Abstract
Human transcription factor IIH (TFIIH) is part of the general transcriptional machinery required by RNA polymerase II for the initiation of eukaryotic gene transcription1. Composed of ten subunits that add up to a molecular mass of about 500 kDa, TFIIH is also essential for nucleotide excision repair1. The seven-subunit TFIIH core complex formed by XPB, XPD, p62, p52, p44, p34, and p8 is competent for DNA repair2, while the CDK-activating kinase subcomplex, which includes the kinase activity of CDK7 as well as the cyclin H and MAT1 subunits, is additionally required for transcription initiation1,2. Mutations in the TFIIH subunits XPB, XPD, and p8 lead to severe premature ageing and cancer propensity in the genetic diseases xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy, highlighting the importance of TFIIH for cellular physiology3. Here we present the cryo-electron microscopy structure of human TFIIH at 4.4 Å resolution. The structure reveals the molecular architecture of the TFIIH core complex, the detailed structures of its constituent XPB and XPD ATPases, and how the core and kinase subcomplexes of TFIIH are connected. Additionally, our structure provides insight into the conformational dynamics of TFIIH and the regulation of its activity.
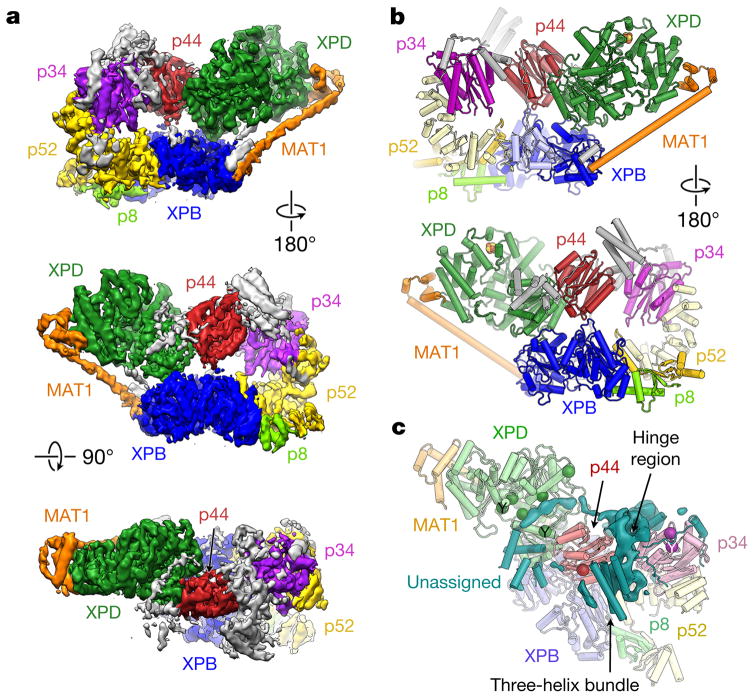
We collected cryo-electron microscopy (cryo-EM) data of human TFIIH, immuno-purified from HeLa cells, and obtained a reconstruction at an overall resolution of 4.4 Å (see Methods and Extended Data Figs 1 and 2). The reconstruction (Fig. 1a) shows a horseshoe-shaped assembly corresponding to the TFIIH core complex, in agreement with previous lower-resolution reconstructions of human TFIIH in isolation4 and in the context of the RNA polymerase II (Pol II) transcription preinitiation complex (PIC) (hereafter, Pol II-PIC)5,6. Secondary structure elements are resolved throughout (Extended Data Fig. 3a–c), and large amino-acid side chains are visible in the best-resolved areas of the map (better than 4 Å; Extended Data Figs 2b and 3d–f). Because the resolution of our cryo-EM map was not sufficient for chain tracing and assignment of the sequence register in several areas, we combined docking and rebuilding of homology models with the placement of secondary structure elements for previously structurally uncharacterized components to generate an atomic model of the TFIIH core–MAT1 complex (Fig. 1b and Extended Data Table 1). This atomic model was refined against the map and fully validated (Extended Data Fig. 3g–i). All protein subunits of the TFIIH core complex except p62, as well as the CDK-activating kinase (CAK) component MAT1, could be unambiguously assigned in our cryo-EM map, while the remainder of the CAK subcomplex is highly flexible and its CDK7 and cyclin H subunits are not resolved (Fig. 1a, b). The architecture of TFIIH is dominated by its ATPase/helicase subunits XPD and XPB, which are situated next to each other at the open end of the horseshoe-shaped structure (green and blue, respectively, in Fig. 1a, b). The arc-like connection between them includes the von Willebrand factor A-like folds of p44 and p34 and the helical domain of p52 (red, magenta, and yellow, respectively, in Fig. 1a, b; domain architectures are shown in Extended Data Fig. 4). Even though p62 could not be unambiguously assigned in our cryo-EM map, data from chemical crosslinking-mass spectrometry (CX-MS) experiments7 suggest that several secondary structure elements built into unassigned density may correspond to parts of p62 (Fig. 1c, Extended Data Fig. 5 and Supplementary Discussion). These α-helical regions and the linkers that seem to connect them localize to distant parts of TFIIH, suggesting that p62 may act as a molecular glue to stabilize the overall assembly of TFIIH.
