Abstract
A century of genetic studies of the meiotic process in Drosophila melanogaster females has been greatly augmented by both modern molecular biology and major advances in cytology. These approaches, and the findings they have allowed, are the subject of this review. Specifically, these efforts have revealed that meiotic pairing in Drosophila females is not an extension of somatic pairing, but rather occurs by a poorly understood process during premeiotic mitoses. This process of meiotic pairing requires the function of several components of the synaptonemal complex (SC). When fully assembled, the SC also plays a critical role in maintaining homolog synapsis and in facilitating the maturation of double-strand breaks (DSBs) into mature crossover (CO) events. Considerable progress has been made in elucidating not only the structure, function, and assembly of the SC, but also the proteins that facilitate the formation and repair of DSBs into both COs and noncrossovers (NCOs). The events that control the decision to mature a DSB as either a CO or an NCO, as well as determining which of the two CO pathways (class I or class II) might be employed, are also being characterized by genetic and genomic approaches. These advances allow a reconsideration of meiotic phenomena such as interference and the centromere effect, which were previously described only by genetic studies. In delineating the mechanisms by which the oocyte controls the number and position of COs, it becomes possible to understand the role of CO position in ensuring the proper orientation of homologs on the first meiotic spindle. Studies of bivalent orientation have occurred in the context of numerous investigations into the assembly, structure, and function of the first meiotic spindle. Additionally, studies have examined the mechanisms ensuring the segregation of chromosomes that have failed to undergo crossing over.
Keywords: meiosis, synapsis, synaptonemal complex, cohesion, double-strand break, meiotic recombination, crossing over, spindle assembly, chromosome segregation, FlyBook
THE process of meiosis is the physical basis of Mendelian genetics. Indeed, it was the analysis of meiotic nondisjunction (missegregation) that allowed Calvin Bridges (Bridges 1916) to validate the chromosome theory of inheritance (Ganetzky and Hawley 2016). Since that time, studies of meiosis in Drosophila melanogaster females have been a leading force in the genetic, genomic, and cytological analysis of the meiotic process.
The first part of our review will focus on the basic genetic characterization of recombination and segregation in Drosophila oocytes. Specifically, we will cover:
The number and position of crossovers (COs).
Processes that control crossing over and CO placement.
The two classes of COs in Drosophila.
The relationship of CO number and position with segregation.
The remainder of our review will focus on several key meiotic events, namely:
The current understanding of meiotic chromosome pairing.
The structure and function of the synaptonemal complex (SC), its assembly and disassembly, and its role in CO maturation.
The mechanisms that control the number and position of COs.
The assembly and function of the meiosis I spindle in terms of the segregation of both exchange and nonexchange chromosomes.
Part I: Recombination as a Read-Out of Meiosis
Over the last century, most genetic studies have used recombination as the primary tool to assess the fidelity of the meiotic process. Such studies have either directly measured changes in recombination frequency or relied upon the fact that genome-wide reductions in exchange can increase the level of nondisjunction, providing an easy read-out for recombination failure. Studies of the number and position of recombination events are also important to the discussion of meiotic cell biology, which comprises the majority of this review. [We would be remiss if we did not note several reviews that have covered many of these subjects in great detail as well; these include: Baker and Hall (1976), Lindsley and Sandler (1977), Hawley et al. (1993), McKim et al. (2002), and Sekelsky (2017).]
The number and position of COs
The standard assay of CO frequency and position in Drosophila involves the use of visible and typically regularly spaced markers along chromosomes [for overviews see: Lindsley and Sandler (1977) and Carpenter (1987, 1988)]. Both recessive and dominant markers can be used, and the resolution of events is limited only by the number of available markers. However, the genetic intervals examined are typically relatively large in terms of megabases of DNA and the exact position of exchanges cannot be determined at the nucleotide level. Moreover, with the exception of one study (Parry 1973), assays have typically been done for only a single chromosome or chromosome arm at a time.
A complementary method of evaluating crossing over by whole-genome sequencing (WGS) has been recently developed (Comeron et al. 2012; Miller et al. 2012, 2016c; Singh et al. 2013). Assaying recombination by WGS uses very large numbers of single-nucleotide polymorphisms (SNPs) to determine the parent of origin for a chromosome (Miller et al. 2012). In the simplest assays, COs are determined by identifying the sites where the SNP profile changes from one parental chromosome to another in females heterozygous for many SNPs. WGS has its own limitations (e.g., cost and adequate SNP density between parents) and this strategy can presently only be applied to a relatively small number of flies. While it initially focused on wild-type flies, WGS provides a method for better understanding recombination-defective mutants, and WGS studies of meiotic mutants have already been initiated.
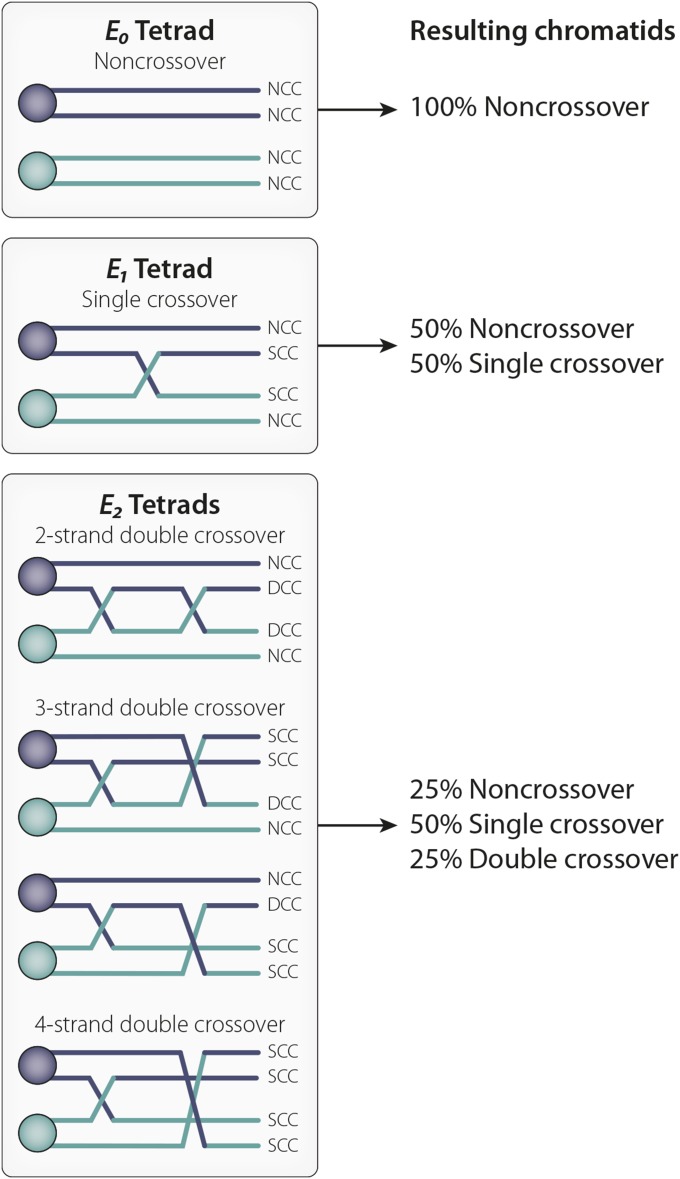
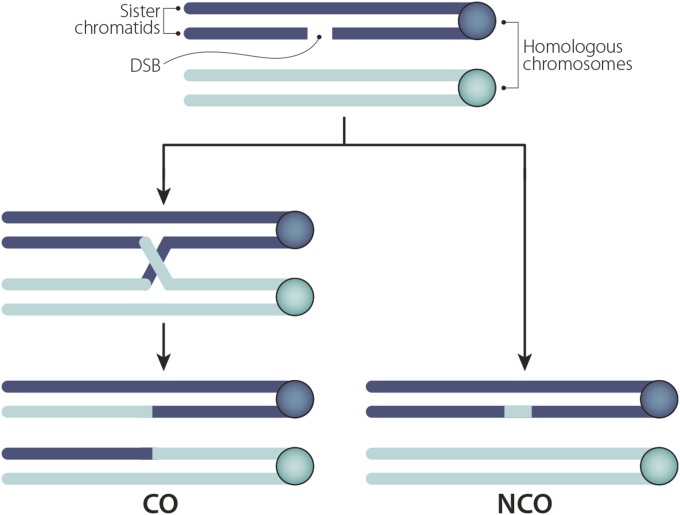
Both types of assays yield the number of progeny bearing chromosomes with zero, one, two, or more CO events. However, as first noted by Weinstein (1918), a parental or “noncrossover” (NCO) chromosome can result not only from a bivalent in which no COs occurred, but also from a bivalent with one, two, or more CO events (Figure 1). This realization forces us to consider our data in terms of the fraction of nonexchange tetrads (E0), single-exchange tetrads (E1), double-exchange tetrads (E2), and so on (En), that produced the observed set of progeny. Discussions of methods for converting progeny counts into tetrad distributions may be found in Merriam and Frost (1964), Zwick et al. (1999), Hawley and Walker (2003), and Ashburner et al. (2005). The most important of these values, the fraction of E0 tetrads, provides us with the probability that a given pair of homologs (for example the X chromosomes) failed to undergo crossing over.
Figure 1.
Tetrad analysis. Tetrad analysis allows us to determine the type and frequency of different kinds of exchange events. An E0 tetrad did not undergo crossing over and thus results in four noncrossover chromatids (NCC). An E1 tetrad experienced one crossover and results in two chromatids that each exhibit a single crossover (SCC) and two NCCs. An E2 tetrad experienced two crossovers; E2 tetrads result in some combination of NCCs, SCCs, or double-crossover chromatids (DCCs), depending on which chromatids were involved in the two crossover events. Although four chromatids are associated with an exchange (or nonexchange) event, only one chromatid will eventually make its way into the Drosophila oocyte nucleus; the remaining three are discarded. Because it is not currently possible in Drosophila to recover full tetrads, the frequency of E0, E1, E2, … En. tetrads must be calculated based on the number and type of exchange chromatids actually observed. This calculation is derived from classic Weinstein algebraic tetrad analysis [Weinstein 1918; Merriam and Frost 1964; adapted from Zwick et al. (1999)].
Based on both classical and WGS studies, we can conclude that in D. melanogaster most chromosomes undergo an average of ∼1.2 CO events per chromosome arm, such that most oocytes experience five to six CO events per meiosis (Lindsley and Sandler 1977; Carpenter 1988; Miller et al. 2016c). The frequency of bivalents without a CO event (E0 bivalents) is much lower than expected. While exchange distributions vary by chromosome arm (Miller et al. 2016c), generally the frequencies of E0, E1, and E2 tetrads are ∼5–10, 60–70, and 30–35%, respectively. E3 and E4 bivalents are quite rare. The relative paucity of bivalents with more than two exchange events reflects the phenomenon of CO interference, which is quite strong in Drosophila (see Interference) (Sturtevant 1913, 1915; Muller 1916).
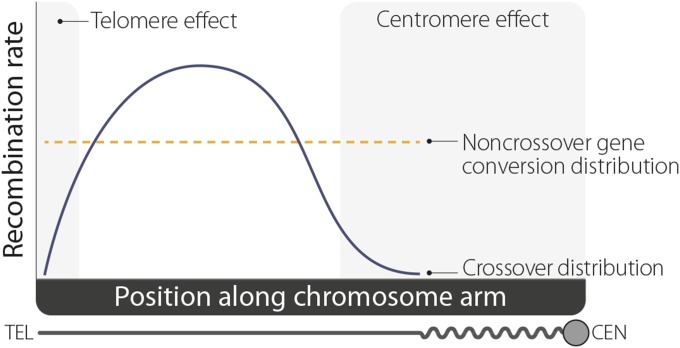
As shown in Figure 2, COs are not distributed randomly, but rather most exchange occurs in the medial and distal euchromatic regions of each chromosome arm. Exchange in the proximal euchromatic regions is strongly suppressed by the centromere effect, a strong polar suppression of exchange emanating from the centromeres. Crossing over does not occur in the heterochromatin or on chromosome 4. Extensive WGS- and population-based assays have failed to find evidence for recombination hotspots in flies (Chan et al. 2012; Comeron et al. 2012; Manzano-Winkler et al. 2013; Smukowski Heil et al. 2015; Miller et al. 2016c); however, additional studies may be required to fully address this absence of hotspots.
Figure 2.
Recombination rates. The centromere effect strongly suppresses crossing over proximal to the centromere (CEN), while a much weaker telomere effect reduces crossovers at the distal tip of the chromosome (TEL). At the same time, interference reduces the likelihood of two crossovers occurring in close proximity. These forces result in the general crossover distribution (solid blue line) depicted here; this is fairly consistent among each arm of the X, second, and third chromosomes. Noncrossover gene conversions, on the other hand, are not subject to the centromere effect nor to interference, thus their distribution (dashed orange line) is more uniformly patterned.
The centromere effect and the absence of crossing over in the pericentric heterochromatin
As noted above, recombination frequency is reduced in the proximal regions of each chromosome (Beadle 1932; Offermann and Muller 1932; Sturtevant and Beadle 1936; Hawley 1980; Miller et al. 2016c). This phenomenon is known as the centromere effect. It was initially unclear whether the centromere effect was caused by a polar suppression of exchange emanating from the centromeres or by the proximity of the proximal euchromatin to large blocks of pericentromeric heterochromatin. Yamamoto and Miklos (1977) examined this question by removing increasingly larger segments of proximal heterochromatin and measuring the effect that the amount of remaining heterochromatin had on the distribution of exchange. They found that such heterochromatic deletions strengthened the polar suppression of exchange, demonstrating that the observed reduction in exchange was due to the proximity of the centromere itself rather than simply the amount of pericentromeric heterochromatin. Alternatively, Sturtevant and Beadle (1936), used the recombinational properties of homozygous inversions to demonstrate that moving proximal regions of euchromatin to a more distal position relieved the exchange suppression generated by the centromere on these regions, while exchange was reduced when normally distal exchanges were moved to a more proximal position. Unfortunately, the mechanisms that mediate the centromere effect are still unclear.
The suppressive effect of the centromeres on crossing over is partially ameliorated by the presence of large blocks of pericentric heterochromatin that surround the centromeres. These blocks of heterochromatin are recombinationally inert. COs do not occur in the heterochromatic regions even when they are moved far away from the centromeres (Sturtevant and Beadle 1936; Szauter 1984). This is likely because double-strand breaks (DSBs), which are required for crossing over, do not typically form in heterochromatic regions. When examined in conjunction with a marker for heterochromatin, the DSB marker γH2AV (a modification to the histone H2A variant made in response to DSBs) failed to identify meiotically-induced DSBs within the labeled heterochromatic regions (Mehrotra and McKim 2006). However, DSBs could be identified in heterochromatin after the application of X-rays. Thus, the absence of heterochromatic γH2AV foci was due not to a failure to modify H2AV histones in response to DSBs in heterochromatin but, more likely, to the presence of mechanisms excluding programmed DSBs from highly repetitive regions (Mehrotra and McKim 2006).
Interference
CO placement is also affected by interference, the phenomenon that acts to ensure that when two COs occur on the same pair of homologs they are widely spaced (Sturtevant 1913, 1915; Muller 1916; Berchowitz and Copenhaver 2010). Interference is a probabilistic rather than an absolute property, with the probability of a second DSB being resolved as a CO increasing the farther away one moves from an existing CO site. Indeed, although the average distance between double-CO events as assayed by WGS is 8–10 Mb, double COs can and do occur closer together, even as close as 1.5 Mb on occasion (Miller et al. 2016c). It appears that the strength of interference may vary among chromosome arms, with the recovery of fewer double COs on the arms of chromosome 2 compared to the X and third chromosomes (Miller et al. 2016c). How interference works remains one of the major unsolved problems of meiotic biology.
CO assurance: is there an obligate CO in Drosophila?
The fact that the observed number of E0 tetrads in Drosophila females is less than expected based on the average number of CO events suggests the existence of a process known as CO assurance. In many organisms, CO assurance acts to ensure that at least one CO occurs per chromosome arm (Martini et al. 2006; Dumont 2017). Recently, Hatkevich et al. (2017) suggested that CO assurance does exist in Drosophila females, but that it is not as strong as in other organisms. However, the lack of COs on the fourth chromosome demonstrates that CO assurance is certainly not absolute. In addition, evidence against CO assurance in Drosophila has been presented by Mehrotra and McKim (2006). This question clearly remains open. As noted by J. J. Sekelsky, “Drosophila females do not have an obligate CO, but more of a highly recommended CO” (J. J. Sekelsky, personal communication). In Drosophila males, COs are completely absent during meiosis, demonstrating the presence of other mechanisms to segregate chromosomes lacking a CO.
Are there two pathways for crossing over in Drosophila?
In many organisms, COs can be produced by two separate pathways, class I and class II (Zalevsky et al. 1999; de los Santos et al. 2003; Hollingsworth and Brill 2004). These two CO pathways are defined both by the different enzymatic functions required to execute them and by how the CO events they produce respond to CO interference. Class I exchanges are highly sensitive to interference, while class II exchanges appear to be placed without regard to the proximity of other CO events. Recent work in Drosophila females indicates that there may be two classes of COs in this organism as well. While the vast majority of COs observed under normal conditions are class I, and thus are highly sensitive to interference, there may be a smaller set of class II COs that are not affected by interference (Miller et al. 2016c). Moreover, Sekelsky and his collaborators have provided strong evidence that the residual COs observed in at least some recombination-deficient mutants are class II COs (Kohl et al. 2012; Hatkevich et al. 2017). It is not yet clear if class II COs in Drosophila are processed by the same enzymatic pathway that produces class II COs in other organisms.
Gene conversion
DSBs created during meiosis can also be repaired as gene conversions, which involve the nonreciprocal transfer of genetic information from one homolog to the other. In other organisms, gene conversions can occur on their own or in association with COs, but it is currently unknown whether CO-associated gene conversion occurs in Drosophila. Gene conversions that are not associated with COs are also called NCO events. Such events are difficult to detect by standard marker-based recombination tests and therefore require large and complex assays to detect them. Early work on identifying NCO events in Drosophila involved assays in which only those progeny that had experienced an NCO within two deleterious alleles of the genes maroon-like or rosy could survive. This worked because an NCO event here would generate a wild-type copy of the gene, allowing the offspring to survive on selective media (Smith et al. 1970; Hilliker and Chovnick 1981; Hilliker et al. 1991). More recent work has used WGS to identify events genome-wide by finding polymorphisms that were copied from one homologous chromosome to another (Comeron et al. 2012; Miller et al. 2012, 2016c). Using both of these approaches, the frequency of NCO in D. melanogaster has been found to be ∼2.1 × 10−8 per bp (Hilliker et al. 1994; Blanton et al. 2005; Miller et al. 2016c). WGS-based assays have also revealed that NCO events are evenly distributed along chromosome arms and are not sensitive to either interference or the centromere effect, nor do they generate interference (Figure 2) (Comeron et al. 2012; Miller et al. 2016c).
Exchange and the fourth chromosome
Under normal laboratory conditions, the fourth chromosome does not undergo crossing over. However, rare CO events may be recovered on the fourth chromosome by placing females at an elevated temperature or by analyzing triploids [reviewed in Hartmann and Sekelsky (2017)]. The fourth chromosome is extremely small, consisting of a large block of centromere-proximal heterochromatin followed by ∼1.5 Mb of euchromatin. Indeed, the centromere effect may largely explain the absence of recombination on the fourth chromosome (Hartmann and Sekelsky 2017).
It remains unclear whether NCOs occur on the fourth chromosome. However, in a study that looked at pooled NCO events after either a single or multiple rounds of meiosis, Comeron et al. (2012) reported several NCO events on the fourth chromosome. Curiously, these authors found both a higher frequency of gene conversion and a longer tract length on the fourth chromosome than was observed for NCO events on the other five chromosome arms.
Balancer chromosomes and the interchromosomal effect
Balancer chromosomes are multiply inverted and rearranged chromosomes that are used in Drosophila either to prevent exchange between a given chromosome and its balancer homolog or, if exchange does occur, to prevent the recovery of recombinant products (Sturtevant and Beadle 1936; Roberts 1976; Ashburner et al. 2005; Hawley and Ganetzky 2016; Miller et al. 2016a,b; Kaufman 2017). Balancers are available in Drosophila for the X, second, and third chromosomes. Each balancer is marked with a dominant visible mutation for easy identification and typically carries a recessive lethal or sterile mutation.
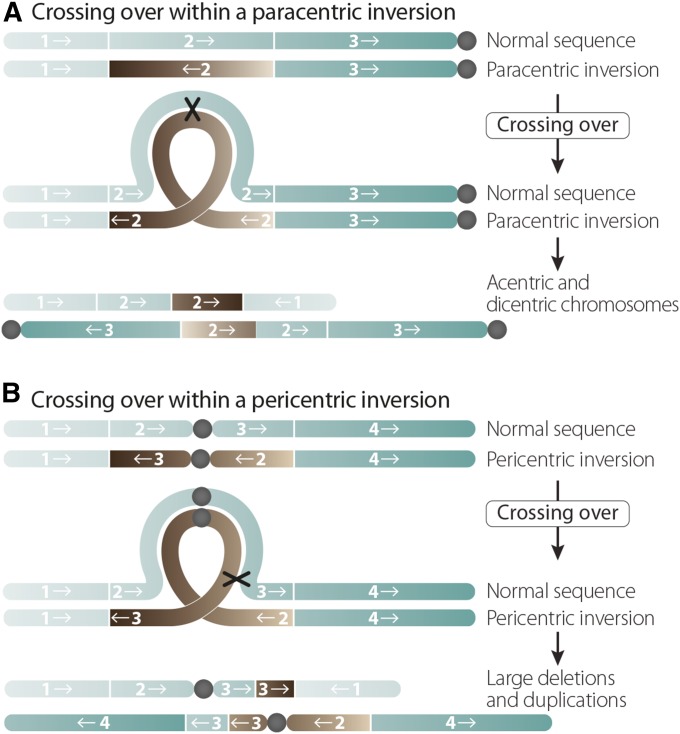
As demonstrated by both genetic and cytological studies, balancers are highly effective at preventing exchange between homologs. This is because, when heterozygous, the multiple inversion breakpoints that form the balancer function as powerful polar suppressors of exchange (Novitski and Braver 1954; Miller et al. 2016a). Exchange events do occasionally occur between a balancer and its normal-sequence homolog, but in these cases the recovery of recombinant products is prevented (Sturtevant and Beadle 1936). In the case of the X chromosome balancers, which are composed of overlapping paracentric inversions (inversions that do not involve the centromere), a single-exchange event within an inverted segment will result in an acentric fragment and a dicentric bridge, neither of which will segregate properly at the first meiotic division (Figure 3). Alternatively, many of the inversions that make up autosomal balancers do involve the centromere and are thus known as pericentric inversions. Single-exchange events within pericentric inversions will result in two chromosomes carrying large deletions and duplications that are lethal to the developing embryo (Figure 3).
