Abstract
Cervical cancer continues to be a leading cancer among women in many parts of the world. Nation-wide screening with the Pap smear has not been implemented in India due to the lack of adequately trained cytologists. Identification of biomarkers to predict malignant potential of the identified low risk lesions is essential to avoid excessive retesting and follow up. The current study analyzed the expression patterns of DNA replication licensing proteins, proliferation inhibitor protein p16INK4A and tumor suppresser protein p63 in cervical tissues and smears to assess the ability of these proteins to predict progression.
Methods:
Cervical smears and corresponding tissues were immunostained using mouse monoclonal antibodies against MCM2, MCM5, CDC6, p16 and p63. Smears were treated with a non-ionic surfactant sodium deoxycholate prior to immuno-cytochemistry. The standard ABC method of immunohistochemistry was performed using DAB as the chromogen. The immunostained samples were scored on a 0-3+ scale and staining patterns of smears were compared with those of tissue sections. Sensitivity and specificity for each of these markers were calculated taking histopathology as the gold standard.
Result:
All the markers were positive in malignant and dysplastic cells. MCM protein expression was found to be up-regulated in LSIL, HSIL and in malignancies to a greater extent than p16 as well as p63. CDC6 protein was preferentially expressed in high grade lesions and in invasive squamous cell carcinomas. A progressive increase in the expression of DNA replication licensing proteins in accordance with the grades of cervical intraepithelial lesion suggests these markers as significant to predict malignant potential of low grade lesions in cervical smears.
Conclusion:
MCMs and CDC6 can be applied as biomarkers to predict malignant potential of low grade lesions identified in screening programmes and retesting / follow up might be confined to those with high risk lesions alone so that overuse of resources can be safely avoided.
Keywords: Cervical cancer, HPV, biomarkers, cervical intraepithelial neoplasia
Introduction
Despite drastic decrease in the incidence of cervical cancer in countries that have implemented systematically organized population screening programmes using Pap smear, this disease is one of the most commonly diagnosed cancer in women in underdeveloped countries. (Siegel et al., 2012). The clinical performance of the cytology-based screening technology has limitations. The sensitivity of the conventional Pap test for the detection of high-grade lesions has a wide range from 30% to 87% (Masoudi et al., 2006). To overcome this limitation, liquid based cytology (LBC) was developed, which was also not found effective. Human Papilloma Virus (HPV) testing is widely being used to improve the accuracy of cervical cancer screening. Pap smear plus HPV DNA test and vaccination against high risk HPV is prevalent in the developed world. India contributes one fifth of the global burden of cervical cancer. None of the screening programmes or vaccination has been implemented in India and similar countries. However, sporadic screening programmes are available in selected areas, particularly in the state of Kerala where, all women with cytological abnormalities are being treated or followed up with repeat Pap smear examination, even though their lesions are likely to revert to normal. An average of 12,000 women is being screened annually in the early cancer detection programmes of the Regional Cancer Centre Thiruvananthapuram, Kerala. About 15% of these cases are being diagnosed to have low-grade squamous intra epithelial lesion LSIL and / Atypical Squamous cells of Undetermined significance (ASCUS) /Atypical glandular cells of undetermined significance (AG-US) and these cases are further evaluated by colposcopy followed by biopsy and treated by cryosurgery or leep, if the abnormality is persistent one. About 80% of the preselected women are being treated unnecessarily causing a heavy over use of resources. It is because of our inability to correctly assess the malignant potential of the preselected lesions. If a biological marker can be characterized to predict high risk lesions, retesting and /follow up can be confined to these lesions alone so that over use of resources can be prevented. The aim of the present study was to evaluate the expression patterns of DNA replication licensing proteins, Mini Chromosome Maintenance proteins 2,5 and Cell Division Control protein (MCM2, MCM5,CDC6) and tumor suppressor proteins p16 and p63 in cervical smears and the corresponding histology sections to see whether any of these markers can be used to assess the malignant potential of the intra epithelial lesions and whether any of these markers can be used as a standalone test for the detection of cervical intra epithelial lesions, so that population screening can be implemented using this marker.
Materials and Methods
The samples for the study were obtained from a cohort of 1850 women attending the population screening programme. It includes 1,650 symptomatic women who have attended the gynecology clinic of Women and Children hospital, Trivandrum, Kerala with some gynecological complaints. The study was approved by the Institute Review Board as well as the Human Ethical Committee of Regional Cancer Centre (RCC). The Study material included cervical smears and tissues samples ranging from negative for intraepithelial lesion (NILM) to invasive squamous cell carcinoma. The cervical scrape smears were obtained from all participants by using Ayers spatula, which were fixed in 95% ethanol and processed in the classical pap staining method. These smears were reported in the Division of Pathology according to the Bethesda system of Pap smear reporting. One separate scraping from each of the participants was collected in the buffer provided by Surepath for LBC preparation. Colposcopic biopsies of the cytologically positive women were obtained in the Community Oncology Division of the RCC.
