Abstract
Background
Intestinal parasitic infections are considered a serious public health problem and widely distributed worldwide, mainly in urban and rural environments of tropical and subtropical countries. Globally, soil-transmitted helminths and protozoa are the most common intestinal parasites. Blastocystis sp. is a highly prevalent suspected pathogenic protozoan, and considered an unusual protist due to its significant genetic diversity and host plasticity.
Methodology/main findings
A total of 294 stool samples were collected from inhabitants of three rural valleys in Rio de Janeiro, Brazil. The stool samples were evaluated by parasitological methods, fecal culture, nested PCR and PCR/Sequencing. Overall prevalence by parasitological analyses was 64.3% (189 out of 294 cases). Blastocystis sp. (55.8%) was the most prevalent, followed by Endolimax nana (18.7%), Entamoeba histolytica complex (7.1%), hookworm infection (7.1%), Entomoeba coli (5.8%), Giardia intestinalis (4.1%), Iodamoeba butchilii (1.0%), Trichuris trichiura (1.0%), Pentatrichomonas hominis (0.7%), Enterobius vermicularis (0.7%), Ascaris lumbricoides (0.7%) and Strongyloides stercoralis (0.7%). Prevalence of IPIs was significantly different by gender. Phylogenetic analysis of Blastocystis sp. and BLAST search revealed five different subtypes: ST3 (34.0%), ST1 (27.0%), ST2 (27.0%), ST4 (3.5%), ST8 (7.0%) and a non-identified subtype.
Conclusions/significance
Our findings demonstrate that intestinal parasite infection rates in rural areas of the Sumidouro municipality of Rio de Janeiro, Brazil are still high and remain a challenge to public health. Moreover, our data reveals significant genetic heterogeneity of Blastocystis sp. subtypes and a possible novel subtype, whose confirmation will require additional data. Our study contributes to the understanding of potential routes of transmission, epidemiology, and genetic diversity of Blastocystis sp. in rural areas both at a regional and global scale.
Introduction
Intestinal parasitic infections (IPIs) are ubiquitous in humans, both in urban and rural environments from tropical and subtropical countries. The poorest and most deprived communities are at increased risk of intestinal parasitic infections, which are present in more than a quarter of the world’s population [1–3]. The frequency of intestinal parasites is an indicator of low socioeconomic development of a population, which is directly associated with educational deficits and poor sanitary conditions [4]. However, in tropical developing countries, rural life is by itself associated with a high risk of infections due to negligible health knowledge, lower socio-economic conditions, inadequate environmental sanitation, insufficient water supply [5, 6] and higher contact rates with wildlife and domestic reservoirs of infection [7–9]. Rural populations experience a vicious cycle of malnutrition and re-infections leading to continuous morbidity and perpetuation of poverty cycles. IPIs are a public health problem caused by helminths and intestinal protozoa [3, 10, 11]. Globally, the soil-transmitted helminthes Ascaris lumbricoides, hookworm and Trichuris trichiura and the protozoan Entamoeba histolytica, Giardia intestinalis and Cryptosporidium sp. are the most common intestinal parasites [12]. Except for E. histolytica, Cryptosporidium spp and Balantidium coli, they are unable to invade the mucosal tissues or other organs. In the recent years, there has been a growing recognition that Blastocystis sp. presents pathogenic potential, although its virulence mechanisms are not understood. They infect a wide range of animals including birds, amphibians, reptiles, insects and mammals, including humans [13–18]. Although the current knowledge of reservoirs for human infection is limited, it is known that a vehicle for Blastocystis sp. transmission is close contact with animals [15, 16, 19]. Blastocystis sp. has potential pandemic distribution, possibly reaching 30% in industrialized countries and up to 76% in developing nations [12, 20, 21].
Analysis of the small-subunit ribosomal RNA of Blastocystis sp. isolates revealed substantial genetic diversity, represented by 17 genetically distinct ribosomal lineages (subtypes/ST1-ST17) [18, 22]. Ten subtypes (ST1-ST9, and ST12) have been isolated from human and animal fecal samples (except ST9) [7, 16, 23–25], whereas others were exclusively found in non-human hosts [14, 26]. The significance of Blastocystis sp. to public health, potential zoonotic differences and clinical outcomes may be related to specific subtype, a hypothesis that has been the topic of recent debate and requires further examination [15, 27].
The molecular epidemiology of Blastocystis sp. infections is still unknown in many parts of the world. Brazil is a continental country and information on the distribution of STs in the country is still incipient, but a high prevalence of Blastocystis sp. has been reported both in urban and rural environments. Moreover, IPIs are still a major public health problem, especially among the impoverished and underprivileged communities living in rural and remote areas, which represent an important target of public health interventions. To address this gap, we investigated the prevalence of intestinal parasites and distribution of Blastocystis sp. STs in inhabitants of rural areas of Rio de Janeiro, Brazil. We report a high prevalence of intestinal parasitic infections and a wide range of Blastocystis sp. STs in the area.
Materials and methods
Study population
A cross-sectional study was carried out from October to December of 2013 in Pamparrão, Porteira Verde and Porteira Verde Alta valleys, in rural area of Sumidouro, State of Rio de Janeiro, Brazil (22° 02’ 46” S; 42° 41’ 21”W). The valleys are populated by small rural properties characterized by vegetable crops and pasture. Total population is around 14,900 inhabitants, and 87.7% of people work in agriculture [28]. The Sumidouro municipality is located in the central mountains bordering the Serra do Mar in Rio de Janeiro, at altitudes varying between 264 and 1,300 meters. The region exhibits marked climatic contrasts due to its peculiar geographical position and humid-mesothermic climate [29], with average rainfall between 153.5 mm and 269.4 mm.
Participants were informed about the study aims, potential risks and benefits, and provided written informed consent followed by signatures of two external witnesses. Relatives or legal tutors provided informed consent for children. Participation was voluntary and people could withdraw from the study at any time without further obligation. To preserve anonymity, each study participant was given a unique identification number. At the end of the study, all participants diagnosed with enteric parasitic pathogens received proper therapy. Treatment was not offered to individuals infected with Blastocystis sp., because its pathogenic potential is still a topic of debate. The study protocol was reviewed and approved by the Human Ethics Committee of the Faculdade Medicina from Universidade Federal Fluminense (CAAE 35028314.2.0000.5 243).
