Abstract
Meiosis is a specific type of cell division that is essential for sexual reproduction in most eukaryotes. Mitochondria are crucial cellular organelles that play important roles in reproduction, though the detailed mechanism by which the mitochondrial respiratory chain functions during meiosis remains elusive. Here, we show that components of the respiratory chain (Complexes I–V) play essential roles in meiosis initiation during the sporulation of budding yeast, Saccharomyces cerevisiae. Any functional defects in the Complex I component Ndi1p resulted in the abolishment of sporulation. Further studies revealed that respiratory deficiency resulted in the failure of premeiotic DNA replication due to insufficient IME1 expression. In addition, respiration promoted the expression of RIM101, whose product inhibits Smp1p, a negative transcriptional regulator of IME1, to promote meiosis initiation. In summary, our studies unveiled the close relationship between mitochondria and sporulation, and uncover a novel meiosis initiation pathway that is regulated by the respiratory chain.
Keywords: sporulation, respiratory chain, meiosis initiation, SMP1, NDI1
MITOCHONDRIA are fundamental organelles in eukaryotic cells that have been reported to be involved in spermatogenesis, oogenesis, and sporulation in yeast (Gorsich and Shaw 2004; Jambhekar and Amon 2008; Huang and Sha 2011). Proteomic and bioinformatic studies have revealed that during oogenesis and yeast sporulation, mitochondrial proteins are closely related to meiosis (Wang et al. 2010; Wen et al. 2016), and defects in mitochondrial dynamics or distribution ultimately results in decreased spore viability and abrogation of spore respiration (Gorsich and Shaw 2004). Moreover, respiration is reported to be essential for entry into the meiotic program and for providing energy for subsequent meiotic processes during sporulation in yeast (Jambhekar and Amon 2008). In addition, mitochondrial dysfunction has been reported to be associated with many diseases causing infertility (Ramalho-Santos et al. 2009; Rajender et al. 2010; Wai and Langer 2016). Thus, active mitochondria participate in multiple processes and are required for the function of the reproductive system (Ramalho-Santos et al. 2009).
Respiration involves a series of metabolic reactions that convert nutrients into adenosine triphosphate (ATP) for cellular utilization; among these reactions, oxidative phosphorylation (OXPHOS) is key for aerobic respiration. During OXPHOS, electrons are transferred through the electron transport chain (ETC), also known as the respiratory chain, to generate a proton gradient and synthesize ATP (Semenza 2007). Most ETC enzymes are large multi-subunit protein assemblages (Complexes I–IV) that contain many redox cofactors (Sazanov 2015). A component of Complex I, Ndi1p, the mitochondrial nicotinamide adenine dinucleotide (NADH) oxidoreductase of Saccharomyces cerevisiae, catalyzes electron transfer from NADH to ubiquinone (De Vries et al. 1992). Ndi1p forms a globular α/β structure and contains two canonical Rossmann domains with a flavin adenine dinucleotide (FAD) molecule buried deeply in the first domain (Feng et al. 2012). In addition to providing energy, mitochondria participate in various cellular functions during gametogenesis, such as hormone synthesis (Ramalho-Santos and Amaral 2013), apoptosis (Mishra et al. 2006; Tiwari et al. 2015), reactive oxygen species production (Lu et al. 2008), and the integration of metabolic to signaling pathways (Amaral et al. 2013; Mishra and Chan 2014; Tiwari et al. 2015).
In response to nitrogen starvation, the budding yeast enters the meiosis process (sporulation) in the presence of a nonfermentable carbon source (Zaman et al. 2008). The utilization of a nonfermentable carbon source requires respiration in mitochondria, and respiration has been reported to be necessary for yeast sporulation (Treinin and Simchen 1993). Moreover, the initiation of meiosis in yeast cells is regulated by multiple signals (Mitchell 1994). These signals converge at the promoter of a master regulator of yeast meiosis, IME1, which encodes a transcription factor that activates the expression of the so-called early meiotic genes (van Werven and Amon 2011). Ime1p in turn promotes the subsequent events of the sporulation program, such as premeiotic DNA replication, meiosis-specific chromosome remodeling, and homologous recombination (Smith et al. 1990; Benjamin et al. 2003). In addition, respiration has been shown to be required for PolII transcription, IME1 expression, DNA replication, and recombination during meiosis (Jambhekar and Amon 2008), and a separate respiration-sensing pathway differing from the energy supply has been proposed to govern meiotic entry (Jambhekar and Amon 2008). A recent study has shown that the expression of IME1 could be induced by inhibiting the protein kinase A (PKA) and target of rapamycin Complex I (TORC1) pathways in respiration-deficient cells (Weidberg et al. 2016). However, the functional role and molecular mechanism underlying respiration in gametogenesis have not been well understood, and whether there is an ATP production independent pathway regulated by respiration and how it works still require further investigation.
Here, we show that components of the respiratory chain (Complexes I–V) play essential roles in meiosis initiation during yeast sporulation. Defects in the Complex I component Ndi1p result in the abolishment of meiosis entry. Artificial induction of IME1 could bypass sporulation defects due to respiration deficiency, suggesting that Ime1p is a key mediator between respiration and meiosis initiation. During meiosis initiation, respiration promotes the expression of RIM101, inhibiting the expression of SMP1, which encodes a transcription repressor of IME1, thus relieving inhibition of IME1 expression to promote the initiation of meiosis. In summary, we dissected the close relationship between mitochondria and meiosis, and our studies uncovered a novel meiosis initiation pathway that is regulated by the respiratory chain.
Materials and Methods
Strains and plasmids
All experiments were performed using diploid SK1 strains produced by mating between appropriate haploids. The genotypes of all strains are listed in Supplemental Material, Table S1 in File S1. Unless otherwise stated, the mutations were homozygous. Strains expressing C-terminal-tagged proteins were constructed using a polymerase chain reaction (PCR)-based method (Longtine et al. 1998). The yeast deletion strains were constructed using a PCR-mediated gene replacement method as previously described (Wach et al. 1994). The truncated and mutant NDI1 expression plasmids were constructed by inserting the PCR products into the yeast vector pADH-YES2 (Cui et al. 2012). The IME1 and RIM101 overexpression plasmids were constructed by inserting the PCR products into YEplac195-CUP1 (Tagwerker et al. 2006).
Sporulation conditions and meiotic nuclear division assays
Sporulation was induced using potassium acetate as previously described (Wen et al. 2016). The strains were grown for 24 hr in YPD medium (1% yeast extract, 2% peptone, and 2% glucose), diluted in liquid YPA medium (1% yeast extract, 2% peptone, and 2% potassium acetate) to OD600 = 0.3, and grown for 10 hr at 30°. Cells were harvested, washed, resuspended in a sporulation medium (SPM, 2% potassium acetate) to OD600 = 1.9, and sporulated at 30° for different durations. The sporulation induced by rapamycin was performed as previously described (Zheng and Schreiber 1997). Wild-type (WT) and respiratory chain-deficient cells were incubated in YPD for 24 hr with vigorous shaking. When the cells reached saturation, and arrested at G1 phase, the cell culture was equally divided into two aliquots. Each aliquot was then incubated with methanol (control) or rapamycin at a final concentration of 100 nM. Meiotic nuclear divisions, representing the sporulation efficiency, were visualized by staining chromosomal DNA with 1 μg/ml 4’,6-diamidino-2-phenylindole (DAPI): the samples were harvested at the indicated times and directly fixed in an equal volume of 100% ethanol for subsequent DAPI staining. Images were recorded and analyzed under a Nikon Eclipse Ti microscope (Eclipse Ti-S; Nikon, Tokyo, Japan).
Intracellular ATP measurement
The concentration of intracellular ATP was measured using an ATP Assay Kit (S0027; Beyotime, Shanghai, China). Briefly, cells were induced to sporulate by adding rapamycin as described above, harvested at the indicated time points, and stored at −80° before detection. Cell pellets were resuspended in 400 μl ATP determination lysis solution and transferred to 2-ml snap-cap tubes with 500 μl of glass beads. The cells were then vortexed for 5 min at full speed at 4° and centrifuged at 12,000 rpm at 4° for 5 min. The supernatants were processed, and intracellular ATP concentrations were determined according to the ATP Assay Kit instructions (Zhang et al. 2014). Luminance (RLU) was detected using a FLUOstar OPTIMA (BMG LABTECH, Offenburg, Germany), and the concentration of ATP was measured. A standard curve representing the ATP concentration (10 nM–10 μM) was prepared from a known amount of ATP; luminescence was then normalized to the protein concentration.