Figure 1. Cryo-EM reconstruction of human TFIIH.
a, Cryo-EM map of TFIIH, colour coded and labelled according to constituent subunits; unassigned density is grey. b, Front and back views of the molecular structure of TFIIH. Protein subunits are labelled. Unassigned secondary structure elements attributed to XPB and p52 are light blue and pale yellow, respectively; remaining unassigned elements, grey. c, Unassigned secondary structure elements and remaining unassigned density in the p34–p44 hinge region (teal) may correspond to p62 and the zinc-binding domains of p34 and p44. Positions of crosslinks between p62 and other TFIIH core proteins7 shown by spheres coloured according to the crosslinked partner; crosslinks from studies of yeast TFIIH indicated by ‘Y’.
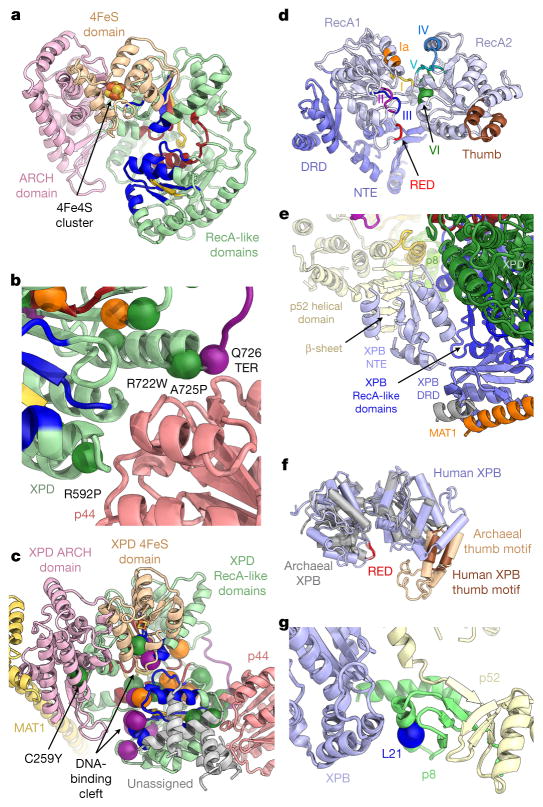
The structures of XPD and XPB (Fig. 2) show that both ATPases are in the apo-state, with unoccupied nucleotide-binding pockets. Human XPD shares its overall architecture with homologous archaeal helicases8–10, with two RecA-like domains (RecA1 and RecA2) harbouring the structurally conserved helicase motifs, and 4FeS and ARCH domain insertions within RecA1 (Fig. 2a and Extended Data Fig. 4). The 4Fe4S cluster is clearly visualized in the cryo-EM map as a strong density peak within the 4FeS domain8,10 (Extended Data Fig. 6a–c). The 4FeS domain is adjacent to the pore, through which single-stranded DNA has been proposed to be threaded before reaching the two RecA-like domains of XPD11,12 (Extended Data Fig. 6c–e). The region around the entrance to the pore in human XPD is narrower than in the archaeal enzymes owing to a nearby insertion segment within the human ARCH domain (Extended Data Fig. 6c, e), although the pore still appears to be accessible along the path proposed for archaeal XPD8,10 (Extended Data Fig. 6e).
