs are known to form pseudopodia. These cellular protrusions may function to both directly contact blood vessels and to increase cell motility to better position endocrine cells near blood vessels for hormone release (Navratil et al. 2007). Pseudopodia may also function in the organization of anterior pituitary cellular networks for regulation of hormone release, allowing a small number of cells to coordinate the timing of hormone release to the bloodstream (Hodson et al. 2012a,b). These functions, while described in other pituitary endocrine cells, may be similar in ACTH-releasing corticotrophs. Notably, upregulation of pseudopodia-associated genes was largely observed in aggressive foxes, which are characterized by higher ACTH blood level relative to tame foxes.
Regulation of exocytosis
cAMP, a major mediator of exocytosis (Seino and Shibasaki 2005), was associated with a variety of DEG in the tame vs. aggressive fox pituitary. Of the 10 most significant GO terms associated with DEG in pituitary tissue, four are associated with regulation of cAMP. Other DEG not included in these GO groups are also associated with cAMP regulation, including two members of the family of phosphodiesterases involved in metabolism of cAMP: PDE4B and PDE7B. Interestingly, while PDE4B is upregulated in the tame pituitary, PDE7B is upregulated in the aggressive pituitary, suggesting a difference in function between the two proteins. Two members of the adenylyl cyclase family of proteins involved with synthesis of cAMP—ADCY7 and ADCY8—are both upregulated in the tame pituitary; along with phosphodiesterases, this family of proteins represents the major regulator of cAMP concentration in the cell. Adenylyl cyclase function is additionally regulated by the DEG CALCRL (a member of the Brown gene coexpression module), P2RY11 (also a member of the Brown module), and RAPGEF4 (Smith et al. 2002; Qi et al. 2001; Anand-Srivastava 2005; Sugawara et al. 2016). A scaffolding protein, AKAP7 (a member of the Pale Turquoise module), is upregulated in the aggressive pituitary. AKAPs serve as anchoring proteins to spatially restrict cAMP activation (Kritzer et al. 2012), and are also known to bind PDE4s to further spatially regulate cAMP (McCahill et al. 2005; Dodge et al. 2001; Taskén et al. 2001). The cAMP regulation pathway therefore appears to be perturbed in tame pituitaries compared to aggressive pituitaries at several different levels. This supports the hypothesis that release of ACTH through cAMP-mediated exocytosis is an important factor in the different ACTH concentrations in tame and aggressive fox circulations.
The JAK-STAT pathway also appeared to be differentially regulated in tame vs. aggressive pituitaries; this pathway has multiple associations with cAMP and cell adhesion. Among the most significant GO terms (Table 1), four GO terms are related to the JAK-STAT pathway. The JAK-STAT pathway, besides its relationship to IL-17 as previously mentioned, is known to regulate cell migration in epithelial tissues (Pocha and Montell 2014). In the pituitary, JAK-STAT signaling operates synergistically with the cAMP pathway by mediating gp130 cytokine genes such as IL-6, LIF, IL-11, and CNTF to regulate POMC expression and ACTH release (Chesnokova and Melmed 2002) even in the absence of CRH (Kariagina et al. 2004). Of the 11 DEG in the tame and aggressive fox pituitaries associated with this term, four (ERBB4, FLRT1, FLRT3, and LRRC4B) are also implicated in cell–cell adhesion and cellular migration (Gambarotta et al. 2004; Jackson et al. 2016; Yamagishi et al. 2011; Wu et al. 2008). All four are also upregulated in the aggressive pituitary. This pathway appears to link IL-17 signaling, associated with the highly significant enriched GO term granulocyte chemotaxis, with cAMP signaling.
Genes associated with secretory vesicles are also implicated in differences between tame and aggressive fox pituitaries. Two members of the synaptotagmin family of membrane trafficking proteins (Südhof 2002) are differentially expressed: SYT10, upregulated in the aggressive pituitary, and SYT13, upregulated in the tame pituitary. SYT10 is a calcium sensor required for exocytosis of secretory vesicles (Brose et al. 1992); this gene is implicated in IGF-1 co-secretion with ACTH (Cao et al. 2011), suggesting IGF-1 functionality in regulation of the stress response at the level of the pituitary (Eppler et al. 2007). SYT10 is also a hub gene in the Antique White 4 module. SYT13 is an atypical synaptotagmin, lacking specific amino acid residues that are conserved in other family members, which may limit its calcium binding activity; though its function is not well understood, the SYT13 protein has been associated with constitutive vesicular transport, and shown to be upregulated in contextual fear conditioning (Han et al. 2012). Another DEG associated with secretory vesicle function is VAMP1 (synaptobrevin), a membrane protein with binding specificity for vesicular proteins (Calakos et al. 1994); this gene is upregulated in the tame pituitary. Overall, genes associated with exocytosis, both at the level of vesicle function and at the level of cAMP, display differences in expression between tame and aggressive fox pituitaries, not just individually, but in enriched GO terms and gene coexpression modules.
Regulation of ACTH release
The most significantly DEG, as well as the gene with the largest log2 fold change, was GRK7—a member of a gene family containing important regulators of CRH receptor function (Grammatopoulos 2012)—upregulated in the aggressive pituitary. Activation of the CRH receptor stimulates ACTH synthesis and release. Notably, however, the G protein-coupled receptor kinase (GRK) family is also associated with regulation of cell motility, including pseudopodium formation and cell adhesion (Chodniewicz and Klemke 2004; Penela et al. 2014). GRK7 represents a potential candidate gene for future studies of the functional differences between the tame and aggressive fox pituitary.
The AVP gene, which codes for the vasopressin protein, is significantly upregulated in the tame fox pituitary. Vasopressin is known to potentiate ACTH release, but not POMC production, in the anterior pituitary (Levin et al. 1989), both in endocrine fashion through release from the posterior pituitary into the systemic circulation, and in paracrine fashion through local synthesis and release (Denef 2008). However, interspecies analysis of adjacent, recently diverged genes such as AVP and OXT (Acher 1980) introduces the potential to conflate paralogy and homology, because sequence divergence has occurred between gene copies both within each species as well as within genes but between species. As a result, some fox OXT reads may have inappropriately mapped to dog AVP, resulting in apparent upregulation due to artifacts of the alignment algorithm rather than biological shifts. Therefore, the apparent upregulation of AVP should be treated cautiously until more work can more clearly define differences between dog and fox AVP sequences.
