Abstract
Background and Aims:
It has been demonstrated that homozygote and heterozygote mutant allele carriers for thiopurine S-methyltransferase (TPMT) are at high risk of developing myelosuppression after receiving standard doses of 6-mercaptopurine (6-MP). The aim of this study was to determine the frequency of TPMT deficient alleles in children with acute lymphoblastic leukemia (ALL) in Jordan and to compare it with other ethnic groups.
Methods:
We included 52 ALL childhood cases from King Hussein Cancer Research Center in Jordan. Genotyping of the rs1800460, rs1800462, and rs1142345 SNPs was performed by polymerase chain reaction (PCR) followed by sequencing. Comparisons were made with historical data for controls and for both volunteers and cases from other middle-eastern countries.
Results:
Mutant TPMT alleles were present in 3.8% (2/52) of patients. Allelic frequencies were 1.0% for both TPMT*B and TPMT*C. None of the patients were heterozygous or homozygous for TPMT*3A or TPMT *2. We did not find statistically significant differences in the distribution of mutant alleles between Jordan and other middle-eastern countries for both healthy volunteers or ALL patients.
Conclusions:
The overall frequency of TPMT mutant alleles was low and did not exhibit differences compared to other middle-eastern countries, including Jordanian studies assessing TPMT mutant alleles in healthy volunteers. The current results question the value of TPMT genotyping in the Jordanian population.
Keywords: Thiopurine methytransferase, childhood acute lymphoblastic leukemia, pharmacogenetics, 6−mercaptopurine
Introduction
Thiopurine drugs, including Azathioprine (AZA) and 6-mercaptopurine (6-MP) are cytotoxic drugs used in the treatment of several serious diseases such as childhood acute lymphoblastic leukemia (ALL) and inflammatory bowel disease (Krynetski and Evans, 2003).
AZA is almost completely converted to 6-MP via a non-enzymatic reaction(Dubinsky, 2004);(Bradford and Shih, 2011). Subsequently, 6-MP is metabolized by three competing enzyme systems (Figure 1), xanthine oxidase (XO), thiopurine S-methyltransferase (TPMT) and hypoxanthine phosphoribosyl transferase (HPRT). The enzyme XO catalyses the transformation of 6-MP into the inactive metabolite 6-thiouric-acid (6-TUA) which is excreted in the urine, whereas the enzyme TPMT methylates 6-MP into 6- methyl mercaptopurine (6-mMPN) which is associated with hepatotoxicity. The HPRT enzyme is responsible for the conversion of 6-MP into 6-thioinosine-monophosphate (6-TIMP) which is converted via multiple steps to 6-thioguanine nucleotides (6-TGN) including 6-thioguanine monophosphate (6-TGMP), diphosphate (6-TGDP) and -triphosphate (6-TGTP) which are the active products that are incorporated into DNA, resulting in cell apoptosis (Louis and Belaiche, 2003; Sahasranaman et al., 2008; Bradford and Shih, 2011). TPMT activity determines the balance between two active metabolites, the myelosuppressive 6-TGNs and the potentially hepatotoxic 6-mMPNs (Derijks et al., 2006). It has been demonstrated that there is a significant negative correlation between 6-TGNS levels in RBCS and TPMT activity(Balis and Adamson, 1999). Therefore, patients with high 6-TGNs levels and low TPMT activity are more susceptible for the myelosuppression or even death in some cases (Katsanos and Tsianos, 2008; Nguyen et al., 2011). TPMT is encoded by a 34 kb gene consisting of ten exons and nine introns and has been known to be polymorphic in the general population (Szumlanski et al., 1996). The distribution of alleles is trimodal with homozygote allele (two low TPMT metabolizing alleles;0.3%), heterozygote allele (one high and one low metabolizing TPMT allele; 11.1%) and wild type with two high metabolizing TPMT allele type; 88.6% (Louis and Belaiche, 2003; Hanai et al., 2010).
