Abstract
There is good laboratory and field evidence that some cattle excrete far more Escherichia coli O157 than others; these are known as 'super-shedders'. Super-shedding has important consequences for the epidemiology of E. coli O157 in cattle - its main reservoir - and for the risk of human infection, particularly via environmental exposure. Ultimately, control measures targeted at super-shedders may prove to be highly effective. At present, there is limited understanding of both the nature of and the determinants of super-shedding. However, super-shedding has been observed to be associated with colonization at the terminal rectum and it may be also more probable with certain pathogen phage types. More generally, epidemiological evidence suggests that super-shedding may turn out to be important in other bacterial and viral infections.
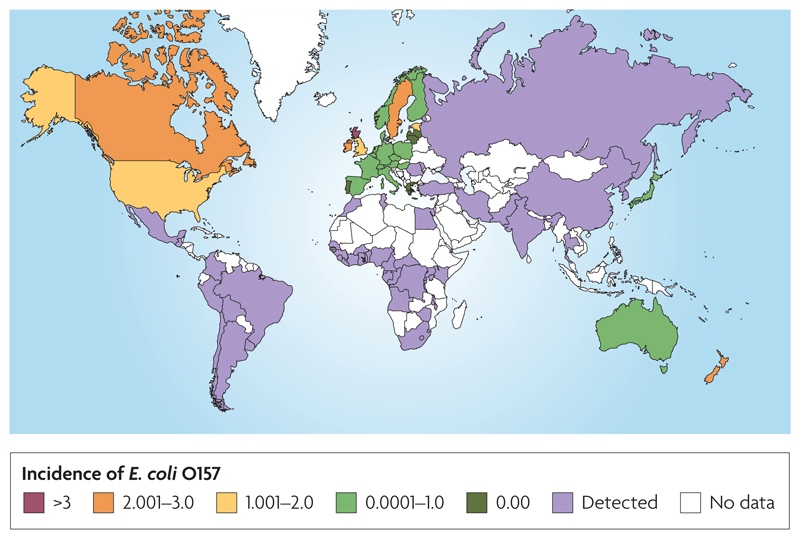
Verocytotoxi-producing Escherichia coli (VTEC), such as E. coli O157, are an important group of zoonotic pathogens of significant worldwide public health concern. E. coli O157 was first recognised as a human pathogen in 1982 when it was detected as the bacteria responsible for food-borne disease outbreaks in North America. Since 1982, infections have been reported in more than 50 countries in every continent other than Antarctica (FIG. 1). Over the past 25 years E. coli O157 has become an important cause of severe gastrointestinal illness, often causing bloody diarrhoea that can progress to more serious illness including haemolytic uraemic syndrome (HUS) and death1. During the 1980’s, most reported outbreaks of E. coli O157 were food-borne, primarily beef products and unpasteurized milk2. Increasingly, however, E. coli O157 food-borne outbreaks are being caused by a much wider variety of food products including unpasteurized apple juice, spinach and salami3,4. The probable or confirmed source of contamination for most of these foods is cross-contamination. This contamination may originate from faeces and arise from the presence of animals on fields or indirectly via irrigation and/or runoff, or through contamination during packaging and/or food preparation. The highest annual incidences of human infection with E. coli O157 during the last two decades have been reported in parts of Canada, the United States, Japan and Scotland5,6. Nationally, the incidence of E. coli O157 infection in Scotland is consistently high (1999-2005 the rate per 100,000 population ranged from 3.0 to 5.73)7, higher than elsewhere in Britain and abroad (FIG. 1).
Figure 1.
Map of the worldwide relative burden of E. coli O157 in humans in 2005 per 100,000 population. Crude rates are presented for countries where there are surveillance programs77,78,79, noting that surveillance and detection methods differ and therefore direct comparisons of burden are problematic. Cross hatching represents the detection of E. coli O157 in that country but no estimate of incidence rate was available. White represents no data available, noting that infections may have occurred in (some of) these countries, especially those without developed E. coli O157 surveillance systems. This figure reflects information in published reports as of August, 2008.
Though E. coli O157 was initially identified as a food-borne pathogen it is increasingly recognised that environmental exposure can also lead to human infection. Cattle are the main reservoir of E. coli O157 in the developed world8, though other animals have been shown as possible carriers. Visits to infected farms, contact with animal excreta and recreational use of animal pasture are risk factors contributing to sporadic cases of E. coli O157 in humans3. In analyses of the spatial distribution of human cases, incidences of human cases were positively associated with indicators such as livestock density and the ratio of cattle to human population9,10,11. There have been several surveys throughout the world of the proportion of cattle farms with E. coli O157 present (TABLE 1). Two of the largest of these were conducted during the past decade in Scotland12,13, both reporting around 20% cattle farms affected.
Table 1.
Examples of published estimates of E. coli O157 farm-level prevalence (proportion of farms tested positive to E. coli O157) from different classes of cattle in different countries. True comparisons are difficult as a result of differences in industry and cattle structure between countries as well as differences in study design and laboratory methods. To reduce some of the bias, all studies presented here used the Immunomagnetic bead Separation (IMS) method to detect E. coli O157 in the faeces. IMS is a more sensitive technique for generating accurate estimates of E. coli O157 prevalence12.
| Country | Cattle Type | N farms | % positive | Ref. |
|---|---|---|---|---|
| UK: | ||||
| England and Wales | Dairy / suckler / fattener | 75 | 38.7 | 80 |
| Scotland | Store / finishing cattle | 952 | 21.7 | 12 |
| Store / finishing cattle | 481 | 18.9 | 13 | |
| Sweden | Dairy cattle | 371 | 8.9 | 81 |
| Denmark | Dairy cattle | 60 | 16.7 | 37 |
| Norway | Heifers and milking cows | 197 | 1.0 | 82 |
| Dairy cattle | 50 | 0.0 | 83 | |
| Spain | Dairy cattle | 124 | 7.0 | 84 |
| Beef cattle | 82 | 1.6 | ||
| Netherlands | Dairy cattle | 678 | 7.2 | 85 |
| Veal calves | 462 | 9.1 | 86 | |
| Dairy cattle | 10 | 70 | 87 | |
| Iran | Dairy cattle | 26 | 3.9 | 88 |
| Canada: | ||||
| Saskatchewan | Feedlot cattle | 20 | 60.0 | 89 |
| Alberta | Feedlot cattle | 84 | 48.0 | 90 |
| USA: | ||||
| Ohio | Dairy cattle | 50 | 8.0 | 83 |
| Midwest | Ranch and feedlot cattle | 29 | 72.0 | 91 |
| Tennessee | Dairy cattle | 30 | 26.7 | 92 |
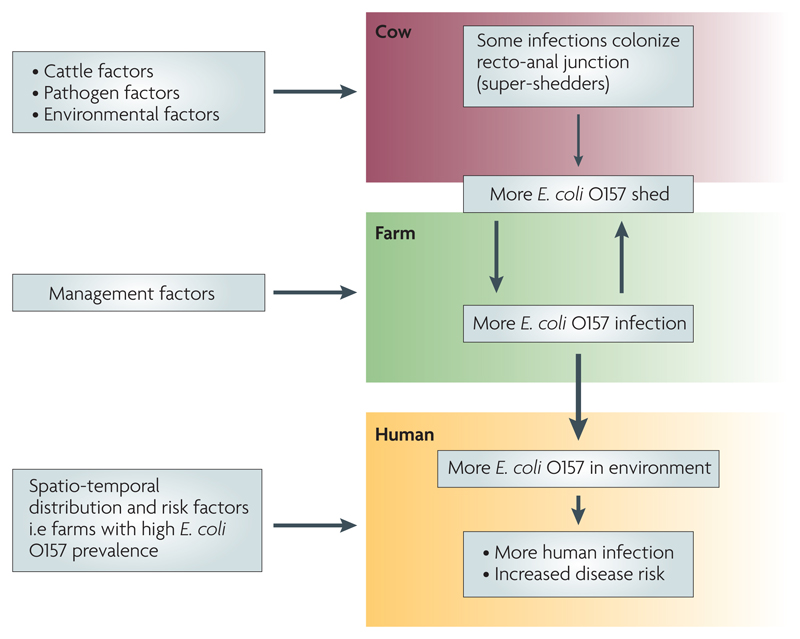
In this article, we focus on a key observation arising from the Scottish studies, namely that some cattle – referred to as ‘super-shedders’ – excrete far more bacteria than others13,14. Our current understanding of the pathogenesis and epidemiology of super-shedding is summarized in FIG. 2. We review the evidence behind this schema and discuss its implications for the transmission dynamics of E. coli O157 in cattle, the risk of human infection (concentrating on environmental rather than food-borne exposure), and the prospects for disease control.
