The model organism Encylopedia of Regulatory Elements (modERN) project was designed to generate genome-wide binding profiles for the majority of transcription...
Keywords: Drosophila, Caenorhabditis elegans, transcription factors, binding sites, regulation
Abstract
To develop a catalog of regulatory sites in two major model organisms, Drosophila melanogaster and Caenorhabditis elegans, the modERN (model organism Encyclopedia of Regulatory Networks) consortium has systematically assayed the binding sites of transcription factors (TFs). Combined with data produced by our predecessor, modENCODE (Model Organism ENCyclopedia Of DNA Elements), we now have data for 262 TFs identifying 1.23 M sites in the fly genome and 217 TFs identifying 0.67 M sites in the worm genome. Because sites from different TFs are often overlapping and tightly clustered, they fall into 91,011 and 59,150 regions in the fly and worm, respectively, and these binding sites span as little as 8.7 and 5.8 Mb in the two organisms. Clusters with large numbers of sites (so-called high occupancy target, or HOT regions) predominantly associate with broadly expressed genes, whereas clusters containing sites from just a few factors are associated with genes expressed in tissue-specific patterns. All of the strains expressing GFP-tagged TFs are available at the stock centers, and the chromatin immunoprecipitation sequencing data are available through the ENCODE Data Coordinating Center and also through a simple interface (http://epic.gs.washington.edu/modERN/) that facilitates rapid accessibility of processed data sets. These data will facilitate a vast number of scientific inquiries into the function of individual TFs in key developmental, metabolic, and defense and homeostatic regulatory pathways, as well as provide a broader perspective on how individual TFs work together in local networks and globally across the life spans of these two key model organisms.
TRANSCRIPTION factors (TFs) play key roles in development and physiology, including sex determination, early pattern formation, organogenesis, and the response to environmental cues. A catalog of genomic sites where TFs bind (regulatory sequences) is perhaps only second in importance to a catalog of genes in understanding how a genome encodes an organism.
The Caenorhabditis elegans and Drosophila melanogaster model organisms have several advantages for global mapping of TF–DNA interactions. Both organisms have extensive comparative genomics resources, both have powerful tools to investigate gene expression, and both are easy to manipulate in the laboratory. Their genomes are among the most thoroughly and meticulously annotated metazoan genomes [a result in part of the transcript identification and annotation efforts of the modENCODE (Model Organism ENCyclopedia Of DNA Elements) project] (Brown and Celniker 2015), thereby providing a stable platform upon which to investigate TF action. At only ∼1/30th the size of the human genome, the 100 Mb worm genome (C. elegans Sequencing Consortium 1998; Hillier et al. 2005) and 143 Mb fly genome (Adams et al. 2000; Hoskins et al. 2015) are compact. Identifying regulatory motifs in these genomes is relatively efficient because they are proportionately high in information content, and regulatory motifs are confined to small regions relatively close to the promoter, when compared to the human genome. Additionally, these compact genomes decrease the cost of chromatin immunoprecipitation sequencing (ChIP-seq) experiments since less sequencing is required to cover the genome, permitting a high level of multiplexing. Also, their reduced complexity increases the likelihood of detecting relatively rare TF binding events occurring in only limited numbers of cells. Most importantly, these model systems provide the opportunity to map TF binding in living organisms and their development stages. Such studies are difficult for human TFs, which must use cell lines and tissues. Finally, many of the TFs in both worms and flies are homologous to human proteins, and both organisms have long been successfully used to investigate the functions of these proteins during development (Lewis 1998). Research on individual fly and worm orthologs has led to important insights into the function of human disease genes and human biology generally (Gehring 1996; Braun and Woollard 2009; Bellen et al. 2010; Kropp and Gannon 2016). Thus, studying key conserved factors in this project will greatly enhance the analysis, interpretation, and the broader relevance of data gathered in the human ENCODE project and in other studies of gene regulation. As we transition into a period where all the “parts lists” in genomes are being defined, it will be crucial to have detailed network maps in model organisms to accelerate the understanding of how the cognate genes function in homologous and analogous networks in humans.
From the previous modENCODE project (Araya et al. 2014; Boyle et al. 2014) and our efforts to date in the modERN (model organism Encyclopedia of Regulatory Networks) project, we generated GFP-tagged strains for 403 worm TFs and 427 fly TFs. From these lines, we successfully obtained ChIP-seq data sets for 219 worm and 267 fly TFs. These data sets define hundreds of thousands of binding sites at diverse stages of development, begin to outline the relationships between various TFs, and identify sets of candidate target genes regulated by these factors across many different cell types. The data are broadly useful to the community for investigating the function of single TFs and for regulatory network analysis.
Materials and Methods
GFP strain production for flies
Recombineering was used to insert a GFP tag into the C-terminus of fly TF genes using the P[acman] (φC31 artificial chromosome for manipulation) system and two P[acman] BAC libraries, one with on average 30-kb and the other with on average 80-kb genomic fragments (Venken et al. 2009). The “GFP.FPTB” tag is a superfolderGFP-FLAG-PreScission-TEV-BLRP tag combination. The tagging cassettes are flanked by 50 nucleotides of PCR-introduced homology arms, and were introduced into the BAC by recombineering using pSIM6 (gift of D. Court) prior to the stop codon of the gene. We verified the tag junctions and GFP sequence for multiple independent clones for each reaction.
To generate tagged TF transgenic fly lines, the tagged P[acman] clone cultures were induced to high plasmid copy number with CopyControl solution (Epicentre) and BAC DNA was isolated using the PureLink HiPure plasmid prep kit (Invitrogen, Carlsbad, CA) with modifications for BAC DNA (Venken et al. 2010). The purified DNA was injected into yw embryos carrying an attP docking site and, on the X chromosome, φC31 integrase driven in the germ line by the vasa promoter. For TFs on the X, second, and fourth chromosomes, we injected into line VK00033 (Bloomington stock 42673), which has an attP docking site on the third chromosome. For TFs on the third chromosome, we injected into a strain with an attP docking site on the second chromosome, either VK00037 (Bloomington stock 24872) or a stock with docking site attP40 [gift of N. Perrimon (Markstein et al. 2008)]. For small BACs (< 50 kb) we injected 100–200 embryos, depending on the docking site used, using a concentration of 150 μg/ml. For large BACs (> 70 kb), we injected 300–600 embryos, depending on the docking site, at a concentration of 50 μg/ml. Hatched larvae were transferred to vials and eclosing G0 adults were crossed to yw flies. The progeny were screened for transformants, identified by w+ eye color. Transformants on the third chromosome were crossed to w1118; TM2/TM6C,Sb (Bloomington stock number 5906), and transformants on the second chromosome were crossed to yw; Sco/Cyo balancer flies to establish balanced lines and remove the integrase-containing X chromosome. Homozygous lines were constructed where possible. For lines that are lethal as homozygotes (∼10%), a balanced stock was generated. The lines were PCR-verified to confirm that they contain the expected TF and that the transgene inserted in the correct attP-landing site.
