Abstract
Efficient phagocytosis of apoptotic cells (efferocytosis) is essential for immune homeostasis. Phospholipids exposed on the surface of apoptotic cells, for example phosphatidylserine, supply important “eat-me” signals. Liposomes are lipid bilayer vesicles that can be generated with one or several phospholipids of interest. Thus, they offer versatility, flexibility and, importantly, a three-dimensional structure, for studying the interaction between lipids and their receptors, and the lipid-receptor interaction-mediated signaling events controlling efferocytosis by cells like professional phagocytes. Here, we describe methods to prepare liposomes, perform liposome-based lipid-receptor binding assays, use liposomes to block efferocytosis, and utilize liposome-coated beads as apoptotic cell surrogates for phagocytosis.
Keywords: lipid-receptor interaction, phospholipids, liposomes, liposome-coated beads, efferocytosis
Introduction
This unit describes four protocols that discuss the preparation of liposomes containing specific phospholipids and liposome-coated beads, analysis of interaction between phospholipids and receptors, and the use of liposomes as blocking reagents, or liposome-coated beads as apoptotic cell surrogate to study phagocytosis.
Basic protocol I describes the preparation of liposomes. This method is based on a combination of sonication (Choi et al., 2011) and the freely available extrusion-based protocols from Avanti Tech Support, and has been optimized to give high-quality liposomes. The protocol also includes the preparation of fluorescent- or biotin-labeled liposomes, which is further discussed in protocol II and III.
Basic protocol II describes how to coat magnetic beads with phospholipid-containing liposomes. Biotin-conjugated phospholipids are incorporated into liposomes, and then the biotinylated liposomes are captured onto streptavidin magnetic beads. The coated liposomes are evaluated for the phospholipid presence with fluorescent-labeled probes, using flow cytometry and confocal microscopy.
Basic protocol III describes two methods to analyze the binding of phospholipids to their receptors. First method describes the analysis of phospholipid-receptor binding kinetics using surface plasmon resonance (SPR). The liposomes are immobilized onto a L1 sensor chip, and the binding of a soluble receptor is recorded. The second method describes the analysis of fluorescent-labeled liposome binding to the cell surface, using flow cytometry.
Basic protocol IV illustrates the use of liposomes as competitive inhibitors to block phagocytosis of apoptotic cells, and the use of liposome-coated beads as apoptotic cell surrogates for phagocytosis assays. The former method utilizes flow cytometry to analyze phagocytosis of pHrodo-labeled apoptotic cells in the presence or absence of phospholipid-containing liposomes. In the latter method, the phagocytosis of liposome-coated beads is analyzed by confocal microscopic imaging via the acquisition of multiple optical sections, and the quantification of the 3D images.
Although these protocols were primarily developed to study to role of synthetic phospholipid-containing liposomes during efferocytosis, they are also applicable to natural phospholipid-containing liposomes in other physiological studies.
Basic Protocol I
PREPARATION OF PHOSPHOLIPID-CONTAINING LIPOSOMES
This protocol describes the preparation of liposomes containing one or more phospholipids. Briefly, liposomes are prepared by evaporating chloroform from the desired phospholipid mixture with argon gas. Then, phospholipids are reconstituted in PBS, pH 7.4, at a concentration of 5–10 mg/ml. Small unilamellar vesicles, composed of synthetic phospholipids, or ceramide are formed by extrusion through filters with 0.1 µm pore size, using an Avanti Mini-Extruder. Using this protocol, unilamellar liposomes of about 100 nm can be generated.
Materials
Degassed phosphate-buffer saline (PBS, without Ca2+ or Mg2+; Lonza, cat. no. 17-516F)
10 mg/ml 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti Polar Lipids, cat. no. 850375C)
10 mg/ml 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS, Avanti Polar Lipids, cat. no. 840035C)
10 mg/ml 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE, Avanti Polar Lipids, cat. no. 850725C)
10 mg/ml 1,2-dielaidoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (DOPG, Avanti Polar Lipids, cat. no. 840477C)
5 mg/ml ceramide (Avanti Polar Lipids, cat. no. 860524P)
Avanti Mini-Extruder (Avanti Polar Lipids, cat. no. 610000)
Nucleopore Track-Etched Polycarbonate 0.1 µm pore size (Whatman, cat. no. 800309)
Filter supports (Avanti Polar Lipids, cat. no. 610014)
Argon gas (Although non-toxic, Argon gas is considered a dangerous asphyxiant and appropriate safety precautions should be considered.)
Ice
Forceps
Glass vials (Thomas Scientific, cat. no. 1234R80)
Glass syringe
Vacuum desiccator
Chloroform (This volatile liquid is a suspected carcinogen and reproductive toxin and appropriate safety precautions should be considered)
Gas dispersion tubes with fritted cylinders (Fisher scientific)
Sonicator
Vacuum
Side-arm flask
Lipid preparation
-
1
Dissolve lipids that come in a powdered form, e.g. ceramide, using chloroform and a glass syringe.
The volume of added chloroform depends on the solubility of the lipid; for example, most listed lipids are soluble at a concentration of 10 mg/ml, while others, like ceramide, have a lower solubility and the maximal concentration is 5 mg/ml or less. Please refer to the provided datasheet from Avanti Polar Lipids to find the solubility limit for a particular lipid of interest.
-
2
Incubate lipid suspensions in glass vials for ~ 10 min at 37°C to facilitate homogenization.
-
3
Mix the prepared homogenized lipid suspensions at a ratio of 80% DOPC and 20% of either DOPS, DOPE, DOPG or ceramide.
Transferring of the lipid suspensions requires the use of glass syringes. Also, rinsing the glass syringes three times with chloroform is strongly recommended when different phospholipids need to be transferred. The lipid ratio of the liposomes dependents on an experimental design; however, it is important to note that each liposome can be made of one type of phospholipid (e.g. 100% DOPC or 100% DOPS).
-
4
Evaporate the solvents from all lipid suspensions, whether freshly made or from vials that already contained the lipid suspensions, for 30 min under argon gas to form a dry film.
-
5
Place all glass vials in a vacuum chamber for at least 12h to remove all remaining traces of the solvent.
Removing all traces of the solvent is critical, as solvent remnants will interfere with the formation of the liposomes.
Liposome formation
-
6
Resuspend the lipids using 1× degassed PBS (pH 7.4) to obtain a 1–10 mg/ml lipid suspension.
Degassing PBS is necessary to remove excess of air from the liquid and reduce the air bubble formation in the mixture. To degas the PBS, transfer PBS to a sterile side-arm flask and seal top of flask with a rubber stopper. Connect the flask’s side-arm to a vacuum pump, and turn on the vacuum at a low rate. When no further bubbling occurs, disconnect the hose and turn off the vacuum. Avoid vigorous shaking of the PBS after degassing, to prevent reintroduction of air to the buffer.
-
7
Incubate lipid suspensions for ~ 10–30 min at 37°C to facilitate homogenization.
The implementation of gentle vortexing every 5 min can help to facilitate the homogenization process.
