Abstract
It is now well established that the environment to which we are exposed during fetal and neonatal life can have a long-term impact on our health. This has been termed the developmental origins of health and disease. Factors known to have such programming effects include intrauterine nutrient availability, (determined by maternal nutrition and placental function), endocrine disruptors, toxins and infectious agents. Epigenetic processes have emerged as a key mechanism by which the early environment can permanently influence cell function and metabolism after multiple rounds of cell division. More recently it has been suggested that programmed effects can be observed beyond the first generation and that therefore epigenetic mechanisms could form the basis of transmission of phenotype from parent to child to grandchild and beyond. Here we review the evidence for such processes.
1. Introduction: Early life programming of future disease risk
Amongst both scientists and laypersons, the notion that a human being is a product of both our genes and our environment is now well accepted. It follows that a person’s health is not necessarily limited to what their DNA permits, but can be modified by lifestyle and environment. In recent years fetal and neonatal life have been highlighted as particularly critical periods of development when the environment can interact with our genotype to have a permanent effect on our phenotype. A strong case for this has been shown recently in a study by Rosenquist and colleagues [1], which found that the impact of the FTO gene variant which has been linked to obesity is largely affected by the year of birth, such that there was no correlation in participants born prior to 1942, whereas there was a far stronger correlation for those born post 1942 (post-World War II). This study was preceded by a number of epidemiological studies showing the effects of historical cases of hunger or malnutrition resulting from wars or natural famine not only immediate effects on the contemporary population, but also that of individuals who were in-utero at the time of these events. The Dutch Hunger Winter [2][3] and the Leningrad Siege [4] were catastrophic periods of hunger and malnutrition during which rations were strictly imposed on all sections of the population including pregnant and nursing mothers. A large number of studies have focused on the malnutrition experienced during these periods of famine and starvation and uncovered associations with chronic adult disease such as cardiovascular disease and metabolic disease in individuals born around the affected periods. As well as long term detrimental effects of under-nutrition in utero, there is now also a wealth of evidence that maternal over nutrition or obesity is also associated with offspring cardio-metabolic disease. This is particularly relevant in Western Societies where a combination of a reduction in physical activity and increased ease of access to highly palatable foods has tilted the balance of energy homeostasis, in favour of energy intake over expenditure, leading to an epidemic of obesity. Studies in animal models have shown that this is a causal relationship between maternal under-nutrition and over-nutrition on offspring metabolic and cardiovascular health that is independent of genotype. Such studies have also highlighted the importance of the pre- and early postnatal environment in growth and development, and that the timing of an insult or deviation from the norm is as important as the insult itself in determining (a) the organ systems affected and (b) the timing of onset and severity of disease outcome. Information on precise mechanisms through which such events in early life program a permanent effect on tissue structure and function, even after numerous rounds of cellular replication during early development and constitutive growth and differentiation, are less well characterized. However, growing evidence to indicate that the programmed phenotype brought about by early environmental insults such toxicants and pollutants, maternal under or over nutrition or parental obesity may extend through more than one generation has led to great interest in the role of epigenetic mechanisms [5][6].
2. Epigenetics and chromatin
The term "epigenetics" was first coined by Conrad Waddington to define the “interactions of genes with their environment which bring the phenotype into being”[7]. It is now used, but not without a great deal of controversy[8], to refer to covalent modifications of DNA and core histones that are heritable and affect genome function without altering the DNA nucleotide sequence. It is however clear that epigenetic information is transmitted from parental cells to daughter cells, and potentially inherited across generations, through the stable perpetuation of chromatin states.
2.1. Chromatin
The genome of eukaryotic cells is packaged into “chromatin”, a structure that comprises the complex of histone proteins and DNA. The nucleosome is the basic unit of chromatin; it contains an octamer of two each of histones H2A, H2B, H3 and H4, or some variant of these canonical core histones, wrapped inside ~147 base pairs (bp) of DNA. Additionally, histone H1 is involved in the compaction of chromatin, functioning as an internucleosome linker. Research over the past decades revealed that chromatin not only provides the scaffold for the packaging of the entire genome, but also plays key roles in both transcriptional regulation and the maintenance of genomic stability.
2.1.1. Chromatin marks
Covalent post-translational modifications of DNA and histone proteins, defined here as “chromatin marks”, can alter the organization and function of chromatin, with implications for the regulation of DNA-based processes, such as DNA repair, replication and transcription. These modifications, or marks, are laid down and removed in a dynamic fashion by specialized enzymes. The characterization of such chromatin-modifying enzymes represented major breakthroughs, as it provided a first handle on how to control the modifications and established the principle of a dynamic system that can respond to cellular stimuli and environmental cues.
Table 1 shows an overview of key (selected) chromatin marks, with information related to proposed function, association with genomic location and annotation of corresponding writers, readers, and erasers of the modification.
Table 1. Chromatin marks.
Listed are chromatin marks discussed in this review
| Chromatin mark | “Writers” | “Reader” domains & proteins | “Erasers” | Genomic distribution | Main function as epigenetic mark | Other features |
|---|---|---|---|---|---|---|
| DNA: | ||||||
| 5mC | DNA methyltransferases (Dnmts 1, 3a, 3b) |
Methyl-CpG Binding Domains (MBDs) (e.g. MeCP2, MBDs1-4) BTB/POZ domains (Kaiso, ZBTB4, ZBTB38) |
Active demethylation (TET 1,2,3 proteins) Passive demethylation processes |
CpG dinucleotides throughout the genome: intergenic regions, gene bodies, heterochromatin, satellite repeats. Excluded from CpG islands associated with promoters |
Repressive mark Cellular memory |
Essential for chromosome stability, silencing of transposons, genomic imprinting and X-inactivation |
| 5hmC, 5fC, 5caC | Ten-eleven translocation enzymes (TET 1, 2, 3) |
Unknown | TDG followed by base excision repair (BER) Passive processes |
Oxidation products of 5mC; abundant levels of 5hmC in brain and ES cells | Unknown | Demethylation intermediates of 5mC; Mutations targeting TET genes frequently observed in human cancers |
| Histone tails: | ||||||
| H3K27me | Histone lysine methyltransferases - HMTs (EZH2) | Chromo and WD40 domains (e.g. Polycomb protein Pc, EED, CBX7) | Histone lysine demethylases- HDMs (KDM6a, KDM6b) |
Repressive domains and silent developmental genes | Repressive mark Cellular memory |
Established by polycomb complex activity; mark associated with bivalent chromatin* |
| H3K4me | Lysine HMT (e.g.SETD1a,b,SETD7, NSD3, MLL, PRDM9) |
Chromo, PHD, Tudor, MBT, Zf-W domains (e.g. CHD1, ING2) | HDMs (e.g.KDM1A,5A,5D, PHF8) |
H3K4me1: enhancers H3K4me2: promoters & enhancers H3K4me3: promoters & transcription start sites |
Activating mark | H3K4me3 mark is enriched at CpG islands devoided of DNA methylation & associated with bivalent chromatin* |
| H3K9me | Lysine HMT (e.g. G9a/KTM1C, PRDM2, SETDB1-2; SUV39H1-H2/KTM1A-B) | Chromo, PHD, Tudor, WD40 domains, Ankyrin repeats (e.g. HP1, CDY, EED) | HDMs (e.g. KDM1A, 1B, 3A, 3B, 4A, 4C, 4D) |
H3K9me2: heterochromatin H3K9me3: constitutive heterochromatin and repetitive elements |
Repressive mark | The chromodomain of all HP1 isoforms binds to H3K9me3 and this interaction is important for the recruitment of HP1 proteins to heterochromatic areas of the genome. |