Figure 2. Structures of human XPB and XPD.
a, Colour-coded domain architecture of XPD: ARCH domain, pink; 4FeS domain, brown; RecA-like domains, green; with helicase motifs distinctively coloured by function (dark red: nucleotide binding and hydrolysis; yellow: coordination between nucleotide hydrolysis and helicase activity; blue: DNA binding)10,16. b, Disease-causing mutations mapped onto the structure of human XPD as solid spheres: xeroderma pigmentosum mutations, purple; TTD mutations, dark green; mutations causing combined xeroderma pigmentosum and Cockayne’s syndrome, orange. XPD mutations near p44 (light red) are labelled and numbered. Q726 is affected by a nonsense mutation in xeroderma pigmentosum; the protein segment truncated thereby is shown in purple. c, XPD in the context of surrounding proteins, with disease-causing mutations mapped onto the structure as in b. d, Structure of XPB (RecA-like domains light, violet; NTE and DRD, blue; RED motif, red; thumb motif, brown; conserved helicase motifs colour coded and labelled)20. e, Secondary structure elements modelled into unassigned density (red in Extended Data Fig. 5b) include the XPB NTE and DRD, and a β-sheet tentatively assigned to p52. f, Superposition of the RecA-like domains of human (violet) and archaeal (grey) XPB (Protein Data Bank (PDB) accession number 2FWR (ref. 20)); domains superposed separately owing to large conformational differences. g, The p8 subunit (light green) bridges the p52 C-terminal domain (pale yellow) with XPB (violet). The L21P mutation (blue sphere) causes TTD.
The helicase activity of XPD is known to be upregulated by interactions with p44, which are also required for assembly of the helicase into the TFIIH complex13. Our structure shows that these interactions with p44 localize near the conserved helicase motifs IV and V in XPD RecA2 (Extended Data Fig. 6f), but on the outside of the RecA-like domain, facing away from the DNA-binding site (Extended Data Fig. 6d, g). Mutations of XPD residues near this interaction surface (Fig. 2b) cause xeroderma pigmentosum or trichothiodystrophy (TTD)1,3,10, highlighting the importance of the XPD–p44 interaction for assembly and function of TFIIH. Unlike proteins that stimulate the activity of DNA and RNA helicases by contacting their N-terminal RecA-like domain and promoting substrate release, such as the translation initiation factor eIF4G and the protein Gle1, which regulate the DEAD-box RNA helicases eIF4A and Dbp5, respectively14,15 (Extended Data Fig. 6g, h), p44 is more likely to regulate XPD through subtle effects on the conformation of the nearby helicase motifs IV and V, which are involved in DNA binding and coupling of ATPase and helicase activity16. This hypothesis agrees with the observation that p44 enhances the helicase activity of XPD, but does not alter its ATPase activity13. For a further discussion of the structural elements of p44 contributing to the XPD-binding interface, see Supplementary Discussion.
Mapping of disease-causing mutations3 onto our structure of human XPD shows that most of the mutations that cause xeroderma pigmentosum or xeroderma pigmentosum combined with Cockayne syndrome cluster near the DNA- or ATP-binding sites of the RecA-like domains of XPD (Fig. 2c), in agreement with their strong effect on helicase activity in both human and archaeal XPD9,10,17. Their position explains the defects they cause in nucleotide excision repair (NER)17 and confirms previous predictions based on the structures of archaeal XPD homologues9,10. While these xeroderma pigmentosum or xeroderma pigmentosum/Cockayne syndrome mutations are mostly confined to the helicase core of the enzyme, TTD-causing mutations also affect peripheral regions important for contacts with the other components of TFIIH (Fig. 2b, c). Some of these TTD mutations are located at the interaction site with p44 (see above) and in the ARCH domain, which is the site of association with MAT1 (see below). TTD-causing mutations have been associated with defects in both NER and basal transcription in vitro17. Because transcription does not depend on the helicase activity of XPD18 and the TTD mutations include sites away from the critical elements in the helicase domain, some of these mutations may cause transcription defects by impairing the ability of XPD to correctly assemble into TFIIH, where it may be required to support the association of TFIIH with the Pol II-PIC and to correctly position the CAK subcomplex near Pol II during transcription initiation (see below).