Parasitological assays
A total of 294 unpreserved stool samples were collected from participants at home, kept at 4°C and transported to the laboratory at the Fundação Oswaldo Cruz in ice packs. Fresh unpreserved stool samples were fractionated into two aliquots upon receipt. Parasitological examination was performed by spontaneous sedimentation technique (also known as Lutz technique or Hoffman, Pons and Janer technique)[30] and flotation in saturated sodium chloride solution density 1.2 g/mL[31]. Direct in vitro xenic cultivation of Blastocystis sp. from an aliquot of stool samples was performed using Pavlova’s medium, supplemented with 10% heat inactivated adult bovine serum and penicillin-streptomycin antibiotics and incubated at 37°C [32]. Screening of Blastocystis sp. cultures was performed using standard light microscopy during the first three days of incubation. When typical forms of the parasite (vacuolar, granular and amoeboid forms) were observed, Blastocystis sp. culture aliquots were frozen at -20°C for subsequent DNA extraction. An aliquot of fresh unpreserved stool samples was stored at -20°C prior to analysis. Statistical analyses were carried out with Epi-Info 3.5.1. Relationships between variables were examined by chi-square tests with significance levels set at 5%.
Molecular analysis
DNA extraction from cultured samples, positive for Blastocystis sp. and from human stools, positive for E. histolytica complex was carried out using the the Qiamp DNA Stool Mini Kit (Qiagen, Valencia, CA) according to manufacturer’s recommendations. Samples exhibiting structures of the E. histolytica complex under microscopic examination were subjected to DNA amplification.
Nested PCR targeting 18S-like ribosomal RNA gene was applied to genetically identify the E. histolytica, E. dispar and E. moshkovskii complex [33]. First-round PCR for detection of Entamoeba genus used the forward primer E-1 (5' TAAGATGCACGAG AGCGAAA3') and reverse primer E-2 (5' GTACAAAG GGCAGGGACGTA 3'). PCR reaction was performed in a 50 μL volume, with the final mix containing 10× PCR buffer, 1.25 mM of dNTPs, 2.5 mM MgCl2, 10 pmoles of each primer, 2.5 U of Taq polymerase (Invitrogen Life Technologies, Carlsbad, CA, USA), and 2.5 μlL of DNA template. PCR cycles consisted of 5 min at 95°C and 40 cycles of 30 s at 95°C, 60 s at 56°C, and 60 s at 72°C, with a final step of 2 min at 72°C. Afterwards, the primary PCR products were subjected to second-round PCRs for Entamoeba species-specific characterization, which had multiple primers sets in the same tube. Amplification was achieved using the following primer sets EH-1 (5’-AAGCATTGTTTCTAGA TCTGAG-3’) and EH-2 (5’-AAGAGGTCTAACCGAAATTAG-3’) to detect E. histolytica (439 bp); ED-1 (5’-TCTAATTTCGATTAGAACTCT-3’) and ED-2 (5’-TCCCTACCTATTAGACATAGC-3’) to detect E. dispar (174 bp); Mos-1 (5’-GAAACCAAGAGTTTCACAAC-3’) and Mos-2 (5’-CAATATAAGGCTTGGATGA T-3’) to detect E. moshkovskii (553 bp) [33]. For the second PCR round, only the annealing temperature was changed to 48°C, while all other amplification cycle parameters were unchanged. Amplified products were visualized with stained with Gelred (Biotium Inc., Hayward, CA, USA) under UV transllumination after electrophoresis on 1.5% agarose gels. PCR amplification of partial SSU rDNA to identify subtypes of Blastocystis sp. was performed as protocol previously described [17], which amplifies a fragment approximately 500 bp long.
Genomic DNA was subjected to PCR analysis using primer pair Blast 505–532 (5’ GGA GGT AGT GAC AAT AAATC 3’) and Blast 998–1017 (5’TGC TTT CGC ACT TGT TCATC 3’). The PCR reaction was performed in a final volume of 50 μL and each reaction contained 100 mM of TrisHCl (pH 9.0), 500 mM of KCl, 1.5 mM of MgCl2, 200 μM of dATP, dGTP, dCTP, and dTTP each, 0.2 μM of primer, 1.5 U of Taq DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA, USA), 0.05% of bovine serum albumin (BSA), and 5 μL of DNA sample. PCR conditions consisted of an initial cycle at 95°C for 5 min, 35 cycles including denaturing at 95°C for 30 s, annealing at 55°C for 30 s, extending at 72°C for 2 min, with a final step of 2 min at 72°C. PCR products were electrophoresed in 1.5% agarose gel in Tris-borate ethylenediamine tetraacetic acid (EDTA) buffer, stained with Gelred (Biotium Inc., Hayward, CA, USA) and visualized under UV transllumination. Amplicons were purified using the Wizard® SV gel and PCR clean up system kit (Promega, Madison, WI, USA) and sequenced in both directions using Big Dye chemistries in an ABI3100 sequencer analyzer (Applied Biosystems, Foster City, CA). Our SSU rDNA partial gene sequences were deposited in GenBank under accession numbers KX523972 to KX524055. They were compared to available GenBank sequences using the BLASTN program on the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov/BLAST). In addition, DNA sequences of ST1 to ST9 of Blastocystis sp. were downloaded from Genbank, and multiple sequence alignment was performed using the ClustalW algorithm of the MEGA software version 6.0 [34].
Phylogenetic analysis
To identify the phylogenetic position of isolates, phylogenetic trees were created using Proteromonas lacertaeas, as the outgroup [35, 36]. Phylogenetic trees were constructed using two probabilistic methods: Maximum Likelihood (ML) and Bayesian Inference (BI), which are based on HKY + G (gamma distribution of rates with four rate categories) model [37]. The best model of nucleotide substitutions was selected based on the Akaike Information Criterion using Jmodeltest [38] for ML, and Mrmodeltest for BI [39]. The ML tree was created using PhyML 3.1 [40] and BI in MrBayes software version 3.2 [41]. Bootstrap values were calculated through the analysis of 1000 replicates for the ML analysis and through Bayesian posterior probability analysis using the MCMC algorithm, with four chains. Chains were run for 107 generations and sampled every 100 generations. Convergence was assessed by evaluating the average standard deviation of split frequencies, which were well below the recommended values (< 0.01). The first 25% of the sampled trees were discarded as burn-in for each data set, and the consensus tree topology and nodal support were estimated from the remaining samples as posterior probability values.