Immunoblotting
Cells were subjected to mild alkaline treatment and then boiled in a standard electrophoresis loading buffer, as previously described (Kushnirov 2000). The samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions and transferred to nitrocellulose membranes. The blots were incubated with a primary antibody and a fluorescent dye-labeled secondary antibody (926-32211; LI-COR Biosciences, Lincoln, NE); the blots were then scanned using an Odyssey infrared 740 imager (9120; LI-COR Biosciences). Myc antibodies were purchased from Abmart (M20002; Abmart, Shanghai, China), the RGS-His antibody was purchased from Qiagen (34610; Qiagen, Hilden, North Rhine-Westphalia, Germany), and the TAP antibody was obtained from Thermo Scientific (MA1-108; Thermo Scientific, Waltham, MA). The Pgk1p polyclonal antibody was generated in rabbits using the corresponding recombinant protein as the antigen. The quantification of immunoblotting was performed using Odyssey imager software and normalized with Pgk1p signal.
Extraction of total RNA from yeast
RNA was isolated from yeast as previously described (Schmitt et al. 1990). Samples were collected and resuspended in 400 μl AE buffer [50 mM Na acetate pH 5.3 and 10 mM ethylenediaminetetraacetic acid (EDTA)], and 40 μl 10% SDS was added. The suspension was vortexed for 5 min, and 400 μl fresh phenol was added. The mixture was vortexed again for 5 min and incubated at 65° for 4 min; the mixture was rapidly chilled on ice for 5 min and then centrifuged for 2 min at 12,000 rpm. The upper aqueous phase was transferred to a fresh tube, after which phenol and chloroform (1:1) were added and incubated for 5 min at room temperature. After centrifuging for 5 min at 12,000 rpm, the upper aqueous phase was again transferred to a fresh tube, and 40 μl 3 M Na acetate and 2.5 vol ethanol were added to precipitate the RNA. After washing with 80% ethanol, the pellet was dried for 5 min, resuspended in 20 μl diethyl pyrocarbonate (DEPC)-treated water and stored at −80°.
Chromatin immunoprecipitation (ChIP) analyses
ChIP analysis of target proteins was performed using antibodies, primarily as described by El Hage et al. (2014). Cells were harvested at the indicated time points after rapamycin treatment. Meiotic cells were cross-linked with formaldehyde (1%) for 25 min at room temperature. The pellets were resuspended in 400 μl FA-1 lysis buffer [50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA at pH 8, 1% Triton X-100, 0.1% w/v sodium deoxycholate, plus CPI-EDTA 1× (11697498001, Protease inhibitor cocktail; Roche)], mixed with 500 μl of glass beads (G8772; Sigma, St. Louis, MO), and vortexed for 45 min at full speed at 4°. The glass beads were removed, and the cross-linked chromatin was recovered by centrifugation at 12,000 rpm for 10 min at 4° (the supernatant was discarded). FA-1 buffer (800 μl) was added to the top of the pellet. The chromatin was sonicated for 2 min (10 sec ON, 15 sec OFF, 20% amplitude) and then centrifuged for 15 min at 12,000 rpm at 4°; glycerol 5% was added to the supernatants. The sonicated chromatin was mixed with Sepharose Cl-4B beads (CL4B200; Sigma) and cleared for 1 hr at 4°. Immunoprecipitation was performed by mixing “cleared-sonicated chromatin” with 35–40 mg IgG2a antibody and a 100-μl bed of Protein A Sepharose CL-4B beads (17-0780-01; GE Healthcare) on a rotating wheel overnight at 4°. The beads were recovered and washed successively with FA-1 buffer (plus CPI-EDTA 1×), FA-2 buffer (as FA-1 buffer but with 500 mM NaCl, plus CPI-EDTA 1×), FA-3 buffer (10 mM Tris-HCl at pH 8, 0.25 M LiCl, 0.5% NP-40, 0.5% w/v sodium deoxycholate, 1 mM EDTA at pH 8, plus CPI-EDTA 1×), and TE 1× (100 mM Tris-Cl at pH 8, 10 mM EDTA at pH 8) at 4°. The cross-link of the sonicated chromatin was reversed by incubating the washed beads with 250 μl TE buffer containing 1% SDS and 1 mg/ml proteinase K overnight at 65°. The DNA was purified using a Qiagen PCR purification kit and eluted with 55 μl of buffer EB containing RNase A (0.5 mg/ml). Quantitative PCR was performed using the immunoprecipitated DNA or whole-cell extracts.
Quantitative real-time PCR
DNA was synthesized using PrimeScript RT Reagent Kit (RR037A; TaKaRa, Kusatsu, Japan). Amplification was performed in a 10-μl reaction with 5 μl 2× EvaGreen mix (MasterMix-S; Applied Biological Materials, Richmond, Canada), 0.8 μl each primer (10 nmol/liter), 2 μl sample complementary DNA (cDNA), and 2.2 μl ddH2O. Real-time PCR was performed using a Roche Light Cycler 480II System (Roche Diagnostics, Mannheim, Germany). The PCR program was initiated at 95° for 10 min, followed by 40 cycles of denaturation for 5 sec at 95°, annealing for 30 sec at 60°, and elongation for 60 sec at 72°. Fluorescence signals were observed at 72° during the elongation step. Each sample was analyzed with at least three biological replicates and normalized to ACT1 or NUP84, respectively. The results were analyzed using Light Cycle 480 SoftWare 1.5.1 in the Roche Light Cycler 480II System.
Yeast growth sensitivity
Yeast strains were grown in YPD or liquid synthetic complete medium with glucose without the corresponding essential amino acid at 30° to an OD600 of 1.0. The cultures were then serially diluted 10 times, and each dilution was spotted onto an auxotrophic plate with glucose or glycerol. The plates were incubated at 30° for 3 days.
Flow cytometry
To analyze the DNA content, 1 × 107 cells were fixed with 1 ml cold 70% ethanol overnight and then resuspended in 1 ml 50 mM sodium citrate. The samples were centrifuged at 2000 rpm for 5 min, and the supernatant was removed. The pellets were digested with RNase A for 2 hr at 37° and then sonicated for 2 sec at 20% power, stained with 1 μM Sytox Green (S-7020; Molecular Probes, Eugene, OR), and analyzed using a BD FACSvantage SE Flow Cytometry System (BD, Franklin Lakes, NJ).
Statistical analysis
All data were performed with at least three biological replicates, and a representative result was shown in Figure 3B. All data were presented as the mean ± SEM. The statistical significance of the differences between the mean values for the different groups was measured by the Student’s t-test with a paired two-tailed distribution. The data were considered significant when the P value was <0.05 (*) or 0.01 (**).
Figure 3.
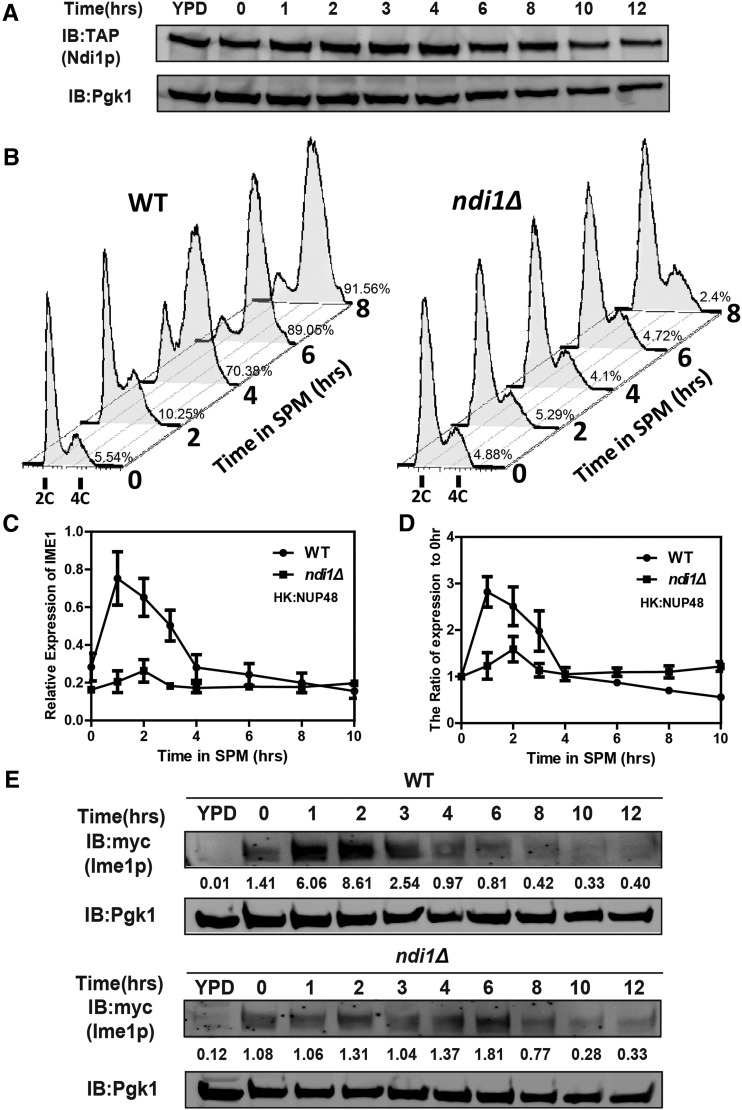
DNA replication and Ime1p induction defects in ndi1Δ cells. (A) Expression of Ndi1p during sporulation. The WT strain expressing the Ndi1p-TAP allele (LW1542) was incubated in SPM, and samples were collected at different times after sporulation induction. Expression of Ndi1p-TAP was analyzed over time by immunoblotting using an anti-TAP antibody. Pgk1p served as a loading control. (B) Premeiotic DNA replication was inhibited in the ndi1Δ strain during sporulation. WT (LW0066) or ndi1Δ strains (LW0323) were incubated in SPM, and samples were collected at different times after induction. The DNA content was analyzed by flow cytometry to detect premeiotic DNA replication (2C–4C transition). The percentage of cells with a 4C DNA content was shown on the right of the corresponding 4C DNA peak. (n = 3). (C) Quantitative PCR analysis of the IME1 expression level in WT (LW0066) and NDI1 deletion strains (LW0323). Cells were harvested at the indicated time points. Total RNA was isolated and reverse transcribed, and IME1 mRNA levels were measured by quantitative PCR. The signals were normalized to the levels of NUP48. HK, Housekeeping gene. Error bars indicate ± SEM (n = 3). (D) Quantification of IME1 by Q-PCR relative to the 0 hr time point shown in (C). (E) Expression of Ime1p was detected in WT and ndi1Δ strains by immunoblotting during sporulation. WT (LW1543) or ndi1Δ (LW1544) cells expressing the IME1-3×Myc allele were incubated in SPM, and samples were collected at different times. Pgk1p served as a loading control. Quantification of Ime1p via western blotting normalized by Pgk1p was shown below the corresponding bands.