GFP strain production for worms
Transgenic strains were generated using fosmids provided by the TransgeneOme Project and were constructed as described (Sarov et al. 2012). These fosmids contain a 35–40 kb section of the C. elegans genome, thus capturing the coding sequence and flanking regulatory elements. The gene of interest was tagged at its C-terminus with an in-frame GFP:3xFLAG tag through recombineering. Cultures of clones were induced with CopyControl solution and DNA was isolated with a FosmidMax DNA purification kit (Epicentre). Integrated strains were generated using microparticle bombardment as previously described (Praitis et al. 2001), with the following exceptions. Particle bombardment of unc-119(tm4063) mutants was performed by using 1100 psi rupture disks and 15–50 μg of total fosmid DNA per transformation. Each 100-mm worm plate was bombarded twice with the same DNA construct using the Bio-Rad (Hercules, CA) Biolistic PDS-1000 with Hepta adapter. After bombardment, worms were transferred to 20 plates (60 mm) seeded with NA22 and then screened for the presence of dauers after 4 weeks. To identify homozygous integrated lines, individuals from presumptive integrated lines were isolated for four generations to confirm the absence of the unc-119 phenotype. Once lines were confirmed to be homozygous they were screened by fluorescent microscopy to determine expression.
Worm growth and ChIP
Embryonic stages were collected after bleaching and arresting in M9 at 20° until the desired stage was visualized. Worm synchronization was achieved by bleaching and L1 starvation. Arrested L1s were plated on peptone-enriched NGM plates seeded with OP50 bacteria and grown for 6 hr at 20° for L1 collection, or grown to the desired stage based on visual examination of their development (Brenner 1974). GFP fluorescent images were collected at this time. ChIP was conducted as previously described (Zhong et al. 2010; Niu et al. 2011; Kasper et al. 2014). Briefly, worm samples were cross-linked with 2% formaldehyde for 30 min at room temperature and then quenched with 1 M Tris pH 7.5. The pelleted worms were subsequently flash frozen in liquid nitrogen and stored at −80°. Samples were sonicated using a microtip to achieve mostly 200–800 bp DNA fragments. For each sample, 2 or 4 mg of protein lysate was immunoprecipitated using anti-GFP antibodies (gifts of Tony Hyman and Kevin White). For a subset of factors (OP662, OP565, OP579, OP638, OP553, OP550, OP658, OP696, OP552, OP563, OP688, OP685, OP707, OR3349, and XIL99), an additional step was added prior to sonication, in which worm pellets were thawed on ice and 750 µl of FA buffer containing protease inhibitors (one Roche Cat#11697498001 cOmplete Protease Inhibitor Cocktail Tablet, 125 μl 100 mM PMSF, and 25 μl 1 M DTT per 25 ml FA buffer) was added, and samples were then transferred to a 2 ml KONTES dounce (Kimble Chase, Vineland, NJ). On ice, samples were dounced 15 times with the small “A” pestle for two cycles with a 1 min hold between each cycle. Samples were then dounced 15 times with the large “B” pestle for four rounds with a 1 min hold between each cycle. Samples were then sonicated and followed the subsequent procedures as described.
Fly growth and ChIP
Transgenic flies were expanded in vials or bottles containing molasses media and stored at 20° with 50% humidity. Postembryonic stages were collected directly from these bottles. For embryos, adult flies were placed in embryo cages at 25° with apple juice plates. Next, 400 mg of flies were collected and divided into four replicates. Embryos were washed with embryo wash buffer (6.8 mM NaCl and 0.003% Triton X-100) before and after dechorination in 50% bleach for 1–2 min. Nonembryonic stages were homogenized first in Broeck-type homogenizers (Wheaton) followed by dounce-type homogenizers (Wheaton), while embryonic stages only required dounce. Organisms were combined with 6 ml A1 buffer (60 mM KCl, 15 mM NaCl, 15 mM HEPES pH 7.6, 0.5% Triton X-100, 4 mM MgCl2, 0.5 mM DTT and Roche protease inhibitor #1873580). Formaldehyde was added to the samples to a final concentration of 1.8%, thoroughly homogenized, and left on ice. Fifteen min after the addition of formaldehyde, 540 μl of 2.5 M glycine was added. Samples were centrifuged at 4000 rpm for 5 min at 4° and pellets washed three times with 3 ml cold A1 buffer. Pellets were then washed once with 3 ml lysis buffer (140 mM NaCl, 15 mM HEPES pH 7.6, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 1% Triton X-100, 500 μM DTT, and Roche protease inhibitor #1873580) and resuspended in 500 μl cold lysis buffer with 0.1% SDS and 0.5% N-lauroylsarcosine. Chromatin extracts were incubated at 4° for 10 min on a rotator prior to sonication.
Extracts were sonicated for 15 min on a Diagenode Bioruptor, with chiller, on high power (30 sec on/off). After sonication, the samples were rotated for another 10 min at 4°. Sheared chromatin was transferred to a microcentrifuge tube and spun at 15,000 rpm at 25° for 3 min. Supernatants were transferred to new tubes. The pellets were resuspended in 500 μl lysis buffer with SDS, rotated at 4°, centrifuged, and the supernatant combined with the first. The samples were spun once more at 15,000 rpm for 7 min, transferred to a new tube with sodium azide, and stored for < 2 months at −80°.
GammaBind G Sepharose beads (GE Healthcare Life Sciences) were washed three times in equal bead volumes of lysis buffer. Beads were blocked for 1 hr with a final concentration of 0.1 mg/ml BSA at 4° while rotating. Beads were again washed three times with cold lysis buffer. Samples were precleared by adding 100 μl of 50/50 bead/buffer solution and rotated at 4° for 4 hr. Samples were centrifuged at max speed for 1 min and supernatants transferred to new tubes. Next, 60 μl from each replicate were removed, pooled to serve as total chromatin input, and stored at 4°. To each replicate, 15 μg of antibody was added and rotated overnight at 4°. Next, 50 μl of 50/50 bead mix was added to the samples, which were rotated for 4 hr at 4°. Immunoprecititates (IPs) were washed four times with cold lysis buffer and twice with cold TE, rotating for 5 min at 4° between washes. Pellets were resuspended in 60 μl elution buffer 1 (10 mM EDTA, 1% SDS, and 50 mM Tris-HCl pH 8) and incubated at 65° for 10 min with mild shaking. Samples were centrifuged and supernatants transferred a fresh tube. Pellets were resuspended again with 60 μl elution buffer 2 (29% TE and 0.67% SDS) and immediately centrifuged. Elution supernatants were combined and incubated at 65° with mild shaking overnight. Chromatin input samples were incubated at 60° with mild shaking overnight, after the addition of Proteinase K and SDS to final concentrations of 0.1 mg/ml and 0.01%, respectively. The next day, inputs were incubated at 70° for 20 min. Proteinase K was added to each IP to a concentration of 4 mg/ml and incubated at 50° for 2 hr. RNaseA was added to the chromatin input to a concentration of 0.017 mg/ml and incubated at 37° for 2 hr. DNA was purified with MinElute columns (QIAGEN, Valencia, CA), eluting in 13 μl (elution buffer provided with MinElute kit). An additional 48 μl EB was added to input samples after purification. Samples were stored at −20°.