Immunohistochemistry
A total of 190 cytologically abnormal samples with histologically confirmed LSIL (n=52), HSIL (n=64), SCC (n=63), and Normal (n=11) were selected for immunocytochemical evaluation. The corresponding tissue blocks for immunohistochemistry were obtained from the department of Pathology, RCC. Among these, 178 samples were included in the statistical analysis and others were omitted due to lack of adequate cellularity. Novolink polymer system was used in the current study. Mouse monoclonal antibody against MCM2, MCM5, CDC6, p16 and P63 were procured from Santa Cruz Biotechnology Inc., USA. Antibodies were diluted with TBS buffer without adding Tween-20. Novolink polymer detection system (Novocastra, Leica bio-systems, New castels limited, UK) was used as the secondary system with DAB as chromogen. Immunostaining was done in Liquid Based Cytology smears prepared according to the instructions provided by the BD Sure path (Tripath Imaging Inc. Burlington NC, 27215, USA). The smears were treated with sodium deoxycholate solution for half an hour before antigen retrieval to increase the permeability of the cell membrane. Further steps were similar to that of immunohistochemistry protocol.
Statistical analysis
Statistical analysis was performed by using SPSS (Statistical Package for the Social Sciences, Version 11) software. The diagnostic accuracy of each marker was calculated by using histopathology as the gold standard. The percentage of positivity in different squamous lesions was assessed in smears and the corresponding histologic sections. Staining in >30% of cells for each marker was considered as the cut off for positivity. Descriptive analysis of the data such as mean, standard deviation and 95% confidence interval was calculated based on mean H- score of all markers in all the lesions. One way analysis of variance (ANOVA) was done to determine whether there were any statistically significant differences between the means of expressions of each marker in different lesions and Bonferroni Multiple Comparison method was used for multiple comparisons. The sensitivity and specificity along with positive (PPV) and negative predictive values (NPV) of each marker was assessed taking histopathology as gold standard. Two sided testing procedure was followed for all statistical analysis and p value <0.05 were considered as significant.
Results
Majority (67%) of the study subjects were in the age group 41- 50 years and the mean age of LSIL, HSIL, and Invasive Squamous Cell Carcinoma were 45, 50, and 57 years respectively. Higher incidence of invasive squamous cell carcinoma was found among postmenopausal women, whereas the low-grade lesions were confined to premenopausal group only.
Strong nuclear expressions of MCM proteins (MCM2 and MCM5) were observed in SCC and HSIL and mild diffuse positivity were observed in LSIL samples (Figure 1 and 2 A-F). Significant difference in expression patterns were noticed for MCM2 and MCM5 in histology and cytology samples in one way ANOVA as well as in multiple comparisons (Table 1 and 2). The sensitivity and specificity of MCM2 for the diagnosis of SCC was 95% and 21% with a positive and negative predictive value of 72% and 67% respectively. For the diagnosis of HSIL, this protein expression showed a sensitivity of 93% with a positive predictive value of 61% and specificity of 28% with a negative predictive value of 78 %. The sensitivity of MCM2 expression for a diagnosis of LSIL was 73% with a positive predictive value of 61% and specificity of 48 % with a negative predictive value of 61% respectively. Similarly MCM5 had a sensitivity of 97% and specificity of 23% in SCC samples and 88%, 22% for HSIL (Figure 3.16). The sensitivity and specificity of MCM5 proteins in low grade lesions (LSIL) were 75% and 20% respectively. The diagnostic accuracy of MCM2 and MCM5 proteins for the identification of SCC were 71% and 78%, and in HSIL 64% and 60%. The diagnostic accuracy of these proteins was 61% and 47% for LSIL respectively.
Figure 1.
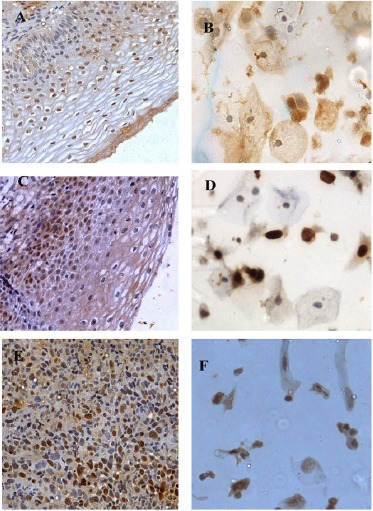
A-F, Expression patterns of MCM2 in histology and corresponding cytology samples; A and B, LSIL 40(X)- Mild nuclear expression; C and D, HSIL (40X)- Moderate nuclear and mild cytoplasmic expression; E and F, SCC (40X)- Intense nuclear expression.