-
8
Sonicate the lipid suspensions for 10 min with one-minute gap intervals.
Mini-Extruder assembly
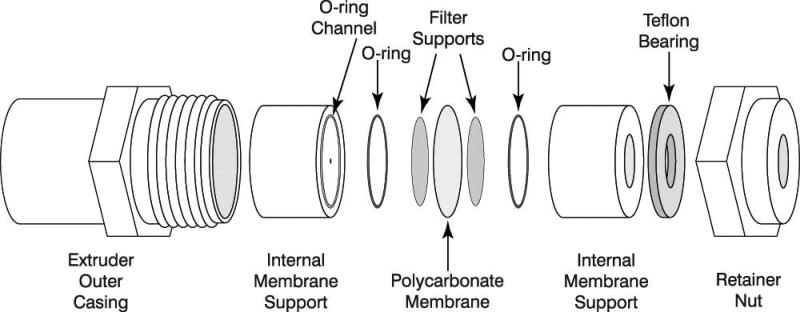
All used parts necessary for the assembly of the Mini-Extruder are highlighted in Figure 1.
-
9
Take two Internal Membrane Supports and place them on a flat surface with the O-rings facing up.
-
10
Take two Filter Supports and pre-wet them in 1× PBS (pH 7.4), and place them on top of the teflon orifice inside the O-ring inner diameter.
-
11
Place the Internal Membrane Support together with the Filter Support into the Extruder Outer Casing with the O-ring facing up.
-
12
Place one Polycarbonate Membrane in the Extruder Outer Casing over the Filter Support and O-ring.
The membrane is protected by blue paper disks, which need to be removed from the Polycarbonate Membrane before installment. Polycarbonate Membrane and Filter Support can only be used once, and should not be reused.
-
13
Take a second pair of Filter Supports, pre-wet them in 1× PBS (pH 7.4), and place them on top of the remaining Internal Membrane Support.
-
14
Gently take the second Internal Membrane Support and place it into the casing with the O-ring facing down.
-
15
Place the teflon bearing into the Retainer Nut.
-
16
Take the Retainer Nut and place it onto the threaded end of the Extruder Outer Casing and gently tighten.
The tighten of the Retainer Nut is accomplished by hand and does not requirement any tools, e.g. a wrench.
Figure 1.
Diagram of the parts involved of the Mini-Extruder assembly (source www.avantilipids.com; used with permission of Avanti Polar Lipids).
Mini-Extruder extrusion
-
17
Load the sonicated sample into one of the gas-tight syringes. Gently take the lipid-containing syringe and insert it into one end of the mini-extruder.
-
18
Take the empty syringe and insert it into the open end of the mini-extruder.
Ensure that the empty syringe plunger is set to zero (i.e., fully inserted), because the syringe will fill as the lipid suspension is pushed (extruded) through the membrane.
-
19
Place the fully assembled extruder apparatus into the stand. Insert the hex nut so that the apex of the hex nut points down and ensure that the swing-arm clips hold the syringes in place (see Figure 2).
-
20
Gently push the plunger of the lipid-containing syringe until the suspension is fully transferred to the other syringe.
-
21
Repeat step 20 by pushing the suspension back to the original syringe.
-
22
Repeat steps 20 and 21 until the lipid suspension changes its color from milky to clear, an indication for liposome formation.
Approximately 20 passes through the membrane should be sufficient to achieve liposome formation.
-
23
Make sure that the final pass of the lipid suspension fills the second syringe (i.e., the one from point 18).
This step is important to avoid any contaminations with larger particles.
-
24
Gently detach the lipid-filled syringe from the extruder and transfer the lipid suspension into a clean glass vial.
Removing of the syringes requires a straight pulling out from the extruder to avoid damaging the syringe.
-
25
To avoid oxidation of the lipid solution the glass vial should be filled with argon gas for at least 2 min and sealed tightly using parafilm.
-
26
Lipid suspensions should be stored at 4°C and should never be frozen.
Using this liposome preparation protocol will result in liposomes of around 100 nm, which is easily measured by dynamic light scattering (DLS), a method previously described by Hupfeld et al., 2006 (Hupfeld, Holsaeter, Skar, Frantzen, & Brandl, 2006).
Lipid suspensions can be used for several months when lipids are kept in a saturated atmosphere with argon gas after each usage. However, it is highly recommended to periodically reevaluate the integrity of the liposome suspensions over time using DLS.
Figure 2.
Fully assembled extruder apparatus (source www.avantilipids.com; used with permission of Avanti Polar Lipids).
Cleaning and maintenance of the Mini-Extruder
-
27
Directly after use, gently rinse both syringes, Internal Membrane Support, Retainer Nut and Extruder Outer Casing with 1× PBS, 100% ethanol, and then with deionized water.
-
28
Air-dry all cleaned materials.
Syringes or other materials should not get in contact with any solvents other than water or alcohol to avoid damaging any parts of the Mini-Extruder. Also, autoclaving of any parts, especially the Teflon inserts, is not recommended.
Basic Protocol 2
PREPARATION OF PHOSPHOLIPID-CONTAINING LIPOSOME-COATED BEADS
This protocol describes the preparation of phospholipid-containing-liposome-coated beads. Briefly, liposomes are prepared as described in basic protocol I, with the exception that biotin-labeled DHPE is incorporated into the liposome. The liposomes are conjugated to M-280 streptavidin-coated Dynabeads. After washing, the phospholipid-coated dynabeads can be stored in argon gas for future use.
Materials
Degassed Phosphate-buffer saline (PBS)
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti Polar Lipids, cat. no. 850375C)
1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS, Avanti Polar Lipids, cat. no. 840035C)
N-(biotinoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine triethylammonium salt (biotin-labeled DHPE, ThermoFisher Scientific, cat. no. B1616)
M-280 streptavidin-coated Dynabeads (ThermoFisher Scientific, cat. no. 11205D)
Bovine Serum Albumin (BSA; Gold Biotech)
Annexin V-APC (BioLegend, cat. no. 640920)
Liposome preparation and formation
Pre-incubate M-280 streptavidin-coated Dynabeads in 0.5 ml of degassed 1× PBS containing 1% BSA at 37 °C for 2 h.
Dissolve biotin-labeled DHPE in chloroform (~ 10 mg/ml).
Incubate suspensions in a glass vial for ~ 10 min at 37°C to facilitate homogenization.
-
Mix the prepared homogenized lipid suspensions at a ratio of 1% biotin-labeled DHPE and 99% DOPC for PC liposomes, and 1% biotin-labeled DHPE with 49% DOPC and 50% of DOPS for PS liposomes.
Of note, all percentages are calculated based on the weight of each liposome.
Evaporate the solvents from all lipid suspensions, whether freshly made or from vials that already contained the lipid suspensions, for 30 min under argon gas to form a dry film.
Resuspend the lipids using degassed 1 × PBS (pH 7.4) to obtain a ~ 10 mg/ml lipid suspension.
Incubate lipid suspensions for 10–30 min at 37°C to facilitate homogenization.