Figure 1.
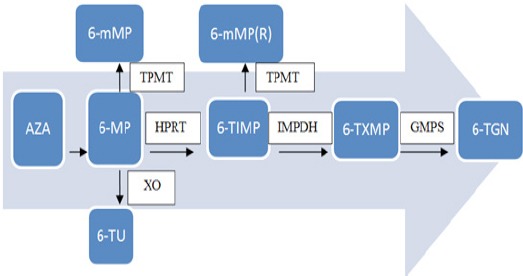
7 Metabolism of Thiopurine Drugs. AZA is converted to 6-MP by a nonenzymatic route. Initial 6-MP transformation occurs via competition of xanthine oxidase (XO) and thiopurine methyltransferase (TPMT) and the hypoxanthine phosphoribosyltransferase (HPRT) enzymatic pathway. Once produced, 6-thiosine 5’-monophosphate (6-TIMP) may be transformed either into 6-TGN or 6-mMP(R) by the rate-limiting enzymatic pathways, inosine monophosphate dehydrogenase (IMPDH) and [guanosine monophosphate synthetase (GMPS). The production of high levels of 6-TGN and 6-mMP (R) increases the risk of myelosuppression and hepatotoxicity associated with therapy. Adapted from Bradford and Shih (2011)
So far, twenty-eight variant alleles have been identified (Garat et al., 2008), however 4 variant alleles [TPMT*2 (c.238G>C), TPMT*3A (c.460G>A and c.719A>G) and TPMT*3C (c.719A>G)] and TPMT*3B (c.460G>A) have been intensively studied both according to their molecular mechanisms and/or clinical implications(Wang et al., 2010). Genotyping for the TPMT*3 family of variant alleles detect more than 92% of deficient activity alleles of TPMT enzyme, addition of TPMT*2 genotyping pushes this to over 95%(Schaeffeler et al., 2004; Wang and Weinshilboum, 2006). On the other hand, concordance between low TPMT activity and a mutant heterozygote phenotype was reported to be 83%(Ford et al., 2009).
In heterozygote individuals with intermediate TPMT activity, the initial doses of thipurines should be reduced by 30-70%, whereas in homozygote non-functional allele carriers the doses of thiopurine drugs should be reduced by 90%, or patients should receive another treatment (Peregud-Pogorzelski et al., 2011). Therefore, analysis of TPMT genotypes can provide an important molecular biomarker that predicts drug response in patients treated from hematological malignancies and autoimmune diseases(Sahasranaman et al., 2008; Budhiraja and Popovtzer, 2011). The aim of this study was to determine the frequency of the most common TPMT gene polymorphisms (238G>C), 460G>A and 719A>G), among children with ALL in Jordan and to compare it with other ethnic groups.
Materials and Methods
Study subjects
Fifty-two children who were diagnosed with ALL, attending the outpatient clinic at King Hussein Cancer Centre (KHCC), and receiving 6-MP were recruited in the study. The study was approved by the research committee in King Hussein Cancer Centre (KHCC), Reference number (15KHCC 57). Patients’ data and blood sample were collected after obtaining a parental consent form. Recruitment was commenced over a period of one year from September, 2015 to September, 2016.
An aliquot of 2 ml blood was obtained from a routine clinical blood sample withdrawn from the patient at the clinic using K3 EDTA coated tubes and stored in ice box during transportation. All blood samples were processed within four hours of blood withdrwal.
Genotyping
Peripheral blood mononuclear cells (PBMC) were extracted from anticoagulated whole blood using “Promega-Wizard genomic DNA purification kit, Promega Corporation, USA” according to manufacturer’s instructions. A total of 52 DNA samples were analyzed. Total genomic DNA extracted blood samples was processed either immediately or stored at -20°C until being used. Four TPMT SNPs (G460A (TPMT*3B), A719G (TPMT*3C), (G460A and A719G (TPMT*3A) andG238C (TPMT*2) were tested by Polymerase chain reaction (PCR) followed by sequencing using the Primers showed in the Table 1. All customized primers were synthesized by Princess Haya Biotechnology Centre (Irbid, Jordan). Sequences were analyzed using the sequencing analysis software (Chromas Lite, version 2.1.1).