Figure 3.
How balancers work. Balancers both suppress recombination and prevent the recovery of crossover events that occur within an inversion. (A) A single crossover within a paracentric inversion (one that does not include the centromere) will result in an acentric fragment, which the cell discards, and a dicentric chromosome, which cannot segregate properly. (B) A single crossover within a paracentric inversion (one that encompasses the centromere) will produce chromosomes with large deletions and duplications, which are inviable to the cell. Balancers that incorporate more inversions are thus more effective [adapted from Miller et al. (2016b)].
Although heterozygosity for a balancer (or inversions) suppresses exchange between those homologs, it increases recombination on the remaining unbalanced chromosomes (Schultz and Redfield 1951; Lucchesi and Suzuki 1968; Ashburner et al. 2005). This phenomenon, known as the interchromosomal effect, can increase the level of exchange by as much as twofold in some intervals, and even in the context of total map length (Schultz and Redfield 1951; Ramel 1962; Ramel and Valentin 1966; Lucchesi and Suzuki 1968; Carpenter and Sandler 1974). The extent of the observed increase appears to intensify with the number of inversion breakpoints, perhaps resulting from a substantial weakening of both interference and the centromere effect.
Exchange and segregation
As meiosis proceeds, COs are matured into chiasmata. The role of chiasmata in ensuring segregation is well documented (and equally well reviewed; see Nicklas (1974, 1977) and Hawley (1988)]. By physically linking two nonsister chromatids, one from each homolog, the bivalent uses sister chromatid cohesion both proximal and distal to the CO event to lock homologs together until anaphase I, at which point sister chromatid cohesion is released along the arms of the chromosomes. It is perhaps no surprise, then, that most of the Drosophila meiotic mutants known to induce the nondisjunction of exchange chromosomes either ablate sister chromatid cohesion or disrupt spindle structure and function [reviewed in Hawley et al. (1993)].
Exchange and nondisjunction in wild-type oocytes:
Wild-type Drosophila females segregate their chromosomes with extremely high fidelity, but spontaneous chromosome nondisjunction does occur at a low level. Spontaneous nondisjunction of the X chromosome is most commonly studied in Drosophila oocytes by assaying the appearance of exceptional progeny derived from females free of both structural aberrations or meiotic mutants. The observed exceptional progeny are referred to as patroclinous males (derived from oocytes lacking an X chromosome) and matroclinous females (derived from oocytes providing two X chromosomes). In some studies, patroclinous and matroclinous exceptions are also known as nullo-X and diplo-X exceptions, respectively. The frequency of spontaneous nondisjunction varies substantially among wild-type stocks in Drosophila (Zeng et al. 2010). However, most estimates are in the range of 0.001–0.0001 exceptional X chromosome progeny per generation. Provided that the centromeres are properly marked on the maternal X chromosomes (usually by appending a wild-type copy of the yellow gene to the right arm of one of the X chromosomes), it is possible to distinguish between those diplo-X exceptions resulting from nondisjunction at meiosis I (reductional exceptions) and those that appear to have occurred at meiosis II (equational exceptions).
The original analysis of spontaneous nondisjunction was performed by Bridges (1916); reviewed in Ganetzky and Hawley (2016). A subsequent study by Koehler et al. (1996) observed that > 76% of reductional exceptions involved nonexchange (E0) X chromosome bivalents. Among the exceptions that were produced by exchange bivalents, all arose from E1 tetrads and most were distal exchanges.* These findings are consistent with similar studies in yeast (Ross et al. 1996) and humans (Lamb et al. 1996), as well as with studies of nondisjunction in Drosophila meiotic mutants (Lindsley and Sandler 1977; Koehler et al. 1996).
Exchange and nondisjunction in meiotic mutants that suppress exchange:
The rules that cover the disjunctional behavior of recombination-defective mutants have been covered in detail by Baker and Hall (1976). Briefly, the vast majority of nondisjunction in such mutants is due to nonhomologous segregation events involving two pairs of nonexchange major chromosomes. For example, X nondisjunction can result from both X chromosomes segregating away from a single second chromosome (with the remaining second chromosome segregating at random) in oocytes in which X, 2L, and 2R are all nonexchange. Because both arms of a major autosome must fail to CO for an E0 X bivalent to nondisjoin, the frequency of X nondisjunction is proportional to E0 cubed (Baker and Hall 1976).
For reasons that remain poorly understood, the frequency of nondisjunction for the obligately nonexchange fourth chromosome rises with the frequency of X chromosome nondisjunction in these mutants as well, usually to approximately one-third to one-half the frequency of X chromosome nondisjunction. This increase in the frequency of fourth chromosome nondisjunction cannot be accounted for by XX ↔ 44 nonhomologous segregations.
Summary
Studies of recombination have taught us a number of critical lessons about CO number and placement:
The average number of COs per chromosome arm is slightly greater than one, such that most oocytes experience 5–6 CO events per meiosis.
The frequency of chromosome arms without at least one CO is much lower than expected.
COs are not distributed randomly, but rather most exchange occurs in the medial and distal regions of each chromosome arm.
Crossing over does not occur in the heterochromatin or on chromosome 4.
CO interference is strong.
NCOs are not sensitive to, nor do they generate, interference, leading to a random distribution along chromosomes.
COs are almost always sufficient to ensure segregation.
Part II: Overview of Drosophila Female Meiosis
Much of our ability to understand the cell biology of meiosis in Drosophila females reflects the major advances in cytology that have developed over the last 20–30 years. These studies have allowed us to reframe the formalisms created by genetic studies in terms of specific cellular events. To provide a context for those cytological studies, we begin our discussion with a summary of the cell biology of meiosis (Lake and Hawley 2012).
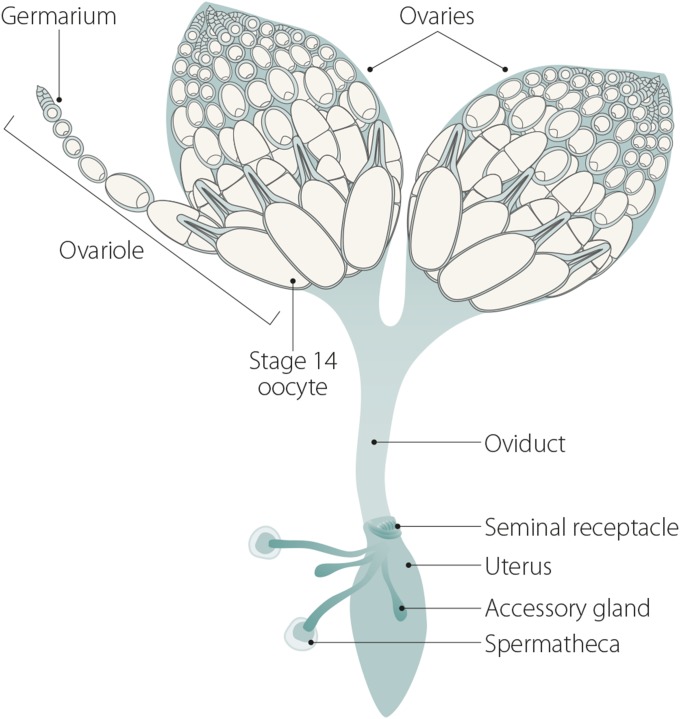
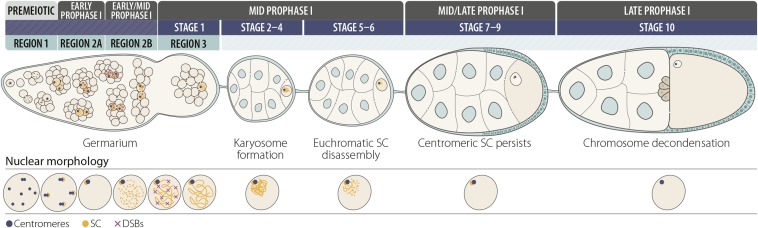
In females, the steps of meiosis can be visualized as a progression of stages in the several ovarioles that make up each ovary (Figure 4). Meiosis begins in the germarium, which is subdivided into regions based on developmental stage (Figure 5). At the tip of the germarium, in region 1, are two to three germline stem cells (GSCs) surrounded by support cells. After a GSC divides, the resulting cystoblast daughter cell and its progenitors undergo a total of four synchronized mitotic cell divisions with incomplete cytokinesis. These divisions generate a 16-cell interconnected cyst that will eventually yield the oocyte. For a review of germarium biology see Kirilly and Xie (2007), and for a review of oogenesis see McLaughlin and Bratu (2015).
Figure 4.
Ovaries. Each Drosophila ovary contains 12–16 ovarioles. Each ovariole in turn consists of a progression of distinct follicles, starting with the germarium at the anterior tip of the ovariole and progressing to a fully developed stage 14 oocyte at the posterior end.
Figure 5.
Oocyte development: stages 1–10. In the premeiotic region 1 of the germarium, a germline stem cell (asterisk) divides to produce a cystoblast, which then undergoes four incomplete mitotic divisions to produce a 16-cell interconnected cyst. Synaptonemal complex (SC) components (orange) begin loading next to unpaired centromeres (blue) in two-cell cysts, and centromeres begin pairing in four-cell cysts and clustering in eight-cell cysts. Prophase I of meiosis begins in 16-cell cysts in region 2A with the initiation of the euchromatic SC in up to four nuclei, followed by double-strand break (DSB) formation (purple). DSB formation and repair are dynamic processes that occur throughout early and early/midprophase. DSBs marked by γ-H2AV are mostly absent by midprophase in region 3 (stage 1), at which time one nucleus has been chosen as the oocyte nucleus. The other cells in the cyst serve as nurse cells. Around stages 2–3, chromosomes are reorganized and condense to form the karyosome. Euchromatic SC begins to disassemble in midprophase around stage 5 and will be completely absent by stages 7–9 during the transition from mid to late prophase (mid/late prophase). Centromeres remain clustered and centromeric SC persists through at least stage 9. In late prophase, chromosomes briefly decondense in stage 10 oocytes and transcription is upregulated before chromosomes recondense in stage 11.
Pairing
Surprisingly, the events that appear to facilitate the pairing of homologous chromosomes are initiated in region 1, while nuclei are still undergoing premeiotic mitotic divisions. Centromeric regions in Drosophila can be identified based on antibodies to Centromere identifier (Cid), a centromere-specific H3 variant (Blower and Karpen 2001). The centromeres of homologous chromosomes first begin to pair in region 1 and are fully paired by the eight-cell cyst (Christophorou et al. 2013; Joyce et al. 2013). By the completion of the last mitotic divisions, the paired centromeres have clustered into an average of two groups. These clusters of paired centromeres persist throughout meiotic prophase (Takeo et al. 2011; Tanneti et al. 2011; Christophorou et al. 2013). Centromere clustering, as well as homologous centromere pairing, requires functional centromeres, as evidenced by the observation that decreased levels of the centromere assembly factors Centromeric protein-C (Cenp-C) and Chromosome alignment defect 1 (Cal1) lead to defects in meiotic centromere pairing and clustering (Unhavaithaya and Orr-Weaver 2013).
During the period of centromere clustering, the nucleus rotates and the centromeres undergo dynamic movements (Christophorou et al. 2015). The dynamic movements of centromeres, and centromere pairing and clustering, are dependent on microtubules as well as the minus-directed motor Dynein, centrosome components, the SUN (Sad1p, UNC-84)-domain protein Klaroid, and the KASH (Klarsicht, ANC-1, Syne homology)-domain protein Klarsicht. Perhaps more surprisingly, centromere clustering (but not homologous centromere pairing) requires components of the SC, the proteinaceous structure that holds homologous chromosomes together (Takeo et al. 2011; Tanneti et al. 2011; Christophorou et al. 2013).
The SC
The first visible cytological sign of meiosis is seen in germarium region 2A, visualized as the loading of the SC along the arms of the chromosomes in up to four nuclei within the 16-cell cyst (Figure 5). As the cyst progresses through the germarium, two of the four nuclei that initially formed euchromatic SC will disassemble their SC and back out of the meiotic program, followed by disassembly of the SC within an additional nucleus in region 2B. This results in a single pro-oocyte by region 3 that maintains full-length SC along the arms of the chromosomes. The remaining 15 cells assume a supporting role as nurse cells.
DSBs and recombination
Studies in multiple organisms have revealed that programmed DSBs are initiated during meiosis. In Drosophila oocytes, DSBs are first visualized in region 2A as the modification of the histone H2A variant γH2AV in response to DSB formation, shortly after the initiation of SC formation along the chromosome arms (Mehrotra and McKim 2006; Lake et al. 2013). A second cellular response to DSB formation is p53 activation, which, based on a p53 reporter construct, is activated in regions 2A and 2B and is dependent on DSB formation (Lu et al. 2010).
A subset of DSBs will result in the formation of around six COs, for an average of 1.2 COs per chromosome arm (Lindsley and Sandler 1977). CO formation is thought to occur at the sites of recombination nodules (RNs), electron-dense structures identified initially by Carpenter in electron microscopy (EM) studies (Carpenter 1975a,b). Those DSBs that do not become COs are repaired as NCO events (Figure 6).
Figure 6.
Homologous recombination. Repair of a double-strand break (DSB) by homologous recombination typically yields either a crossover (CO) or a noncrossover gene conversion (NCO). COs exhibit a Mendelian, or 2:2, segregation pattern of alleles, while NCOs exhibit a 1:3 segregation pattern of the alleles covered by the NCO. In flies, NCOs may be formed by synthesis-dependent strand annealing (SDSA) or by a modified version of SDSA in which both ends of the break engage with the homolog. Other, more complex outcomes of repair by gene conversion have also been observed in Drosophila (Crown et al. 2014).
DSBs are repaired progressively as the cyst moves through the germarium, and although few γH2AV foci are visible in the oocyte by region 3, it is unknown when the final stages of repair are complete (Mehrotra and McKim 2006). COs are matured into chiasmata that physically interlock homologous chromosomes together. These chiasmata are crucial for proper biorientation of homologs on the meiotic spindle, thus ensuring that they segregate away from each other at the first meiotic division.
If DSBs are not repaired when the cyst buds off from the germarium and enters the vitellarium at stage 2, as occurs in mutants such as spindle-B (XRCC3 homolog), spindle-D (Rad51C homolog), and okra (Rad54L homolog), a checkpoint is initiated [reviewed in Ghabrial et al. (1998) and Sekelsky (2017)]. This checkpoint, which is dependent on Mei-41 (ATR homolog) and Loki (Chk2 homolog, also known as Mnk) (Abdu et al. 2002), initiates a cascade that leads to defects in chromosome condensation, abnormal egg chamber development, and ultimately sterility (Ghabrial et al. 1998).
Segregation
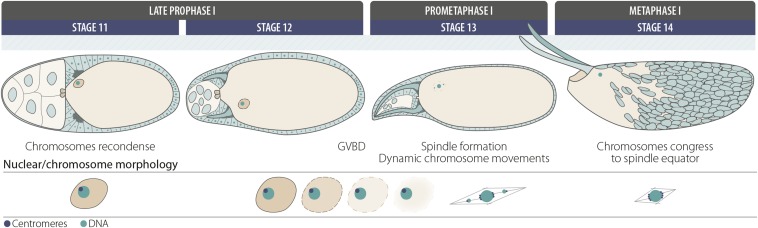
In terms of meiotic progression, only a few notable events have been documented during stages 2–12 of oocyte development, namely karyosome formation and SC disassembly. Around stages 12–13, germinal vesicle breakdown (GVBD) and spindle assembly occur (Figure 7). After homologous chromosomes have bioriented on the meiosis I spindle, they are then segregated to opposite spindle poles at anaphase I. Sister chromatids are subsequently segregated at meiosis II to form four haploid products.
Figure 7.
Oocyte development: stages 11–14. In stage 11, the karyosome recondenses and undergoes preparations during stage 12 for germinal vesicle breakdown (GVDB), which occurs at approximately the start of stage 13. After GVBD, tubulin is recruited to the chromosomes and then organized into a bipolar spindle. Achiasmate chromosomes undergo dynamic movements toward the spindle poles during prometaphase I, and by stage 14 the achiasmate chromosomes congress to join the chiasmate chromosomes. At this stage, homologous chromosomes have bioriented toward opposite spindles poles and formed a compact structure at the metaphase plate of the spindle. Oocytes will maintain this metaphase I arrest configuration until activation.
Part III: Chromosome Pairing and Synapsis
Pairing, SC assembly, DSB formation/resolution, and the segregation of homologous chromosomes at meiosis I will serve both as our central areas of concentration and as our landmarks in the discussion of meiotic biology that follow. This discussion will primarily be in the context of genes (and their protein products) that were identified in numerous screens for meiotic mutants (Box 1).
Box 1. Screening for Meiotic Mutants.
Much of our knowledge of the proteins involved in female Drosophila meiosis has come through the analysis of mutations that disrupt various aspects of meiosis, many of which were isolated through forward genetic screens for mutations that cause chromosome nondisjunction. The screens have varied in their choice of mutagen and utilized different strategies for recovering and identifying the mutation of interest. These efforts have identified not only mutations that decrease recombination, which leads to chromosome nondisjunction, but also mutations that directly disrupt chromosome segregation during the two meiotic divisions.
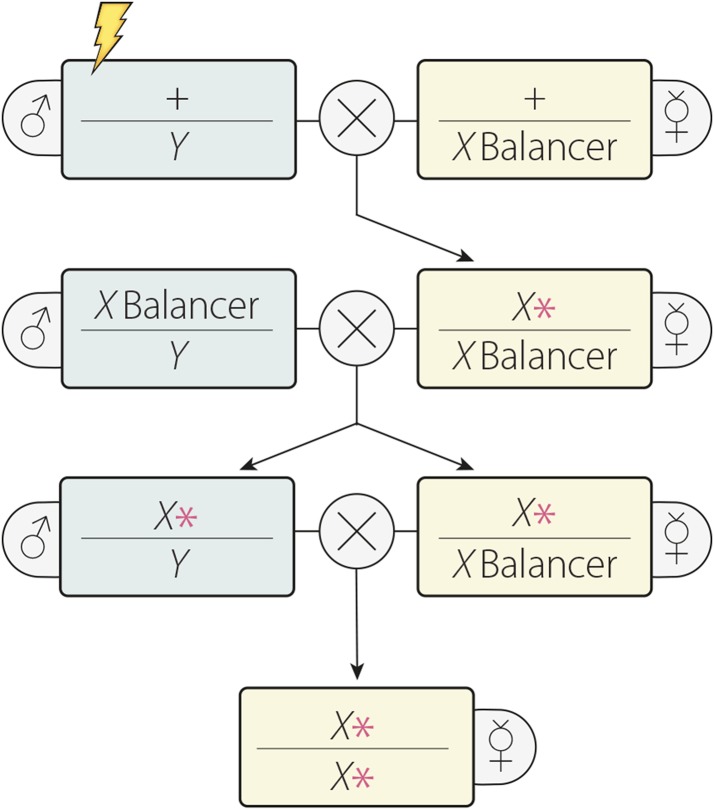
The first meiotic mutations identified were found fortuitously: a mutation in crossover suppressor on 3 of Gowen (c(3)G) (Gowen and Gowen 1922; Gowen 1933), and a mutation in D. simulans that later was found to affect the homolog of the D. melanogaster meiotic gene nonclaret disjunctional (ncd) as well as the gene claret (Davis 1969; Sequeira et al. 1989). The c(3)G mutant provided the first insights into the genetic control of female meiosis. Later, Sandler et al. (1968) undertook a groundbreaking effort to systematically screen for genetic mutations that alter meiosis in natural fly populations by looking for lines with increased chromosome nondisjunction in females and males, which yielded several strong meiotic mutants. Baker and Carpenter (1972) used a similar approach to identify new female meiotic mutants induced by EMS (see figure).
Genetic screens continue to be used to identify new genes that play a role in meiotic processes, using sex chromosome nondisjunction as the assay. Traditional screens included using P-elements as the mutagen (Sekelsky et al. 1999), an EMS screen of the X chromosomes (Liu et al. 2000), and the examination of already existing highly EMS-mutagenized fly lines (Giunta et al. 2002). More recently, advances in screening methods by two labs employed a FLP-FRT system to screen for recessive meiotic mutants in heterozygous F1 females, with a requirement for nondisjunction of an autosome to select for strong female meiotic mutants and to potentially allow for the recovery of recessive lethal mutations (Fedorova et al. 2001; Page et al. 2007; Collins et al. 2012).
Additional methods to identify genes that play roles in female meiosis have included further analyzing mutants that originally were identified for their effects on male meiosis (Yan and McKee 2013; Krishnan et al. 2014), analyzing female-sterile collections (Pearson et al. 2005), screening deficiencies (Harris et al. 2003; Sousa-Guimaraes et al. 2011; Von Stetina et al. 2011), and, more recently, candidate RNAi approaches (Radford et al. 2012b; Hughes and Hawley 2014). The genes defined by these mutations have provided us with insights into the mechanisms that control SC formation, DSB formation, CO maturation and distribution, spindle formation, and chromosome segregation events.
Screening for new meiotic mutants on the X chromosome. Shown is a simple example of a forward genetic screen for new meiotic mutations on the X chromosome. Most commonly, females homozygous for the mutagenized chromosome have been tested by scoring the level of X chromosome nondisjunction among the progeny, but autosomal nondisjunction, recombination, or cytological parameters can also be examined. For more detailed descriptions of forward genetic screens of the X chromosome, including more complicated selection-type screens, see Baker and Carpenter (1972), Liu et al. (2000), Collins et al. (2012).