Figure 2.
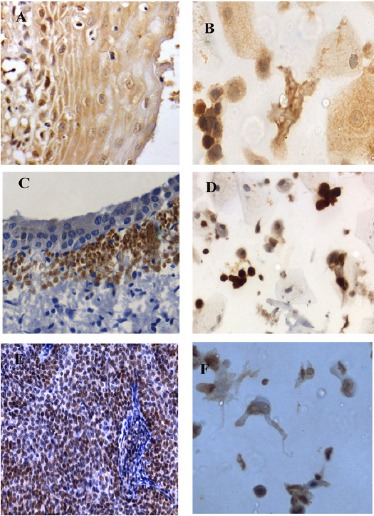
A-F, Expression patterns of MCM5 in histology and corresponding cytology samples; A and B, LSIL 40(X)- Mild nuclear and cytoplasmic expression; C and D, HSIL (40X)- Intense nuclear expression; E and F, SCC (40X)- Intense nuclear expression.
Table 1.
One way ANOVA and Bonferroni Multiple Comparison of MCM2 Expression in Tissues and their Corresponding Cytology Samples
| Lesion (n) | Positive Cases (%) | Mean | Std. Deviation | Range | p-value | |
|---|---|---|---|---|---|---|
| Histology Samples | ||||||
| SCC (63) | 57 (90.5) | 142.06 | 61.017 | 0-240 | ||
| HSIL (55) | 46 (83.6) | 119.27 | 71.151 | 0-240 | ||
| LSIL (49) | 37 (75.5) | 99.59 | 56.235 | 0-190 | ||
| Normal (11) | 1 (9.1) | 16.36 | 24.606 | 0-80 | ||
| Cytology Samples | <0.001 | |||||
| SCC (63) | 43 (68.3) | 83.85 | 54.778 | 0-180 | ||
| HSIL (55) | 30 (54.5) | 70.1 | 62.809 | 0-190 | ||
| LSIL (49) | 26 (53.1) | 49.25 | 46.043 | 0-140 | ||
| Normal (11) | 0 (0) | 9.09 | 10.445 | 0-30 | ||
| Bonferroni Multiple Comparison | ||||||
| Lesion | p-value (Histology) | p-value (Cytology) | ||||
| SCC | HSIL | 0.281 | 1 | |||
| LSIL | 0.002 | 0.095 | ||||
| Normal | 0.0001 | 0.002 | ||||
| LSIL | 0.637 | 0.296 | ||||
| HSIL | ||||||
| Normal | 0.0001 | 0.005 | ||||
| LSIL | Normal | 0.0001 | 0.227 | |||
Table 2.
One way ANOVA and Bonferroni Multiple Comparison of MCM5 Expression in Tissues and their Corresponding Cytology Samples
| Lesion (n) | Positive Cases (%) | Mean | Std. Deviation | Range | p-value |
|---|---|---|---|---|---|
| Histology Samples | |||||
| SCC (63) | 58 (92.1) | 152.7 | 72.45 | 0-270 | |
| HSIL (55) | 46 (83.6) | 127.64 | 69.361 | 0-240 | |
| LSIL (49) | 40 (81.6) | 104.49 | 49.374 | 0-190 | |
| Normal (11) | 2 (18.2) | 16.36 | 24.196 | 0-80 | |
| Cytology Samples | <0.001 | ||||
| SCC (63) | 46 (73) | 85.77 | 69.292 | 0-210 | |
| HSIL (55) | 32 (58.2) | 77.92 | 59.604 | 0-190 | |
| LSIL (49) | 23 (46.9) | 55.5 | 48.089 | 0-190 | |
| Normal (11) | 6 (54.5) | 25.45 | 21.616 | 0-60 | |
| Bonferroni Multiple Comparison | |||||
| Lesion | p-value (Histology) | p-value (Cytology) | |||
| SCC | HSIL | 0.208 | 1 | ||
| LSIL | 0.001 | 0.224 | |||
| Normal | 0.0001 | 0.023 | |||
| HSIL | LSIL | 0.399 | 0.225 | ||
| Normal | 0.0001 | 0.026 | |||
| LSIL | Normal | 0.0001 | 0.75 | ||
Figure 3.