Table 1.
Primers Used for the DNA Sequencing of TPMT SNPs
| Gene | Rs | Forward primer | Reverse primer |
|---|---|---|---|
| TPMT G460A | 1800460 | AGGCCACACAGCTTGAAAGT | CCCAGGTCCACACATTCCTC |
| TPMT A719G | 1142345 | AATCTGCAAGACACATAGGCA | AGGTTGATGCTTTTGAAGAACGA |
| TPMT G238C | 1800462 | ACCTTAAATACTTTGGTTCCAGG | GCTTACTCTAATATAACCCTCT |
PCR amplifications were done in a reaction volume of 20 µL containing 50-200ng of genomic DNA, final concentrations of 0.5 µMforward and reverse primers, 10 µL of 2X KAPA2 Fast ReadyMix (Kapabiosystem, USA).
All PCR reactions started with denaturation for 5 minutes at 95, followed by 39 cycles of 30 seconds at 94ºC, then 30 seconds at 58ºC annealing temperature, and finally for 30 seconds at 72ºC as extension temperature.
PCR products were analyzed and resolved using agarose gel (2 %). The resolved DNA bands are detected by staining the gel with safe-red dye, followed by photographing under ultraviolet (UV) illumination. A 50-bp ladder was used as a convenient marker to estimate the size of the amplified product.
Samples with sharp PCR products are good candidates for sequencing. The Sanger sequencing technique was used in our study by GENEWIZ Technical Support Group, USA (http://www.genewiz.com).
Statistical analysis
Online Encyclopedia for Genetic Epidemiology studies (http://www.oege.org/software/hwe-mr-calc.shtml) was used to test Hardy-Weinberg equilibrium (HWE) for genotype distributions in ALL cases. Data was analysed using Fisher’s exact test to evaluate the difference in allele frequencies between populations. Differences were considered significant if p-value <0.05. SPSS, version 22.0, was used for the analysis of the statistical data.
Results
Genotype and allele frequencies
In this study, the frequencies of TPMT variants were investigated in 52 patients with ALL. Mean age of children was 8.90 years (SD= 4.44) and 40.4% of patients were female.
Table 2 shows the studied genotypes and allele frequencies. The most predominant genotype of TPMT (rs1800460) was the homozygous wild type genotype GG (N=51, 98.08%) followed by the heterozygous GA (N=1, 1.92%), whereas the homozygous variant AA was not detected. Figure 2 shows chromatograms of the wild and heterozygous variant. For TPMT (rs1142345), the vast majority of patients were carriers for the homozygous wild type genotype AA (N=47, 98%), while one patient was a carrier for the heterozygous AG (N=1, 2%) Figure 3 shows chromatograms of the wild and heterozygous variant.
Table 2.
Genotypes and Minor Allele Frequencies among Recruited ALL Patients
| rs | Variant | Genotype / Allele | Observed frequency N(%) | Expected frequency* N(%) | P- value | MAF (%) |
|---|---|---|---|---|---|---|
| rs1800460 | (*3B) c.460G > A | GG | 51 (98.08%) | 51 (98.08%) | 0.99 | 1.00% |
| GA | 1 (1.92 %) | 1 (1.92 %) | ||||
| AA | 0 (0.0 %) | 0 (0.0 %) | ||||
| rs1142345 | (*3C) c.719A > G | AA | 47 (98%) | 47 (98%) | 0.99 | 1.00% |
| AG | 1 (2%) | 1 (2%) | ||||
| GG | 0 (0.0%) | 0 (0.0%) | ||||
| rs1800462 | (*2) c.238G>C | GG | 51 (100%) | 51 (100%) | 1 | 0.00% |
| GC | 0 (0.0%) | 0 (0.00%) | ||||
| CC | 0 (0.0%) | 0 (0.00%) | ||||
| rs1800460 & rs1142345 | TPMT*3A (c.460G>A and c.719A>G) | Carriers of both TPMT*3B and TPMT*3C alleles | 0 (0.0%) | 0 (0.00%) | - | - |
, Using Hardy-Weinberg Equilibrium; MAF, minor allele frequency; N, numbe; %, percentage
Figure 2.