Figure 2.
Diagrammatic representation of the chain of events linking super-shedding cattle and the risk of human infection with E. coli O157.
What are super-shedders?
Within the past decade researchers have adopted the terms ‘super-shedding’ and ‘super-spreading’ to describe heterogeneities in the epidemiology of infectious diseases. Although the terms are often used interchangeably we do not regard them as equivalent (Box 1).
Box 1. What is a super-shedder?
(i). Super-shedders or super-spreaders?
The terms ‘super-shedding’ and ‘super-spreading’ are not always clearly distinguished in the existing literature we therefore recommend the following definitions.
Super-shedder: is a term applied to an individual who for a period yields many more infectious organisms of a particular type than the majority of the same host species. Typically this would mean that many more infectious units are released from a super-shedder. The term is most useful when there is a clear biological basis for the distinction between super-shedders and non-super-shedders (such as host genetic differences, host immune suppression, type differences in the infectious organism, presence or absence of co-infections).
Super-spreader: Super-spreading is a term applied to an individual who has more opportunities to infect other hosts with a given pathogen type than do the majority of individuals of the same host type. Typically this would mean that an individual had many more contacts, where ‘contact’ is defined by the route of transmission of the pathogen (e.g. direct contact through proximity, physical contact, sexual contact, or indirect contact by contamination of food, being bitten by a large number of vectors etc.)58.
Thus defined, super-shedding and super-spreader are independent traits: super-shedding reflects the interaction of the host with the pathogen whereas super-spreading reflects the interaction of the host with other hosts.
(ii). E. coli O157 super-shedders
For E. coli O157, several working definitions of a super-shedder can be found in the literature. The most basic of these are derived from single direct counts of E. coli O157 in faecal samples. Cut-offs for super-shedding have been suggested at counts ≥103 or ≥104 colony forming units (CFU) g-1 faeces15,17,59,60, or the simple identification of outlying counts has been used61.
Such measures are simple to use but do not directly indicate whether an animal is colonized at the terminal rectum, nor do they allow for (possibly substantial) sample-to-sample variation in bacteria counts during the course of an infection. A recent longitudinal study20 using recto-anal mucosal swabs (RAMS) defined a super-shedder on the basis of both mean concentration (≥104 CFU/RAMS) and persistent colonization (≥ 4 consecutive positive RAMS, where sampling was carried out twice per week for 14 weeks).
The above definition appears ideal as it encompasses both properties of a super-shedder (high excretion and colonization of the terminal rectum), however, longitudinal examination is not always possible. Hence, for the purpose of identifying super-shedders (for control purposes, for example) we need a suitable working definition that is readily employable in field situations. A recent study20 observed the concentration of E. coli O157 in the faeces was positively associated with the estimated duration of culture-positive status. This is consistent with the findings of a study of cattle at slaughter where shedding >103 CFU g-1 faeces was associated with bacterial carriage close to the terminal rectum whereas shedding <103 was not17.
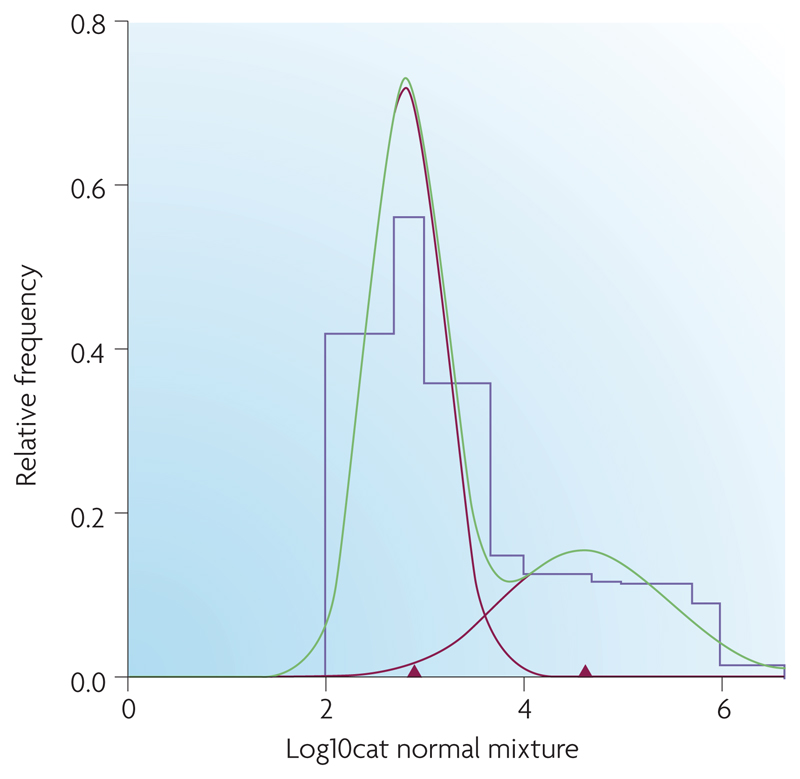
A more formal analysis of E. coli O157 faecal counts was performed in a recent paper13. Mixture distribution analysis has suggested a cut-off of 3,135 CFU g-1 faeces (low and high confidence threshold: 1,658 and 10,395)13 (FIG 3). This figure formally confirms the approximated estimates of >103 to >104 in the current literature.
On the basis of the available evidence we propose a working definition of an E. coli O157 super-shedder as an animal excreting >104 CFU g-1 faeces. Although less stringent that previous recommendations19, we consider that such high shedding levels are unlikely to occur without colonization. We note, however, that at times during the course of infection or due to sampling variation even colonized animals may excrete at levels below this threshold. In other words, our working definition has high specificity but somewhat lower sensitivity for detecting super-shedders.
