Abstract
Moths can biosynthesize sex pheromones in the female sex pheromone glands (PGs) and can distinguish species-specific sex pheromones using their antennae. However, the biosynthesis and transportation mechanism for Type II sex pheromone components has rarely been documented in moths. In this study, we constructed a massive PG transcriptome database (14.72 Gb) from a moth species, Ectropis grisescens, which uses type II sex pheromones and is a major tea pest in China. We further identified putative sex pheromone biosynthesis and transportation-related unigenes: 111 cytochrome P450 monooxygenases (CYPs), 25 odorant-binding proteins (OBPs), and 20 chemosensory proteins (CSPs). Tissue expression and phylogenetic tree analyses showed that one CYP (EgriCYP341-fragment3), one OBP (EgriOBP4), and one CSP (EgriCSP10) gene displayed an enriched expression in the PGs, and that EgriOBP2, 3, and 25 are clustered in the moth pheromone-binding protein clade. We considered these our candidate genes. Our results yielded large-scale PG sequence information for further functional studies.
Keywords: transcriptome, sex pheromone biosynthesis, tissue expression pattern, Ectropis grisescens
Reproductive isolation in moths relies on species-specific and multiple sex pheromone components, which are diverse in structure and ratio (Ando et al. 2004). Sex pheromone components are divided into three types according to their structure: type I (75%), type II (15%), and miscellaneous type (10%) (Ando et al. 2004; Löfstedt et al. 2016). Type I sex pheromone components comprise a C10–C18 straight chain with unsaturated aliphatic compounds, and different terminal functional groups making them an alcohol, aldehyde, or acetate. Type II consists of one to three cis double bonds separated by methylene groups of C17–C23 straight chains, in addition to zero, one, or two epoxide functions (Millar 2000; Ando et al. 2004).
Type I sex pheromone components are biosynthesized from saturated fatty acids, typically palmitic acid, through many enzymatic reactions (Jurenka 2004a; He et al. 2017), whereas type II sex pheromone components are derived through decarboxylation and epoxidation from dietary linoleic or linolenic acid (Jurenka 2004a; Tillman et al. 1999; Millar 2000). Production sites also differ between type I and type II. Type I pheromone components are usually produced and released from female extrudable glands (pheromone glands, PGs) located between the eighth and ninth abdominal segments. However, the hydrocarbon precursor of type II pheromone components is expected to be produced from dietary linolenic acid by chain elongation and decarboxylation in oenocytes and then transported to the PG in which it is epoxidized and released (Millar 2000; Jurenka 2003, 2004b). The proteins that are responsible for transportation and epoxidation of the type II sex pheromone precursor are not yet well documented.
Odorant-binding proteins (OBPs) and chemosensory proteins (CSPs) are two types of soluble protein that are well known as odor transporters (including transporters of sex pheromones) in insect antennal lymph (Vogt et al. 2015; Pelosi et al. 2014). Numerous insect OBPs and CSPs have been identified recently by the blooming sequencing techniques (X.-M. Li et al. 2015; He and He 2014; He et al. 2011; Xu et al. 2009). Insect OBPs can be divided into five subfamilies: classic, minus-C, plus-C, atypical, and dimer. Classic OBPs possess six conserved cysteines (C1–C6), paired to form three complete salt bridges (Leal et al. 1999). By contrast, minus-C lacks two conserved cysteines (C2 and C5) (Hekmat-Scafe et al. 2002), and plus-C OBPs have two additional conserved cysteines and a proline. Atypical and dimer OBPs are rare; the former possess 12 conserved cysteine residues (Graham and Davies 2002), and the latter have an additional conserved cysteine in an extended C-terminal region, compared with classic OBPs (Xu et al. 2003).
In the Lepidoptera, OBPs are divided, according to differences in odorant substrate, into pheromone-binding proteins (PBPs) and general OBPs (GOBPs). Generally, PBPs possess three paralog genes (PBP1, PBP2, and PBP3) and GOBPs contain two (GOBP1 and GOBP2). It was thought that the paralog genes in the PBPs bound pheromone components and those in GOBPs bound general odors (Jacquin-Joly et al. 2000; Vogt et al. 2015). However, a recent study stated that GOBP2 assists moth larvae to find better food though sex pheromone cues (J. Zhu et al. 2016).
In insects, CSPs are more conserved than OBPs (Xu et al. 2009). CSPs have two disulfide bonds formed by four conserved cysteines (Angeli et al. 1999; Leal et al. 1999). The existence of some CSPs in subsets of chemosensory sensilla suggests a potential olfactory function, which has been confirmed by several odor-binding experiments with CSPs (Briand et al. 2002; Ozaki et al. 2005; González et al. 2009; Guo et al. 2011). However, CSPs also have nonolfactory functions because of their varied expression patterns, such as being involved in leg regeneration (Kitabayashi et al. 1998) and insecticide resistance (G. X. Liu et al. 2014). Some OBP and CSP members have been found to be abundant in antennae and enriched in other tissues, such as the female sex PGs (Dani et al. 2011; Jacquin-Joly et al. 2001). These results strengthen our assertion that OBPs and CSPs do not just play a part in olfaction, but may participate in other crucial physical functions (Sun et al. 2012).