Protein targeting
Four highly significantly enriched GO terms (see Table 1) are related to protein targeting and signal recognition proteins (SRP), and genes in these terms appear to be upregulated in tame rather than aggressive pituitary tissues (see Table S10). These terms all describe processes controlling the translocation of proteins during translation to the organelle where they will ultimately reside. Notably, however, all but one of the DEG associated with these terms are associated with ribosomal proteins, i.e., with protein elongation rather than protein translocation. The remaining gene, ZFAND2B, is associated with protein folding as well as translocation (Glinka et al. 2014). Interestingly, all of these genes are up regulated in the tame fox. This finding is surprising, as the aggressive pituitary is believed to release more ACTH and therefore might be expected to synthesize a greater number of proteins related to hormone synthesis and release.
Complexity of behavioral regulation
The observed differences between DEG in pituitaries from the tame and aggressive lines were less than twofold. A relatively small magnitude of changes was expected due to the outbred nature of these lines of foxes and complex genetic inheritance of their behavioral differences (Kukekova et al. 2012, Johnson et al. 2015; Nelson et al. 2017). Additionally, differences in gene expression between the two lines may not be a precise indicator of differences in protein expression, or of protein activity. A comparison of intraspecific differences in gene and protein between humans and chimpanzees suggested that protein expression levels are under stronger evolutionary constraints than are gene expression levels, and, therefore, a change in gene expression may not always indicate a corresponding change in protein expression (Khan et al. 2013). Protein activity, moreover, may be modified not just by gene or protein expression changes, but by translational regulation or, post-translationally, of protein modifications such as phosphorylation, glycosylation, lipidation, ubiquitination, and acetylation. Such modifications may affect protein stabilization, activation, folding, and interaction with other proteins (Deribe et al. 2010), and, thereby, may potentially significantly affect cellular processes without an accompanying perturbation of gene expression. Therefore, the transcriptome study presented here identified genes and gene pathways likely implicated in behavioral and physiological differences between tame and aggressive foxes, but further proteomic studies are needed to elucidate post-transcriptional differences between the two lines.
Conclusions
Experimentally domesticated foxes provide a novel model for the study of the behavioral phenotype of domesticated animals. An attenuated ACTH response to psychogenic stressors has been shown in the tame foxes, but the cellular mechanisms resulting in the inhibition of this response are not understood. Differences in ACTH concentrations in tame and aggressive fox blood have been demonstrated (Trut et al. 2004), but both POMC mRNA and ACTH protein concentrations in tame and aggressive fox pituitaries are similar (Gulevich et al. 2004). These results suggest that differences between the two lines may lie in regulation of ACTH release through exocytosis rather than regulation of ACTH synthesis and processing or in corticotroph cell number. In this study, examination of gene expression through sequencing of mRNA demonstrated differences in formation of pseudopodia and in signaling by cAMP. Pseudopodia formation have been implicated in release of pituitary hormones, such as lutenizing hormone, into blood vessels and in the formation of regulatory networks for coordinating hormone release by endocrine cells, while cAMP is a major regulator of exocytosis.
We have demonstrated here for the first time the possibility that genes in networks related to cAMP regulation, cell adhesion, and pseudopod formation may have been targeted in selection for the domesticated phenotype. Although our findings are in the experimentally domesticated fox, the similarity of phenotypes across domesticated species suggest the possibility of selection on a shared set of gene pathways. Therefore, our findings may provide new avenues for investigation of the biological differences between domesticated and wild species.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300508/-/DC1.
Acknowledgments
We thank Irina V. Pivovarova, Tatyana I. Semenova, Eugene A. Martinov, and all the animal keepers at the Institute of Cytology and Genetics (ICG) experimental farm for research assistance. We are grateful for the insights of Andrew G. Clark, Xu Wang, Lisa Stubbs, Michael Saul, Jonathan E. Beever, and Waqar Yarif. This project was supported by National Institutes of Health (grant number GM120782), the United States Department of Agriculture Federal Hatch Project (grant number 538922), a Campus Research Board grant from the University of Illinois at Urbana-Champaign, the Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences (grant number 0324-2018-0016; animal maintenance), the Russian Scientific Foundation (grant number 16-14-10216; data collection and analysis), the Jonathan Baldwin Turner Scholarship Program (to J.P.H.), and personal support from Temple Grandin (to J.P.H.).
Footnotes
Communicating editor: D. Bannasch
Literature Cited
- Acher R., 1980. Molecular evolution of biologically active polypeptides. Proc. R. Soc. Lond. B Biol. Sci. 210: 21–43. 10.1098/rspb.1980.0116 [DOI] [PubMed] [Google Scholar]
- Adams J. C., 2001. Thrombospondins: multifunctional regulators of cell interactions. Annu. Rev. Cell Dev. Biol. 17: 25–51. 10.1146/annurev.cellbio.17.1.25 [DOI] [PubMed] [Google Scholar]
- Albert F. W., Shchepina O., Winter C., Römpler H., Teupser D., et al. , 2008. Phenotypic differences in behavior, physiology and neurochemistry between rats selected for tameness and for defensive aggression towards humans. Horm. Behav. 53: 413–421. 10.1016/j.yhbeh.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Anand-Srivastava M. B., 2005. Natriuretic peptide receptor-C signaling and regulation. Peptides 26: 1044–1059. 10.1016/j.peptides.2004.09.023 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S., 2010. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc Accessed December 1, 2017.