Homolog pairing
Genetic studies over the last century have attempted to provide insight into the pairing process by analyzing the effects of structural aberrations on meiotic recombination (Dobzhansky 1930, 1931; E. H. Grell 1964, R. F. Grell 1964, 1976; Roberts 1972; Hawley 1980; Craymer 1984). Unfortunately, in the absence of cytology it proved difficult—indeed, impossible—to make inferences about pairing based on a readout of CO frequency and distribution. The process of understanding meiotic pairing was further complicated (if not entirely stymied) by repeated assertions that meiotic pairing in Drosophila was simply an extension of the somatic pairing that is so ubiquitously observed in this organism [for review, see Joyce et al. (2013)]. However, this long-lived notion has recently been dispelled by advances in cytology that have allowed the direct observation of the process of meiotic pairing in the female germline (Box 2).
Box 2. The Three Peculiarities of Drosophila Female Meiosis.
We are often asked why meiosis is different in flies compared to in yeast or worms. It would be convenient if, as we presume is true for things like gene regulation, there were only one mechanism for executing meiosis—a mechanism that is absolutely conserved in all species. Unfortunately, meiosis appears to be soft clay for the evolutionary process, resulting in quite impressive variation in many aspects of the meiotic process. This variation does make the study of meiosis more challenging.
There are three significant ways in which meiosis in Drosophila differs from meiosis in other well-characterized systems (i.e., budding yeast, mice, and humans).
First, meiotic pairing begins during the premeiotic mitoses (Christophorou et al. 2013; Joyce et al. 2013).
Second, although in most other organisms, DSBs are required to initiate SC formation, complete SC assembly can occur in flies even in mutants that fully block DSB formation (McKim et al. 1998). Studies of a mutant that makes only a partial SC suggest that, under normal conditions, SC assembly results both from the extension of pericentromeric SC and from extension of the SC initiated from additional sites along the arms (Manheim and McKim 2003).
Lastly, Drosophila oocytes do not have a canonical diplotene–diakinesis in which homologs fully repel each other. Rather, homolog pairing is maintained in the heterochromatin that surrounds the centromere until prometaphase I (Dernburg et al. 1996).
We find the unique aspects of the Drosophila meiotic process, most notably premeiotic pairing, to be among the most fascinating problems for further study. Our rationale for focusing on what others may dismiss as “some odd fly thing” is that regardless of the exact route, flies still achieve the fundamental goals of pairing, synapsis, CO control, and segregation. Insights into any process that facilitates an end such as pairing will provide critical understanding of what it means to be “paired” or to be selected as a DSB that will become a CO.
We now know that in GSCs, both euchromatic and heterochromatic regions of autosomes are unpaired, and thus meiotic pairing is not simply an extension of somatic pairing (Christophorou et al. 2013; Joyce et al. 2013; Rubin et al. 2016). Euchromatic regions begin to show low levels of pairing in cystoblasts, but heterochromatic regions remain mostly unpaired (Christophorou et al. 2013; Joyce et al. 2013). During the premeiotic mitotic divisions, both euchromatic and heterochromatic regions progressively pair to maximum levels by the 8–16-cell cyst stages, and this pairing is maintained until SC disassembly (Sherizen et al. 2005; Joyce et al. 2013). Homologous chromosome pairing thus initiates during the mitotic divisions that precede meiosis. At the end of pachytene, heterochromatic regions remain paired while euchromatic regions begin to unpair, with over half of oocytes showing two distinct foci for the euchromatic histone locus (Dernburg et al. 1996).
The timing of X chromosomal euchromatic pairing is similar to the pairing of autosomal euchromatic regions (Joyce et al. 2013). Euchromatic regions of the X chromosome remain highly paired in zygotene and in early pachytene, even in oocytes heterozygous for a highly rearranged balancer chromosome, which would require the pairing of homologous loci despite structural rearrangement of one of the two X chromosomes. (Gong et al. 2005; Sherizen et al. 2005). However, pairing of the heterochromatic 359-bp repeat region of the X chromosome is unusual. For reasons that remain unclear, the 359-bp repeat region on the X chromosome, which lies adjacent to the X chromosomal rDNA, is already paired in GSCs. Heterochromatic regions on the X chromosome remain highly paired throughout prophase, even in oocytes heterozygous for a multiply inverted balancer chromosome (Dernburg et al. 1996; Joyce et al. 2013).
The SC
The SC runs the full length of each set of paired homologous chromosomes. At least within the euchromatin, the tripartite structure of the SC can be compared to a railroad track or a zipper. The basic structure of the Drosophila SC was explored in detail by serial reconstruction EM performed by Carpenter and others (Carpenter 1975b; von Wettstein et al. 1984; Schmekel et al. 1993; Schmekel and Daneholt 1995). These studies have been reviewed previously (Carpenter 1988; Hawley et al. 1993; Ashburner et al. 2005).
Composition and structure of the SC:
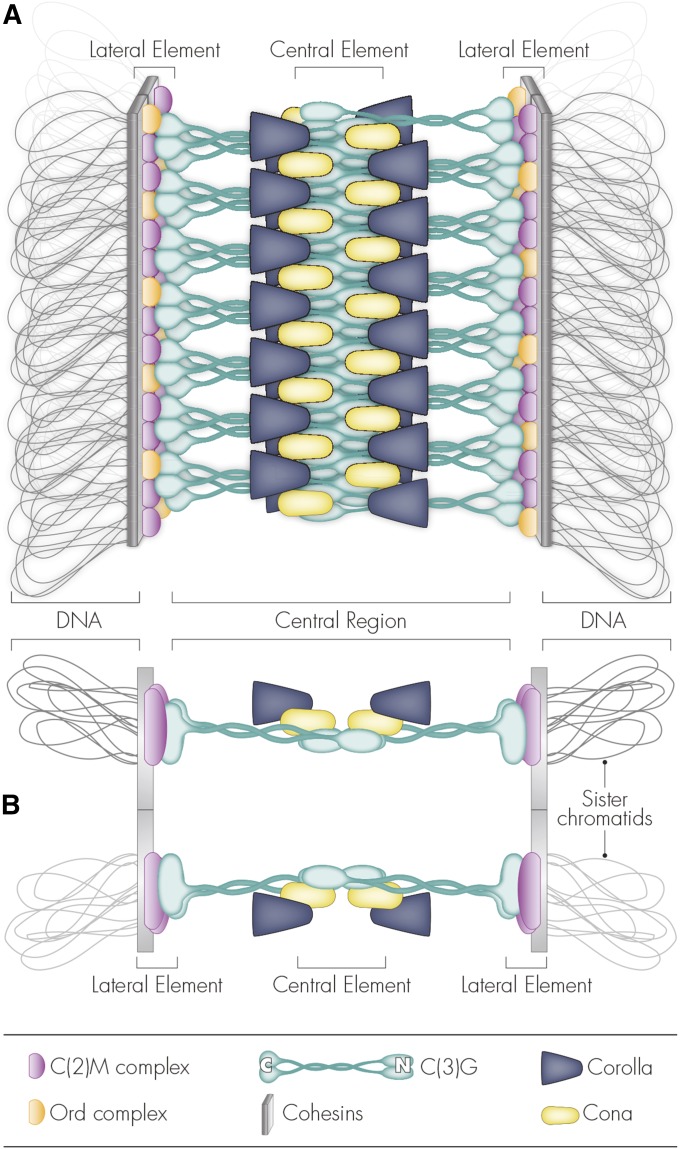
As shown in Figure 8A, the SC comprises three components:
Figure 8.
The synaptonemal complex (SC). (A) Along the chromosome arms, the SC consists of two axial/lateral elements and a central region that spans the distance between lateral elements. The axial elements serve as a scaffold that both connects sister chromatids and provides a link between chromatin and the SC in an unknown fashion. In Drosophila, axial element proteins include the cohesins Smc1/3, as well as Ord, Solo, Sunn, and the cohesin loader Nipped-B. These proteins localize along chromosome arms as well as at the centromere. C(2)M is a cohesin-like protein, generally described as a lateral element protein, that is found only along chromosome arms. The central region proteins include Corolla, the transverse filament C(3)G, and the central element protein Cona. (B) The SC forms in two distinct layers that mirror one another. These layers are known to contain C(2)M, C(3)G, Corolla, and Cona.
Two lateral elements (LEs), derived from the axial cores of each of the two homologs, run along the chromosome arms.
The central region (CR) is composed of proteins extending between and connecting the LEs and, ultimately, homologous chromosomes.
The central element (CE) runs down the middle of the CR.
Although multiple LE proteins have been identified (Table 1), the internal architecture of the LE remains poorly defined in Drosophila, but has been explored in other organisms (Kohler et al. 2017). The LE links the chromosome cores to CR components of the SC and appears to be composed primarily of cohesin and cohesin-related complexes. Cohesin complexes are multiunit complexes that contain the core components Smc1 and Smc3 (Smc1/3) that hold sister chromatids together during critical stages of both mitosis and meiosis (Losada and Hirano 2005). In Drosophila, Smc1/3 are thought to form at least two different complexes in the female germline (Khetani and Bickel 2007; Gyuricza et al. 2016). These complexes are defined based on shared protein localization patterns and mutant phenotypes. The first of these complexes includes three proteins: Orientation disrupter (Ord), a protein required for proper cohesion but whose exact function in cohesion remains elusive (Khetani and Bickel 2007); the stromalin-related protein Sisters unbound (Sunn); and Sisters on the loose (Solo) (Yan and McKee 2013; Krishnan et al. 2014). These proteins localize to both the centromeres and the chromosome arms (Yan and McKee 2013; Krishnan et al. 2014). For simplicity, we will refer to this complex as the Ord complex.
Table 1. Genes involved in pairing and synapsis.
| Gene symbol | Common name (homolog) | Functiona | References |
|---|---|---|---|
| klaroid | SUN domain protein | Inner nuclear membrane protein | Christophorou et al. (2015) |
| klarsicht | KASH domain | Outer nuclear member protein | Christophorou et al. (2015) |
| dhc | Dynein heavy chain | Minus-end-directed motor | Christophorou et al. (2015) |
| Chromosome axis | |||
| smc1 | Smc1 | Core cohesion component | Khetani and Bickel (2007); Gyuricza et al. (2016) |
| smc3 | Smc3 | Core cohesion component | Khetani and Bickel (2007); Gyuricza et al. (2016) |
| ord | Cohesion: Ord complex | Bickel et al. (1996), (2002); Gyuricza et al. (2016) | |
| sunn | Stromalin-related | Cohesion: Ord complex | Krishnan et al. (2014); Gyuricza et al. (2016) |
| solo | Cohesion: Ord complex | Yan and McKee (2013); Gyuricza et al. (2016) | |
| SA | Stromalin | Cohesion: C(2)M complex | Gyuricza et al. (2016) |
| NipB | SCC2 | Cohesion: C(2)M complex | Gyuricza et al. (2016) |
| rad21/vtd | SCC1 | Cohesion | Urban et al. (2014) |
| SC lateral element | |||
| c(2)M | Kleisin-like protein; cohesion: C(2)M complex | Manheim and McKim (2003); Heidmann et al. (2004); Gyuricza et al. (2016); Cahoon et al. (2017) | |
| SC central region | |||
| c(3)G | Zip1/SCP1 | Transverse filament | Hall (1972); Page and Hawley (2001); Cahoon et al. (2017) |
| corolla | Transverse filament-like protein | Collins et al. (2014); Cahoon et al. (2017) | |
| cona | Central element | Page et al. (2008); Lake and Hawley (2012); Cahoon et al. (2017) | |
SUN, Sad1p, UNC-84; KASH, Klarsicht, ANC-1, Syne homology; SC, synaptonemal complex.
Most likely function based on genetic, biochemical, or cytological data.
The second proposed cohesin complex, referred to here as the C(2)M complex, consists of the cohesin component Stromalin (SA), the SCC2 homolog Nipped-B, and the kleisin-like protein C(2)M (Heidmann et al. 2004; Gyuricza et al. 2016). C(2)M, which has homology to kleisin-family proteins and has been to shown to interact with Smc3 (Manheim and McKim 2003; Heidmann et al. 2004), is loaded along the chromosome arms but is absent from the centromeres (Manheim and McKim 2003; Gyuricza et al. 2016). The mitotic α-kleisin Rad21 also interacts with C(2)M, and targeted cleavage of Rad21 in the ovary leads to an early disassembly of the SC (Urban et al. 2014).
The CR of the SC appears to be composed primarily of transverse filament proteins (TFs), which are characterized by their central coiled-coil domains flanked by globular domains at their N- and C-termini. In flies, the primary TF protein is encoded by the c(3)G gene. C(3)G forms a homodimer (Page and Hawley 2001) and, based on immuno-EM, the C-terminus of the dimer is located within the LE adjacent to C(2)M, while the N-terminus of the dimer overlaps with a second C(3)G dimer at the middle of the CR (Anderson et al. 2005) (Figure 8A). It is the C(3)G protein, then, that forms the zipper-like component of the SC to span the distance between LEs (Page and Hawley 2001). Two additional CR proteins have been identified: Corolla, which forms two tracks through the CR, and Corona (Cona), which also appears to reside as two tracks in or beside the CE (Page et al. 2008; Collins et al. 2014; Cahoon et al. 2017).
Recent work using more advanced microscopy techniques has provided insight into the three-dimensional architecture of the Drosophila SC as well. Based on this study, the SC forms as two mirrored layers, with C(2)M, Corolla, and Cona all lying slightly above/below each layer of C(3)G (Figure 8B) (Cahoon et al. 2017).
Assembly and function of the SC at the centromeres:
The SC shows a highly regulated pattern of assembly. During the mitotic divisions to create the 16-cell cyst, the cohesin proteins Smc1/3 and the cohesin-related Ord complex proteins localize to the centromeres in all 16 nuclei of the cyst (Webber et al. 2004; Khetani and Bickel 2007; Yan and McKee 2013; Krishnan et al. 2014). The localization of Smc1/3 to the centromeres is dependent on Ord complex proteins (Khetani and Bickel 2007; Yan and McKee 2013; Krishnan et al. 2014), and the Ord complex proteins are mutually dependent on one another for loading to the centromeres (Webber et al. 2004; Khetani and Bickel 2007; Yan and McKee 2013; Krishnan et al. 2014; Gyuricza et al. 2016).
The CR proteins C(3)G and Corolla first load near the centromeres before the cyst has exited the mitotic divisions of region 1 (Christophorou et al. 2013). The loading of CR proteins to the centromeres has been shown to be dependent on Ord and Solo, and mutation of these cohesion proteins results in a failure to load centromeric SC (Yan and McKee 2013; Gyuricza et al. 2016). Loss of the CR proteins [either C(3)G or Cona] leads to defects in the pairing of pericentric heterochromatin during the mitotic divisions of the cyst (Christophorou et al. 2013). CR mutants also display greatly decreased levels of heterochromatic pairing in pachytene when compared to wild-type flies (Christophorou et al. 2013). The absence of any one of the three known CR proteins [Cona, Corolla, or C(3)G] also leads to a defect in centromere clustering (Takeo et al. 2011; Collins et al. 2014). This requirement for the CR proteins to cluster the centromeres is consistent with the role of the SC in mediating centromere clustering in several other organisms (Da Ines and White 2015; Kurdzo and Dawson 2015).
Loss of the Ord complex proteins also leads to defects in both centromere clustering and centromere pairing (Khetani and Bickel 2007; Takeo et al. 2011; Tanneti et al. 2011; Yan and McKee 2013; Krishnan et al. 2014; Gyuricza et al. 2016). These oocytes display up to eight centromeric foci in region 2A, suggesting that the centromeres of homologous chromosomes are unpaired, while the centromeres of sister chromatids remain associated (Takeo et al. 2011; Tanneti et al. 2011; Yan and McKee 2013; Krishnan et al. 2014). However, pro-oocytes with more than eight Cid foci can be observed at later stages (Takeo et al. 2011; Yan and McKee 2013; Krishnan et al. 2014; Gyuricza et al. 2016), indicating a progressive loss of sister chromatid cohesion at the centromeres. These observations demonstrate a critical role for the Ord complex in meiotic centromere clustering and sister chromatid cohesion.
Unlike the Ord Complex, C(2)M complex proteins are not essential for centromere clustering. Defects in centromere clustering are not seen in ovaries with decreased levels of the C(2)M complex proteins, consistent with C(2)M’s failure to show localization to the centromeres (Takeo et al. 2011; Tanneti et al. 2011; Gyuricza et al. 2016). The loss of C(2)M, SA, or Nipped-B also has no effect on Smc1/3 retention at the centromeres (Gyuricza et al. 2016).
Assembly of the SC along the arms:
After centromeres have clustered, the zipper-like SC will then assemble along the euchromatic arms in up to four nuclei of the 16-cell cyst in region 2A (Figure 5). As the cyst matures, this euchromatic SC then quickly disassembles in all but the pro-oocyte, leaving two nuclei with mostly full-length tracts of the SC at region 2B and only the single pro-oocyte with full-length SC at region 3. The cyst will exit the germarium and enter the vitellarium at stage 2, when the oocyte nucleus is reorganized into the karyosome. Full-length euchromatic SC persists in the oocyte nucleus until approximately stage 5, when it is progressively disassembled from along the chromosome arms. By approximately stage 7–8, the SC is completely disassembled along the chromosome arms but remains at the centromeres for additional stages.
Functional dependencies for LE assembly:
The initial loading of Smc1/3 to the chromosome arms is dependent on the C(2)M complex, and the LE protein C(2)M localizes only to the arms of the chromosomes (Manheim and McKim 2003; Gyuricza et al. 2016). Loss of either SA or Nipped-B leads to a failure of C(2)M to load along the chromosome arms, and loss of any member of the complex also leads to a loss of Smc1/3 along chromosome arms (Gyuricza et al. 2016).
The initial loading of Smc1/3 along the chromosome arms does not appear to be dependent on the Ord complex (Webber et al. 2004; Khetani and Bickel 2007; Yan and McKee 2013; Krishnan et al. 2014; Gyuricza et al. 2016). However, although Smc1/3 is initially loaded to the chromosome arms in solo and ord mutants, it is prematurely lost from the chromosome cores in older cysts, resulting in the eventual separation of sister chromatids (Khetani and Bickel 2007; Yan and McKee 2013; Krishnan et al. 2014).
The two cohesion complexes, C(2)M and Ord, appear to be interdependent in terms of their localization. In c(2)M mutants expressing a GFP-tagged version of Ord, the localization of Ord-GFP was patchy along the chromosome arms but Ord-GFP was loaded to the centromeres (Khetani and Bickel 2007). In ord mutants, C(2)M initially loads to the chromosomes arms but, like Smc1/3, the C(2)M protein is then precociously lost from the chromosome arms (Webber et al. 2004).
Finally, although the loss of LE proteins strongly affects the localization of CR proteins either along the arms or to the centromeres, the loss of CR proteins does not appear to affect the localization of any of the LE proteins thus far tested, based on immunofluorescence analyses (Page et al. 2008).
Functional dependencies for CR assembly:
Loss of the C(2)M complex proteins has strong effects on elongation of the CR along the chromosome arms (Manheim and McKim 2003; Gyuricza et al. 2016). Specifically, in c(2)M mutants, the CR protein C(3)G can still load near the centromeres but loads only to several small sites along the chromosome arms (Manheim and McKim 2003; Gyuricza et al. 2016). RNA interference (RNAi) knockdown of Nipped-B or SA leads to a similar phenotype, with C(3)G loading only to the centromeres and several discreet sites on the chromosomes, and a failure to extend these sites into full-length SC (Gyuricza et al. 2016). The loading of C(3)G only to several discreet sites suggests that C(2)M, Nipped-B, and SA are required not for the initiation of SC assembly, but for its elongation into a full-length SC.
Based on immunofluorescence, CR proteins of the SC are still initially loaded along the chromosome arms in region 2A in Ord complex mutants (Webber et al. 2004; Yan and McKee 2013; Krishnan et al. 2014). However, although the CR initially appears wild-type in ord mutants by immunofluorescence, EM studies reveal the CR to be abnormal (Webber et al. 2004). Additionally, the CR progressively fragments and disassembles much earlier than in wild-type flies and is completely disassembled before the cyst exits the germarium (Khetani and Bickel 2007; Yan and McKee 2013).
Cona, Corolla, and C(3)G are mutually dependent on one another for loading to the CR of the SC (Page et al. 2008; Collins et al. 2014), and loss of any of these proteins leads to a complete failure to form the CR of the SC. As a consequence of a failure to assemble the SC, CR mutants also display greatly decreased levels of both heterochromatic and euchromatic pairing in pachytene when compared to wild-type flies (Sherizen et al. 2005; Page et al. 2008; Christophorou et al. 2013).
Role of SC proteins in facilitating meiotic recombination:
All known null alleles of CR-encoding genes completely suppress recombination (Page and Hawley 2001; Page et al. 2008; Collins et al. 2014). CR mutants do still form DSBs, although at a reduced level (Mehrotra and McKim 2006; Collins et al. 2014), indicating that the CR of the SC is required for the maturation of DSBs into COs.
Null alleles of ord and solo cause large decreases in recombination, particularly in distal regions of the chromosomes, and a subset of the residual COs appear to be between sister chromatids (Mason 1976; Webber et al. 2004; Yan and McKee 2013). These studies indicate that the Ord complex may play a role in promoting exchange between homologs over sister chromatid exchange (Webber et al. 2004; Yan and McKee 2013). These defects in recombination lead to chromosome missegregation at both meiotic divisions for these mutants (Bickel et al. 1997; Yan and McKee 2013; Krishnan et al. 2014).
Mutants in c(2)M show a decrease in recombination to 25% of wild-type levels (Manheim and McKim 2003). Moreover, the residual COs show an altered distribution that is more proportional to physical distance, indicating a decrease in CO interference. Due to the strong decreases in recombination, c(2)M mutations cause increased chromosome missegregation. Approximately 10% of the X chromosome nondisjunction events observed by Manheim and McKim (2003) appeared to be sister chromatid nondisjunction, suggesting that C(2)M plays a moderate role in sister chromatid cohesion. The short stretches of residual SC observed in c(2)M mutant oocytes frequently colocalize with DSBs, suggesting that these short SC segments are sufficient for the maturation of COs in some instances (Tanneti et al. 2011).