| H3K36me3 | Lysine HMT (SETD2, NSD2) |
Chromo, PWWP Domains (e.g. EAF3, MRG15, N-PAC) |
HDMs (KDM4A, NO66) |
Associated with transcribed portions of genes, with preference for 3’regions after intron1 | Elongation mark associated with transcription | H3K36me3 regulates DNA mismatch repair (MMR) in human cells, being required to recruit MMR recognition proteins onto chromatin |
| H3K4ac | Histone acetyltransferases – HATs (KAT2A/GCN5) |
Bromo Domains | Histone deacetylases-HDAC’s (HDAC3) | Enriched at transcription start sites (TSS) and along gene bodies | Activating mark (“Open” chromatin) |
|
| H3K9ac | HATs (e.g. KAT2A/GCN5, ELP3) | Bromo Domains (e.g. BRG1, BRD4, TAF1) |
HDAC’s (SIRT1, SIRT6) | Preference for promoters (near TSS is related to transcriptional activation) |
Activating mark (“Open” chromatin) |
Mark of active regulatory elements |
| H3K27ac | HATs (P300, CREBBP/CBP) |
Bromo Domains (e.g. BRG1, BRD4, TAF1) |
HDACs (e.g. HDAC1) |
Active regulatory elements (may distinguish between active promoters and enhancers from inactive ones) | Activating mark (“Open” chromatin) |
Distinguishes active enhancers from inactive/poised enhancer elements containing H3K4me1 alone |
contains both activating and repressing marks in the same domain of chromatin; these bivalent domains are considered to “poise” expression of developmental genes, allowing timely activation while maintaining repression in the absence of differentiation signals; for extended information on the chromatin marks listed see review articles: Huang H et al. (2004) Snapshot: Histone modifications Cell, 159:p458; Li E and Zhang Y (2014) DNA methylation in mammals. CSH Perspect Biol 6:a019133; Yun M et al. (2015) Readers of histone modifications. Cell Research 21:564-578; see also main text for details)
2.1.2. Histone marks
Histone marks occur in the N-terminal tail domains of the core histones that protrude out from the nucleosome, but also in the core histone domains and in newly synthesized histones. Histone tails contain an extraordinary number of sites that can be subjected to post-translational modifications. Some of these modifications, such as acetylation and phosphorylation, can alter the charge of the tails and, thus, have the potential to influence chromatin through electrostatic mechanisms. However, the primary mechanism by which tail modifications act seems to be through their function as “docking” sites for chromatin “readers” that specifically recognize these modifications, and in turn recruit additional chromatin modifiers and remodeling enzymes. Chromatin readers include large families of proteins containing domains such as bromodomain, chromodomain, Tudor domains, plant homeodomain (PHD), PWWP domains, YEATS domains to effect diverse downstream chromatin-based processes (reviewed in Yun et al. 2011; Eberl et al. 2013; Li et al. 2014)[9]–[11]. Recent studies suggest that core domain modifications may also function through distinct mechanisms involving structural alterations to the nucleosome (reviewed in Tessarz and Kourzarides, 2014)[12].
Vincent Allfrey and colleagues[13] were the first to propose that post-translational modification in histones (i.e. histone acetylation) may provide ‘a dynamic and reversible mechanism for activation as well as repression of RNA synthesis’. This hypothesis was validated several decades later with the finding that transcriptionally active genes carry acetylated core histones and the establishment of causal links between histone acetylation and transcriptional regulation. A major breakthrough in this regard was the discovery of the enzymes that acetylate or de-acetylate histones (histone acetyltransferases - HATS and deacetylases- HDACS, respectively) (reviewed in Verdin and Ott, 2015)[14]. It is now well recognized that acetylation of histones inhibits the folding of nucleosome arrays into secondary and tertiary structure formation, thus resulting in chromatin decondensation (thus allowing access to transcription factors and co-activators of transcription).
Lysine methylation of histones, like acetylation of histones, was first described in the 1960’s but decade passed without much insight into the functional significance of this modification being offered. Histone methylation can affect higher order chromatin structure directly as shown recently for methylation of H4 at lysine 20 (H4K20), which enhanced the ability of nucleosomal arrays to fold and condense in vitro[15] It has also become apparent that lysine methylation can alter the local properties of chromatin for transcription by creating binding sites for reader proteins (Table 1). H3K4 methylation, for example, is generally associated with active transcription (H3K4 dimethylation broadly associated with active and potentially active genes, while H3K4 trimethylation is a mark associated with the start site of transcription) (Table 1).
These known modifications may however represent just the tip of the iceberg. Recent work of Tan and colleagues, biochemically identified 67 novel histone marks[16] including histone tyrosine hydroxylation and lysine crotonylation (Kcr). In particular, the authors focused on the significance of Kcr by demonstrating that this mark is a robust indicator of active cellular genes (marking either active promoters or potential enhancers) and that it is likely an important histone mark for sperm cell differentiation. The functional significance of this observation was provided this year by Sabari and coworkers showing that histone crotonylation by the coactivator p300 was able to stimulate transcription to a greater degree than histone acetylation [17]. However there is still much to be done in defining the precise role and relative importance of these marks.
2.1.3. DNA marks
The DNA of vertebrates can be covalently modified by methylation of the cytosine base in the dinucleotide sequence CpG (p is an abbreviation for phosphate, which links the cystosine and guanine nucleotides together in DNA). Until recently, DNA methylation (abbreviated to 5mC) was the only covalent DNA modification known. This situation changed with the identification of 5-hydroxymethylation (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) as bona fide bases of DNA. However, these marks are generated by oxidation of 5mC by the TET family of dioxygenases as part of DNA demethylation pathways (5fC/5caC are then later processed by the DNA repair mechanism enzyme TDG). It remains unclear if 5hmC, given its accumulation in certain cell types and tissues, have specific functions as an “epigenetic” mark or is simply an intermediate for DNA demethylation[18].
DNA methylation (5mC) patterns are established during embryonic development by the de novo methylating enzymes Dnmt3a and Dnmt3b. These patterns are then maintained when cells divide by a “maintenance methyltransferase” Dnmt1, that copies the parental pattern onto the progeny strand during DNA replication (thus exclusively methylating CpGs base-paired with a methylated parental CpG). This mechanism ensures that patterns are replicated semiconservatively like the base sequence of DNA itself.
5mC is associated with gene silencing and it plays an important role in developmental processes such as genomic imprinting and X-inactivation. Regions of CpG methylation either prevent binding of certain transcription factors, thereby preventing transcription, or they attract or repel numerous DNA binding proteins. Methyl-CpG binding domain proteins (or MBDs) are a family of proteins that recognize methylated DNA and recruit repressor complexes to methylated promoter regions, thus contributing to transcriptional silencing. These chromatin-inactivating complexes often include histone deacetylases and histone methyltransferases.
Certain regions of the genome are DNA methylation free, called CpG islands or CGIs, which are clusters of CpG sequences mostly found directly upstream of gene promoters. A major question in the field is how CGI is protected from DNA methylation when most CpGs are methylated[19]. CpGs are enriched of the H3K4me3 mark, through the action of the enzyme that writes this mark, Setd1, a member of the MLL family of H3K4 methyltransferases, which is recruited to CGIs. This enrichment is likely to prevent CGs from being methylated as this modification can repulse de novo methyltransferases in vitro. Another possible factor at play being the enrichment of Tet1 protein, which is capable of removing “accidental” methylation at CGIs through oxidation of 5mC, followed by base excision repair mechanisms. Furthermore, two proteins, Cfp1 and Kdm2a, can then bind specifically to non-metylated CpGs via the CXXC domain, and contribute to creating a transcriptionally competent chromatin configuration (reviewed by Li and Zhang, 2014[18]).