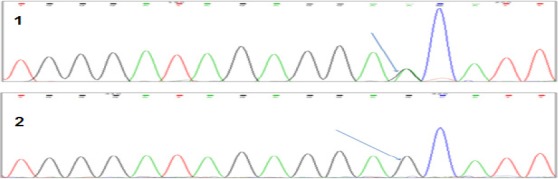
Chromatograms of (*3B) c.460G > A(rs1800460): (1) Heterozygote Genotype (GA). (2) Wild Type Genotype (GG)
Figure 3.
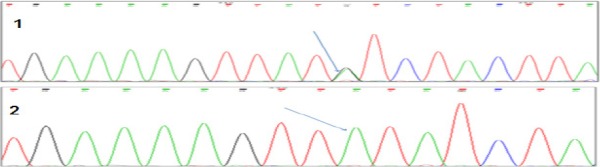
Chromatogram(*3C) c.719A > G (rs1142345): (1) Heterozygote Genotype (AG). (2) Wild Type Genotype (AA)
None of the patients carried the TPMT*3A or TPMT *2 allele. Genotype and allele frequencies in our study were matched to expectation by Hardy-Weinberg Equilibrium of the tested polymorphisms.
Discussion
Interracial variations in the frequency of genes encoding for drug metabolizing enzymes, transports and receptors are well documented in the literature (Phan et al., 2009). Genetic variations can account for a large proportion of the variability observed in drug response, thus elucidating these variations and understanding their roles in drug disposition is of paramount importance for individualizing patient care. Among various races, the Arab race, due to significant genetic admixture, is considered one of the distinct races (Hamdy et al., 2003). Among Arabs, we focused on the present study on the Jordanian population, and we aimed to investigate the frequency of the TPMT deficient alleles in children with ALL in Jordan and to compare with other ethnic groups.
None of the ALL children included in the present study carried the TPMT*2 allele. This finding comes in line not only with the results of previous Jordanian studies conducted in healthy volunteers (Hakooz et al., 2010; Elawi et al., 2013), but also with the results of all studies conducted in Asian countries and the majority of studies conducted at Middle-eastern countries such as Palestine and Turkey (Table 3). Interestingly, a higher frequency of this variant has been reported by an Iranian study (3.9%) (Azad et al., 2009), this frequency was statistically different than our frequency (p<0.05). Furthermore, two later larger sample-sized studies conducted in South Iran reported lower frequency of this variant allele 0.1% and 2.2% (Bahari et al., 2010; Moini et al., 2012). Finally, as shown in Table 3 there was no significant difference in the frequency of the TPMT*2 between our study and studies conducted in Europe and Africa.
Table 3.