Part IV: DSB Formation and Recombination
In Drosophila females, meiotic DSBs are initiated after the formation of the SC. DSBs can be visualized cytologically in the Drosophila ovary with an antibody recognizing γH2AV, a modification that occurs in response to DSBs (Mehrotra and McKim 2006; Lake et al. 2013). In the first 16-cell cyst of the germarium there are almost no γH2AV foci, and after SC formation, a few γH2AV foci are observed (Mehrotra and McKim 2006). The number of γH2AV foci steadily increases with each older cyst until it reaches a maximum of ∼15 γH2AV foci and then steadily drops until few or no foci are observed by region 3 (Mehrotra and McKim 2006).
In mutants that are unable to complete repair of DSBs (Table 2), such as spn-A (Rad51 homolog), spn-B (Xrcc3 homolog), or okra (Rad54L homolog), the average number of γH2AV foci in region 3 is 21.3, 24.3, and 20.6, respectively (Mehrotra and McKim 2006). Since DSBs are not being repaired in these mutants, it is believed that these numbers would represent the upper limit of meiotic DSBs that occur. Observations in wild-type oocytes have shown that an average of 15 DSBs can be observed in a single region of the germarium, but DSBs may be induced and repaired in several regions so this number may be on the lower limit of the average number of meiotic DSBs created (Mehrotra and McKim 2006). The wild-type number of DSBs is two to three times higher than the around six COs per genome, or 1.2 COs per arm, observed by genetic and WGS techniques (Lindsley and Sandler 1977; Miller et al. 2016c), indicating that the remaining DSBs should be recovered as NCOs. Indeed, a recent study using WGS to identify CO and NCO events observed an average of 11.2 NCOs during a single meiosis (Miller et al. 2016c).
Table 2. Genes involved in DSBs and recombination.
MCM, mini-chromosome maintenance; DSB, double-strand break.
Most likely function based on genetic, biochemical, or cytological data.
Genes involved in the induction of meiotic DSBs
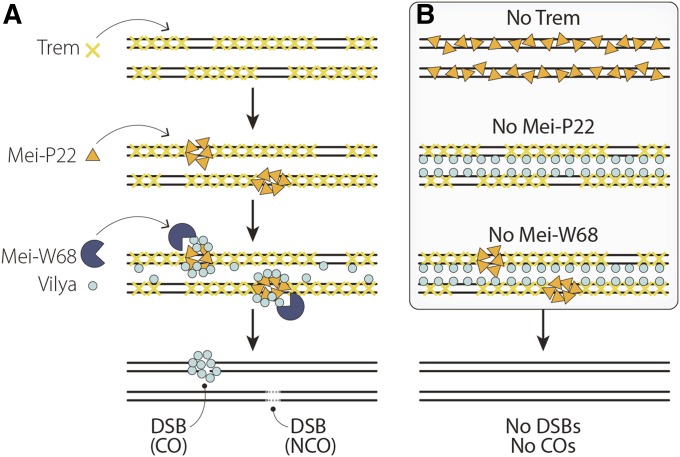
Four genes are known whose products are required for the induction of meiotic DSBs: mei-W68, mei-P22, vilya, and trade embargo (trem) (Figure 9 and Table 2). The Mei-W68 protein, a homolog of yeast SPO11, catalyzes meiotic DSBs (McKim and Hayashi-Hagihara 1998) and is related to a subunit of TopoVI DNA topoisomerase (TopoVIA) (Bergerat et al. 1997). In mei-W68 mutants, recombination and gene conversion are absent and γH2AV foci fail to be detected (McKim and Hayashi-Hagihara 1998; Mehrotra and McKim 2006).
Figure 9.
Double-strand break (DSB) formation. (A) Before DSB formation, Trem localizes to the chromosomes and is required for the recruitment of Mei-P22 to discreet foci. Mei-W68 and Vilya are recruited to sites of Mei-P22, leading to the formation of DSBs. (B) Without Trem, Mei-P22 localizes along chromosome arms rather than to discreet foci. Without either Mei-P22 or Mei-W68, Trem still localizes to chromosome arms, but Vilya does not form discreet foci. If any of these proteins are missing, DSBs are not made. Adapted from Lake et al. (2015). CO, crossover; NCO, noncrossover.
Like Mei-W68, Mei-P22 is also required for the induction of meiotic DSBs as assessed by γH2AV foci, and recombination is eliminated in these mutants (Liu et al. 2002). Recently, Mei-P22 was found to have homology to the TopoVIB family, which works with A subunits as a heterotetramer to catalyze DSBs (Robert et al. 2016), suggesting that Mei-P22 and Mei-W68 may work as a complex to initiate meiotic DSBs. Although no antibody is available for localizing Mei-W68, a tagged rescue construct of Mei-P22 shows that Mei-P22 foci first appear just prior to the appearance of γH2AV foci (DSBs). Moreover, the two types of foci are highly colocalized, consistent with Mei-P22’s role in DSB formation (Liu et al. 2002; Mehrotra and McKim 2006).
The localization of Mei-P22 to discreet foci requires the protein Trem. The Trem protein contains a zinc finger-associated domain and five C2H2 zinc fingers. Mutation of trem leads to a strong decrease in the number of meiotic DSBs as based on γH2AV foci and recombination is strongly reduced, which results in high levels of chromosome missegregation (Lake et al. 2011). The Trem protein appears to be chromatin-associated, leading to the hypothesis that Trem may be required to alter chromatin structure to allow the binding of Mei-P22 or may assist directly in bringing Mei-P22 to DNA (Lake et al. 2011) (Figure 9).
Finally, Vilya is a protein with homology to the yeast E3 ligase Zip3. Mutations in vilya eliminate recombination, display a strong reduction in the number of γH2AV foci, and cause high levels of chromosome nondisjunction (Lake et al. 2015). The strong reduction in γH2AV foci indicates that one role of Vilya is to promote DSB formation. Indeed, Vilya strongly interacts with Mei-P22 in yeast two-hybrid assays. Prior to the appearance of γH2AV foci, an epitope-tagged version of Vilya localizes along the length of the SC. In the presence of DSBs, Vilya then concentrates into around six foci in early/midpachytene, believed to be the sites of COs. Vilya foci are not formed in mutants that do not make DSBs (Lake et al. 2015). This localization strongly suggests that Vilya may play an additional role after DSB formation (described in RNs).
Genes involved in DSB fate determination
Once formed, a meiotic DSB can be repaired either as a CO or as an NCO. Because only six COs are formed per meiosis and approximately three times as many DSBs are observed by cytological methods, the majority of DSBs are repaired as NCOs (Miller et al. 2016c). We currently understand the process of DSB maturation and fate choice at two levels: (1) the enzymes that process the DSB itself to create an NCO or CO event, and (2) the formation and function of cytologically visible RNs that mark the physical sites of CO formation.
The molecular events that underlie the production of CO vs. NCO resolution events in Drosophila have been thoroughly reviewed by Kohl and Sekelsky (2013) and Sekelsky (2017). In other organisms, NCOs are thought to form through a process called synthesis-dependent strand annealing (SDSA) (Figure 6), as described in Haber (2013). In Drosophila females, many NCOs also appear to be formed via a modified version of the SDSA pathway. Evidence suggests that instead of only one end of the DSB engaging with the homolog to prime synthesis, about half of the time both ends of the break interact with the homolog in some manner (Figure 6) (Crown et al. 2014).
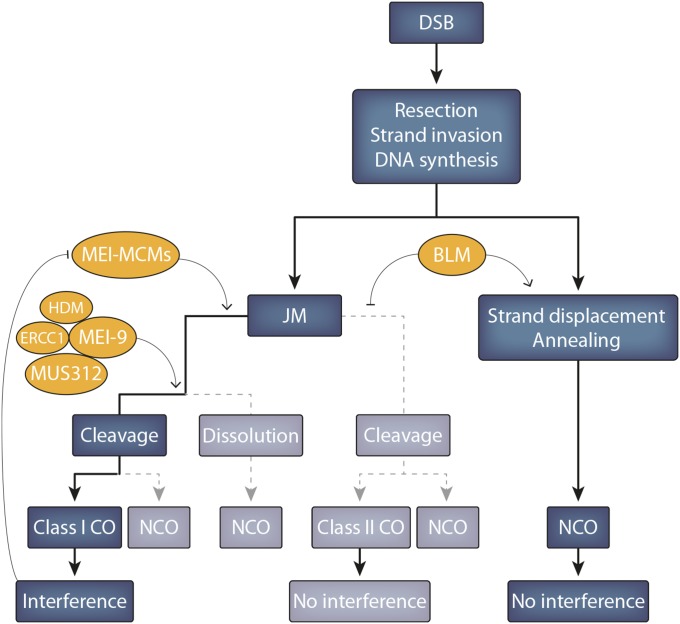
We will organize our further description of the enzymology of Drosophila meiotic recombination into three sets of functions: (1) a group of proteins known collectively as the Mei-MCM (mini-chromosome maintenance) proteins, (2) the Mei-9 resolvase complex, and (3) the Bloom syndrome helicase (Blm helicase) (Table 2). An oversimplified model that explains the roles of these proteins is presented in Figure 10. Briefly, the Mei-MCM proteins are thought to act to promote the resolution of DSBs along the interference-sensitive class I CO pathway. The Mei-9 resolvase complex then acts to facilitate the processing of recombination intermediates that have been matured in the presence of the Mei-MCM complex into class I COs. Blm helicase functions as an anti-CO protein that inhibits DSBs from being processed by the interference-insensitive class II pathway (see section Are there two pathways for crossing over in Drosophila? for a discussion of class I vs. class II COs).
Figure 10.
Double-strand break (DSB) repair. A DSB is typically repaired as either a crossover (CO) or a noncrossover gene conversion (NCO). Two classes of COs can occur, each of which follows the formation of a joint molecule (JM). Class I COs, which are sensitive to interference, are by far the most common, while noninterfering class II COs happen infrequently under normal circumstances (rarely, a JM may be dissolved into an NCO.) The Mei-MCM proteins (Rec, Mei-217, Mei-218, and MCM5) are thought to stabilize those JMs designated to become class I COs, and the Mei-9 resolvase likely functions to cleave double Holliday junctions into COs. Under wild-type conditions, the anticrossover helicase Bloom (BLM) both inhibits class II crossovers and promotes the formation of NCOs by synthesis-dependent strand annealing.
The Mei-MCM proteins:
The Mei-MCM proteins are related to the MCM family of proteins, and these proteins are hypothesized to work as a complex (Kohl et al. 2012). The Mei-MCM complex is thought to include Rec/Mcm8, Mei-217, Mei-218, and likely Mcm5 proteins, all of which appear to act in place of the Msh4 and Msh5 proteins [in other organisms Msh4 and Msh5 play critical roles in promoting the repair of DSBs into COs, but these proteins appear to have been lost in the evolution of Drosophila (Sekelsky et al. 1998; Sekelsky 2017)]. Each of these genes is briefly described below.
Null alleles of rec display high rates of chromosome nondisjunction (Grell 1984) and recombination is strongly decreased. The reduction in recombination is polar, with the reduction in COs strongest in centromere-distal regions and less severe nearer the centromeres (Grell 1984; Blanton et al. 2005). In addition, NCO frequencies are increased, but NCO tract length is shortened (Grell 1984; Blanton et al. 2005).
Mutations in mei-217 and mei-218 reduce exchange to < 10% of wild-type levels, with residual COs occurring more in proportion to physical distance (Sandler et al. 1968; Baker and Carpenter 1972; Carpenter and Sandler 1974; McKim et al. 1996; Liu et al. 2000). Mei-217 and Mei-218 proteins have been shown to interact by yeast two-hybrid assay, and sequence analysis of each protein led to the proposal that they both evolved from a single MCM-like protein (Kohl et al. 2012). Mei-217 also physically interacts with Rec (Kohl et al. 2012). The physical interaction of Mei-217 with both Mei-218 and Rec, as well as the similarity of the phenotypes of mei-217/mei-218 mutants with rec mutants, led to the classification of Mei-217, Mei-218, and Rec as Mei-MCM proteins. Mei-218 and Rec may also have additional roles in blocking nonhomologous end-joining and in the DSB repair checkpoint during female meiosis (Joyce et al. 2012).
Lake et al. (2007) characterized a meiosis-specific allele of mcm5 (null alleles are lethal) that caused increased chromosome missegregation and a 10-fold reduction in crossing over. The reduction in crossing over was not uniform, with CO reductions being strongest in intervals distal to the centromere. Since DSBs were both induced and repaired (based on γH2AV foci and a lack of checkpoint activation associated with failure to repair DSBs), it was concluded that loss of mcm5 function results in the repair of DSBs by NCO or sister-chromatid exchange over COs (Lake et al. 2007). The similarity of the mcm5 mutant phenotype with mei-217, mei-218, and rec mutants suggests that Mcm5 functions in the Mei-MCM complex as well.
The observation that the residual COs observed in Mei-MCM mutants are distributed more in proportion to physical distance suggests that one of the primary roles of the Mei-MCM complex is to promote class I COs. Based on the model proposed by Blanton et al. (2005), loss of Mei-MCM proteins leads to those DSBs designated to mature as class I COs being repaired instead by SDSA. Models suggest that the remaining COs in Mei-MCM mutants are class II COs that are not influenced by interference (Carpenter 1982; Liu et al. 2000; Blanton et al. 2005; McVey et al. 2007; Kohl et al. 2012).
The Mei-9 complex:
Recombination intermediates must be fully resolved to complete repair. Evidence suggests that Mei-9 acts in concert with Mus312, Ercc1, and Hold’em (Hdm) to resolve such intermediates that have been directed down the class I CO pathway by the Mei-MCM complex (Hatkevich et al. 2017). Mei-9, the Drosophila homolog of the Rad1/XPF/ERCC4 single-strand endonuclease, is thought to play a critical role in resolving recombination intermediates into COs during meiotic recombination (Sekelsky et al. 1995). Although mutations in mei-9 do not appear to reduce the frequency of NCOs (at least at the rosy locus), crossing over is reduced to < 10% of wild-type (Baker and Carpenter 1972; Carpenter and Sandler 1974; Carpenter 1982). Importantly, the remaining COs display a wild-type distribution, unlike the mutations in Mei-MCM protein-coding genes, which show an abnormal distribution of COs (Yildiz et al. 2004).
Mei-9 physically interacts by yeast two-hybrid assay with Mus312, which is required for repair of interstrand cross-links, and mus312 mutants display strong recombination and nondisjunction defects similar to mei-9 mutants (Yildiz et al. 2002, 2004). A point mutation in mei-9 that abrogates the physical interaction with Mus312 causes strong meiotic phenotypes, indicating that Mei-9 functions with Mus312 to facilitate resolution of recombination intermediates into class I COs (Yildiz et al. 2002). Mei-9 also physically interacts by yeast two-hybrid assay with the protein Ercc1, using a different part of the protein than the interaction with Mus312 (Yildiz et al. 2002; Radford et al. 2007). Ercc1 mutants display only about half the levels of nondisjunction of mei-9 mutants, indicating that Mei-9 is dependent on Ercc1 for only a portion of its role in resolving meiotic COs (Yildiz et al. 2002).
The Mei-9 complex likely utilizes the single-stranded binding protein Hdm to resolve the remaining COs. Hdm interacts with both Mei-9 and Ercc1 by yeast two-hybrid assay (Joyce et al. 2009), and mutations in hdm reduce recombination and increase chromosome nondisjunction to levels comparable to ercc1 mutations (Joyce et al. 2009). More importantly, double mutants for ercc1 and hdm affect recombination and nondisjunction as strongly as mei-9 mutants, indicating that the Mei-9 complex may utilize Ercc1 and Hdm to resolve different COs (Joyce et al. 2009).
The Blm helicase:
Studies of Blm (or Mus309) provide more insight into the designation of class I vs. class II COs. Both hypomorphic and null alleles of blm cause a moderate reduction in meiotic recombination (McVey et al. 2007; Kohl et al. 2012). The decrease in recombination is not uniform across the chromosome arms, with COs distributed in proportion to physical distance on the chromosomes (McVey et al. 2007; Hatkevich et al. 2017).
Mutants in blm produce enough COs to directly examine interference, which is strongly reduced in these mutants (Hatkevich et al. 2017). Additionally, COs in blm mutants can be identified on the fourth chromosomes, which normally fail to cross over in wild-type flies (Hatkevich et al. 2017). The lack of interference suggests that the remaining COs in blm mutants are generated via the class II CO pathway. To further address this idea, mutations in blm were combined with either mei-218 or rec mutations (Mei-MCM mutants) (Kohl et al. 2012). The strong reduction in recombination and high X chromosome nondisjunction that is characteristic of both mei-218 and rec mutants was substantially alleviated, resulting in nearly wild-type levels of recombination in the mei-MCM blm double mutants (Kohl et al. 2012). blm rec double mutants also display even higher levels of crossing over on the fourth chromosome than blm mutants alone (Hatkevich et al. 2017). Moreover, the observed CO events display a distribution similar to that of blm mutants alone. These results suggest that, in the absence of Blm function, DSBs can be directed into a CO pathway (class II) that does not require Mei-218 and Rec (Kohl et al. 2012).
Double mutants of blm and mei-9 display recombination levels similar to blm mutants, indicating that the COs remaining in blm mutants are not dependent on mei-9 (Hatkevich et al. 2017). Since the Mei-9 complex is thought to be required for class I COs, these results lend further support to the hypothesis that the noninterfering COs observed in blm mutants must be class II COs. (Hatkevich et al. 2017).
Taken together, these studies indicate that the role of Blm helicase is to inhibit the repair of DSBs by the class II pathway and instead direct DSBs down the path to be repaired as class I COs (Hatkevich et al. 2017). There is still much to be learned about the mechanisms determining the designation of DSBs into NCOs vs. class I or II COs, but the intent of the model shown in Figure 10 is to provide a framework to drive future studies.
RNs
As DSBs are made and their fates designated, some DSB sites appear to be associated with large proteinaceous structures known as RNs. Using EM, Carpenter first observed these electron-dense spherical structures in a distribution consistent with the number of COs in Drosophila females (Carpenter 1975a,b, 1979). Thus, RNs, which associate with the SC, appear to mark the sites of crossing over. Despite their obvious association with sites of crossing over, very little is known about how RNs function. The most obvious possibility seems to lie in a role for RNs in the modification of chromosome axes to accommodate the DNA interchange associated with crossing over.
To date, only one RN component (Vilya) has been definitively identified. An epitope-tagged version of the Vilya protein was observed to first localize in tracts along the SC and then concentrate into discreet foci along the chromosome arms in midpachytene (Figure 9). The localization of the epitope-tagged Vilya foci showed a strong correlation with the number and distribution of the COs observed by genetic crosses (Lake et al. 2015). More importantly, immuno-EM demonstrated Vilya localization to the RNs. By immunofluorescence, Mre11 localizes to foci in a pattern indicating that it may also be a component of the RN, but Mre11 localization has not yet been examined by EM (Kusch 2015).
Part V: Stages 2–14; Oocyte Development, Spindle Assembly, and Segregation
The events detailed thus far all occur primarily within the germarium of oocyte development (Figure 5). Once DSB formation and resolution has concluded, a number of additional steps must still be completed for the successful production of a haploid oocyte. The remainder of this review will describe our current understanding of these later stages of meiosis in Drosophila (Table 3).
Table 3. Genes involved in oocyte development.
| Gene symbol | Common name (homolog) | Functiona | References |
|---|---|---|---|
| nhk-1 | Vrk family | Karysome formation | Cullen et al. (2005); Ivanovska et al. (2005) |
| baf | Banf | Karysome formation; links nuclear envelope and chromatin | Lancaster et al. (2007) |
| SRPK | SRPK | Karysome formation; kinase | Loh et al. (2012) |
| kdm5/lid | Kdm5 | Karysome formation; trimethyl H3K4 histone demethylase | Zhaunova et al. (2016) |
| Su(var)205 | HP1a | Heterochromatin-binding protein | Giauque and Bickel (2016) |
| piwi | Argonaute superfamily | piRNA binding | Giauque and Bickel (2016) |
| Su(var)3-9 | SUV39 | H3K9 methyltransferase | Giauque and Bickel (2016) |
| eggless | SetDB | H3K9 methyltransferase | Giauque and Bickel (2016) |
| polo | Plk1-4 | Spindle assembly; chromosome alignment; GVBD | Xiang et al. (2007); Das et al. (2016) |
| mtrm | Inhibitor of Polo kinase | Harris et al. (2003); Xiang et al. (2007), Bonner et al. (2013); Whitfield et al. (2013) | |
| endos | Ensa | GVDB; spindle formation; chromosome alignment | Von Stetina et al. (2008) |
| twe | Germline-specific version of CDC25 | GVBD; chromosome alignment; spindle assembly; cell cycle | Alphey et al. (1992); Courtot et al. (1992), White-Cooper et al. (1993); Xiang et al. (2007) |
| gwl | MastL | Polo regulator | Archambault et al. (2007) |
| elgi | Nrdp1 | GVDB; predicted E3 ubiquitin ligase | Von Stetina et al. (2008) |
| Wispy | Gld-2 | Cytoplasmic poly(A) polymerase; mRNA regulation | Brent et al. (2000); Cui et al. (2008) |
GVBD, germinal vesicle breakdown.
Most likely function based on genetic, biochemical, or cytological data.
Karyosome formation
After the designation of a single pro-oocyte in region 3 or stage 1, the 16-cell cyst buds off from the germarium. In this stage 2–3 oocyte, the nucleus reorganizes to form a compact structure called the karyosome (King 1970; Spradling 1993). The histone H2A kinase Nhk-1 (Nucleosomal histone kinase-1) appears to mediate karyosome formation by phosphorylating the protein Baf (Barrier to autointegration factor), which links chromatin to the nuclear envelope (Cullen et al. 2005; Lancaster et al. 2007). The conserved kinase SRPK (SR protein kinase) also appears to be required to establish and maintain the karyosome (Loh et al. 2012).
Mutants in nhk-1 and SRPK show severe defects in maintaining a single-chromosome mass, and the multiple chromosome masses remain attached to the nuclear envelope (Cullen et al. 2005; Ivanovska et al. 2005; Loh et al. 2012). Heterochromatic regions are also disrupted in SRPK mutants (Loh et al. 2012). nhk-1 mutants display both a delay in the disassembly of euchromatic SC and a failure to properly load condensin components to the chromosomes (Ivanovska et al. 2005). The dispersed chromosomes go on to nucleate separate spindles after meiotic spindle assembly in both nhk-1 and SRPK mutants, indicating that the condensation of the chromosomes into a single karyosome is an important prerequisite for later meiotic events (Cullen et al. 2005; Ivanovska et al. 2005; Loh et al. 2012).