Worm and fly library preparation and sequencing
The enriched DNA fragments and input control (genomic DNA from the same sample) for two biological replicates for worm or three for fly were used for library preparation and sequencing, as previously described for modENCODE samples (Zhong et al. 2010; Nègre et al. 2011). Briefly, modERN samples were libraried and multiplexed as described (Kasper et al. 2014), using the Ovation Ultralow DR Multiplex Systems 1–8 and 9–16 (NuGEN Technologies, San Carlos, CA) following the manufacturer’s protocol, except that QIAGEN MinElute PCR purification kits were used to isolate the DNA. Briefly, 1 μl of input DNA and 10 μl of IP DNA was used to prepare sequencing libraries using NuGEN Ultralow library kits. Samples were prepared according to the manufacturer’s protocol with the following modifications. After adapter ligation, samples were purified with MinElute with two elutions in 18.5 μl EB. MinElute columns were also used after amplification, eluting with 21 μl EB. Samples were subsequently run on an Agilent Bioanalyzer DNA1000 chip. Samples showing appropriate library concentrations were size-selected, targeting a range between 200–1000 bp. Initially, libraries were size-selected using eGELs (Invitrogen). However, for the majority of libraries, we used SPRIselect beads (Beckman, Fullerton, CA). Sample volumes were increased with EB to 50 μl and combined with 42.5 μl beads for left-sided selection. Samples were eluted in 50 μl EB and subjected to right-sided selection using first 28 μl beads and then aspirating 73 μl, which was combined with 90.5 μl beads. Next, 23 μl EB was added to the washed beads and 21 μl removed as the final library sample. Library quality was assayed again on a DNA1000 chip as well as an Agilent bioanalyzer high-sensitivity chip. Sequencing was performed on the Illumina HiSequation 2000/2500/4000.
Peak calling/bioinformatics analysis
The Illumina sequencing data were aligned to the reference genome using the Burrows–Wheeler Aligner (BWA) (Li and Durbin 2009). Fly data were aligned to genome version dm6 and worm data were aligned to genome version WS245. Tools for converting sequence coordinates between different versions are available at FlyBase (http://flybase.org/static_pages/downloads/COORD.html) and WormBase (http://www.wormbase.org/wiki/index.php/Converting_Coordinates_between_releases). In addition to using aligned reads from each biological replicate, a pooled replicate was generated using aligned reads from each replicate. Furthermore, for each biological replicate, aligned reads were randomly divided into two pseudoreplicates. Peak regions significantly enriched in aligned reads were called by ChIP-seq processing pipeline (SPP) following the standard ENCODE/modENCODE pipeline (Kharchenko et al. 2008). Only data with strong peak concordance between pseudoreplicates, as well as between replicates and the pooled replicate, were used. Peaks above an irreproducibility discovery rate (IDR) of 0.1% were used to generate final peak sets (Li et al. 2011; Landt et al. 2012; Yue et al. 2014). The peaks per experiment were plotted in R using the violplot library and function.
Peak clustering
We clustered peaks from TFs using standard approaches, but noticed that with the high numbers of peaks in promoter regions this clustering often resulted in merging of what appeared to be distinct regions, as defined by the positions of the peak summits (for the worm, where some stages were assayed multiple times, testing different protocols, only the experiment using the standard protocol was kept and included in the counts and analysis). To distinguish close but distinct segments more effectively, we clustered peaks based on their summits, placing peak summits that lay no more than x bases apart. After evaluating clusters obtained with various values of x by visual inspection of the clusters in the browser, we selected 60 bases as the cutoff for x. This yielded 59,163 clusters of one or more sites in the worm, including 29,114 singletons, and 114,593 clusters in the fly, including 65,937 singletons.
Motif analysis in TF binding sites
From the Cis-BP database (Weirauch et al. 2014), we collected motifs determined by systematic evolution of ligands by exponential enrichment (SELIX), protein binding microarray (PBM), and yeast one-hybrid (Y1H) experiments. Position weight matrix files (PWM) of the motifs were used to search against the genomes by Fimo (Grant et al. 2011). A genomic region contains a motif if they are significantly similar (P-value < 10−4 by Fimo). For a binding site, we define its core region as 100 bp around its summit, and thus each core binding site is a 200-bp genomic region. All binding sites from this project and their motifs in the core regions are listed in Supplemental Material, Table S3 and Table S4. For each motif hit, we report its motif identifier in Cis-BP, P-value by Fimo, and genomic coordinates. Moreover, we also state whether a motif is enriched in the corresponding ChIP-seq data. To calculate this, we first counted the numbers of binding sites with and without motifs in the total binding sites of the TF. Second, to generate the same two quantities from random binding sites, we divided the genome into 50-bp bins and shuffled the sequence within each bin. Starting with this random genome, we repeated the above analysis to get the same quantities as references. At last, the two quantities and their references were compared with Fisher’s exact test, and a P-value < 0.05 indicates that the motif is significantly enriched in the binding sites of the TF, compared to random binding sites. A TF may have multiple motifs, and the representative motif is the one with lowest enrichment P-value. The logos of representative motifs are also from Cis-BP and listed together with their TFs in Table S3 and Table S4.
Global pairwise TF coassociations
Using methods similar to those described (Araya et al. 2014), we calculated a coassociation strength between all ChIP-seq experiments to define the level of similarity between binding sites identified. The analysis was confined to only those sites that fell outside of clusters of > 40 sites. Further, the peak interval was defined as the region ± 25 bases from the summit of the peak. The interval statistics methods (Chikina and Troyanskaya 2012) used calculates directional exact P-values for proximity between binding sites. We confined our analyses to possible promoter regions in the C. elegans genome by masking bases in the genome from the second exon to the last exon (including introns), and defining all remaining regions as possible promoter regions. P-values were calculated using IntervalStats (Chikina and Troyanskaya 2012), restricting comparisons to the current chromosome when calculating the numerator and denominator for the P-value. After performing all pairwise comparisons in both directions, we computed the fraction of significant (P < 0.05) proximal binding events in promoter domains. We reported the mean values of the complementary (inverted query and reference) comparisons as the coassociation between ChIP-Seq experiments. In the resulting matrix, rows were excluded that had no coclusters (e.g., no clusters with ≤ 40 sites) before clustering and plotting the data. The raw cluster score matrix was hierarchically clustered using (in scipy version 0.17.1 with numpy version 1.13.1) the scipy.spatial.distance.pdist function with the “cosine” distance metric to generate a distance matrix, and the scipy.cluster.hierarchy.linkage function to do the clustering with the “average” cluster joining method. The dendrogram was plotted using scipy.cluster.hierarchy.dendrogram, and the clustered data were plotted with the matplotlib.pyplot.matshow function (matplotlib version 1.5.1).
Data availability
Strains are available from the Caenorhabditis Genetics Center (CGC) (worm) and the Bloomington Drosophila Stock Center (BDSC) (fly) public repositories, and identifying information is given in Table S3 and Table S4. Complete ChIP-seq data sets and all metadata are available at http://encodeproject.org and via http://epic.gs.washington.edu/modERN. All accession and identifying information for each data set is listed in Table S5. Supplemental Material is available on Figshare at https://figshare.com/articles/Supplemental_Data_for_Kudron_et_al_2018_The_modERN_Resource_Genome-wide_Binding_Profiles_for_Hundreds_of_Drosophila_and_C_elegans_Transcription_Factors_/5729667.