Results
A total of 294 individuals participated in this study, 145 (49.3%) males and 149 (50.7%) females (age range = 2–87 years, mean age = 38.3 years). Participants were stratified by age and gender (Table 1). There were 21 (7.1%) participants aged 1–5 years, 24 (8.2%) aged 6–14 years, 190 (64.6%) aged 15–55 years and 59 (20.1%) participants aged 55 years and over. A total of 126 individuals (42.8%) provided three stool samples, 99 (33.6%) two samples and 69 (23.4%) one sample. The study population was characterized predominantly by adults (81.2%). Overall, 189 of participants out of 294 (64.3%) were infected with at least one enteric parasite, with gender specific prevalence being 54.8% in males and 45.2% females. Prevalence was significantly different between the genders (χ2 = 6.82; p = 0.009). The total prevalence of protozoan infections was 60.2%. The most predominant species was Blastocystis sp., followed by Endolimax nana, E. histolytica complex, Entamoeba coli, and Giardia intestinalis. The total prevalence of helminth infections was 13.7%. Hookworm was the most prevalent helminth specie, followed by Trichuris trichiura (Table 1).
Table 1. Distribution of intestinal parasites according to age groups and gender.
| Age group, in years | Gender* | ||||||
|---|---|---|---|---|---|---|---|
| 1–5 | 6–14 | 15–55 | 55 y plus | M | F | Total (%) | |
| N° (%) | 21(7.1) | 24(8.2) | 190(64.6) | 59(20.1) | 145(49.3) | 149(50.7) | 294(100) |
| Pos | 8 (38.0) | 12 (50.0) | 128 (67.3) | 41 (69.5) | 103 (71.0) | 86 (57.7) | 189 (64.3) |
| Neg | 13 (62.0) | 12 (50.0 | 62 (32.7) | 18 (30.5) | 42 (29.0) | 63 (42.3) | 105 (35.7) |
| Blast | 7 (33.3) | 10 (41.7) | 110 (57.9) | 37 62.7) | 92 (63.4) | 72 (48.3) | 164 (55.8) |
| Ec | 2 (9.5) | 1 (4.2) | 9 (4.7) | 5 (8.5) | 10 (6.9) | 7 (4.7) | 17 (5.8) |
| Eh complex | 2 (9.5) | 1 (4.2) | 14 (7.4) | 4 (6.8) | 12 (8.3) | 9 (6.0) | 21 (7.1) |
| En | 2 (9.5) | 0 (0.0) | 44 (23.2) | 9 (15.3) | 33 (22.8) | 22 (14.8) | 55 (18.7) |
| Gi | 2 (9.5) | 3 (12.5) | 5 (2.6) | 2 (3.4) | 9 (6.2) | 3 (2.0) | 12 (4.1) |
| Ib | 0 (0.0) | 0 (0.0) | 1 (0.5) | 2 (3.4) | 2 (1.4) | 1 (0.7) | 3 (1.0) |
| Hook | 0 (0.0) | 0 (0.0) | 19 (10.0) | 2 (3.4) | 13 (9.0) | 8 (5.4) | 21 (7.1) |
| Ss | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 1 (0.7) | 0 (0.0) | 1 (0.3) |
| Ph | 0 (0.0) | 1 (4.2) | 1 (0.5) | 0 (0.0) | 0 (0.0) | 2 (1.3) | 2 (0.7) |
| Al | 1 (4.8) | 0 (0.0) | 0 (0.0) | 1 (1.7) | 1 (0.7 | 1 (0.7) | 2 (0.7) |
| Tt | 0 (0.0) | 0 (0.0) | 3 (1.6) | 0 (0.0) | 1 (0.7) | 2 (1.3) | 3 (1.0) |
| Ev | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (3.4) | 1 (0.7) | 1 (0.7) | 2 (0.7) |
Pos = positive samples; neg = negative samples; Blast = Blastocystis sp; Ec = Entamoeba coli; En = Endolimax nana; Eh complex = E. histolytica, E. dispar, or E. moshkovskii Gi = Giardia intestinalis; Ib = Iodamoeba butschlii; Hook = hookworm; Ss: Strongyloides stercoralis; Tt = Trichiuris trichiura; Ph = Pentatrichomonas hominis; Al = Ascaris lumbricoides; EV = Enterobius vermicularis. F = female; M = male; y = years.
*(p = 0.009)
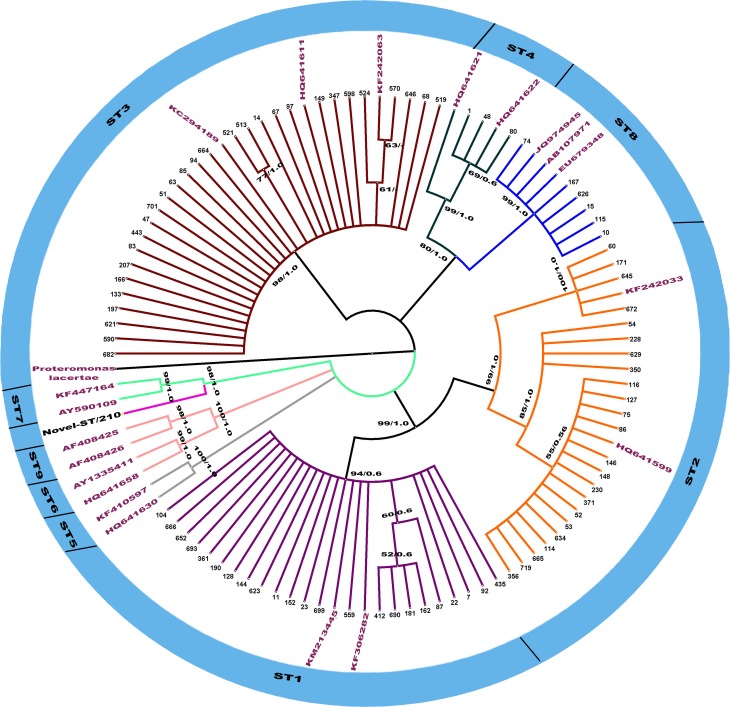
All 21 microscopically samples positive for E. histolytica complex were successfully amplified by nested PCR. Of those samples, 20 (95%) were positive for E. dispar and one (5.0%) had a mixed infection of E. histolytica and E. dispar. Culture and Hoffman techniques, both used for diagnosis of Blastocystis sp., exhibited very similar sensibility: 118 of 164 (71.9%) samples were diagnosed as positive by both techniques, 24 only by the use of culture and 22 only by the Hoffman technique. There was no statistical difference between techniques (p = 0.888). Of the 164 samples positive for Blastocystis sp., a subset of 85 (51.8%) was successfully sequenced. The sequencing of partial SSU rDNA gene indicated the presence of subtypes: ST3 (34.0%), ST1 (27.0%), ST2 (27.0%), ST4 (3.5%), ST8 (7.0%) and a non-identified subtype (ID 210). Sequences showed high identity (98–100%) to sequences available at the NCBI database, while sample ID 210 returned a BLAST identity of 93%. Topology of the constructed phylogenetic tree revealed that ST1, ST2, ST3, ST4, ST8 and the non-identified ST clustered into well-supported clades (with grouped bootstrap values over 70.0% and Bayesian posterior probabilities) (Fig 1). The non-identified Blastocystis sp. ST is evolutionarily closer to ST7. Intra-ST diversity was observed in ST1, ST2 and ST3 as indicated by their small clades.