It is also possible that wildlife reservoirs and other livestock species play a role as sheep have been reported to excrete concentrations of bacteria comparable with super-shedder cattle24. In a study of sheep at slaughter in Scotland, the counts of E. coli O157 in faeces were occasionally >107 CFU ml-1, suggestive of the existence of other host species as super-shedders of the organism (unpublished data). In addition, a high prevalence of E. coli O157 was observed24 in a sheep flock in Scotland with individuals in the flock shedding up to 106 CFU.g-1.
Despite an increasing number of published articles on E. coli O157 super-shedders, no formal definition has been provided (see Box 1). The results of both field and experimental studies highlight the heterogeneity of E. coli O157 carriage and excretion rates. Field studies have demonstrated that the majority (75%) of positive faecal samples contain <102 colony forming units (CFU) CFU.g-1 faeces of E. coli O15713. This is above the detection threshold for immunomagnetic separation (IMS) techniques but below the threshold for accurate enumeration13. By comparison, a minority of animals may be excreting E. coli O157 at levels up to >107 CFU.g-1 faeces13. Such a variation in levels of E. coli O157 excretion cannot be explained by a single distribution representing one homogeneous population (FIG 3). The consequences of this are substantial; one study reported that high shedders (there defined as ≥104 CFU.g-1 faeces) made up 9% of a sample of slaughter cattle but were responsible for >96% of all E. coli O157 bacteria shed15. Similar results were observed in data from the Scottish study13 where high shedders (there defined as ≥3 x 103 CFU.g-1 faeces) comprised 8% of the cattle but 99% of the bacteria shed.
Figure 3.
Results of mixture distribution analysis on a histogram of E. coli O157 counts (log transformed) gathered from faecal pats collected during a cross-sectional survey of Scottish store and finishing cattle (2002-2004)13. This data was analyzed using MIX (Ichthus Data Systems, Hamilton, Ontario, Canada), a program that analyzes histograms as mixtures of statistical distributions. Parameters of the mixture distribution (mean, standard deviation, proportion) were calculated as maximum likelihood estimates (MLE) using a combination of the Expectation-Maximization (EM) algorithm and Newton-type methods. Final models were fitted assuming two normal components. This figure shows the original histogram of E. coli O157 counts (purple line) fitted with 2 component normal distributions (red lines). The green line is their sum, the mixture distribution, which matches the histogram as closely as possible. The red triangles mark the mean E. coli O157 count within each component distribution. The point estimate of threshold for a super- shedder was chosen as the point of overlap of the two unscaled distributions, approximately 3135 C.F.U. g-1; 95% CI for threshold estimate, 1658 and 10395. Note that this does not match the value which might be inferred from this figure, where the components are scaled relative to the sizes of the observed populations. Reproduced with permission from REF 13 © 2007American society of Microbiology.
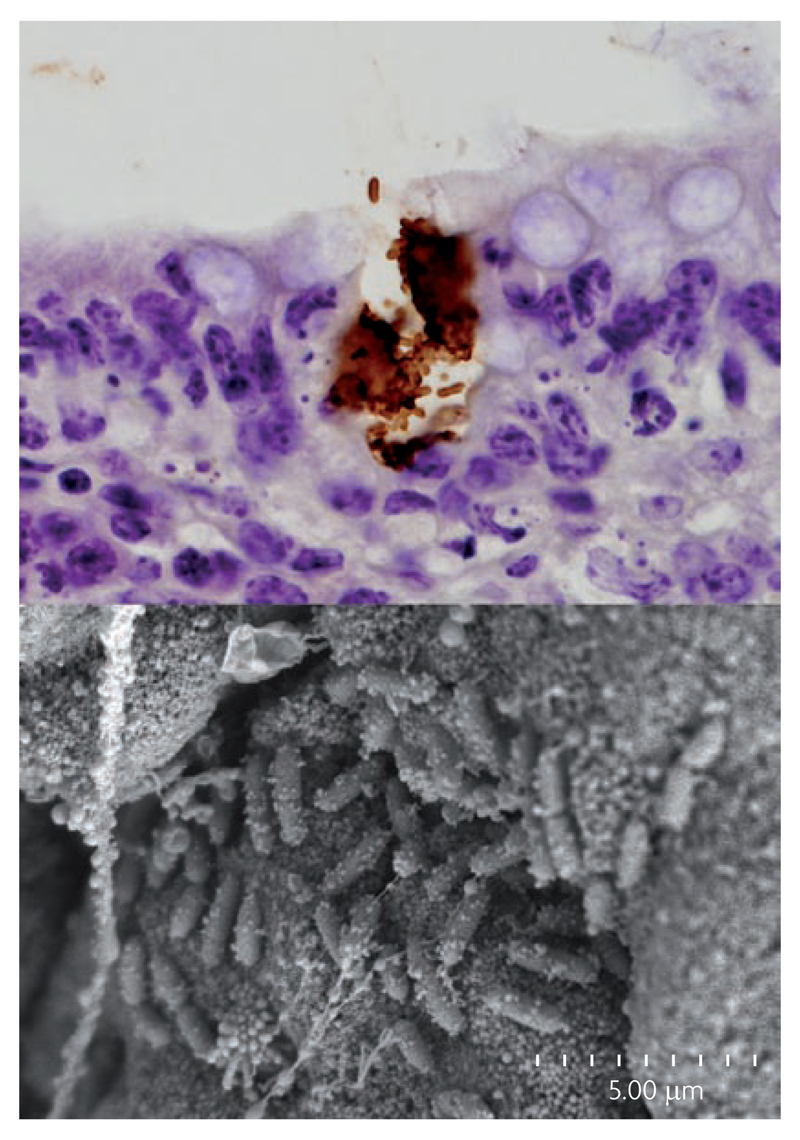
In 2003 it was shown that some E. coli O157 infections of cattle colonize the mucosal epithelium at the terminal rectum16 (FIG. 4). Subsequently, both field and experimental studies have indicated that cattle colonized at this site are associated with the high levels of faecal excretion and longer-duration faecal shedding17,18,19. Non-colonized cattle may only shed bacteria over a few days whereas colonized cattle may shed bacteria for weeks or months18,20. The positive correlation between the concentration of E. coli O157 in the faeces and the duration of carriage17,20 can be exploited to suggest a working definition of a super-shedder based on a threshold excretion rate of ≥104 CFU.g-1 faeces (Box 1). We propose that super-shedders are the subset of animals that are colonised at the terminal rectum and that replication at this site leads to excretion in the faeces at levels greater than this threshold. By contrast, we suggest that the majority of positive animals shedding at lower levels may be the result of amplification in the faeces during transient passage through the animal host or the result of colonisation with lower level replication at sites other than the terminal rectum. Based on examination of clonal similarity of E. coli O157 isolates using PFGE analysis it has been suggested21 that E. coli O157 in faeces came from two sources: colonized at the terminal rectum or transient in the gastrointestinal tract. This result is confirmed in a study of long-duration E. coli O157 colonization18 where the bacteria were cultured only from the terminal rectum and not from any other location in the gastrointestinal tract.
Figure 4.
Colonization of cattle terminal rectum by E. coli O157. The top panel shows immuno-peroxidase staining of a microcolony of E. coli O157 at the rectal epithelium of an experimentally-colonized calf. The brown staining is of the surface O157 LPS and shows that the E. coli O157 micro-colony has eroded the epithelium (x2000). The bottom panel shows a scanning electron micrograph of a bacterial micro-colony from the terminal rectum of a calf colonized with E. coli O157 (scale indicated). Images provided courtesy of Pablo Nart, University of Edinburgh.