Prepare the PC- or PS-containing liposomes using the Mini-Extruder protocol (see Basic Protocol I)
Remove supernatant from the M-280 streptavidin-coated Dynabeads and mix beads with either PC or PS liposomes for 2 h at 37 °C in degassed 1× PBS containing 1% BSA.
Remove supernatants and wash 2 times with 1× PBS.
-
Remove supernatants and resuspend liposome-bead mixtures in 1× PBS.
To verify the coating of PS onto the beads, a small aliquot of liposomes-coated beads can be stained with an Annexin V-APC antibody and analyzed by flow cytometry. Also, like liposomes, liposome-coated beads need to be stored in argon gas to prevent lipid oxidization.
Basic Protocol 3
IDENTIFICATION OF LIPID-RECEPTOR INTERACTIONS BY SURFACE PLASMON RESONANCE
This protocol describes the interaction analysis between lipids and receptors using surface plasmon resonance (SPR). The lipid-containing liposomes are coated onto a sensor chip L1 followed by the injection of various concentrations of Fc-tagged extracellular domains of the receptor (lipid receptor-Fc fusion). The binding signals are recorded and the association kinetics are analyzed using the BIAevaluation Software.
Materials
BIAcore system (Biacore Inc.)
BIAevaluation Software (Biacore Inc.)
Series S Sensor chip L1 (GE Healthcare, cat. no. 29-1049-93)
1 mM liposome solutions (DOPC, DOPS, DOPE, DOPG, or ceramide, see Basic Protocol I)
Running buffer/protein dilution buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2)
Dissociation/Regeneration buffer: running buffer containing 2.5 M NaCl, 5 mM EDTA but no CaCl2)
Degassed Phosphate-buffer saline (PBS)
HEPES
5 M Sodium chloride
2 M Calcium chloride
0.5 M EDTA
0.05 M Sodium citrate, pH 3
2 M Tris-HCL, pH 8.8
Isopropanol
Sodium hydroxide solution
lipid receptor Fc-fusion proteins (e.g. murine CD300s) at various concentrations (10 µg/ml is a good starting point)
HEK293T cells
T175 tissue culture flasks
DMEM medium
Fetal bovine serum (FBS) (Sigma)
Transfection reagent PolyJet (SignaGen, cat.no. SL100688)
(Lipofectamine 2000 or others are equal applicable)
Protein A-Sepharose (GE, cat.no. GHC-17-1279-01)
Econo-column (Bio-Rad, cat.no. 7371012)
Amicon Ultra-4 Filter Units (Millipore, cat.no. UFC801096)
Eppendorf tubes
Production of lipid receptor-Fc fusion proteins
-
1
Seed HEK293T cells into T175 flasks the day prior to the transfection (~ 20 ml of DMEM + 10% FBS is sufficient to cover the cells in the flask).
The number of flasks required to obtain ~ 0.5 mg of lipid receptor-Fc fusion proteins depends strongly on the expression level of the construct (pCDNA3.1 is the most commonly used expression vector) and has to be empirically tested. In general, a good expression construct would require 10 flasks to obtain this amount.
-
2
On the day of transfection, replace the old media from the flask with 20 ml of fresh DMEM + 10% FBS.
-
3
Prepare two transfection tubes. Tube 1 should contain 1 ml of DMEM (no serum) + 20 µg of plasmid DNA (e.g. pCDNA3.1 vector containing the gene encoding the lipid receptor-Fc DNA of interest), while tube 2 should contain 1 ml of DMEM (no serum) + 60 µl of PolyJet (Transfection reagent)
This reaction setup is enough for one T175 flask.
-
4
Mix the content of both tubes by gentle vortexing.
-
5
Transfer reaction from tube 1 into tube 2, and mix gently by vortexing.
-
6
Incubate the mixture for 15 min at room temperature.
-
7
Add the transfection mixture (from step 6) to the T175 flask with HEK293T cells (from step 2), and mix gently by hand. Place the cells in a humidified incubator with 5% CO2 at 37°C.
-
8
After 12h, replace the old media with 20 ml of fresh DMEM (no serum) and incubate the cells for additional 48h before collecting the cell culture medium.
Supernatants from cells transfected with the same plasmid DNA can be combined.
-
9
Centrifuge the cell culture media at 1000 × g for 20–30 min at 4°C to remove cell debris.
Purification of lipid receptor-Fc fusion proteins from cell culture medium
-
10
Load 2 ml of protein A-Sepharose into a fast flow column and wash for about 30 min with 1× PBS.
-
11
Load the entire volume of cell culture media supernatant (step 9) onto the column.
-
12
Wash again for 60 min with 1× PBS.
-
13
Elute the bound proteins with sodium citrate solution (pH 3), by adding it stepwise to the column, use 0.9 ml for each elution step, collect fraction into Eppendorf tubes and repeat this step ~ 8–10. Keep tubes on ice after eluent is collected.
-
14
Add 0.1 ml of 2 M Tris-HCL to each tube and mix gently.
This step is required to neutralize the very acetic elution condition into a more physiological buffer to avoid potential problems with protein functionality.
-
15
Analyze the collected fractions for the presence of the lipid receptor-Fc fusion proteins by SDS-page and Coomassie Blue staining.
A small aliquot (20–30 µl) of each eluted fraction should be sufficient to detect the fusion protein.
-
16
Combine all the fractions containing Fc-fusion protein, and concentrate the protein by centrifugation using the Amicon Filter Units.
-
17
Centrifuge the combined fractions at 1500 × g, 4°C until the solution reaches a volume of about 0.5 ml.
Time of centrifugation will vary dependent on the rotor size, temperature of the centrifuge, etc., and therefore should be tested prior to concentrating your protein.
-
18
Add 4.5 ml of 1× PBS to the concentrated Fc-fusion protein solution (from step 17), and centrifuge again until the solution reaches about 0.5 ml. Repeat this step twice.
This step is required to completely exchange the elution buffer to a more physiological buffer (i.e., PBS), and to avoid potential problems with protein functionality. PBS alone as protein storage buffer was sufficient due to the high stability of our proteins, however the addition of 10% glycerol is recommended for proteins with lower stability for the freezing and thawing process.
-
19
Determine the protein concentration, using your preferred method (e.g. the colorimetric Bradford protein assay at absorbance of 595 nm).
Protein concentration should be in the range of 1 – 5 mg/ml. If the protein concentration is lower, further centrifugation is recommended to concentrate the protein solution. Of note, this concentration step could lead to the aggregation of the protein when the concentration of the solution is too high.
-
20
Store proteins at −80°C for future use.
Analyses of the lipid receptor Fc-fusion protein-liposome interaction by SPR Conditioning and liposome binding to the Sensor Chip L1
-
21
Dock chip to the Biacore instrument.
Follow the chip docking instructions for the available BIAcore system.
-
22
Prepare liposome solution (0.5 – 1 mM) using the running buffer (see Materials for recipe).
Of note, liposome concentrations were determined using the Stewart assay as described by Stewart et al., 1980.
-
23
Precondition the chip surface with two 30 sec injections of Isopropanol/NaOH solution (2 parts Isopropanol and 3 parts 50 mM NaOH in the running buffer).