Frequency of Selected TPMT Variant Allels Across the World in Healthy or ALL Patients
| Country | Total number of alleles | Frequency of TPMT *2 | Frequency of TPMT *3B | Frequency of TPMT *3C | Frequency of TPMT *3A | Healthy or patients | Reference |
|---|---|---|---|---|---|---|---|
| Jordan | 96-104 | 0.00% | 1.00% | 1.00% | 0.00% | ALL patients | This study |
| Jordan | 338 | 0.00% | 0.00% | 0.30% | 0.59% | Healthy | (Hakooz et al., 2010) |
| Jordan | 500 | 0.00% | 0.40% | 0.00% | 0.40% | Healthy | (Elawi et al., 2013) |
| Middle eastern | |||||||
| Egypt | 400 | 0.00% | 0.00% | 1.30% | 0.30% | Healthy (Students and staff at Cairo University) | (Hamdy et al., 2003) |
| Iran | 1000 | 0.10% | 0.00% | 2.50% | 0.00% | Healthy | (Moini et al., 2012) |
| Iran | 1664 | 2.16% | 1.62% | 0.54% | 1.68% | Healthy | (Bahari et al., 2010) |
| Iran | 254 | 3.93%* | 0.00% | 1.57% | 0.79% | Healthy | (Azad et al., 2009) |
| Turkey | 212 | 0.00% | 0.00% | 0.90% | 0.90% | ALL patients | (Tumer et al., 2007) |
| Palestine | 112 | 0.00% | 0.00% | 0.00% | 0.89% | ALL patients | (Ayesh et al., 2013) |
| European countries | |||||||
| British- Caucasians | 2298 | 0.22% | 0.00%* | 0.70% | 4.50%* | ALL patients | (Lennard et al., 2013) |
| French - Caucasians | 938 | 0.70% | 0.00% | 0.40% | 3.00% | n=304 healthy, n=147 children hospitalized for day care surgery n=18 neonates (cord bloods) | (Ganiere-Monteil et al., 2004) |
| Italian | 1886 | 0% | 0.32% | 0.32% | 2.20% | Healthy | (Serpe et al., 2009) |
| German- Caucasians | 2428 | 0.20% | 0.00%* | 0.40% | 4.40%* | Healthy | (Schaeffeler et al., 2004) |
| Russia | 1990 | 0.10% | 0.00%* | 0.40% | 2.30% | n=446 children with malignant diseases, n= 549 children and adults without malignant disease | (Samochatova et al., 2009) |
| Spain (Spanish) | 276 | - | 1.45% | 1.45% | 3.26% | Healthy | (Corominas et al., 2006) |
| Sweden | 1600 | 0.06% | 0.13% | 0.44% | 3.75%* | n=800 DNA samples obtained from a data bank | (Haglund et al., 2004) |
| Countries of North, Central and South America | |||||||
| Argentina | 294 | 0.70% | 0.00% | 0.00% | 3.06% | Healthy | (Larovere et al., 2003) |
| Brazil | 408 | 2.20% | 0.20% | 1.00% | 1.50% | ALL n=2, non-ALL n=202 | (Boson et al., 2003) |
| Bolivia | 230 | 0.00% | 0.00% | 0.00% | 6.52%* | NS | (Lu et al., 2005) |
| Mexico | 216 | 0.90% | 2.30% | 1.40% | 3.24% | Healthy | (Taja-Chayeb et al., 2008) |
| Mexico | 78 | 2.70% | 0.00% | 2.50% | 7.60%* | ALL | (Taja-Chayeb et al., 2008) |
| USA-African | 496 | 0.40% | 0.00% | 2.42% | 0.81% | n=196 healthy, n=52 ALL | (Hon et al., 1999) |
| USA-Caucasian | 564 | 0.17% | 0.00% | 0.18% | 3.19% | unrelated white subjects | (Hon et al., 1999) |
| African countries | |||||||
| Ghana | 434 | 0.00% | 0.00% | 7.60%* | 0.00% | Healthy | (Ameyaw et al., 1999) |
| Ghana | 232 | 0.00% | - | 6.47%* | 0.00% | Healthy | (Schaeffeler et al., 2008) |
| Kenyan | 202 | 0.00% | 0.00% | 5.45%* | 0.00% | Healthy | (McLeod et al., 1999) |
| Libya | 492 | 0.00% | 0.00% | 1.02% | 0.61% | Healthy | (Zeglam et al., 2015) |
| Mozambique | 500 | 0.00% | - | 3.80% | 0.20% | Healthy | (Alves et al., 2004) |
| Angola | 206 | 0.00% | 0.00% | 3.90% | 0.00% | Healthy | (Oliveira et al., 2007) |
| Asian countries | |||||||
| Chinese | 1402 | 0.00% | 0.00% | 1.05% | 0.42% | Healthy | (Zhang et al., 2006) |
| Chinese | 426 | 0.00% | 0.00% | 0.23% | 0.00% | Healthy | (Zhang et al., 2004) |
| Japanese | 302 | 0.00% | 0.00% | 0.33% | 0.00% | Healthy | (Kubota and Chiba, 2001) |
| Korean | 1800 | 0.00% | 0.00% | 1.40% | 0.00% | Patients (type of disease not reported) | (Kim et al., 2015) |
Significant p-value<0.05