The cytochrome P450 monooxygenases (P450s, CYPs) are a large and complex superfamily found across life forms, from prokaryotes to eukaryotes, and are responsible for the oxidative metabolism of many diverse compounds (Nebert and Gonzalez 1987). In insects, the major role of CYPs is to catalyze the synthesis of endogenous, physiologically crucial chemical compounds, such as juvenile hormones, odors, and ecdysteroids. Despite this, most research has focused on their role in the detoxification of pesticides and plant allelochemicals (N. Liu et al. 2015). In the fall webworm, Hyphantria cunea, one P450 gene (CYP341B14) has been characterized and found to be able to epoxidize a specific (Z)-double bond at the ninth position in its type II pheromone precursor: (3Z,6Z,9Z)-3,6,9-henicosatriene (Rong et al. 2014).
Ectropis grisescens is a major tea pest that has spread to most tea fields in China. It was recently distinguished from its sibling species, Ectropis obliqua, by cytochrome oxidase I sequencing (Nan et al. 2014; Yu et al. 2014). The moth sex pheromone components have been characterized as two type II compounds: Z3, Z6, Z9-18:H and Z3, epo6, Z9-18:H, and they trigger a strong gas chromatography–electroantennogram detection reaction in male moths (Ma et al. 2016). Moreover, a 1:4 ratio of the two compounds gives the most attractive effect in tea fields (Ma et al. 2016). However, the biosynthesis mechanism for sex pheromone components remains unclear. We aimed to find CYP, OBP, and CSP members potentially involved in sex pheromone biosynthesis and transportation. We sequenced the PG transcriptome of E. grisescens and then analyzed the phylogenetic tree and tissue expression patterns of these three gene types.
Materials and Methods
Insect samples and tissue collection
The E. grisescens colony used in this study was originally collected from the experimental tea plantation in the Tea Research Institute of the Chinese Academy of Agricultural Sciences (Hangzhou, Zhejiang, China). Newly hatched larvae were transferred onto fresh tea shoots in enclosed nylon mesh cages (70 × 70 × 70 cm). They were kept in a climate-controlled room at 25 ± 1° with 70 ± 5% relative humidity under a photoperiod of 14:10 (light:dark). After pupation, male and female pupae were separated based on their body size and morphological characters and kept in darkness until eclosion. After emergence, adult moths were given a 10% honey solution. For transcriptome sequencing, each biological replicate was made up of 40 female PGs from unmated females collected 2–3 d after eclosion. For the quantitative polymerase chain reaction (qPCR), a different sample of 20 female adults was used to collect antennae, proboscises, heads without antennae and proboscises, thoraxes, sex PGs, abdomens without sex PGs, legs, and wings, as well as 20 male adults to collect antennae, proboscises, heads without antennae or proboscises, thoraxes, abdomens, legs, and wings. All tissues were immediately snap-frozen in liquid nitrogen and stored at −80° until extraction. Total RNA was extracted using an SV Total Isolation System (Promega, Madison, WI). The integrity of RNA samples was evaluated by gel electrophoresis and the quantification determined using a NanoDrop 2000 spectrophotometer (NanoDrop, Wilmington, DE).
cDNA library construction and Illumina sequencing
The cDNA library construction and Illumina sequencing of the samples were performed by Novogene Bioinformatics Technology Co. Ltd., Beijing, China. Poly-adenylated RNA was isolated from 20 µg of the total pooled RNA using oligo (dT) magnetic beads. The mRNA was then fragmented into short pieces in the presence of divalent cations in fragmentation buffer at 94° for 5 min. Using the cleaved fragments as templates, random hexamer primers were used to synthesize first-strand cDNA. Second-strand cDNA was generated using the buffer, dNTPs, RNAse H, and DNA polymerase I. Following end repair and adaptor ligation, short sequences were amplified by PCR and purified with a QIAquick PCR extraction kit (Qiagen, Venlo, The Netherlands), then sequenced on a HiSeq 2000 platform (Illumina, San Diego, CA).
Assembly and annotation
The PG transcriptomes were assembled de novo using the short-read assembly program Trinity (version r20140413p1) based on the paired-end reads. Transcripts larger than 150 bp were compared using BLASTX to existing sequences in the protein databases, including the NCBI NR, NT, KO, Swiss-Prot, PFAM, and KOG. We then used the Blast2GO program for gene ontology (GO) annotation of the transcripts and WEGO software to plot the GO annotation results.
Analysis of transcript expression in the pheromone glands
Transcript expression abundances were calculated by the FPKM (reads per kilobase per million mapped reads) method, which can eliminate the influence of different transcript lengths and sequencing discrepancies in the calculation of expression abundance. FPKM was calculated using equation (1):
| (1) |
where FPKM (A) is the expression of transcript A; C is the number of reads uniquely aligned to transcript A; N is the total number of fragments uniquely aligned to all transcripts; and L is the number of bases in transcript A.
