Abstract

The most promising F508del-CFTR corrector, VX-809, has been unsuccessful as an effective, stand-alone treatment for CF patients, but the rescue effect in combination with other drugs may confer an acceptable level of therapeutic benefit. Targeting cellular factors that modify trafficking may act to enhance the cell surface density of F508-CFTR with VX-809 correction. Our goal is to identify druggable kinases that enhance F508del-CFTR rescue and stabilization at the cell surface beyond that achievable with the VX-809 corrector alone. To achieve this goal, we implemented a new high-throughput screening paradigm that quickly and quantitatively measures surface density and total protein in the same cells. This allowed for rapid screening for increased surface targeting and proteostatic regulation. The assay utilizes fluorogen-activating-protein (FAP) technology with cell excluded and cell permeant fluorogenic dyes in a quick, wash-free fluorescent plate reader format on live cells to first measure F508del-CFTR expressed on the surface and then the total amount of F508del-CFTR protein present. To screen for kinase targets, we used Dharmacon’s ON-TARGETplus SMARTpool siRNA Kinase library (715 target kinases) with and without 10 μM VX-809 treatment in triplicate at 37 °C. We identified several targets that had a significant interaction with VX-809 treatment in enhancing surface density with siRNA knockdown. Select small-molecule inhibitors of the kinase targets demonstrated augmented surface expression with VX-809 treatment.
Keywords: cystic fibrosis, VX-809, high-throughput siRNA kinase library screen, fluorgen-activating-protein (FAP)
Introduction
The cystic fibrosis transmembrane conductance regulator (CFTR) is finely controlled in its bioavailability at the apical surface of epithelial cells, where it plays a significant role as a chloride channel. Aberrant cellular trafficking and dysfunction of CFTR in the lung leads to decreased chloride transport, resulting in cystic fibrosis (CF) symptoms. These arise from a thickened mucus layer in lung epithelia, which causes airway obstruction, inflammation, chronic infection, and severe reduction in life expectancy. CF remains the most common fatal genetic disease among Caucasians.1
The disease is most frequently caused by the deletion of phenylalanine at position 508 of the CFTR gene (F508del-CFTR). F508del-CFTR is misfolded and becomes trafficking defective, where it is largely degraded by endoplasmic reticulum associated degradation pathways involving the proteasome.2 Rescuing trafficking to increase bioavailability would help restore anion channel function, where recovering only 30% of wild-type activity can provide a therapeutic benefit to CF patients.3 Partial rescue of F508del-CFTR trafficking to the plasma membrane (PM) has been shown with low temperature incubation that promotes proper folding of the channel, and through small molecule correctors, which can either act by direct participation in stabilizing CFTR conformation, or indirectly through interactions with quality control machinery or other cellular components involved in CFTR folding and trafficking.4 F508del-CFTR that is rescued to the cell surface shows a moderate functional defect due to its inherent altered protein conformation and faces significant stability problems, where the peripheral protein quality control system redirects mutant CFTR from recycling endosomes toward degradation.5 Hence, rescue of CFTR to the surface, combined with decreased degradation of rescued CFTR may deliver improved therapeutic effects.
Identified correctors have been unsuccessful at achieving clinical relief as stand-alone treatments. The corrector, VX-809, promotes stabilization of the disrupted F508del-CFTR conformation, but that alone is insufficient for therapeutic effect.6 The combination of VX-809 and VX-770, a CFTR potentiator that increases channel activity at the PM, has been approved as a treatment strategy (Orkambi) for improving lung function in specific patient populations. Recently, two phase 3 clinical trials with VX-770 in combination with a different candidate corrector, VX-661, resulted in significant improvements in lung function.7 However, there have been concerns regarding the use of VX-770 for treatment of F508del-CFTR mutations, because it appears to accelerate F508del-CFTR degradation from the PM after rescue with VX-809 or VX-661 correctors.8,9
Repairing trafficking, the implicit defect of F508del-CFTR, has been a difficult objective for F508del-CFTR directed therapies. Currently, new-generation correctors, VX-152 and VX-440, are undergoing clinical development alone and in a triple combination with VX-661/VX-770.10 Combinations of CFTR modulators have the potential to confer an acceptable therapeutic benefit.11,12 Despite VX-809’s stand-alone clinical shortcomings and low therapeutic effect in combination with VX-770, it still remains an encouraging corrector since its discovery in 2011. A synergy-based small molecule screen for enhancing VX-809 correction efficacy was previously conducted to search for molecular correctors that restore F508del-CFTR structure stability that is not fully accomplished through VX-809 interaction alone. Identifying second-generation correctors can result in further rescue when applied with first generation correctors like VX-809.13 Combinational therapies that utilize different possible methods of correction have the potential to be the tipping point for a powerful treatment strategy for CF. Thus, identification of druggable kinase targets that, when inhibited, enhance the rescuing effect of VX-809 could lead to improved therapeutic strategies.
In screening for F508del-CFTR correction, kinases have been investigated through drug inhibition and RNA interference libraries.14,15 However, such kinase centered high-throughput screens (HTS) have not been conducted in tandem with corrector treatment. Our goal is to develop an assay to identify druggable kinases that enhance F508del-CFTR rescue and stabilization at the cell surface beyond that achievable with the VX-809 corrector alone at physiological temperature. We developed a screening method to rapidly and simply detect surface expression and total F508del-CFTR protein, independently assessing trafficking and proteostatic effects of each treatment.