The structure of human XPB (Fig. 2d) encompasses the two RecA-like domains, a DNA-damage recognition domain (DRD)-like domain, and a set of secondary structure elements corresponding to the N-terminal extension domain (NTE). The NTE is important for anchoring and activity of XPB within TFIIH and is affected by disease mutations3,7,19, but is not present in archaeal XPB. Although a precise delineation of p52 and the XPB NTE at their contact site is not possible at the present resolution, our structure suggests this functionally important interaction may involve regions of predicted β-strands within both proteins (Fig. 2e; for further discussion of the DRD and NTE see Extended Data Figs 5 and 7 and Supplementary Discussion). The overall structure of the two RecA-like domains is similar to the previously reported archaeal XPB structure, including the signature RED and thumb motifs, even though the latter is substantially shortened relative to its archaeal counterpart20 (Fig. 2d, f). XPB also shares structural homology with the archaeal ssoRad54 SWI2/SNF2-type ATPase21 and the eukaryotic Rad54 SWI2/SNF2 chromatin remodeller22, including the presence of a domain insertion at the site of the XPB thumb domain (Extended Data Fig. 6i) and conserved motifs in the RecA-like domains16 (Extended Data Fig. 6j). The relative orientation of the two RecA-like domains observed in XPB within TFIIH is similar to the arrangement in the structure of eukaryotic Rad54 (ref. 22), which lacks bound substrate, but highly dissimilar to the conformations of both the archaeal ssoRad54 SWI2/SNF2 ATPase in complex with DNA21 and archaeal XPB20 as reported previously5. Comparison with human TFIIH within a promoter-bound Pol II-PIC5 shows that DNA binding does not induce major inter-domain rearrangements in XPB within TFIIH. In fact, the conformations observed in DNA-bound ssoRad54 (ref. 21) or in free archaeal XPB would cause extensive steric hindrance or rearrangement of large interaction interfaces (Extended Data Fig. 6k). It seems that the structure of XPB within TFIIH is more restrained than for homologous enzymes that are not part of this large assembly and that the TFIIH scaffold may act to prearrange the otherwise flexible RecA-like domains of XPB in a conformation that is suitable for substrate binding and catalytic activity, in agreement with previous biochemical data23.
Similarly to the p44–XPD interaction, XPB RecA2 interacts with p52 and p8, interactions that are required for XPB assembly and function within TFIIH7,19. The L21P mutation in p8 that causes the disease TTD in humans24 is located in an α-helix that is sandwiched between the fold of p8 and XPB RecA2 (Fig. 2g). The presence of a proline at this site probably distorts or unfolds the affected helix, thereby destabilizing the interactions between XPB, p8, and p52 in this region or reducing p8 stability25.
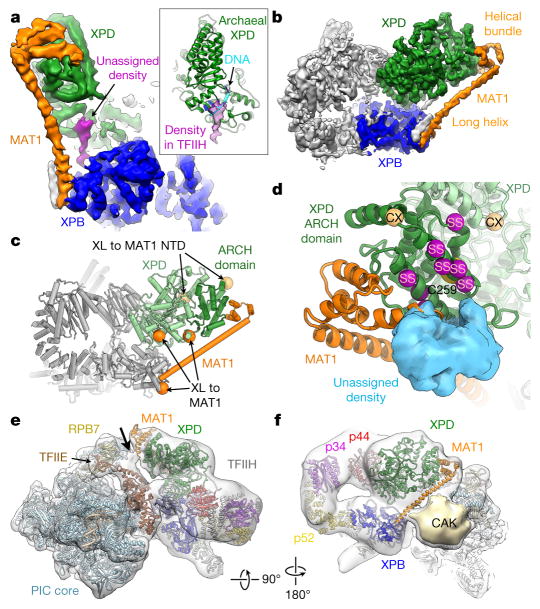
In our cryo-EM map, an elongated density connects the DRD-like domain of XPB with a region of XPD RecA2 that is inferred to be part of the DNA substrate-binding site on the basis of the structures of archaeal XPD–DNA complexes11,12 (Fig. 3a). While this density could be due to residual DNA not removed during TFIIH purification from native source, an alternative possibility is that it corresponds to an unassigned protein region that could potentially act to regulate the interaction of XPD with substrate.