Phylogenetic analysis
To investigate the phylogenetic relationships between the CYPs, OBPs, and CSPs of E. grisescens and those of some other insect genes, we compared them using MAFFT with default settings. The phylogenetic tree was constructed using PhyML 3.0 with default settings and 1000 bootstrap replicates.
Quantitative real-time PCR and data analysis
Quantitative real-time PCR (qRT-PCR) was conducted according to guidelines for the minimum information required for publication of such experiments (Bustin et al. 2009). A blank control without template cDNA (replacing cDNA with H2O) served as the negative control. Each reaction had three independent biological replicates and was repeated three times (technical replicates). Relative expression levels were calculated using the comparative 2−△△Cq method. Total RNA was isolated using the SV Total Isolation System (Promega, Madison, WI) according to the manufacturer’s instructions, including a step of DNAse treatment to avoid genome contamination. Single-stranded cDNA templates were synthesized using 1 µg of total RNA from 15 moth body and 15 PG samples using a Reverse Transcription System (Promega) following the instructions in the manual. qRT-PCR was performed in a Mastercycler ep realplex (Eppendorf, Hamburg, Germany) with primers designed using Beacon Designer 7.7 and based on the E. grisescens nucleotide sequences from the Illumina data. The E. grisescens GTP-binding protein and glyceraldehyde-3-phosphate dehydrogenase genes were used as references, as they had been identified as having stable expression across tissues in this species. Expression levels of the tested mRNA were determined using GoTaq qPCR Master Mix (Promega) according to the manufacturer’s instructions. The primers are listed in Supplemental Material, File S2.
Data availability
The datasets of PG transcriptomes used in this study are available in the NCBI SRA database (http://trace.ncbi.nlm.nih.gov/Traces/sra/), under accession numbers SRR5571992; SRR5571993; SRR5571994 and SRR5571995.
Results
Overview of the PG transcriptomes
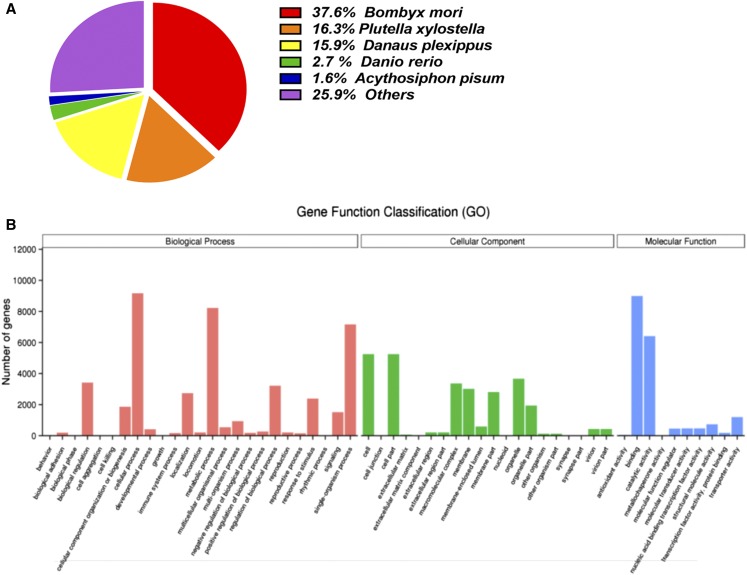
The female PG transcriptome was sequenced using the Illumina HiSeq platform and assembled with the program Trinity (version r20140413p1). Two biological replicates were sequenced, yielding 7.31 and 7.41 Gb each. We performed a de novo assembly of the PG transcriptomes, yielding 101,632 transcripts with N50 lengths of 1491 nt (Figure S1 and Table 1). BLASTx searches of all 76,074 unigenes showed that 28.56% were homologous to proteins in several other insect species. The highest level of sequence identities (37.6%) was with Bombyx mori sequences, followed by sequences from Plutella xylostella (16.3%), Danaus plexippus (15.9%), and Acythosiphon pisum (1.6%) (Figure 1A).
Table 1. Summary of Ectropis grisescens de novo PG transcriptome assembly.
| Ectropis grisescens | ||
|---|---|---|
| PG-1 | PG-2 | |
| Total Number of Raw Reads | 51,184,742 | 51,899,126 |
| Total Number of Clean Reads | 48,724,984 | 49,372,382 |
| Total Number of Clean Nucleotides (nt) | 7.31 Gb | 7.41 Gb |
| Q20 Percentage | 96.94% | 96.83% |
| GC Percentage | 46.64% | 46.16% |
| Total Number of Transcripts | 76,074 | |
| N50 (nt) | 1491 | |
| Percentage of Transcripts Annotated by NCBI NR Database | 28.56% | |
| Percentage of Transcripts Annotated by Swiss-Prot Database | 19.05% | |
| Percentage of Transcripts Annotated by PFAM Database | 21.63% | |
| Percentage of Transcripts Annotated by KOG Database | 13.35% | |
| Percentage of Transcripts Annotated by GO Database | 21.88% | |
Figure 1.
Annotation summaries for E. grisescens unigenes. (A) Species distribution of unigenes with the best-hit annotation terms in the nonredundant (NR) database. (B) Gene ontology (GO) classifications of E. grisescens unigenes.