A majority of F508del-CFTR corrector screens have been based on functional rescue. Such HTS activity assays include the yellow fluorescent protein based halide sensor, or the use of a voltage-sensitive membrane dye.16−19,14,15 In contrast, there have been several F508del-CFTR correction screens developed that measure rescued trafficking directly by detecting the presence of CFTR at the plasma membrane in nonpermeabilized cells using immunodetection of epitope tags (HA and FLAG) with fluorescent antibody labeling, or quantifying horseradish peroxidase (HRP)-tagged protein for surface specific luminescence.20,21,13 These surface-selective assays have several drawbacks, involving multiple wash steps and lengthy incubation periods, which limits processing speed and introduces potential variability. The HRP method allows for the interrogation of surface or total protein, in different assay wells, depending on permeabilization. There is a need for single well measurements of surface and total protein pools in a fast, no-wash whole-well format, consistent with high-throughput screening workflows.
A fluorogen-activating-protein (FAP)-based platform was previously introduced and validated as an alternative method for selective and sensitive F508del-CFTR surface fluorescence measurements.22 More recently this platform was utilized for high-content imaging of F508del-CFTR detection.23 Here, we extended the FAP-F508del-CFTR platform to establish a new high-throughput screening paradigm that quickly and quantitatively measures surface expression and total protein at 37 °C in a plate reader format, enabled by new quantum-yield matched cell permeant and cell-excluded dyes. Using this assay, we performed a siRNA screen targeting kinases to identify kinase targets that enhanced surface trafficking, and enhanced stability of F508del-CFTR. From these results, we identified targets that increased VX-809-mediated rescue, and several showed a significant positive interaction with VX-809. Plate reader results were recapitulated in high-throughput flow cytometry with individual siRNA treatments, and the influence of two identified target kinases were validated using specific inhibitors with single cell (flow cytometry) measurements. Specific kinase suppression resulted in significant and substantial increases in VX-809-mediated rescue at 37 °C.
Experimental Section
Materials
Dulbecco’s Modified Eagle’s Medium (DMEM) was obtained from Hyclone (SH30022.01). OptiMEM reduced serum with no phenol red was from Thermo Fisher Scientific (11058201). Hanks balanced salt solution (HBSS) with calcium and magnesium was from Life Technologies (14025134). High optical quality 96-well cell culture-treated plates were obtained from Ibidi (89626). Poly-l-lysine for coating 96-well plates was from Sigma (P4707). The ON-TARGETplus SMARTpool siRNA Kinase library, single ON-TARGETplus siRNAs (CAMKK1 and RAF1), DharmaFect1 transfection reagent (T-2001-02), the positive controls (ON-TARGET PLUS SMART POOL siRNA CFTR, L-006425-00-0005), and negative controls (ON-TARGET Plus Nontargeting pool, D-001810-10) were from GE Heathcare Dharmacon. The kinase inhibitors were purchased from Cayman Chemicals and Selleck chemicals. VX-809 was purchased from Selleck chemicals. MG dyes were synthesized at Carnegie Mellon University, and Hoechst 33342 cell stain was from Thermo Fisher Scientific.
Cell Line Generation and Cell Culture
ΔF508-CFTR and WT CFTR were fused with FAP (dL5**) at the N-terminus through an added membrane-spanning segment (Figure 1). The fusion constructs were made with a pBabeSacLac2 plasmid and expressed in HEK-293 cells for stable cell lines, described previously.24 Clonal FAP expressing cell lines were generated by BD FACS Diva through selecting cells with the brightest fluorescence after MG-B-Tau dye surface labeling. The FAP-CFTR ΔF508 cell lines were sorted with the BD FACS Diva for the enrichment of highest responders to 24 h treatment of 10 μM VX-809 at 27 °C. The enriched population was expanded and cryopreserved for use at the same passage for each screening experiment. HEK-293 cells were maintained in DMEM with 10% FBS, 100 units ml–1 penicillin, and 100 μg mL–1 streptomycin in a humidified atmosphere of 5% CO2 at 37 °C. Antibiotics were absent during transfection and the 24 h incubation of VX-809/DMSO treatment.
-
1.
After plate treatment, the wells were aspirated.
-
2.
HBSS (100 μL) with Hoechst33342 (1 μg/mL) were added to the wells. Immediately afterward, 50 μL of MG-B-Tau was added to the plate at a final concentration of 500 nM. The plate was scanned on a M1000 Tecan Plate reader at 640/680 nm, 10 nm width, 250 gain, from the bottom, and 16 multiple reads of distinct areas in each well. The plate was scanned 3×.
-
3.
Cell permeable dye (50 μL), MGnBu, was added at a final concentration of 200 nM and incubated for 20 min at 37 °C. It was then scanned using the same parameters as step 2.
-
4.
After an hour incubation with Hoechst 33342 (1 μg/mL), the plate was scanned at 362/492 nm, 5 nm width, and with 150 gain.
Figure 1.

FAP-CFTR construct. An N-terminal fusion of the dL5** fluorogen-activating-protein (FAP) with a PDGFR transmembrane spanning segment was used to express the FAP at the extracellular face of the plasma membrane.