Figure 3. MAT1 forms a physical connection between all ATP consuming TFIIH moieties.
a, b, A long α-helical density and a helical bundle (orange) connect XPB (blue) and XPD (green). Unassigned density (purple in a) connects the XPB DRD with the DNA-binding site of XPD. Inset: unassigned density superimposed on the structure of archaeal XPD11 (PDB accession number 5H8W; green) bound to DNA (cyan). c, Locations of CX-MS crosslinks (XL, spheres)7 of human MAT1 (orange) and the N-terminal domain of the yeast MAT1 homologue (light orange) on XPD (green; ARCH domain dark green). d, Interaction region between MAT1 and the XPD ARCH domain. Mutations of C259 (partly occluded yellow sphere) impair the MAT1–XPD interaction. Site-specific crosslinks (SS) between RAD3 and TFB3 (yeast homologues of XPD and MAT1)29 shown as purple spheres, CX-MS crosslinks (CX; same as in c) light orange. Unassigned density that may correspond to the N-terminal RING domain of MAT1 (Extended Data Fig. 8d, e) shown as blue surface. e, Docking of the structure of TFIIH into the density of the Pol II-PIC (Electron Microscopy Data Bank (EMDB) accession number EMD-8134)5. Density for the MAT1 helical bundle is visible near a contact site (arrow) of TFIIH with the PIC core near TFIIE and RPB7. f, As e, but viewing TFIIH from the side. Density for the long MAT1 α-helix is not clear, possibly because of flexibility or structural rearrangement. A large but relatively weak density contacting Pol II probably corresponds to the cyclin H and Cdk7 subunits of the CAK5,6 (Extended Data Fig. 8f, g).
We assigned the CAK subunit MAT1 to a very long α-helix and a helical bundle that form interactions with both XPB and the XPD ARCH domain (Fig. 3a, b). This density localizes near the region of attachment of the CAK subcomplex to the TFIIH core, as visualized in 2D class averages and 3D reconstructions of negatively stained TFIIH (Extended Data Fig. 8a–c and refs 6, 26), and reconstructions of TFIIH in the context of the assembled Pol II-PIC (see below and refs 5, 27). Previous data additionally show that MAT1 interacts with the XPD ARCH domain, and that the C259Y mutation in the ARCH domain impairs binding28. Combined with secondary structure prediction showing a very long α-helix in MAT1 (Extended Data Fig. 4) and CX-MS and site-specific crosslinking data showing proximity of MAT1 to XPB and XPD7,29 (Fig. 3c, d), these observations strongly support our assignment of MAT1. At the end of the MAT1 helical bundle, there is weak density that could accommodate the N-terminal RING domain of MAT1 (blue in Fig. 3d and Extended Data Fig. 8d, e); however, unambiguous assignment is currently not possible.
MAT1 connects all three ATP-dependent moieties of TFIIH—the CAK subcomplex and the XPB and XPD ATPases—and may be involved in the regulation and coordination of these functional centres of TFIIH. Interestingly, the presence of the CAK subcomplex, which includes MAT1, is known to inhibit the activity of the XPD helicase28,30. Accordingly, the CAK subcomplex is present during transcription initiation, when the XPD helicase is inactive, and removed during NER, when XPD helicase activity is required2,18,19,31. The interactions between MAT1 and XPD observed in our cryo-EM map might be involved in mediating this inhibition, possibly by limiting the conformational freedom of the ARCH domain, which has been implicated in DNA substrate loading by XPD11. In the context of the human Pol II-PIC5, MAT1 may also be involved in forming contacts between TFIIH and the PIC core (Fig. 3e). Furthermore, MAT1 is in close proximity to a density element located between TFIIH and the Pol II-PIC core that probably corresponds to the remainder of the CAK subcomplex5,6 (Fig. 3f), according to our negative-stain reconstructions of apo-TFIIH (Extended Data Fig. 8f, g and ref. 6) and in agreement with recent studies in yeast27.
The XPD C259Y mutation, which is found in patients with TTD3 and impairs both NER and transcription activity in vitro28, probably destabilizes the structure of the entire ARCH domain8,9. Our structure now indicates that perturbations of the structure of the ARCH domain may prevent functional incorporation of MAT1 into TFIIH, which will impair the proper placement of the remaining components of the CAK subcomplex within the Pol II-PIC, thus explaining the defects in Pol II-CTD phosphorylation and transcription observed in the presence of the C259Y mutant TFIIH in vitro28.