We used Blast2GO to annotate the unigenes into functional groups based on GO. In the molecular function category, the genes expressed were mostly enriched for catalytic activity (e.g., hydrolase and oxidoreductase) and binding (e.g., nucleotide, ion, and odor binding). In the biological process category, cellular and metabolic processes were the most common. In the cellular component category, the terms cell (GO:0005623) and cell part (GO:0044464) were represented the most (Figure 1B).
Identification of putative OBP, CSP, and CYP genes
To identifiy putative OBP, CSP, and CYP genes, we used a local blast program against the OBP and CSP genes of H. cunea (Zhang et al. 2016) and the CYP genes of Operophtera brumata, since its genome is available and it also belongs to the Geometridae (Derks et al. 2015). One hundred and eleven CYP transcripts were identified throughout the PG transcriptome, and they showed high levels of identity (from 39.96 to 92.82%) with other insect CYP genes (File S1). Helix-C (WxxxR) and helix-K (ExxR), the conservative domains of CYP genes, were more conserved than other domains, such as the heme-binding domain (PFxxGxRxCxG/A), the PERF motif (PxxFxPE/DRF), and helix-I (GxE/DTT/S). Additionally, 25 OBPs and 20 CSPs were identified, all of which displayed conservative cysteine domains and high levels of identity with OBPs and CSPs that were previously identified in other insect species: 25–95% for OBPs and 31–90% for CSPs (File S1).
Phylogenetic tree analysis
E. grisescens CYPs were named according to the CYP Gene Family Nomenclature Committee (Dr D. Nelson, University of Tennessee, Memphis, TN). We constructed a phylogenetic tree using O. brumata and B. mori CYP genes (Figure 2). The 111 CYPs were clearly distributed in all four CYP clans: CYP2, CYP3, CYP4, and mitochondrial CYP. Most of them were distributed in the CYP3 clan (59 genes) and the CYP4 clan (33 genes). The rest were clustered within the CYP2 (7 genes) and mitochondrial CYP (12 genes) clans. For the OBP phylogenetic tree (Figure 3), we used four previously identified groups: plus-C, minus-C, PBP, and GOBP. We found four OBPs in plus-C (EgriOBP4, 5, 6, and 7), three in minus-C (EgriOBP13, 14, and 15), three in PBP (EgriOBP2, 3, and 25), and one in GOBP (EgriOBP1). EgriOBP8 was not clustered in any group, as was the case for several BmorOBPs (22, 23, and 28). For CSPs (Figure 4), one group included the most (10) EgriCSPs (EgriCSP3, 4, 5, 6, 7, 8, 9, 10, 17, and 18), as well as CSPs from the two other lepidopteran species: seven from B. mori and four from H. cunea. Another group contained only lepidopteran CSPs and one Apis mellifera CSP: AmelCSP1. In this group, there were five EgriCSPs: 11, 12, 14, 15, and 19. EgriCSP13 and its orthologous genes, BmorCSP10 and HyphCSP11, clustered within a clade of Locusta migratoria CSPs. EgriCSP2 did not cluster in any group, and another 19 CSPs were distributed in four other groups.
Figure 2.
Phylogenetic analysis of CYPs in E. grisescens, O. brumata, and B. mori. The phylogenetic tree was constructed in PhyML3.0 using the maximum likelihood method.
Figure 3.
Phylogenetic analysis of EgriOBPs with some other insect OBPs. The phylogenetic tree was constructed in PhyML3.0 using the maximum likelihood method.
Figure 4.
Phylogenetic analysis of EgriCSPs with some other insect CSPs. The phylogenetic tree was constructed in PhyML3.0 using the maximum likelihood method.
Tissue expression profile and mRNA abundance of the CYP, OBP, and CSP genes
We further characterized the expression levels and tissue expression patterns of the transcripts of the putative CYP, OBP, and CSP genes by qPCR. The aim was to find the genes with expression predominantly in the PG, which may be involved in pheromone biosynthesis or transportation. Transcript abundance in the PG was also calculated as FPKM (File S1). One hundred of the 111 CYPs were successfully amplified by qPCR (Figure 5). Of them, 30 CYPs presented a PG-enriched expression pattern. The mRNA abundance showed that seven EgriCYPs had expression levels >10 times higher in the PG than in the body: EgriCYP340BD1 (149.05-fold), EgriCYP367A1_Ortholog (74.16-fold), EgriCYP4AU2 (42.53-fold), EgriCYP9A116 (28.16-fold), EgriCYP340BC1 (22.58-fold), EgriCYP9CP1 (22.54-fold), and EgriCYP341U1 (16.12-fold). Five of the 100 CYPs were clearly expressed at higher levels than the rest: EgriCYP9A116 (FPKM = 940.07 ± 98.71), EgriCYP340BD1 (929.68 ± 0.71), EgriCYP4L-fragment2 (250.21 ± 9.52), EgriCYP4G174 (202.27 ± 33.27), and EgriCYP6AB120 (153.37 ± 10.59) (Figure 5).
Figure 5.