Fig 1. Maximum likelihood (ML) and Bayesian inference (BI) trees based on the partial sequences of the partial SSU rDNA (437 bp) of Blastocystis sp. isolates of the present study.
The high genetic divergence of rDNA gene revealed six clusters (denoted by ST1, 2, 3, 4, 8 and Novel-ST). The numbers along branches correspond to bootstrap values and Bayesian posterior probability values (values above 50% are shown) in the order: MP/BI.
Discussion
We assessed the prevalence of IPIs and identity of Blastocystis sp. subtypes in a rural community of Rio de Janeiro, Brazil. IPIs have been described as the greatest worldwide cause of diseases [42]. Current epidemiological evaluations suggest that 450 million people, especially in developing countries, may host multiple parasite species [1]. Our results showed that 64.3% of participants presented IPIs, with the most common human parasite being Blastocystis sp., in accordance to previous work across Brazilian regions [12, 43–46]. Although IPIs had received attention in Sumidouro as early as 1995, most of these efforts targeted Schistosomiasis. Gonçalves et al. [47] previously reported a prevalence of 13% of hookworm, 5% of Ascaris lumbricoides, 20% of Entamoeba coli and 10% of Giardia intestinalis in the Sumidouro population. The prevalence of protist parasites that we are reporting is much higher than of helminths, which may be explained by self-medication (antihelminths) practiced by the population, or by the use of pesticides in agriculture above recommended doses, which may have a significant effect on soil-transmitted helminths survival. However, these hypotheses still require investigation.
We also identified statistically significant differences in infection rates between men and women. Several investigations have reported a higher prevalence of parasitic infection in males than females [48, 49]. A possible explanation is the higher engagement of males in farming activities. Most of the Sumidouro population is rural and their most important economic activities are cattle breeding and vegetable cultivation. People from rural communities usually eat fresh food, drink untreated water and disposal of sewage is generally inadequate [50], resulting in increased infection risk. The diversity of protist parasites reveled by our analyses suggest that their distribution depends on factors such as environmental conditions, lack of sanitation and health education, confirming previous studies in rural communities [51, 52]. It was shown that short-term xenic in vitro culture (XIVC) is a more sensitive diagnostic method than parasitological examination [53]. Contrary to the expectations, our findings showed that the proportion of specimens negative by parasitological examination but positive by xenic in vitro culture, and vice-versa, were similar. The likely explanation is that the fecal samples did not contain viable Blastocystis sp. or alternatively cultured organisms were scant and imperceptible thereby avoiding detection by microscopy. Moreover, the screening of Blastocystis sp. cultures was performed in the first three days of incubation, when parasite count was low due to slow growth rates. Screening after a week of cultivation for example might reveal additional positives missed by the analyses performed herein.
The predominant Blastocystis sp. subtype was ST3, ST1 and ST2. STs 1, 2 and 3 had already been identified as the most abundant subtypes in previous studies [53–56]. Recently, Ramirez et al. [21] have reported percentages ranging from 15 to 92% regarding these subtypes. Other investigations have singled out ST3 as the most frequent subtype [18, 57–64]. Subtypes 1 and 2 have been identified both in humans and a wide range of synanthropic animals such as pigs, rats, dogs, horses, monkeys, cattle and chickens [15, 16, 65]. A recent study using animal models revealed that various human Blastocystis sp. subtypes, including ST1 and ST2, could infect chickens and rats [66, 67], suggesting the zoonotic potential of human Blastocystis sp. isolates. We also observed the occurrence of ST4 in only three residents, although this subtype is the second most common in the UK and commonly found across Europe [15, 27, 68–70]. In Colombia, ST4 was identified in a few humans and non-human primates [21]. Rodents were also shown to be reservoir hosts of ST4 [23, 71]. Brazil was the only country in South America where ST8 has been identified [21], although it was also found in Denmark, the UK, Italy and Australia [62, 65, 72, 73]. Large American opossums and non-human primates can be infected by ST8, which is rarely found in other hosts [18]. We could not identify the subtype from sample 210. However, our phylogenetic tree shows that the sample is closely related to ST7. A similar case was reported by Ramirez et al. [21] who reported a novel ST closely related to ST1 and found in six South American countries including Brazil. Further analyses and full-length sequencing of the SSU-rDNA gene are required to truly confirm the existence of the novel ST.
Conclusion
Our study revealed the presence of IPIs in a rural community in Brazil, with high prevalence and a wide diversity of Blastocystis sp. subtypes, and possibly a new subtype of the protozoan. Our findings reinforce the need for additional studies of Blastocystis sp. subtype diversity, which may contribute to more efficient identification of infection sources and potential transmission routes under various ecological conditions, as well as a re-evaluation of control strategies by public health services.
Supporting information
(DOCX)
Acknowledgments
The authors thank Marisa Soares for her significant contributions given to the study and the staff from the DNA sequencing platform/PDTIS Fiocruz.