HTS Plate Reader Surface and Total Expression Assay
siRNA Screen
HEK-293 cells expressing FAP-F508del-CFTR were seeded at a density of 3 × 104 cells/well in a 96-well plate. Transfection was performed following Dharmacon’s Library transfection protocol, using 25 nM siRNA. One-day post transfection, cells were transferred to two poly-l-lysine-coated ibidi 96-well plates at 5 × 105 cells/well. Two days post transfection, the media was treated with either 10 μM VX-809 or DMSO for 24 h. After 24 h of incubation, cells were processed on a plate reader as described in Figure 2.
Figure 2.

Stepwise plate reader fluorescence measurements.
Kinase Drug Target Validation
Cell were plated at 5 × 105 cells/well in a poly-l-lysine-coated ibidi 96-well plate, dosed with GW 5074 (RAF1) or STO-609 (CAMKK1) kinase inhibitors, and were treated in combination with either DMSO or 10 μM VX-809 for 24 h. After 24 h of incubation, cells were processed on a plate reader as described in Figure 2.
Flow Cytometry
Asssessing Relative Brightness of MG Fluorogens
FAP-WT-CFTR cells were plated in 35 mm dishes and grown to 80% confluency. Cells were incubated with 500 nM MG-B-Tau, MG-Ester, or MGnBu in PBS for 15 min, and then they were suspended and analyzed for surface (MG-B-Tau) or total fluorescence via BD Accuri flow cytometer. For the remaining cells/PBS/MG-B-Tau mix, 500 nM MGnBu or MG-Ester was added and incubated for 15 min to label intracellular protein, and then they were measured for total protein fluorescence.
SiRNA Target Validation
Cells were plated at 3 × 104 cells/well and dosed identical to the HTS plate reader method for siRNA screening, using single CAMKK1 and RAF1 siRNAs at 25 nM in a 96-well plate. After VX-809 24 h incubation, media was removed, 100 μL of cell stripper was added to detach cells, and the cells were incubated for 20 min. Cell stripper was gently removed and 150 μL of HBSS with 500 nM MG-B-Tau dye was added to suspend cells and transfer them to a U-bottom 96-well plate, where they were allowed to incubate for 15 min in dye before being read on an Intellicyt HTFC BD Accuri flow cytometer.
Kinase Drug Target Validation
Cells were plated at 5 × 105 cells/well in a 96-well plate, dosed with GW 5074 (RAF1) or STO-609 (CAMKK1) kinase inhibitors, and were treated in combination with either DMSO or 10 μM VX-809 for 24 h. After VX-809 24 h incubation, media was removed, 100 μL of cell stripper was added to detach cells, and the cells were incubated for 20 min. Cell stripper was gently removed and 150 μL of HBSS with 500 nM MG-B-Tau dye was added to suspend cells and transfer them to a U-bottom 96-well plate, where they were allowed to incubate for 15 min in dye before being read on an Intellicyt HTFC BD Accuri flow cytometer.
Data Analysis and Statistics
Hit scoring metrics were calculated according to established high-throughput RNAi screening data analysis and are described in the Supporting Information.
Pre-Processing Screen Data
Normalize fluorescence to Hoechst (cell count) and transform to log scale.
Remove intrawell outlier measurements.
Average intrawell measurements.
Screen Hit Scoring Metrics Using Strictly Standardized Mean Difference (SSMD)
Calculate SSMD-based assay quality score for a moderate control.
Calculate paired SSMD score.
Threshold paired SSMD score based on strong effect (≥2).
Calculate average % increase in surface and total protein.
Calculate siRNA kinase target + VX-809 hit interaction via two-way ANOVA.
Results
High-Throughput Assay Development
FAP-CFTR Platform
FAP technology is a fluorogenic labeling approach that uses high specificity binding to activate the fluorescence of a “dark” dye molecule. The FAP platform consists of a single chain antibody fragment, the size of GFP, fused to the protein of interest. The dL5** FAP complex has subnanomolar affinity binding toward malachite green (MG)-based fluorogens. The fluorogenic MG analog dyes become fluorescent only once bound to the dL5**FAP, which eliminates the need for wash steps. This tool system is dynamic due to the ability to create different compositions of MG fluorogen analogs (Scheme 1), allowing for selective labeling of different subcellular pools of FAP-fused proteins.25 F508del-CFTR and WT CFTR were fused with FAP (dL5**) at the N-terminus through an added membrane-spanning segment (PDGFR-TM, derived from pDisplay (Life Technologies, Inc.)), and they were stably expressed in HEK293 cells, illustrated in Figure 1. These constructs and cell populations have been previously established and validated.24
Scheme 1. Chemical Structure of MG-B-Tau (1), MG-2p (2), MG-11p (3), MG-Ester (4), and MGnBu (5).