Tissue expression profile of selected EgriCYP genes based on relative mRNA quantity and the FPKM values of EgriCYPs with PG-enriched expression. The level of EgriCYPs expression in the abdomen was set at 1. B, abdomen; Pg, female pheromone glands.
For the OBPs (Figure 6), 25 EgriOBPs presented distinct expression patterns. The OBPs with greater levels of expression in male antennae (EgriOBP2, 3, 5, 9, 12, 14, and 25) had ratios of male:female antennae expression of 14.42, 4.79, 2.37, 8.51, 6.73, 4184.72, and 13.28, respectively. Five OBPs were expressed equally in male and female antennae (EgriOBP1, 11, 13, 16, and 17). Only one OBP (EgriOBP4) presented a female PG-enriched expression pattern, with its expression level in the PG being at least four times higher than that in other tissues. Additionally, three OBPs (EgriOBP10, 18, and 19) showed a pattern of abundant expression in female antennae, with the ratios of female:male antennae being 2.25, 2.26, and 5.64, respectively. Of the other OBPs, EgriOBP8 and 22 were highly expressed in male heads, EgriOBP6 and 15 had enriched expression in the male abdomen, EgriOBP20 presented predominant expression in the male leg, and EgriOBP7 and 21 were highly expressed in proboscises. Two OBPs (EgriOBP23 and 24) had ubiquitous expression patterns, with no significant differences between the tissues tested.
Figure 6.
Tissue expression profile of selected EgriOBP genes based on relative mRNA quantity. The level of EgriOBPs expression in the female antennae was set at 1. A, antennae; Ab, abdomen; H, heads; L, legs; Pg, female pheromone glands; Pr, proboscises; T, thoraxes; W, wings.
As for CSPs, seven EgriCSPs (5, 8, 11, 12, 13, 14, and 16) were found to be expressed at high levels in the antennae. EgriCSP5, 11, 12, 13, 14, and 16 showed dominant expression in the female antennae, which was above their expression levels in other tissues that were tested, whereas EgriCSP8 showed equal levels in both sexes (Figure 7). Two other EgriCSPs, EgriCSP3 and 18, showed a proboscis expression pattern indicating their gustatory function. Furthermore, EgriCSP9, 10, and 20 were expressed at obviously raised levels in legs, PGs, and wings, respectively. No other EgriCSPs had significantly raised levels of expression in any of our tissues tested. Among the 20 EgriCSPs, EgriCSP5, 13, 15, 17, and 19 ranked as having the top five expression levels (Figure S2).
Figure 7.
Tissue expression profile of selected EgriCSP genes based on relative mRNA quantity. The level of EgriCSPs expression in the female antennae was set at 1. A, antennae; Ab, abdomen; H, heads; L, legs; Pg, female pheromone glands; Pr, proboscises; T, thoraxes; W, wings.
Discussion
The sex pheromone components of E. grisescens, Z3, Z6, Z9-18:H and Z3, epo6, Z9-18:H (Ma et al. 2016), belong to the type II group. It is expected that triene, a type II pheromone component, is synthesized in epidermal oenocytes before being transported to the PG, where it is oxidized to epoxydiene and then emitted into the atmosphere. We were unable to dissect and extract RNA from oenocytes; instead, we selected PG, since it is an important tissue responsible for the synthesis and release of type I sex pheromones. CYP might be involved in this oxidation process (Rong et al. 2014). OBP and CSP are thought to have a transportation function for sex pheromones in the type I group (Liu et al. 2013; Zhang et al. 2014; Chang et al. 2015; Z.-Q. Li et al. 2015; N. Y. Liu et al. 2015; G.-H.Zhu et al. 2016) and may, therefore, also participate in the transportation of type II sex pheromone components.
Our study aimed to identify the genes that are potentially involved in the biosynthesis (CYPs) and transportation (OBPs and CSPs) of type II sex pheromone components. We constructed a massive PG transcriptome database, yielding 14.72 Gb from two biological replicates of E. grisescens. The result was 111 CYPs in total. Twenty-five of the OBPs and 20 of the CSPs were first identified in this species. We further analyzed the phylogenetic and tissue expression patterns and found that 30 EgriCYPs, EgriOBP4, and EgriCSP10 had enriched expression patterns in the PG, and that EgriOBP2, 3, and 25 clustered in the moth PBP clade. We therefore considered these our candidate genes.
CYP is involved in the epoxidation of triene to epoxydiene in the PGs. It follows that CYPs that are distinctly expressed at higher levels in PGs than in adult somatic cells, and that are more abundant than the other CYPs in the PGs, may be involved in sex pheromone biosynthesis. Five CYPs displayed PG-predominant expression, had far higher abundance than other CYPs in the PGs: EgriCYP9A116, EgriCYP340BD1, EgriCYP4L-fragment2, EgriCYP4G174, and EgriCYP6AB120. Therefore, these five CYP genes might function in sex pheromone biosynthesis. However, previous research stated that in the fall webworm H. cunea, one CYP member (CYP341B14) was able to epoxidize a specific (Z)-double bond in a pheromone precursor, (3Z,6Z,9Z)-3,6,9-henicosatriene (Rong et al. 2014). One of the type II sex pheromone components in E. grisescens is Z3, epo6, Z9-18:H, which contains an epoxide group at the sixth position. We found that its orthologous gene is also a CYP341 member (EgriCYP341B-fragment3) with a dominant expression pattern in the PG. It showed high levels of identity (77%) with CYP341B14 of H. cunea. Further studies will focus on full-length cloning and functional characterization of these six CYP genes.