Data availability
The authors declare that all other data supporting the findings are available within the article. Table S1 in File S1 contains all strains used in this study. Table S2 in File S1 contains all oligonucleotide sequence information.
Results
The functional role of the respiratory chain in sporulation
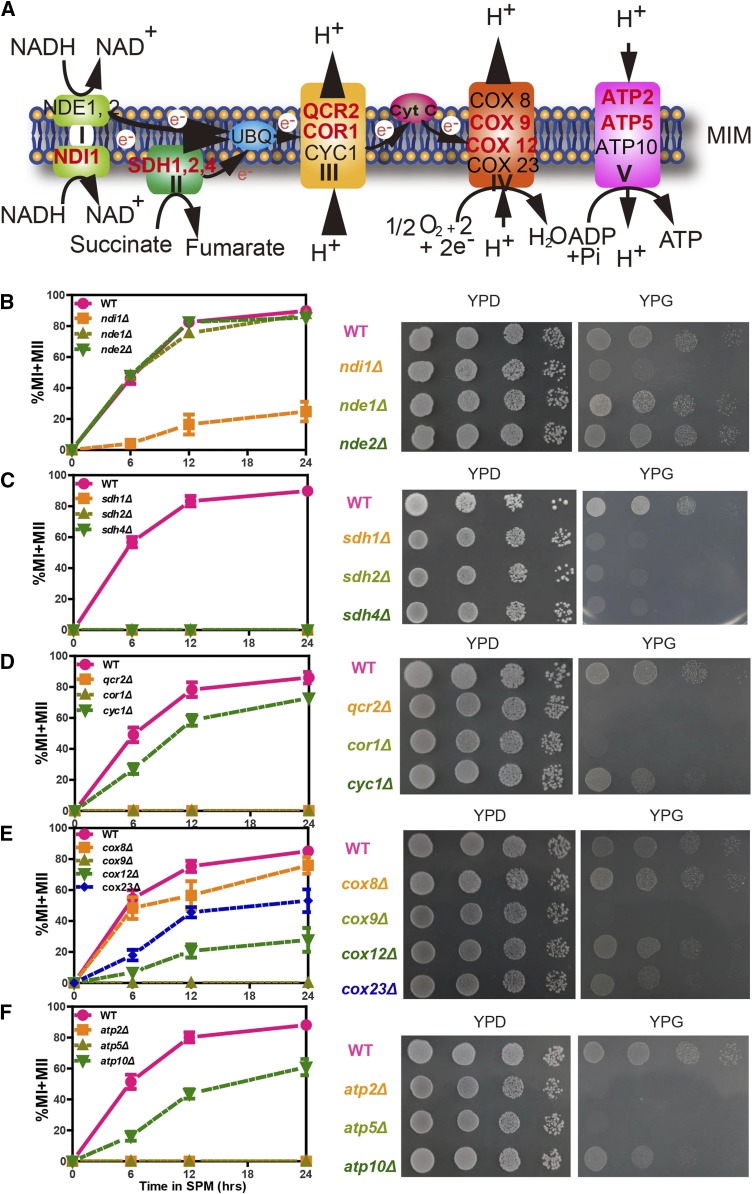
The respiratory chain, which consists of five large multi-subunit protein complexes (Complexes I–V), is a core “machine” in mitochondria that is vital for OXPHOS in electron transfer, proton gradient generation, and ATP synthesis (Figure 1A) (De Vries et al. 1992; Sazanov 2015). To study the functional role of the respiratory chain in meiosis, small-scale screening of the respiratory chain enzymes in Complexes I–V was performed. We selected several key enzymes from each complex and deleted their corresponding genes in the S. cerevisiae SK1 background using homologous recombination. We found that some mutant strains, including ndi1Δ (Complex I), sdh1Δ, sdh2Δ, sdh4Δ (Complex II), cor1Δ, qcr2Δ (Complex III), cox9Δ (Complex IV), atp2Δ and atp5Δ (Complex V), exhibited significantly lower sporulation efficiency than the WT strain (Figure 1, B–F, left panel). However, some mutant strains, including nde1Δ, nde2Δ (Complex I), cyc1Δ (Complex III), cox8Δ, cox12Δ, cox23Δ (Complex IV), and atp10Δ (Complex V), did not show obvious defects in sporulation (Figure 1, B–F, left panel). To further confirm the relationship between the respiratory chain and sporulation, we cultured cells on YPG (yeast extract, peptone, glycerol) medium, which contains a nonfermentable carbon source on which respiration-deficient cells fail to grow, to examine the respiratory activities of individual mutant strains (Figure 1, B–F, right panel). As the mutants with impaired respiratory ability (slow growth on YPG) displayed decreased sporulation efficiency, our results indicate that respiratory activity in mitochondria is essential for yeast sporulation.
Figure 1.
The ETC is essential for sporulation and respiration. (A) Schematic representation of Complexes I–V in the ETC. (B–F) Effects on sporulation and respiration of the deletion of key enzymes from Complexes I–V. Left panel: WT and mutant strains were induced to sporulate by transfer to SPM and analyzed at different time points. The divided nucleates, representing the sporulation efficiency, were stained with DAPI, visualized and counted under a microscope. Error bars indicate ± SEM (n = 3). Right panel: Yeast cells were spotted onto corresponding positions on YPD or YPG (a nonfermentable carbon source medium) plates by serial 10-fold dilutions (n = 4). WT: LW0066; ndi1Δ: LW0323; nde1Δ: LW0326; nde2Δ: LW0329; sdh1Δ: LW1327; sdh2Δ: LW1330; sdh4Δ: LW1333; qcr2Δ: LW1336; cor1Δ: LW1339; cyc1Δ: LW1342; cox8Δ: LW1345; cox9Δ: LW1348; cox12Δ: LW1351; cox23Δ: LW1354; atp2Δ: LW1357; atp5Δ: LW1360; atp10Δ: LW1363.