Defects in karyosome formation are also seen with the loss of the histone demethylase Kdm5/Lid, possibly through defects in chromatin architecture during meiotic prophase (Zhaunova et al. 2016). Kdm5/Lid mutants also cause defects in the maintenance of the SC along the chromosome arms, as well as in the maintenance of centromere pairing and clustering during midprophase (Zhaunova et al. 2016).
SC disassembly
Around stages 4–5 of oocyte development, the SC along the chromosome arms begins to progressively disassemble, and it is completely disassembled by stages 6–8. This loss of the SC along the arms coincides with the loading of condensin complex components (Resnick et al. 2009). Interestingly, at least some SC components remain associated with the centromeres beyond euchromatic SC disassembly (Takeo et al. 2011).
The maintenance of heterochromatic associations
After SC disassembly, chromosomes remain linked by heterochromatic associations throughout prophase (Dernburg et al. 1996). These associations occur regardless of whether chiasmata have formed. This was an important observation, as it revealed how those chromosomes that fail to undergo exchange during meiosis are still able to properly segregate during anaphase I (Grell 1976). The vital role that heterochromatic pairing plays in ensuring the proper segregation of nonexchange chromosomes became apparent through studies examining the consequences of heterochromatic duplications and deletions on both the X and fourth chromosomes (Hawley et al. 1992). Furthermore, experiments using a series of heterochromatic deletions have shown that the rate of missegregation is directly associated with the size of a heterochromatic deletion (Karpen et al. 1996). Heterochromatic associations are seen after SC disassembly between nonexchange chromosomes carrying whole-arm inversions (Dernburg et al. 1996), suggesting that it is not simply the maintenance of centromeric SC that facilitates or helps to maintain these associations. Decreasing the level of the heterochromatin-binding protein HP1a [Su(var)205], the H3K9 methyltransferases Su(var)3-9 and Eggless, or the piRNA-binding protein PIWI all lead to defects in the association of heterochromatic regions of the X chromosomes when X chromosomes are heterozygous for the FM7a balancer (Giauque and Bickel 2016).
Chromosome decondensation
At approximately stage 9–10, the chromosomes undergo a temporary decondensation that is associated with an upregulation in transcription by the oocyte nucleus (Mahowald and Tiefert 1970). Whether this decondensation and transcription is important for meiotic progression is unknown since mutations that solely affect this process have not been identified.
GVBD
The chromatin in the oocyte nucleus recondenses after stage 10 in preparation for GVBD, which begins after the end of stage 12. Polo kinase is a protein shown to have roles in mitotic cell cycle entry (Archambault and Glover 2009), and research on two presumed regulators of Polo (Mtrm and Endos) suggests that Polo is likely a regulator of GVBD in Drosophila oocytes. Specifically, reducing the copy number of the Polo inhibitor Matrimony (Mtrm) causes precocious GVBD that is suppressed by a corresponding reduction in Polo levels (Xiang et al. 2007); GVBD is accelerated when Polo is overexpressed (Xiang et al. 2007). The loss of Endosulfine (Endos), a positive regulator of Polo, leads to a delay in GVBD (Von Stetina et al. 2008). Likewise, mutation of the Polo target Twine, a Cdc25 homolog, also leads to a delay in GVBD (Xiang et al. 2007).
Mutation of the Endos-binding protein Early girl (Elgi), a predicted E3 ubiquitin ligase, leads to precocious GVBD by a mechanism separate from Polo kinase regulation (Von Stetina et al. 2008). Additional mechanisms of regulating GVBD and the meiotic cell cycle in Drosophila oocytes likely exist, but because some GVBD regulators likely also play roles in the mitotic cell cycle, approaches that circumvent earlier lethality will be required to examine the function of these players.
Meiosis I spindle assembly
Upon GVBD, tubulin is recruited to the chromosomes (Matthies et al. 1996; Skold et al. 2005; Hughes et al. 2009). Meiotic spindles are anastral (acentriolar) in Drosophila oocytes, thus it is the chromosomes that recruit tubulin and organize the direction of the developing bipolar spindle (Theurkauf and Hawley 1992; Matthies et al. 1996). The spindle is composed of antiparallel microtubules at the central spindle that interact laterally with the chromosomes, kinetochore microtubules that connect kinetochores to the spindle poles, and additional interpolar microtubules that overlap in the central spindle region (Jang et al. 2005).
Multiple kinesins regulate spindle assembly:
Several proteins that play a role in meiotic spindle formation in Drosophila have been identified (Figure 11 and Table 4). One of the best characterized is Nonclaret disjunctional (Ncd), a minus-directed kinesin protein required for proper bipolar spindle assembly and chromosome movement (Endow et al. 1990; McDonald et al. 1990; Skold et al. 2005). It is thought that Ncd moves along microtubules toward the spindle poles to cross-link and bundle microtubules into focused spindle poles (Hatsumi and Endow 1992a,b; Matthies et al. 1996). Mutations in ncd cause misaligned chromosomes; achiasmate (nonrecombinant) chromosomes to be lost from the primary spindle; frayed, unfocused, or absent spindles poles; and chromosome missegregation (Kimble and Church 1983; Hatsumi and Endow 1992b). Live imaging of ncd mutant oocytes reveals a delay in spindle formation, a defect in microtubules interacting laterally with the chromosomes, and abnormal spindle conformations (Matthies et al. 1996; Skold et al. 2005).
Figure 11.
Spindle assembly. Because meiotic spindles in Drosophila are acentriolar, spindle assembly is organized by the chromosomes. The chromosomal passenger complex proteins Aurora-B and Incenp (teal), as well as Subito (green) and the Centralspindlin complex (blue), localize to the central spindle around the DNA and function in chromosome movement and/or spindle assembly/stabilization. Nod (brown arrowhead) acts as the polar ejection force to push chromosomes away from the spindle poles. The proteins MSPS and D-TACC (pink) function at the spindle poles to maintain spindle bipolarity. The Augmin complex (purple) is also located predominately at the spindle poles, where it recruits γ-tubulin and promotes spindle assembly. Both γ-tubulin (dashed lines) and α/β-tubulin (dotted lines) localize along the entire meiotic spindle. The kinesins Ncd and KLP10A, as well as additional kinesins, help regulate the assembly (Ncd) and disassembly (KLP10A) of the bipolar spindle. Mei-38 promotes the assembly or stabilization of kinetochore microtubules (solid lines).
Table 4. Genes involved in spindle assembly and segregation.
| Gene symbol | Common name (homolog) | Functiona | References |
|---|---|---|---|
| ncd | Kinesin family member C1 | Bundles microtubules | Hatsumi and Endow (1992a); Matthies et al. (1996) |
| asp | Aspm | Microtubule-associated protein | Saunders et al. (1997); Riparbelli et al. (2002) |
| Klp54D | Kinesin family member 12 | Spindle symmetry | Radford et al. (2017) |
| Klp61F | Kinesin family member 11 | Spindle symmetry | Radford et al. (2017) |
| Klp10A | Kinesin-13 | Spindle assembly | Zou et al. (2008); Radford et al. (2012a); Do et al. (2014) |
| Eb1 | Mapre family | Microtubule binding | Do et al. (2014) |
| aurB | Aurora Kinase B | Chromosomal passenger complex; spindle midzone | Radford et al. (2012b) |
| Incenp | Incenp | Chromosomal passenger complex; spindle midzone | Colombie et al. (2008); Radford et al. (2012b) |
| sub | MKLP-2/kinesin 6 | Spindle midzone | Giunta et al. (2002); Jang et al. (2005); Radford et al. (2017) |
| tum | RacGAP50C | Centralspindlin complex; spindle midzone | Das et al. (2016) |
| mei-38 | TPX2 | Promotes kinetochore microtubules | Baker and Carpenter (1972); Wu et al. (2008) |
| msps | XMAP215/TOG | Maintains bipolarity | Cullen and Ohkura (2001) |
| cks30A | CKs/Suc1 | Spindle assembly | Cullen et al. (2005) |
| d-tacc | D-TACC | Maintains bipolarity | Cullen and Ohkura (2001) |
| α-Tub67C | Maternally loaded α-tubulin | Microtubule nucleation | Matthies et al. (1999) |
| γ-Tub37C | γ-tubulin | Microtubule nucleation | Tavosanis et al. (1997); Hughes et al. (2011) |
| Augmin complex | Chromosome alignment and movement | Meireles et al. (2009); Colombie et al. (2013) | |
| Grip71 | γTurc complex | Microtubule nucleation | Reschen et al. (2012) |
| Sentin/ ssp2 | Kinetochore attachment to microtubules | Gluszek et al. (2015) | |
| rod | Kntc1 | Rzz complex; spindle assembly checkpoint | Gluszek et al. (2015) |
| Top2 | Topoisomerase 2 | Chromosome orientation | Hughes and Hawley (2014) |
| nod | Nonmotile member of kinesin family | Polar ejection force; chromosome alignment | Baker and Carpenter (1972); Zhang and Hawley (1990); Zhang et al. (1990); Theurkauf and Hawley (1992); Hughes et al. (2009) |
| Axs | Ano family | Spindle width; achiasmate chromosome segregation | Zitron and Hawley (1989); Kramer and Hawley (2003a) |
| mps1/ald | Mps1 | Meiotic spindle assembly checkpoint; chromosome segregation | O’Tousa (1982); Gilliland et al. (2005), (2007) |
| cnn | Centrosomin | Central aster of meiosis II spindles | Riparbelli and Callaini (2005) |
| Grip75 | γTurc Complex | Meiosis II spindle assembly | Vogt et al. (2006) |
| Grip128 | γTurc Complex | Meiosis II spindle assembly | Vogt et al. (2006) |
| Mud | NuMa | Meiosis II central aster | Yu et al. (2006) |
| sra | RCAN | Meiosis II entry; oocyte activation | Horner et al. (2006); Takeo et al. (2006); (2010) |
| canB2 | calcineurin regulatory subunit B | Meiosis II entry; oocyte activation | Takeo et al. (2010) |
| mei-S332 | Shugoshin | Protection of centromeric cohesion | Kerrebrock et al. (1992), (1995) |
| cort | Cortex | APC; Meiosis II progression; egg activation | Page and Orr-Weaver (1996); Whitfield et al. (2013) |
| grau | Grauzone | Meiosis II progression; egg activation; destruction of proteins | Page and Orr-Weaver (1996) |
| fzy | Cdc20 (APC) | Meiosis II progression; destruction of proteins | Swan and Schupbach (2007) |
APC, anaphase-promoting complex.
Most likely function based on genetic, biochemical, or cytological data.
The maintenance of both spindle and centromere symmetry at meiosis I requires the kinesin-5 protein KLP61F (Radford et al. 2017). The meiosis I spindle develops asymmetrically in klp61F mutants and this spindle asymmetry of klp61F mutants depends on Ncd, the microtubule-associated Abnormal spindle protein, and the kinesin-12 KLP54D (Radford et al. 2017).
Another kinesin, the depolymerizing kinesin-13 KLP10A, has also been shown to have a role in meiotic spindle assembly (Zou et al. 2008; Radford et al. 2012a). KLP10A localizes to the meiotic spindle, and loss of KLP10A results in meiosis I spindles that are extremely elongated, with frayed or absent poles (Zou et al. 2008; Radford et al. 2012a; Do et al. 2014). klp10A mutant oocytes also display a failure of homologous chromosome centromeres to biorient and properly attach to spindle microtubules, and a dominant-negative form of KLP10A results in short meiosis I spindles (Zou et al. 2008). Evidence suggests that KLP10A regulates the rate of microtubule disassembly and the interaction of the microtubule-interacting protein EB1 with the ends of microtubules (Radford et al. 2012a; Do et al. 2014).
Central spindle proteins:
As it does in mitosis, the chromosomal passenger complex (CPC) plays crucial roles in meiosis I spindle assembly. Both Aurora B kinase and Incenp, two components of the CPC, localize to the central region of the meiosis I spindle as a ring around the DNA (Jang et al. 2005; Radford et al. 2012b). This localization is dependent on Subito, a homolog of MKLP-2/kinesin 6 (Jang et al. 2005). Subito also localizes as a ring around the DNA on the meiosis I spindle, and is involved in the formation and maintenance of the central spindle (Jang et al. 2005; Radford et al. 2012b). Like KLP61F, Subito promotes spindle and chromosome symmetry (Radford et al. 2017). The Centralspindlin complex and some downstream effectors of this complex also localize to the central spindle and interact with Subito to regulate central spindle assembly and chromosome orientation (Jang et al. 2005; Das et al. 2016).
Mutations in central spindle proteins cause a variety of defects. Hypomorphic alleles of incenp result in the formation of ectopic or split spindle poles, as well as aberrant chromosome orientation and alignment (Colombie et al. 2008; Resnick et al. 2009). Live analysis of the hypomorphic incenp mutants reveals that spindles are unstable, and assembly is delayed, indicating that Incenp is required for spindle formation and stability (Colombie et al. 2008). Stronger knockdown of either Aurora B or incenp results in strong spindle assembly defects (Radford et al. 2012b). Meanwhile, subito mutations cause the formation of monopolar and tripolar spindles and increased chromosome nondisjunction (Giunta et al. 2002). Spindles in subito mutants are also unstable, although the timing of spindle formation is not delayed as it is in some incenp mutants (Colombie et al. 2008).
Spindle pole proteins:
The Mini-spindles (Msps; XMAP215/TOG homolog) and Transforming acidic coiled-coil (D-TACC) proteins localize to the poles of the acentrosomal meiosis I spindle. Mutations in these proteins cause the formation of tripolar spindles, indicating that these proteins are important for maintaining spindle bipolarity (Cullen and Ohkura 2001). Mutants in cks30A, a homolog of Cks/Suc1 and a subunit of the Cdc2 (Cdk1)-cyclin B complex, cause chromosome misalignment and spindle defects in meiosis I (Cullen et al. 2005). These spindles display ectopic poles near the spindle equator, most likely due to mislocalization of Msps and D-TACC to the spindle equator.
Proteins involved in microtubule nucleation and stability:
Another mutant that affects spindle formation, mei-38, still forms bipolar spindles, but the spindles have decreased microtubule density near the kinetochores (Wu et al. 2008). The localization of central spindle proteins is mostly unaffected in mei-38 mutants, and a tagged version of Mei-38 localizes to microtubules other than those in the central spindle, suggesting that the decreased spindle density is due to loss of kinetochore microtubules (Wu et al. 2008). This indicates that Mei-38 is required for the stability of the kinetochore microtubules. Mutation of mei-38 also leads to chromosomes that are disorganized on the aberrant spindles and causes chromosome nondisjunction, with especially high levels of achiasmate chromosome missegregation (Wu et al. 2008). This increased susceptibility of achiasmate chromosomes to missegregate is a common phenotype of mutations that affect spindle formation.
Not surprisingly, tubulin mutants also display meiotic spindle defects. For example, oocytes heterozygous for a mutation in the maternally loaded α-Tubulin 67c (αTub67C) display shorter meiosis I spindles, and chromosomes fail to stretch toward the spindle poles (Matthies et al. 1999). Centromeric regions of the chromosomes do not orient toward opposite spindle poles and achiasmate chromosomes nondisjoin (Matthies et al. 1999). γ-tubulins are important in the nucleation of microtubules, and γTubulin37C, the female germline version of γ-Tubulin in Drosophila, localizes to the entire meiotic spindle (Hughes et al. 2011). Mutations in γTub37c lead to abnormal spindle formation, with highly disorganized prometaphase I spindles exhibiting untapered poles, chromosomes that fail to align properly on the spindle, and centromeres that fail to biorient toward opposite spindle poles (Tavosanis et al. 1997; Hughes et al. 2011). Additionally, D-TACC localization to the spindle is aberrant in γTub37c mutants, and live imaging of mutant γTub37c oocytes revealed spindles that underwent dynamic movements and changes in shapes, as well as chromosome fluctuations (Hughes et al. 2011).
It is believed that γ-Tubulin functions in a complex called the γ-Tubulin ring complex (γTuRC), which is thought to be recruited to some microtubules by the protein complex Augmin (Meireles et al. 2009; Reschen et al. 2012). In meiosis I, Augmin components localize predominantly to the spindle poles, where they likely promote microtubule assembly (Meireles et al. 2009; Colombie et al. 2013). Loss of the Augmin complex component wee Augmin (wac) in oocytes results in increased chromosome movement on the meiosis I spindle and a failure of chromosomes to achieve proper alignment (Meireles et al. 2009; Colombie et al. 2013). Furthermore, mutation of the protein Dgp71WD, which functions with the γTuRC complex, results in meiosis I spindles that are narrower than wild-type, with reduced or absent D-TACC localization (Reschen et al. 2012).
Mutations in many spindle assembly proteins lead to sterility due to failures in meiosis and the early mitotic divisions in the embryo. This sterility has made the study of many of these proteins during meiosis difficult until the recent use of stage-specific RNAi lines.
Chromosome movement, biorientation, and the polar ejection force
While a proper bipolar spindle is necessary for proper biorientation and segregation of homologous chromosomes, it is not sufficient to ensure that these processes occur correctly. For correct chromosome alignment and biorientation on the spindle, kinetochores must also properly attach to the kinetochore microtubules during acentrosomal spindle assembly, and inappropriate kinetochore–microtubule attachments must be destabilized. The EB1 effector protein Sentin appears to be important for this process in Drosophila oocytes (Gluszek et al. 2015). Mutation of Sentin results in a decrease in chromosome movement in prometaphase I and a failure of homologous centromeres to separate and biorient on the bipolar spindle (Gluszek et al. 2015). Although the meiotic spindle is mostly normal in sentin mutant oocytes, localization of the RZZ (Rod-Zw10-Zwilch) complex component Rough deal (Rod), which is part of the spindle assembly checkpoint, indicates that kinetochores become stably attached to spindle microtubules too quickly after spindle assembly, often to the same pole. Indeed, deficiencies in kinetochore components also cause a failure in chromosome orientation and movement (Unhavaithaya and Orr-Weaver 2013; Radford et al. 2015).
In addition, as the spindle elongates during prometaphase I, chiasmata hold exchange chromosomes together in the middle of the spindle while achiasmate chromosomes move toward the poles (Figure 7). The rate of movement of achiasmate chromosomes to the poles is size-dependent, as evidenced by the smaller fourth chromosome proceeding to the pole before the larger X chromosome. The size-dependent nature of migration location was confirmed by Sullivan and Karpen, who observed that a small X chromosome duplication—smaller in size than a fourth chromosome—migrated farther to the pole than the fourth chromosome itself (W. Sullivan and G. Karpen, personal communication). Chromosomes are pushed away from the poles and toward the spindle equator by the polar ejection force. No distributive disjunction (Nod), a kinesin-like protein (discussed in detail in Distributive mutants), helps to ensure that achiasmate chromosomes do not migrate off the spindle by acting as a brake on chromosome movement toward the spindle poles (Theurkauf and Hawley 1992; Afshar et al. 1995). The elimination of this brake is clearly seen in nod mutant oocytes, where the inherently achiasmate fourth chromosome is essentially always found in the cytoplasm (Theurkauf and Hawley 1992; Hughes et al. 2009).
Meiosis I segregation
In mitosis as well as meiosis II, sister chromatids segregate away from each other at anaphase, but in meiosis I, homologous chromosomes must biorient toward opposite spindle poles and segregate away from each other at anaphase I. While correct positioning and orientation on the meiotic spindle are essential for proper homologous chromosome segregation at the first meiotic division, crossing over and the formation of chiasmata provide the force needed to hold bivalents together. This force balances the tension on the kinetochores of homologous chromosomes from the opposing spindle poles to properly biorient homologous centromeres at the metaphase plate until chromosome segregation occurs at anaphase I. Indeed, chromosomes that fail to recombine often fail to properly segregate away from each other. Additionally, at least one pair of homologous chromosomes must form a chiasma to generate the tension required for oocytes to maintain metaphase I arrest (Jang et al. 1995). In the absence of chiasmata, oocytes precociously enter anaphase I. Finally, when chromosomes fail to become balanced at the middle of the meiosis I spindle, the forces of the microtubules, kinesins, and other proteins progressively move the chromosomes apart. The chromosomes can move so far apart, in fact, that they may nucleate multiple, separate spindles. Thus, although a byproduct of recombination is to increase genetic diversity, its primary function is to ensure proper chromosome segregation at anaphase I.
The distributive system
Although recombination is the primary system for ensuring segregation of the autosomes in Drosophila females, the fourth chromosomes never undergo recombination in wild-type females and the X chromosomes fail to recombine in 8–10% of oocytes (Hawley et al. 1993). Additionally, when a normal-sequence X chromosome is heterozygous to a balancer X chromosome, the X’s will fail to undergo recombination. Nonetheless, such achiasmate X and fourth homologs still segregate faithfully to opposite poles in > 99% of wild-type oocytes using what has been termed the distributive system (Hawley et al. 1993). This mechanism of segregating achiasmate chromosomes differs from the segregation of achiasmate chromosomes in male meiosis, where recombination never occurs.
Heterochromatic sequences on the X and fourth chromosomes are found to be both necessary and sufficient for the distributive system to properly segregate achiasmate chromosomes (Hawley et al. 1992; Karpen et al. 1996). More importantly, heterochromatic associations have been shown to exist between X and fourth homologs during prophase and these associations persist until just prior to GVBD (Dernburg et al. 1996). Additionally, heterochromatic DNA threads have been shown to connect achiasmate chromosomes even after spindle assembly, as discussed in the next section (Hughes et al. 2009).
Achiasmate chromosomes undergo dynamic movements:
Upon assembly of the bipolar spindle, the achiasmate chromosomes undergo dynamic movements toward the spindle poles and back toward the center of the spindle (Figure 7). These dynamic movements, during which homologs may at times associate on the same side of the spindle, continue until all achiasmate homologs have properly bioriented and have congressed to join the chiasmate autosomes that are properly balanced at the central spindle (Gilliland et al. 2009; Hughes et al. 2009). After achiasmate chromosomes have congressed, the chromosomes form a compact lemon-shaped structure with the centromeres of homologous chromosomes bioriented toward opposite spindle poles (Figure 7) (Gilliland et al. 2009).