- Baltz A. G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., et al. , 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46: 674–690. 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Bänziger C., Soldini D., Schütt C., Zipperlen P., Hausmann G., et al. , 2006. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125: 509–522. 10.1016/j.cell.2006.02.049 [DOI] [PubMed] [Google Scholar]
- Benjannet S., Rondeau N., Day R., Chrétien M., Seidah N. G., 1991. PC1 and PC2 are proprotein convertases capable of cleaving proopiomelanocortin at distinct pairs of basic residues. Proc. Natl. Acad. Sci. USA 88: 3564–3568. 10.1073/pnas.88.9.3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besco J. A., Hooft van Huijsduijnen R., Frostholm A., Rotter A., 2006. Intracellular substrates of brain-enriched receptor protein tyrosine phosphatase rho (RPTPρ/PTPRT). Brain Res. 1116: 50–57. 10.1016/j.brainres.2006.07.122 [DOI] [PubMed] [Google Scholar]
- Bi S., Hong P. W., Lee B., Baum L. G., 2011. Galectin-9 binding to cell surface protein disulfide isomerase regulates the redox environment to enhance T-cell migration and HIV entry. Proc. Natl. Acad. Sci. USA 108: 10650–10655. 10.1073/pnas.1017954108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., et al. , 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093. 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfiglio J. J., Inda C., Refojo D., Holsboer F., Arzt E., et al. , 2011. The corticotropin-releasing hormone network and the hypothalamic-pituitary-adrenal axis: molecular and cellular mechanisms involved. Neuroendocrinology 94: 12–20. 10.1159/000328226 [DOI] [PubMed] [Google Scholar]
- Brose N., Petrenko A. G., Südhof T. C., Jahn R., 1992. Synaptotagmin: a calcium sensor on the synaptic vesicle surface. Science 256: 1021–1025. 10.1126/science.1589771 [DOI] [PubMed] [Google Scholar]
- Budry L., Balsalobre A., Gauthier Y., Khetchoumian K., L’honoré A., et al. , 2012. The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 26: 2299–2310. 10.1101/gad.200436.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R. D., Morgan A., 2003. Secretory granule exocytosis. Physiol. Rev. 83: 581–632. 10.1152/physrev.00031.2002 [DOI] [PubMed] [Google Scholar]
- Calakos N., Bennett M. K., Peterson K. E., Scheller R. H., 1994. Protein–protein interactions contributing to the specificity of intracellular vesicular trafficking. Science 263: 1146–1149. 10.1126/science.8108733 [DOI] [PubMed] [Google Scholar]
- Campler M., Jöngren M., Jensen P., 2009. Fearfulness in red junglefowl and domesticated White Leghorn chickens. Behav. Processes 81: 39–43. 10.1016/j.beproc.2008.12.018 [DOI] [PubMed] [Google Scholar]
- Cao P., Maximov A., Südhof T. C., 2011. Activity-dependent IGF-1 exocytosis is controlled by the Ca(2+)-sensor synaptotagmin-10. Cell 145: 300–311. 10.1016/j.cell.2011.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caohuy H., Srivastava M., Pollard H. B., 1996. Membrane fusion protein synexin (annexin VII) as a Ca2+/GTP sensor in exocytotic secretion. Proc. Natl. Acad. Sci. USA 93: 10797–10802. 10.1073/pnas.93.20.10797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesné J., Braza F., Mahay G., Brouard S., Aronica M., et al. , 2014. IL-17 in severe asthma. Where do we stand? Am. J. Respir. Crit. Care Med. 190: 1094–1101. [DOI] [PubMed] [Google Scholar]
- Chesnokova V., Melmed S., 2002. Minireview: neuro-immuno-endocrine modulation of the hypothalamic-pituitary-adrenal (HPA) axis by gp130 signaling molecules. Endocrinology 143: 1571–1574. 10.1210/endo.143.5.8861 [DOI] [PubMed] [Google Scholar]
- Chodniewicz D., Klemke R. L., 2004. Guiding cell migration through directed extension and stabilization of pseudopodia. Exp. Cell Res. 301: 31–37. 10.1016/j.yexcr.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Chrétien M., Mbikay M., 2016. 60 YEARS OF POMC: from the prohormone theory to pro-opiomelanocortin and to proprotein convertases (PCSK1 to PCSK9). J. Mol. Endocrinol. 56: T49–T62. 10.1530/JME-15-0261 [DOI] [PubMed] [Google Scholar]
- Ciccotosto G. D., Schiller M. R., Eipper B. A., Mains R. E., 1999. Induction of integral membrane PAM expression in AtT-20 cells alters the storage and trafficking of POMC and PC1. J. Cell Biol. 144: 459–471. 10.1083/jcb.144.3.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin J. L., 1968. The development of the human fetal adenohypophysis. Anat. Rec. 160: 79–91. 10.1002/ar.1091600108 [DOI] [PubMed] [Google Scholar]
- Cool D. R., Loh Y. P., 1998. Carboxypeptidase E is a sorting receptor for prohormones: binding and kinetic studies. Mol. Cell. Endocrinol. 139: 7–13. 10.1016/S0303-7207(98)00081-1 [DOI] [PubMed] [Google Scholar]
- Cua D. J., Tato C. M., 2010. Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 10: 479–489 (erratum: Nat. Rev. Immunol. 10: 611). 10.1038/nri2800 [DOI] [PubMed] [Google Scholar]
- Cuajungco M. P., Podevin W., Valluri V. K., Bui Q., Nguyen V. H., et al. , 2012. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. 114: 705–712. 10.1016/j.acthis.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Abaco G. M., Ng K., Paradiso L., Godde N. J., Kaye A., et al. , 2006. ADAM22, expressed in normal brain but not in high-grade gliomas, inhibits cellular proliferation via the disintegrin domain. Neurosurgery 58: 179–186. 10.1227/01.NEU.0000192363.84287.8B [DOI] [PubMed] [Google Scholar]