Results
Project overview
The ultimate goal of the modERN project is to generate genome-wide binding profiles for the vast majority of TFs in Drosophila and C. elegans. Drosophila has 703 sequence-specific predicted TF genes, based on having at least one of the 73 identified DNA-binding domains (Hammonds et al. 2013) (Table S1). A large number of these predicted TFs, 215, still remain largely unstudied and are known only by a curated gene identifier (CG) in FlyBase (Gramates et al. 2017). Over half of these 215 uncharacterized predicted factors contain a zf-C2H2 DNA-binding domain and some of these may be involved in DNA binding, RNA binding, or both. Other proteins containing zinc finger domains, such as zf-CCCH and zf-DHHC, were not included in our overall list, since they are associated with protein–protein interactions and palmitoyltransferase activity, respectively. Here, we report results for 38% (267/708) of candidate TFs that were targeted for analysis in the modERN project.
C. elegans has 958 predicted TF genes [pseudogenes removed from Reece-Hoyes et al. (2005) and additional factors from Narasimhan et al. (2015)]. However, several of these have now been classified as RNA-binding or chromatin-remodeling factors, leaving 892 sequence-specific candidate TFs. Of these, 284 represent an expanded family of nuclear hormone receptor genes. A small fraction of these (Antebi 2015) have orthologs outside of nematodes, but most are nematode-specific and are poorly characterized; therefore, all but a representative sample of the nematode-specific nuclear receptor genes are excluded from this project, leaving 685 C. elegans TFs that are candidates for analysis (Table S1). For both organisms, the factors to be analyzed are members of the major conserved families of TFs, including homeobox, GATA, ETS, winged helix, high-mobility group, basic leucine zipper, zinc finger, and T-box DNA-binding domain-containing proteins. Here, we report results for 31% (216/685) of candidate TFs (23% overall).
Our general strategy (Figure 1) for both worms and flies is to create stably integrated transgenic lines expressing individual TFs tagged at the C-terminal end with GFP. A validated goat anti-GFP antibody is then used for IP of chromatin associated with each factor, followed by sequencing. We perform ChIP-seq on whole animals at specific stages, using RNA expression profile data and phenotypic data, where available, to determine the optimal developmental stage for ChIP. Generally, only one stage is assayed, but in some cases we examine additional stages. Typically, for each experiment, two (worm) or three (fly) biological replicates are assayed along with an input control to assess reproducibility using parameters established by the ENCODE and modENCODE projects (Landt et al. 2012).
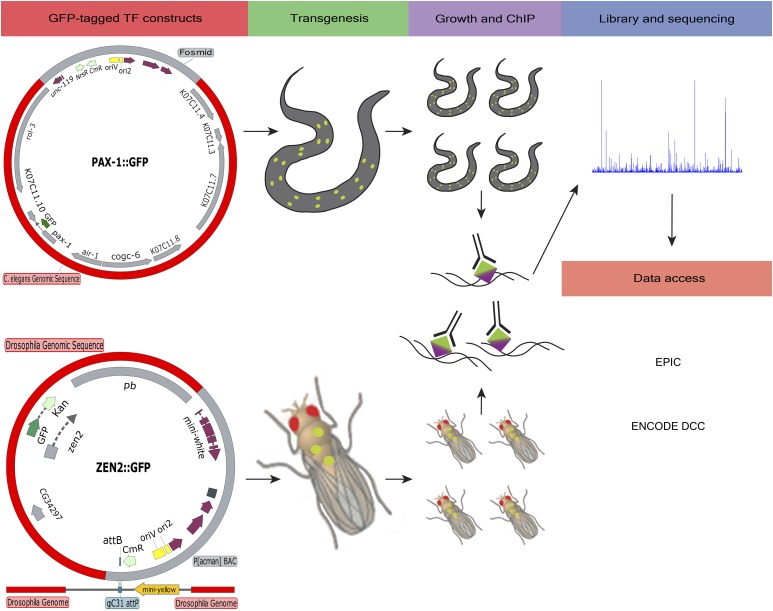
Figure 1.
Schematic of the modERN ChIP-seq pipeline. Example TF-tagged constructs for worm and fly are shown. Transgenic worms were generated by bombardment of fosmid constructs containing a single TF with dual GFP and 3xFLAG tags into unc-119 mutants. For fly, recombineered BACs containing a GFP-tagged TF were injected into embryos expressing ϕ31 integrase to target genomic integration of the entire BAC into well-characterized engineered docking sites. Integration of the BAC was confirmed by PCR. Worms and flies expressing the GFP-tagged TF were grown, fixed, homogenized, and/or sheared to obtain chromatin for immunoprecipitation. The same GFP antibody was used for ChIP in both organisms. All libraries and sequencing were conducted at the same site. Access to all of the modENCODE and modERN ChIP-seq data can be found at either the EPIC modERN website (http://epic.gs.washington.edu/modERN/) or the ENCODE DCC site (http://encodeproject.org). ChIP-seq, chromatin immunoprecipitation sequencing; DCC, Data Coordinating Center; EPIC, European Photonics Industry Consortium; TF, transcription factor; modENCODE, Model Organism ENCyclopedia Of DNA Elements; modERN; model organism Encyclopedia of Regulatory Networks.
Strain construction and resource
Strain construction differs in detail for the two species. For flies, recombineering is used to introduce C-terminal GFP tags into clones from one of two P[acman] libraries (average insert sizes of 30 and 80 kb, respectively). BACs are selected to ensure that the tagged BAC includes the DNA between the two closest predicted insulators (Nègre et al. 2010, 2011), or extends to cover the nearest genes upstream and downstream of the TF. The resultant clones are introduced into flies using the ϕC31 integrase system and attP docking sites on the second or third chromosomes. After phenotypic selection and outcrossing, the transgenic lines are PCR-verified to ensure the that inserts are integrated into the right place in the genome and that they contain the targeted TF. A pilot study showed that 12/16 BACs rescued the corresponding mutant phenotype (Venken et al. 2009). These factors include a variety of DNA-binding domains, involving several different organs or tissues (Table S2).
For worms, we have exploited the tagged protein resource created by Sarov and Hyman, in conjunction with the modENCODE project, where recombineering was used to insert a GFP tag and a 3x FLAG tag at the stop codon of genes residing in fosmids (Sarov et al. 2012). Generally, the gene of interest is flanked on either side by at least one other gene, making it highly likely that the regulatory sequences are present. The fosmid DNA is introduced into adult worms by particle bombardment and stably integrated, expressing lines are selected. Copy number is generally low (1–10) and is verified from a slight increase in the input signal on the ChIP experiment over the TF locus. This signal also confirms the identity of the tagged TF in the transgenic strain.
The transparency of the worm allows us to check the GFP expression patterns directly and document those patterns with fluorescent micrographs. For > 200 factors, detailed embryonic expression data are already available in the European Photonics Industry Consortium database (Murray et al. 2012). The large majority of the 403 strains exhibit nuclear-localized signals in a pattern that is consistent with patterns described in WormBase. For 50 nominal TFs, we find the fusion protein to be predominantly cytoplasmic, suggesting either that the worm lacks the signal by which the TF is localized to the nucleus, or that it is not a TF, but perhaps an RNA-binding protein. Another 19 TFs are nuclear localized but are expressed at such low levels in so few cells that, based on our experience, they will not give a reliable ChIP signal. Thus, these 69 lines (Table S4) have been excluded from ChIP analysis for the present, along with 25 nuclear hormone receptor genes. To date, nine strains have been tested for the ability to rescue a mutant phenotype, and all nine have demonstrated strong rescue. As with fly, these factors include a variety of DNA-binding domains, involving several different organs or tissues (Table S2). Additionally, the strains provide a ready way of identifying and recovering the tagged cells.