It is not fully clear why chromosomes undergo dynamic movements on the spindle during prometaphase I. One possibility is that chromosomes make improper centromere attachments to the kinetochore microtubules at spindle assembly, and these chromosome movements can facilitate the shedding of incorrect attachments and allow for new, correct attachments to form. This idea is supported by data from sentin mutants (described in Chromosome movement, biorientation, and the polar ejection force). Oocytes mutant for sentin display decreased chromosome movement, precocious stable attachment of kinetochore microtubules to centromeres (as based on Rod localization), and, ultimately, centromeres that remain maloriented (Gluszek et al. 2015). Chromosomes undergoing movements would need a mechanism to help ensure that achiasmate homologs end up on opposite half-spindles and biorient in the absence of chiasmata.
During the chromosome movements, DNA threads composed of heterochromatin connect achiasmate homologs (Hughes et al. 2009). These threads may allow achiasmate chromosomes to maintain contact with their homolog in the absence of chiasmata and thus facilitate proper orientation toward opposite spindle poles (Hughes et al. 2009). These DNA threads are likely resolved by Topoisomerase 2 (Top2). Knockdown of Top2 by RNAi results in chromosomes that fail to properly separate and biorient on the meiosis I spindle, and causes the heterochromatic regions of chromosomes to become highly stretched out in many prometaphase I oocytes (Hughes and Hawley 2014). These oocytes fail to properly separate homologs at anaphase I when Top2 is decreased, indicating that the heterochromatic connections must be resolved before homologous chromosome segregation.
Heterologous segregation:
While heterochromatic homology appears to be the primary mechanism for segregating achiasmate homologs, other mechanisms likely influence the segregation of chromosomes that do not have a homolog or heterologous segregation (Table 5). In D. melanogaster, flies with compound chromosomes carrying two arms of the same chromosome attached together are viable. Flies have been generated carrying two sets of compound chromosomes, such as an attached X chromosome and an attached fourth chromosome, or compound 2L and compound 2R chromosomes. In these instances, the compounds segregate away from each other with extremely high fidelity, based on both genetics and the orientation of chromosomes after spindle assembly (Grell 1963, 1970; Gilliland et al. 2014). Thus, a second mechanism may exist that orients chromosomes lacking homologs to ensure balanced chromosome segregation. Interestingly, reduced achiasmate chromosome movement is observed when an attached fourth chromosome is present without a normal fourth chromosome, indicating that the fourth chromosomes influence chromosome movement (Gilliland et al. 2014).
Table 5. Glossary of terms.
| Term | Definition |
|---|---|
| Acentric fragment | A chromosome fragment that does not contain a centromere |
| Acentriolar spindle | A spindle lacking centrioles, most commonly found during meiosis I in oocytes; also known as an anastral spindle |
| Achiasmate chromosome | A chromosome that does not form a chiasma (CO) |
| Balancer chromosome | A chromosome that is multiply inverted and/or rearranged in comparison to the normal chromosome and that, when heterozygous with a normal-sequence chromosome, suppresses exchange and/or prevents the recovery of recombinant products; typically carries both a dominant visible marker and a homozygous lethal or sterile mutation |
| Biorientation | The alignment of homologous chromosomes toward opposite spindle poles during meiosis I |
| Bivalent | A pair of homologous chromosomes held together by at least one chiasma |
| Class I CO | The predominant type of exchange event whose placement is influenced by interference and the centromere effect |
| Class II CO | A CO that is placed in a manner independent of other COs or the centromere |
| Central element (CE) | As observed by electron microscopy, the electron-dense region of the SC that lies within the CR, midway between the lateral elements |
| Central region (CR) | The portion of the SC between the lateral elements that encompasses the space between two homologous chromosomes; includes the transverse filament and CE proteins |
| Centromere clustering | The association of the paired homologous centromeres of one chromosome with the paired centromeres of other, nonhomologous chromosomes |
| Centromere effect | The strong suppression of exchange events in proximity to the centromere |
| Centromere pairing | The association, mediated by SC proteins, of the centromeres from two homologous chromosomes |
| Chiasma (pl. chiasmata) | A mature CO that physically links two nonsister chromatids; required for accurate meiotic chromosome segregation |
| Crossing over | The process that exchanges genetic material between homologous chromosomes, leading to the formation of chiasmata and recombinant chromosomes |
| Crossover (CO) | Event marking the location in the DNA where two nonsister chromatids exchange their genetic material; the physical outcome of the process of crossing over |
| CO assurance | The observed phenomenon where each chromosome is guaranteed at least one CO during meiosis |
| Diakinesis | In most organisms, the last phase of meiotic prophase I, during which individual chromosomes further condense in preparation for germinal vesicle breakdown and spindle assembly; Drosophila lacks a canonical diakinesis |
| Dicentric bridge | An aberrant chromosome with two centromeres that is pulled apart or broken during segregation, resulting in two unstable chromosomes |
| Diplotene | In most organisms, the phase of meiotic prophase I in which the SC disassembles and chromosomes become individualized; Drosophila lacks a canonical diplotene |
| Distributive system | Term applied to the mechanism for properly segregating chromosomes that failed to undergo exchange; denotes two very separate processes: the mechanism that ensures the segregation of nonexchange homologs and a poorly understood process that can segregate nonhomologous chromosomes |
| Double-strand break (DSB) | A break that involves both of the Watson and Crick strands of a DNA molecule |
| E0 tetrad | A tetrad having no CO events; the fraction of E0 tetrads is used to calculate the likelihood that a pair of homologous chromosomes did not undergo crossing over |
| Equational exception | Exceptional progeny resulting from nondisjunction at meiosis II |
| Euchromatin | The more lightly-condensed portion of a chromosome that contains the majority of an organism’s genes |
| Gene conversion | The nonreciprocal exchange of a small amount of genetic information from one homologous chromosome to the other, resulting in a 3:1 allele ratio at that locus; can occur in association with a CO or independently of crossing over |
| Germarium | The structure at the tip of the ovary where egg chambers are formed and meiosis is initiated |
| Germinal vesicle breakdown (GVBD) | The breakdown of the nuclear envelope at the end of stage 12 of oocyte development |
| Heterochromatin | Tightly condensed chromatin that contains few genes and often contains repetitive DNA sequences |
| Heterologous segregation | The segregation of nonhomologous chromosomes away from one another |
| Interchromosomal effect | The phenomenon whereby heterozygosity for chromosome aberrations, especially balancer chromosomes, suppresses crossing over between those homologs but increases crossing over on the other unbalanced chromosomes |
| Interference | The phenomenon in which a CO in one interval decreases the likelihood of a CO in an adjacent interval |
| Karyosome | The condensed oocyte nucleus that forms in midprophase |
| Lateral element (LE) | The outermost components of the SC that run along the axis of two homologous chromosomes |
| Marker | A mutation that produces an innocuous, easily identifiable visible phenotype, such as eye color, wing attribute, or bristle quality |
| Matroclinous females | Exceptional progeny derived from XX oocytes, also known as diplo-X exceptions |
| Meiotic mutant | A stock bearing a mutation in a gene involved in a meiotic process |
| Missegregation | See Nondisjunction |
| Noncrossover (NCO) event | A gene conversion that is not associated with a CO event |
| Nondisjunction | The aberrant segregation of chromosomes during meiosis, such that both homologs (meiosis I) or sister chromatids (meiosis II) go to the same daughter nucleus |
| Nurse cells | Polyploid, interconnected support cells that produce proteins and RNAs needed by the developing oocyte |
| Ovariole | One of ∼16 tubules in each Drosophila ovary that contains a germarium and developing egg chambers connected sequentially from stage 1 through stage 14 |
| Paracentric inversion | A chromosome inversion that does not include the centromere |
| Pachytene | Phase of meiotic prophase I during which paired chromosomes recombine and full-length SC is established; in flies, this begins in region 2A in the germarium |
| Patroclinous males | Exceptional progeny derived from oocytes lacking an X; also known as nullo-X exceptions |
| Pericentric heterochromatin | Recombinationally inert portion of a chromosome located near the centromere |
| Pericentric inversion | A chromosome inversion that encompasses the centromere |
| Polar ejection force | A force that pushes chromosomes away from the spindle poles and toward the central spindle during prometaphase |
| Prometaphase I | An intermediate period between prophase and metaphase I during which the nuclear envelope breaks down, the bipolar spindle elongates, and chromosomes undergo dynamic movements on the spindle |
| Recombination hotspot | A location or particular DNA sequence in the genome that exhibits a higher rate of meiotic DSB formation and/or recombination than expected by chance |
| Recombination nodule (RN) | Protein structure associated with the SC that mediates CO/chiasma formation |
| Reductional exception | Exceptional progeny resulting from nondisjunction at meiosis I |
| Sister chromatid exchange | Meiotic exchange that occurs between sister chromatids rather than homologous chromosomes |
| Synapsis | The full-length alignment of homologous chromosomes by the SC in preparation for recombination |
| Transverse filament (TF) | Coiled-coil-containing SC protein that functions to span the distance between the two lateral elements |
| Triploid | An individual with three sets of each chromosome |
| Synaptonemal complex (SC) | The proteinaceous, zipper-like structure that connects homologous chromosomes during prophase |
| Vitellarium | The posterior portion of an ovariole that contains oocytes in developmental stages 2–14 |
| Zygotene | Phase of meiotic prophase I during which homologous chromosomes begin to pair and synapse; characterized in flies by the observation of short patches of synaptonemal complex in early region 2A of the germarium |
Another example where a chromosome must segregate in the absence of a homolog is in XXY females. Bridges (1916) [reviewed in Ganetzky and Hawley (2016)] observed that in XXY females where the X chromosomes recombine, the X chromosomes segregate away from each other with the Y segregating at random. When the X chromosomes are achiasmate, the Y chromosome associates with the X chromosomes, resulting in their aberrant coorientation after spindle assembly and ultimately in their missegregation (Bridges 1916; Xiang and Hawley 2006). This suggests that the segregation of both achiasmate chromosomes and chromosomes without homologs may be complex.
Distributive mutants:
Some mutants display a stronger effect on the segregation of achiasmate chromosomes than on the segregation of chiasmate chromosomes. This can be viewed as a defect in the distributive system, thus these mutants are termed distributive mutants. As mentioned above, mutations that affect spindle assembly cause defects in proper chromosome biorientation during meiosis I. Chiasmata can partially compensate for a poor meiotic spindle or aberrant forces on the chromosomes. However, chromosomes that are achiasmate, and therefore not held together by chiasmata, are especially prone to biorientation errors when the spindle fails to properly form. Chromosomes that depend solely on heterochromatic associations for accurate segregation require tapered, bipolar spindles and balanced forces to maintain proper alignment. The genes most strongly associated with distributive segregation are nod, Aberrant X segregation (Axs), monopolar spindle 1 [mps1, originally known as altered disjunction (ald)], and mtrm.
One of the first identified distributive mutants was nod (Baker and Carpenter 1972). The nod gene encodes a kinesin-like protein believed to act as the polar ejection force, which is the force that pushes chromosomes back to the metaphase plate (Theurkauf and Hawley 1992; Afshar et al. 1995; Cochran et al. 2009). The polar ejection force acts against the forces of the kinetochore microtubules that pull centromeres of chromosomes toward the spindle poles. Studies of Nod, including the determination of its crystal structure, reveal that while Nod has homology to kinesins, it is nonmotile (Afshar et al. 1995; Cochran et al. 2009). Instead, it binds to microtubules and chromosomes to act as a brake for chromosome movement (Afshar et al. 1995; Cochran et al. 2009). Analysis of fixed and live nod mutant oocytes shows that during the dynamic movements achiasmate chromosomes undergo on the meiosis I spindle, achiasmate chromosomes are often ejected from the primary spindle containing the chiasmate chromosomes (Theurkauf and Hawley 1992; Hughes et al. 2009). Although these lost chromosomes can nucleate their own mini-spindles, they may not always participate in the normal segregation of chromosomes at anaphase of meiosis I and II. Because achiasmate chromosomes are lost from the primary spindle in nod mutant oocytes, mutations in nod result in achiasmate chromosome nondisjunction values > 50% (Carpenter 1973; Rasooly et al. 1994). In the absence of Nod, chiasmata are still able to hold chiasmate chromosomes at the metaphase plate, which allows for the normal segregation of these chromosomes (Carpenter 1973).
The meiosis I spindle is surrounded by a membranous sheath to which the protein Axs localizes (Kramer and Hawley 2003a). A dominant mutation in Axs, AxsD, causes the formation of improperly tapered meiosis I spindles (Kramer and Hawley 2003a). Yet, unlike other spindle mutants, AxsD appears to disrupt only the segregation of achiasmate chromosomes (Zitron and Hawley 1989; Kramer and Hawley 2003a). Spindles in AxsD flies are still bipolar, but they are much broader than in wild-type flies (Kramer and Hawley 2003a). Additionally, overexpression of the AxsD mutant protein leads to a failure to maintain metaphase I arrest in some oocytes (Kramer and Hawley 2003a). Axs has homology to a family of calcium-activated chloride channels (Kramer and Hawley 2003b; Hartzell et al. 2009), but further investigation is needed to fully understand the wild-type function of Axs, as well as the mutant phenotype of AxsD, during meiosis.
Achiasmate chromosome segregation is strongly affected by the haplo-insufficient mutation mtrm (Harris et al. 2003). While heterozygosity for mtrm mutations leads to near random segregation of achiasmate chromosomes, homozygosity leads to sterility due to meiotic catastrophe after meiosis I spindle assembly (Xiang et al. 2007; Bonner et al. 2013). The Mtrm protein appears to be a physical inhibitor of Polo kinase (described in GVBD), which regulates a number of processes in mitosis in addition to cell cycle entry (Archambault and Glover 2009). Strong knockdown of polo leads to spindle and chromosome alignment defects (Das et al. 2016), while the probable increase in unregulated Polo in mtrm mutant oocytes likely leads to the failure of achiasmate chromosomes to properly biorient. A mutation of greatwall (gwl) that causes decreased Polo activity in meiosis can suppress the nondisjunction of mtrm heterozygotes, further illustrating the interaction of Mtrm and Polo (Archambault et al. 2007).
Another example of distributive mutants that undergo improper cell-cycle regulation and highly increased achiasmate chromosome nondisjunction are female germline-specific alleles of mps1/ald. Mutations in mps1 appear to alter the timing of meiosis I events, with chromosomes undergoing anaphase I-like separation very quickly after meiosis I spindle assembly (Gilliland et al. 2005, 2007). Achiasmate chromosomes undergoing movements to biorient on the bipolar spindle likely become trapped on the same side of the meiotic spindle at the time of the precocious chromosome segregation, thus leading to achiasmate chromosome nondisjunction (Gilliland et al. 2007).
The mutants affecting achiasmate chromosome segregation disrupt proteins that appear to play diverse roles. This indicates that the distributive system may be complex, with multiple components.
Meiosis II
Drosophila oocytes can remain arrested at metaphase I for up to 2 days (King 1970). In Drosophila oocytes, it is passage through the oviduct, rather than fertilization, that leads to oocyte activation and the resumption of meiosis (Doane 1960; Mahowald et al. 1983; Page and Orr-Weaver 1997; Heifetz et al. 2001; Horner and Wolfner 2008). After activation, anaphase I and all of meiosis II can be completed within ∼20 min, making analysis of the events after metaphase I difficult (Riparbelli and Callaini 1996). Upon activation, there is a wave of Ca2+ in the oocyte indicating that, like in other organisms, Ca2+ signaling plays an important role in activation (Horner and Wolfner 2008; Kaneuchi et al. 2015). Consistent with Ca2+ playing a role in activation, the progression from anaphase I to meiosis II is dependent on the calcineurin pathway (Horner et al. 2006; Takeo et al. 2006, 2010, 2012). Mutation in either calcineurin B2 (canB2) or sarah (sra), which interacts with and regulates calcineurin activity, leads to an arrest at anaphase I, with chromosomes stopped before reaching the spindle poles (Horner et al. 2006; Takeo et al. 2006, 2010). Both failure to translate bicoid mRNA and elevated cyclin B levels, which are indicative of a failure to enter meiosis II, support that Sarah is required for full egg activation (Horner et al. 2006).
Upon activation, the spindle rotates to align at a right angle with the anterior–posterior axis of the oocyte and the CR of the spindle elongates (Endow and Komma 1998; Riparbelli and Callaini 1996). As the chromosomes move toward the spindle poles in anaphase I, the center of the spindle pinches in between the chromosomes and an aster of microtubules forms between the separating chromosomes (Endow and Komma 1998; Riparbelli and Callaini 1996). This central aster of microtubules progresses to form a central spindle pole body composed of centrosomal components (Riparbelli and Callaini 1996, 2005; Endow and Komma 1998; Llamazares et al. 1999). Loss of the centrosomal protein Centrosomin leads to an aberrant central aster between the developing prometaphase II spindles, illustrating the requirement for centrosomal proteins for the formation of the central aster (Riparbelli and Callaini 2005). After prometaphase II, the sister chromatids segregate away from each other on the twin spindles to rapidly complete the second meiotic division (Riparbelli and Callaini 1996; Endow and Komma 1998).
The γTuRC binds to microtubule minus ends and stimulates microtubule-nucleating activity in vitro (Zheng et al. 1995; Moritz and Agard 2001). In mutants affecting two components of the γTuRC (Grip75 and Grip128), meiosis I spindle formation appears mostly normal, but meiosis II spindles are severely disrupted (Vogt et al. 2006). The central aster of microtubules between the two tandem spindles is absent, the two spindles fail to remain properly aligned with each other and/or with the cortex, and spindles are often disorganized. Similar phenotypes are observed at meiosis II in oocytes mutant for mushroom body defect (mud) (Yu et al. 2006). The Mud protein localizes to the region between the tandem meiosis II spindles.
A hypomorphic allele of polo kinase, polo1, which can complete meiosis I, causes defects in meiosis II spindle formation (Riparbelli et al. 2000). The twin spindles are frequently misaligned, and the central array of microtubules improperly organized, and the chromosomes fail to stay properly aligned on the defective spindles (Riparbelli et al. 2000). Polo kinase appears to play roles in both meiotic divisions of Drosophila oocytes.
In meiosis I, cohesion between the arms of the homologs is cleaved to allow separation of homologs at anaphase I, but cohesion at sister centromeres is maintained until meiosis II by the Shugoshin homolog Mei-S332 (Kerrebrock et al. 1992). Mutation of mei-S332 causes precocious loss of centromeric cohesion and the eventual random segregation of sister chromatids in meiosis II (Davis 1971; Kerrebrock et al. 1992). Mei-S332 localizes near centromeres until the metaphase II/anaphase II transition, consistent with its role in protecting centromeric cohesion (Kerrebrock et al. 1995).
Mutation of cortex or its transcriptional activator grauzone primarily results in meiotic arrest at anaphase II, but both mutants also display defects in anaphase I chromosome segregation (Page and Orr-Weaver 1996). A smaller proportion of oocytes arrest in meiosis I or exhibit aberrant spindle and chromosome configurations, and cortical microtubules fail to rearrange in response to activation. Cortex is a Cdc20-related protein that, with the mitotic Cdc20 homolog Fizzy (Fzy), works in the anaphase-promoting complex (APC) to destroy Cyclins A, B, and B3 in female meiosis to allow for proper cell cycle progression (Swan and Schupbach 2005, 2007; Pesin and Orr-Weaver 2007). Cks30A promotes the activation of the APC that utilizes Cortex (Swan and Schupbach 2007), and Cortex is also required for the destruction of the Polo inhibitor Mtrm during meiosis II to allow proper entry into the embryonic mitoses (Whitfield et al. 2013). These studies demonstrate the importance of the degradation of meiotic proteins before the onset of embryonic mitoses.
Because meiosis II is completed so rapidly, much of our knowledge is based on a limited set of mutants that cause arrest at various stages of meiosis II. There is still much to learn about the regulation of meiosis II.
Conclusions
A century of genetic studies in Drosophila are now embedded in superb cytological techniques. More than ever, we can imagine taking the genetic formalisms that resulted from the first 100 years of meiotic mutant analysis and inserting them in the realities of meiotic cell biology. Indeed, studies examining the genetics and biology of female meiosis have revealed:
The SC is assembled and disassembled in a highly organized fashion.
Components of the SC play important roles in regulating CO formation, centromere clustering, and pairing.
There are two classes of COs and multiple proteins play roles promoting the formation of class I COs (interference-sensitive) over class II COs (interference-insensitive).
Formation of a tapered, bipolar meiosis I spindle is important for proper segregation of homologous chromosomes.
The distributive system helps ensure the segregation of achiasmate chromosomes.
Meiosis II occurs rapidly with sister chromatids segregating away from each other on twin spindles.
However, many important questions remain to be fully answered. For example, how are the number and placement of DSBs and COs chosen? What regulates the assembly and disassembly of the SC? What proteins play a role in the RN? How are meiosis I and II entry and exit controlled? As new advances in imaging, sequencing, and biochemistry are developed, our understanding of the processes regulating meiosis will only deepen.
Acknowledgments
We are grateful to be a part of the supportive, collaborative environment that is the fly community. In particular, we thank our colleagues Jeff Sekelsky, Nicole Crown, Talia Hatkevich, and Sarah Radford for their helpful comments and discussions regarding aspects of this review. We also thank the members of the Hawley lab, especially Cathy Lake and Amanda Bonner, who assisted us during this process. Finally, our apologies to the authors of the many fine studies we were unable to discuss here; we simply ran out of time. R.S.H. is an American Cancer Society Research Professor.
Footnotes
This review is dedicated to the memory of professor Robert C. King, whose early studies on the development and morphology of Drosophila oocytes were critical to the development of the field.
Curiously, a study published by Merriam and Frost (1964) attributed only 25% of reductional exceptions to the failure of exchange. Indeed, Merriam and Frost observed that nearly two-thirds of the exchange bivalents doomed to nondisjunction were E2 tetrads (asterisks). Moreover, they did not observe a difference in the positioning of CO events between bivalents that segregated properly and those undergoing reductional nondisjunction. There does not appear to be a straightforward way to reconcile these findings with those of Koehler et al. (1996)].