-
24
Inject liposome solutions (1 mM concentration is recommended) using a flow rate of 10 µl/min, and select the desired immobilization level (RUs).
Only three different lipid solutions can be compared on a single chip, because the first chamber of each chip is used as a negative control for binding. Also, it is recommended to use lower amounts of RUs of about 100–500 to quantitatively assess the binding between the lipid receptor Fc-fusion protein (or any other recombinant protein) and the liposomes.
-
25
When the desired immobilization level is reached, one 30 sec injection with the regeneration solution (see Materials) is applied to remove unbound vesicles, and to stabilize the base line prior to interaction analysis.
Interaction analysis between Fc-lipid receptor proteins and liposomes
-
26
Prepare various concentrations of each lipid receptor Fc-fusion protein in running buffer (see Materials). Perform serial dilutions starting from 1 µM down to 1 nM.
-
27
Select binding program (see instructions for your available BIAcore system); choose a flow rate of 10 µl/min, and a contact time of about 1 min for the injected lipid receptor Fc-fusion protein solution.
-
28
Select a regeneration phase of about 2–5 min between each binding analysis.
Ensure that the injection of the regeneration buffer allows for the RU levels to reach baseline. If regeneration conditions do not reach baseline, a stronger regeneration solution should be considered.
-
29
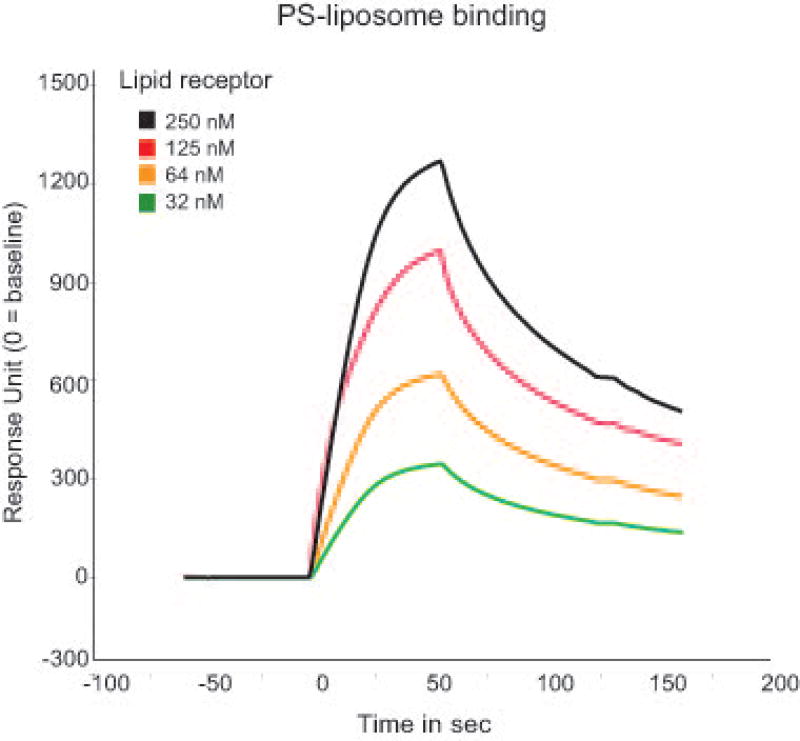
Analyze the association kinetics between lipid receptor Fc-fusion proteins and the different liposomes using the BIAevaluation Software (see Figure 3).
Of note, reliable kinetics and affinity analysis depend on various parameters, such as data acquisition of sufficient analyte concentrations, buffer composition, avidity effects etc. Therefore, experiments should be designed in such a way that the data can be interpreted in the simplest interaction model possible. A detailed review regarding these parameters is provided by Jason-Moller et al. 2006.
Figure 3.
Representative association kinetic profile between one lipid receptor protein at various concentrations (250 nM-32 nM) and PS liposomes using SPR.
Basic Protocol 4
FLUORESCENCE-LABELED LIPOSOME BINDING TO CELL SURFACE RECEPTORS
This protocol describes the analysis of binding of fluorescently-labeled liposome to a cell surface receptor. NPD-PC is incorporated into liposomes containing various phospholipids, and the binding between a liposome and a cell surface receptor is detected as the fluorescence signal from NPD, using the flow cytometry.
Materials
L929 cells
HEK 293T cells
Empty Lentiviral vector (e.g., pCDH-EF1-T2A-puro vector from System Biosciences)
Lentiviral vector encoding the receptor (e.g., pCDH-EF1-T2A-puro vector encoding murine CD300f)
Lentiviral packing helper plasmids (e.g., psPAX2 and pMD2G plasmids from System Biosciences)
PEG-it™ virus precipitation solution (System Biosciences, cat. no. LV810A-1)
10 cm tissue culture dishes (Denville)
Cell culture media: DMEM medium (Lonza)
Fetal bovine serum (FBS) (Sigma)
Transfection reagent PolyJet (SignaGen, cat.no. SL100688)
(Lipofectamine 2000 or others are equal applicable)
Puromycin (Sigma)
Trypsin
6 and 24-well plates
FACS tubes
Sterile 15 ml tubes
Bleach
Protamine sulfate (Sigma)
Antibody against the lipid-recognizing receptor (e.g., AF488-labeled anti-murine CD300f antibody)
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti Polar Lipids, cat. no. 850375C)
1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS, Avanti Polar Lipids, cat. no. 840035C)
1-Oleoyl-2-[12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl]-sn-Glycero-3-Phosphocholine (NBD PC, Avanti Polar Lipids, cat. no. 810133)
Phosphate-buffer saline (PBS)
FBS (sigma)
Flow cytometer (e.g. BD FACSCalibur)
Prepare empty lentivirus or viral particles encoding the receptor
-
1
Seed HEK293T cells in 10 ml DMEM medium with 10% FBS into 10 cm dishes the day prior to the transfection.
-
2
On the day of transfection, replace the old media with 5 ml of fresh DMEM + 10% FBS.
-
3
Prepare the transfection mixture. For one 10 cm dish of cells, add 250 µl DMEM containing 15 µl of PolyJet transfection reagent into 250 µl DMEM containing 5 µg of expression and helper plasmids (3 µg pCDH empty vector or pCDH encoding the receptor + 1.25 µg psPAX2 + 0.75 µg pMD2G). Incubate the mixture for 15 min at room temperature, and then add the transfection mixture dropwise to the 10 cm culture dish; next, swirl the media gently to disperse it evenly throughout the dish.
-
4
Change the medium 18 hrs after transfection, and add 10 ml fresh DMEM medium + 10% FBS.
-
5
At 72 hrs after transfection, collect the medium into a 15 ml sterile, capped conical centrifuge tube. Centrifuge at 500 × g for 10 min at room temperature to pellet cell debris. Transfer the cell culture supernatant (containing the lentiviral particles) into a new tube.
-
6
Add 1 volume of cold PEG-it virus precipitation solution to every 4 volumes of lentivirus-containing supernatant. Keep the mixture at 4°C for 4–5 days.