At present, we have limited knowledge of what leads to rectal colonization and the generation of a super-shedder infection but risk factors may be related to the host (phenotype or genotype), the pathogen (strain) and/or other, ‘environment’ factors (e.g. route of transmission or exposure dose). One epidemiological study has identified a pathogen-related risk factor for super-shedding13. There, super-shedding (defined in terms of faecal bacteria counts – see above) was associated (odds ratio >2) with infections with a particular E. coli O157 phage type (PT), PT 21/28. The biological significance of these findings is unclear. It is possible that PT 21/28 may be a marker for some genetic difference or altered gene expression of E. coli O157 resulting in that particular subgroup being shed from cattle at higher levels (BOX 2) and therefore more likely to be associated with super-shedding animals.
Box 2. The association between phage type and super-shedding.
One explanation for an association of phage type (PT) 21/28 with super-shedder infections could be that PT 21/28 is a marker for some key genetic difference or altered gene expression in E. coli O157. It has been suggested recently that altered regulation of the type III secretion system (T3S) in PT 21/28 strains compared to other phage type strains may enable the bacteria to colonize and be excreted at higher levels. There is variation in the expression of T3S proteins by different E. coli O157 strains that could impact on colonization62. It is known that T3S is essential for bovine colonization of cattle at the terminal rectum and therefore the establishment of super-shedders16,63,64. Macro-sequence differences in E. coli strains occur as a result of the insertion and deletion of horizontally-acquired genomic ‘O’ islands (OIs) as well as the acquisition of plasmids65. Many of the larger OIs were originally inserted bacteriophage genomes and much of the variation between E. coli O157 strains is the result of differences in the OI/phage complement66,67,68. There are different shiga-toxin encoding lambdoid phages and these carry an assortment of accessory genes along with those required for lysogeny/lysis. In addition, their integration sites into the genome can also vary69. Phage typing will be responsive to the phage constitution or insertion history as the presence of different phage, or remnants thereof, can provide resistance to infection with another. Hence the association between phage type and super-shedders is likely to represent differences in the OI/phage repertoire of the strain. OIs contain effector proteins that are secreted by the T3S70, regulators that control T3S71 as well as adhesins72,73 that may also be directly relevant to bovine colonization. Future work should now correlate the presence of particular OIs with phenotypic and epidemiological data.
In addition, a recent study in Canada has also implicated pathogen-related factors. This study revealed strong correlations between the lineage-specific polymorphism assay (LSPA6) genotype and phage type of E. coli O157 strains examined22. Previous research had suggested possible genotypic differences may underlie phenotypic differences between LSPA6 lineage I and II genotypes22. These two lineages differ in their frequency of association with human disease. However, further study of E. coli O157 strains from other geographic regions is needed to determine whether the association observed is widely applicable22.
The possibilities that super-shedding is also influenced by host genotype or phenotype or environmental factors have yet to be investigated but these may well turn out to be significant.
Super-shedding and within-farm transmission
Natural transmission of E. coli O157 between cattle is thought to be largely via the faecal-oral route, although transmission may be indirect via an environmental reservoir23. It is also possible that wildlife reservoirs and other livestock species play a role as sheep have been reported to excrete concentrations of bacteria comparable with super-shedder cattle24 (see Box 1).
Cattle excreting large numbers of bacteria may be expected to pose a greater risk to other cattle than cattle excreting at low levels25. This is substantiated by data generated from both modelling and experimental studies that have quantified E. coli O157 shedding. Mathematical modelling has suggested that the observed distribution of prevalence of E. coli O157 infection at the farm level (most O157-positive farms having low prevalence but a few having very high prevalence) is best explained when a small proportion of cattle are assumed to have much higher transmission rates than the others26,14. Associations have also been observed between the presence of a super-shedder and a high prevalence of low-level shedders on a farm17,19,13 – these results are consistent with higher transmission rates where there are super-shedders, although they could simply reflect that (rare) super-shedders are more likely to be found where there are many exposed cattle. However, the same association has also been found in a longitudinal study19 where the risk of infection was much higher for cattle in a pen with a super-shedder, with the supporting observation that these infections tended to have similar DNA pulsed-field gel electrophoresis (PFGE) type to the super-shedder. In the field, there is often limited PFGE diversity on individual farms27 and on three-quarters of farms with PT 21/28 present no other phage types were found28.
Higher transmission rates on farms with super-shedders could reflect either or both of two effects. First, susceptible cattle are more likely to encounter positive faecal pats and local environmental contamination. Second, when they are exposed they are more likely to be exposed to higher numbers of bacteria. There have been few studies of the effect of dose on E. coli O157 infection of cattle, although it is known that oral exposure to <300 CFU can result in infection and that the probability of infection increases with dose29. We do not know the relationship between dose and the production of super-shedders but this could have important effects on the transmission dynamics (BOX 3).
Box 3. A simple theoretical framework.
A number of simulation models of the within-herd dynamics of E. coli O157 transmission have been developed, but these have not incorporated super-shedders74,75,76. Here, we consider the dynamics of E. coli O157 infection in a local, ‘well-mixed’ population of cattle, represented by a modified version of the standard SIS (susceptible-infected-susceptible) model which allows two kinds of infection (W for super-shedder; V for non-super-shedder) and an environmental reservoir (E, measured in ‘units’ of infection).
The model is:
where S=1-V-W is the fraction uninfected, βVE and βWE are the per capita rates of infection from the reservoir, γV and γW are the per capita recovery rates, λV and λW are the per capita rates infectious units are added to the reservoir, and 1/σ is the mean survival time of an infectious unit in the environment.
This simple model regards all E. coli O157 infections as equivalent, but can readily be adapted to be strain specific. Distinct dynamics for different strains would be represented by variation in any or all of the seven parameters in the model. In particular, for some strains βW may be very low or even zero, i.e. they rarely or never generate super-shedders. Previously published work13 has shown different phage-types to have different propensities to be super-shedders: in this case we might expect the ratio βW/ βV to be greater for the phage-type 21/28 than other phage-types such as PT32.
Viewing the population as a whole, experimental evidence suggests that γV > γW, perhaps by an order of magnitude or more. However, the steady state prevalence of super-shedders in Scottish cattle is lower than the prevalence of non-super-shedders (W*≈1% and V*≈5%). This implies that βV >> βW, i.e. that it is very much easier to generate a non-super-shedder than a super-shedder. One possible explanation is that to generate a super-shedder requires an orders of magnitude higher exposure dose. In the case of PT 21/28, for example, a higher prevalence of infection than observed for other PTs, could mean that cattle have an increased likelihood of encountering a higher dose.
The model predicts a steady state level for the environmental reservoir of
By definition we expect λV << λW, perhaps by two or three orders of magnitude. This difference is much greater than the difference between V* and W*, implying that super-shedders make the greatest contribution to the reservoir and so to the risk of subsequent infection of both cattle and humans. If super-shedders could be rapidly identified and removed (i.e. γW becomes very large) then the prevalence of infection will decrease and if
then E. coli O157 will be eradicated. However, if σ is small the decline in prevalence will be slow without additional measures directed at the environment reservoir. It is also possible that the reservoir is self-sustaining, either by growth of the bacteria in the environment or if the ‘environment’ includes alternative host species. Again, this will slow any decrease in prevalence following the removal of super-shedders and could prevent eradication.