Scheme adapted from Yan, Q. et al., 2015.26
Cell Surface Protein and Total Protein Labeling
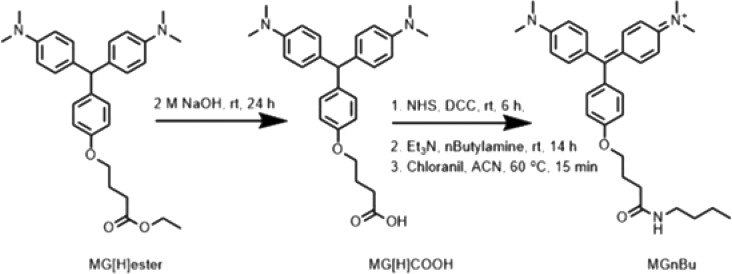
With FAP-CFTR expressing cells, we used MG-B-Tau, a cell excluded sulfonated MG analog, as a method to label extracellular FAP-F508del-CFTR. MG-B-Tau has been reported previously for its selective and rapid surface labeling.26 After labeling and measuring surface protein, we are able to sequentially label intracellular protein and measure total protein in the same cells. Measuring both surface and total protein allows us to consider the % protein present at the cell surface. For labeling intracellular protein, MG-Ester has been the standard cell permeable dye commonly used in FAP technology.27 However, MG-Ester has a lower quantum yield in comparison to MG-B-Tau (Table 1). To achieve an accurate representation of cellular total protein, the cell excluded dye and cell permeable dye (both 640 nm ex/680 nm em) must have matched quantum yields and spectral properties. To label intracellular CFTR protein, we prepared and validated a new dye, MGnBu (Scheme 1, Scheme 2, Scheme S1, and Figures S1–S5), which has a similar quantum yield to MG-B-Tau. We screened various MG variants where the fluorogen had a different functional tail distal to the fluorogenic moiety, which structural studies indicated was oriented outside the binding cleft.28 MGnBu, containing a butanamide linker through conjugating a n-butylamine functional group to the carboxylic acid on the MG fluorogen, showed a 50% increase in the quantum yield compared to that of MG-Ester and was identical in brightness and spectral properties to MG-B-Tau (Table 1). In addition, the overall positive charge of MGnBu causes the fluorogen to be cell-permeable. These properties, enhanced quantum yield and cell-permeability, make this dye an ideal cell-permeant chase reagent, allowing initial cell surface quantitation with the cell excluded MG-B-Tau, followed by total protein measurement by using a spectrally and quantum-yield matched cell-permeant dye to label remaining sites. The final complete dye-protein labeled cell reports total protein through measuring MG-B-Tau (surface) + MGnBu (inside) 680 nm emission. Sequential reads are used to establish the relative fraction of protein at the surface and within intracellular biosynthetic and endocytic compartments. In addition, the amide moiety is less susceptible to cleavage by esterases or media conditions, and may be useful for more reliable labeling in complex specimens or thick tissues.
Table 1. Dye Comparisons (1–5) with Reported Quantum Yield and Kda.
| Quantum Yield | Kd | |
|---|---|---|
| Dye | Φf | (nM) |
| MG-B-Tau (1) | 0.19 | 0.54 ± 0.04 |
| MG-2p (2) | 0.20 | 0.50 ± 0.35 |
| MG-11p (3) | 0.08 | 0.11 ± 0.02 |
| MG-Ester (4) | 0.12 | 0.42 ± 0.05 |
| MGnBu (5) | 0.20 | 0.40 ± 0.35 |
1–4 dye data adapted from Yan et al.26
Scheme 2. Synthesis of MGnBu (5).
In addition to spectral equivalence, MG-B-Tau and MGnBu must also have equal brightness in cellular labeling. Their equivalent brightness permits sequential labeling with cell impermeant and cell permeant dyes to measure surface and total protein, as qualitatively demonstrated previously with two-color fluorogen labeling.29 Flow cytometry-based fluorescence measurements with HEK293 cells expressing FAP-WT-CFTR demonstrated the identical brightness of MG-B-Tau and MGnBu, and the comparable challenges with MG-ester. Cells were first measured for surface fluorescence by MG-B-Tau labeling, followed by remaining protein labeling with MG-ester or MGnBu. Sequentially labeled samples were compared to samples labeled exclusively with 500 nM MGnBu dye or 500 nM MG-Ester alone which labeled both surface and internal pools of protein, at a brightness indicative of the cell permeant dye (Figure 3). There was no significant difference between the two methods of measuring the total protein using MGnBu, validating that in vivo the brightness of the two dyes are matched. However, MG-Ester showed a significantly reduced signal relative to the combined labeling, confirming that the higher intrinsic brightness of MG-B-Tau relative to MG-Ester compromises quantitative assessment of surface and total pools.
Figure 3.

Cell permeable dye, MGnBu, has the same quantum yield as the cell impermeable dye, MG-B-Tau, in FAP-WT-CFTR cells. Fluorescence signal is normalized to surface fluorescence (MG-B-Tau labeling). Total expression is shown in two different ways: (1) Measuring total expression through only labeling with cell permeable dyes, MG-Ester, or MGnBu. (2) Sequential labeling with first 500 nM MG-B-Tau (surface) for 15 min followed by another 15 min incubation with 500 nM MGnBu or MG-Ester (intracellular). MGnBu has the same quantum yield as the cell impermeable dye, MG-B-Tau, at 37 °C in FAP-WT-CFTR due to nonsignificant differences in labeling methods, however MG-Ester shows significant differences in the measurement of total protein. Data are expressed as the mean ± SD (4 replicates). One-way ANOVA with multiple comparisons. **P ≤ 0.01; *P ≤ 0.05; ns > 0.05.