-
7
Centrifuge the lentivirus-containing supernatant at 1500 × g for 30 min at 4°C; the lentiviral particles appear as a white pellet at the bottom of the tube.
-
8
Discard the supernatant and resuspend the lentiviral pellets in 500 µl cold, sterile PBS.
-
9
Aliquot the lentiviral-containing solution to cryogenic vials, and store at −70°C until ready for use.
All dry waste contacting the viral particles should be contained in the double plastic bags (e.g., Ziploc) before discarded into a biohazard box for incineration. The liquid waste should be disinfected with 1% bleach for 20–30 min before discarding.
Make L929 control cells and cells expressing the receptor
-
10
Seed 8 × 104 L929 cells in 500 µl DMEM + 10% FBS in 24-well plates the day before the transduction.
-
11
On the day of transduction, add 50 µl of viral particle solution (empty virus for control cells, virus encoding the receptor for the receptor-expressing cells) and 8 µg/ml protamine sulfate to the cells. Centrifuge the cells at 1500 × g at room temperature for 45 min and return the cells to humidified incubator with 5% CO2 at 37°C.
-
12
Forty-eight hours after the transduction, detach the cells with trypsin and re-seed all the cells to 6-well plate in 2 ml DMEM + 10% FBS and add 20 µg/ml puromycin for selection of the transduced cells.
Please note that puromycin is a selection drug for pCHD constructs. If using a different expression vector, make sure that selection drug appropriate for the vector of choice is used.
-
13
Forty-eight hours after puromycin selection, the control cells or cells transduced to express the receptor are detached for the analysis of cell surface receptor expression with the fluorescence-labeled antibody against the receptor, or the unlabeled primary antibody and fluorescence-labeled anti-species secondary antibody.
Preparation of NBD-liposome
-
14
Mix the lipid suspensions in chloroform at a ratio of 1% NBD PC and 99% DOPC for PC liposomes, or 1% NBD PC, 49% DOPC and 50% of DOPS for PS liposomes.
NBD-PC liposomes and NBD-PS liposomes are prepared here as examples to analyze the binding of PS to PS-recognizing receptors (e.g. murine CD300f).
-
15
Follow the Basic Protocol I to prepare the NBD-PC and NBD-PS liposome. Cover the tubes with aluminum foil to protect from light.
Binding of NBD-liposomes to cell surface receptor
-
16
Resuspend 1 × 106 control cells, or cells expressing the receptor of interest in 100 µl PBS + 2% FBS in FACS tubes, with or without the addition of 10 µM NBD-liposomes.
-
17
Incubate the cells on ice for 30 min with protection from light.
-
18
Add 2 ml PBS + 2% FBS to wash the cells.
-
19
Centrifuge the cells at 500 × g for 5 min at 4°C, and discard the supernatant.
-
20
Repeat steps 18–19 one more time.
-
21
Resuspend the cells in 300 µl PBS + 2% FBS.
-
22
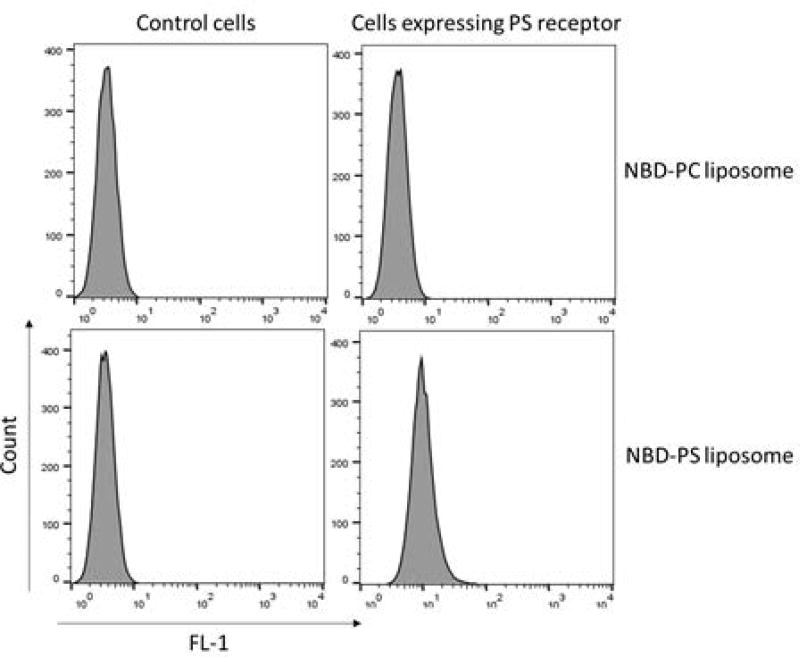
Analyze the fluorescence in the FL-1 channel using a flow cytometer (Figure 4).
Figure 4.
Representative graph showing NBD-PS liposome binds cells expressing PS receptor but not control cells.
Basic Protocol 5
EFFEROCYTOSIS BY PS-CONTAINING LIPOSOMES
The following protocol describes the use of phospholipid-containing liposomes as competitive inhibitors to elucidate if efferocytosis is mediated by the recognition of phospholipids exposed on the surface of the apoptotic cell through the receptor of interest. In this protocol, bone marrow-derived macrophages are used as a model for the efferocytosis assay, but alternatively any professional or non-professional phagocyte can be used (e.g. bone marrow-derived dendritic cells, or L929 cells-expressing the receptor).
Materials
C57BL/6 mice
CO2 chamber
70% ethanol
Scissors
Forceps
L929 cells overexpressing macrophage colony-stimulating factor (M-CSF)
RPMI medium
DMEM medium
Fetal Bovine Serum (FBS)
Phosphate-buffered saline (PBS)
pHrodo™ Red, succinimidyl ester (pHrodo™ Red, SE; Molecular probes)
Sterile 15mL tubes
FACS tubes
ACK lysing buffer (Lonza)
Accutase (Innovative Cell Technologies)
75 cm2 flasks
6 and 24-well plates
10 cm petri dishes
3 mL syringe plungers
20 mL syringes
27-gauge needles
Hemocytometer
70-µm cell strainers
Liposomes (see basic protocol I)
APC anti-mouse F4/80 Antibody (Biolegend)
Basic buffer: 50 mM Tris-HCl (pH 8.8), 153.8 mM NaCl
The humidified incubator with 5% CO2 at 37°C
Irradiator
Flow cytometer (e.g. BD FACSCalibur)
Preparation of L929-conditioned medium
-
1
Plate 7 × 106 L929 cells overexpressing M-CSF in a 75 cm2 flask containing 10 mL of DMEM medium with 10% FBS.
-
2
Culture cells in a humidified incubator with 5% CO2 at 37°C for 96 hrs.
-
3
Collect the supernatant and store L929-conditioned medium at 4°C (for immediate use) or −80°C (for long-term storage).
Bone marrow (BM) isolation and bone marrow-derived macrophage (BMM) differentiation
Start BM isolation and BMM differentiation one week before the efferocytosis assay
-
4
Euthanize the mouse by CO2 asphyxiation.