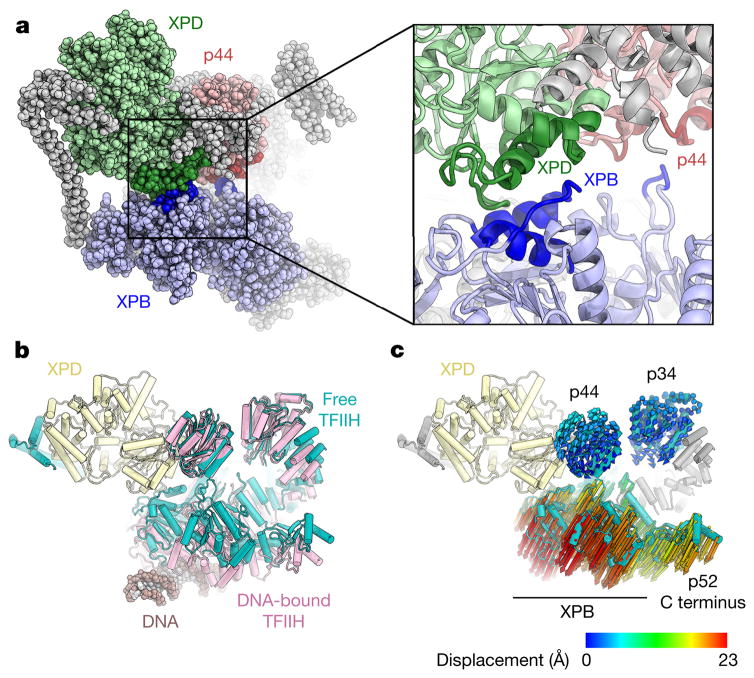
Conformational changes in molecular machines are often associated with functional transitions. In free TFIIH, an interaction between XPB and XPD, possibly supported by additional XPB–p44 contacts, brings together the two ends of the horseshoe-shaped TFIIH core complex (Fig. 4a and Extended Data Fig. 9a). In the Pol II-PIC5, TFIIH undergoes a conformational change during which the XPD–XPB interaction breaks as the distance between them is increased relative to free TFIIH (Fig. 4b, c and Extended Data Fig. 9b–f). Overall, the RecA-like domains of XPB move away from XPD when engaging DNA. Because they are bound to XPB RecA2, p8 and the C-terminal domain of p52 are also relocated by this motion (Fig. 4c). These rearrangements may be required for the activation of the DNA translocase function of XPB within the Pol II-PIC that has been proposed to aid the melting of the promoter DNA and the formation of the transcription bubble5,6,32. Additionally, the MAT1 contact site on the XPD ARCH domain is clearly resolved in the reconstruction of the Pol II-PIC5 (Fig. 3e, f), while the density for the connection to XPB is not clear. This might indicate that the MAT1–XPB contact is released in the Pol II-PIC to facilitate conformational changes that enable the remainder of the CAK subcomplex to access and phosphorylate its targets in the Pol II-PIC. Complete release of the CAK during NER2,31 may not only de-repress the helicase activity of XPD28,30, but might also allow for more conformational flexibility of TFIIH to possibly facilitate the recruitment of downstream NER factors.
Figure 4. Conformational rearrangements of TFIIH.
a, XPD–XPB and p44–XPB contacts in free TFIIH. Interacting regions coloured dark blue on XPB, dark green on XPD, and dark red on p44. b, Free TFIIH (teal) and TFIIH bound to DNA in the Pol II-PIC (pink; subunits of free TFIIH were fitted into the cryo-EM map of TFIIH in the PIC5) superimposed on XPD (light yellow). A conformational change in TFIIH separates the XPB and XPD ATPases when XPB is bound to DNA. c, Analysis of XPB and XPD Cα displacement during the conformational change from free to PIC-bound TFIIH shows a downward movement of XPB, p8, and the p52 C terminus, and slight rotations of p34 and p44.
Acknowledgments
We thank S. Zheng and D. King for providing peptides and XPB monoclonal antibody, P. Grob for cryo-EM support, and T. Houweling and A. Chintangal for computing support. We thank A. B. Patel and R. K. Louder for discussions and help with data collection, and J. H. D. Cate for providing PyMOL scripts. We acknowledge the use of the LAWRENCIUM computing cluster at Lawrence Berkeley National Laboratory and the resources of the National Energy Research Scientific Computing Center, a Department of Energy Office of Science user facility supported by the Office of Science of the US Department of Energy under contract number DE-AC02-05CH11231. This work was funded through NIGMS grants R01-GM63072 to E.N. and P01-GM063210 to P.D.A. B.J.G. was supported by fellowships from the Swiss National Science Foundation (projects P300PA_160983, P300PA_174355), and T.H.D.N. is a University of California, Berkeley Miller Fellow. E.N. is a Howard Hughes medical investigator.