The dynamics of E. coli O157 infection would be considerably more complex (and the effectiveness of control potentially greater) if there were non-linear effects of exposure dose on the probability of infection, requiring that the term βWE be replaced by a function where incidence is no longer simply proportional to E. If transmission increases more rapidly than linearly with E, then simple models demonstrate that the steady state prevalence responds more readily to reductions in shedding. Such a non-linear effect would arise, for example, if there were a minimum dose needed to generate a super-shedder. The relationship between dose and super-shedding in vivo has yet to be systematically investigated.
Between-farm transmission
Some studies30 have suggested frequent between-farm transmission on the basis of finding subtypes of E. coli O157 that could not be distinguished using macro-restriction fragment analysis of PFGE. Through examination of PFGE types it has been suggested that E. coli O157 isolates are frequently transferred over short31 as well as long32 and sometimes global distances33. Data from Scotland show that 15 of 105 PFGE types were found on more than one farm (out of 91 positive farms), consistent with the results of other studies34 and with the suggestion that between farm transmission does occur.
The routes of between-farm transmissions are not clearly understood. Possibilities include the movement of contaminated faeces or soil via farm personnel or equipment, the movement of contaminated feed33 or the movement of wildlife (e.g. wild birds) onto farms30. Another potentially important contributor is the movement of cattle between farms. There is much evidence in the literature to support an association between cattle movement and the farm level risk of E. coli O157 shedding13,35,36,37,38. Exploratory mathematical models39 have shown that cattle movement can make a significant contribution to the observed prevalence of E. coli O157 positive farms but may not be sufficient by itself to maintain E. coli O157 in the population40. In practice, even closed herds (i.e. those not importing any cattle) are not fully protected against the introduction of E. coli O15730.
The role of super-shedders in between-farm transmission and the long term persistence of E. coli O157 in the cattle population have yet to be formally investigated. However, since super-shedding is associated with higher on-farm prevalence there will be a greater chance of transmission via movement (and perhaps via other routes too) from a farm with super-shedders (noting that not all cattle are equally likely to be moved to another farm), and presumably a higher chance of establishing infection on an uninfected farm if a super-shedder is imported.
Carriage by cattle and the risk to humans
Environmental exposure is now recognized as an important route for human infection with E. coli O15740, an observation supported by case-control studies41,42,43 and spatial analysis9,10,11. Oral exposure to cattle faeces is significant, but more general environmental exposure is also likely as the bacteria are capable of long-term survival in manure, pasture and soil44. A dose-response model has been used to estimate an ID50 for environmental exposure of approximately 102 to 105 bacteria45. Exposure doses are typically much lower than this so only a small proportion of exposed individuals will go on to develop an infection46. Importantly, there is expected to be a close relationship between numbers of E. coli O157 shed by cattle and the proportion of exposed humans infected46, implying that environmental contamination by super-shedding cattle results in a much (up to 100-fold or 1000-fold) higher risk.
Sporadic human infections have been related to the presence of cattle, even without knowledge of their status as carriers of E. coli O157. Significant clustering of human infection was found in Scotland in regions where the ratio of the cattle population to human population was high11, echoing similar associations reported by studies elsewhere9,10. Recently, it was observed that HUS paediatric incidence was associated with dairy cattle density and the ratio of calves to children <15 years old47.
A direct link between cattle and human infections has also been suggested on the basis of subtyping, primarily phage typing and PFGE48. In many countries the most common phage types among bovine isolates are also common among human isolates, supporting the idea that cattle are a principal reservoir49. In Scotland, PT 21/28 was by far the most common phage type found, accounting for 50% of the cattle isolates28 and also for 72% of isolates obtained from human infections over the same period (unpublished data). PT 21/28 was also by far the most widespread phage type, found on >50% of all positive farms across Scotland, although with a slight bias toward the north of the country compared to other phage types28.
PT 21/28 is of particular concern because of its association with more severe human morbidity. In Scotland from 1997 to 2001, 61% of HUS cases in individuals less than 16 years old were attributable to infection with PT 21/2850. In the UK and Ireland (1997-2001), the risk of developing diarrhoea-associated HUS was significantly higher in children infected with PT 21/28 compared to other phage types50. There are two (not mutually exclusive) hypotheses to explain this observation: (i) PT 21/28 is a marker for (so far unidentified) human virulence factors; and (ii) PT 21/28 is typically ingested in higher doses than other phage types. Here, we are particularly concerned with the second hypothesis, which seems plausible given the greater abundance of PT 21/28.
Conclusions
The preceding sections all support the schema shown in Figure 2. It suggests that the epidemiology of E. coli O157 may be driven largely by a small number of colonized cattle that for a period are shedding large numbers of bacteria, so called super-shedders (see BOX 1). There is evidence that not all E. coli O157 are equally likely to cause super-shedder infections. In Scotland this trait is often associated (though not always) with a particular phage type, PT 21/28 and this has two important implications.
First, there is obviously a need for greater research effort directed at identifying factors that generate a super-shedder. The association with PT 21/28 implicates pathogen factors, and there are a number of candidates for further investigation (see BOX 2). Host genetic factors have yet to be studied; but some aspects of cattle phenotype could be investigated through epidemiological studies of individual-level risk factors for super-shedding. An additional factor that merits attention is the relationship between super-shedding and exposure dose - the form of this relationship has important implications for the population dynamics of super-shedding (see BOX 3).
Second, there are practical implications for the control of E. coli O157 and reducing the risk of human infection. To date, consideration of control options for E. coli O157 in cattle has not taken super-shedding into account51. However, the development of a method for identifying super-shedders would allow control strategies to be targeted at removing or eliminating high-level faecal excretion and so to greatly reduce the prevalence of the organism in the host and the risk of human infection52. Possible strategies include the detection and removal of super-shedding cattle, testing prior to movement of individual animals, intervention at slaughterhouses and, in the longer term, vaccination to restrict the likelihood of colonization53 or direct treatment of colonized animals52. Mathematical models will be useful for exploring the effectiveness of control measures targeted at super-shedders both for reducing bacterial excretion levels in cattle and the risk of infection in humans (both through environmental and food-borne exposure). Ultimately, however, it will be necessary to implement field trials, and we are optimistic that these will feasible within the next few years.
In this article we have reviewed evidence surrounding our proposed understanding of the pathogenesis and epidemiology of E. coli O157 and its implications for transmission within and among farms, as well as to humans. Central to this article has been the concept of super-shedding cattle. The biology and epidemiology of super-shedding is understood far better for E. coli O157 than any other system. Although we have focused on E. coli O157 in this article, there is now recognition that super-shedding may be a feature of other bacterial infections, such as Mycobacterium avium subsp. paratuberculosis (MAP)54 and Salmonella enterica Typhimurium55.
Analysis of outbreak data has suggested that, in general, a minority of individuals are responsible for the majority of infections in a population14,56. If super-shedders are a common phenomenon of most infections, this will have profound implications for our understanding of the epidemiology and control of many infectious diseases. In the case of E. coli O157 in cattle, this could potentially lead to a reduction in the risk of human infections. Although we have focussed on environmental exposure, super-shedding does affect carcass contamination and so is likely to be important for food contamination as well57.