-
5
Sterilize the hind legs by spraying with 70% ethanol.
-
6
Using scissors, remove skin and muscle tissue from the bones. Separate the femur and tibia by breaking the knee joint and cut the bones at both ends. Place the bones into a 6-well plate containing PBS on ice.
-
7
Flush the bones with PBS using a 20-mL syringe and a 27-gauge needle until the bones turn white.
-
8
After centrifugation at 500 × g for 5 min at 4°C, discard the supernatant.
-
9
Remove red blood cells using ACK buffer for 5 min on ice. Add FBS to stop red blood cell lysis.
-
10
Pass the cells through a 70-µm cell strainer. Repeat step 8.
-
11
Wash cells using PBS for complete removal of ACK buffer. Repeat step 8.
-
12
Resuspend cells using complete RPMI medium with 10% FBS.
-
13
Count BM cells using a hemocytometer. Adjust the concentration to 2 × 106 cells/mL in RPMI medium with 10% FBS.
Alternatively, an automated cell counter of choice can be used to quantify cell numbers.
-
14
Plate 10 × 106 cells (5 mL) in a 10-cm petri dish containing 2 mL of RPMI medium with 10% FBS and 3 mL of L929-conditioned medium (final concentration: 30%).
-
15
Differentiate BM cells into BMMs in a humidified incubator with 5% CO2 at 37°C for 7 days. If cells reach 80% confluency (on day 4), detach cells from the plate using Accutase and split them into two petri dishes with fresh RPMI medium containing 10% FBS and 30% L929-conditioned medium.
-
16
On day 7, after removing non-adherent cells by PBS washing, detach BMMs from the plate using Accutase and use for the further experiments.
Preparation of Apoptotic thymocytes
-
17
On the day of the efferocytosis assay, euthanize mice by CO2 asphyxiation.
-
18
Wet the fur by spraying with 70% ethanol.
-
19
Dissect mice and collect the thymus.
-
20
Place the thymus into a 6-well plate containing PBS on ice.
-
21
Place a 70 µm cell strainer onto a 6-well plate containing 5 mL of PBS and transfer the thymus to the cell strainer.
-
22
Disrupt the thymus using a 3 mL syringe plunger.
-
23
Transfer the cells to a 15 mL tube.
-
24
After centrifugation at 500 × g for 5 min at 4°C, discard the supernatant.
-
25
Remove red blood cells using ACK buffer for 5 min on ice. Add FBS to stop red blood cell lysis.
-
26
Remove the cell debris by passing through a 70-µm cell strainer. Repeat step 24.
-
27
Wash cells using PBS for complete removal of ACK buffer. Repeat step 24.
-
28
Resuspend cells using RPMI medium with 1% FBS.
-
29
Count thymocytes using a hemocytometer (or an automated cell counter). Adjust the concentration to 1 × 106 cells/mL in RPMI medium with 1% FBS.
-
30
Transfer 10 mL of cell suspension into 15 mL tubes.
-
31
Irradiate cells with 2000 rad.
-
32
Incubate cells in a humidified incubator with 5% CO2 at 37°C for 6 h.
Labelling apoptotic thymocytes with pHrodo™ Red
-
33
After 6 h incubation, centrifuge apoptotic thymocytes at 500 × g for 5 min at 4°C, and discard the supernatant.
-
34
Resuspend apoptotic thymocytes in PBS.
-
35
Count apoptotic thymocytes using a hemocytometer (without trypan blue staining). Adjust the concentration to 4 × 106 cells/mL in PBS.
Trypan blue staining is not necessary, but can be used to verify the loss of irradiated cell viability.
-
36
Transfer 2.5 mL of cell suspension (10 × 106 cells) into a 15-mL tube.
-
37
Add pHrodo™ Red SE, a pH-sensitive fluorescent dye, as a final concentration 100–1000 ng/mL.
-
38
Incubate cells in a humidified incubator with 5% CO2 at 37°C for 30 min.
-
39
Stop the staining by adding an equal volume (2.5 mL) of FBS. Incubate for 2 min at room temperature to allow binding of excess dye, and then add 5 mL of PBS.
-
40
Centrifuge the cells at 500 × g for 5 min at 4°C, and discard the supernatant.
-
41
Wash cells with 10 mL of PBS. Repeat step 40.
-
42
Repeat step 41.
-
43
Resuspend pHrodo™ Red-labelled apoptotic thymocytes in RPMI medium containing 10% FBS.
-
44
Count cells using a hemocytometer (without trypan blue staining). Adjust the concentration to 0.5 × 106 cells/mL in RPMI medium with 10% FBS.
pHrodo™ Red-labelled apoptotic thymocytes need to be used immediately.
Efferocytosis blocked by PS-containing liposomes
-
45
On day 7 of BMM differentiation (the day before efferocytosis assay), plate 0.8 × 105 BMMs in a 24-well plate containing 500 µL of RPMI medium with 10% FBS. Incubate in a humidified incubator with 5% CO2 at 37°C for overnight.
-
46
On the day of efferocytosis assay, remove the medium from the plate and add 500 µL of the fresh RPMI medium with 10 µM liposomes (e.g. liposomes containing 80% DOPC and 20% DOPS).
-
47
Incubate BMMs in the medium with liposomes in a humidified incubator with 5% CO2 at 37°C for 30 min.
-
48
Add 500 µL of pHrodo™ Red-labelled apoptotic thymocytes (2.5 × 105 cells) to BMMs (0.8 × 105 cells) in the medium with liposomes (the ratio of BMM to apoptotic thymocytes is 1:3).
-
49
Coculture BMMs and apoptotic thymocytes in a humidified incubator with 5% CO2 at 37°C for 1 h.
-
50
Remove non-engulfed apoptotic cells by washing twice with PBS.
-
51
Detach BMMs from the plate using Accutase, and collect cells in the FACS tubes.
-
52
After centrifugation at 500 × g for 5 min at 4°C, discard the supernatant.
-
53
Resuspend the cells in 100 µL of PBS containing 2 % FBS
-
54
Add 1 µL of APC-conjugated anti-mouse F4/80 antibodies to the cell suspension. Stain for 30 min on ice, protected from light.
-
55
Wash cells with 2mL of PBS containing 2 % FBS.
-
56
After centrifugation at 500 × g for 5 min at 4°C, discard the supernatant.
-
57
Resuspend the cells in 250 µL of basic buffer (pH 8.8).
This step is to quench fluorescence of any non-engulfed cells. Only engulfed apoptotic thymocytes will display pHrodo™ fluorescence.