Ndi1p is essential for the ability of respiration to regulate yeast sporulation
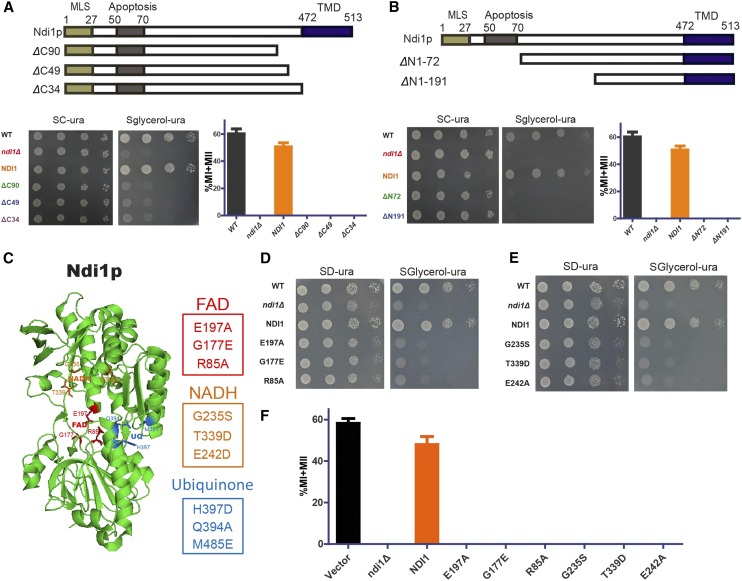
As NADH-Q oxidoreductase is the first enzyme in the respiratory chain, Ndi1p could catalyze electron transfer from NADH to ubiquinone (De Vries et al. 1992). To study the detailed relationship between the respiratory chain and sporulation, Ndi1p was selected as a representative for further investigation. First, a NDI1 expression vector under the control of its own promoter was generated and transformed into a ndi1∆ strain. We found that compared with the control strain, the NDI1 expression in the ndi1Δ strain rescued its sporulation defect (Figure 2A), indicating that NDI1 indeed plays a very important role in yeast sporulation. Previous studies have identified the following three main domains in Ndi1p: an MLS (mitochondria localization signal) domain at the N-terminus, an apoptosis-related domain in the middle, and a TMD (transmembrane domain) at the C-terminus (Yang et al. 2011; Iwata et al. 2012) (Figure 2A, top panel). To determine which domain is necessary for the involvement of Ndi1p in yeast sporulation, we generated a series of truncations (Figure 2, A and B and Figure S1, A and B in File S1) to delete the above-mentioned domains, and then transformed these mutants into ndi1Δ cells. The TMD domain was important for the localization of Ndi1p in the inner mitochondrial membrane, and the truncations that deleted the TMD domains could not rescue the sporulation and respiration deficiency (Figure 2A). Next, we detected the functional role of the MLS domain of Ndi1p in the N-terminus, and found that following the deletion of the MLS domain, the cells did not show growth defects on the YPG plate and maintained high sporulation efficiency (Figure S1, A and B in File S1). However, cells did not display respiratory activity and sporulation competence following deletion of both the MLS and apoptosis-related domains (Figure 2B). We also generated a series of point-mutations in NDI1 (Cui et al. 2012) (Figure 2C) disrupting the FAD-binding pocket, NADH-binding pocket, and ubiquinone-binding sites, as based on structural and functional information (Feng et al. 2012). Our data indicated that cells with a disrupted FAD- or NADH-binding pocket exhibited slow growth on YPG medium and could not sporulate in SPM (Figure 2, D and E). In contrast, cells lacking the ubiquinone-binding pocket showed no growth defects on YPG medium and maintained high sporulation efficiency (Figure S1C in File S1). In addition, these results were not due to an expression defect in the mutants (Figure S1D in File S1). Our results suggest that the functional role of Ndi1p in sporulation depends on the binding of FAD and NADH, but not ubiquinone, and that sporulation processes are closely related to respiration.
Figure 2.
Effects of Ndi1p on sporulation and respiration. (A and B, top panel) Schematic representation of the domains of Ndi1p, including ΔC90 (Δ424–513 aa, LW1522), ΔC49 (Δ465–513 aa, LW1523), ΔC34 (Δ480–513 aa, LW1524), ΔN1–72 (Δ1–72 aa, LW1531), and ΔN1–191 (Δ1–191 aa, LW1532). MLS indicates the mitochondria localization signal; apoptosis indicates the apoptosis-related region; TMD indicates the transmembrane domain. (A and B, bottom panel) Effects of different Ndi1p truncations on respiration and sporulation. Left panel, yeast cells were spotted onto corresponding positions on SD-ura or SG-ura plates by serial 10-fold dilutions (n = 4). Right panel shows that the indicated strains were induced to sporulate by transfer to SPM, and the percentages of MI and MII cells were measured after 24 hr. Error bars indicate ± SEM (n = 3). (C) Schematic representation of key residues of the functional sites in Ndi1p, including the FAD-binding pocket (R85, G177, and E197), NADH-binding pocket (G235, E242, and T339), and ubiquinone (UQ)-binding site (Q394, H397, and M485). (D) Interaction between Ndi1p and FAD is essential for respiration. Yeast cells (WT: LW1399; ndi1Δ with an empty vector: LW1398; Ndi1p: LW1521; G177E: LW1534; and E197A: LW1533; R85A: LW1535) were established as described in A. (E) The NADH-binding pocket of Ndi1p is essential for respiration. Yeast cells (WT; ndi1Δ with an empty vector; Ndi1p; G235S: LW1536; T339D: LW1537 and E242A: LW1538) were established as described in A. (F) The FAD- and NADH-binding pockets of Ndi1p are essential for sporulation. The yeast cells shown in F were established as described in A. Error bars indicate ± SEM (n = 3).
IME1 induction defect in ndi1Δ cells
To further investigate the mechanism by which meiosis is regulated by respiration, we determined the phases of sporulation that were affected in the NDI1-deleted strain. First, we detected expression of Ndi1p during sporulation by generating a tandem affinity purification (TAP) tag at the C-terminus. Based on our results, Ndi1p was continuously expressed throughout the sporulation program (Figure 3A), suggesting that the protein may have an ongoing function in sporulation, which is consistent with the requirement of mitochondrial activity throughout meiosis (Jambhekar and Amon 2008). To further identify the major meiotic process controlled by Ndi1p, we compared premeiotic DNA synthesis in the ndi1Δ strain with that in the WT strain by performing flow cytometry analysis. In WT cells, DNA replication began at 2 hr and was completed at ∼6 hr after the induction of sporulation (Figure 3B), whereas ndi1Δ cells did not complete DNA replication, even by 8 hr after induction (Figure 3B).
Yeast meiosis is initiated by the expression of IME1, which serves as a master regulatory switch in meiosis (Kassir et al. 1988; Vershon and Pierce 2000). Thus, we detected the transcription level of IME1 in the WT and ndi1Δ cells by real-time PCR. In WT cells, IME1 was highly expressed during the early phase of sporulation and subsequently decreased during the middle phases at ∼4 hr (Figure 3, C and D and Figure S2, A and B in File S1). In contrast, the expression of IME1 in ndi1Δ cells was maintained at a very low level throughout the sporulation process (Figure 3, C and D and Figure S2, A and B in File S1). We also measured the Ime1p level in the ndi1∆ strain by adding a 3×Myc tag at the C-terminus of Ime1p. The western blotting results showed that the protein levels were consistent with the corresponding messenger RNA (mRNA) levels (Figure 3E). These results indicate that Ndi1p participates in meiosis initiation by regulating the IME1 expression.
Energy supply-independent function of respiration during sporulation
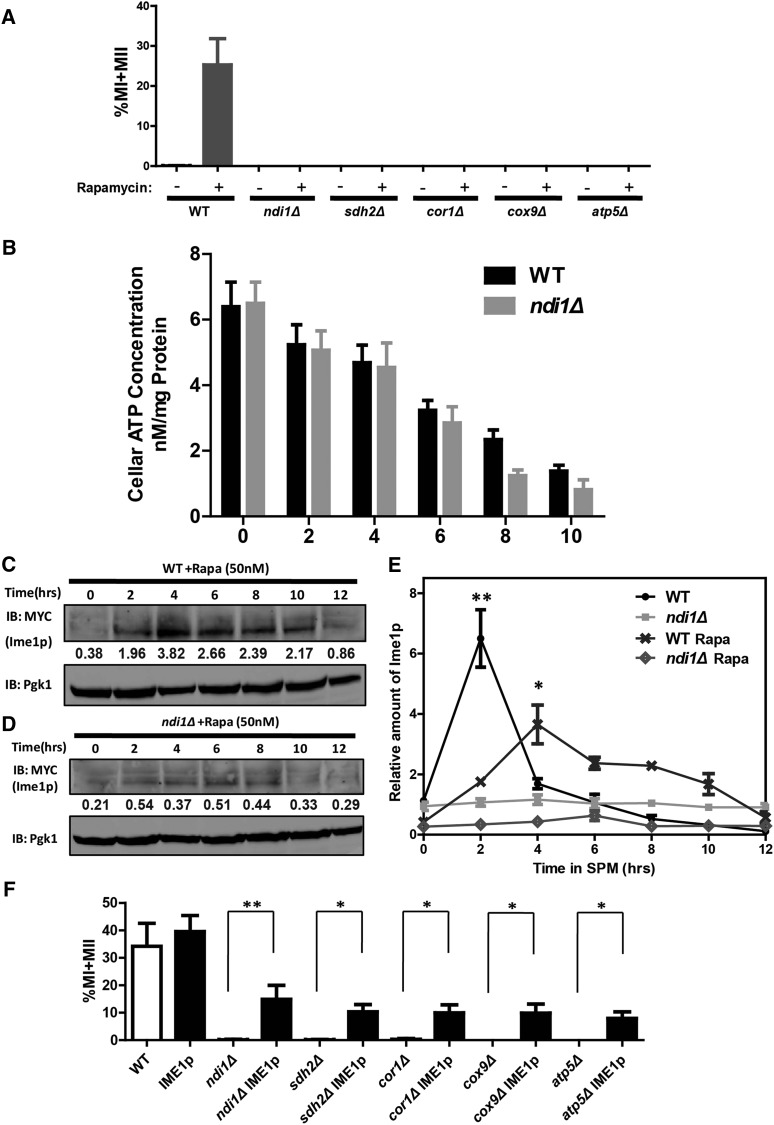
As the major energy source during yeast sporulation, the utilization of nonfermentable carbon source requires respiration in mitochondria (Neiman 2011). Usually, yeast sporulation is induced by nutritional starvation. The nitrogen source and fermentable carbon source (glucose) are lacking in the sporulation induction medium, whereas acetate, which is a nonfermentable carbon source, is added to the medium. ETC-deficient cells cannot efficiently utilize acetate to generate ATP because of the deficiency in respiration, but glucose is a fermentable carbon source that could be utilized by ETC-deficient cells to supply energy. Rapamycin treatment in the presence of glucose has been reported to induce yeast sporulation (Zheng and Schreiber 1997). Therefore, when the sporulation is induced under this condition, the energy levels of ETC-deficient cells could be restored during meiosis (Jambhekar and Amon 2008). If the major role of Ndi1p in yeast sporulation is to provide ATP, the sporulation defect observed in ndi1Δ cells might be rescued by a rapamycin treatment. We found that following treatment with rapamycin, WT cells could be induced to sporulate, whereas ndi1Δ and the other respiratory chain-deficient cells did not undergo meiosis (Figure 4A). To assess the energy supply in these strains, we measured intracellular ATP levels in WT and ETC-deficient cells during sporulation after rapamycin treatment, and no obvious differences were observed at the early meiotic stage (Figure 4B). These results suggest that Ndi1p and other respiratory chain components must have other functional roles in addition to supplying energy during sporulation.