Data Availability
All relevant data are within the paper and its Supporting Information files. In this study, SSU rDNA partial gene sequences were deposited in GenBank under accession numbers KX523972 to KX524055 and MG309719.
Funding Statement
Financial support was provided by Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq) and Fundação Oswaldo Cruz (PAPES-VI/FIOCRUZ) number (407768/2012-2) to HLCS and MMB and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro to CMd. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Robertson LJ, van der Giessen JW, Batz MB, Kojima M, Cahill S. Have foodborne parasites finally become a global concern? Trends Parasitol. 2013;29(3):101–3. Epub 2013/02/05. doi: 10.1016/j.pt.2012.12.004 . [DOI] [PubMed] [Google Scholar]
- 2.de Silva NR, Brooker S, Hotez PJ, Montresor A, Engels D, Savioli L. Soil-transmitted helminth infections: updating the global picture. Trends Parasitol. 2003;19(12):547–51. Epub 2003/12/03. . [DOI] [PubMed] [Google Scholar]
- 3.Alum A, Rubino JR, Ijaz MK. The global war against intestinal parasites—should we use a holistic approach? Int J Infect Dis. 2010;14(9):e732–8. Epub 2010/04/20. doi: 10.1016/j.ijid.2009.11.036 . [DOI] [PubMed] [Google Scholar]
- 4.Agudelo-Lopez S, Gomez-Rodriguez L, Coronado X, Orozco A, Valencia-Gutierrez CA, Restrepo-Betancur LF, et al. Prevalence of intestinal parasitism and associated factors in a village on the Colombian Atlantic Coast. Rev Salud Publica (Bogota). 2008;10(4):633–42. Epub 2009/04/11. . [DOI] [PubMed] [Google Scholar]
- 5.Abdulsalam AM, Ithoi I, Al-Mekhlafi HM, Ahmed A, Surin J, Mak JW. Drinking water is a significant predictor of Blastocystis infection among rural Malaysian primary schoolchildren. Parasitology. 2012;139(8):1014–20. Epub 2012/03/27. doi: 10.1017/S0031182012000340 . [DOI] [PubMed] [Google Scholar]
- 6.Speich B, Croll D, Furst T, Utzinger J, Keiser J. Effect of sanitation and water treatment on intestinal protozoa infection: a systematic review and meta-analysis. Lancet Infect Dis. 2016;16(1):87–99. Epub 2015/09/26. doi: 10.1016/S1473-3099(15)00349-7 . [DOI] [PubMed] [Google Scholar]
- 7.Yoshikawa H, Wu Z, Pandey K, Pandey BD, Sherchand JB, Yanagi T, et al. Molecular characterization of Blastocystis isolates from children and rhesus monkeys in Kathmandu, Nepal. Vet Parasitol. 2009;160(3–4):295–300. Epub 2009/01/13. doi: 10.1016/j.vetpar.2008.11.029 . [DOI] [PubMed] [Google Scholar]
- 8.Helenbrook WD, Shields WM, Whipps CM. Characterization of Blastocystis species infection in humans and mantled howler monkeys, Alouatta palliata aequatorialis, living in close proximity to one another. Parasitol Res. 2015;114(7):2517–25. Epub 2015/04/11. doi: 10.1007/s00436-015-4451-x . [DOI] [PubMed] [Google Scholar]
- 9.Yoshikawa H, Tokoro M, Nagamoto T, Arayama S, Asih PB, Rozi IE, et al. Molecular survey of Blastocystis sp. from humans and associated animals in an Indonesian community with poor hygiene. Parasitol Int. 2016;65(6 Pt B):780–4. Epub 2016/04/16. doi: 10.1016/j.parint.2016.03.010 . [DOI] [PubMed] [Google Scholar]
- 10.Turkeltaub JA, McCarty TR 3rd, Hotez PJ. The intestinal protozoa: emerging impact on global health and development. Curr Opin Gastroenterol. 2015;31(1):38–44. Epub 2014/11/14. doi: 10.1097/MOG.0000000000000135 . [DOI] [PubMed] [Google Scholar]
- 11.Hotez PJ, Bottazzi ME, Strych U, Chang LY, Lim YA, Goodenow MM, et al. Neglected tropical diseases among the Association of Southeast Asian Nations (ASEAN): overview and update. PLoS Negl Trop Dis. 2015;9(4):e0003575 Epub 2015/04/17. doi: 10.1371/journal.pntd.0003575 ; PubMed Central PMCID: PMCPMC4400050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David EB, Guimaraes S, de Oliveira AP, Goulart de Oliveira-Sequeira TC, Nogueira Bittencourt G, Moraes Nardi AR, et al. Molecular characterization of intestinal protozoa in two poor communities in the State of Sao Paulo, Brazil. Parasit Vectors. 2015;8:103 Epub 2015/04/19. doi: 10.1186/s13071-015-0714-8 ; PubMed Central PMCID: PMCPMC4335703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abe N. Molecular and phylogenetic analysis of Blastocystis isolates from various hosts. Vet Parasitol. 2004;120(3):235–42. Epub 2004/03/26. doi: 10.1016/j.vetpar.2004.01.003 . [DOI] [PubMed] [Google Scholar]
- 14.Yoshikawa H, Abe N, Wu Z. PCR-based identification of zoonotic isolates of Blastocystis from mammals and birds. Microbiology. 2004;150(Pt 5):1147–51. Epub 2004/05/11. doi: 10.1099/mic.0.26899-0 . [DOI] [PubMed] [Google Scholar]
- 15.Stensvold CR, Alfellani MA, Norskov-Lauritsen S, Prip K, Victory EL, Maddox C, et al. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int J Parasitol. 2009;39(4):473–9. Epub 2008/08/30. doi: 10.1016/j.ijpara.2008.07.006 . [DOI] [PubMed] [Google Scholar]
- 16.Parkar U, Traub RJ, Vitali S, Elliot A, Levecke B, Robertson I, et al. Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet Parasitol. 2010;169(1–2):8–17. Epub 2010/01/22. doi: 10.1016/j.vetpar.2009.12.032 . [DOI] [PubMed] [Google Scholar]