-
58
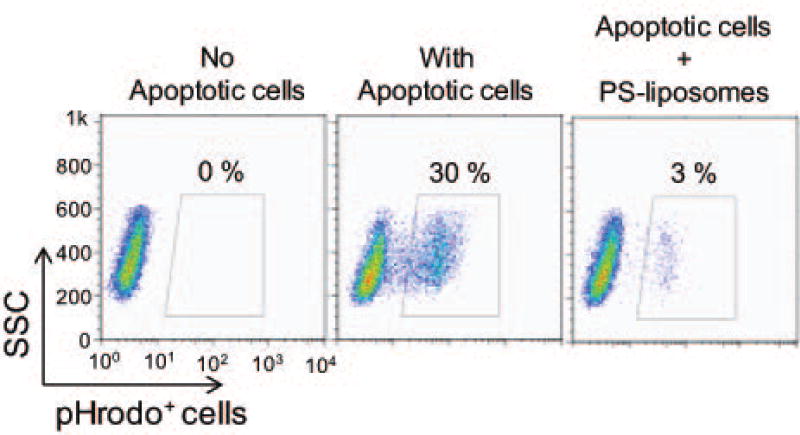
Perform flow cytometry analysis immediately (Figure 5).
Figure 5.
Representative flow cytometry data showing the percentage of engulfment pHrodo-labeled apoptotic cells in PS receptor-expressing L929 cells after 2 h in the presence or absence of PS-containing liposomes (10 µM).
Basic Protocol 6
PHAGOCYTOSIS OF LIPOSOME-COATED DYNABEADS
The following protocol describes how to use liposome-coated beads as apoptotic cell surrogates in a phagocytosis assay. It is used to identify the essential phospholipid(s) required for the uptake of apoptotic cells by phagocytes.
Materials
Liposome-coated dynabeads (see Basic Protocol II)
Phagocytes (e.g. bone marrow-derived macrophages)
RPMI medium
Fetal Bovine Serum (FBS)
Phosphate-buffered saline (PBS)
Accutase (Lonza)
AF647-Phalloidin (Invitrogen)
Saponin (Sigma)
4% paraformaldehyde
Prolong Gold Medium (Invitrogen)
Glass bottom microwell dishes (35 mm petri dish, 10 mm microwell and 0.16–0.19 mm coverglass)
Sterile 15 mL tube
Hemacytometer
Fluorescence microscope (e.g. Zeiss LSM 780 laser scanning confocal microscope)
Phagocytosis of liposome-coated dynabeads
Plate 0.5 × 105 BMMs in a glass bottom microwell dish containing 100 µL of RPMI medium with 10% FBS. Incubate in a humidified incubator with 5% CO2 at 37°C.
Six hours later (once BMMs attach to the glass bottom of microwell dish), add 2 mL of RPMI medium with 10% FBS. Incubate in a humidified incubator with 5% CO2 at 37°C for overnight.
On the day of phagocytosis assay, remove the medium from the plate and add 100 µL of the fresh RPMI medium with liposome-coated dynabeads (the ratio of BMM to liposome-coated dynabeads is 1:3).
Incubate in a humidified incubator with 5% CO2 at 37°C for 10, 30 or 60 min.
Remove non-engulfed liposome-coated dynabeads by washing twice with PBS.
Fix the cells with 1 ml of 4% paraformaldehyde in PBS for 10 min.
Wash the cells in 1 ml of PBS 3 times for each 5 min.
Permeabilize cells with 1 ml of 0.4% saponin in PBS for 15 min.
Wash the cells in 1 ml of PBS once and block nonspecific binding with 1% BSA in PBS for 30 min.
Wash the cells in 1 ml of PBS once and stain the cells with AF647-phalloidin (1:200 dilution in 500 µl PBS) for 30 min at room temperature.
Wash cells with 1 ml of PBS for 5 min, repeat this step 2 more times
Mounted washed cells with ProLong Gold mounting medium.
-
Examine the cells under a confocal fluorescence microscope using an appropriate filter setup (490 nm Ex/525 nm Em for the auto-fluorescence of Dynabeads, 650 nm Ex/665 nm Em for Phalloidin-stained F-actin). Acquire several optical sections (z-stacks) in order to reconstruct the three-dimensional images required to evaluate the number of the engulfed liposome-coated Dynabeads per BMM cell.
0.5 or 1 µm intervals in the z-plane should be sufficient for the 3D reconstruction; please, make sure to acquire enough z-sections to capture the entire cell.
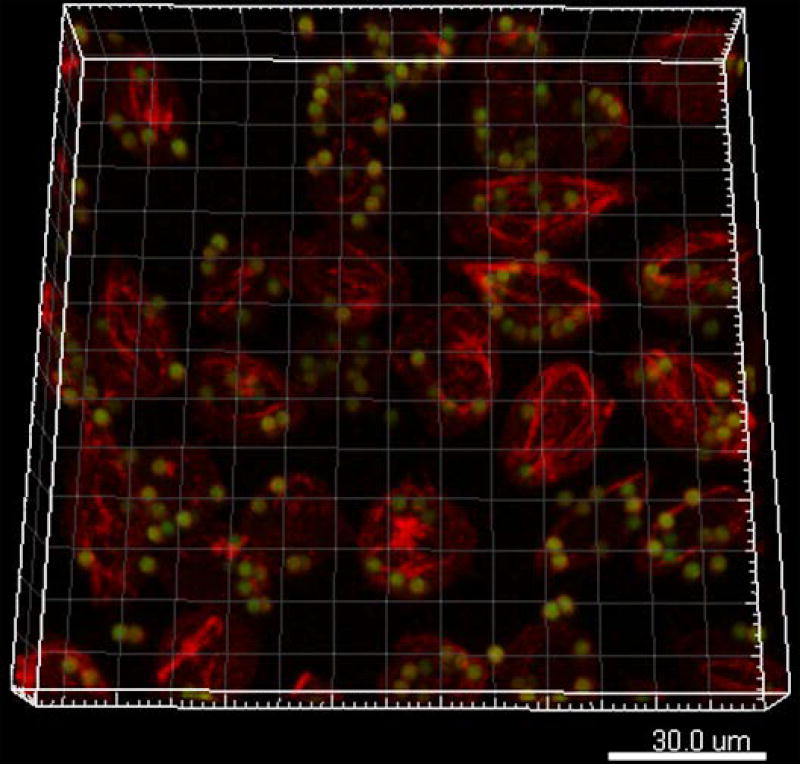
Generate 3D reconstructions of BMM cells to visualize the engulfed pHrodo+ liposome-coated dynabeads. Quantify the number of engulfed beads using a software of choice (e.g., Image J, IMARIS). See Figure 6.
Figure 6.
Representative 3D image showing the engulfment of PS liposome-coated beads (green) by phagocytes (e.g. PS receptor-expressing L929 cells in red).
Commentary
Background Information
Phospholipids consist of a hydrophilic head and hydrophobic fatty acid tails. Due to the amphipathic nature and insolubility in water, phospholipids are ideal compounds for forming biological membranes. Phospholipids in the context of the cell membrane are recognized by the interacting cells via their cell surface receptors, and therefore play important roles in regulating the immune response, including cytokine production, efferocytosis and antigen processing (Birge et al., 2016; Lee et al., 2017; Murakami et al., 2014; Tian et al., 2016; Tian et al., 2014). A bilayer membrane is formed spontaneously when phospholipids are dispersed in an aqueous solution. One form of the lipid bilayer is the liposome. Based on the number of lamellae, liposome can be categorized as multilamellar or unilammellar vesicles. Multilamellar vesicles are usually heterogeneous in size from 200 nm to 3 µm; while unilamellar vesicles are prepared in a narrow size range, including small unilamellar vesicles (SUV, 20 nm-100 nm), large unilamellar vesicles (LUV, 100 nm-400 nm), or giant unilamellar vesicles (GUV, over 1 µm). The vesicle size is an important parameter in determining the circulation half-life of liposomes, and both size and number of bilayers affect the encapsulation volume in the liposomes. Liposomes have been utilized as drug carriers for cellular delivery in disease treatments, and as a tool in the studies of cell interaction and recognition processes (Akbarzadeh et al., 2013).