Figure 4.
Respiration regulates meiosis initiation via Ime1p. (A) ETC-deficient cells could not be induced to sporulate by rapamycin in YPD medium. Indicated cells were grown to saturation in YPD for 24 hr at 30° and then treated with methanol (control) or rapamycin. The ratio of divided nuclei in cells was analyzed at 24 hr after induction using DAPI staining. Error bars indicate ± SEM (n = 3). WT: LW0066; ndi1Δ: LW0323; sdh2Δ: LW1330; cor1Δ: LW1339; cox9Δ: LW1348; atp5Δ: LW1360. (B) The intracellular ATP level of WT (LW0066) and ndi1Δ strains (LW0323) during rapamycin-induced sporulation. The cells were harvested at indicated time points and lysised by ATP determination lysis solution. The supernatants were processed and intracellular ATP concentrations were determined as described in the protocol of the ATP Assay Kit. Error bars indicate ± SEM (n = 5). (C and D) Expression of Ime1p was detected in WT and ndi1Δ strains by immunoblotting during rapamycin-induced sporulation. WT (LW1543) or ndi1Δ (LW1544) expressing the IME1-3×Myc allele were incubated in YPD and induced to sporulate with rapamycin; samples were collected at different times. Pgk1p served as a loading control. The relative amount of Ime1p was normalized with Pgk1p. (E) Quantitative results of Figure 4, C and D. * P < 0.05. (F) Overexpression of Ime1p rescued the sporulation defect in ETC-deficient strains (ndi1Δ, sdh2Δ, cor1Δ, cox9Δ, and atp5Δ). An Ime1p expression vector under control of the CUP1 promoter was generated and transformed into ETC-deficient strains. Cells were induced to sporulate as described in A and stained with DAPI after 24 hr. Error bars indicate ± SEM (n = 3). * P < 0.05 or ** P < 0.01. WT, ndi1Δ, sdh2Δ, cor1Δ, cox9Δ, and atp5Δ strains were the same as those shown in A; IME1: LW1563; ndi1ΔIME1: LW1564; sdh2ΔIME1: LW1565; cor1ΔIME1: LW1566; cox9ΔIME1: LW1567; atp5ΔIME1: LW1568.
Under sporulation conditions, IME1 was strongly induced by rapamycin treatment, while deletion of NDI1 severely impaired such induction (Figure 4, C–E). These results were consistent with the previous results: IME1 cannot be induced by rapamycin treatment in respiration-defective mutants (Jambhekar and Amon 2008). Thus, rapamycin treatment mimicked the normal sporulation process, and this approach can be used to investigate additional roles of Ndi1p and respiratory chain components in sporulation. To further study the downstream response of the respiratory chain in meiosis initiation, we generated an IME1 overexpression construct under the control of the CUP1 promote and transformed this plasmid into either WT or respiratory chain-deficient strains. We found that appropriate copper ion concentrations were necessary for IME1 induction and yeast sporulation, and that higher concentrations of copper ion might be toxic to cells and inhibit sporulation (Figure S3, A and B in File S1); therefore, we chose 100 mM copper to induce IME1 expression. After treatment, we detected the sporulation efficiency in these strains and found that expression of Ime1p in the respiratory chain-deficient strains could partially rescue the sporulation defects to a level greater than half of that observed in WT (Figure 4F and Figure S3C in File S1). These results suggest that in addition to providing energy, the respiratory chain regulates meiosis initiation by inducing IME1 expression.
Rim101p participates in meiosis initiation
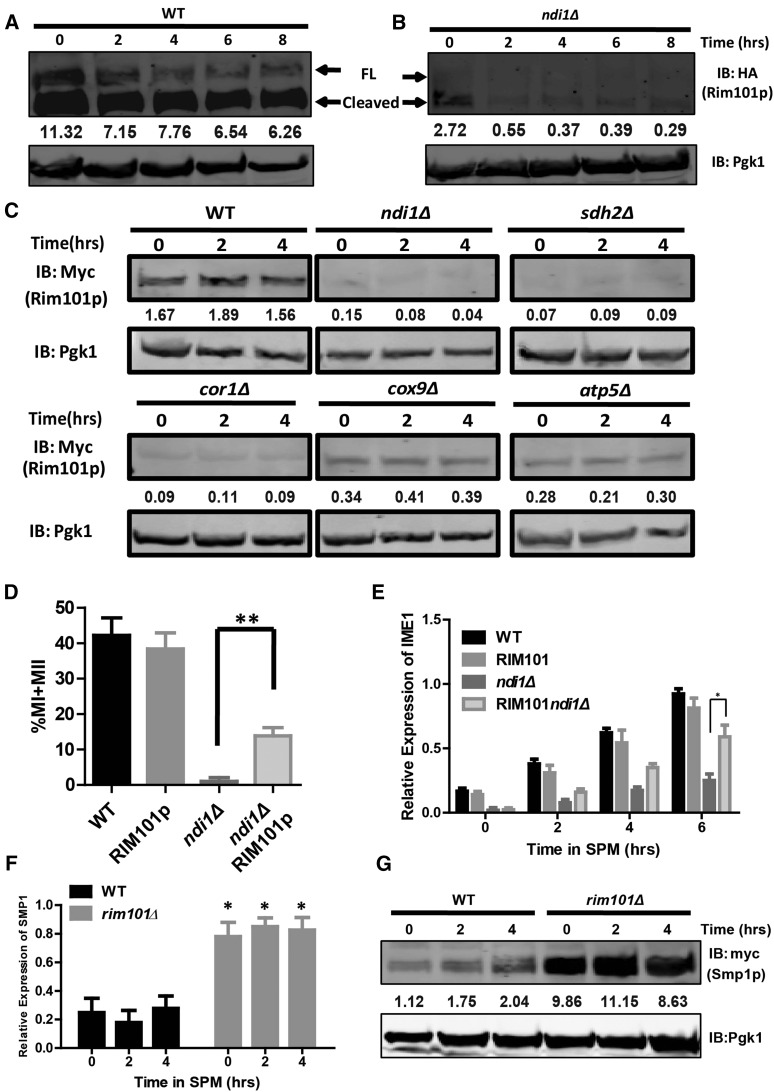
The transcription repressor RIM101 has been reported to participate in IME1 regulation (Su and Mitchell 1993; Li and Mitchell 1997; Epstein et al. 2001), and the expression level of RIM101 varies in response to different types of carbon utilization conditions and is reduced in mitochondrial dysfunction cells (DeRisi et al. 1997; Gasch et al. 2000). To investigate the potential role of Rim101p, we assessed cellular Rim101p level and its activation in WT and respiratory chain-deficient cells. First, we inserted a hemagglutinin (HA) tag into the middle of the coding sequence (CDS) of RIM101 as previously reported (Li and Mitchell 1997) to examine its proteolytic processing. In WT cells, Rim101p levels remained high from 0 to 8 hr, and the proteolytic process was efficient (Figure 5A). In ndi1Δ cells, Rim101p was processed during sporulation although its protein level was low (Figure 5B).
Figure 5.