DNA methylation patterns, although they can be transmitted from cell to cell, are not permanent. Changes to DNA methylation may arise as a physiological response to environmental changes, while other changes might be associated with aging or disease such as cancer, diabetes, cardiovascular disease. DNA methylation marks can be removed by either a passive mechanism of demethylation by inhibition of the maintenance methyltransferase Dnmt1, or an active mechanism involving the family of Tet proteins that can oxidise 5mC to 5hmC and further to 5fC and 5caC, the latter being excised by glycosylases such as TDG, followed by DNA repair to generate C.
The epigenetic processes that affect genome function are now recognized to also include the regulatory effects of non-coding RNAs (such as microRNAs, miRNAs, and long non-coding RNAs, lncRNAs). The majority of lncRNAs are nuclear and their most common mechanism of action is the modification of chromatin structures, via recruitment of chromatin modifiers to DNA. These modifiers can be activating (such as the WDR5-MLL complex) or repressive (such as the LSD1-CoREST complexes, or PRC1/2)[20].
In addition to epigenetic regulation of transcription, variability in protein expression forms yet another layer of organismal and tissue functional complexity. This can be attributed to post-transcriptional events such as binding of existing transcripts to ribosomal complexes of the translational machinery, transcript half-life or the binding and interaction of microRNAs which are now commonly included as an epigenetic component. MicroRNAs are 21-24 nucleotides in length and bind sequence specifically to the 3' untranslated regions of mRNA transcripts and subsequently interact with the Dicer complex and sequester the bound transcript for degradation, or interfere with transcript binding to the translational machinery. In the cytosol, lncRNAs can also act as as sponges for miRNAs, thus inhibiting the actions of miRNAs on mRNAs (there are also few examples of lncRNAS that affect the half life of mRNAs by either stabilizing or destabilizing specific subsets of mRNAs)[20].
2.2. Chromatin marks as carriers of epigenetic information during development
Histone modifications are often referred to as epigenetic marks. However, for that to be the case, at least according to certain definitions, the modification should be stably inherited through cell divisions in the absence of the initiating event (and the mechanisms by which this might occur, if it occurs, are at present unknown). Equally controversial is the notion that histone marks act sequentially or in combination to signal downstream events, as if there were following a pre-determined “code”. Although it is clear that histone marks often function as short-term “signalling” molecules, it is also evident that histones can perpetuate chromatin states together with their “writer” and “reader” complexes. Indeed, PcG and TrX protein reader complexes (Table 1) are major players in chromatin-based memory strategies for maintaining gene activity in somatic cell lineages. Maintenance of cellular memory by chromatin-based epigenetic mechanisms, including DNA methylation, is essential for cellular differentiation processes and the perpetuation of cell-specific functions. Indeed, there is growing evidence that epigenetic regulators play a key role in very early development at the point of cell differentiation when cells become committed to extra-embryonic tissues vs embryonic tissues, and ectoderm, endoderm and mesoderm lineages. This lineage commitment is mediated by these stable epigenetic marks that are inherited through several rounds of proliferation and are retained throughout life. As a result, somatic tissues have distinct epigenetic signatures that they acquire during development and that can be modified, to certain extent, by the environment.
In mammals, the developmentally acquired epigenetic signatures, including those induced by the environment, will be erased in the early embryo and in the germline. These two rounds of epigenetic erasure, so called epigenetic reprogramming, occur to restore totipotency of the zygote and leave little chance for inheritance of epigenetic marks, whether programmed, environmentally induced or accidental. Indeed, if germline reprogramming fails, epigenetic marks could be retained and potentially transmitted from one generation to the next. Recent genome-wide DNA methylation profiling confirms that the bulk of the genome becomes demetylated in primordial germ cells but there are a number of loci (>4,500), predominantly repeat associated, that escape reprogramming[21], [22]. Those loci could represent prime candidates for possible transgenerational epigenetic inheritance in mammals.
It is important to distinguish intergenerational effects (or parental) from truly transgenerational effects. The former include effects such as the impact of in utero exposure to particular stress, toxic, nutritional, hormonal environments on the developing embryo and its germline, as opposed to the latter, which refers to effects that are found in generations that were not exposed to the initial exposure. Parental or intergenerational programming effects certainly occur in mammals and may have an epigenetic basis, which will be explored below.
3. Maternal Effects
3.1. Epigenetic effects - DNA methylation and Histone Modifications
3.1.1. Evidence from Humans
The Dutch Hunger Winter famine of 1944 is regarded as a ‘natural experiment’ for the study of the prenatal environment in relation to metabolic disease in later life. Individuals who were exposed to this famine in utero can be traced from birth records and the timing of the exposure can be established. Several DNA methylation studies have been reported using whole blood from individuals that were exposed in early gestation (representing the developmental window of extensive epigenetic reprogramming) and late gestation, compared with same-sex unexposed siblings. Initial reports using gene candidate approaches found evidence for differential methylation at promoters and imprinted regions regulating genes involved in growth and metabolism, including IGF2 and LEP[23], [24]. More recently, a genome-scale DNA methylation analysis of 24 exposed individuals and 24 same-sex sibilings controls, by Reduced Representation Bisulphite Sequencing (RRBS; with a coverage of 1.2M individual CpG dinucleotides), led to the identification of 181 P-DMRs (Pre-natal malnutrition-associated Differentially Methylated Regions)[25]. The majority of P-DMRs occurred in gene bodies, and were associated in pathways involved in development and metabolic regulation. An important recent study by Guenard et al (2013) [26] utilised a mass-spectrometry approach (EpiTYPER, Sequenom) to investigate CpG methylation changes in blood of offspring born to mothers before and after bariatric surgery to reduce obesity. The authors reported differential methylation in glucoregulatory genes and genes involved in diabetes-related cardiometabolic pathways. This supports the role of the maternal metabolic state in the aetiology of offspring disease through dysregulation of methylation signals and the efficacy of bariatric surgery as a treatment for the amelioration of future offspring cardiometabolic disease. Altogether these studies suggest that the phenotypic associations between prenatal exposure to famine and adverse metabolic profile, i.e. suboptimal glucose handling, higher BMI, elevated total and LDL cholesterol, may have an underlying epigenetic basis.
One limitation of epigenetic studies in humans is that they are usually restricted to clinically accessible tissues such as white blood cells. Although identification of epigenetic changes in such tissues may be useful from a biomarker perspective, most (but not all) epigenetic changes in these tissues will not be reflective of changes present in more metabolically relevant tissues. It is therefore of relevance to focus on regions within the genome where epigenetic differences may be conserved between tissues. In 2002, Rakyan and co-workers [27] first coined the term “metastable epialleles” (MEs) to describe such regions within the genome where DNA methylation is established in the early embryo, and then is stably maintained in differentiated tissues, leading to epigenetic variation that affects multiple cell types. The term “metastable” refers to the labile nature of the epigenetic mark, while “epiallele” defines their potential to maintain epigenetic marks transgenerationally. A recent study by Dominguez-Salas and colleagues[28] therefore focused on these regions of the genome in a human population in the Gambia. This population is affected by pronounced naturally occurring seasonal variations in diet availability and substrate utilization and therefore season of birth has been shown to have a major impact on the long-term health of an individual. Candidate methylation analysis of white blood cells and hair follicle samples (representing mesodermal and ectodermal tissues, respectively) from the offspring of these mothers born in the rainy season had increased methylation of six metastable alleles.