The phylogenetic tree analysis of EgriOBPs showed that EgriOBP2, 3, and 25 cluster in a moth PBP clade. Tissue expression pattern results showed that the three OBPs highly expressed in male antennae correspond to expression patterns in other moth PBPs (Liu et al. 2013; Sun et al. 2013; Jin et al. 2014; N. Y. Liu et al. 2015). Notably, we found two PBP1 analogs (EgriOBP2 and 25), and one PBP2 analog (EgriOBP3), but no PBP3 analog. Another moth, H. cunea, has the miscellaneous pheromone type and full PBP1, 2, and 3 genes (Zhang et al. 2016). It may be that the moths with type II pheromones and PBP3 genes have not separated from PBP1. Only the female PG transcriptome database has been built, so the antennae transcriptome and genome of this species is needed to further confirm our results and conclusions. Similarly, in the GOBP clade, GOBP1 could not be found in our database; only the GOBP2 analog was identified. However, among 25 EgriOBPs, the mRNA expression of four genes (EgriOBP1, 2, 3, and 25) did not rank highly among all EgriOBPs. There are some olfactory sensilla distributed on the ovipositor (Faucheux 1988). EgriOBP4 is the only OBP gene to display a PG-enriched expression pattern, which ranked it sixth in expression level of the OBPs in the PG. We propose that, in E. grisescens, EgriOBP2, 3, 4, and 25 are type II sex pheromone transporters and that further functional characterization is needed.
Unlike the EgriOBPs, the expression patterns of EgriCSPs are similar to those of other insect CSPs (Z.-Q. Li et al. 2015; X.-M. Li et al. 2015; Zhang et al. 2016). Interestingly, we did not find any male antennae with enriched CSP expression levels, but found three female antennae enriched with EgriCSPs: 5, 11, 12, 14, and 16. These may participate in female-specific physiological behaviors in moths, such as locating ovipositor sites. We found high levels of EgriCSP3 and 18 expression in proboscises, suggesting that they may have a role in moth tasting, as was found in previous functional characterizations of CSPs from Helicoverpa armigera and Helicoverpa assulta (Y. L. Liu et al. 2014). Three BmorCSPs (6, 11, and 15) of B. mori were detected by protein sequencing in PGs but not in antennae (Dani et al. 2011). Their analogs are EgriCSP6, 8, and 18, respectively, but these are not expressed at high levels in PGs. In this study, EgriCSP10 was the only EgriCSP expressed at high levels in PGs, and we thus propose that they are involved in sex pheromone transportation in this moth.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300543/-/DC1.
Acknowledgments
This study was funded by National Key Research & Development (R&D) Plan (2016YFD0200900), the National Natural Science Foundation of China (31701795 and 31601892), and the Modern Agricultural Industry Technology System (CARS-23).
Author contributions: Z.-Q.L. and Z.-M.C. conceived and designed the experiments; Z.-Q.L. performed the experiments; Z.-Q.L., X.-M.C., L.M., Q.Y., Z.-X.L., Z.J.-X., L.B., and P.H. analyzed the data; and Z.-Q.L. and P.H. wrote the manuscript. All authors reviewed the final manuscript. The authors declare no conflict of interests.