Rim101p participates in meiosis initiation. (A and B) Expression and activation of Rim101p were detected in WT (LW1571) and ndi1Δ strains (LW1572) by immunoblotting during rapamycin-induced sporulation. FL, full-length Rim101p; Cleaved, cleaved Rim101p. Pgk1p was used as a loading control. The relative amount of Rim101p was normalized with Pgk1p and is shown below the corresponding band. (C) Expression of Rim101p was lower in ETC-deficient strains (ndi1Δ, sdh2Δ, cor1Δ, cox9Δ, and atp5Δ) than that in WT during meiosis. Pgk1p served as a loading control. The relative amount of Rim101p was normalized with Pgk1p and is shown below the corresponding band. WT: LW1545; ndi1Δ: LW1546; sdh2Δ: LW1547; cor1Δ: LW1548; cox9Δ: LW1549; atp5Δ: LW1550. (D) Overexpression of Rim101p rescued the sporulation defect in the NDI1 deletion strain. RIM101 was expressed in ndi1Δ cells, which were then induced to sporulate as described in A; the cells were stained with DAPI after 24 hr. Error bars indicate ± SEM (n = 3). ** P < 0.01. WT: LW0066; RIM101: LW1551; ndi1Δ: LW1552; ndi1ΔRIM101: LW1553. (E) Overexpression of Rim101p increased the transcription level of IME1 in ndi1Δ cells. The cells and procedures used to induce sporulation were the same as those described in D. The IME1 expression level in WT and NDI1 deletion strains was analyzed by real-time PCR. The signals were normalized to ACT1 levels. Error bars indicate ± SEM (n = 3). * P < 0.05. (F) Real-time PCR analysis of the SMP1 expression level in WT (LW0066) and RIM101 deletion strains (LW1560). The procedure was the same as that described in E. Error bars indicate ± SEM (n = 3). * P < 0.05. (G) The protein level of Smp1p was higher in rim101Δ cells (LW1573) than in WT cells (LW1366) during meiosis. Cells expressing the Smp1p-9×Myc allele were induced to sporulate with rapamycin, and samples were collected at different times. Pgk1p served as a loading control. The relative amount of Smp1p was normalized with Pgk1p and is shown below the corresponding band.
High external pH activates a conserved pH sensing pathway that includes some cell surface receptors and Rim101p (Hayashi et al. 2005; Baek et al. 2006; Lorenz and Cohen 2014). However, we found that the sporulation defect in ndi1Δ cells could not be rescued by increasing the pH of the medium (Figure S4, A and B in File S1). Although high external pH induced Rim101p activation in ndi1Δ cells (Figure S4C in File S1), the Ime1p levels were still very low in various alkaline media (Figure S4D in File S1). These results suggest that RIM101 expression alone is not sufficient to induce sporulation.
Next, we measured Rim101p levels in several respiratory chain-deficient strains and found that the Rim101p level was very low or undetectable (Figure 5C). To further confirm the function of Rim101p in sporulation, we constitutively overexpressed RIM101 in ndi1Δ cells (Figure S3D in File S1). After the induction of RIM101 for 2–6 hr, the IME1 transcript level was significantly higher in the RIM101 overexpressed strain than that in the ndi1Δ strain (Figure 5E). Most importantly, RIM101 overexpression improved the sporulation efficiency in the ndi1Δ strain from <2 to >15% (Figure 5D), suggesting that Ndi1p might regulate sporulation by up-regulating RIM101.
Smp1p directly regulates the transcription of IME1
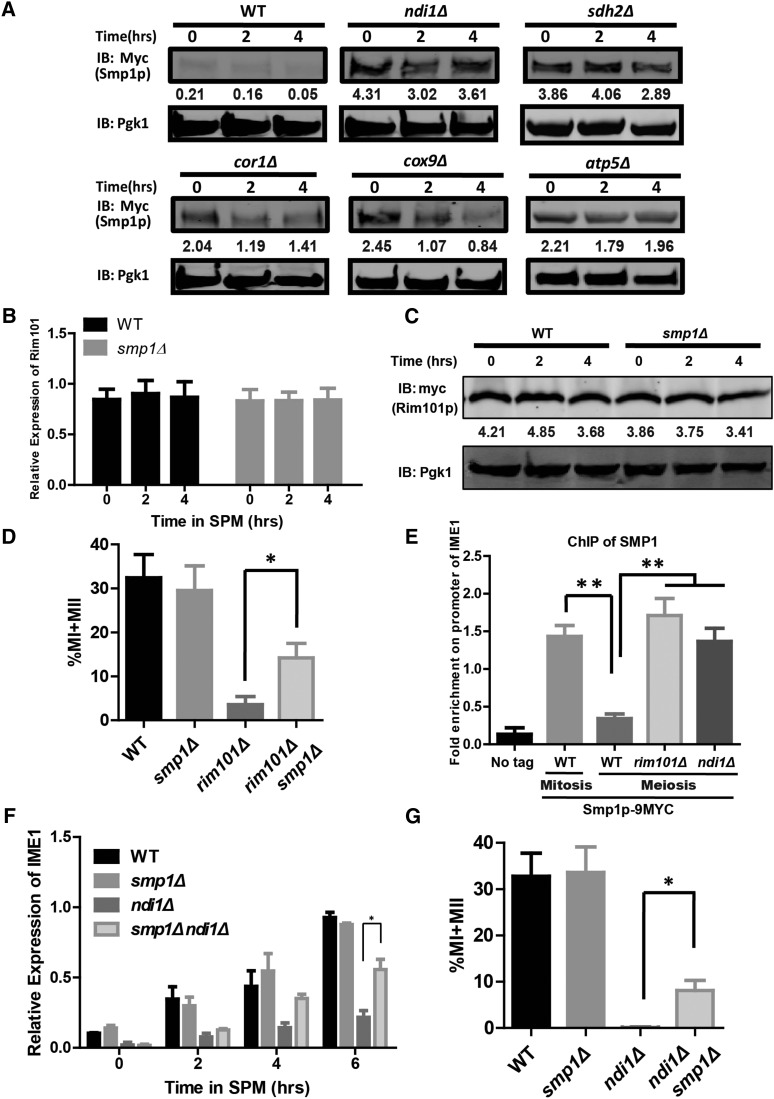
We then explored the mechanism by which Rim101p activates the downstream cascade to induce sporulation. Smp1p is a transcription factor negatively controlled by Rim101p and has been implicated in yeast sporulation (Lamb and Mitchell 2003). To determine whether Rim101p modulates sporulation by down regulating SMP1, we generated C-terminal 9 × Myc tagged Smp1p in both the WT and respiratory chain-deficient strains and then compared protein levels of Smp1p by western blotting. Compared to WT cells, all respiratory chain-deficient cells displayed high levels of Smp1p after sporulation induction (Figure 6A).
Figure 6.
The IME1 repressor Smp1p is down-regulated by Rim101p during meiosis. (A) Repression of Smp1p was impaired in ETC-deficient strains (ndi1Δ, sdh2Δ, cor1Δ, cox9Δ, and atp5Δ) during meiosis. Cells expressing Smp1p-9×Myc were induced to sporulate with rapamycin, and samples were collected at different times. Pgk1p served as a loading control. The relative amount of Smp1p was normalized with Pgk1p and is shown below the corresponding band. WT: LW1366; ndi1Δ: LW1554; sdh2Δ: LW1555; cor1Δ: LW1556; cox9Δ: LW1557; atp5Δ: LW1558. (B) Real-time PCR analysis of the RIM101 expression level in WT (LW0066) and SMP1 deletion (LW1559) strains. The procedure was the same as that described in Figure 5F. Error bars indicate ± SEM (n = 3). (C) The protein level of Rim101p did not differ markedly between smp1Δ cells (LW1574) and WT cells (LW1545) during meiosis. The procedure was the same as that described in Figure 5G. The relative amount of Rim101p was normalized with Pgk1p and is shown below the corresponding band. (D) SMP1 deletion rescued the sporulation defect in rim101Δ cells. The procedure used to induce sporulation was the same as that described in Figure 5A, and cells were stained with DAPI after 24 hr. Error bars indicate ± SEM (n = 3). * P < 0.05. WT: LW0066; smp1Δ: LW1559; rim101Δ: LW1560; smp1Δrim101Δ: LW1561. (E) The different binding activities of Smp1p to the IME1 promoter in WT (LW1366), rim101Δ (LW1573), and ndi1Δ (LW1554) cells. Mitotic cells (in YPD medium) and meiotic cells (in rapamycin + YPD medium) were cross-linked with formaldehyde, and Smp1p was immunoprecipitated from chromatin extracts. The recovered DNA was quantified by real-time PCR using primers (IME1-P3-F/R) corresponding to the IME1 promoter. The signals were normalized to whole-cell DNA. Error bars indicate ± SEM (n = 3). * P < 0.05. (F) Deletion of SMP1 increased the transcription level of IME1 in ndi1Δ cells. The IME1 expression level in WT and NDI1 deletion strains was analyzed by real-time PCR. The cells and procedures used to induce sporulation were the same as those described in Figure 4A. Error bars indicate ± SEM (n = 3). * P < 0.05. (G) SMP1 deletion rescued the sporulation defect in ndi1Δ cells. Cells were induced to sporulate as described in Figure 4A and stained with DAPI after 24 hr. Error bars indicate ± SEM (n = 3). * P < 0.05. WT: LW0066; smp1Δ: LW1559; ndi1Δ: LW0323; smp1Δndi1Δ: LW1562.