3.1.2. Evidence from Animal Models
Animal models have been key in demonstrating causal effects of changes in the maternal environmental factors on epigenetic modifications in offspring tissue. A wide range of metabolically relevant tissues has been observed to be epigenetically modified in response to the early environment.
Liver
Regardless of species, many models of maternal feeding of a high fat or highly palatable diet during pregnancy have shown that maternal obesity during pregnancy results in development of fatty liver in the offspring. This is associated with changes in histone modifications and DNA methylation patterns. In a primate model of maternal obesity, Japanes macaques were fed a high-fat (35% fat) breeder diet and mated. This resulted in a three-fold increase in fetal liver triglycerides, which was accompanied by a hyperacetylation at H3K14 in this tissue, although this did not associate with gene repression. Fetal histone deacetylase 1 (HDAC1) expression was however reduced, which correlated with depleted HDAC1 protein levels and in-vitro functional activity[29]. In mice fed a high fat diet over 3 consecutive generations (F0, F1 and F2), it was shown that the onset of obesity in succeeding generations occurred earlier and increased in severity. An increase in steatosis accompanied this intergenerational high fat feeding, which was apparently compounded by the obesity in previous generations, with the highest degree of steatosis observed in the F2 generation. Leptin and insulin levels were also the highest in the F2 mice. There was increased lipogenesis in their livers, which correlated to a progressive reduction in histone methylation in the LXRα and ERO1-α gene promoters[30].
Persistent metabolic changes in the offspring accompanied by epigenetic changes are also observed in models of maternal protein restriction. For example, hypomethylation of GR and PPAR alpha promoters is induced in the livers of juvenile and adult offspring whose mothers were fed a PR diet[31]. This is associated with increased levels of the expression of those genes and in the metabolic processes under their control. Interestingly, epigenetic changes at these two key metabolic genes are reversed in the liver of offspring of mothers fed a global dietary restriction, i.e. promoter hypermethylation rather than hypomethylation [32]. Perhaps not surprisingly, the nature of the maternal nutritional challenge is an important determinant of the adaptive response on the epigenome of the offspring.
Pancreatic Islets
Transcription factors in pancreatic islet have been shown to be particularly vulnerable to the epigenetic changes in response to a suboptimal environment in utero. In a model of intrauterine growth restriction (IUGR) following uterine artery ligation, Park et al (2008) [33] observed decreased H3 and H4 acetylation and loss of binding of USF-1 to the proximal promoter of Pdx1, resulting in markedly reduced Pdx1 transcription. In a model of maternal protein restriction it was observed that there was reduced expression of Hnf4a in offspring pancreatic islets in adulthood. This was associated with a small increase in DNA methylation at the active HNF4a promoter (P2) in the low protein offspring islets. However, more notably, there were substantial changes in histone marks specifically at the enhancer region, with an excess of the repressive mark H3K9me2 and loss of the active mark H3K4me1[34]. Consistent with these epigenetic changes, a significant reduction of the P2–enhancer interaction in LP offspring islets was observed, providing a mechanistic basis for the reduction in HNF4a expression. This study also highlighted the importance of carrying out epigenetic studies across the life course. It was observed that exposure to the low protein diet in early life modulated the dynamics of epigenetic changes with age. There was a greater age-dependent accumulation of the repressive histone mark H3K27me3 in the low protein offspring islets.
Epigenetic changes involving the imprinted Igf2/H19 loci in pancreatic islets was recently reported in a mouse model of intergenerational transmission of glucose intolerance induced by intra-uterine hyperglycemia[35]. In this study, F0 females were randomly divided into GDM and control groups and injected with a single injection of streptozotocin (STZ) or vehicle control, respectively. F1 adult offspring from the Control and GDM groups were then intercrossed to obtain F2 offspring of four groups (C-C; GDM-C; GDM-C; GDM-GDM) for metabolic and epigenetic analyses. Intra-uterine hyperglycemia in F0 induced impaired glucose tolerance in F1-GDM and F2-GDM groups, which was more pronounced in males, and resulted in altered birth weight in F2-GDM but not F1-GDM offspring. The expression of the imprinted Igf2 and H19 genes was reduced in islets of F1 and F2-GDM, which was associated with hypermethylation at Igf2 DMR2 and H19 DMR regions, with expression of these genes also downregulated in sperm of F1-GDM. The authors speculate that intrauterine hyperglycemia can alter imprinted gene expression in germ cells and contribute, by yet unknown mechanisms, to transgenerational transmission of the metabolic phenotype.
Muscle
Histone code modifications have also been shown to be involved in the repression of glucose transporter expression in IUGR rat offspring. Raychaudhuri and colleagues showed that de-acetylation and di-methylation of specific amino acid residues in the N-tail of histone 3 had a putative role in co-repressor complex formation, and therefore interfere with formation of a co-activator complex. Both at birth and persisting in the adult, these epigenetic changes decreased GLUT4 transcription, the major insulin responsive glucose transporter[36]. This epigenetically programmed reduction in GLUT 4 may therefore explain the increased susceptibility to diabetes in these animals.
Sperm
Recent studies have demonstrated that as well as effects on somatic tissue, maternal diet can also impact on methylation of offspring germ cells. Using an established model of maternal under-nutrition, which leads to low birth weight and glucose intolerance in male and female F1 offspring, Radford et al showed that DMRs in sperm DNA of the F1 males were hypomethylated and enriched in nucleosome-retaining regions [37]. Phenotypically, this hypomethylation in the DMRs was associated with transmission of the low birth weight and glucose intolerance in to the F2 offspring. Radford argued that although these differences were not retained in late-gestation somatic tissues of the F2 offspring the alterations seen in the F1 sperm could provide a mechanism for paternal transmission. Luciferase studies suggest that these DMRs are enriched in regulatory elements and therefore potently involved in transcriptional regulation. As DMRs are late to regain methylation after erasure in normal primordial germ cells, they may be susceptible to environmental changes that delay or impair re-methylation later in gestation. Park et al showed in porcine zygotes, that the methylation mark of the paternal allele Igf2/H19 DMR3 is erased by active demethylation, whereas that of the maternal allele is de novo methylated [38]. Furthermore, they showed that the hemimethylated pattern in zygotes fertilized in vitro was present up to the 4-cell embryo stage and then exclusively demethylated at the 8-cell stage and finally restored at the morula stage. These dynamic methylation changes during early embryonic development allow flexibility to primordial germ cells, which render them sensitive to the prevailing maternal environment.