Critical parameters and troubleshooting
Liposome preparation
Plastic pipettes and tips can be dissolved by chloroform and thereby can lead to contaminations. Glass syringe must be used to transfer or dissolve the phospholipids in chloroform. Since chloroform interferes with the formation of liposomes, glass vials after evaporating the bulk of chloroform with argon gas are put in a vacuum chamber for at least 12 hrs to remove all remaining traces of the solvent. All buffers must be filtered through a 0.22 µm filter to avoid accumulation of particles, which can clog the membrane in the extrusion step. Phospholipids with unsaturated bonds in fatty acid chains are subject to oxidation. To avoid oxidation, all the buffers must be degassed and the liposome must be stored in tubes filled with argon gas.
Efferocytosis
The ratio of target cells to phagocytes and the incubation time are critical factors for the phagocytosis background. With a high phagocytic background, one should try a lower ratio of apoptotic cells to phagocytes, and/or decrease the incubation time (e.g., 15–30 min). The higher background may also come from tethering of targets to the phagocyte cell surface. To decrease the tethering effects, the addition of basic buffer (pH 8.8) to quench the pHrodo signal of the tethered apoptotic cells is critical.
Biosafety
Use a fume hood to work with chloroform and used chloroform is disposed in an organic waste container. Virus particles must be prepared in the Biological Safety Cabinet. All dry waste containing the viral particles should be stored in double plastic bags and are discarded into a biohazards box. The liquid waste should be disinfected with 1% bleach for 20–30 min before being poured down the sink. Wearing safety clothing, and exercising proper care and common sense during the experiments is highly recommended to avoid any hazardous situations.
Anticipated results
The protocols described here will allow investigators to characterize lipid-receptor interactions. The main advantage of the assays described here is that they are performed in a three-dimensional space, thereby minimizing the effect of spatial and steric hindrances that may be common for the two-dimensional approaches utilized in other commonly used assays (e.g., ELISA, lipid strips). Moreover, these protocols will enable evaluation of the effect of particular receptors, and/or phospholipids on efferocytic cell ability to phagocytose apoptotic cells. Such characterization may help to uncover new receptor and ligand interactions, as well as highlight differences in contribution of phospholipid-recognizing receptors to efferocytosis.
Time considerations
For the detailed time frames, please refer to a particular protocol. The Basic Protocol I requires about 14 hrs of total time, including 12 hrs incubation that can be done overnight. Basic Protocol II can be completed in 5–6 hrs. In the Basic Protocol III, the significant variables in total experimental time include cell transduction (3 days total), protein purification (half a day to one day), lentiviral particle preparation (about a week), and cell infection (2–3 days). Once the prerequisite conditions are met (e.g., cells are transduced and selected, Fc-fusion proteins are available), the assays should be completed within 4 to 8 hrs, depending on the assay. For the Basic Protocol IV, the biggest time considerations are preparation of conditioned media (4 days), isolation of BM and BMM differentiation (7–8 days total time), BMM plating and culture (6 hrs for plating and an overnight incubation) and apoptotic thymocyte preparation and labeling (7–8 hrs); these are the longest steps. The efferocytosis assay should take about 2–3 hrs, depending on the number of samples analyzed. The liposome-coated bead phagocytosis assay should take 2–3 hrs and additional 3–4 hrs for image acquisition, depending on the number of samples and time points; 3D image reconstruction, microscopic analysis and engulfed bead quantification likely will require at least several hrs depending on the number of images acquired.
Acknowledgments
This work was supported by the Intramural Programs of the National Institute of Allergy and Infectious Diseases.
Footnotes
Conflict of Interest
None of the authors has a conflict of interest to disclose.
Literature Cited
- Akbarzadeh A, Rezaei-Sadabady R, Davaran S, Joo SW, Zarghami N, Hanifehpour Y, Nejati-Koshki K. Liposome: classification, preparation, and applications. Nanoscale Res Lett. 2013;8(1):102. doi: 10.1186/1556-276X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge RB, Boeltz S, Kumar S, Carlson J, Wanderley J, Calianese D, Herrmann M. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 2016;23(6):962–978. doi: 10.1038/cdd.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SC, Simhadri VR, Tian L, Gil-Krzewska A, Krzewski K, Borrego F, Coligan JE. Cutting edge: mouse CD300f (CMRF-35-like molecule-1) recognizes outer membrane-exposed phosphatidylserine and can promote phagocytosis. J Immunol. 2011;187(7):3483–3487. doi: 10.4049/jimmunol.1101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupfeld S, Holsaeter AM, Skar M, Frantzen CB, Brandl M. Liposome size analysis by dynamic/static light scattering upon size exclusion-/field flow-fractionation. J Nanosci Nanotechnol. 2006;6(9–10):3025–3031. doi: 10.1166/jnn.2006.454. [DOI] [PubMed] [Google Scholar]
- Jason-Moller L, Murphy M, Bruno J. Overview of Biacore Systems and Their Applications. Curr Protoc Protein Sci. 2006;(Unit 19.13) doi: 10.1002/0471140864.ps1913s45. Chapter 19. [DOI] [PubMed] [Google Scholar]
- Lee HN, Tian L, Bouladoux N, Davis J, Quinones M, Belkaid Y, Krzewski K. Dendritic cells expressing immunoreceptor CD300f are critical for controlling chronic gut inflammation. J Clin Invest. 2017;127(5):1905–1917. doi: 10.1172/JCI89531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Tian L, Voss OH, Margulies DH, Krzewski K, Coligan JE. CD300b regulates the phagocytosis of apoptotic cells via phosphatidylserine recognition. Cell Death Differ. 2014;21(11):1746–1757. doi: 10.1038/cdd.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Choi SC, Lee HN, Murakami Y, Qi CF, Sengottuvelu M, Coligan JE. Enhanced efferocytosis by dendritic cells underlies memory T-cell expansion and susceptibility to autoimmune disease in CD300f-deficient mice. Cell Death Differ. 2016;23(6):1086–1096. doi: 10.1038/cdd.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Choi SC, Murakami Y, Allen J, Morse HC, 3rd, Qi CF, Coligan JE. p85alpha recruitment by the CD300f phosphatidylserine receptor mediates apoptotic cell clearance required for autoimmunity suppression. Nat Commun. 2014;5:3146. doi: 10.1038/ncomms4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JC. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal Biochem. 1980;104(1):10–4. doi: 10.1016/0003-2697(80)90269-9. [DOI] [PubMed] [Google Scholar]