It seemed that Smp1p cannot be suppressed in respiratory chain-deficient cells due to the low level of Rim101p and ultimately blocked meiosis initiation. To study the relationship among Rim101p, Smp1p, and Ime1p, we first detected the expression of RIM101 and SMP1 in the deletion strains. In rim101Δ cells, the mRNA and protein levels of SMP1 were higher than those in the WT strain during sporulation (Figure 5, F and G). However, in smp1Δ cells, RIM101 mRNA and protein levels were not significantly changed (Figure 6, B and C). Moreover, we generated two double mutant strains, rim101Δ smp1Δ and ndi1Δ smp1Δ, and then assessed their sporulation efficiencies. After 24-hr induction, the sporulation efficiency of the rim101Δ smp1Δ strain was markedly higher than that of the rim101Δ strain (Figure 6D), suggesting that Smp1p is a negative regulator of meiosis that must be suppressed by Rim101p. Furthermore, the ndi1Δsmp1Δ strain transcribed more IME1 mRNA and exhibited a sporulation efficiency that was higher than that of the ndi1Δ strain (Figure 6, F and G), indicating that Ndi1p might regulate IME1 expression by up-regulating Rim101p and then inhibiting Smp1p expression. Additionally, Tup1p is another repressor of IME1 promoter (Weidberg et al. 2016). We found that the deletion of TUP1 enhanced IME1 expression in ndi1Δ cells (Figure S6 in File S1), suggesting that the repression of the IME1 promoter by Tup1p might not be dependent on respiration completely.
Next, ChIP experiments were performed to investigate the direct target gene of Rim101p and Smp1p during meiosis initiation. Although we did not detect a direct interaction between Rim101p and IME1 or SMP1 promoter (Figure S5, C–E in File S1), we found that Smp1p was highly enriched on the IME1 promoter in the region ∼1000 base pairs upstream of the transcription start site (Figure S5, A and B in File S1). Moreover, the ChIP results showed that Smp1p could directly bind to the IME1 promoter during mitosis, and this binding was weakened during meiosis (Figure 6E). This result suggested that Smp1p represses IME1 transcription during mitosis and is detached from the IME1 promoter during meiosis to induce IME1 expression. In rim101Δ cells, the interaction between Smp1p and the IME1 promoter was much stronger than that in WT cells, suggesting that Rim101p governs IME1 expression by regulating SMP1 expression during meiosis (Figure 6E). In addition, deletion of NDI1 also repressed detachment of Smp1p from the IME1 promoter (Figure 6E). These results suggest that IME1 is directly regulated by Smp1p during meiosis initiation.
Discussion
During mitosis, respiratory function is dispensable because yeast can utilize fermentation for growth (Chacinska and Boguta 2000). Nonetheless, respiration is essential for meiosis, and the mechanisms underlying respiration during meiosis remains poorly understood. To obtain comprehensive insight into this regulation and examine the functional role of the respiratory chain in meiosis, we generated a series of strains harboring deletions in different respiratory chain complexes (Complexes I–V). We found that not all deletion strains had sporulation defects, and only those deletions that disrupted respiratory function could abolish the meiotic process (Figure 1). Genes whose deletion did not affect both sporulation and respiration might be due to their redundant roles in these processes. Furthermore, a detailed examination of the function of Ndi1p confirmed the relationship between respiratory efficiency and sporulation (Figure 2).
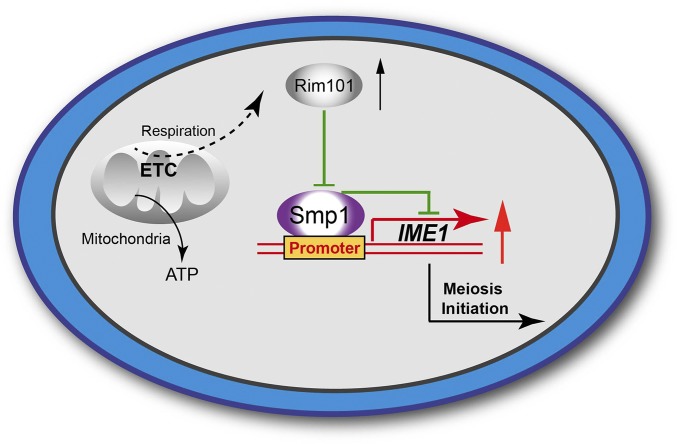
Mitochondrial function has been reported to be essential for meiosis initiation during yeast sporulation (Treinin and Simchen 1993; Gorsich and Shaw 2004). The primary function of mitochondria is ATP synthesis by oxidative phosphorylation via the respiratory chain. However, meiotic program entry is not only dependent on energy produced by respiration but is also controlled by a respiration-associated signaling pathway (Jambhekar and Amon 2008). To elucidate the detailed mechanism underlying this process, we generated a serial of respiratory chain-deficient strains, and induced them to sporulate in glucose-rich medium containing rapamycin. Our results suggested that the respiratory chain regulates meiosis initiation using a Rim101p-Smp1p-Ime1p cascade that is independent of the mitochondrial energy supply (Figure 7).
Figure 7.
Proposed model for the functional role of respiration during meiosis initiation. In addition to providing energy, respiration is involved in meiosis initiation during sporulation in yeast. Respiration stimulates expression of RIM101, followed by suppression of SMP1. Abolishment of Smp1p binding to the IME1 promoter facilitates IME1 transcription. The high expression level of IME1 initiates premeiotic DNA replication, ultimately initiating the sporulation process. The gray areas indicate known results from previous studies. The colored areas indicate the new findings from the current study.
In a previous study, the mutant strain pet100Δ was used to explore the relationship between respiration and meiosis, and the authors found that the overexpression of IME1 could not rescue the sporulation defect (Weidberg et al. 2016). Because pet100-null mutant cells accumulate cytochrome c oxidase due to the failure of cytochrome c oxidase assembly (Church et al. 2005), the pet100 null may not only have impaired respiratory activity, but also other mitochondrial dysfunctions, which could explain the discrepancy between their and our observations. Interestingly, despite IME1 overexpression promoting meiosis initiation, the sporulation defect in the respiratory chain-deficient cells could not be fully rescued (Figure 4F). Because some mitochondrial functions are essential for nuclear divisions (Jambhekar and Amon 2008), we propose that respiration affects not only meiosis initiation but also other aspects of sporulation. Future studies are needed to decipher the mechanism by which respiration regulates downstream meiotic events, such as nuclear division and spore packaging.
In addition to regulating IME1 expression, Rim101p is also involved in the alkalization response (Su and Mitchell 1993; Li and Mitchell 1997), and Rim101p is activated by alkaline pH-stimulated C-terminal proteolytic cleavage (Peñalva et al. 2008). Although the level of sporulation medium alkalization of respiration-deficient cells was actually lower than that of WT cells (Jambhekar and Amon 2008), we found that increasing the pH of the medium could not rescue the sporulation defect in ndi1Δ cells (Figure S4, A and B in File S1). These results suggest that RIM101 expression is insufficient to induce sporulation and must be combined with other condition(s). We favor the idea that an unknown respiration-dependent mechanism exists that regulates RIM101 expression.
A previous study showed that inhibiting two highly conserved, nutrient-sensing pathways, protein kinase A (PKA) and target of rapamycin Complex I (TORC1), mimics starvation-induced sporulation and drives cells to induce IME1, and to enter into the meiotic process under nutrient-rich conditions (Weidberg et al. 2016). Ime1p could be strongly induced in respiration-deficient cells by blocking the PKA and TORC1 pathways using mutagenesis, the ATP analog 1NM-PP1, and high concentrations of rapamycin (1000 ng/μl, ∼1 mM) (Weidberg et al. 2016). In addition, the authors proposed that glucose had already been consumed and was insufficient to support IME1 expression when respiration was blocked (Weidberg et al. 2016). However, our data reveal no obvious difference in the energy levels between WT and ETC-deficient cells during the early stage of sporulation induced by rapamycin (Figure 4A). In respiration-deficient cells, Ime1p expression could not be sufficiently induced to promote meiosis initiation by inhibiting the TORC1 pathway using low concentrations of rapamycin (100 nM). Tup1p is a key repressor of IME1, while PKA and TORC1 promote Tup1p binding to the IME1 promoter (Weidberg et al. 2016). Since the deletion of TUP1 enhanced the transcription of Ime1p in ndi1Δ cells (Figure S6 in File S1), the repression of the IME1 promoter by Tup1p may not be solely controlled by respiration. Therefore, we propose that respiration, PKA, and TORC1 pathways cooperatively control the expression of IME1, but do not work in a linear pathway.