3.2. Evidence for programming of miRNAs
Adipose tissue
Adipose tissue appears to be an early site of programming effects. There is good evidence to suggest that the early environment can impact on adipocyte cell size that in term can influence insulin sensitivity. In an IUGR model using maternal protein restriction, it has been shown that the F1 offspring have persistently smaller and more numerous adipocytes [39]. This phenotype was associated with an increased abundance of miR-483-3p that directly regulates translation of growth differentiation factor (GDF)-3, a determinant of cell size. Similar effects on miR-483-3p (an increase) and GDF3 (a decrease) were observed in adipose tissue biopsies from young low birth weight men. These findings suggest that these effects are conserved between species and therefore likely to be fundamentally important.
miRNAs have also been shown to be dysregulated in models of maternal obesity. For example, miR-126 levels are elevated in epidydymal adipose tissue of offspring of obese mouse dams. This miRNA directly regulates IRS-1 and therefore the programmed change in the miR could explain the programmed reduction in IRS-1 observed in adipose tissue from the offspring of obese dams. Importantly, these effects on miR-126 and IRS-1 were cell autonomous and were retained following in vitro differentiation of programmed preadipocytes [40]
Skeletal muscle
Skeletal muscle is another tissue in which dysregulation of miRs is manifest as a consequence of low birth weight. In monozygotic twins, the expression of miR-15b and miR-16 in skeletal muscle biopsies of the diabetic twin was found to be higher than that of the non-diabetic twin, and this was negatively associated with the direct targets of these miRs targets, the insulin receptor and IRS-1. Furthermore, the expression of miR-15b was also elevated in skeletal muscle of rats that were protein-restricted in-utero, again highlighting the conservation of programming of miRs between species [41]. Maternal obesity has also been shown to affect miR expression levels in skeletal muscle. In an ovine model of maternal obesity there was reduced expression of let-7g fetal skeletal muscle [42]. The let-7g downregulation was proposed to enhance intramusclular adipogenesis during fetal muscle development. Since let-7g has been shown to be secreted as a pre-miR in microparticles from mesenchymal stem cells (MSC) [43], the reduction in let-7g might reflect a reduction in the proportion of MSCs to committed myoblasts. The importance of let-7, its relationship with Lin28a and their roles in regulating glucose tolerance and insulin sensitivity was elegantly dissected in mice overexpressing these two miRs, and established their interacting pathways as central to the regulation of mammalian glucose metabolism[44].
Heart
The first demonstration of altered miR expression in cardiac tissue was in relation to miR-133, which was up-regulated in heart tissue of young offspring exposed to maternal obesity [45]. In this model the offspring develops cardiac hypertrophy very early in life (3 weeks of age)[46] that is associated with increased stimulation of the MAPK pathways[45].
4. Paternal Effects
Early evidence from human epidemiological studies suggested a link between paternal grandfather's food supply and grandchild's risk of diabetic death and cardiovascular diseases. In the Överkalix cohort in Sweden [47] and in the ALSPAC (Avon Longitudinal Study of Parents and Children) cohort in the UK [48], paternal grandfathers food supply was linked to the cardiovascular and diabetes mortality of grandsons, while paternal grandmother food supply was only associated with granddaughters mortality, although it is critical to note that the exposure had to have occurred during the slow growth period or fetal/infant life.
In spite of this evidence, until recently, nearly all studies into the programming of health and disease were focused on the maternal line. The role of paternal factors in programming of offspring health has only in recent years, become a focus of study. It has now been established that both paternal under-nutrition (Carone et al., 2010) and over-nutrition (Ng et al., 2010) can have an effect on the next generation. Carone et al demonstrated a modest increase in methylation in an intergenic CpG island between PPARα and Wnt7b in offspring of males fed a low-protein diet [49]. The importance and susceptibility of this locus to low-protein diet exposure in the preceding generation is implied since differential methylation was also observed at the PPARα promoter in offspring of female rats fed a low-protein diet during pregnancy [50]. Paternal over-nutrition has been shown to programme beta cell dysfunction in female offspring of male rats fed a high fat diet[51]. More recently, Wei et al showed that paternal diabetes resulted in reduced expression of Pik3ca, Pik3r1 and Ptpn1 in offspring pancreatic islets, and consistent with this, they found increases in methylation at intragenic regions of Pik3r1 and Pik3ca [52]. A large proportion of these differentially methylated genes were also differentially methylated in the fathers' sperm. Moreover, when F1 male mice were mated with normal females, their offspring (F2) also developed impaired glucose tolerance and the methylation status of Pik3r1, Pik3ca, and Ptpn1 in the F2 pancreatic islets was similarly perturbed as with the F1 generation.
In a drosophila model of paternal programming, acute low- or high-sugar feeding to the fathers (2 days) was found to increase offspring F1 triglyceride content when challenged by an obesogenic high-sugar diet [53]. These findings are consistent with evidence from mammalian models showing that suboptimal nutrition at either end of the spectrum (i.e., parental over-nutrition or parental under-nutrition) causes an increased risk of metabolic dysfunction in the offspring [54]. Furthermore, the authors identified requirements for H3K9/K27me3-dependent reprogramming of metabolic genes in two distinct germline and zygotic windows, and in effect, they identified a clear and conserved epigenetic signature that is associated with obesity in mammals as well as flies.
As well as paternal effects on offspring DNA methylation and histone modifications, there is also evidence that they can impact on miRNA levels. F1 male offspring of prenatally stressed dams were observed to develop dysmasculinization and this phenotype was transmitted to the F2 males via the paternal lineage. This was associated with reduced anogenital distance and significant reductions in miR-322, miR-574, and miR-873 in the F2 stressed male brains [55].
5. Programming: potential for transgenerational inheritance?
There is now undisputable evidence that paternal or maternal exposures can influence the epigenotype of the F1 offspring. However less well established is if the epigenotype can be transmitted to the F2 generation and beyond. As highlighted by Skinner[56], the exposure of a gestating female (F0), to a nutritional, hormonal or toxic insult, would affect not only her, but her F1 generation as well as the germ cells that will form the F2 offspring (which develop very early in development of F1). Thus, the effects of the initial exposure should cease to have effect on the F3 (i.e. the first generation to be free of exposure). However, if phenotypic changes are present in the F3 (mother exposed) this can be defined as a transgenerational effect mediated by epigenetic processes[57]–[59]. In the case of inheritance via the male germ line, in which an epigenetic change is induced in males only, the individual (F0) and his germline (F1) are exposed, which signifies that only F2 and subsequent generations can be considered for evidence of transgenerational inheritance.
There is significant evidence that exposure to endocrine disruptors (EDs) in utero significantly modifies male offspring digits lengths, promoting a more feminized digit ratio [60]. Auger et al showed that similar effects were carried through in the next generation of unexposed males sired by exposed fathers and unexposed mothers. In parallel, the same demasculinizing agents methoxychlor and vinclozin, and the estrogenic compound bisphenol A administered during gestation were shown to disrupt the development of the male reproductive tract and spermatogenesis. The result was a decrease in sperm counts and methylation pattern changes in a selection of paternally and maternally expressed imprinted genes. Furthermore, the damaging effects of the EDs were specific to gamete cells and transmitted to F3[61].
Some of the strongest evidence for transgenerational effects of a nutritional insult came from a study in C. elegans. This demonstrated that starvation (known to lead to an increase in lifespan) led to the induction of expression of small RNAs with gene targets involved in nutritional regulation and that these differences were maintained into the third generation. It was established that this response was dependent on the germline-expressed nuclear argonaute HRDE-1 [62]. Moreover, the F3 offspring of starved animals showed an increased lifespan, thus demonstrating a transgenerational memory of past conditions. There is limited evidence for effects of early nutritional manipulation in rats to the F3 generation. One study using a maternal protein restriction model (F0) showed this dietary manipulation during pregnancy and lactation led to altered glucose metabolism in the F1 and F2 generation, and a more modest effect in the F3 generation [63]. Thus, it is possible the effects of nutritional exposure may be transmitted transgenerationally but the effect size becomes reduced with successive generations.