Supplementary Material
Acknowlegdements
This study was a part of the International Partnership Research Award in Veterinary Epidemiology (IPRAVE), Epidemiology and Evolution of Enterobacteriaceae Infections in Humans and Domestic Animals, funded by the Wellcome Trust. DG is funded by DEFRA under the Veterinary Training and Research Initiative and by LK0006. The authors would like to thank all members of the IPRAVE consortium.
References
- 1.Karmali MA. Infection by shiga toxin-producing Escherichia coli. An overview. Mol Biotechnol. 2004;26:117–122. doi: 10.1385/MB:26:2:117. [DOI] [PubMed] [Google Scholar]
- 2.Griffin PM, Tauxe RV. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli and the associated haemolytic uremic syndrome. Epidemiol Rev. 1991;30:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 3.Caprioli A, Morabito S, Brugère H, Oswald E. Enterohaemorragic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 Infections associated with consumption of fresh spinach---United States, September 2006. MMWR Weekly. 2006;55:1045–1046. [PubMed] [Google Scholar]
- 5.Mead PS, Griffin PM. Escherichia coli O157:H7. Lancet. 1998;352:1207–1212. doi: 10.1016/S0140-6736(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 6.Sakuma M, Urashima M, Okabe N. Verocytotoxin producing Escherichia coli, Japan, 1999-2004. Emerg Infect Dis. 2006;12:323–325. doi: 10.3201/eid1202.050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollock KGJ, Beattie TJ, Reynolds B, Stewart A. Clinical management of children with suspected or confirmed E. coli O157 infection. S.M.J. 2007;52:5–7. doi: 10.1258/rsmsmj.52.3.5. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong GL, Hollingsworth J, Morris JG. Emerging food pathogens: Escherichia coli O157:H7 as a model entry of a new pathogen into the food supply of the developed world. Epidemiol Rev. 1996;18:29–51. doi: 10.1093/oxfordjournals.epirev.a017914. [DOI] [PubMed] [Google Scholar]
- 9.Michel P, et al. Temporal and geographic distributions of reported cases of Escherichia coli O157:H7 infection in Ontario. Epidemiol Infect. 1999;122:193–200. doi: 10.1017/s0950268899002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valcour JE, Michel P, McEwen SA, Wilson JB. Associations between indicators of livestock farming intensity and incidence of human shiga toxin-producing Escherichia coli infection. Emerg Infect Dis. 2002;8:252–257. doi: 10.3201/eid0803.010159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Innocent GT, et al. Spatial and temporal epidemiology of sporadic human cases of Escherichia coli O157 in Scotland 1996-1999. Epidemiol Infect. 2005;153:1033–1041. doi: 10.1017/S0950268805003687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunn GJ, et al. An investigation of factors associated with the prevalence of verocytotoxin producing Escherichia coli O157 shedding Scottish beef cattle. Vet J. 2007;174:554–564. doi: 10.1016/j.tvjl.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 13.Chase-Topping ME, et al. Risk factors for the presence of high-level shedders of Escherichia coli O157 on Scottish farms. J Clin Microbiol. 2007;45:1594–1603. doi: 10.1128/JCM.01690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews L, et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc Nat Acad Sci USA. 2006;103:547–552. doi: 10.1073/pnas.0503776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omisakin F, MacRae M, Ogden ID, Strachan NJC. Concentration and prevalence of Escherichia coli O157 in cattle faeces at slaughter. Appl Environ Microbiol. 2003;69:2444–2447. doi: 10.1128/AEM.69.5.2444-2447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naylor SW, et al. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonisation of enterohaemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun. 2003;71:1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Low JC, et al. Rectal Carriage of Enterohemorrhagic Escherichia coli O157 in Slaughtered cattle. Appl Environ Microbiol. 2005;71:98–7. doi: 10.1128/AEM.71.1.93-97.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim JY, Li J, Sheng H, Besser TE, Potter K, Hodve CJ. Escherichia coli O157:H7 colonization at the rectoanal junction of long duration culture positive cattle. Appl Environ Microbiol. 2007;73:1380–1382. doi: 10.1128/AEM.02242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobbold RN, et al. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl Environ Microbiol. 2007;73:1563–1568. doi: 10.1128/AEM.01742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis MA, et al. Comparison of cultures from rectoanal-junction mucosal swabs and feces for detection of Escherichia coli O157 in dairy herds. Appl Environ Microbiol. 2006;72:3766–3770. doi: 10.1128/AEM.72.5.3766-3770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox JT, Shi X, Nagaraja TG. Escherichia coli O157 in the rectoanal mucosal region of cattle. Foodborne Pathog Dis. 2008;5:69–77. doi: 10.1089/fpd.2008.0042. [DOI] [PubMed] [Google Scholar]
- 22.Ziebell K, et al. Genotypic characterization and prevalence of virulence factors among Canadian Escherichia coli O157:H7 strains. Appl Environ Microbiol. 2008;74:4314–4323. doi: 10.1128/AEM.02821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancock DD, et al. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the northwestern United States. Prev Vet Med. 1998;35:11–19. doi: 10.1016/s0167-5877(98)00050-6. [DOI] [PubMed] [Google Scholar]
- 24.Ogden ID, MacRae M, Strachan NJC. Concentration and prevalence of Escherichia coli O157 in sheep faeces at pasture in Scotland. J Appl Microbiol. 2005;98:646–651. doi: 10.1111/j.1365-2672.2004.02493.x. [DOI] [PubMed] [Google Scholar]
- 25.Berg J, et al. Escherichia coli O157:H7 excretion by commercial feedlot cattle fed either barley- or corn-based finishing diets. J Food Protect. 2004;67:666–671. doi: 10.4315/0362-028x-67.4.666. [DOI] [PubMed] [Google Scholar]
- 26.Matthews L, et al. Super-shedding cattle and the transmission dynamics of Escherichia coli O157. Epidemiol Infect. 2006a;134:131–142. doi: 10.1017/S0950268805004590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vali L, et al. Comparison of diversities of Escherichia coli O157 shed from a cohort of spring-born beef calves at pasture and in housing. Appl Environ Microbiol. 2005;71:1648–1652. doi: 10.1128/AEM.71.3.1648-1652.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halliday JEB, et al. Herd-level factors associated with the presence of Phage type 21/28 E. coli O157 on Scottish farms. BMC Microbiol. 2006;6:99. doi: 10.1186/1471-2180-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besser TE, Richards BL, Rice DH, Hancock DD. Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiol Infect. 2001;127:555–560. doi: 10.1017/s095026880100615x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wetzel AN, LeJeune JT. Clonal dissemination of Escherichia coli O157:H7 subtypes among dairy farms in northeast Ohio. Appl Environ Microbiol. 2006;72:2621–2626. doi: 10.1128/AEM.72.4.2621-2626.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vali L, et al. High-level genotypic variation and antibiotic sensitivity among Escherichia coli O157 strains isolated from two Scottish beef cattle farms. Appl Environ Microbiol. 2004;70:5947–5954. doi: 10.1128/AEM.70.10.5947-5954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renter DG, Sargeant JM, Hungerford LL. Distribution of Escherichia coli O157:H7 within and among cattle operations in pasture-based agricultural areas. AJVR. 2004;65:1367–1376. doi: 10.2460/ajvr.2004.65.1367. [DOI] [PubMed] [Google Scholar]