Footnotes
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Author Contributions E.N. directed the study. J.F. performed HeLa cell culture and prepared TFIIH. B.J.G. performed cryo-EM specimen preparation, data collection, data processing, and initial model building. B.J.G. and T.H.D.N. built the final model. B.J.G. and T.H.D.N. performed coordinate refinement, supported by P.V.A. in the laboratory of P.D.A. B.J.G. wrote the initial draft of the manuscript and all authors contributed to the final version.
References
- 1.Compe E, Egly JM. Nucleotide excision repair and transcriptional regulation: TFIIH and beyond. Annu Rev Biochem. 2016;85:265–290. doi: 10.1146/annurev-biochem-060815-014857. [DOI] [PubMed] [Google Scholar]
- 2.Svejstrup JQ, et al. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 3.Cleaver JE, Thompson LH, Richardson AS, States JC. A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum Mutat. 1999;14:9–22. doi: 10.1002/(SICI)1098-1004(1999)14:1<9::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 4.Schultz P, et al. Molecular structure of human TFIIH. Cell. 2000;102:599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 5.He Y, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016;533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo J, et al. Architecture of the human and yeast general transcription and DNA repair factor TFIIH. Mol Cell. 2015;59:794–806. doi: 10.1016/j.molcel.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolski SC, et al. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, et al. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan L, et al. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Constantinescu-Aruxandei D, Petrovic-Stojanovska B, Penedo JC, White MF, Naismith JH. Mechanism of DNA loading by the DNA repair helicase XPD. Nucleic Acids Res. 2016;44:2806–2815. doi: 10.1093/nar/gkw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuper J, Wolski SC, Michels G, Kisker C. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 2012;31:494–502. doi: 10.1038/emboj.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coin F, et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20:184–188. doi: 10.1038/2491. [DOI] [PubMed] [Google Scholar]
- 14.Schütz P, et al. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montpetit B, et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubaele S, et al. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 18.Kuper J, et al. In TFIIH, XPD helicase is exclusively devoted to DNA repair. PLoS Biol. 2014;12:e1001954. doi: 10.1371/journal.pbio.1001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coin F, Oksenych V, Egly JM. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell. 2007;26:245–256. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Fan L, et al. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol Cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Dürr H, Körner C, Müller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 22.Thomä NH, et al. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 23.Grünberg S, Warfield L, Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol. 2012;19:788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giglia-Mari G, et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36:714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 25.Kainov DE, Vitorino M, Cavarelli J, Poterszman A, Egly JM. Structural basis for group A trichothiodystrophy. Nat Struct Mol Biol. 2008;15:980–984. doi: 10.1038/nsmb.1478. [DOI] [PubMed] [Google Scholar]
- 26.Gibbons BJ, et al. Subunit architecture of general transcription factor TFIIH. Proc Natl Acad Sci USA. 2012;109:1949–1954. doi: 10.1073/pnas.1105266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai KL, et al. Mediator structure and rearrangements required for holoenzyme formation. Nature. 2017;544:196–201. doi: 10.1038/nature21393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdulrahman W, et al. ARCH domain of XPD, an anchoring platform for CAK that conditions TFIIH DNA repair and transcription activities. Proc Natl Acad Sci USA. 2013;110:E633–E642. doi: 10.1073/pnas.1213981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warfield L, Luo J, Ranish J, Hahn S. Function of conserved topological regions within the Saccharomyces cerevisiae basal transcription factor TFIIH. Mol Cell Biol. 2016;36:2464–2475. doi: 10.1128/MCB.00182-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandrock B, Egly JM. A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. J Biol Chem. 2001;276:35328–35333. doi: 10.1074/jbc.M105570200. [DOI] [PubMed] [Google Scholar]
- 31.Coin F, et al. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol Cell. 2008;31:9–20. doi: 10.1016/j.molcel.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 32.Fishburn J, Tomko E, Galburt E, Hahn S. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci USA. 2015;112:3961–3966. doi: 10.1073/pnas.1417709112. [DOI] [PMC free article] [PubMed] [Google Scholar]