- Dantzer R., O’Connor J. C., Freund G. G., Johnson R. W., Kelley K. W., 2008. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 9: 46–56. 10.1038/nrn2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy A., Robbins S. M., 2000. Ephrin-A5 modulates cell adhesion and morphology in an integrin-dependent manner. EMBO J. 19: 5396–5405. 10.1093/emboj/19.20.5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Valle-Pérez B., Martínez V. G., Lacasa-Salavert C., Figueras A., Shapiro S. S., et al. , 2010. Filamin B plays a key role in vascular endothelial growth factor-induced endothelial cell motility through its interaction with Rac-1 and Vav-2. J. Biol. Chem. 285: 10748–10760. 10.1074/jbc.M109.062984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef C., 2008. Paracrinicity: the story of 30 years of cellular pituitary crosstalk. J. Neuroendocrinol. 20: 1–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deribe Y., Pawson T., Dikic I., 2010. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17: 666–672. 10.1038/nsmb.1842 [DOI] [PubMed] [Google Scholar]
- Diebold M.-L., Loeliger E., Koch M., Winston F., Cavarelli J., et al. , 2010. Noncanonical tandem SH2 enables interaction of elongation factor Spt6 with RNA polymerase II. J. Biol. Chem. 285: 38389–38398. 10.1074/jbc.M110.146696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge K. L., Khouangsathiene S., Kapiloff M. S., Mouton R., Hill E. V., et al. , 2001. mAKAP assembles a protein kinase A/PDE4 phosphodiesterase cAMP signaling module. EMBO J. 20: 1921–1930. 10.1093/emboj/20.8.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M., Roehr J. T., Ahmed R., Dieterich C., 2012. FLEXBAR—flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 1: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell M. I., Aguilar E., Friedlander M., 2002. Retinal vascular development is mediated by endothelial filopodia, a preexisting astrocytic template and specific R-cadherin adhesion. Invest. Ophthalmol. Vis. Sci. 43: 3500–3510. [PubMed] [Google Scholar]
- Driscoll C. A., Macdonald D. W., O’Brien S. J., 2009. From wild animals to domestic pets, an evolutionary view of domestication. Proc. Natl. Acad. Sci. USA 106(Suppl. 1): 9971–9978. 10.1073/pnas.0901586106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin J., 2016. 60 YEARS OF POMC: transcriptional and epigenetic regulation of POMC gene expression. J. Mol. Endocrinol. 56: T99–T112. 10.1530/JME-15-0289 [DOI] [PubMed] [Google Scholar]
- Endris V., Haussmann L., Buss E., Bacon C., Bartsch D., et al. , 2011. SrGAP3 interacts with lamellipodin at the cell membrane and regulates Rac-dependent cellular protrusions. J. Cell Sci. 124: 3941–3955. 10.1242/jcs.077081 [DOI] [PubMed] [Google Scholar]
- Eppler E., Jevdjovic T., Maake C., Reinecke M., 2007. Insulin-like growth factor I (IGF-I) and its receptor (IGF-1R) in the rat anterior pituitary. Eur. J. Neurosci. 25: 191–200. 10.1111/j.1460-9568.2006.05248.x [DOI] [PubMed] [Google Scholar]
- Florio T., 2011. Adult pituitary stem cells: from pituitary plasticity to adenoma development. Neuroendocrinology 94: 265–277. 10.1159/000330857 [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Ali S., 2008. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem. Soc. Trans. 36: 609–612. 10.1042/BST0360609 [DOI] [PubMed] [Google Scholar]
- Gaffen S. L., 2009. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 9: 556–567 (erratum: Nat. Rev. Immunol. 9: 747). 10.1038/nri2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambarotta G., Garzotto D., Destro E., Mautino B., Giampietro C., et al. , 2004. ErbB4 expression in neural progenitor cells (ST14A) is necessary to mediate neuregulin-1β1-induced migration. J. Biol. Chem. 279: 48808–48816. 10.1074/jbc.M408374200 [DOI] [PubMed] [Google Scholar]
- Gibson M. A., Leavesley D. I., Ashman L. K., 1999. Microfibril-associated glycoprotein-2 specifically interacts with a range of bovine and human cell types via αVβ3 integrin. J. Biol. Chem. 274: 13060–13065. 10.1074/jbc.274.19.13060 [DOI] [PubMed] [Google Scholar]
- Glinka T., Alter J., Braunstein I., Tzach L., Sheng C. W., et al. , 2014. Signal-peptide-mediated translocation is regulated by a p97–AIRAPL complex. Biochem. J. 457: 253–261. 10.1042/BJ20130710 [DOI] [PubMed] [Google Scholar]
- Grammatopoulos D. K., 2012. Insights into mechanisms of corticotropin-releasing hormone receptor signal transduction. Br. J. Pharmacol. 166: 85–97. 10.1111/j.1476-5381.2011.01631.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Sumida Y., Sanzen N., Sekiguchi K., 2001. Laminin-10/11 and fibronectin differentially regulate integrin-dependent Rho and Rac activation via p130Cas-CrkII-DOCK180 pathway. J. Biol. Chem. 276: 27090–27097. 10.1074/jbc.M102284200 [DOI] [PubMed] [Google Scholar]
- Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N. V., et al. , 2009. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138: 990–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulevich R. G., Oskina I. N., Shikhevich S. G., Fedorova E. V., Trut L. N., 2004. Effect of selection for behavior on pituitary–adrenal axis and proopiomelanocortin gene expression in silver foxes (Vulpes vulpes). Physiol. Behav. 82: 513–518. 10.1016/j.physbeh.2004.04.062 [DOI] [PubMed] [Google Scholar]
- Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N. V., et al. , 2009. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138(5): 990–1004. 10.1016/j.cell.2009.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Hong S., Lee D., Lee M.-H., Choi J.-S., et al. , 2012. Altered expression of synaptotagmin 13 mRNA in adult mouse brain after contextual fear conditioning. Biochem. Biophys. Res. Commun. 425: 880–885. 10.1016/j.bbrc.2012.07.166 [DOI] [PubMed] [Google Scholar]
- Hare B., Wobber V., Wrangham R., 2012. The self-domestication hypothesis: evolution of bonobo psychology is due to selection against aggression. Anim. Behav. 83: 573–585. 10.1016/j.anbehav.2011.12.007 [DOI] [Google Scholar]