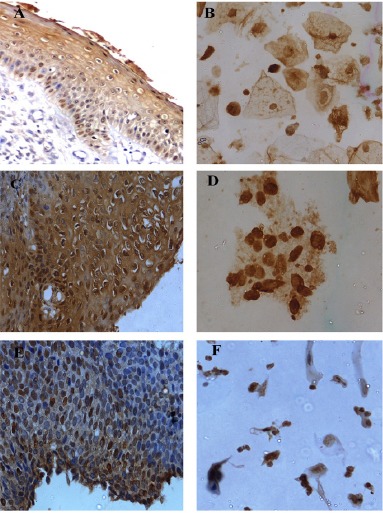
A-F, Expression patterns of CDC6 in histology and corresponding cytology samples; A and B, LSIL 40(X)- Mild nuclear and diffuse cytoplasmic expression; C and D, HSIL (40X)- Moderate nuclear and cytoplasmic expression; E and F, SCC (40X)- Moderate to Intense nuclear expression.
Immunoreactivity of CDC6 was observed mainly in the dysplastic nucleus. Mild diffuse expression was found in the cytoplasm also (Figure 3 A-F). Moderate to intense expression of CDC6 was noted in 89% of SCC in histology and 87% in cytology samples. Significant difference was observed for CDC6 expression patterns in the different lesions in one-way analysis (ANOVA) (p<0.001) as well as in Bonferroni Multi Comparison in both histology and cytology (Table 3). In multiple comparisons, only SCC showed significance with respect to normal (p= 0.028) in cytology, but in histology samples SCC (p= 0.001) and HSIL (p= 0.001) found significant with respect to normal. The sensitivity of CDC6 for SCC was 94%, with a positive predictive value of 80% and a specificity of 27% with a negative predictive value of 57% whereas, it was 89% and 67% for a diagnosis of HSIL with a specificity of 25% and negative predictive value of 56 %. The sensitivity of CDC6 for LSIL was 62% with a positive predictive value of 57% and a specificity of 38% with a negative predictive value of 53%.
Table 3.
One way ANOVA and Bonferroni Multiple Comparison of CDC6 Expression in Tissues and their Corresponding Cytology Samples
| Lesion (n) | Positive Cases (%) | Mean | Std. Deviation | Range | p-value |
|---|---|---|---|---|---|
| Histology Samples | |||||
| SCC (63) | 56 (88.9) | 133.49 | 66.385 | 0-270 | |
| HSIL (55) | 46 (83.6) | 110 | 65.064 | 0-260 | |
| LSIL (49) | 31 (63.3) | 72.65 | 52.471 | 10-170 | |
| Normal (11) | 1 (9.1) | 20.91 | 9.439 | 0-40 | |
| Cytology Samples <0.001 | |||||
| SCC (63) | 48 (76.2) | 79.23 | 48.409 | 0-180 | <0.001 |
| HSIL (55) | 35 (63.6) | 67.33 | 59.243 | 0-260 | |
| LSIL (49) | 26 (53.1) | 58.25 | 52.667 | 0-190 | |
| Normal (11) | 4 (36.4) | 22.73 | 25.334 | 0-60 | |
| Bonferroni Multiple Comparison | |||||
| Lesion | p-value (Cytology) | ||||
| SCC | HSIL | 0.219 | 1 | ||
| LSIL | 0.001 | 0.786 | |||
| Normal | 0.001 | 0.028 | |||
| HSIL | LSIL | 0.012 | 1 | ||
| Normal | 0.001 | 0.068 | |||
| LSIL | Normal | 0.067 | 0.354 | ||
Expression of p16INK4A in both LBC smears and tissues samples showed strong nuclear positivity in the squamous cell carcinoma samples compared to other lesions (Figure 4 A-F). Significant differences (p<0.001) were observed for p16 expression pattern with respect to the different lesions in one way analysis (ANOVA). Bonferroni Multiple Comparison also showed significance for p16 to differentiate lesions in histology with a p value <0.005 for both SCC and HSIL with respect to other lesions (Table 4). The sensitivity of p16 for the identification of SCC was 96% with a positive predictive value of 81%. The specificity was only 68% with a negative predictive value of 42%. The sensitivity and specificity of p16 for the diagnosis of HSIL was 94%, and 42%, and for LSIL it was 53 and 24 % respectively. The diagnostic accuracy of p16 for LSIL was only 53%. The diagnostic accuracy of p16 for SCC and HSIL were 76% and 62%
Figure 4.
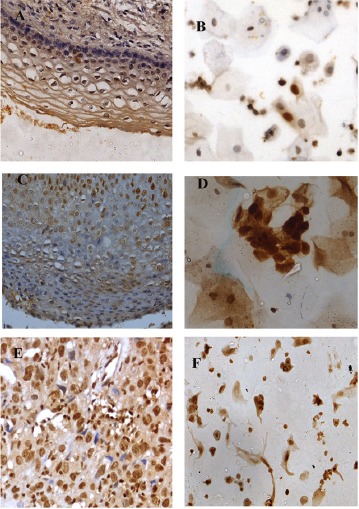
Expression patterns of p16 in histology and corresponding cytology samples; A and B, LSIL 40(X)- Mild nuclear expression; C and D, HSIL (40X)- Moderate nuclear expression; E and F, SCC (40X)- Dense nuclear expression.