Another study demonstrated the transmission of liver lipid metabolism defects through altered Lxra methylation in the 3rd generation via the paternal line. Male mice whose mothers were 50% calorie restricted during gestation were found to have a low birth weight and developed obesity and glucose intolerance by 4-6 months of age[64]. The reduced birth weight phenotype was observed in the F2 generation from the paternal line, although obesity was only transmitted through the maternal line, whereas impaired glucose tolerance progressed through both parents[65]. They proposed therefore that DNA methylation contributed to the metabolic dysfunction in the 3rd generation via the paternal lineage. They subsequently demonstrated that lipogenic gene expression was reduced, in part, by reduced expression of Lxra and Srebf1, and that methylation at the Lxra locus was reduced both in sperm of the F1 and livers of the F2 offspring[66].
The epigenetic state of sperm and oocyte are considered to be the primary mechanisms that mediate paternal and maternal programming effects. It is now clear that exposure of either male or female gametes can lead to changes in their epigenetic stage and potentially lead to phenotypic changes in the offspring that develop from these gametes. A major barrier for propagation of the history of environmental exposures across generations is the significant epigenetic mark erasure that occurs in the germline and early embryo, but recent studies, highlighted in previous sections, show that this reprogramming is not completely reset. Future studies looking at mechanisms of epigenetic transmission of parental exposures are likely to focus on those genomic sequences and the associated epigenetic marks that resist re-programming. Non-coding RNAs are also emerging as potential mechanisms for transgenerational transmission of phenotypes, as potential mediators of the response to environmental signals. It is now known that oocytes and sperm produce (and deliver at fertilization) a vast array of small non-coding RNAs that have been proposed to aid in multiple functions, ranging from degradation of maternal mRNA, to regulation of the epigenetic state. Such small RNAs (18-24 bp long) include for example, piwi-interacting RNAs (piRNAs), mature-sperm-enriched tRNA derived small RNAs (mse-tsRNAs), miRNAs, small nuclear RNAs (snRNAs), YRNAs. Recently, it was shown that paternal changes to sperm RNAs are implicated in the transmission of behavioral and metabolic responses to the next generation in a model of early-life traumatic stress in mice[67]. Formal proof that sperm RNA mediates the intergenerational transmission was provided by showing that injecting sperm RNAs from traumatized males into fertilized wild-type oocytes mimics the alterations in behavior and metabolism in the resulting offspring[67]. It is tempting to speculate that RNAs are initial signals that result in chromatin mark changes in the gametes. In that context, the work establishing links between the piwi (piRNA) pathway and CpG methylation is particularly relevant. The piwi pathway is a well-established mechanism for retrotransposon silencing in the genome that was recently implicated in the regulation of the de novo DNA methylation at the imprinted, paternally expressed, Rasgrf1 locus[68]. Although the physiological function of endogenous small RNAs in epigenetic regulation is well described in many other organisms, little is known in mammals, and this is undoubtedly a hot area for research in the future, in particular, for maternal and paternal gamete-based programming.
Elaborating on the regulatory role of noncoding RNAs in RNA-mediated hereditary variation (reviewed by Mercer & Mattick, 2013[69], Kiani and co-workers questioned the role of methyltransferases and other interacting molecules by using a mouse paramutation in the white tail phenotype Kit, to show that the loss of function of the RNA methyltransferase DNMT2 was able to override the paternal transmission of the paramutation in sperm [70]. Their experiments therefore strongly suggest that DNMT2 methyltransferase activity also has an important role in the stable transmission of sperm-borne transgenerational effects and epigenetic heredity. It also highlights the potential contribution of methylating/demethylating enzymes in fine-tuning the heritability of phenotypes through sperm.
Summary
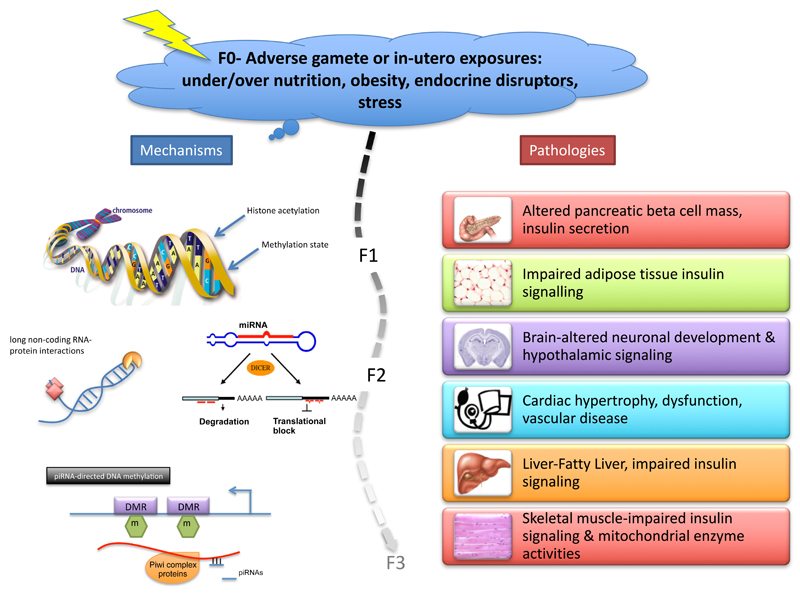
The field of developmental programming has progressed substantially since its conception around twenty-five years ago. Epigenetic processes have emerged as an attractive mechanism to explain how suboptimal exposures at critical times of development can have a long term consequence on the function of a tissue that persist following multiple rounds of cell division. Table 2 summarizes the identification of some of these epigenetic alterations in models of developmental programming, the tissues affected and the generations affected. Studies to date show a causative effect of the exposure on the epigenetic mark (i.e. DNA methylation, histone acetylation, miRNA levels and latterly piRNA directed DNA methylation), as depicted in Figure 1. However, these studies merely demonstrate an association of the epigenetic modulation with a change in cell function. Therefore the challenge remains to show a causative relationship between programmed epigenetic changes and the phenotype of the organism.
Table 2. Parental exposures and intergenerational epigenetic alterations in models of programming of adult metabolic disease.
| Parental exposure | Metabolic organ/Gamete | Nature of epigenetic modification or MiRs | Association with disease | Physiological function of the altered gene | Generation affected |
|---|---|---|---|---|---|
| maternal high fat feeding[27] | Liver | reduced histone methylation | increased lipogenesis, steatosis | LXRα and ERO1-α gene promoters | F1, F2 |
| Maternal protein restriction[28] | Liver | hypomethylation of PPAR alpha promoters | impaired glucose tolerance, obesity | increased expression of GR and PPARα | F1 |
| Global maternal calorie restriction[29] | Liver | hypermethylation of GR and PPAR alpha | impaired glucose tolerance, obesity | altered expression of GR and PPARα | F1 |
| Uterine artery ligation[30] | pancreatic islets | deacetylation of histones H3 and H4 in PDX1 proximal promoter | type-2 diabetes | reduced PDX1 expression | F1 |
| Maternal protein restriction[31] | pancreatic islets | excess of repressive H3K9me2; loss of active H3K4me1 | type-2 diabetes | reduced interaction with P2–enhancer of HNF4a and its expression | F1 |
| Gestational diabetes[32] | pancreatic islets | abnormal methylation at DMR | impaired glucose tolearance | downregulation of Igf2 and H19 | F1, F2 |
| 50% calorie restriction[33] | muscle | H3.K14 de-acetylation | insulin resistance | diminished GLUT4 | F1 |
| undernourished dams[34] | liver, sperm | hypomethylated DMRs | metabolic defects | increased expression of genes involved in lipid oxidation, i.e PPARα, Pgc1α,and Pgc1β, and a trend toward down-regulation of genes involved in lipid synthesis, including Scd1, Srebp1,and Dgat1 | F1, F2 |
| maternal protein restriction[36] | adipose tssue | overexpression of miR483-3p | impaired glucose tolerance | reduced levels of target protein GDF3 | F1 |
| maternal obesity[37] | adipose tissue | overexpression of mir-126-3p | impaired glucose tolerance, obesity | reduced levels of target protein IRS1 | F1 |
| twin pregnancies[38] | skeletal muscle | increased miR-15b, miR-16 in the diabetic twin | diabetes | reduced levels of target proteins InsR and IRS1 | F1 |
| maternal obesity[40] | skeletal muscle | reduced expression of let-7g | obesity | reduced mesenchymal stem cell to myoblast commitment | F1 |
| maternal obesity [42, 43] | heart | overexpression of miR-133 | cardiac hypertrophy | impaired cardiac function | F1 |
Figure 1.