- Hirsch D. S., Pirone D. M., Burbelo P. D., 2001. A new family of Cdc42 effector proteins, CEPs, function in fibroblast and epithelial cell shape changes. J. Biol. Chem. 276: 875–883. 10.1074/jbc.M007039200 [DOI] [PubMed] [Google Scholar]
- Hodson D. J., Romanò N., Schaeffer M., Fontanaud P., Lafont C., et al. , 2012a Coordination of calcium signals by pituitary endocrine cells in situ. Cell Calcium 51: 222–230. 10.1016/j.ceca.2011.11.007 [DOI] [PubMed] [Google Scholar]
- Hodson D. J., Schaeffer M., Romanò N., Fontanaud P., Lafont C., et al. , 2012b Existence of long-lasting experience-dependent plasticity in endocrine cell networks. Nat. Commun. 3: 605 10.1038/ncomms1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houslay M. D., Adams D. R., 2003. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 370: 1–18. 10.1042/bj20021698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu D. K., Chernyavsky A. I., Chen H.-Y., Yu L., Grando S. A., et al. , 2009. Endogenous galectin-3 is localized in membrane lipid rafts and regulates migration of dendritic cells. J. Invest. Dermatol. 129: 573–583. 10.1038/jid.2008.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince-Dunn G., Okano H. J., Jensen K. B., Park W.-Y., Zhong R., et al. , 2012. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 75: 1067–1080. 10.1016/j.neuron.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Hatayama M., Tohmonda T., Itohara S., Aruga J., et al. , 2004. Mouse Zic5 deficiency results in neural tube defects and hypoplasia of cephalic neural crest derivatives. Dev. Biol. 270: 146–162. 10.1016/j.ydbio.2004.02.017 [DOI] [PubMed] [Google Scholar]
- Ivanov D., Philippova M., Tkachuk V., Erne P., Resink T., 2004. Cell adhesion molecule T-cadherin regulates vascular cell adhesion, phenotype and motility. Exp. Cell Res. 293: 207–218. 10.1016/j.yexcr.2003.09.030 [DOI] [PubMed] [Google Scholar]
- Jackson V. A., Mehmood S., Chavent M., Roversi P., Carrasquero M., et al. , 2016. Super-complexes of adhesion GPCRs and neural guidance receptors. Nat. Commun. 7: 11184 10.1038/ncomms11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarjour A. A., Durko M., Luk T. L., Marçal N., Shekarabi M., et al. , 2011. Autocrine netrin function inhibits glioma cell motility and promotes focal adhesion formation. PLoS One 6: e25408 10.1371/journal.pone.0025408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Wittgenstein H., Mitchell S. E., Hyma K. E., Temnykh S. V., et al. , 2015. Genotyping-by-sequencing (GBS) detects genetic structure and confirms behavioral QTL in tame and aggressive foxes (Vulpes vulpes). PLoS One 10: e0127013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M. S., Licklider L. J., Gygi S. R., Grigorieff N., Moore M. J., 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439. 10.1017/S1355838202021088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S., Hennessy M. B., Sachser N., 2015. Domestication affects the structure, development and stability of biobehavioural profiles. Front. Zool. 12(Suppl. 1): S19 10.1186/1742-9994-12-S1-S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariagina A., Romanenko D., Ren S.-G., Chesnokova V., 2004. Hypothalamic-pituitary cytokine network. Endocrinology 145: 104–112. 10.1210/en.2003-0669 [DOI] [PubMed] [Google Scholar]
- Kasamatsu A., Uzawa K., Nakashima D., Koike H., Shiiba M., et al. , 2005. Galectin-9 as a regulator of cellular adhesion in human oral squamous cell carcinoma cell lines. Int. J. Mol. Med. 16: 269–273. [PubMed] [Google Scholar]
- Khan Z., Ford M. J., Cusanovich D. A., Mitrano A., Pritchard J. K., et al. , 2013. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 342: 1100–1104. 10.1126/science.1242379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-G., Layman L. C., 2011. The role of CHD7 and the newly identified WDR11 gene in patients with idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Mol. Cell. Endocrinol. 346: 74–83. 10.1016/j.mce.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T., Remm M., 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289–1291. 10.1093/bioinformatics/btm091 [DOI] [PubMed] [Google Scholar]
- Krause M., Gautreau A., 2014. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15: 577–590. 10.1038/nrm3861 [DOI] [PubMed] [Google Scholar]
- Kritzer M. D., Li J., Dodge-Kafka K., Kapiloff M. S., 2012. AKAPs: the architectural underpinnings of local cAMP signaling. J. Mol. Cell. Cardiol. 52: 351–358. 10.1016/j.yjmcc.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukekova A. V., Temnykh S. V., Johnson J. L., Trut L. N., Acland G. M., 2012. Genetics of behavior in the silver fox. Mamm. Genome 23: 164–177. 10.1007/s00335-011-9373-z [DOI] [PubMed] [Google Scholar]
- Kukekova A. V., Trut L. N., Acland G. M., 2014. Genetics of domesticated behavior in dogs and foxes, pp. 361–396 in Genetics and the Behavior of Domestic Animals, Ed. 2, edited by Grandin T., Deesing M. Academic Press, Cambridge, MA: 10.1016/B978-0-12-394586-0.00010-X [DOI] [Google Scholar]
- Langfelder P., Horvath S., 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.-C., Kolkman A., van Schaik F. M. A., Mulder K. W., Pim W. W., et al. , 2009. Human Ccr4–Not complexes contain variable deadenylase subunits. Biochem. J. 422: 443–453. 10.1042/BJ20090500 [DOI] [PubMed] [Google Scholar]