Footnotes
Communicating editor: R. Anholt
Literature Cited
- Ando T., Inomata S., Yamamoto M., 2004. Lepidopteran sex pheromones. Top. Curr. Chem. 239: 951–961. [DOI] [PubMed] [Google Scholar]
- Angeli S., Ceron F., Scaloni A., Monti M., Monteforti G., et al. , 1999. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 262: 745–754. [DOI] [PubMed] [Google Scholar]
- Briand L., Swasdipan N., Nespoulous C., Bezirard V., Blon F., et al. , 2002. Characterization of a chemosensory protein (ASP3c) from honeybee (Apis mellifera L.) as a brood pheromone carrier. Eur. J. Biochem. 269: 4586–4596. [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., et al. , 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Chang H., Liu Y., Yang T., Pelosi P., Dong S., et al. , 2015. Pheromone binding proteins enhance the sensitivity of olfactory receptors to sex pheromones in Chilo suppressalis. Sci. Rep. 5: 13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani F. R., Michelucci E., Francese S., Mastrobuoni G., Cappellozza S., et al. , 2011. Odorant-binding proteins and chemosensory proteins in pheromone detection and release in the silkmoth Bombyx mori. Chem. Senses 36: 335–344. [DOI] [PubMed] [Google Scholar]
- Derks M. F. L., Smit S., Salis L., Schijlen E., Bossers A., et al. , 2015. The genome of winter moth (Operophtera brumata) provides a genomic perspective on sexual dimorphism and phenology. Genome Biol. Evol. 7: 2321–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucheux M. J., 1988. Multiporous sensilla on the ovipositor of Monopis crocicapitella clem. (Lepidoptera: Tineidae). Int. J. Insect Morphol. Embryol. 17: 473–475. [Google Scholar]
- González D., Zhao Q., McMahan C., Velasquez D., Haskins W. E., et al. , 2009. The major antennal chemosensory protein of red imported fire ant workers. Insect Mol. Biol. 18: 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham L. A., Davies P. L., 2002. The odorant-binding proteins of Drosophila melanogaster: annotation and characterization of a divergent gene family. Gene 292: 43–55. [DOI] [PubMed] [Google Scholar]
- Guo W., Wang X., Ma Z., Xue L., Han J., et al. , 2011. CSP and Takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genet. 7: e1001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., He P., 2014. Molecular characterization, expression profiling, and binding properties of odorant binding protein genes in the whitebacked planthopper, Sogatella furcifera. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 174: 1–8. [DOI] [PubMed] [Google Scholar]
- He P., Zhang J., Liu N.-Y., Zhang Y.-N., Yang K., et al. , 2011. Distinct expression profiles and different functions of odorant binding proteins in Nilaparvata lugens stål. PLoS One 6: e28921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Zhang Y.-F., Hong D.-Y., Wang J., Wang X.-L., et al. , 2017. A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genomics 18: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe D. S., Scafe C. R., McKinney A. J., Tanouye M. A., 2002. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 12: 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquin-Joly E., Bohbot J., Francois M. C., Cain A. H., Nagnan-Le Meillour P., 2000. Characterization of the general odorant-binding protein 2 in the molecular coding of odorants in Mamestra brassicae. Eur. J. Biochem. 267: 6708–6714. [DOI] [PubMed] [Google Scholar]
- Jacquin-Joly E., Vogt R. G., Francois M. C., Nagnan-Le Meillour P., 2001. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. Senses 26: 833–844. [DOI] [PubMed] [Google Scholar]
- Jin J.-Y., Li Z.-Q., Zhang Y.-N., Liu N.-Y., Dong S.-L., 2014. Different roles suggested by sex-biased expression and pheromone binding affinity among three pheromone binding proteins in the pink rice borer, Sesamia inferens (Walker) (Lepidoptera: Noctuidae). J. Insect Physiol. 66: 71–79. [DOI] [PubMed] [Google Scholar]
- Jurenka R., 2003. Biochemistry of female moth sex pheromones, in Insect Pheromone Biochemistry and Molecular Biology, edited by Blomquist G. J., Vogt R. Elsevier Academic Press, London. [Google Scholar]
- Jurenka R., 2004a Insect pheromone biosynthesis, pp. 97–132 in The Chemistry of Pheromones and Other Semiochemicals I, edited by Schulz S. Springer, Berlin Heidelberg. [DOI] [PubMed] [Google Scholar]
- Jurenka R., 2004b Insect pheromone biosynthesis. Top. Curr. Chem. 239: 97–132. [DOI] [PubMed] [Google Scholar]
- Kitabayashi A. N., Arai T., Kubo T., Natori S., 1998. Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach). Insect Biochem. Mol. Biol. 28: 785–790. [DOI] [PubMed] [Google Scholar]
- Leal W. S., Nikonova L., Peng G., 1999. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 464: 85–90. [DOI] [PubMed] [Google Scholar]
- Li X.-M., Zhu X.-Y., Wang Z.-Q., Wang Y., He P., et al. , 2015. Candidate chemosensory genes identified in Colaphellus bowringi by antennal transcriptome analysis. BMC Genomics 16: 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.-Q., Zhang S., Luo J.-Y., Zhu J., Cui J.-J., et al. , 2015. Expression analysis and binding assays in the chemosensory protein gene family indicate multiple roles in Helicoverpa armigera. J. Chem. Ecol. 41: 473–485. [DOI] [PubMed] [Google Scholar]