- 33.Davis MA, et al. Correlation between geographic distance and genetic similarity in an international collection of bovine faecal Escherichia coli O157:H7 isolates. Epidemiol Infect. 2003;131:923–930. doi: 10.1017/s0950268803008884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sargeant JM, Shi X, Sanderson MW, Renter DG, Nagaraja TG. Pulsed-field gel electrophoresis patterns of Escherichia coli O157 isolates from Kansas feedlots. Foodborne Pathog Dis. 2006;3:251–258. doi: 10.1089/fpd.2006.3.251. [DOI] [PubMed] [Google Scholar]
- 35.Hancock DD, et al. The prevalence of Escherichia coli O157:H7 in dairy and beef cattle in Washington State. Epidemiol Infect. 1994;113:199–207. doi: 10.1017/s0950268800051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson JB, et al. Risk factors for bovine infection with verocytotoxigenic Escherichia coli in Ontario. Can J of Prev Vet Med. 1993;16:159–170. doi: 10.1016/s0167-5877(97)00010-x. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen EM, Tegtmeier C, Andersen HJ, Gronbaek C, Andersen JS. Influence of age, sex and herd characteristics on the occurrence of verocytotoxin-producing Escherichia coli O157 in Danish farms. Vet Microbiol. 2002;88:245–257. doi: 10.1016/s0378-1135(02)00108-6. [DOI] [PubMed] [Google Scholar]
- 38.Schouten JM, et al. Risk factor analysis of verocytotoxin producing Escherichia coli O157 on Dutch dairy farms. Prev Vet Med. 2004;64:49–61. doi: 10.1016/j.prevetmed.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Lui W-c, et al. Metapopulation dynamics of Escherichia coli O157 in cattle: an explatory model. J Roy Soc Interface. 2007;4:917–924. doi: 10.1098/rsif.2007.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strachan NJC, Dunn GM, Locking ME, Reid TMS, Ogden ID. Escherichia coli O157: Burger bug or environmental pathogen. Int J of Food Microbiol. 2006;112:129–137. doi: 10.1016/j.ijfoodmicro.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Locking ME, et al. Risk factors for sporadic cases of Escherichia coli O157 infection: the importance of contact with animal excreta. Epidemiol Infect. 2001;127:215–220. doi: 10.1017/s0950268801006045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Brien SJ, Adak GK, Gilham C. Contact with farming environment as a major risk factor for shiga toxin (verocytotoxin)-producing Escherichia coli O157 infection in humans. Emerg Infect Dis. 2001;7:1049–1051. doi: 10.3201/eid0706.010626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willshaw GA, Evans J, Cheasty T, Cummins A, Pritchard GC. Verocytotoxin-producing Escherichia coli infection and private farm visits. Vet Rec. 2003;153:365–366. doi: 10.1136/vr.152.12.365. [DOI] [PubMed] [Google Scholar]
- 44.Ogden ID, et al. Long term survival of Escherchia coli O157 on pasture following an outbreak associated with sheep at a scout camp. Lett Appl Microbiol. 2002;34:100–104. doi: 10.1046/j.1472-765x.2002.01052.x. [DOI] [PubMed] [Google Scholar]
- 45.Cassin MH, Lammerding AM, Ross W, McColl RS. Quantitative Risk Assessment of Escherichia coli O157:H7 in ground beef hamburgers. Int J of Food Microbiol. 1998;41:21–44. doi: 10.1016/s0168-1605(98)00028-2. [DOI] [PubMed] [Google Scholar]
- 46.Strachan NJC, Fenlon DR, Ogden ID. Modelling the vector pathway and infection of humans in an environmental outbreak of Escherichia coli O157. FEMS Microbiol Lett. 2001;203:69–73. doi: 10.1111/j.1574-6968.2001.tb10822.x. [DOI] [PubMed] [Google Scholar]
- 47.Haus-Cheymol R, et al. Association between indicators of cattle density and incidence of paediatric haemolytic-uraemic syndrome (HUS) in children under 15 years of age in France between 1996 and 2001: an ecological study. Epidemiol Infect. 2006;134:712–718. doi: 10.1017/S095026880500542X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mora A, et al. Phage types and genotypes of shiga toxin-producing Escherichia coli O157:H7 isolates from humans and animals in Spain: identification and characterization of two predominating phage types (PT2 and PT8) J Clin Microbiol. 2004;42:4007–4015. doi: 10.1128/JCM.42.9.4007-4015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lahti E, et al. Use of phenotyping and genotyping to verify transmission of Escherichia coli O157:H7 from dairy farms. Eur J Clin Microbiol Infect Dis. 2002;21:189–195. doi: 10.1007/s10096-001-0682-0. [DOI] [PubMed] [Google Scholar]
- 50.Lynn RM, et al. Childhood haemolytic Uremic Syndrome, United Kingdom and Ireland. Emerg Infect Dis. 2005;11:590–596. doi: 10.3201/eid1104.040833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callaway TR, et al. What are we doing about Escherichia coli O157 in cattle? J Anim Sci. 2004;82:E93–E99. doi: 10.2527/2004.8213_supplE93x. [DOI] [PubMed] [Google Scholar]
- 52.Naylor SW, et al. Impact of the direct application of therapeutic agents to the terminal recta of experimentally colonised calves on Escherichia coli O157:H7 shedding. Appl Environ Microbiol. 2007;73:1493–1500. doi: 10.1128/AEM.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potter AA, et al. Decreased shedding of Escherichia coli O157:H7 by cattle following vaccination with type III secreted proteins. Vaccine. 2004;22:362–369. doi: 10.1016/j.vaccine.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 54.Whitlock RH, et al. Cattle shedding MAP: A new paradigm. Lansing, Michigan: 2008. [Google Scholar]
- 55.Lawley TD, et al. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigeneous intestinal microbiota. Infect Immum. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fox JT, Renter DG, Sanderson MW, Nutsch AL, Shi X, Nagaraja TG. Associations between the presence and magnitude of Escherichia coli O157 in faeces at harvest and contamination of preintervention beef carcasses. J Food Prot. 2008;71:1761–1767. doi: 10.4315/0362-028x-71.9.1761. [DOI] [PubMed] [Google Scholar]
- 58.Woolhouse MEJ, et al. Heterogeneities in the transmission of infectious agents: Implications for the design of control programs. Proc Nat Acad Sci USA. 1997;94:338–342. doi: 10.1073/pnas.94.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson SE, Wright EJ, Hart CA, Bennett M, French NP. Intermittent and persistent shedding of Escherchia coli O157 in cohorts of naturally infected calves. J Appl Microbiol. 2004;94:1045–1053. doi: 10.1111/j.1365-2672.2004.02390.x. [DOI] [PubMed] [Google Scholar]
- 60.Ogden ID, MacRae M, Strachan NJC. Is the prevalence and shedding concentrations of E. coli O157 in beef cattle in Scotland seasonal? FEMS Microbiol Lett. 2004;233:297–300. doi: 10.1016/j.femsle.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 61.Bach SJ, Selinger LJ, Stanford K, McAllister TA. Effect of supplementing corn- or barley-based feedlot diets with canola oil on faecal shedding of Escherichia coli O157:H7 by steers. J Appl Microbiol. 2005;98:464–475. doi: 10.1111/j.1365-2672.2004.02465.x. [DOI] [PubMed] [Google Scholar]