Table 4.
One way ANOVA and Bonferroni Multiple Comparison of p16 Expression in Histopathology and their Corresponding Cytology Lesions of the Cervix
| Lesion (n) | Positive Cases (%) | Mean | Std. Deviation | Range | p-value |
|---|---|---|---|---|---|
| Histology Samples | |||||
| SCC (63) | 58 (92.1) | 165.4 | 78.572 | 0-280 | |
| HSIL (55) | 48 (87.3) | 128.73 | 68.369 | 0-240 | |
| LSIL (49) | 32 (65.3) | 70.41 | 46.724 | 0-180 | |
| Normal (11) | 1 (9.1) | 11.82 | 14.709 | 0-40 | |
| Cytology Samples <0.001 | |||||
| SCC (63) | 51 (81.0) | 110.77 | 62.86 | 0-180 | <0.001 |
| HSIL (55) | 31 (56.4) | 69.01 | 61.652 | 0-190 | |
| LSIL (49) | 24 (49) | 46.25 | 39.851 | 0-120 | |
| Normal (11) | 1 (9.1) | 9.09 | 13.751 | 0-40 | |
| Bonferroni Multiple Comparison | |||||
| Lesion | p-value (Cytology) | ||||
| SCC | HSIL | 0.016 | 0.005 | ||
| LSIL | 0.0001 | 0.0001 | |||
| Normal | 0.0001 | 0.0001 | |||
| HSIL | LSIL | 0.0001 | 0.183 | ||
| Normal | 0.0001 | 0.005 | |||
| LSIL | Normal | 0.047 | 0.314 | ||
The staining pattern of p63 was predominantly in the nucleus and found to be highly expressed in majority of SCC in tissue and cytology samples compared to other lesions (fig. 5 A-F). The difference in expression patterns of p63 in the different lesions analyzed was found to be significant in one-way analysis (ANOVA) (p<0.001). But in Bonferroni Multiple Comparison SCC (p= 0.001) and HSIL (p= 0.001) was found significant with respect to normal in histology and in cytology samples. HSIL showed significance with respect to normal (p= 0.032) and LSIL (p= 0.001) (Table 5). The diagnostic sensitivity of p63 for SCC samples was 97% with a specificity of 33%. The sensitivity and specificity for HSIL were 92% and 24% and for LSIL 54% and 33% respectively.
Figure 5.
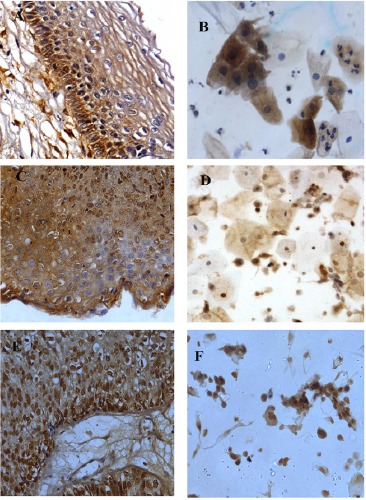
A-F, Expression patterns of p63 in histology and corresponding cytology samples; A and B, LSIL 40(X)- Mild nuclear and cytoplasmic expression; C and D; HSIL (40X)- Moderate nuclear expression and diffuse mild cytoplasmic expression; E and F, SCC (40X)- Dense nuclear expression.
Table 5.