A measurable and quantifiable difference in surface expression of FAP-F508del-CFTR can be obtained using a whole-well plate-reader format assay with fluorogen detection. HEK293 cells expressing FAP-F508del-CFTR were plated in a 96-well high optical quality ibidi plate. The following day cells were treated with or without 10 μM VX-809 for 24 h at 27 or 37 °C. For surface labeling, 500 nM MG-B-Tau in HBSS was added to the cells followed immediately by fluorescence measurement on a plate reader (Figure 4). Incubation (37 °C) with VX-809 treatment showed a small, but significant increase in fluorescence (∼20%). As previously shown, 27 °C incubation with VX-809 induced a dramatic increase in surface signal.
Figure 4.

Measurable surface protein fluorescence differences with ΔF508-CFTR in a 96-well plate reader format. Cells were either incubated at 37 and 27 °C, with or without 10 μM VX-809, for 24 h. Fluorescence was normalized to 37 °C ΔF508-CFTR treated with vehicle control, DMSO. Data are expressed as the mean ± SD of 6 replicates. One-way ANOVA with multiple comparisons. ****P ≤ 0.0001; ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
Hoechst 33342 nuclear stain was added to normalize for any potential differences in cell confluency across wells that may arise from transfection or the action of kinases on cell proliferation. Surface and total measurements were normalized to Hoechst stain. This high-throughput assay for conducting fluorescence measurements is illustrated and described in Figure 2. This simple plate reader-based, 1 h assay was applied as a method for quickly processing 96-well plates in a kinase knockdown screen using an siRNA library.
Identification of Kinases Whose Repression Promotes Surface Expression of ΔF508-CFTR Beyond That of VX-809 Alone
Using the high-throughput FAP-based assay, we screened the Dharmacon ON-TARGETplus SMARTpool siRNA Kinase library (715 target kinases) with and without 10 μM VX-809 treatment in triplicate at 37 °C to determine surface and total cellular expression of the F508del-CFTR construct. Using this data, we identified kinase targets whose suppression results in increases of surface F508del-CFTR above the level obtained by VX809 alone.
SiRNA Screen Quality Assessment
The quality control (QC) of the siRNA screen was assessed through a strictly standardized mean difference (SSMD) score.30,31 This scoring method is favorable due to its ability to report consistent quality control results with positive controls that are not necessarily very strong. In our system, the positive control for transfection is siRNA that targets CFTR, with success resulting in a clear reduction in total fluorescence signal measured upon addition of MGnBu. The total amount of CFTR, however, is reduced due to the F508del mutation, and thus even with VX-809 treatment, total protein expression is low, making knockdown to background levels only a moderate change in total fluorescence. Our QC criterion was based off of a moderate control effect, assessing the VX-809-treated plate of a transfection pair, and using the CFTR siRNA wells as a positive control for total protein knockdown and scrambled NC siRNA wells as a negative control. The siRNA kinase library was supplied in a 96-well format consisting of 9 plates total, where each library plate was utilized in true biological triplicate assays for both treatment conditions (with DMSO and VX-809). A plate passes QC if it has SSMD < −1 (good or excellent) quality. The QC results are presented in Figure 5, showing that all replicates passed our QC criterion for effective transfection.
Figure 5.

Quality assessment of transfection throughout siRNA screen. The assay quality scores for each 96-well library plate and their respective replicates. Thresholds for a moderate control: excellent: SMMD ≤ −2; good: −2 < SMMD ≤ −1; inferior: −1< SMMD ≤ −0.5; poor: SSMD>-0.5. Only one replicate of plate 9 borders between good and inferior at −1.
Initial Hit Selection
Paired SSMD and mean fold change in CFTR surface expression were used to rank initial hits, identifying the kinase targets whose suppression increases surface expression of F508del-CFTR. The paired SSMD score was implemented from Zhang,30 and is ideal for processing RNAi screens with replicates. The SSMD score is the average log fold change penalized by variability. Similar to QC, a threshold of SSMD ≥ 2 (strong positive effect) was set to identify kinase target candidates whose knockdown increased CFTR surface levels. The SSMD score vs average log fold change of each target can be visualized in a flashlight plot (Figure 6) to show the spread of effect size and assay statistical quality.
Figure 6.
Flashlight plot thresholds of kinase screen surface florescence change. SSMD thresholds are set at 2 and the area below SSMD score 2 is grayed out. The negative control (nontargeted, scrambled siRNA) in each plot is represented at 0. The line on the mean fold change axis represents the threshold that yields a significant positive change from the neg. ctrl., the area below this threshold is grayed out. (A) VX-809 flashlight plot where interesting targets are those above the visualized thresholds set from the VX-809 neg. ctrl. (B) DMSO (vehicle treated) flashlight plot shows that some siRNA treatments without VX-809 treatment may increase F508del-CFTR surface expression.