- Levin N., Blum M., Roberts J. L., 1989. Modulation of basal and corticotropin-releasing factor-stimulated proopiomelanocortin gene expression by vasopressin in rat anterior pituitary. Endocrinology 125: 2957–2966. 10.1210/endo-125-6-2957 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Yang L., Yang J. J., Huang Y., Liu Z.-R., 2005. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol. Cell. Biol. 25: 7484–7493. 10.1128/MCB.25.17.7484-7493.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Grant W. M., Persky D., Latham V. M., Singer R. H., et al. , 2002. Interactions of elongation factor 1α with F-Actin and β-Actin mRNA: implications for anchoring mRNA in cell protrusions. Mol. Biol. Cell 13: 579–592. 10.1091/mbc.01-03-0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. P., Robinson P. J., Funder J. W., Engler D., 1994. A comparative study of the role of adenylate cyclase in the release of adrenocorticotropin from the ovine and rat anterior pituitary. Mol. Cell. Endocrinol. 101: 173–181. 10.1016/0303-7207(94)90232-1 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNair L., Xiao S., Miletic D., Ghani M., Julien J.-P., et al. , 2016. MTHFSD and DDX58 are novel RNA-binding proteins abnormally regulated in amyotrophic lateral sclerosis. Brain 139: 86–100. 10.1093/brain/awv308 [DOI] [PubMed] [Google Scholar]
- Malik M., Chen Y.-Y., Kienzle M. F., Tomkowicz B. E., Collman R. G., et al. , 2008. Monocyte migration and LFA-1-mediated attachment to brain microvascular endothelia is regulated by SDF-1α through Lyn kinase. J. Immunol. 181: 4632–4637. 10.4049/jimmunol.181.7.4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell K. J., Babbin B. A., Nusrat A., Parkos C. A., 2005. Junctional adhesion molecule 1 regulates epithelial cell morphology through effects on β1 integrins and Rap1 activity. J. Biol. Chem. 280: 11665–11674. 10.1074/jbc.M412650200 [DOI] [PubMed] [Google Scholar]
- Matsunaga K., Taoka M., Isobe T., Izumi T., 2017. Rab2a and Rab27a cooperatively regulate the transition from granule maturation to exocytosis through the dual effector Noc2. J. Cell Sci. 130: 541–550. 10.1242/jcs.195479 [DOI] [PubMed] [Google Scholar]
- McCahill A., McSorley T., Huston E., Hill E. V., Lynch M. J., et al. , 2005. In resting COS1 cells a dominant negative approach shows that specific, anchored PDE4 cAMP phosphodiesterase isoforms gate the activation, by basal cyclic AMP production, of AKAP-tethered protein kinase A type II located in the centrosomal region. Cell. Signal. 17: 1158–1173. 10.1016/j.cellsig.2005.04.003 [DOI] [PubMed] [Google Scholar]
- McLaughlin F., Ludbrook V. J., Cox J., von Carlowitz I., Brown S., et al. , 2001. Combined genomic and antisense analysis reveals that the transcription factor Erg is implicated in endothelial cell differentiation. Blood 98: 3332–3339. 10.1182/blood.V98.12.3332 [DOI] [PubMed] [Google Scholar]
- Mehul B., Hughes R. C., 1997. Plasma membrane targetting, vesicular budding and release of galectin 3 from the cytoplasm of mammalian cells during secretion. J. Cell Sci. 110: 1169–1178. [DOI] [PubMed] [Google Scholar]
- Melmed S. (Editor), 2010. The Pituitary. Academic Press, Cambridge, MA. [Google Scholar]
- Mercurio A. M., Rabinovitz I., Shaw L. M., 2001. The α6β4 integrin and epithelial cell migration. Curr. Opin. Cell Biol. 13: 541–545. 10.1016/S0955-0674(00)00249-0 [DOI] [PubMed] [Google Scholar]
- Mosher D. F., Adams J. C., 2012. Adhesion-modulating/matricellular ECM protein families: a structural, functional and evolutionary appraisal. Matrix Biol. 31: 155–161. 10.1016/j.matbio.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Movilla N., Bustelo X. R., 1999. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol. Cell. Biol. 19: 7870–7885. 10.1128/MCB.19.11.7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K., Koyabu Y., Aruga J., Mikoshiba K., 2000. A novel member of the Xenopus Zic family, Zic5, mediates neural crest development. Mech. Dev. 99: 83–91. 10.1016/S0925-4773(00)00480-9 [DOI] [PubMed] [Google Scholar]
- Nakata Y., Tomkowicz B., Gewirtz A. M., Ptasznik A., 2006. Integrin inhibition through Lyn-dependent cross talk from CXCR4 chemokine receptors in normal human CD34+ marrow cells. Blood 107: 4234–4239. 10.1182/blood-2005-08-3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumenko E. V., Popova N. K., Nikulina E. M., Dygalo N. N., Shishkina G. T., et al. , 1989. Behavior, adrenocortical activity, and brain monoamines in Norway rats selected for reduced aggressiveness towards man. Pharmacol. Biochem. Behav. 33: 85–91. 10.1016/0091-3057(89)90434-6 [DOI] [PubMed] [Google Scholar]
- Navratil A. M., Knoll J. G., Whitesell J. D., Tobet S. A., Clay C. M., 2007. Neuroendocrine plasticity in the anterior pituitary: gonadotropin-releasing hormone-mediated movement in vitro and in vivo. Endocrinology 148: 1736–1744. 10.1210/en.2006-1153 [DOI] [PubMed] [Google Scholar]
- Nelson R. M., Temnykh S. V., Johnson J. L., Kharlamova A. V., Vladimirova A. V., et al. , 2017. Genetics of interactive behavior in silver foxes (Vulpes vulpes). Behav. Genet. 47: 88–101. 10.1007/s10519-016-9815-1 [DOI] [PubMed] [Google Scholar]
- Nobumoto A., Nagahara K., Oomizu S., Katoh S., Nishi N., et al. , 2008. Galectin-9 suppresses tumor metastasis by blocking adhesion to endothelium and extracellular matrices. Glycobiology 18: 735–744. 10.1093/glycob/cwn062 [DOI] [PubMed] [Google Scholar]
- Ogi T., Lehmann A. R., 2006. The Y-family DNA polymerase [kappa](pol [kappa]) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 8: 640–642. 10.1038/ncb1417 [DOI] [PubMed] [Google Scholar]
- Oskina I. N., Plyusnina I. Z., 2000. The pituitary-adrenal axis at selection for domesticated behavior, pp. 327–333 in Current Concepts of Evolutionary Genetics, edited by Schumny V. K., Markel A. L. Institute of Cytology and Genetics, Novosibirsk, Russia. [Google Scholar]
- Parnet P., Kelley K. W., Bluthé R. M., Dantzer R., 2002. Expression and regulation of interleukin-1 receptors in the brain. Role in cytokines-induced sickness behavior. J. Neuroimmunol. 125: 5–14. 10.1016/S0165-5728(02)00022-X [DOI] [PubMed] [Google Scholar]