- 17.Santin M, Gomez-Munoz MT, Solano-Aguilar G, Fayer R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol Res. 2011;109(1):205–12. Epub 2011/01/07. doi: 10.1007/s00436-010-2244-9 . [DOI] [PubMed] [Google Scholar]
- 18.Alfellani MA, Taner-Mulla D, Jacob AS, Imeede CA, Yoshikawa H, Stensvold CR, et al. Genetic diversity of blastocystis in livestock and zoo animals. Protist. 2013;164(4):497–509. Epub 2013/06/19. doi: 10.1016/j.protis.2013.05.003 . [DOI] [PubMed] [Google Scholar]
- 19.Rajah Salim H, Suresh Kumar G, Vellayan S, Mak JW, Khairul Anuar A, Init I, et al. Blastocystis in animal handlers. Parasitol Res. 1999;85(12):1032–3. Epub 1999/12/22. . [DOI] [PubMed] [Google Scholar]
- 20.Balint A, Doczi I, Bereczki L, Gyulai R, Szucs M, Farkas K, et al. Do not forget the stool examination!-cutaneous and gastrointestinal manifestations of Blastocystis sp. infection. Parasitol Res. 2014;113(4):1585–90. Epub 2014/02/21. doi: 10.1007/s00436-014-3805-0 . [DOI] [PubMed] [Google Scholar]
- 21.Ramirez JD, Sanchez LV, Bautista DC, Corredor AF, Florez AC, Stensvold CR. Blastocystis subtypes detected in humans and animals from Colombia. Infect Genet Evol. 2014;22:223–8. Epub 2013/07/28. doi: 10.1016/j.meegid.2013.07.020 . [DOI] [PubMed] [Google Scholar]
- 22.Stensvold CR, Alfellani M, Clark CG. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol. 2012;12(2):263–73. Epub 2011/11/26. doi: 10.1016/j.meegid.2011.11.002 . [DOI] [PubMed] [Google Scholar]
- 23.Noel C, Dufernez F, Gerbod D, Edgcomb VP, Delgado-Viscogliosi P, Ho LC, et al. Molecular phylogenies of Blastocystis isolates from different hosts: implications for genetic diversity, identification of species, and zoonosis. J Clin Microbiol. 2005;43(1):348–55. Epub 2005/01/07. doi: 10.1128/JCM.43.1.348-355.2005 ; PubMed Central PMCID: PMCPMC540115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkar U, Traub RJ, Kumar S, Mungthin M, Vitali S, Leelayoova S, et al. Direct characterization of Blastocystis from faeces by PCR and evidence of zoonotic potential. Parasitology. 2007;134(Pt 3):359–67. Epub 2006/10/21. doi: 10.1017/S0031182006001582 . [DOI] [PubMed] [Google Scholar]
- 25.Yan Y, Su S, Ye J, Lai X, Lai R, Liao H, et al. Blastocystis sp. subtype 5: a possibly zoonotic genotype. Parasitol Res. 2007;101(6):1527–32. Epub 2007/08/01. doi: 10.1007/s00436-007-0672-y . [DOI] [PubMed] [Google Scholar]
- 26.Cian A, El Safadi D, Osman M, Moriniere R, Gantois N, Benamrouz-Vanneste S, et al. Molecular Epidemiology of Blastocystis sp. in Various Animal Groups from Two French Zoos and Evaluation of Potential Zoonotic Risk. PLoS One. 2017;12(1):e0169659 Epub 2017/01/07. doi: 10.1371/journal.pone.0169659 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stensvold CR, Christiansen DB, Olsen KE, Nielsen HV. Blastocystis sp. subtype 4 is common in Danish Blastocystis-positive patients presenting with acute diarrhea. Am J Trop Med Hyg. 2011;84(6):883–5. Epub 2011/06/03. doi: 10.4269/ajtmh.2011.11-0005 ; PubMed Central PMCID: PMCPMC3110361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.BRASIL. Contagem Populacional. Censo Demográfico 2010. Instituto Brasileiro de Geografia e Estatística (IBGE) 2010.
- 29.Nimer E. Climatologia do Brasil 2.ed ed: IBGE, Departamento de Recursos Naturais e Estudos Ambientais; 1989. [Google Scholar]
- 30.Hoffman WA, Pons JA, Janer JL. The sedimentation concentration method in schistosomiasis mansoni. P R J Publ Health Trop Med. 1934;9: 283–91. [Google Scholar]
- 31.Willis H H. A simple levitation method for the detection of hook-worm ova. Medicine Journal of Australia. 1921; 29: 375–376 [Google Scholar]
- 32.Zerpa LRH, L.; Naquira C.; Espinoza I. A simplified culture method for Blastocystis hominis. Revista Mexicana de Patología Clínica. 2000;47(1):17–9. [Google Scholar]
- 33.Khairnar K, Parija SC. A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples. BMC Microbiol. 2007;7:47 Epub 2007/05/26. doi: 10.1186/1471-2180-7-47 ; PubMed Central PMCID: PMCPMC1888694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. Epub 2013/10/18. doi: 10.1093/molbev/mst197 ; PubMed Central PMCID: PMCPMC3840312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leipe DD TS, Goggin CL, Slemenda SB, Pieniazek NJ, Sogin ML. 16S-like rDNA sequences from Developayella elegans, Labyrinthuloides haliotidis, and Proteromonas lacerate confirm that the stramenopiles are a primarily heterotrophic group. European Journal of Protistology. 19916;32(4):449–58. doi: 10.1016/S0932-4739(96)80004-6 [Google Scholar]
- 36.Silberman JD, Sogin ML, Leipe DD, Clark CG. Human parasite finds taxonomic home. Nature. 1996;380(6573):398 Epub 1996/04/04. doi: 10.1038/380398a0 . [DOI] [PubMed] [Google Scholar]
- 37.Hasegawa M, Iida Y, Yano T, Takaiwa F, Iwabuchi M. Phylogenetic relationships among eukaryotic kingdoms inferred from ribosomal RNA sequences. J Mol Evol. 1985;22(1):32–8. Epub 1985/01/01. . [DOI] [PubMed] [Google Scholar]
- 38.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53(5):793–808. Epub 2004/11/17. doi: 10.1080/10635150490522304 . [DOI] [PubMed] [Google Scholar]