For both worms and flies, the inclusion of a wider genomic context increases the likelihood that spatial and temporal transcriptional regulation reflects the native gene. Further, the presence of all introns and intact 5′- and 3′-UTR regions facilitates faithful post-transcriptional control. To date, we have generated fly and worm strains for 429 and 403 TFs, respectively (Table S3 and Table S4). In addition, for the worm we generated strains for 17 DNA-associated or nuclear-localized factors, and for the fly we generated strains for five chromatin or TF-associated cofactors. As shown in these tables, the vast majority of these strains are available through the BDSC (fly) or the CGC (worm).
ChIP-seq resource
Prior to performing ChIP-seq on the strains expressing nuclear-localized GFP-tagged TFs, we gathered information on the RNA profiles for each TF from WormBase, FlyBase, modENCODE, and the literature. Based on these data, we selected a primary developmental stage when the factor has maximal expression and/or function, and performed ChIP-seq using whole-animal chromatin preparations. We also selected secondary stages, if warranted. We assessed whether the transgenic strain exhibited any features that possibly indicated an overexpression phenotype or a disruption of an important gene by transgene insertion, and might thus preclude analysis. Very few lines have exhibited visible phenotypes. For selected strains, we cultured animals in a synchronized fashion to the desired developmental stage(s), and then collected and fixed them with formaldehyde. These samples were then lysed and sonicated (sometimes with an intervening douncing step) to shear chromatin, and subjected to immunoprecipitation with a validated anti-GFP antibody. A small fraction was reserved prior to immunoprecipitation to serve as a whole-genome input control.
Our ChIP-seq data-processing pipeline closely mirrors that of ENCODE (ENCODE Project Consortium 2012). Raw fastq files are aligned to the reference genome (for fly: Release 6; for worm: WS245) using BWA (Li and Durbin 2009). Aligned reads are scored for mappability to the target genome and PCR duplication rate. High-quality, unique reads are fed into SPP (Kharchenko et al. 2008) to call peaks; with rare exception, MACS2 has been used when broad peaks are expected (Zhang et al. 2008). Significant peaks are identified using an IDR with a threshold of 0.01. Data sets are evaluated using the self-consistency ratio and rescue ratio metrics set by ENCODE. The self-consistency ratio is a measure of how similar replicates are to one another. Pseudoreplicates are generated by randomly splitting the sequencing reads within a replicate. Common peaks between each pseudoreplicate, and above an IDR threshold of 0.01, are considered significant. Replicates must have less than a twofold difference in their numbers of significant pseudoreplicate peaks. The rescue ratio is calculated in a similar manner and measures the similarity of each replicate to the entire data set. Significant peaks (IDR < 0.01) are called on two pooled pseudoreplicates generated from every replicate’s reads. Likewise, pairwise significant peaks (IDR < 0.02) are called by comparing the peaks called on each individual replicate. Valid data sets must have less than a twofold difference between all sets of pairwise replicate peaks, as well as between each set of pairwise replicate peaks and the number of pooled pseudoreplicate peaks. Upon consulting ENCODE project members, we no longer use normalized strand coefficient minimum and relative strand correlation score as additional metrics. The thresholds previously used by modENCODE and ENCODE were subjectively chosen based on human ChIP-seq data, and do not translate well to worm and fly due to their smaller genomes.
To date, we have completed ChIP-seq experiments for 262 TFs in the fly and 217 TFs in the worm, along with 35 and 18 DNA-associated proteins, respectively (these include data sets generated previously for the modENCODE project). Some TFs have been assayed at multiple stages or in different backgrounds, with a total of 302 and 366 experiments summarized here. Total data sets by stage are summarized in Table 1 and the full list is presented in Table S5. The factors assayed in each organism include representatives of all the major DNA-binding domains (see Table S3 and Table S4 for specifics). The assayed TFs have 1035 orthologs in human (Table S6), as identified using Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool (DIOPT) (Hu et al. 2011), forming 1786 orthologous pairs. As expected, almost all the pairs are one-to-many (28%) or many-to-many (70%) orthologous types because numerous gene duplications occurred due to the extended evolution after speciation. The surviving TF duplicates, i.e., the ones we observe today, are potentially subject to neo/subfunctionalization. Moreover, the assayed TFs of fly have 436 orthologs from worm, forming 697 orthologous pairs, while the assayed worm TFs have 366 orthologs, forming 576 pairs with fly. Taken together, these abundant TFs with homologs within and between species provide an opportunity to study regulatory network expansion and rewiring by gene duplication.
Table 1. Experiments by stage.
| Fly stage | Factors assayed | Worm stage | Factors assayed |
|---|---|---|---|
| Embryo | 200 | Embryo | 91 |
| W3L | 37 | L1 larva | 72 |
| WPP | 52 | L2 | 47 |
| PUP | 1 | L3 | 46 |
| Adult | 14 | L4 | 64 |
| Kc167 | 5 | Young adult | 51 |
| Total | 309 | Total | 371 |
In total, for the fly we detect 1,232,334 peaks across 302 data sets for 262 factors (Table S7). Similarly, for the worm we detect 667,924 peaks (TF binding sites) across the 366 data sets for the 217 factors (Table S8). The number of sites detected per experiment varied considerably with 5–95% quantiles of 226 and 8708 for the fly and of 98 and 6036 for the worm (see Figure S1 for the full distribution). Very few examples exist in which GFP-tagged TF ChIP profiles can be compared directly to endogenous TF profiles that used an antibody with ENCODE-level validation, but one such instance is worm EFL-1. EFL-1:GFP was compared directly to endogenous EFL-1 performed at the same L1 developmental stage and analyzed with the same pipeline. There was a 98% agreement between the two data sets (Kudron et al. 2013). As a further test of the binding sites, we found factors with prior experimentally determined motifs in Cis-Bp (http://cisbp.ccbr.utoronto.ca/) and asked if the motifs were enriched in the binding sites of that factor. Of 118 fly TFs with prior experimentally determined motifs, 71 had at least one motif enriched in our data, and of 54 worm factors, 27 had at least one enriched motif (P < 0.05).
The binding sites often lie close to one another, with tens to hundreds of peaks from multiple factors lying within a few 100 bp, so called high occupancy target (HOT) sites (Moorman et al. 2006; Gerstein et al. 2010). Such HOT sites are often associated with broadly and highly expressed genes such as ribosomal protein genes, and are often presumed to represent open chromatin. We grouped the ChIP-seq sites into clusters, using the peak summit rather than the whole binding site, so that we could resolve apparently distinct clusters (see the Materials and Methods). We detected 88,507 clusters in the fly and 59,136 clusters in the worm (Table S9 and Table S10). Of these, 48,604 and 29,103 are singleton sites, respectively, and the remainder represent a continuum from two to hundreds of sites. This tight clustering of sites means that the ∼1,232,000 and to ∼667,000 worm binding sites span just 8.7 and 5.77 Mb in the clusters, even allowing for 25 bases on each side of the peak summit.