Communicating editor: T. Orr-Weaver
Literature Cited
- Abdu U., Brodsky M., Schupbach T., 2002. Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol. 12: 1645–1651. [DOI] [PubMed] [Google Scholar]
- Afshar K., Barton N. R., Hawley R. S., Goldstein L. S., 1995. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell 81: 129–138. [DOI] [PubMed] [Google Scholar]
- Alphey L., Jimenez J., White-Cooper H., Dawson I., Nurse P., et al. , 1992. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell 69: 977–988. [DOI] [PubMed] [Google Scholar]
- Anderson L. K., Royer S. M., Page S. L., McKim K. S., Lai A., et al. , 2005. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. USA 102: 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V., Glover D. M., 2009. Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 10: 265–275. [DOI] [PubMed] [Google Scholar]
- Archambault V., Zhao X., White-Cooper H., Carpenter A. T., Glover D. M., 2007. Mutations in Drosophila greatwall/scant reveal its roles in mitosis and meiosis and interdependence with Polo kinase. PLoS Genet. 3: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Golic K. G., Hawley R. S., 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Baker B. S., Carpenter A. T., 1972. Genetic analysis of sex chromosomal meiotic mutants in Drosophilia melanogaster. Genetics 71: 255–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., Hall J. C., 1976. Meiotic mutants: genic control of meiotic recombination and chromosome segregation, pp. 351–434 in The Genetics and Biology of Drosophila, Vol. 1a, edited by Ashburner M., Novitski E. Academic Press, New York. [Google Scholar]
- Beadle G. W., 1932. A possible influence of the spindle fibre on crossing over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., Copenhaver G. P., 2010. Genetic interference: don’t stand so close to me. Curr. Genomics 11: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A., de Massy B., Gadelle D., Varoutas P. C., Nicolas A., et al. , 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417. [DOI] [PubMed] [Google Scholar]
- Bickel S. E., Wyman D. W., Miyazaki W. Y., Moore D. P., Orr-Weaver T. L., 1996. Identification of ORD, a Drosophila protein essential for sister chromatid cohesion. EMBO J. 15: 1451–1459. [PMC free article] [PubMed] [Google Scholar]
- Bickel S. E., Wyman D. W., Orr-Weaver T. L., 1997. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics 146: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel S. E., Orr-Weaver T. L., Balicky E. M., 2002. The sister-chromatid cohesion protein ORD is required for chiasma maintenance in Drosophila oocytes. Curr. Biol. 12: 925–929. [DOI] [PubMed] [Google Scholar]
- Blanton H. L., Radford S. J., McMahan S., Kearney H. M., Ibrahim J. G., et al. , 2005. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 1: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M. D., Karpen G. H., 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner A. M., Hughes S. E., Chisholm J. A., Smith S. K., Slaughter B. D., et al. , 2013. Binding of Drosophila Polo kinase to its regulator matrimony is noncanonical and involves two separate functional domains. Proc. Natl. Acad. Sci. USA 110: E1222–E1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent A. E., MacQueen A., Hazelrigg T., 2000. The Drosophila wispy gene is required for RNA localization and other microtubule-based events of meiosis and early embryogenesis. Genetics 154: 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges C. B., 1916. Non-disjunction as proof of the chromosome theory of heredity (concluded). Genetics 1: 107–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon C. K., Yu Z., Wang Y., Guo F., Unruh J. R., et al. , 2017. Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. USA 114: E6857–E6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. T., 1973. A meiotic mutant defective in distributive disjunction in Drosophila melanogaster. Genetics 73: 393–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. T., 1975a Electron microscopy of meiosis in Drosophila melanogaster females: II: the recombination nodule—a recombination-associated structure at pachytene? Proc. Natl. Acad. Sci. USA 72: 3186–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. T., 1975b Electron microscopy of meiosis in Drosophila melanogaster females. I. Structure, arrangement, and temporal change of the synaptonemal complex in wild-type. Chromosoma 51: 157–182. [DOI] [PubMed] [Google Scholar]
- Carpenter A. T., 1979. Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma 75: 259–292. [DOI] [PubMed] [Google Scholar]
- Carpenter A. T., 1982. Mismatch repair, gene conversion, and crossing over in two recombination-defective mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 79: 5961–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. T., 1987. Gene conversion, recombination nodules, and the initiation of meiotic synapsis. Bioessays 6: 232–236. [DOI] [PubMed] [Google Scholar]
- Carpenter A. T. C., 1988. Thoughts on recombination nodules, meiotic recombination, and chiasmata, pp. 529–548 in Genetic Recombination, edited by Kucherlapati R., Smith G. R. American Society for Microbiology, Washington, DC. [Google Scholar]
- Carpenter A. T., Sandler L., 1974. On recombination-defective meiotic mutants in Drosophila melanogaster. Genetics 76: 453–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. H., Jenkins P. A., Song Y. S., 2012. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet. 8: e1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou N., Rubin T., Huynh J. R., 2013. Synaptonemal complex components promote centromere pairing in pre-meiotic germ cells. PLoS Genet. 9: e1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou N., Rubin T., Bonnet I., Piolot T., Arnaud M., et al. , 2015. Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nat. Cell Biol. 17: 1388–1400. [DOI] [PubMed] [Google Scholar]
- Cochran J. C., Sindelar C. V., Mulko N. K., Collins K. A., Kong S. E., et al. , 2009. ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement. Cell 136: 110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. A., Callicoat J. G., Lake C. M., McClurken C. M., Kohl K. P., et al. , 2012. A germline clone screen on the X chromosome reveals novel meiotic mutants in Drosophila melanogaster. G3 (Bethesda) 2: 1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. A., Unruh J. R., Slaughter B. D., Yu Z., Lake C. M., et al. , 2014. Corolla is a novel protein that contributes to the architecture of the synaptonemal complex of Drosophila. Genetics 198: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombie N., Cullen C. F., Brittle A. L., Jang J. K., Earnshaw W. C., et al. , 2008. Dual roles of incenp crucial to the assembly of the acentrosomal metaphase spindle in female meiosis. Development 135: 3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombie N., Gluszek A. A., Meireles A. M., Ohkura H., 2013. Meiosis-specific stable binding of augmin to acentrosomal spindle poles promotes biased microtubule assembly in oocytes. PLoS Genet. 9: e1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot C., Fankhauser C., Simanis V., Lehner C. F., 1992. The Drosophila cdc25 homolog twine is required for meiosis. Development 116: 405–416. [DOI] [PubMed] [Google Scholar]
- Craymer L., 1984. Techniques for manipulating chromosomal rearrangements and their application to Drosophila melanogaster. II. Translocations. Genetics 108: 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown K. N., McMahan S., Sekelsky J., 2014. Eliminating both canonical and short-patch mismatch repair in Drosophila melanogaster suggests a new meiotic recombination model. PLoS Genet. 10: e1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Sackton K. L., Horner V. L., Kumar K. E., Wolfner M. F., 2008. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics 178: 2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen C. F., Ohkura H., 2001. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3: 637–642. [DOI] [PubMed] [Google Scholar]
- Cullen C. F., Brittle A. L., Ito T., Ohkura H., 2005. The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J. Cell Biol. 171: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ines O., White C. I., 2015. Centromere associations in meiotic chromosome pairing. Annu. Rev. Genet. 49: 95–114. [DOI] [PubMed] [Google Scholar]
- Das A., Shah S. J., Fan B., Paik D., DiSanto D. J., et al. , 2016. Spindle assembly and chromosome segregation requires central spindle proteins in Drosophila oocytes. Genetics 202: 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. K., 1971. Genetic analysis of a meiotic mutant resulting in precocious sister-centromere separation in Drosophila melanogaster. Mol. Gen. Genet. 113: 251–272. [DOI] [PubMed] [Google Scholar]
- Davis D. G., 1969. Chromosome behavior under the influence of claret-nondisjunctional in Drosophila melanogaster. Genetics 61: 577–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., et al. , 2003. The Mus81/Mms4 endonuclease acts independently of double-holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., Sedat J. W., Hawley R. S., 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 135–146. [DOI] [PubMed] [Google Scholar]
- Do K. K., Hoang K. L., Endow S. A., 2014. The kinesin-13 KLP10A motor regulates oocyte spindle length and affects EB1 binding without altering microtubule growth rates. Biol. Open 3: 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doane W. W., 1960. Completion of meiosis in uninseminated eggs of Drosophila melanogaster. Science 132: 677–678. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T., 1930. Translocations involving the third and the fourth chromosomes of Drosophila melanogaster. Genetics 15: 347–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1931. Translocations involving the second and the fourth chromosomes of Drosophila melanogaster. Genetics 16: 629–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont B. L., 2017. Variation and evolution of the meiotic requirement for crossing over in mammals. Genetics 205: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S. A., Komma D. J., 1998. Assembly and dynamics of an anastral:astral spindle: the meiosis II spindle of Drosophila oocytes. J. Cell Sci. 111: 2487–2495. [DOI] [PubMed] [Google Scholar]
- Endow S. A., Henikoff S., Soler-Niedziela L., 1990. Mediation of meiotic and early mitotic chromosome segregation in Drosophila by a protein related to kinesin. Nature 345: 81–83. [DOI] [PubMed] [Google Scholar]
- Fedorova S. A., Nokkala S., Omel’ianchuk L. V., 2001. Genetic screening of meiotic mutations in mosaic clones of the Drosophila melanogaster female germline. Genetika 37: 1621–1631. [PubMed] [Google Scholar]
- Ganetzky B., Hawley R. S., 2016. The centenary of GENETICS: bridges to the future. Genetics 202: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A., Ray R. P., Schupbach T., 1998. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 12: 2711–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giauque C. C., Bickel S. E., 2016. Heterochromatin-associated proteins HP1a and piwi collaborate to maintain the association of achiasmate homologs in Drosophila oocytes. Genetics 203: 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland W. D., Wayson S. M., Hawley R. S., 2005. The meiotic defects of mutants in the Drosophila mps1 gene reveal a critical role of Mps1 in the segregation of achiasmate homologs. Curr. Biol. 15: 672–677. [DOI] [PubMed] [Google Scholar]
- Gilliland W. D., Hughes S. E., Cotitta J. L., Takeo S., Xiang Y., et al. , 2007. The multiple roles of Mps1 in Drosophila female meiosis. PLoS Genet. 3: e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland W. D., Hughes S. E., Vietti D. R., Hawley R. S., 2009. Congression of achiasmate chromosomes to the metaphase plate in Drosophila melanogaster oocytes. Dev. Biol. 325: 122–128. [DOI] [PubMed] [Google Scholar]
- Gilliland W. D., Colwell E. M., Lane F. M., Snouffer A. A., 2014. Behavior of aberrant chromosome configurations in Drosophila melanogaster female meiosis I. G3 (Bethesda) 5: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta K. L., Jang J. K., Manheim E. A., Subramanian G., McKim K. S., 2002. subito encodes a kinesin-like protein required for meiotic spindle pole formation in Drosophila melanogaster. Genetics 160: 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluszek A. A., Cullen C. F., Li W., Battaglia R. A., Radford S. J., et al. , 2015. The microtubule catastrophe promoter sentin delays stable kinetochore-microtubule attachment in oocytes. J. Cell Biol. 211: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W. J., McKim K. S., Hawley R. S., 2005. All paired up with no place to go: pairing, synapsis, and DSB formation in a balancer heterozygote. PLoS Genet. 1: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen J. W., 1933. Meiosis as a genetic character in Drosophila melanogaster. J. Exp. Zool. 65: 83–106. [Google Scholar]
- Gowen M. S., Gowen J. W., 1922. Complete linkage in Drosophila melanogaster. Am. Nat. 61: 286–288. [Google Scholar]
- Green M. M., 1981. mus(3)312D1, a mutagen sensitive mutant with profound effects on female meiosis in Drosophila melanogaster. Chromosoma 82: 259–266. [DOI] [PubMed] [Google Scholar]
- Grell E. H., 1963. Distributive pairing of compound chromosomes in females of Drosophila melanogaster. Genetics 48: 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell E. H., 1964. Influence of the location of a chromosome duplication on crossing over in Drosophila melanogaster. Genetics 50: 251–252. [Google Scholar]
- Grell E. H., 1970. Distributive pairing: mechanism for segregation of compound autosomal chromosomes in oocytes of Drosophila melanogaster. Genetics 65: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell R. F., 1964. Chromosome size at distributive pairing in Drosophila melanogaster females. Genetics 50: 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell R. F., 1976. Distributive pairing, pp. 435–486 in The Genetics and Biology of Drosophila. Vol. 1a, edited by Ashburner M., Novitski E. Academic Press, New York. [Google Scholar]
- Grell R. F., 1984. Time of recombination in the Drosophila melanogaster oocyte. III. Selection and characterization of temperature-sensitive and -insensitive, recombination-deficient alleles in Drosophila. Genetics 108: 425–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell R. F., Generoso E. E., 1980. Time of recombination in the Drosophila melanogaster oocyte. II. Electron microscopic and genetic studies of a temperature-sensitive recombination mutant. Chromosoma 81: 339–348. [DOI] [PubMed] [Google Scholar]
- Gyuricza M. R., Manheimer K. B., Apte V., Krishnan B., Joyce E. F., et al. , 2016. Dynamic and stable cohesins regulate synaptonemal complex assembly and chromosome segregation. Curr. Biol. 26: 1688–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., 2013. Genome Stability: DNA Repair and Recombination. Garland Science, New York. [Google Scholar]
- Hall J. C., 1972. Chromosome segregation influenced by two alleles of the meiotic mutant c(3)G in Drosophila melanogaster. Genetics 71: 367–400. [DOI] [PubMed] [Google Scholar]
- Harris D., Orme C., Kramer J., Namba L., Champion M., et al. , 2003. A deficiency screen of the major autosomes identifies a gene (matrimony) that is haplo-insufficient for achiasmate segregation in Drosophila oocytes. Genetics 165: 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M. A., Sekelsky J., 2017. The absence of crossovers on chromosome 4 in Drosophila melanogaster: imperfection or interesting exception? Fly (Austin) DOI: 10.1080/19336934.2017.132118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell H. C., Yu K., Xiao Q., Chien L. T., Qu Z., 2009. Anoctamin/TMEM16 family members are Ca2+-activated Cl– channels. J. Physiol. 587: 2127–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkevich T., Kohl K. P., McMahan S., Hartmann M. A., Williams A. M., et al. , 2017. Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr. Biol. 27: 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsumi M., Endow S. A., 1992a The Drosophila ncd microtubule motor protein is spindle-associated in meiotic and mitotic cells. J. Cell Sci. 103: 1013–1020. [DOI] [PubMed] [Google Scholar]
- Hatsumi M., Endow S. A., 1992b Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J. Cell Sci. 101: 547–559. [DOI] [PubMed] [Google Scholar]
- Hawley R. S., 1980. Chromosomal sites necessary for normal levels of meiotic recombination in Drosophila melanogaster. I. Evidence for and mapping of the sites. Genetics 94: 625–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R. S., 1988. Exchange and chromosomal segregation in eukaryotes, pp. 497–528 in Genetic Recombination, edited by Kucherlapati R., Smith G. R. American Society for Microbiology, Washington, D. C. [Google Scholar]
- Hawley R. S., Ganetzky B., 2016. Alfred Sturtevant and George Beadle untangle inversions. Genetics 203: 1001–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R. S., Walker M., 2003. Advanced Genetic Analysis. Blackwell Science Ltd, Malden, MA. [Google Scholar]
- Hawley R. S., Irick H., Zitron A. E., Haddox D. A., Lohe A., et al. , 1992. There are two mechanisms of achiasmate segregation in Drosophila females, one of which requires heterochromatic homology. Dev. Genet. 13: 440–467. [DOI] [PubMed] [Google Scholar]
- Hawley R. S., McKim K. S., Arbel T., 1993. Meiotic segregation in Drosophila melanogaster females: molecules, mechanisms, and myths. Annu. Rev. Genet. 27: 281–317. [DOI] [PubMed] [Google Scholar]
- Heidmann D., Horn S., Heidmann S., Schleiffer A., Nasmyth K., et al. , 2004. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma 113: 177–187. [DOI] [PubMed] [Google Scholar]
- Heifetz Y., Yu J., Wolfner M. F., 2001. Ovulation triggers activation of Drosophila oocytes. Dev. Biol. 234: 416–424. [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Chovnick A., 1981. Further observations on intragenic recombination in Drosophila melanogaster. Genet. Res. 38: 281–296. [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Clark S. H., Chovnick A., 1991. The effect of DNA sequence polymorphisms on intragenic recombination in the rosy locus of Drosophila melanogaster. Genetics 129: 779–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A. J., Harauz G., Reaume A. G., Gray M., Clark S. H., et al. , 1994. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics 137: 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Brill S. J., 2004. The Mus81 solution to resolution: generating meiotic crossovers without holliday junctions. Genes Dev. 18: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner V. L., Wolfner M. F., 2008. Mechanical stimulation by osmotic and hydrostatic pressure activates Drosophila oocytes in vitro in a calcium-dependent manner. Dev. Biol. 316: 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner V. L., Czank A., Jang J. K., Singh N., Williams B. C., et al. , 2006. The Drosophila calcipressin Sarah is required for several aspects of egg activation. Curr. Biol. 16: 1441–1446. [DOI] [PubMed] [Google Scholar]
- Hughes S. E., Hawley R. S., 2014. Topoisomerase II is required for the proper separation of heterochromatic regions during Drosophila melanogaster female meiosis. PLoS Genet. 10: e1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. E., Gilliland W. D., Cotitta J. L., Takeo S., Collins K. A., et al. , 2009. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genet. 5: e1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. E., Beeler J. S., Seat A., Slaughter B. D., Unruh J. R., et al. , 2011. Gamma-tubulin is required for bipolar spindle assembly and for proper kinetochore microtubule attachments during prometaphase I in Drosophila oocytes. PLoS Genet. 7: e1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I., Khandan T., Ito T., Orr-Weaver T. L., 2005. A histone code in meiosis: the histone kinase, NHK-1, is required for proper chromosomal architecture in Drosophila oocytes. Genes Dev. 19: 2571–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. K., Messina L., Erdman M. B., Arbel T., Hawley R. S., 1995. Induction of metaphase arrest in Drosophila oocytes by chiasma-based kinetochore tension. Science 268: 1917–1919. [DOI] [PubMed] [Google Scholar]
- Jang J. K., Rahman T., McKim K. S., 2005. The kinesinlike protein subito contributes to central spindle assembly and organization of the meiotic spindle in Drosophila oocytes. Mol. Biol. Cell 16: 4684–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Tanneti S. N., McKim K. S., 2009. Drosophila Hold’em is required for a subset of meiotic crossovers and interacts with the DNA repair endonuclease complex subunits MEI-9 and ERCC1. Genetics 181: 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Paul A., Chen K. E., Tanneti N., McKim K. S., 2012. Multiple barriers to nonhomologous DNA end joining during meiosis in Drosophila. Genetics 191: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Apostolopoulos N., Beliveau B. J., Wu C. T., 2013. Germline progenitors escape the widespread phenomenon of homolog pairing during Drosophila development. PLoS Genet. 9: e1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneuchi T., Sartain C. V., Takeo S., Horner V. L., Buehner N. A., et al. , 2015. Calcium waves occur as Drosophila oocytes activate. Proc. Natl. Acad. Sci. USA 112: 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G. H., Le M. H., Le H., 1996. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science 273: 118–122. [DOI] [PubMed] [Google Scholar]
- Kaufman T. C., 2017. A short history and description of Drosophila melanogaster classical genetics: chromosome aberrations, forward genetic screens, and the nature of mutations. Genetics 206: 665–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock A. W., Miyazaki W. Y., Birnby D., Orr-Weaver T. L., 1992. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics 130: 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock A. W., Moore D. P., Wu J. S., Orr-Weaver T. L., 1995. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 83: 247–256. [DOI] [PubMed] [Google Scholar]
- Khetani R. S., Bickel S. E., 2007. Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J. Cell Sci. 120: 3123–3137. [DOI] [PubMed] [Google Scholar]
- Kimble M., Church K., 1983. Meiosis and early cleavage in Drosophila melanogaster eggs: effects of the claret-non-disjunctional mutation. J. Cell Sci. 62: 301–318. [DOI] [PubMed] [Google Scholar]
- King R. C., 1970. Ovarian Development in Drosophila melanogaster. Academic Press, New York. [Google Scholar]
- Kirilly D., Xie T., 2007. The Drosophila ovary: an active stem cell community. Cell Res. 17: 15–25. [DOI] [PubMed] [Google Scholar]
- Koehler K. E., Boulton C. L., Collins H. E., French R. L., Herman K. C., et al. , 1996. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat. Genet. 14: 406–414. [DOI] [PubMed] [Google Scholar]
- Kohl K. P., Sekelsky J., 2013. Meiotic and mitotic recombination in meiosis. Genetics 194: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Jones C. D., Sekelsky J., 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S., Wojcik M., Xu K., Dernburg A. F., 2017. Superresolution microscopy reveals the three-dimensional organization of meiotic chromosome axes in intact Caenorhabditis elegans tissue. Proc. Natl. Acad. Sci. USA 114: E4734–E4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J., Hawley R. S., 2003a The spindle-associated transmembrane protein Axs identifies a membranous structure ensheathing the meiotic spindle. Nat. Cell Biol. 5: 261–263. [DOI] [PubMed] [Google Scholar]
- Kramer J., Hawley R. S., 2003b The spindle-associated transmembrane protein Axs identifies a new family of transmembrane proteins in eukaryotes. Cell Cycle 2: 174–176. [PubMed] [Google Scholar]
- Krishnan B., Thomas S. E., Yan R., Yamada H., Zhulin I. B., et al. , 2014. Sisters unbound is required for meiotic centromeric cohesion in Drosophila melanogaster. Genetics 198: 947–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdzo E. L., Dawson D. S., 2015. Centromere pairing–tethering partner chromosomes in meiosis I. FEBS J. 282: 2458–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T., 2015. Brca2-Pds5 complexes mobilize persistent meiotic recombination sites to the nuclear envelope. J. Cell Sci. 128: 717–727. [DOI] [PubMed] [Google Scholar]
- Lake C. M., Hawley R. S., 2012. The molecular control of meiotic chromosomal behavior: events in early meiotic prophase in Drosophila oocytes. Annu. Rev. Physiol. 74: 425–451. [DOI] [PubMed] [Google Scholar]
- Lake C. M., Hawley R. S., 2016. Becoming a crossover-competent DSB. Semin. Cell Dev. Biol. 54: 117–125. [DOI] [PubMed] [Google Scholar]