- Penela P., Nogués L., Mayor F., Jr, 2014. Role of G protein-coupled receptor kinases in cell migration. Curr. Opin. Cell Biol. 27: 10–17. 10.1016/j.ceb.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Piesse C., Tymms M., Garrafa E., Gouzy C., Lacasa M., et al. , 2002. Human aminopeptidase B (rnpep) on chromosome 1q32.2: complementary DNA, genomic structure and expression. Gene 292: 129–140. 10.1016/S0378-1119(02)00650-9 [DOI] [PubMed] [Google Scholar]
- Pocha S. M., Montell D. J., 2014. Cellular and molecular mechanisms of single and collective cell migrations in Drosophila: themes and variations. Annu. Rev. Genet. 48: 295–318. 10.1146/annurev-genet-120213-092218 [DOI] [PubMed] [Google Scholar]
- Prasolova L. A., Gerbek Y. E., Gulevich R. G., Shikhevich S. G., Konoshenko M. Yu., et al. , 2014. The effects of prolonged selection for behavior on the stress response and activity of the reproductive system of male grey mice (Rattus norvegicus). Russ. J. Genet. 50: 846–852. 10.1134/S1022795414080031 [DOI] [PubMed] [Google Scholar]
- Price E. O., 2008. Principles and Applications of Domestic Animal Behavior: An Introductory Text. CABI, Wallingford, United Kingdom: 10.1079/9781845933982.0000 [DOI] [Google Scholar]
- Pulichino A.-M., Vallette-Kasic S., Tsai J. P.-Y., Couture C., Gauthier Y., et al. , 2003. Tpit determines alternate fates during pituitary cell differentiation. Genes Dev. 17: 738–747. 10.1101/gad.1065703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi A.-D., Kennedy C., Harden T. K., Nicholas R. A., 2001. Differential coupling of the human P2Y11 receptor to phospholipase C and adenylyl cyclase. Br. J. Pharmacol. 132: 318–326. 10.1038/sj.bjp.0703788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisine T., Rougon G., Barbet J., Affolter H. U., 1985. Corticotropin-releasing factor-induced adrenocorticotropin hormone release and synthesis is blocked by incorporation of the inhibitor of cyclic AMP-dependent protein kinase into anterior pituitary tumor cells by liposomes. Proc. Natl. Acad. Sci. USA 82: 8261–8265. 10.1073/pnas.82.23.8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribon V., Herrera R., Kay B. K., Saltiel A. R., 1998. A role for CAP, a novel, multifunctional Src homology 3 domain-containing protein in formation of actin stress fibers and focal adhesions. J. Biol. Chem. 273: 4073–4080. 10.1074/jbc.273.7.4073 [DOI] [PubMed] [Google Scholar]
- Ridley A. J., 1994. Membrane ruffling and signal transduction. BioEssays 16: 321–327. 10.1002/bies.950160506 [DOI] [PubMed] [Google Scholar]
- Rizzoti K., 2010. Adult pituitary progenitors/stem cells: from in vitro characterization to in vivo function. Eur. J. Neurosci. 32: 2053–2062. 10.1111/j.1460-9568.2010.07524.x [DOI] [PubMed] [Google Scholar]
- Russo A., Siciliano G., Catillo M., Giangrande C., Amoresano A., et al. , 2010. hnRNP H1 and intronic G runs in the splicing control of the human rpL3 gene. Biochim. Biophys. Acta 1799: 419–428. 10.1016/j.bbagrm.2010.01.008 [DOI] [PubMed] [Google Scholar]
- Sachdev P., Zeng L., Wang L. H., 2002. Distinct role of phosphatidylinositol 3-kinase and Rho family GTPases in Vav3-induced cell transformation, cell motility, and morphological changes. J. Biol. Chem. 277: 17638–17648. 10.1074/jbc.M111575200 [DOI] [PubMed] [Google Scholar]
- Sano H., Hsu D. K., Yu L., Apgar J. R., Kuwabara I., et al. , 2000. Human galectin-3 is a novel chemoattractant for monocytes and macrophages. J. Immunol. 165: 2156–2164. 10.4049/jimmunol.165.4.2156 [DOI] [PubMed] [Google Scholar]
- Sansone P., Bromberg J., 2012. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J. Clin. Oncol. 30: 1005–1014. 10.1200/JCO.2010.31.8907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax J. K., El-Deiry W. S., 2003. Identification and characterization of the cytoplasmic protein TRAF4 as a p53-regulated proapoptotic gene. J. Biol. Chem. 278: 36435–36444. 10.1074/jbc.M303191200 [DOI] [PubMed] [Google Scholar]
- Segovia M., Louvet C., Charnet P., Savina A., Tilly G., et al. , 2014. Autologous dendritic cells prolong allograft survival through Tmem176b-dependent antigen cross-presentation. Am. J. Transplant. 14: 1021–1031. 10.1111/ajt.12708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino S., Shibasaki T., 2005. PKA-dependent and PKA-independent pathways for cAMP-regulated exocytosis. Physiol. Rev. 85: 1303–1342. 10.1152/physrev.00001.2005 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Park J. W., Lu Z.-X., Lin L., Henry M. D., et al. , 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 111: E5593–E5601. 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikhevich S. G., Oskina I. N., Pliusnina I. Z., 2002. Effect of stress and immune stimulus on the pituitary-adrenal axis in gray rats selected for behavior. Ross. Fiziol. Zh. Im. I M Sechenova 88: 781–789. [PubMed] [Google Scholar]
- Sindrilaru A., Peters T., Schymeinsky J., Oreshkova T., Wang H., et al. , 2009. Wound healing defect of Vav3−/− mice due to impaired β2-integrin–dependent macrophage phagocytosis of apoptotic neutrophils. Blood 113: 5266–5276. 10.1182/blood-2008-07-166702 [DOI] [PubMed] [Google Scholar]
- Skalsky Y. M., Ajuh P. M., Parker C., Lamond A. I., Goodwin G., et al. , 2001. PRCC, the commonest TFE3 fusion partner in papillary renal carcinoma is associated with pre-mRNA splicing factors. Oncogene 20: 178–187. 10.1038/sj.onc.1204056 [DOI] [PubMed] [Google Scholar]
- Smith D. M., Coppock H. A., Withers D. J., Owji A. A., Hay D. L., et al. , 2002. Adrenomedullin: receptor and signal transduction. Biochem. Soc. Trans. 30: 432–437. 10.1042/bst0300432 [DOI] [PubMed] [Google Scholar]