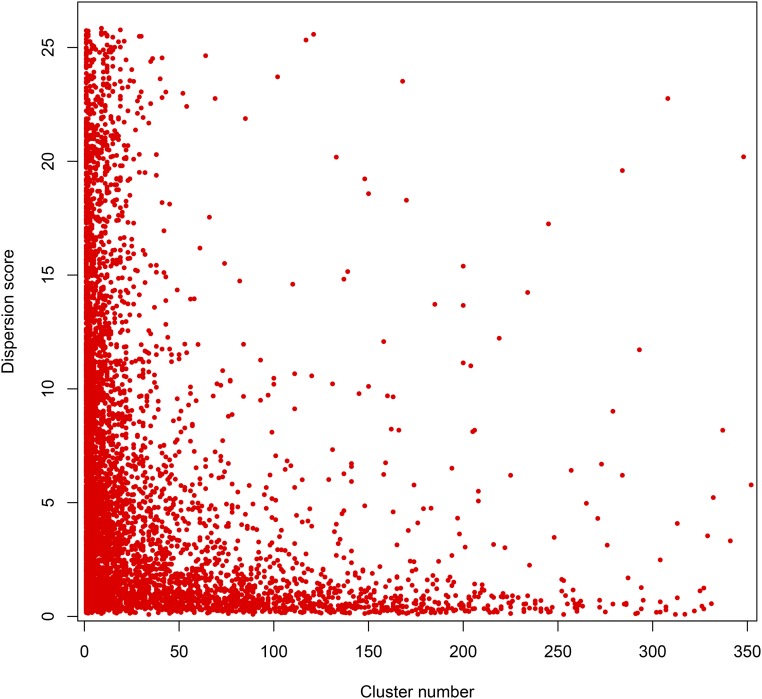
Exploiting recently produced single-cell RNA sequencing (RNA-seq) data that defines expression profiles for 28 cell types in the L2 worm (Cao et al. 2017), we examined the relationship between the number of binding sites near a given gene and the cell type-specificity of gene expression in the worm data. Clusters containing many binding sites (HOT sites) were primarily found associated with broadly expressed genes, whereas regions containing relatively few sites were generally associated with genes expressed in specific cell types (Figure 2). Even though about one-third of all worm TFs have been assayed, 35% of genes fail to have any upstream TF binding site (considering only genes that do not potentially “share” regulatory regions). However, ∼23% of these genes are primarily expressed in males, which we have not assayed. Additionally, on average, genes without TF binding sites, and with maximal expression in hermaphrodites, are expressed at only one-fifth of the level of genes with sites (Boeck et al. 2016), suggesting that the expression of many genes without binding sites is limited to very few cells or a brief window of time.
Figure 2.
Cell type specificity of expression reflects the number of binding sites in promoters. The dispersion score (a measure of how broadly or specifically expressed a gene is, with increasing score representing increasing specificity) of each of 5401 expressed genes is plotted against the number of binding sites in the largest cluster of sites upstream of the gene. Genes with high dispersion scores overwhelmingly have < 30 binding sites in the largest upstream cluster, whereas genes with low dispersion scores (< 3) can have very large clusters of sites upstream. Dispersion scores for 14,535 protein-coding genes were obtained from the L2 single-cell combinatorial indexing RNA sequencing data set (Cao et al. 2017) using the estimateDispersions function in Monocle2. Dispersion scores > 10 show expression predominantly in a single cell type. Of these, 7503 had the upstream gene in the same orientation; all binding sites in the intergenic space plus 200 bases downstream of the transcript start site were accordingly assigned to the downstream gene. Of these, 5401 had at least one binding site. The cluster with the maximum number of sites was used for plotting.
Extending the previous analysis of coassociation (Araya et al. 2014), we looked for coassociations of TFs in this larger worm data set (Figure 3). We continue to see the many previously reported associations, along with new ones such as UNC-120 with RNT-1, CEH-18, UNC-62, and HLH-1 in the embryo, and XND-1, F49E8.2, EFL-1, and DPL-1 in young adults. The coexpression of these groups of genes in muscle and gonad, respectively, suggest that they may cooperate in regulating expression in these cell types. Reinforcing these associations, Cao et al. (2017), found that binding sites from similar combinations of factors in small clusters could predict cell-specific expression patterns. This coassociation analysis is also useful for identifying possible functions for relatively unknown factors. One example is F16B12.6, an essentially unstudied AT-hook-containing TF. This factor groups quite strongly with a discrete cluster of TFs in larval animals that includes EFL-1, DPL-1, and LIN-15, all of which are part of the synthetic multivulva (SynMuv) pathway, which acts to repress gene expression during somatic development (e.g., Cui et al. 2006).
Figure 3.
Global pairwise transcription factor coassociation matrix (NT = 155,630) as defined by promoter interval statistics (Chikina and Troyanskaya 2012), followed by clustering of factors based on those scores. Coassociation scores are scaled by the SD (uncentered) for visualization purposes. Clusters with mutually high-scoring coassociations are apparent both along and off the diagonal. Several clusters that contain transcription factors of known specificity are outlined and enlarged to show the various factors and stages involved.
Accessing the data
Two avenues are available to access the data produced by the modENCODE and modERN consortia. After passing QC filters, the ChIP-seq data from both the modENCODE and modERN projects are submitted to the ENCODE Data Coordinating Center (DCC), where it is available to the public (http://encodeproject.org). Users accessing the DCC ENCODE site can directly enter a TF of interest into the search bar in the upper right-hand corner and then choose the ChIP-seq experiment from the data types listed. The input control is listed as a separate experiment for each stage assayed. The experiment summary page provides all necessary information for the TF and the ChIP-seq experiment, such as the strain genotype, antibody information, library, and sequencing platform information, and all associated images, documents, and files.