VX-809 + kinase targets above the selected threshold score were ranked based on their observed average % increase in surface expression from the VX-809 negative control. All hits that were significantly higher than the VX-809 negative control were selected (one-way ANOVA, P ≥ 0.05). The highest scoring, significant hits are shown in Figure 7. Surface signal of target-specific siRNA transfected cells treated with VX-809 are shown in red, and siRNA-transfected cells treated with DMSO vehicle control are shown in black. The negative controls were treated with nontargeted, scrambled siRNA. The screen had a primary focus for identifying VX-809 + siRNA kinase target hits that increased surface CFTR expression significantly above VX-809 treatment alone. Any kinase target + VX-809 treatment that produced a significant interaction, where the combination is higher than the sum of their individual treatments, are identified alongside hits as P-value symbols in blue. We also assessed DMSO + siRNA kinase targets and classified hits compared to the DMSO negative control. These effects were generally weaker, suggesting that many may be below statistical significance, but targeting the FGFR4 kinase in both the VX-809-treated and DMSO-treated cell screens showed a significant increase in surface expression. These results are represented in Figure 8, depicting the distribution of these kinase targets.
Figure 7.

Ranked surface signal increase of identified kinase targets + VX-809 hits. SiRNA kinase targets + VX-809 with a score of SSMD ≥ 2, and are significantly higher than VX-809 control (one-way ANOVA, P ≥ 0.05), are ranked based on their average % surface increase from DMSO negative control. SiRNA targets treated with VX-809 are shown in red, and targets treated with DMSO vehicle control are shown in black. Kinase targets in combination with VX-809 treatment that promote surface rescue, beyond VX-809 alone, are shown with blue asterisks. Error bars are shown as mean ± SEM (3 replicates). Two-way ANOVA for enhancing interaction (blue P-values). ****P ≤ 0.0001; ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
Figure 8.

Pie-graph illustrating independent and overlapping hits. VX-809 + siRNA target hits are those showing a strong increase in surface expression from VX-809 negative control, and DMSO + siRNA target hits are those showing a strong, significant increase in surface expression from DMSO negative control.
Identified targets can be further evaluated by comparing surface vs total CFTR protein in respect to basal F508del-CFTR expression (Figure 9). The total protein measurements provide information on overall protein expression levels in addition to showing if hits shifted distribution of protein. Overall, the surface vs total protein plot shows a loose, positive linear trend. Targets above the diagonal may represent kinases whose inhibition would increase surface targeting of the F508del-CFTR protein without a concomitant increase in overall protein expression.
Figure 9.

Average % surface vs total signal in VX-809-treated siRNA kinase target knockdown. Average % surface and % total protein increase are normalized to F508del-CFTR DMSO negative control. Listing top VX-809 + siRNA % surface increase hits. Targets above the grayed area are those that increased the amount of total protein expressed at the surface from DMSO negative controls. Mean values from screening (3) replicates are plotted.
Select Kinase Target Validation
Single siRNA CAMKK1 and RAF1 Validation in Flow Cytometry
Following the screen, the two kinases, CAMKK1 and RAF1, were selected for further validation. These targets were chosen based upon their significant enhancement of VX-809 and the availability of specific inhibitors. We confirmed the select kinase knockdown in the SMARTpool siRNA library by cherry picking those wells for transfection and performing immunofluorescence (Figure S6). To demonstrate agreement with the plate-reader results, which measures on a population scale, we performed confirmation experiments on a BD Accuri flow cytometer with Intellicyt HTFC for single-cell quantitative measurements. For flow-cytometry assays, after treatment, cells are plated on a 96-well plate and treated with 500 nM MG-B-Tau in HBSS for 15 min before measuring.
The initial validation experiment utilized single Dharmacon ON-TARGETplus siRNA vs the SMARTpool siRNA used in the screening process. The individual siRNA from the pools were used to confirm the significant increase in surface expression upon siRNA knockdown of CAMKK1 and RAF1. The SMARTpool is composed of 4 siRNAs, and the 4 individual siRNA results are shown in Figure 10, assessed by flow cytometry. All four CAMKK1 siRNA targets and one siRNA targeting RAF1, combined with VX-809, significantly increased surface expression relative to the VX-809-scrambled siRNA control.
Figure 10.

RAF1 and CAMKK1 single siRNA validation in flow cytometry. RAF1 and CAMKK1 single Dharmacon ON-TARGETplus siRNAs with or without VX-809 treatment. Measured surface CFTR fluorescence with a BD Accuri flow cytometer with Intellicyt HTFC. The grayed out region represents VX-809s average surface fluorescence. VX-809 + siRNA significance is shown from VX-809 negative control. Data are shown as mean ± SEM (3–4 replicates). One-way ANOVA with multiple comparison for significant changes from VX-809 neg. ctrl (0 nM) (red P-values). ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
CAMKK1 and RAF1 Drug Inhibition Validation in Flow Cytometry
Using an independent flow cytometry method, individual siRNAs showed agreement with screening results. Further validation was conducted with specific kinase inhibitors (Figure 11). For these studies, we used GW 5074, a drug that acts on Raf1, previously shown to enhance function of F508del-CFTR14 and STO-609, a known selective inhibitor of CAMKK1. Kinase inhibitor experiments were carried out via flow cytometry (Figure 12). The flow cytometry fluorescence surface measuring assay produced confirmatory results. In combination with VX-809, the CAMKK1 inhibitor, STO-609, and RAF1 inhibitor, GW 5074, both showed a significant increase in surface signal from the VX-809 only control with a significant interaction.
Figure 11.