- 62.Roe AJ, et al. Co-ordinate single-cell expression of LEE4-and LEE5-encoded proteins of Escherichia coli O157:H7. Mol Microbiol. 2004;54:337–352. doi: 10.1111/j.1365-2958.2004.04277.x. [DOI] [PubMed] [Google Scholar]
- 63.Dziva F, van Diemen PM, Stevens MP, Smith AJ, Wallis TS. Identification of Escherichia coli O157:H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiol-SGM. 2004;150:3631–3645. doi: 10.1099/mic.0.27448-0. [DOI] [PubMed] [Google Scholar]
- 64.Naylor SW, et al. E. coli O157:H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiol. 2005;151:2773–2781. doi: 10.1099/mic.0.28060-0. [DOI] [PubMed] [Google Scholar]
- 65.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nature Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 66.Ohnishi M, et al. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc Natl Acad Sci USA. 2002;99:17043–17048. doi: 10.1073/pnas.262441699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kudva IT, et al. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J Bacteriol. 2002;184:1873–1879. doi: 10.1128/JB.184.7.1873-1879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steele M, et al. Identification of Escherichia coli O157:H7 genomic regions conserved in strains with a genotype associated with human infection. Appl Environ Microbiol. 2007;73:22–31. doi: 10.1128/AEM.00982-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Besser TE, et al. Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl Environ Microbiol. 2007;73:671–679. doi: 10.1128/AEM.01035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tobe T, et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. PNAS. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abe H, et al. Global Regulation by Horizontally Transferred Regulators Establishes the Pathogenicity of Escherichia coli. DNA Res. 2008;15:25–38. doi: 10.1093/dnares/dsm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Low AS, et al. Cloning, expression, and characterization of fimbrial operon F9 from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2006;74:2233–44. doi: 10.1128/IAI.74.4.2233-2244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spears KJ, Roe AJ, Gally DL. A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol Lett. 2006;255:187–202. doi: 10.1111/j.1574-6968.2006.00119.x. [DOI] [PubMed] [Google Scholar]
- 74.Turner J, Bowers RG, Begon M, Robinson SE, French NP. A semistochastic model of the transmission of Escherichia coli O157 in a typical dairy herd: dynamics, sensitivity analysis and intervention / prevention strategies. J Theor Biol. 2006;241:806–822. doi: 10.1016/j.jtbi.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 75.Turner J, Bowers RG, Clancy D, Behnke MC, Christley RM. A network model of E. coli transmission within a typical UK dairy herd: the effect of heterogeneity and clustering on the prevalence of infection. J Theor Biol. 2008;254:45–54. doi: 10.1016/j.jtbi.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 76.Wood JC, Spiers DC, Naylor SW, Gettinby G, McKendrick IJ. A continuum model of the within-animal population dynamics of E. coli O157. J Biol Sys. 2006;14:425–443. [Google Scholar]
- 77.European Centre for Disease Provention and Control (ECDC) Annual Epidemiological Report on Communicable Diseases in Europe. 2005 [Online] http://ecdc.europa.eu/pdf/ECDC_epi_report_ 2007.pdf.
- 78.Health protection Agency, Northern Ireland addition. Gastrointestinal infections: 2005. Monthly Report. 2006;15:2–13. [Online] http://www.cdscni.org.uk/publications/MonthlyReports/Volume_15_2006/MonthlyReportVol15No4.pdf. [Google Scholar]
- 79.The Centers for Disease Control and Prevention (CDC) Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food – 10 sites, United States 2005. MMWR. 2006;55:392–395. [Online] http://www.cdc.gov//mmwr/preview/mmwrhtml/mm5414a2.htm. [PubMed] [Google Scholar]
- 80.Paiba GA, et al. Prevalence of faecal excretion of verocytotoxogenic Escherichia coli O157 in cattle in England and Wales. Vet Rec. 2003;153:347–353. doi: 10.1136/vr.153.12.347. [DOI] [PubMed] [Google Scholar]
- 81.Eriksson E, Aspan A, Gunnarsson A, Vågsholm I. Prevalence of verotoxin-producing Escherichia coli (VTEC) O157 in Swedish dairy herds. Epidemiol Infect. 2005;133:349–358. doi: 10.1017/s0950268804003371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vold L, Klungseth B, Kruse H, Skjerve E, Wasteson Y. Occurrence of shigatoxinogenic Escherichia coli O157 in Norwegian cattle herds. Epidemiol Infect. 1998;120:21–28. doi: 10.1017/s0950268897008364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.LeJeune JT, Hancock D, Wasteson Y, Skjerve E, Urdahl AM. Comparison of E. coli O157 and shiga toxin encoding genes (stx) prevalence between Ohio, USA and Norwegian dairy cattle. Int J Food Microbiol. 2006;109:19–24. doi: 10.1016/j.ijfoodmicro.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 84.Oporto B, Esteban JI, Aduriz G, Juste RA, Hurtado A. Escherichia coli O157:H7 and Non-O157 shiga toxin-producing E. Coli in healthy cattel, sheep and swine herds in northern Spain. Zoonoses Public health. 2008;55:73–81. doi: 10.1111/j.1863-2378.2007.01080.x. [DOI] [PubMed] [Google Scholar]
- 85.Schouten JM, et al. Prevalence estimation and risk factors for Escherichia coli on Dutch dairy farms. Prev Vet Med. 2004;64:49–61. doi: 10.1016/j.prevetmed.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 86.Schouten JM, van de Giessen AW, Frankena K, De Jong MCM, Graat EAM. Escherichia coli O157 prevalence in Dutch poultry, pig finishing and veal herds and risk factors in Dutch veal herds. Prev Vet Med. 2005;70:1–15. doi: 10.1016/j.prevetmed.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 87.Heuvelink AE, et al. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J Clin Microbiol. 1998;36:3480–3487. doi: 10.1128/jcm.36.12.3480-3487.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sami M, Firouzi R, Shekarforoush SS. Prevalence of Escherichia coli O157:H7 on dairy farms in Shiraz, Iran by immunomagnetic separation and multiplex PCR. Iran. J Vet Res. 2007;8:319–324. [Google Scholar]
- 89.Vidovic S, Korber DR. Prevalence of Escherichia coli O157 in Saskatchewan cattle: characterization of isolates by using random amplified polymorphic DNA PCR, Antibiotic Resistance Profiles and Pathogenicity Determinants. Appl Environ Microbiol. 2006;72:4347–4355. doi: 10.1128/AEM.02791-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Renter DG, et al. Detection and determinants of Escherichia coli O157:H7 in Alberta feedlot pens immediately prior to slaughter. Can J Vet Res. 2008;72:217–227. [PMC free article] [PubMed] [Google Scholar]
- 91.Elder RO, et al. Correlation of enterohemorragic Escherichia coli O157 prevalence in feces, hides and carcasses of beef cattle during processing. PNAS. 2000;97:2999–3003. doi: 10.1073/pnas.060024897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Murinda SE, et al. Prevalence and molecular characterization of Escherichia coli O157:H7 in bulk tank milk survey of dairy farms in east Tennessee. J Food Prot. 2002;65:752–759. doi: 10.4315/0362-028x-65.5.752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.