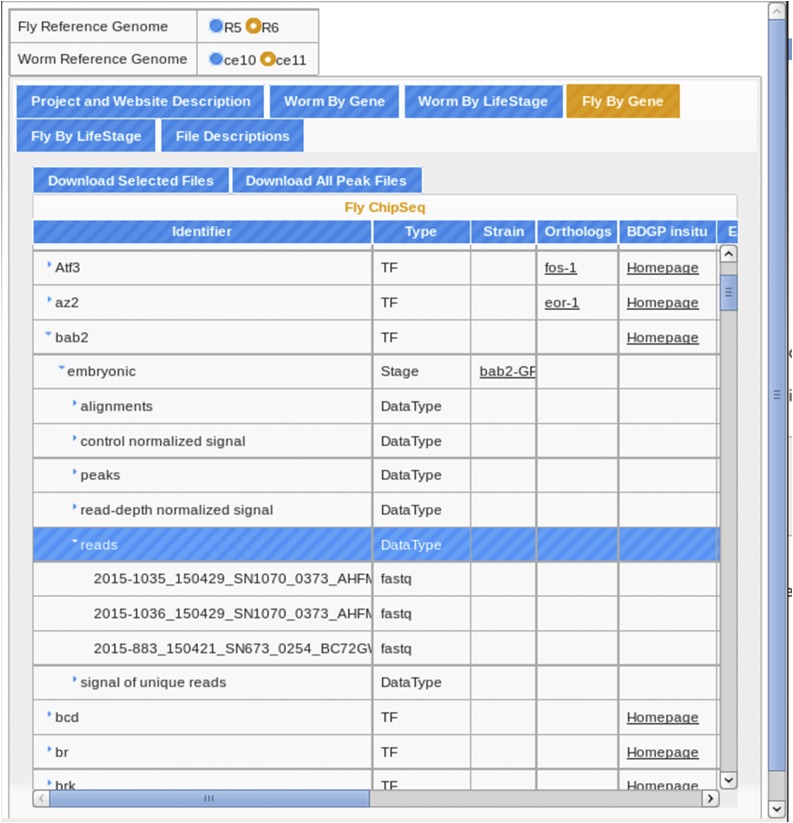
To provide intuitive, direct access to our data with additional information, we created a website, http://epic.gs.washington.edu/modERN/, which organizes all the ChIP-seq files generated for TFs in worm and fly for both modENCODE and modERN data (Figure 4) (Figure S2 provides a tutorial). Users can search for data sets in worm and fly by TF or by life-stage in their chosen reference genome. Individual or groups of TFs can be easily accessed simply by using the drop-down arrow for each chosen TF. Individual files or groups of files can be selected and downloaded. A document describing available raw and processed file types is also accessible (rightmost button at http://epic.gs.washington.edu/modERN/). We also provide links to the DCC ENCODE site, the University of California, Santa Cruz (UCSC) Genome Browser, and WormBase/FlyBase for each TF/data set. The UCSC browser links provide overviews of the data sets for all features of the genome, updated daily.
Figure 4.
Screenshot of the EPIC modERN database. All worm and fly data from both the modERN and modENCODE consortiums can be accessed at (http://epic.gs.washington.edu/modERN/). See Figure S2 for a tutorial on how to navigate the site. BDGP, Berkeley Drosophila Genome Project; ChIP, chromatin immunoprecipitation; EPIC, European Photonics Industry Consortium; TF, transcription factor; modENCODE, Model Organism ENCyclopedia Of DNA Elements; modERN; model organism Encyclopedia of Regulatory Networks.
Discussion
Here, we describe the efforts of the modERN consortium to date, in which, combined with data from modENCODE, we have defined binding sites for about one-third of targeted TFs in two key model organisms. The binding sites cluster tightly in both organisms, both in HOT regions and also in other regions with distinctly fewer sites. As compared to the worm, the fly has about double the number of peaks per factor. The larger fly genome size could partially account for this difference, as could the greater complexity of the fly body plan, which utilizes essentially the same number of TFs in the genome as the simpler worm. The use of the same methods, including peak calling, for both worm and fly rules out many possible artifactual causes of the difference, but not organism-specific differences, such as cross-reactivity of endogenous proteins with the GFP antibody. The genomic regions with 2–40 sites in the worm are greatly enriched for genes that exhibit cell- or tissue-specific expression. Also, pairs of worm TFs show significant coassociations in these smaller clusters, suggesting likely interactions in regulating the nearby genes. Indeed, in recent work examining cell type expression using single-cell combinatorial indexing RNA-seq, the combinations of TF binding sites in small clusters were highly predictive of the observed expression patterns (Cao et al. 2017). The models also suggest extensive regulatory networks.
Obtaining a comprehensive picture of the binding sites for all factors will be challenging. Our strategy of performing whole-animal ChIP-seq using a GFP-tagged protein on the stage where the factor is most highly expressed has limitations. The anti-GFP antibody may have artifactual binding, possibly contributing to broadly bound regions. Focusing analyses on sites with relatively few factors and the development of modified peak callers may partially ameliorate any such problem. Factors may bind at different sites in stages that we have not assayed. Factors expressed in a very limited number of cells may not yield signals above background for many sites. Factors may also target different sites in different tissues, again complicating their detection. To deal with issues of sensitivity and tissue specificity, we are exploring more sensitive methods, such as Cut & Run (Skene and Henikoff 2017), that may allow the detection of binding sites in flow-sorted cells. However, assaying additional stages for a given factor would necessarily come in exchange for assays of new factors. Instead, we expect that the data we provide on a single stage will provide the community with the leads they need to explore these factors further, with the advantage that tagged strains are already available.
The GFP-tagged strains are themselves of wide utility to the community and are in high demand from the respective stock centers. Because they are embedded in large segments of DNA containing flanking genes and are integrated into the genome, the expression patterns generally are faithful representations of when and where those TFs are expressed (micrographs illustrating the expression patterns based on the GFP tag or in situ hybridization are readily accessible through our web interface: http://epic.gs.washington.edu/modERN/). For worm genes expressed in the first half of embryogenesis, expression patterns for these TFs are being determined systematically at the single-cell level with high temporal resolution (Murray et al. 2012), and of course experts in worm anatomy can determine expression patterns for genes of interest. Once patterns are defined, these strains can serve as a means to retrieve those cells by FACS for RNA-seq analysis and other assays.
The analyses we have done to date only begin to tap the utility of these data sets. We expect that the modERN ChIP-seq resource will facilitate many additional scientific inquiries into the function of individual TFs and combinations of TFs in key conserved and species-specific regulatory pathways. These data will be essential to understand how individual TFs work together in both cell type-specific and global networks across development and homeostasis. One of the outstanding questions in the field of TF mapping is how frequently (or infrequently) binding leads to regulation, and data sets like these will help to clarify these issues. Buttressed with the classical experimental strengths of worms and flies, including sophisticated genetic and cell biological approaches, the genomic regulatory data that we generate should be leveraged highly efficiently and extensively by the larger community into a sophisticated understanding of how complex regulatory systems are integrated in the living animal.
Moreover, our project will complement efforts in humans, both those already underway and planned for the near future. The challenge to define regulatory networks in humans is much greater than worms and flies, with ∼1500 TFs needed to be assayed across multiple cell lines. With our data deposited in the ENCODE DCC, links between our results and human projects should be easier to establish.
Acknowledgments
The authors thank the ENCODE Data Coordinating Center for providing data access; Elise Feingold for her support during the project; members of the Berkeley Drosophila Genome Project for their input, especially Erwin Frise for GFP image analysis; members of the University of Chicago core facilities HGAC = High-throughput Genome Analysis Core (HGAC) and Genomics Facility (GF) (P30 CA014599), especially Adam Dedier and Jigyasa Tuteja, for sequencing; Stacy Holtzman and Thomas C. Kaufman for their production of 12 tagged transcription factor (TF) lines: bab1, Bro, CG7839, Eip74EF, ERR, Ets21C, gcm, lola, polybromo, pros, Sp1, and ss made during the bridge period between modENCODE and modERN; Rebecca Spokony for her input and suggestions; the Bloomington Drosophila Stock Center (BDSC) for distributing the fly GFP-tagged TF strains; and the Caenorhabditis Genetics Center (CGC) for distributing the worm GFP-tagged TF strains. The BDSC and CGC are funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs P40 OD-018537 and P40 OD-010440, respectively. This work was supported by NIH grants U41-HG-007355 (R.H.W.) and R01-GM-076655 (S.E.C.), and a William Gates III Endowed Chair in Biomedical Sciences (R.H.W.).