Chemical structures of inhibitors of two identified kinase targets, RAF1 and CAMKK1.
Figure 12.

Measuring surface fluorescence changes using select kinase drug inhibitors with a plate reader and flow cytometry method. Kinase inhibitors, GW 5074 and STO-609, with VX-809 treatment showed an increase in surface fluorescence using a BD Accuri flow cytometer with Intellicyt HTFC. VX-809 + kinase inhibitor is shown normalized to DMSO-treated control cells. The grayed out region represents VX-809s average surface fluorescence. Each drug showed significant interaction effects. Data are shown as mean ± SEM (3 or more replicates). One-way ANOVA with multiple comparisons for significant changes from VX-809 control (0 nM) (red P-values). Two-way ANOVA was used for measuring interaction. ****P ≤ 0.0001; **P ≤ 0.01.
Discussion
We have developed a new assay that selectively and quantitatively assesses the cell surface expression and overall protein content of membrane proteins in the same population of cells, and applied this assay to identify potential kinase targets that may enhance F508del- CFTR rescue to the plasma membrane in combination with VX-809 treatment. To determine the overall total protein levels, we have introduced a new cell-permeant fluorogenic dye, MGnBu, that possesses the same quantum yield and spectral properties as the surface labeling dye, MG-B-Tau, to determine the ratio of surface exposed protein to total expressed protein. This assay utilizes a simple plate-reader format for accelerated screen processing, where surface and total protein measurements are readily transitioned to other instruments, such as flow cytometry or microscopy. The ability to readily collect complementary fluorescence measurements across different devices, while using a single established labeling approach, corroborates results readily. Through our developed screening assay, we identified several targets whose inhibition showed increased CFTR rescue with VX-809 treatment using a SSMD scoring metric. Of these hits, we used a two-way ANOVA interaction test to determine targets that resulted in a significantly higher CFTR surface localization than that of either siRNA or VX-809 treatment alone, or the expected sum of the treatments. Kinase targets, CAMKK1 and RAF1, VX-809 enhancing effects were further validated using single siRNA knockdown and kinase inhibitors, and were evaluated utilizing flow cytometric measurements. We have shown a high-throughput approach that uses both surface and total measurements that was applied to screening potential kinase targets that show an enhancement of VX-809 F508del-CFTR rescue. This platform could also be used for other cell surface protein trafficking related screens that require understanding the fraction of total protein at the surface and robust measurements to assess or control for proteostatic effects.
Acknowledgments
This work was supported in part by a pilot grant from the Cystic Fibrosis Research Center at University of Pittsburgh (5P30DK072506), the Shurl and Kay Curci Foundation, and NIH grant (R01EB017268). L.A.P. was supported by a Presidential Fellowship at CMU. We would like to thank Gunnar Atli Sigurdsson for statistical advice, Professor Jon Jarvik for the FAP-CFTR cell line, and Professor Ray Frizzell for extensive discussion and advice about CFTR assays and trafficking.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.molpharmaceut.7b00928.
Synthesis and characterization of MGnBu, MGnBu photophysical properties, analysis and statistics, and select siRNA kinase knock-down immunofluorescence (PDF)
The authors declare the following competing financial interest(s): Marcel Bruchez is a founder in Sharp Edge Labs, a company using FAP technology for drug discovery.
Supplementary Material
References
- Ratjen F.; Döring G. Cystic fibrosis. Lancet 2003, 361, 681–689. 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- Cheng S. H.; et al. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 1990, 63, 827–834. 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Kerem E. Pharmacologic therapy for stop mutations: how much CFTR activity is enough?. Curr. Opin. Pulm. Med. 2004, 10, 547–552. 10.1097/01.mcp.0000141247.22078.46. [DOI] [PubMed] [Google Scholar]
- Amaral M. D.; Farinha C. M. Rescuing mutant CFTR: a multi-task approach to a better outcome in treating cystic fibrosis. Curr. Pharm. Des. 2013, 19, 3497–3508. [DOI] [PubMed] [Google Scholar]
- Okiyoneda T.; et al. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 2010, 329, 805–10. 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy J. P.; et al. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax 2012, 67, 12–18. 10.1136/thoraxjnl-2011-200393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Two Phase 3 Studies of the Tezacaftor/Ivacaftor Combination Treatment Met Primary Endpoints with Statistically Significant Improvements in Lung Function (FEV1) in People with Cystic Fibrosis. Business Wire, http://www.businesswire.com/news/home/20170328006545/en/.
- Cholon D. M.; Quinney N. L.; Fulcher M. L.; Esther C. R.; Das J.; Dokholyan N. V.; Randell S. H.; Boucher R. C.; Gentzsch M. Potentiator Ivacaftor Abrogates Pharmacological Correction of ΔF508 CFTR in Cystic Fibrosis. Sci. Transl. Med. 2014, 6, 246ra96. 10.1126/scitranslmed.3008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit G.; et al. Some gating potentiators, including VX-70, diminish DeltaF508-CFTR functional expression. Sci. Transl. Med. 2014, 6, 246ra97. 10.1126/scitranslmed.3008889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertex Announces Significant Progress in Its Development Efforts to Treat the Cause of Cystic Fibrosis in the Vast Majority of People with the Disease. Business Wire, http://www.businesswire.com/news/home/20151008005754/en/Vertex-Announces-Significant-Progress-Development-Efforts-Treat.