- 39.Nylander JAA. Mrmodeltest 2.3. Program Distributed by the Author. Uppsala University; Uppsala, Switzerland: 2004. [Google Scholar]
- 40.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. Epub 2003/10/08. . [DOI] [PubMed] [Google Scholar]
- 41.Huelsenbeck JP, Ronquist F. Bayesian Analysis of Molecular Evolution Using MrBayes 2005. 183–226 p. [Google Scholar]
- 42.Steketee RW. Pregnancy, nutrition and parasitic diseases. J Nutr. 2003;133(5 Suppl 2):1661S–7S. Epub 2003/05/06. . [DOI] [PubMed] [Google Scholar]
- 43.Aguiar JI, Goncalves AQ, Sodre FC, Pereira Sdos R, Boia MN, de Lemos ER, et al. Intestinal protozoa and helminths among Terena Indians in the State of Mato Grosso do Sul: high prevalence of Blastocystis hominis. Rev Soc Bras Med Trop. 2007;40(6):631–4. Epub 2008/01/18. . [DOI] [PubMed] [Google Scholar]
- 44.Gil FF, Busatti HG, Cruz VL, Santos JF, Gomes MA. High prevalence of enteroparasitosis in urban slums of Belo Horizonte-Brazil. Presence of enteroparasites as a risk factor in the family group. Pathog Glob Health. 2013;107(6):320–4. Epub 2013/10/05. doi: 10.1179/2047773213Y.0000000107 ; PubMed Central PMCID: PMCPMC4001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rebolla MF, Silva EM, Gomes JF, Falcao AX, Rebolla MV, Franco RM. High prevalence of Blastocystis spp. infection in children and staff members attending public urban schools in Sao Paulo state, Brazil. Rev Inst Med Trop Sao Paulo. 2016;58:31 Epub 2016/04/14. doi: 10.1590/S1678-9946201658031 ; PubMed Central PMCID: PMCPMC4826084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabrine-Santos M, Cintra Edo N, do Carmo RA, Nascentes GA, Pedrosa AL, Correia D, et al. Occurrence of Blastocystis spp. in Uberaba, Minas Gerais, BRAZIL. Rev Inst Med Trop Sao Paulo. 2015;57(3):211–4. Epub 2015/07/23. doi: 10.1590/S0036-46652015000300005 ; PubMed Central PMCID: PMCPMC4544244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goncalves MM, Barreto MM, Maldonado A Jr., Maione VR, Rey L, Soares Mda S. [Socio-cultural and ethical factors involved in the diagnosis of schistosomiasis mansoni in an area of low endemicity]. Cad Saude Publica. 2005;21(1):92–100. Epub 2005/02/05. doi: /S0102-311X2005000100011. PubMed . [DOI] [PubMed] [Google Scholar]
- 48.Goncalves AQ, Junqueira AC, Abellana R, Barrio PC, Terrazas WC, Sodre FC, et al. Prevalence of intestinal parasites and risk factors forspecific and multiple helminth infections in a remote city of the Brazilian Amazon. Rev Soc Bras Med Trop. 2016;49(1):119–24. Epub 2016/05/11. doi: 10.1590/0037-8682-0128-2015 . [DOI] [PubMed] [Google Scholar]
- 49.Nasiri V, Esmailnia K, Karim G, Nasir M, Akhavan O. Intestinal parasitic infections among inhabitants of Karaj City, Tehran province, Iran in 2006–2008. Korean J Parasitol. 2009;47(3):265–8. Epub 2009/09/03. doi: 10.3347/kjp.2009.47.3.265 ; PubMed Central PMCID: PMCPMC2735692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boonjaraspinyo S, Boonmars T, Kaewsamut B, Ekobol N, Laummaunwai P, Aukkanimart R, et al. A cross-sectional study on intestinal parasitic infections in rural communities, northeast Thailand. Korean J Parasitol. 2013;51(6):727–34. Epub 2014/02/12. doi: 10.3347/kjp.2013.51.6.727 ; PubMed Central PMCID: PMCPMC3916464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valverde JG, Gomes-Silva A, De Carvalho Moreira CJ, Leles De Souza D, Jaeger LH, Martins PP, et al. Prevalence and epidemiology of intestinal parasitism, as revealed by three distinct techniques in an endemic area in the Brazilian Amazon. Ann Trop Med Parasitol. 2011;105(6):413–24. Epub 2011/11/29. doi: 10.1179/1364859411Y.0000000034 ; PubMed Central PMCID: PMCPMC4100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Damazio SM, Lima Mde S, Soares AR, Souza MA. Intestinal parasites in a quilombola community of the Northern State of Espirito Santo, Brazil. Rev Inst Med Trop Sao Paulo. 2013;55(3). Epub 2013/06/07. doi: 10.1590/S0036-46652013000300007 . [DOI] [PubMed] [Google Scholar]
- 53.Stensvold CR, Arendrup MC, Jespersgaard C, Molbak K, Nielsen HV. Detecting Blastocystis using parasitologic and DNA-based methods: a comparative study. Diagn Microbiol Infect Dis. 2007;59(3):303–7. Epub 2007/10/05. doi: 10.1016/j.diagmicrobio.2007.06.003 . [DOI] [PubMed] [Google Scholar]
- 54.Popruk S, Udonsom R, Koompapong K, Mahittikorn A, Kusolsuk T, Ruangsittichai J, et al. Subtype distribution of Blastocystis in Thai-Myanmar border, Thailand. Korean J Parasitol. 2015;53(1):13–9. Epub 2015/03/10. doi: 10.3347/kjp.2015.53.1.13 ; PubMed Central PMCID: PMCPMC4384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malheiros AF, Stensvold CR, Clark CG, Braga GB, Shaw JJ. Short report: Molecular characterization of Blastocystis obtained from members of the indigenous Tapirape ethnic group from the Brazilian Amazon region, Brazil. Am J Trop Med Hyg. 2011;85(6):1050–3. Epub 2011/12/07. doi: 10.4269/ajtmh.2011.11-0481 ; PubMed Central PMCID: PMCPMC3225150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rene BA, Stensvold CR, Badsberg JH, Nielsen HV. Subtype analysis of Blastocystis isolates from Blastocystis cyst excreting patients. Am J Trop Med Hyg. 2009;80(4):588–92. Epub 2009/04/07. . [PubMed] [Google Scholar]