One way ANOVA and Bonferroni Multiple Comparison of p63 Expression in Histopathology and their Corresponding Cytology Lesions of the Cervix
| Lesion (n) | Positive Cases (%) | Mean | Std. Deviation | Range | p-value |
|---|---|---|---|---|---|
| Histology Samples | |||||
| SCC (63) | 55 (87.3) | 154.76 | 78.758 | 0-270 | |
| HSIL (55) | 48 (87.3) | 156.18 | 75.216 | 0-270 | |
| LSIL (49) | 38 (77.6) | 118.57 | 69.761 | 0-190 | |
| Normal (11) | 5 (45.5) | 39.09 | 38.589 | 0-90 | |
| Cytology Samples <0.001 | |||||
| SCC (63) | 38 (60.3) | 63.46 | 54.548 | 0-170 | <0.001 |
| HSIL (55) | 35 (64.8) | 80.3 | 69.623 | 0-210 | |
| LSIL (49) | 19 (39.8) | 35.75 | 41.068 | 0-170 | |
| Normal (11) | 3 (27.3) | 26.36 | 24.606 | 0-70 | |
| Bonferroni Multiple Comparison | |||||
| Lesion | p-value (Histology) | p-value (Cytology) | |||
| SCC | HSIL | 1 | 1 | ||
| LSIL | 0.063 | 0.415 | |||
| Normal | 0.001 | 0.529 | |||
| HSIL | LSIL | 0.06 | 0.001 | ||
| Normal | 0.001 | 0.032 | |||
| LSIL | Normal | 0.008 | 1 | ||
Discussion
Management of precursor lesions identified in cervical cancer screening programme is difficult for low grade squamous intraepithelial lesions as 70-80% of low grade lesions spontaneously regress without any intervention. So follow up studies and treatment for these lesions is unnecessary. Thus, the identification of biomarkers to select women who are truly at risk of lesion progression will lead to reduction in the incidence of invasive squamous cell carcinoma as well as it will prevent over use of resources and eliminate patient anxiety. Usefulness of a wide array of molecular markers to identify progressive precancerous lesions of the cervix was reviewed and we have selected a panel of markers which have repeatedly been reported in several meta-analysis. Also the well-established putative role of these proteins in cervical carcinogenesis (Yim 2006) formed the rationale for selection of these markers.
Majority of the women diagnosed as SCC and HSIL were presented with some clinical symptoms and abnormalities in cervix on clinical examination, which supports our earlier suggestion that visual examination plus symptom history should be tried as a pre-selection criterion for cervical cancer screening in low resource countries (Sujathan et al., 1994). Similarly, most of the participants with low grade squamous intraepithelial lesion were in the premenopausal group and showed clinically healthy cervix whereas majority of the participants diagnosed as high-grade lesion and malignancy were in the post-menopausal group, which has already been reported in several similar studies (Murthy et al., 1990). Immuno expression pattern of MCM2, MCM5, CDC6, p16, and p63, were observed mainly on nucleus. A very few normal cells in abnormal samples showed positive staining with these markers. All these selected protein markers were found positive in all the dysplastic cells of more than 88% of HSIL cases and more than 93% of the squamous cell carcinoma samples in moderate to intense pattern whereas in the samples of low-grade lesions, about 70% of the dysplastic cells only showed these proteins in mild to moderate fashion in few studies (Murphy et al., 2005). Nearly 100% of HSIL and SCC have been reported to show overexpression of p16 protein but negative in non-dysplastic lesions in a few of the studies (Kalof et al., 2007). In a Spanish study, 91.7% of invasive carcinoma samples showed over expression of p16 protein (Perez et al., 2014). The current study also observed p16 over expression in 92.1% of SCC in histology samples whereas in cytology smears, 81% only showed over expression. Sixty five percentage of LSIL in the current study showed immunopositivity in tissue samples whereas it was only 49 % in cytology samples and is not found sensitive for a diagnosis of LSIL. Moreover it was found positive in immature metaplastic cells and reserve cells both in smears as well as in tissue sections. So this protein expression cannot be suggested as a screening test for cervical cancer.
MCM proteins are important regulators in the process of DNA replication (Borlado et al., 2008). These MCM proteins are detectable in the dividing phase and lost in differentiation stage of cell cycle (Madine et al., 2000). The MCM proteins have been considered as useful biomarkers that reflect the stages of the cell cycle (proliferative phase), but they are degraded in cells that have exited the cell cycle (quiescent). An increased expression of MCM proteins has been observed not only in the proliferative phase but also in pre malignant cells (Freeman et al., 1999; Rodins et al., 2002). A few studies have reported an increased expression of MCM5 proteins in tissue samples with features of HPV and this might be due to release of Rb inhibition on E2F by binding of HPV E7 oncoproteins. The E2F is the transcription factor that facilitates an increased transcription of MCM5 by binding to MCM5 promoter sites (Murphy et al., 2005). The current study also observed an increased immunopositivity of MCM2 and MCM5 in high grade dysplasic cells of HSIL and SCC than LSIL. An increased expression pattern of CDC6 protein was observed in both nucleus and cytoplasm of SCC and HSIL samples compared to LSIL samples. Like minichromosome maintenance proteins, the CDC6 proteins are also involved in the process of DNA replication (Borlado et al., 2008). A few cell line studies also have reported an increased expression of CDC6 in cervical carcinoma, both in nucleus and in cytoplasm (Fujita et al., 1999). The reason for this pattern of expression is, after phosphorylation of cyclin A-cyclin2 the CDC6 protein is translocated from its chromatin sites to the cytoplasm during the replication phase (S phase) of the cell cycle (Saha et al.,1998). CDC6 is then degraded by ubiquitin dependent proteolysis by the anaphase promoting complex/cyclosme (Biermann et al., 2002). Re-localization of CDC6 into the cytoplasm prevents re-initiation of replication and is necessary for coupling phase with the following mitosis. The over expression of CDC6 protein in cytoplasm of high grade lesion and SCC may be due to the accumulation of CDC6 protein in the cytoplasm after repeated and prolonged S phase in dysplastic cells (Clay-Farrace et al., 2003; Coverley et al., 2000). We found progressive over expression in CIN III, and SCC respectively in tissue samples. So the present study suggests CDC6 to be tried as biomarkers for progressive dysplastic lesions.