Acknowledgements
This work was supported by the MRC Metabolic Diseases Unit award: MC_UU_12012/4 (all authors). S.E.O. is a British Heart Foundation Senior Fellow and a member of the MRC Metabolic Diseases Unit.
Footnotes
Disclosure Summary
The authors have nothing to disclose.
References
- [1].Rosenquist JN, Lehrer SF, O’Malley aJ, Zaslavsky AM, Smoller JW, Christakis Na. Cohort of birth modifies the association between FTO genotype and BMI. Proc Natl Acad Sci. 2014 Dec; doi: 10.1073/pnas.1411893111. 201411893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: An overview. Reprod Toxicol. 2005;20:345–352. doi: 10.1016/j.reprotox.2005.04.005. [DOI] [PubMed] [Google Scholar]
- [3].Ravelli A, van der Meulen J, Michels R, Osmond C, Barker D, Hales C, Bleker O. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998 Jan;351(9097):173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- [4].Stanner SA, Bulmer K, Andrès C, Lantseva OE, Borodina V, Poteen VV, Yudkin JS. Does malnutrition in utero determine diabetes and coronary heart disease in adulthood? Results from the Leningrad siege study, a cross sectional study. BMJ Br Med J. 1997 Nov;315(7119):1342–1348. doi: 10.1136/bmj.315.7119.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Heerwagen MJR, Miller MR, Barbour LA, Friedman JE. Maternal obesity and fetal metabolic programming: a fertile epigenetic soil. Am J Physiol - Regul Integr Comp Physiol. 2010 Sep;299(3):R711–R722. doi: 10.1152/ajpregu.00310.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nilsson EE, Anway MD, Stanfield J, Skinner MK. Transgenerational epigenetic effects of the endocrine disruptor vinclozolin on pregnancies and female adult onset disease. Reproduction. 2008;135:713–721. doi: 10.1530/REP-07-0542. Corwin 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Waddington C. Organizers and Genes. Cambridge University Press; 1940. [Google Scholar]
- [8].Deans C, Maggert Ka. What Do You Mean, ‘Epigenetic’? Genetics. 2015;199(4):887–896. doi: 10.1534/genetics.114.173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21(4):564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. A Map of General and Specialized Chromatin Readers in Mouse Tissues Generated by Label-free Interaction Proteomics. Mol Cell. 2013;49(2):368–378. doi: 10.1016/j.molcel.2012.10.026. [DOI] [PubMed] [Google Scholar]
- [11].Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SYR, Li W, Li H, et al. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell. 2014 Oct;159(3):558–571. doi: 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014 Nov;15(11):703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- [13].Allfrey V, Faulkner R, Mirsky A. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. … Sci United States …. 1964;315(1938):786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015 Apr;16(4):258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- [15].Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol. 2008;15(10):1122–1124. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011 Sep;146(6):1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, Dai L, Shimada M, Cross JR, Zhao Y, Roeder RG, et al. Intracellular Crotonyl-CoA Stimulates Transcription through p300-Catalyzed Histone Crotonylation. Mol Cell. 2015 Mar;58(2):203–15. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014;6(5) doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schubeler D. Function and information content of DNA methylation. Nature. 2015 Jan;517(7534):321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- [20].Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol. 2015 Mar;11(3):151–160. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA, Pringle JE. Germline DNA Demethylation Dynamics and Imprint Erasure Through 5-Hydroxymethylcytosine. Sci. 2013 Jan;339(6118):448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The Dynamics of Genome-wide DNA Methylation Reprogramming in Mouse Primordial Germ Cells. Mol Cell. 2012;48(6):849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009 Nov;18(21):4046–53. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, Slieker RC, Stok AP, Thijssen PE, Müller F, van Zwet EW, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun. 2014 Nov;5 doi: 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Guénard F, Deshaies Y, Cianflone K, Kral JG, Marceau P, Vohl M-C. Differential methylation in glucoregulatory genes of offspring born before vs. after maternal gastrointestinal bypass surgery. Proc Natl Acad Sci U S A. 2013 Jul;110(28):11439–11444. doi: 10.1073/pnas.1216959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18(7):348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- [28].Dominguez-Salas P, Moore SE, Baker MS, Bergen AW, Cox SE, Dyer Ra, Fulford AJ, Guan Y, Laritsky E, Silver MJ, Swan GE, et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun. 2014 Jan;5:3746. doi: 10.1038/ncomms4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Aagaard-tillery KM, Grove K, Bishop J, Ke X, Fu Q, Mc Knight R, Lane RH. Developmental origins of disease and determinant of chromatin structure: Maternal Diet Modifies the Primate Fetal Epigenome. J Mol Endocrinol. 2008;41(2):91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li J, Huang J, Li J-S, Chen H, Huang K, Zheng L. Accumulation of endoplasmic reticulum stress and lipogenesis in the liver through generational effects of high fat diets. J Hepatol. 2012 Apr;56(4):900–907. doi: 10.1016/j.jhep.2011.10.018. [DOI] [PubMed] [Google Scholar]
- [31].Lillycrop Ka, Phillips ES, Torrens C, Hanson Ma, Jackson Aa, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br J Nutr. 2008;100:278–282. doi: 10.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lillycrop KA, Burdge GC. Maternal diet as a modifier of offspring epigenetics. J Dev Orig Health Dis. 2015 Apr;6(2):88–95. doi: 10.1017/S2040174415000124. [DOI] [PubMed] [Google Scholar]
- [33].Park JH, Stoffers Da, Nicholls RD, Simmons Ra. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118(6):2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sandovici I, Smith NH, Nitert MD, Ackers-Johnson M, Uribe-Lewis S, Ito Y, Jones RH, Marquez VE, Cairns W, Tadayyon M, O’Neill LP, et al. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci U S A. 2011;108(13):5449–5454. doi: 10.1073/pnas.1019007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ding GL, Wang FF, Shu J, Tian S, Jiang Y, Zhang D, Wang N, Luo Q, Zhang Y, Jin F, Leung PCK, et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61(5):1133–1142. doi: 10.2337/db11-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Raychaudhuri N, Raychaudhuri S, Thamotharan M, Devaskar SU. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J Biol Chem. 2008;283(20):13611–13626. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Radford EJ, Ito M, Shi H, Corish Ja, Yamazawa K, Isganaitis E, Seisenberger S, Hore Ta, Reik W, Erkek S, Peters aHFM, et al. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science (80-. ) 2014 Jul; doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Park C-H, Kim H-S, Lee S-G, Lee C-K. Methylation status of differentially methylated regions at Igf2/H19 locus in porcine gametes and preimplantation embryos. Genomics. 2009 Feb;93(2):179–186. doi: 10.1016/j.ygeno.2008.10.002. [DOI] [PubMed] [Google Scholar]