Author contributions: R.H.W., K.P.W., V.R., and S.E.C. designed and managed the project. L.G. and J.P. helped develop the modERN website. A.V., J.M., D.S., M.S., S.S., M.Ka., and N.J. recombineered the fly BACs. W.W.F. and S.P. made the fly transgenic lines. A.S.H. imaged GFP-expressing fly embryos. M.Ki., J.G., H.A., and M.L.W. performed fly ChIP. D.V., R.T., and M.C. made the worm transgenic lines. M.M.K., S.S., and M.H. grew and imaged worm strains. M.M.K. and S.S. performed worm ChIP. M.Ki., H.A., and A.V. built libraries for worm and fly samples. A.V. analyzed the fly ChIP data and M.M.K. analyzed the worm ChIP data. R.H.W., T.J.D., and L.W.H. did additional analyses on the ChIP data. J.X., K.-K.Y., and M.G. helped on orthology. M.M.K., A.V., R.H.W., V.R., S.E.C., and A.S.H. wrote the manuscript. L.G., J.P., J.X., K.-K.Y., K.P.W., L.M., and M.G. provided input on the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online at https://figshare.com/articles/Supplemental_Data_for_Kudron_et_al_2018_The_modERN_Resource_Genome-wide_Binding_Profiles_for_Hundreds_of_Drosophila_and_C_elegans_Transcription_Factors_/5729667.
Communicating editor: O. Hobert
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila Melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Antebi, A., 2015 Nuclear receptor signal transduction in C. elegans (June 9, 2015), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.64.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Araya C. L., Kawli T., Kundaje A., Jiang L., Wu B., et al. , 2014. Regulatory analysis of the C. Elegans genome with spatiotemporal resolution. Nature 512: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Tong C., Tsuda H., 2010. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat. Rev. Neurosci. 11: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck M. E., Huynh C., Gevirtzman L., Thompson O. A., Wang G., et al. , 2016. The time-resolved transcriptome of C. Elegans. Genome Res. 26: 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. P., Araya C. L., Brdlik C., Cayting P., Cheng C., et al. , 2014. Comparative analysis of regulatory information and circuits across distant species. Nature 512: 453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Woollard A., 2009. RUNX factors in development: lessons from invertebrate model systems. Blood Cells Mol. Dis. 43: 43–48. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis Elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. B., Celniker S. E., 2015. Lessons from modENCODE. Annu. Rev. Genomics Hum. Genet. 16: 31–53. [DOI] [PubMed] [Google Scholar]
- Cao J., Packer J. S., Ramani V., Cusanovich D. A., Huynh C., et al. , 2017. Comprehensive single cell transcriptional profiling of a multicellular organism. Science 357: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998 Genome sequence of the nematode C. Elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed]
- Chikina M. D., Troyanskaya O. G., 2012. An effective statistical evaluation of ChIPseq dataset similarity. Bioinformatics 28: 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Chen J., Myers T. R., Hwang B. J., Sternberg P. W., et al. , 2006. SynMuv genes redundantly inhibit Lin-3/EGF expression to prevent inappropriate vulval induction in C. Elegans. Dev. Cell 10: 667–672. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W. J., 1996. The master control gene for morphogenesis and evolution of the eye. Genes Cells 1: 11–15. [DOI] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis Elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates L. S., Marygold S. J., Dos Santos G., Urbano J.-M., Antonazzo G., et al. , 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45: D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. E., Bailey T. L., Noble W. S., 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammonds A. S., Bristow C. A., Fisher W. W., Weiszmann R., Wu S., et al. , 2013. Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol. 14: R140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L. W., Coulson A., Murray J. I., Bao Z., Sulston J. E., et al. , 2005. Genomics in C. Elegans: so many genes, such a little worm. Genome Res. 15: 1651–1660. [DOI] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I., et al. , 2015. The Release 6 reference sequence of the Drosophila Melanogaster genome. Genome Res. 25: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., et al. , 2011. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper D. M., Wang G., Gardner K. E., Johnstone T. G., Reinke V., 2014. The C. Elegans SNAPc component SNPC-4 coats piRNA domains and is globally required for piRNA abundance. Dev. Cell 31: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P. V., Tolstorukov M. Y., Park P. J., 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 26: 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropp P. A., Gannon M., 2016. Onecut transcription factors in development and disease. Trends Dev. Biol. 9: 43–57. [PMC free article] [PubMed] [Google Scholar]
- Kudron M., Niu W., Lu Z., Wang G., Gerstein M., et al. , 2013. Tissue-specific direct targets of Caenorhabditis elegans Rb/E2F dictate distinct somatic and germline programs. Genome Biol. 14: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt S. G., Marinov G. K., Kundaje A., Kheradpour P., Pauli F., et al. , 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22: 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1998. The bithorax complex: the first fifty years. Int. J. Dev. Biol. 42: 403–415. [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Brown J. B., Huang H., Bickel P. J., 2011. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5: 1752–1779. [Google Scholar]
- Moorman C., Sun L. V., Wang J., de Wit E., Talhout W., et al. , 2006. Hotspots of transcription factor colocalization in the genome of Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 103: 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. I., Boyle T. J., Preston E., Vafeados D., Mericle B., et al. , 2012. Multidimensional regulation of gene expression in the C. Elegans embryo. Genome Res. 22: 1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan K., Lambert S. A., Yang A. W. H., Riddell J., Mnaimneh S., et al. , 2015. Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. Elife 4: e06967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A., et al. , 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6: e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Ma L., Bristow C. A., Miller S. W., et al. , 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W., Lu Z. J., Zhong M., Sarov M., Murray J. I., et al. , 2011. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. Elegans. Genome Res. 21: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis Elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes J. S., Deplancke B., Shingles J., Grove C. A., Hope I. A., et al. , 2005. A compendium of Caenorhabditis Elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 6: R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M., Murray J. I., Schanze K., Pozniakovski A., Niu W., et al. , 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. Elegans. Cell 150: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene P. J., Henikoff S., 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6: e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., Carlson J. W., Schulze K. L., Pan H., He Y., et al. , 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila Melanogaster. Nat. Methods 6: 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., Popodi E., Holtzman S. L., Schulze K. L., Park S., et al. , 2010. A molecularly defined duplication set for the X chromosome of Drosophila Melanogaster. Genetics 186: 1111–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch M. T., Yang A., Albu M., Cote A. G., Montenegro-Montero A., et al. , 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., et al. , 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M., Niu W., Lu Z. J., Sarov M., Murray J. I., et al. , 2010. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis Elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 6: e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available from the Caenorhabditis Genetics Center (CGC) (worm) and the Bloomington Drosophila Stock Center (BDSC) (fly) public repositories, and identifying information is given in Table S3 and Table S4. Complete ChIP-seq data sets and all metadata are available at http://encodeproject.org and via http://epic.gs.washington.edu/modERN. All accession and identifying information for each data set is listed in Table S5. Supplemental Material is available on Figshare at https://figshare.com/articles/Supplemental_Data_for_Kudron_et_al_2018_The_modERN_Resource_Genome-wide_Binding_Profiles_for_Hundreds_of_Drosophila_and_C_elegans_Transcription_Factors_/5729667.