In summary, our data show that respiration stimulates the expression of RIM101 and that Rim101p inhibits the expression of SMP1, a negative regulator of IME1. This situation promotes the expression of IME1 to start premeiotic DNA replication, and ultimately initiates the sporulation process. In respiratory chain-deficient cells, the expression of SMP1 cannot be suppressed with low levels of Rim101p and finally blocks meiosis initiation.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300689/-/DC1.
Acknowledgments
We thank Bing Zhou, Maojun Yang, and Yixian Cui at Tsinghua University for providing NDI1 constructs and its variants. We thank Tracey Baas for her critical reading and editing of the manuscript. This work was supported by the National key R&D program of China (2016YFA0500901) and the National Natural Science Foundation of China (grant No. 31171374, 31471277, 31271228). The authors declare no conflicts of interest with the contents of this article.
Author contributions: Conceived and designed the experiments: WL and HZ. Performed the experiments: HZ, QW, CL, F. Wen, and F. Wang. Analyzed the data: HZ, QW, CL, WL, YS, and WX. Contributed reagents/materials/analysis tools: HZ, QW, WL, and WX. Wrote the paper: HZ and WL.
Footnotes
Communicating editor: O. Cohen-Fix
Literature Cited
- Amaral A., Lourenco B., Marques M., Ramalho-Santos J., 2013. Mitochondria functionality and sperm quality. Reproduction 146: R163–R174. [DOI] [PubMed] [Google Scholar]
- Baek Y. U., Martin S. J., Davis D. A., 2006. Evidence for novel pH-dependent regulation of Candida albicans Rim101, a direct transcriptional repressor of the cell wall beta-glycosidase Phr2. Eukaryot. Cell 5: 1550–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin K. R., Zhang C., Shokat K. M., Herskowitz I., 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17: 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., Boguta M., 2000. Coupling of mitochondrial translation with the formation of respiratory complexes in yeast mitochondria. Acta Biochim. Pol. 47: 973–991. [PubMed] [Google Scholar]
- Church C., Goehring B., Forsha D., Wazny P., Poyton R. O., 2005. A role for Pet100p in the assembly of yeast cytochrome c oxidase: interaction with a subassembly that accumulates in a pet100 mutant. J. Biol. Chem. 280: 1854–1863. [DOI] [PubMed] [Google Scholar]
- Cui Y., Zhao S., Wu Z., Dai P., Zhou B., 2012. Mitochondrial release of the NADH dehydrogenase Ndi1 induces apoptosis in yeast. Mol. Biol. Cell 23: 4373–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J. L., Iyer V. R., Brown P. O., 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686. [DOI] [PubMed] [Google Scholar]
- De Vries S., Van Witzenburg R., Grivell L. A., Marres C. A., 1992. Primary structure and import pathway of the rotenone-insensitive NADH-ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur. J. Biochem. 203: 587–592. [DOI] [PubMed] [Google Scholar]
- El Hage A., Webb S., Kerr A., Tollervey D., 2014. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 10: e1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein C. B., Waddle J. A., Hale W., Dave V., Thornton J., et al. , 2001. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 12: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Li W., Li J., Wang J., Ge J., et al. , 2012. Structural insight into the type-II mitochondrial NADH dehydrogenases. Nature 491: 478–482. [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsich S. W., Shaw J. M., 2004. Importance of mitochondrial dynamics during meiosis and sporulation. Mol. Biol. Cell 15: 4369–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Fukuzawa T., Sorimachi H., Maeda T., 2005. Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol. Cell. Biol. 25: 9478–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Y., Sha J. H., 2011. Proteomics of spermatogenesis: from protein lists to understanding the regulation of male fertility and infertility. Asian J. Androl. 13: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M., Lee Y., Yamashita T., Yagi T., Iwata S., et al. , 2012. The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proc. Natl. Acad. Sci. USA 109: 15247–15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A., Amon A., 2008. Control of meiosis by respiration. Curr. Biol. 18: 969–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y., Granot D., Simchen G., 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52: 853–862. [DOI] [PubMed] [Google Scholar]
- Kushnirov V. V., 2000. Rapid and reliable protein extraction from yeast. Yeast 16: 857–860. [DOI] [PubMed] [Google Scholar]
- Lamb T. M., Mitchell A. P., 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Mitchell A. P., 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lorenz K., Cohen B. A., 2014. Causal variation in yeast sporulation tends to reside in a pathway bottleneck. PLoS Genet. 10: e1004634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Poirier C., Gaspar T., Gratzke C., Harrison W., et al. , 2008. A mutation in the inner mitochondrial membrane peptidase 2-like gene (Immp2l) affects mitochondrial function and impairs fertility in mice. Biol. Reprod. 78: 601–610. [DOI] [PubMed] [Google Scholar]
- Mishra D. P., Pal R., Shaha C., 2006. Changes in cytosolic Ca2+ levels regulate Bcl-xS and Bcl-xL expression in spermatogenic cells during apoptotic death. J. Biol. Chem. 281: 2133–2143. [DOI] [PubMed] [Google Scholar]
- Mishra P., Chan D. C., 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15: 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., 1994. Control of meiotic gene-expression in Saccharomyces-cerevisiae. Microbiol. Rev. 58: 56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M., 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189: 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva M. A., Tilburn J., Bignell E., Arst H. N., Jr., 2008. Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 16: 291–300. [DOI] [PubMed] [Google Scholar]
- Rajender S., Rahul P., Mahdi A. A., 2010. Mitochondria, spermatogenesis and male infertility. Mitochondrion 10: 419–428. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos J., Amaral S., 2013. Mitochondria and mammalian reproduction. Mol. Cell. Endocrinol. 379: 74–84. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos J., Varum S., Amaral S., Mota P. C., Sousa A. P., et al. , 2009. Mitochondrial functionality in reproduction: from gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 15: 553–572. [DOI] [PubMed] [Google Scholar]
- Sazanov L. A., 2015. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 16: 375–388. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L., 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18: 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. L., 2007. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem. J. 405: 1–9. [DOI] [PubMed] [Google Scholar]
- Smith H. E., Su S. S., Neigeborn L., Driscoll S. E., Mitchell A. P., 1990. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 6103–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S. S., Mitchell A. P., 1993. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 21: 3789–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagwerker, C., K. Flick, M. Cui, C. Guerrero, Y. Dou et al., 2006 A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics 5: 737–748. [DOI] [PubMed] [Google Scholar]
- Tiwari, M., S. Prasad, A. Tripathi, A. N. Pandey, I. Ali et al., 2015 Apoptosis in mammalian oocytes: a review. Apoptosis 20: 1019–1025. [DOI] [PubMed] [Google Scholar]
- Treinin, M., and G. Simchen, 1993 Mitochondrial activity is required for the expression of IME1, a regulator of meiosis in yeast. Curr. Genet. 23: 223–227. [DOI] [PubMed] [Google Scholar]
- van Werven F. J., Amon A., 2011. Regulation of entry into gametogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3521–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon A. K., Pierce M., 2000. Transcriptional regulation of meiosis in yeast. Curr. Opin. Cell Biol. 12: 334–339. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wai T., Langer T., 2016. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 27: 105–117. [DOI] [PubMed] [Google Scholar]
- Wang S., Kou Z., Jing Z., Zhang Y., Guo X., et al. , 2010. Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. USA 107: 17639–17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Moretto F., Spedale G., Amon A., van Werven F. J., 2016. Nutrient control of yeast gametogenesis is mediated by TORC1, PKA and energy availability. PLoS Genet. 12: e1006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F. P., Guo Y. S., Hu Y., Liu W. X., Wang Q., et al. , 2016. Distinct temporal requirements for autophagy and the proteasome in yeast meiosis. Autophagy 12: 671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yamashita T., Nakamaru-Ogiso E., Hashimoto T., Murai M., et al. , 2011. Reaction mechanism of single subunit NADH-ubiquinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae: evidence for a ternary complex mechanism. J. Biol. Chem. 286: 9287–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Zhao X., Broach J. R., 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42: 27–81. [DOI] [PubMed] [Google Scholar]
- Zhang X. R., Zuo X. X., Yang B., Li Z. R., Xue Y. C., et al. , 2014. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. F., Schreiber S. L., 1997. Target of rapamycin proteins and their kinase activities are required for meiosis. Proc. Natl. Acad. Sci. USA 94: 3070–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all other data supporting the findings are available within the article. Table S1 in File S1 contains all strains used in this study. Table S2 in File S1 contains all oligonucleotide sequence information.