- Lin S.; et al. Identification of synergistic combinations of F508del cystic fibrosis transmembrane conductance regulator (CFTR) modulators. Assay Drug Dev. Technol. 2010, 8, 669–684. 10.1089/adt.2010.0313. [DOI] [PubMed] [Google Scholar]
- Cholon D. M.; Esther C. R.; Gentzsch M. Efficacy of lumacaftor-ivacaftor for the treatment of cystic fibrosis patients homozygous for the F508del-CFTR mutation. Expert Review of Precision Medicine and Drug Development 2016, 1, 235. 10.1080/23808993.2016.1175299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuan P.-W.; et al. Synergy-based Small-Molecule Screen Using a Human Lung Epithelial Cell Line Yields F508-CFTR Correctors that Augment VX-809 Maximal Efficacy. Mol. Pharmacol. 2014, 86, 42–51. 10.1124/mol.114.092478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzcinska-Daneluti A. M.; et al. Use of kinase inhibitors to correct ΔF508-CFTR function. Mol. Cell. Proteomics 2012, 11, 745–57. 10.1074/mcp.M111.016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzcińska-Daneluti A. M.; et al. RNA Interference Screen to Identify Kinases That Suppress Rescue of ΔF508-CFTR. Mol. Cell. Proteomics 2015, 14, 1569–83. 10.1074/mcp.M114.046375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedemonte N.; Zegarra-moran O.; Galietta L. J. V. High-Throughput Screening of Libraries of Compounds to Identify CFTR Modulators. Methods Mol. Biol. 2011, 741, 13–21. 10.1007/978-1-61779-117-8_2. [DOI] [PubMed] [Google Scholar]
- Pedemonte N.; et al. Small-molecule correctors of defective DeltaF508-CFTR cellular processing identified by high-throughput screening. J. Clin. Invest. 2005, 115, 2564–71. 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor F.; Straley K.; et al. Rescue of DF508-CFTR trafficking and gating in human cystic fibrosis airway primary cultures by small molecules. Am. J. Physiol Lung Cell Mol. Physiol 2006, 290, 1117–1130. 10.1152/ajplung.00169.2005. [DOI] [PubMed] [Google Scholar]
- Trzcinska-Daneluti A. M.; et al. High-content functional screen to identify proteins that correct F508del-CFTR function. Mol. Cell. Proteomics 2009, 8, 780–90. 10.1074/mcp.M800268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile G. W.; et al. Correctors of protein trafficking defects identified by a novel high-throughput screening assay. ChemBioChem 2007, 8, 1012–20. 10.1002/cbic.200700027. [DOI] [PubMed] [Google Scholar]
- Botelho H. M.; et al. Protein Traffic Disorders: an Effective High-Throughput Fluorescence Microscopy Pipeline for Drug Discovery. Sci. Rep. 2015, 5, 9038. 10.1038/srep09038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran J. P.; et al. Pharmacological rescue of the mutant cystic fibrosis transmembrane conductance regulator (CFTR) detected by use of a novel fluorescence platform. Mol. Med. 2012, 18, 685–696. 10.2119/molmed.2012.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. B.; Hu J.; Frizzell R. A.; Watkins S. C. Simple image-based no-wash method for quantitative detection of surface expressed CFTR. Methods 2016, 96, 40–45. 10.1016/j.ymeth.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran J.; et al. Fluorogen-Activating Proteins as Biosensors of Cell-Surface Proteins in Living Cells. Cytometry, Part A 2011, 76, 1358–1375. 10.1002/cyto.a.20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi C.; et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat. Biotechnol. 2008, 26, 235–240. 10.1038/nbt1368. [DOI] [PubMed] [Google Scholar]
- Yan Q.; et al. Near-instant surface-selective fluorogenic protein quantification using sulfonated triarylmethane dyes and fluorogen activating proteins. Org. Biomol. Chem. 2015, 13, 2078–2086. 10.1039/C4OB02309A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telmer C. A.; et al. Rapid, specific, no-wash, far-red fluorogen activation in subcellular compartments by targeted fluorogen activating proteins. ACS Chem. Biol. 2015, 10, 1239–46. 10.1021/cb500957k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi C.; et al. Malachite Green Mediates Homodimerization of Antibody VL Domains to Form a Fluorescent Ternary Complex with Singular Symmetric Interfaces. J. Mol. Biol. 2013, 425, 4595–4613. 10.1016/j.jmb.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt C. P.; He J.; Wang Y.; Barth A. L.; Bruchez M. P. Fluorogenic Green-Inside Red-Outside (GIRO) Labeling Approach Reveals Adenylyl Cyclase-Dependent Control of BKα Surface Expression. Bioconjugate Chem. 2015, 26, 1963–1971. 10.1021/acs.bioconjchem.5b00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. D. Illustration of SSMD, z Score, SSMD*, z* Score, and t Statistic for Hit Selection in RNAi High-Throughput Screens. J. Biomol. Screening 2011, 16, 775–785. 10.1177/1087057111405851. [DOI] [PubMed] [Google Scholar]
- Zhang X. D. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J. Biomol. Screening 2011, 16, 775–85. 10.1177/1087057111405851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.