- 57.Scicluna SM, Tawari B, Clark CG. DNA barcoding of blastocystis. Protist. 2006;157(1):77–85. Epub 2006/01/25. doi: 10.1016/j.protis.2005.12.001 . [DOI] [PubMed] [Google Scholar]
- 58.Yan Y, Su S, Lai R, Liao H, Ye J, Li X, et al. Genetic variability of Blastocystis hominis isolates in China. Parasitol Res. 2006;99(5):597–601. Epub 2006/05/12. doi: 10.1007/s00436-006-0186-z . [DOI] [PubMed] [Google Scholar]
- 59.Ozyurt M, Kurt O, Molbak K, Nielsen HV, Haznedaroglu T, Stensvold CR. Molecular epidemiology of Blastocystis infections in Turkey. Parasitol Int. 2008;57(3):300–6. Epub 2008/03/14. doi: 10.1016/j.parint.2008.01.004 . [DOI] [PubMed] [Google Scholar]
- 60.Jones MS, Whipps CM, Ganac RD, Hudson NR, Boorom K. Association of Blastocystis subtype 3 and 1 with patients from an Oregon community presenting with chronic gastrointestinal illness. Parasitol Res. 2009;104(2):341–5. Epub 2008/10/17. doi: 10.1007/s00436-008-1198-7 . [DOI] [PubMed] [Google Scholar]
- 61.Souppart L, Moussa H, Cian A, Sanciu G, Poirier P, El Alaoui H, et al. Subtype analysis of Blastocystis isolates from symptomatic patients in Egypt. Parasitol Res. 2010;106(2):505–11. Epub 2009/12/03. doi: 10.1007/s00436-009-1693-5 . [DOI] [PubMed] [Google Scholar]
- 62.Meloni D, Sanciu G, Poirier P, El Alaoui H, Chabe M, Delhaes L, et al. Molecular subtyping of Blastocystis sp. isolates from symptomatic patients in Italy. Parasitol Res. 2011;109(3):613–9. Epub 2011/02/23. doi: 10.1007/s00436-011-2294-7 . [DOI] [PubMed] [Google Scholar]
- 63.Bart A, Wentink-Bonnema EM, Gilis H, Verhaar N, Wassenaar CJ, van Vugt M, et al. Diagnosis and subtype analysis of Blastocystis sp. in 442 patients in a hospital setting in the Netherlands. BMC Infect Dis. 2013;13:389 Epub 2013/08/27. doi: 10.1186/1471-2334-13-389 ; PubMed Central PMCID: PMCPMC3765316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pandey PK, Verma P, Marathe N, Shetty S, Bavdekar A, Patole MS, et al. Prevalence and subtype analysis of Blastocystis in healthy Indian individuals. Infect Genet Evol. 2015;31:296–9. Epub 2015/02/24. doi: 10.1016/j.meegid.2015.02.012 . [DOI] [PubMed] [Google Scholar]
- 65.Roberts T, Stark D, Harkness J, Ellis J. Subtype distribution of Blastocystis isolates identified in a Sydney population and pathogenic potential of Blastocystis. Eur J Clin Microbiol Infect Dis. 2013;32(3):335–43. Epub 2012/09/22. doi: 10.1007/s10096-012-1746-z . [DOI] [PubMed] [Google Scholar]
- 66.Iguchi A, Ebisu A, Nagata S, Saitou Y, Yoshikawa H, Iwatani S, et al. Infectivity of different genotypes of human Blastocystis hominis isolates in chickens and rats. Parasitol Int. 2007;56(2):107–12. Epub 2007/01/26. doi: 10.1016/j.parint.2006.12.004 . [DOI] [PubMed] [Google Scholar]
- 67.Hussein EM, Hussein AM, Eida MM, Atwa MM. Pathophysiological variability of different genotypes of human Blastocystis hominis Egyptian isolates in experimentally infected rats. Parasitol Res. 2008;102(5):853–60. Epub 2008/01/15. doi: 10.1007/s00436-007-0833-z . [DOI] [PubMed] [Google Scholar]
- 68.Forsell J, Granlund M, Stensvold CR, Clark CG, Evengard B. Subtype analysis of Blastocystis isolates in Swedish patients. Eur J Clin Microbiol Infect Dis. 2012;31(7):1689–96. Epub 2012/02/22. doi: 10.1007/s10096-011-1416-6 . [DOI] [PubMed] [Google Scholar]
- 69.Poirier P, Wawrzyniak I, Albert A, El Alaoui H, Delbac F, Livrelli V. Development and evaluation of a real-time PCR assay for detection and quantification of blastocystis parasites in human stool samples: prospective study of patients with hematological malignancies. J Clin Microbiol. 2011;49(3):975–83. Epub 2010/12/24. doi: 10.1128/JCM.01392-10 ; PubMed Central PMCID: PMCPMC3067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dominguez-Marquez MV, Guna R, Munoz C, Gomez-Munoz MT, Borras R. High prevalence of subtype 4 among isolates of Blastocystis hominis from symptomatic patients of a health district of Valencia (Spain). Parasitol Res. 2009;105(4):949–55. Epub 2009/05/28. doi: 10.1007/s00436-009-1485-y . [DOI] [PubMed] [Google Scholar]
- 71.Yoshikawa H, Nagano I, Wu Z, Yap EH, Singh M, Takahashi Y. Genomic polymorphism among Blastocystis hominis strains and development of subtype-specific diagnostic primers. Mol Cell Probes. 1998;12(3):153–9. Epub 1998/07/17. doi: 10.1006/mcpr.1998.0161 . [DOI] [PubMed] [Google Scholar]
- 72.Stensvold R, Brillowska-Dabrowska A, Nielsen HV, Arendrup MC. Detection of Blastocystis hominis in unpreserved stool specimens by using polymerase chain reaction. J Parasitol. 2006;92(5):1081–7. Epub 2006/12/13. doi: 10.1645/GE-840R.1 . [DOI] [PubMed] [Google Scholar]
- 73.Clark CG. Extensive genetic diversity in Blastocystis hominis. Mol Biochem Parasitol. 1997;87(1):79–83. Epub 1997/07/01. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. In this study, SSU rDNA partial gene sequences were deposited in GenBank under accession numbers KX523972 to KX524055 and MG309719.