- [39].Ferland-McCollough D, Fernandez-Twinn DS, Cannell IG, David H, Warner M, Vaag AA, Bork-Jensen J, Brøns C, Gant TW, Willis AE, Siddle K, et al. Programming of adipose tissue miR-483-3p and GDF-3 expression by maternal diet in type 2 diabetes. Cell Death Differ. 2012 Jul;44:1–10. doi: 10.1038/cdd.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fernandez-Twinn DS, Alfaradhi MZ, Martin-Gronert MS, Duque-Guimaraes DE, Piekarz A, Ferland-McCollough D, Bushell M, Ozanne SE. Downregulation of IRS-1 in adipose tissue of offspring of obese mice is programmed cell-autonomously through post-transcriptional mechanisms. Mol Metab. 2014;3:325–333. doi: 10.1016/j.molmet.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bork-jensen J, Scheele C, Christophersen DV, Nilsson E, Friedrichsen M, Fernandez-twinn DS, Grunnet LG, Litman T, Holmstrøm K, Vind B, Højlund K. Glucose tolerance is associated with differential expression of microRNAs in skeletal muscle: results from studies of twins with and without type 2 diabetes. 2015:363–373. doi: 10.1007/s00125-014-3434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yan X, Huang Y, Zhao J-X, Rogers CJ, Zhu M-J, Ford SP, Nathanielsz PW, Du M. Maternal obesity downregulates microRNA let-7g expression, a possible mechanism for enhanced adipogenesis during ovine fetal skeletal muscle development. Int J Obes. 2012 May;:568–575. doi: 10.1038/ijo.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen TS, Lai RC, Lee MM, Choo ABH, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2009;38(1):215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhu H, Ng SC, Segr AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fernandez-Twinn DS, Blackmore HL, Siggens L, Giussani DA, Cross CM, Foo R, Ozanne SE. The Programming of Cardiac Hypertrophy in the Offspring by Maternal Obesity Is Associated with Hyperinsulinemia, AKT, ERK, and mTOR Activation. Endocrinology. 2012 Oct;153(12):5961–5971. doi: 10.1210/en.2012-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Blackmore H, Niu Y, Fernandez-Twinn DS, Tarry-Adkins JL, Giussani DA, Ozanne SE. Maternal diet-induced obesity programmes cardiovascular dysfunction in adult male mouse offspring independent of current body weight. Endocrinology. 2014 Oct;155:3970–3980. doi: 10.1210/en.2014-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents' slow growth period. Eur J Hum Genet. 2002;10(11):682–688. doi: 10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- [48].Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjöström M, Golding J. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14(2):159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- [49].Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143(7):1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPARα promoter of the offspring. Br J Nutr. 2008;100(2):278–282. doi: 10.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ng S-F, Lin RCY, Laybutt DR, Barres R, Owens Ja, Morris MJ. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467(7318):963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- [52].Wei Y, Yang C-R, Wei Y-P, Zhao Z-A, Hou Y, Schatten H, Sun Q-Y. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A. 2014 Feb;111(5):1873–8. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Öst A, Lempradl A, Casas E, Weigert M, Tiko T, Deniz M, Pantano L, Boenisch U, Itskov PM, Stoeckius M, Ruf M, et al. Paternal Diet Defines Offspring Chromatin State and Intergenerational Obesity. Cell. 2014 doi: 10.1016/j.cell.2014.11.005. [DOI] [PubMed] [Google Scholar]
- [54].Fernandez-Twinn DS, Ozanne SE. Early life nutrition and metabolic programming. Ann N Y Acad Sci. 2010;1212:78–96. doi: 10.1111/j.1749-6632.2010.05798.x. [DOI] [PubMed] [Google Scholar]
- [55].Morgan CP, Bale TL. Early Prenatal Stress Epigenetically Programs Dysmasculinization in Second-Generation Offspring via the Paternal Lineage. J Neurosci. 2011;31(33):11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Skinner MK. Environmental stress and epigenetic transgenerational inheritance. 2014;12(7):153. doi: 10.1186/s12916-014-0153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13(3):153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- [58].Lim J, Brunet A. Bridging the transgenerational gap with epigenetic memory. Changes. 2013;29(3):176–186. doi: 10.1016/j.tig.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ferguson-Smith AC, Patti ME. You are what your dad ate. Cell Metab. 2011;13(2):115–117. doi: 10.1016/j.cmet.2011.01.011. [DOI] [PubMed] [Google Scholar]
- [60].Auger J, Le Denmat D, Berges R, Doridot L, Salmon B, Canivenc-Lavier MC, Eustache F. Environmental levels of oestrogenic and antiandrogenic compounds feminize digit ratios in male rats and their unexposed male progeny. Proc Biol Sci. 2013;280(1768):20131532. doi: 10.1098/rspb.2013.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Paoloni-Giacobino A. Chapter Eight - Epigenetic Effects of Methoxychlor and Vinclozolin on Male Gametes. In: L G, editor. Endocrine Disrupters. Vol. 94. Academic Press; 2014. pp. 211–227. [DOI] [PubMed] [Google Scholar]
- [62].Rechavi O, Houri-Ze’Evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158(2):277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Benyshek DC, Johnston CS, Martin JF. Glucose metabolism is altered in the adequately-nourished grand-offspring (F3 generation) of rats malnourished during gestation and perinatal life [5] Diabetologia. 2006;49(5):1117–1119. doi: 10.1007/s00125-006-0196-5. [DOI] [PubMed] [Google Scholar]
- [64].Jimenez-Chillaron JC, Hernandez-Valencia M, Reamer C, Fisher S, Joszi A, Hirshman M, Oge A, Walrond S, Przybyla R, Boozer C, Goodyear LJ, et al. {b}-Cell Secretory Dysfunction in the Pathogenesis of Low Birth Weight – Associated Diabetes: A Murine Model. Diabetes. 2005 Mar;54:702–711. doi: 10.2337/diabetes.54.3.702. [DOI] [PubMed] [Google Scholar]
- [65].Jimenez-chillaron JC, Isganaitis E, Charalambous M, Gesta S, Pentinat-Pelegrin T, Faucette RR, Otis JP, Chow A, Diaz R, Ferguson-Smith A, Patti M-E. Intergenerational Transmission of Glucose Intolerance and Obesity by In Utero Undernutrition in Mice. Diabetes. 2009 Feb;58:460–468. doi: 10.2337/db08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Martínez D, Pentinat T, Ribó S, Daviaud C, Bloks VW, Cebrià J, Villalmanzo N, Kalko SG, Ramón-Krauel M, Díaz R, Plösch T, et al. In utero undernutrition in male mice programs liver lipid metabolism in the second-generation offspring involving altered Lxra DNA methylation. Cell Metab. 2014;19:941–951. doi: 10.1016/j.cmet.2014.03.026. [DOI] [PubMed] [Google Scholar]
- [67].Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17(5):667–9. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, et al. Role for piRNAs and Noncoding RNA in de Novo DNA Methylation of the Imprinted Mouse Rasgrf1 Locus. Sci. 2011 May;332(6031):848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013 Mar;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- [70].Kiani J, Grandjean V, Liebers R, Tuorto F, Ghanbarian H, Lyko F, Cuzin F, Rassoulzadegan M. RNA-Mediated Epigenetic Heredity Requires the Cytosine Methyltransferase Dnmt2. PLoS Genet. 2013;9(5) doi: 10.1371/journal.pgen.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]