- Soulavie F., Piepenbrock D., Thomas J., Vieillard J., Duteyrat J.-L., et al. , 2014. hemingway is required for sperm flagella assembly and ciliary motility in Drosophila. Mol. Biol. Cell 25: 1276–1286. 10.1091/mbc.E13-10-0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed E., Balda M. S., Matter K., 2010. Dynamics and functions of tight junctions. Trends Cell Biol. 20: 142–149. 10.1016/j.tcb.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Stillman B. N., Hsu D. K., Pang M., Brewer C. F., Johnson P., et al. , 2006. Galectin-3 and galectin-1 bind distinct cell surface glycoprotein receptors to induce T cell death. J. Immunol. 176: 778–789. 10.4049/jimmunol.176.2.778 [DOI] [PubMed] [Google Scholar]
- Subramaniam S. V., Cooper R. S., Adunyah S. E., 1999. Evidence for the involvement of JAK/STAT pathway in the signaling mechanism of interleukin-17. Biochem. Biophys. Res. Commun. 262: 14–19. 10.1006/bbrc.1999.1156 [DOI] [PubMed] [Google Scholar]
- Südhof T. C., 2002. Synaptotagmins: why so many? J. Biol. Chem. 277: 7629–7632. [DOI] [PubMed] [Google Scholar]
- Sugawara K., Shibasaki T., Takahashi H., Seino S., 2016. Structure and functional roles of Epac2 (Rapgef4). Gene 575: 577–583. 10.1016/j.gene.2015.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton K. A., Jungnickel M. K., Wang Y., Cullen K., Lambert S., et al. , 2004. Enkurin is a novel calmodulin and TRPC channel binding protein in sperm. Dev. Biol. 274: 426–435. 10.1016/j.ydbio.2004.07.031 [DOI] [PubMed] [Google Scholar]
- Taskén K. A., Collas P., Kemmner W. A., Witczak O., Conti M., et al. , 2001. Phosphodiesterase 4D and protein kinase A type II constitute a signaling unit in the centrosomal area. J. Biol. Chem. 276: 21999–22002. 10.1074/jbc.C000911200 [DOI] [PubMed] [Google Scholar]
- Teufel M., Saudek V., Ledig J.-P., Bernhardt A., Boularand S., et al. , 2003. Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J. Biol. Chem. 278: 6521–6531. 10.1074/jbc.M209764200 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., et al. , 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta C. R., Miao F., Arn E. A., Stevens S. W., Ho C. K., et al. , 1997. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89: 849–858. 10.1016/S0092-8674(00)80270-6 [DOI] [PubMed] [Google Scholar]
- Trut L., Oskina I., Kharlamova A., 2009. Animal evolution during domestication: the domesticated fox as a model. BioEssays 31: 349–360. 10.1002/bies.200800070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trut L. N., Plyusnina I. Z., Oskina I. N., 2004. An experiment on fox domestication and debatable issues of evolution of the dog. Russ. J. Genet. 40: 644–655. 10.1023/B:RUGE.0000033312.92773.c1 [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., et al. , 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40: e115 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada J., Ota K., Kumar A., Wallner E. I., Kanwar Y. S., 1997. Developmental regulation, expression, and apoptotic potential of galectin-9, a beta-galactoside binding lectin. J. Clin. Invest. 99: 2452–2461. 10.1172/JCI119429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak M., Martens S., 2013. Dissecting the role of the Atg12–Atg5-Atg16 complex during autophagosome formation. Autophagy 9: 424–425. 10.4161/auto.22931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Huang J., Hu Z., 2012. RNA helicase DDX5 regulates microRNA expression and contributes to cytoskeletal reorganization in basal breast cancer cells. Mol. Cell. Proteomics 11: M111.011932 10.1074/mcp.M111.011932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne R. K., 1993. Molecular evolution of the dog family. Trends Genet. 9: 218–224. 10.1016/0168-9525(93)90122-X [DOI] [PubMed] [Google Scholar]
- Weinstein L. S., Liu J., Sakamoto A., Xie T., Chen M., 2004. Minireview: GNAS: normal and abnormal functions. Endocrinology 145: 5459–5464. 10.1210/en.2004-0865 [DOI] [PubMed] [Google Scholar]
- Wilkins A. S., Wrangham R. W., Fitch W. T., 2014. The “domestication syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics 197: 795–808. 10.1534/genetics.114.165423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. K., Spence H. J., Rodgers R. R., Ozanne B. W., Fitzgerald U., et al. , 2005. Role of Mayven, a kelch-related protein in oligodendrocyte process formation. J. Neurosci. Res. 81: 622–631. 10.1002/jnr.20588 [DOI] [PubMed] [Google Scholar]
- Wu M., Huang C., Li X., Li X., Gan K., et al. , 2008. LRRC4 inhibits glioblastoma cell proliferation, migration, and angiogenesis by downregulating pleiotropic cytokine expression and responses. J. Cell. Physiol. 214: 65–74. 10.1002/jcp.21163 [DOI] [PubMed] [Google Scholar]
- Yamagishi S., Hampel F., Hata K., Del Toro D., Schwark M., et al. , 2011. FLRT2 and FLRT3 act as repulsive guidance cues for Unc5-positive neurons. EMBO J. 30: 2920–2933. 10.1038/emboj.2011.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T., Calaminus S. D. J., Caswell P., Spence H. J., Carnell M., et al. , 2011. The Arp2/3 activator WASH regulates α5β1-integrin-mediated invasive migration. J. Cell Sci. 124: 3753–3759. 10.1242/jcs.080986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo D.-X., Zhang X.-W., Jin B., Zhang Z., Xie B.-S., et al. , 2013. CSTP1, a novel protein phosphatase, blocks cell cycle, promotes cell apoptosis, and suppresses tumor growth of bladder cancer by directly dephosphorylating Akt at Ser473 site. PLoS One 8: e65679 10.1371/journal.pone.0065679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock L., Ancuta P., Crowe S., Dalod M., Grau V., et al. , 2010. Nomenclature of monocytes and dendritic cells in blood. Blood 116: e74–e80. 10.1182/blood-2010-02-258558 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The primary sequencing data have been deposited in the GenBank Sequence Read Archive (SRA) under accession number SRP107226.