The p63 protein belongs to the family of p53, the most widely studied tumor suppressor protein and over expression of this protein has been reported in several human cancers (Rivlin et al., 2011). Like p53, aberrant expression of p63 has also been found in several human cancers in association with specific cell type differentiation and progression (Urist et al., 2002; Inoue 2014). The immunoexpression pattern of p63 on CIN samples has been reported to typically localize in the basal and parabasal cells in low-grade lesions and found extended to the middle and upper layers in samples of high-grade lesions (Quade et al., 2001). Our results are also similar in histology samples, but in cytology samples we couldn’t find much difference between normal and abnormal samples, because metaplastic squamous cells also showed positive immunoexpression for p63 protein. Also p63 has been suggested as a marker to distinguish squamous lesions (Garcia et al., 2007) from glandular lesions in cervical samples due to their negative expression in glandular lesions. Our observations are contradictory to this report. We found the atypical glandular cells adjacent to the SCC lesions expressing p63 protein. The link between p63 over expression and HPV associated oncoprotein expression is controversial. The E6 and E7 genes of high risk HPV types are essential for the inactivation of the p53 and retinoblastoma proteins for the deregulation of the tumor suppressor genes. Teissier et al., reported that the p63 target genes could be activated by suppressing the E6/E6AP pathway (Teissier, 2007). Also the protein p63 is reported as a marker of reserve cells or stem like cells (Witkiewicz et al., 2005). Reserve cells are seen in the basal layers of the epithelium. We found p63 to express in a homogenous pattern in the basal layer of the tissue samples of LSIL, HSIL and in the reserve cell population in the cytology samples. The reserve cells are reported as the target cell for HPV infection (Smedts et al., 1992). The cytology samples often contain reserve cells and immature metaplastic cells, particularly in the child baring age group. Hence this protein expression cannot be considered as a significant marker for screening purpose.
While analyzing the sensitivity and specificity of these markers individually, it was observed that, all these five markers have high sensitivity (>90%) for identifying SCC and HSIL, but in LSIL sensitivity of MCM proteins and CDC6 was >70% but the sensitivity of p16 and p63 for the identification of LSIL was less than <65%. It was found positive in metaplastic cells both in smears as well as in tissue sections. The diagnostic accuracy of p16 for the identification of low grade lesions was very low compared to SCC and HSIL. MCM proteins and cdc6 were found to have highest diagnostic accuracy for identifying low grade dysplastic cells. The progressive increase in the expression patterns of these proteins in low grade to high grade lesions suggests its significance in predicting malignant potential of LSILs. Follow up studies of these lesions and quantitative assessment of these proteins is being evaluated to establish the same.
In conclusion, even though several reports are available for the significance of MCM2, MCM5, CDC6, p16 and p63 for identification of cervical intraepithelial lesions in tissue samples, none of them has been validated as a standalone test for cervical cancer screening in smears as biopsy collection or any such surgical procedures are not acceptable as screening test. The current study suggests MCM2 MCM5 and CDC6 should be tried as markers for identifying progressive precancerous lesions.
Conflict of interest
Authors have no Conflict of interest.
Acknowledgements
Authors wish to acknowledge Kerala State Council for Science Technology and Environment, Govt. of Kerala for funding.