- Liu G. X., Xuan N., Chu D., Xie H. Y., Fan Z. X., et al. , 2014. Biotype expression and insecticide response of Bemisia tabaci chemosensory protein-1. Arch. Insect Biochem. Physiol. 85: 137–151. [DOI] [PubMed] [Google Scholar]
- Liu N., Li M., Gong Y., Liu F., Li T., 2015. Cytochrome P450s – their expression, regulation, and role in insecticide resistance. Pestic. Biochem. Physiol. 120: 77–81. [DOI] [PubMed] [Google Scholar]
- Liu N. Y., Liu C. C., Dong S. L., 2013. Functional differentiation of pheromone-binding proteins in the common cutworm Spodoptera litura. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 165: 254–262. [DOI] [PubMed] [Google Scholar]
- Liu N. Y., Yang F., Yang K., He P., Niu X. H., et al. , 2015. Two subclasses of odorant-binding proteins in Spodoptera exigua display structural conservation and functional divergence. Insect Mol. Biol. 24: 167–182. [DOI] [PubMed] [Google Scholar]
- Liu Y. L., Guo H., Huang L. Q., Pelosi P., Wang C. Z., 2014. Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J. Exp. Biol. 217: 1821–1826. [DOI] [PubMed] [Google Scholar]
- Löfstedt C., Wahlberg N., Millar J. G., 2016. Evolutionary patterns of pheromone diversity in Lepidoptera, pp. 401 in Pheromone Communication in Moths: Evolution, Behavior, and Application, edited by Allison J. D., Cardé R. T. University of California Press, Oakland, CA. [Google Scholar]
- Ma T., Xiao Q., Yu Y. G., Wang C., Zhu C. Q., et al. , 2016. Analysis of tea geometrid (Ectropis grisescens) pheromone gland extracts using GC-EAD and GCxGC/TOFMS. J. Agric. Food Chem. 64: 3161–3166. [DOI] [PubMed] [Google Scholar]
- Millar J. G., 2000. Polyene hydrocarbons and epoxides: a second major class of lepidopteran sex attractant pheromones. Annu. Rev. Entomol. 45: 575–604. [DOI] [PubMed] [Google Scholar]
- Nan J., Shu-Xian L., Da-Yong X., Mei-Jun T., Qiang X., et al. , 2014. External morphology and molecular identification of two tea Geometrid moth from southern China. Yingyong Kunchong Xuebao 51: 987–1002. [Google Scholar]
- Nebert D. W., Gonzalez F. J., 1987. P450 genes: structure, evolution, and regulation. Annu. Rev. Biochem. 56: 945–993. [DOI] [PubMed] [Google Scholar]
- Ozaki M., Wada-Katsumata A., Fujikawa K., Iwasaki M., Yokohari F., et al. , 2005. Ant nestmate and non-nestmate discrimination by a chemosensory sensillum. Science 309: 311–314. [DOI] [PubMed] [Google Scholar]
- Pelosi P., Iovinella I., Felicioli A., Dani F. R., 2014. Soluble proteins of chemical communication: an overview across arthropods. Front. Physiol. 5: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y., Fujii T., Katsuma S., Yamamoto M., Ando T., et al. , 2014. CYP341B14: a cytochrome P450 involved in the specific epoxidation of pheromone precursors in the fall webworm Hyphantria cunea. Insect Biochem. Mol. Biol. 54: 122–128. [DOI] [PubMed] [Google Scholar]
- Sun M., Liu Y., Wang G., 2013. Expression patterns and binding properties of three pheromone binding proteins in the diamondback moth, Plutella xyllotella. J. Insect Physiol. 59: 46–55. [DOI] [PubMed] [Google Scholar]
- Sun Y. L., Huang L. Q., Pelosi P., Wang C. Z., 2012. Expression in antennae and reproductive organs suggests a dual role of an odorant-binding protein in two sibling Helicoverpa species. PLoS One 7: e30040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillman J. A., Seybold S. J., Jurenka R. A., Blomquist G. J., 1999. Insect pheromones–an overview of biosynthesis and endocrine regulation. Insect Biochem. Mol. Biol. 29: 481–514. [DOI] [PubMed] [Google Scholar]
- Vogt R. G., Grosse-Wilde E., Zhou J.-J., 2015. The Lepidoptera odorant binding protein gene family: gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 62: 142–153. [DOI] [PubMed] [Google Scholar]
- Xu P. X., Zwiebel L. J., Smith D. P., 2003. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 12: 549–560. [DOI] [PubMed] [Google Scholar]
- Xu Y. L., He P., Zhang L., Fang S. Q., Dong S. L., et al. , 2009. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genomics 10: 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Kun-Shan Y., Mei-Jun T., Qiang X., 2014. Geographic populations of the tea geometrid, Ectropis obliqua (Lepidoptera: Geometridae) in Zhejiang, eastern China have differentiated into different species. Acta Entomologica Sinica 57: 1117–1122. [Google Scholar]
- Zhang L.-W., Kang K., Jiang S.-C., Zhang Y.-N., Wang T.-T., et al. , 2016. Analysis of the antennal transcriptome and insights into olfactory genes in Hyphantria cunea (Drury). PLoS One 11: e0164729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-N., Ye Z.-F., Yang K., Dong S.-L., 2014. Antenna-predominant and male-biased CSP19 of Sesamia inferens is able to bind the female sex pheromones and host plant volatiles. Gene 536: 279–286. [DOI] [PubMed] [Google Scholar]
- Zhu G.-H., Xu J., Cui Z., Dong X.-T., Ye Z.-F., et al. , 2016. Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome editing. Insect Biochem. Mol. Biol. 75: 1–9. [DOI] [PubMed] [Google Scholar]
- Zhu J., Ban L., Song L.-M., Liu Y., Pelosi P., et al. , 2016. General odorant-binding proteins and sex pheromone guide larvae of Plutella xylostella to better food. Insect Biochem. Mol. Biol. 72: 10–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets of PG transcriptomes used in this study are available in the NCBI SRA database (http://trace.ncbi.nlm.nih.gov/Traces/sra/), under accession numbers SRR5571992; SRR5571993; SRR5571994 and SRR5571995.