- Lake C. M., Teeter K., Page S. L., Nielsen R., Hawley R. S., 2007. A genetic analysis of the Drosophila mcm5 gene defines a domain specifically required for meiotic recombination. Genetics 176: 2151–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Nielsen R. J., Hawley R. S., 2011. The Drosophila zinc finger protein trade embargo is required for double strand break formation in meiosis. PLoS Genet. 7: e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Holsclaw J. K., Bellendir S. P., Sekelsky J., Hawley R. S., 2013. The development of a monoclonal antibody recognizing the Drosophila melanogaster phosphorylated histone H2A variant (gamma-H2AV). G3 (Bethesda) 3: 1539–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Nielsen R. J., Guo F., Unruh J. R., Slaughter B. D., et al. , 2015. Vilya, a component of the recombination nodule, is required for meiotic double-strand break formation in Drosophila. Elife 4: e08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb N. E., Freeman S. B., Savage-Austin A., Pettay D., Taft L., et al. , 1996. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat. Genet. 14: 400–405. [DOI] [PubMed] [Google Scholar]
- Lancaster O. M., Cullen C. F., Ohkura H., 2007. NHK-1 phosphorylates BAF to allow karyosome formation in the Drosophila oocyte nucleus. J. Cell Biol. 179: 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., Sandler L., 1977. The genetic analysis of meiosis in female Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci. 277: 295–312. [DOI] [PubMed] [Google Scholar]
- Liu H., Jang J. K., Graham J., Nycz K., McKim K. S., 2000. Two genes required for meiotic recombination in Drosophila are expressed from a dicistronic message. Genetics 154: 1735–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jang J. K., Kato N., McKim K. S., 2002. Mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamazares S., Tavosanis G., Gonzalez C., 1999. Cytological characterisation of the mutant phenotypes produced during early embryogenesis by null and loss-of-function alleles of the γTub37C gene in Drosophila. J. Cell Sci. 112: 659–667. [DOI] [PubMed] [Google Scholar]
- Loh B. J., Cullen C. F., Vogt N., Ohkura H., 2012. The conserved kinase SRPK regulates karyosome formation and spindle microtubule assembly in Drosophila oocytes. J. Cell Sci. 125: 4457–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano T., 2005. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 19: 1269–1287. [DOI] [PubMed] [Google Scholar]
- Lu W. J., Chapo J., Roig I., Abrams J. M., 2010. Meiotic recombination provokes functional activation of the p53 regulatory network. Science 328: 1278–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J. C., Suzuki D. T., 1968. The interchromosomal control of recombination. Annu. Rev. Genet. 2: 53–86. [Google Scholar]
- Mahowald A. P., Tiefert M., 1970. Fine structural changes in the Drosophila oocyte nucleus during a short period of RNA synthesis: an autoradiographic and ultrastructural study of RNA synthesis in the oocyte nucleus of Drosophila. Wilhelm Roux’. Arch. Entwickl. Mech. Org. 165: 8–25. [DOI] [PubMed] [Google Scholar]
- Mahowald A. P., Goralski T. J., Caulton J. H., 1983. In vitro activation of Drosophila eggs. Dev. Biol. 98: 437–445. [DOI] [PubMed] [Google Scholar]
- Manheim E. A., McKim K. S., 2003. The synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr. Biol. 13: 276–285. [DOI] [PubMed] [Google Scholar]
- Manzano-Winkler B., McGaugh S. E., Noor M. A., 2013. How hot are Drosophila hotspots? Examining recombination rate variation and associations with nucleotide diversity, divergence, and maternal age in Drosophila pseudoobscura. PLoS One 8: e71582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E., Diaz R. L., Hunter N., Keeney S., 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., 1976. Orientation disruptor (ord): a recombination-defective and disjunction-defective meiotic mutant in Drosophila melanogaster. Genetics 84: 545–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H. J., McDonald H. B., Goldstein L. S., Theurkauf W. E., 1996. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 134: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H. J., Messina L. G., Namba R., Greer K. J., Walker M. Y., et al. , 1999. Mutations in the alpha-tubulin 67C gene specifically impair achiasmate segregation in Drosophila melanogaster. J. Cell Biol. 147: 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald H. B., Stewart R. J., Goldstein L. S., 1990. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell 63: 1159–1165. [DOI] [PubMed] [Google Scholar]
- McKim K. S., Hayashi-Hagihara A., 1998. Mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12: 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Dahmus J. B., Hawley R. S., 1996. Cloning of the Drosophila melanogaster meiotic recombination gene mei-218: a genetic and molecular analysis of interval 15E. Genetics 144: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Green-Marroquin B. L., Sekelsky J. J., Chin G., Steinberg C., et al. , 1998. Meiotic synapsis in the absence of recombination. Science 279: 876–878. [DOI] [PubMed] [Google Scholar]
- McKim K. S., Jang J. K., Manheim E. A., 2002. Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36: 205–232. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. M., Bratu D. P., 2015. Drosophila melanogaster oogenesis: an overview. Methods Mol. Biol. 1328: 1–20. [DOI] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S., McKim K. S., 2006. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles A. M., Fisher K. H., Colombie N., Wakefield J. G., Ohkura H., 2009. Wac: a new augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J. Cell Biol. 184: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriam J. R., Frost J. N., 1964. Exchange and nondisjunction of the X chromosomes in female Drosophila melanogaster. Genetics 49: 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Takeo S., Nandanan K., Paulson A., Gogol M. M., et al. , 2012. A whole-chromosome analysis of meiotic recombination in Drosophila melanogaster. G3 (Bethesda) 2: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Cook K. R., Arvanitakis A. V., Hawley R. S., 2016a Third chromosome balancer inversions disrupt protein-coding genes and influence distal recombination events in Drosophila melanogaster. G3 (Bethesda) 6: 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Cook K. R., Yeganeh Kazemi N., Smith C. B., Cockrell A. J., et al. , 2016b Rare recombination events generate sequence diversity among balancer chromosomes in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 113: E1352–E1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Smith C. B., Kazemi N. Y., Cockrell A. J., Arvanitakas A. V., et al. , 2016c Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect. Genetics 203: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Agard D. A., 2001. Gamma-tubulin complexes and microtubule nucleation. Curr. Opin. Struct. Biol. 11: 174–181. [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1916. The mechanism of crossing over. Am. Nat. 50: 193–221. [Google Scholar]
- Nicklas R. B., 1974. Chromosome segregation mechanisms. Genetics 78: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R. B., 1977. Chromosome distribution: experiments on cell hybrids and in vitro. Philos. Trans. R. Soc. Lond. B Biol. Sci. 277: 267–276. [DOI] [PubMed] [Google Scholar]
- Novitski E., Braver G., 1954. An analysis of crossing over within a heterozygous inversion in Drosophila melanogaster. Genetics 39: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermann, C. A., and H. J. Muller, 1932 Regional differences in crossing over as a function of the chromosome structure. Proc. Sixth Int. Congress Genet. 2: 143–145. [Google Scholar]
- O’Tousa J., 1982. Meiotic chromosome behavior influenced by mutation-altered disjunction in Drosophila melanogaster females. Genetics 102: 503–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. W., Orr-Weaver T. L., 1996. The Drosophila genes grauzone and cortex are necessary for proper female meiosis. J. Cell Sci. 109: 1707–1715. [DOI] [PubMed] [Google Scholar]
- Page A. W., Orr-Weaver T. L., 1997. Activation of the meiotic divisions in Drosophila oocytes. Dev. Biol. 183: 195–207. [DOI] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Nielsen R. J., Teeter K., Lake C. M., Ong S., et al. , 2007. A germline clone screen for meiotic mutants in Drosophila melanogaster. Fly (Austin) 1: 172–181. [DOI] [PubMed] [Google Scholar]
- Page S. L., Khetani R. S., Lake C. M., Nielsen R. J., Jeffress J. K., et al. , 2008. Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes. PLoS Genet. 4: e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D. M., 1973. A meiotic mutant affecting recombination in female Drosophila melanogaster. Genetics 73: 465–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson N. J., Cullen C. F., Dzhindzhev N. S., Ohkura H., 2005. A pre-anaphase role for a Cks/Suc1 in acentrosomal spindle formation of Drosophila female meiosis. EMBO Rep. 6: 1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesin J. A., Orr-Weaver T. L., 2007. Developmental role and regulation of cortex, a meiosis-specific anaphase-promoting complex/cyclosome activator. PLoS Genet. 3: e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. J., McMahan S., Blanton H. L., Sekelsky J., 2007. Heteroduplex DNA in meiotic recombination in Drosophila mei-9 mutants. Genetics 176: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. J., Harrison A. M., McKim K. S., 2012a Microtubule-depolymerizing kinesin KLP10A restricts the length of the acentrosomal meiotic spindle in Drosophila females. Genetics 192: 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. J., Jang J. K., McKim K. S., 2012b The chromosomal passenger complex is required for meiotic acentrosomal spindle assembly and chromosome biorientation. Genetics 192: 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. J., Hoang T. L., Gluszek A. A., Ohkura H., McKim K. S., 2015. Lateral and end-on kinetochore attachments are coordinated to achieve bi-orientation in Drosophila oocytes. PLoS Genet. 11: e1005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. J., Go A. M., McKim K. S., 2017. Cooperation between kinesin motors promotes spindle symmetry and chromosome organization in oocytes. Genetics 205: 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel C., 1962. Interchromosomal effects of inversions in Drosophila melanogaster. Hereditas 48: 1–58. [DOI] [PubMed] [Google Scholar]
- Ramel C., Valentin J., 1966. Interchromosomal effects of the Curly inversions on crossing over in Drosophila melanogaster. Hereditas 54: 307–313. [DOI] [PubMed] [Google Scholar]
- Rasooly R. S., Zhang P., Tibolla A. K., Hawley R. S., 1994. A structure-function analysis of NOD, a kinesin-like protein from Drosophila melanogaster. Mol. Gen. Genet. 242: 145–151. [DOI] [PubMed] [Google Scholar]
- Reschen R. F., Colombie N., Wheatley L., Dobbelaere J., St Johnston D., et al. , 2012. Dgp71WD is required for the assembly of the acentrosomal meiosis I spindle, and is not a general targeting factor for the gamma-TuRC. Biol. Open 1: 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick T. D., Dej K. J., Xiang Y., Hawley R. S., Ahn C., et al. , 2009. Mutations in the chromosomal passenger complex and the condensin complex differentially affect synaptonemal complex disassembly and metaphase I configuration in Drosophila female meiosis. Genetics 181: 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli M. G., Callaini G., 1996. Meiotic spindle organization in fertilized Drosophila oocyte: presence of centrosomal components in the meiotic apparatus. J. Cell Sci. 109: 911–918. [DOI] [PubMed] [Google Scholar]
- Riparbelli M. G., Callaini G., 2005. The meiotic spindle of the Drosophila oocyte: the role of Centrosomin and the central aster. J. Cell Sci. 118: 2827–2836. [DOI] [PubMed] [Google Scholar]
- Riparbelli M. G., Callaini G., Glover D. M., 2000. Failure of pronuclear migration and repeated divisions of polar body nuclei associated with MTOC defects in polo eggs of Drosophila. J. Cell Sci. 113: 3341–3350. [DOI] [PubMed] [Google Scholar]
- Riparbelli M. G., Callaini G., Glover D. M., Avides Mdo C., 2002. A requirement for the abnormal spindle protein to organise microtubules of the central spindle for cytokinesis in Drosophila. J. Cell Sci. 115: 913–922. [DOI] [PubMed] [Google Scholar]
- Robert T., Nore A., Brun C., Maffre C., Crimi B., et al. , 2016. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351: 943–949. [DOI] [PubMed] [Google Scholar]
- Roberts P. A., 1972. Differences in synaptic affinity of chromosome arms of Drosophila melanogaster revealed by differential sensitivity to translocation heterozygosity. Genetics 71: 401–415. [DOI] [PubMed] [Google Scholar]
- Roberts P. A., 1976. The genetics of chromosome aberration, pp. 68–184 in The Genetics and Biology of Drosophila. Vol. 1a, edited by Ashburner M., Novitski E. Academic Press, London. [Google Scholar]
- Ross L. O., Maxfield R., Dawson D., 1996. Exchanges are not equally able to enhance meiotic chromosome segregation in yeast. Proc. Natl. Acad. Sci. USA 93: 4979–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin T., Christophorou N., Huynh J. R., 2016. How to pre-pair chromosomes for meiosis. Cell Cycle 15: 609–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L., Lindsley D. L., Nicoletti B., Trippa G., 1968. Mutants affecting meiosis in natural populations of Drosophila melanogaster. Genetics 60: 525–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders R. D., Avides M. C., Howard T., Gonzalez C., Glover D. M., 1997. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 137: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmekel K., Daneholt B., 1995. The central region of the synaptonemal complex revealed in three dimensions. Trends Cell Biol. 5: 239–242. [DOI] [PubMed] [Google Scholar]
- Schmekel K., Skoglund U., Daneholt B., 1993. The three-dimensional structure of the central region in a synaptonemal complex: a comparison between rat and two insect species, Drosophila melanogaster and Blaps cribrosa. Chromosoma 102: 682–692. [DOI] [PubMed] [Google Scholar]
- Schultz J., Redfield H., 1951. Interchromosomal effects on crossing over in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 16: 175–197. [DOI] [PubMed] [Google Scholar]
- Sekelsky J., 2017. DNA repair in Drosophila: mutagens, models, and missing genes. Genetics 205: 471–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., McKim K. S., Chin G. M., Hawley R. S., 1995. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., Burtis K. C., Hawley R. S., 1998. Damage control: the pleiotropy of DNA repair genes in Drosophila melanogaster. Genetics 148: 1587–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., McKim K. S., Messina L., French R. L., Hurley W. D., et al. , 1999. Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics 152: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira W., Nelson C. R., Szauter P., 1989. Genetic analysis of the claret locus of Drosophila melanogaster. Genetics 123: 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherizen D., Jang J. K., Bhagat R., Kato N., McKim K. S., 2005. Meiotic recombination in Drosophila females depends on chromosome continuity between genetically defined boundaries. Genetics 169: 767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. D., Stone E. A., Aquadro C. F., Clark A. G., 2013. Fine-scale heterogeneity in crossover rate in the garnet-scalloped region of the Drosophila melanogaster X chromosome. Genetics 194: 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skold H. N., Komma D. J., Endow S. A., 2005. Assembly pathway of the anastral Drosophila oocyte meiosis I spindle. J. Cell Sci. 118: 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. D., Finnerty V. G., Chovnick A., 1970. Gene conversion in Drosophila: non-reciprocal events at the maroon-like cistron. Nature 228: 442–444. [DOI] [PubMed] [Google Scholar]
- Smukowski Heil C. S., Ellison C., Dubin M., Noor M. A., 2015. Recombining without hotspots: a comprehensive evolutionary portrait of recombination in two closely related species of Drosophila. Genome Biol. Evol. 7: 2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Guimaraes S., Sunkel C., Malmanche N., 2011. Polo is identified as a suppressor of bubR1 nondisjunction in a deficiency screen of the third chromosome in Drosophila melanogaster. G3 (Bethesda) 1: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., 1993 Developmental genetics of oogenesis, pp. 1–70 in The Development of Drosophila melanogaster, edited by M. Bate, and A. M. Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sturtevant A. H., 1913. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zool. 14: 43–59. [Google Scholar]
- Sturtevant A. H., 1915. The Behavior of the Chromosomes as Studied Through Linkage. Gerb. Bortraeger, Berlin. [Google Scholar]
- Sturtevant A. H., Beadle G. W., 1936. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics 21: 554–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan A., Schupbach T., 2005. Drosophila female meiosis and embryonic syncytial mitosis use specialized Cks and CDC20 proteins for cyclin destruction. Cell Cycle 4: 1332–1334. [DOI] [PubMed] [Google Scholar]
- Swan A., Schupbach T., 2007. The Cdc20 (Fzy)/Cdh1-related protein, Cort, cooperates with Fzy in cyclin destruction and anaphase progression in meiosis I and II in Drosophila. Development 134: 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szauter P., 1984. An analysis of regional constraints on exchange in Drosophila melanogaster using recombination-defective meiotic mutants. Genetics 106: 45–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo S., Tsuda M., Akahori S., Matsuo T., Aigaki T., 2006. The calcineurin regulator Sra plays an essential role in female meiosis in Drosophila. Curr. Biol. 16: 1435–1440. [DOI] [PubMed] [Google Scholar]
- Takeo S., Hawley R. S., Aigaki T., 2010. Calcineurin and its regulation by Sra/RCAN is required for completion of meiosis in Drosophila. Dev. Biol. 344: 957–967. [DOI] [PubMed] [Google Scholar]
- Takeo S., Lake C. M., Morais-de-Sa E., Sunkel C. E., Hawley R. S., 2011. Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr. Biol. 21: 1845–1851. [DOI] [PubMed] [Google Scholar]
- Takeo S., Swanson S. K., Nandanan K., Nakai Y., Aigaki T., et al. , 2012. Shaggy/glycogen synthase kinase 3β and phosphorylation of Sarah/regulator of calcineurin are essential for completion of Drosophila female meiosis. Proc. Natl. Acad. Sci. USA 109: 6382–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanneti N. S., Landy K., Joyce E. F., McKim K. S., 2011. A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr. Biol. 21: 1852–1857. [DOI] [PubMed] [Google Scholar]
- Tavosanis G., Llamazares S., Goulielmos G., Gonzalez C., 1997. Essential role for γ-tubulin in the acentriolar female meiotic spindle of Drosophila. EMBO J. 16: 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf W. E., Hawley R. S., 1992. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J. Cell Biol. 116: 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y., Orr-Weaver T. L., 2013. Centromere proteins CENP-C and CAL1 functionally interact in meiosis for centromere clustering, pairing, and chromosome segregation. Proc. Natl. Acad. Sci. USA 110: 19878–19883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban E., Nagarkar-Jaiswal S., Lehner C. F., Heidmann S. K., 2014. The cohesin subunit Rad21 is required for synaptonemal complex maintenance, but not sister chromatid cohesion, during Drosophila female meiosis. PLoS Genet. 10: e1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N., Koch I., Schwarz H., Schnorrer F., Nusslein-Volhard C., 2006. The gammaTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133: 3963–3972. [DOI] [PubMed] [Google Scholar]
- Von Stetina J. R., Tranguch S., Dey S. K., Lee L. A., Cha B., et al. , 2008. Alpha-endosulfine is a conserved protein required for oocyte meiotic maturation in Drosophila. Development 135: 3697–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina J. R., Lafever K. S., Rubin M., Drummond-Barbosa D., 2011. A genetic screen for dominant enhancers of the cell-cycle regulator alpha-endosulfine identifies matrimony as a strong functional interactor in Drosophila. G3 (Bethesda) 1: 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D., Rasmussen S. W., Holm P. B., 1984. The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 18: 331–413. [DOI] [PubMed] [Google Scholar]
- Webber H. A., Howard L., Bickel S. E., 2004. The cohesion protein ORD is required for homologue bias during meiotic recombination. J. Cell Biol. 164: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein A., 1918. Coincidence of crossing over in Drosophila melanogaster (Ampelophila). Genetics 3: 135–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H., Alphey L., Glover D. M., 1993. The cdc25 homologue twine is required for only some aspects of the entry into meiosis in Drosophila. J. Cell Sci. 106: 1035–1044. [DOI] [PubMed] [Google Scholar]
- Whitfield Z. J., Chisholm J., Hawley R. S., Orr-Weaver T. L., 2013. A meiosis-specific form of the APC/C promotes the oocyte-to-embryo transition by decreasing levels of the Polo kinase inhibitor matrimony. PLoS Biol. 11: e1001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Singaram V., McKim K. S., 2008. Mei-38 is required for chromosome segregation during meiosis in Drosophila females. Genetics 180: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Hawley R. S., 2006. The mechanism of secondary nondisjunction in Drosophila melanogaster females. Genetics 174: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Takeo S., Florens L., Hughes S. E., Huo L. J., et al. , 2007. The inhibition of Polo kinase by matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol. 5: e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Miklos G. L., 1977. Genetic dissection of heterochromatin in Drosophila: the role of basal X heterochromatin in meiotic sex chromosome behaviour. Chromosoma 60: 283–296. [DOI] [PubMed] [Google Scholar]
- Yan R., McKee B. D., 2013. The cohesion protein SOLO associates with SMC1 and is required for synapsis, recombination, homolog bias and cohesion and pairing of centromeres in Drosophila meiosis. PLoS Genet. 9: e1003637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O., Majumder S., Kramer B., Sekelsky J. J., 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell 10: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O., Kearney H., Kramer B. C., Sekelsky J. J., 2004. Mutational analysis of the Drosophila DNA repair and recombination gene mei-9. Genetics 167: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. X., Guan Z., Nash H. A., 2006. The mushroom body defect gene product is an essential component of the meiosis II spindle apparatus in Drosophila oocytes. Genetics 173: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., MacQueen A. J., Duffy J. B., Kemphues K. J., Villeneuve A. M., 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Li H., Schweppe N. M., Hawley R. S., Gilliland W. D., 2010. Statistical analysis of nondisjunction assays in Drosophila. Genetics 186: 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Hawley R. S., 1990. The genetic analysis of distributive segregation in Drosophila melanogaster. II. Further genetic analysis of the nod locus. Genetics 125: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Knowles B. A., Goldstein L. S., Hawley R. S., 1990. A kinesin-like protein required for distributive chromosome segregation in Drosophila. Cell 62: 1053–1062. [DOI] [PubMed] [Google Scholar]
- Zhaunova L., Ohkura H., Breuer M., 2016. Kdm5/Lid regulates chromosome architecture in meiotic prophase I independently of its histone demethylase activity. PLoS Genet. 12: e1006241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wong M. L., Alberts B., Mitchison T., 1995. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378: 578–583. [DOI] [PubMed] [Google Scholar]
- Zitron A. E., Hawley R. S., 1989. The genetic analysis of distributive segregation in Drosophila melanogaster. I. Isolation and characterization of Aberrant X segregation (Axs), a mutation defective in chromosome partner choice. Genetics 122: 801–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Hallen M. A., Yankel C. D., Endow S. A., 2008. A microtubule-destabilizing kinesin motor regulates spindle length and anchoring in oocytes. J. Cell Biol. 180: 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick M. E., Cutler D. J., Langley C. H., 1999. Classic Weinstein: tetrad analysis, genetic variation and achiasmate segregation in Drosophila and humans. Genetics 152: 1615–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]