References
- Biermann E, Baack M, Kreitz S, et al. Synthesis and turn-over of the replicative Cdc6 protein during the HeLa cell cycle. Eur J Biochem. 2002;269:1040–6. doi: 10.1046/j.0014-2956.2001.02746.x. [DOI] [PubMed] [Google Scholar]
- Borlado LR, Mendez J. CDC6:from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis. 2008;29:237–3. doi: 10.1093/carcin/bgm268. [DOI] [PubMed] [Google Scholar]
- Clay-Farrace L, Pelizon C, Santamaria D, et al. Human replication protein Cdc6 prevents mitosis through a checkpoint mechanism that implicates Chk1. EMBO J. 2003;22:704–2. doi: 10.1093/emboj/cdg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Pelizon C, Trewick S, et al. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J Cell Sci. 2000;113:1929–8. doi: 10.1242/jcs.113.11.1929. [DOI] [PubMed] [Google Scholar]
- Freeman A, Morris LS, Mills AD, et al. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res. 1999;5:2121–2. [PubMed] [Google Scholar]
- Fujita M, Yamada C, Goto H. Cell cycle regulation of human Cdc6 protein. J Biol Chem. 1999;274:25927–2. doi: 10.1074/jbc.274.36.25927. [DOI] [PubMed] [Google Scholar]
- Garcia MT, Acar MD, Jorda M, et al. Use of p63 for distinction of glandular versus squamous lesions in cervicovaginal specimens. Cancer Cytopathol. 2007;111:54–7. doi: 10.1002/cncr.22419. [DOI] [PubMed] [Google Scholar]
- Inoue K, Fry EA. Alterations of p63 and p73 in human cancers. Subcell Biochem. 2014;85:17–40. doi: 10.1007/978-94-017-9211-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalof AN, Cooper K. Our approach to squamous intraepithelial lesions of the uterine cervix. J Clin Pathol. 2007;60:449–5. doi: 10.1136/jcp.2005.036426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine MA, Swietlik M, Pelizon C. The roles of the MCM, ORC and Cdc6 proteins in determining the replication competence of chromatin in quiescent cells. J Struct Biol. 2000;129:198–10. doi: 10.1006/jsbi.2000.4218. [DOI] [PubMed] [Google Scholar]
- Masoudi H, Van Niekerk DJ, Gilks CB, et al. Loss of p16 INK4 expression in invasive squamous cell carcinoma of the uterine cervix is an adverse prognostic marker. Histopathology. 2006;49:542–5. doi: 10.1111/j.1365-2559.2006.02510.x. [DOI] [PubMed] [Google Scholar]
- Murphy N, Ring M, Heffron CC, et al. p16INK4A, CDC6, and MCM5:predictive biomarkers in cervical preinvasiveneoplasia and cervical cancer. J Clin Pathol. 2005;58:525–4. doi: 10.1136/jcp.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy NS, Sehgal A, Satyanarayana L, et al. Risk factors related to biological behaviour of precancerous lesions of the uterine cervix. Br J Cancer. 1990;61:732–6. doi: 10.1038/bjc.1990.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quade BJ, Yang A, Wang Y, et al. Expression of the p53 homologue p63 in early cervical neoplasia. Gynecol Oncol. 2001;80:24–9. doi: 10.1006/gyno.2000.5953. [DOI] [PubMed] [Google Scholar]
- Rivlin N, Brosh R, Oren M, et al. Mutations in the p53 tumor suppressor gene important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466–4. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodins K, Cheale M, Coleman N, et al. Minichromosome maintenance protein2 expression in normal kidney and renal cell carcinoma:relationship to tumor dormancy and potential clinical utility. Clin Cancer Res. 2002;8:1075–1. [PubMed] [Google Scholar]
- Saha P, Chen J, Thome KC, et al. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol. 1998;18:2758–7. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarawardana P, Singh M, Shroyer KR. Dual stain immunohistochemical localization of p16INK4A and ki-67:a synergistic approach to identify clinically significant cervical mucosal lesions. Appl Immuno histochem Mol Morphol. 2011;19:514–8. doi: 10.1097/PAI.0b013e3182167c66. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Smedt F, Ramaekers F, Troyanovsky S, et al. Basal-cell keratins in cervical reserve cells and a comparison to their expression in cervical intraepithelial neoplasia. Am J Pathol. 1992;140:601–2. [PMC free article] [PubMed] [Google Scholar]
- Sujathan K, Kannan S, Pillai KR, et al. Implications of Gynaecological abnormalities in the pre-selection Criteria for cervical cancer screening:preliminary evaluation of 3602 subjects in South India. Cytopathol. 1994;6:75–7. doi: 10.1111/j.1365-2303.1995.tb00451.x. [DOI] [PubMed] [Google Scholar]
- Urist MJ, Como CJ, Lu ML, et al. Loss of p63 expression is associated with tumor progression in bladder cancer. Am J Pathol. 2002;161:1199–6. doi: 10.1016/S0002-9440(10)64396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Putte G, Holm R, Lie AK, et al. Expression of p27, p21, and p16 protein in early squamous cervical cancer and its relation to prognosis. Gynecol Oncol. 2003;89:140–7. doi: 10.1016/s0090-8258(03)00010-6. [DOI] [PubMed] [Google Scholar]
- Witkiewicz AK, Hecht JL, Cviko A, et al. Microglandular hyperplasia:a model for the de novo emergence and evolution of endocervical reserve cells. Hum Pathol. 2005;36:154–1. doi: 10.1016/j.humpath.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Yim EK, Park JS. Biomarkers in cervical cancer. Biomark Insights. 2006;1:215–5. [PMC free article] [PubMed] [Google Scholar]