The TPMT*3A allele, which contains two genetic variations [G460A in exon 7 and A719G in exon 10], has not been detected in the present study. The *3A is a common variant allele in Caucasians (Barik et al., 2017). Many white/Caucasian populations have reported a significantly higher frequency of this allele compared to this study. For instance, the *3A allele has been found in a frequency of 4.5% in British Caucasians and a similar frequency has been also reported in German-Caucasians. Furthermore, compared to our results the frequency of the *3A was also significantly higher in certain populations of the Americas (Table 3). On the other hand, the frequency of the *3A allele was not significantly different than our frequency in Asian, African and other Jordanian studies in healthy volunteers.
TPMT*3B is a rare allele that is usually absent in most populations. The Mexican population has been reported to harbor a high frequency of this variant allele, as its frequency has mounted to 2.3% in healthy volunteers (Rossino et al., 2006). Studies conducted in the Jordanian population reported a frequency of this allele ranging between 0.0% and 1% (Table 3). Amongst the European countries, a relatively high frequency has been reported in the Spanish population (1.5%), and a statistically significant lower frequency of this variant has been detected in the Russian (0/1990 alleles), the British population (0/2298 alleles) and German population (0/2428 alleles) compared to our population.
The early work of Yates et al., (1997) suggested that the TPMT*3C allele might be more prevalent in black subjects as opposed to white subjects, with the accumulation of knowledge from subsequent investigations this information has been repetitively confirmed, as noted in Table 3. The TPMT*3C allele accounted for more than 60% up to 100% of the TPMT *2 *3A*3B and *3C alleles in African studies, and its frequency in the Ghanian and Kenyan population was significantly higher than its frequency at our population. In lower frequencies, this variant has been also detected in Asian populations (0.23% - 1.44%), European populations (0.32% - 1.45%), and the Jordanian population (0.0%-1.0%) (Table 3).
Individuals who are homozygous for the TPMT variant alleles or who are compound heterozygotes (i.e. carriers of two different alleles; TPMT*2/3C, TPMT*2/*3B or TPMT*2/*3A), have been shown to be associated with low TPMT activity and severe bone marrow toxicity when treated with mercaptopurine (Brouwer et al., 2001; Belen et al., 2014). We could not detect any individual carrying these genotypes; however, due to their rarity, our statistical power is inadequate to preclude their presence in the Jordanian population.
The present study is limited by the small sample size, which is related to the small pediatric ALL population at KHCC. Nevertheless, it provides the first insight of TPMT variations in ALL patients. Furthermore, the current results, pose a serious question regarding the value of initiationTPMT genotyping service in Jordan for patients initiating the thiopurine drugs. This question can be only answered by conducting a large scale genetic study combined with a pharmacoeconomic study.
In conclusion, in this study wedetermined, for the first time, the frequency of four major TPMT mutant alleles in a Jordainan children with ALL. The overall frequency of TPMT mutant alleles was low (3.8%) of patients, with equal distribution of the two mutant alleles TPMT*3B and TPMT*3C, (1%). Nevertheless, it did not show differential distribution compared to Jordainian healthy population or other middle-eastern countries. The presentstudy open new horizons for investigating the value of TPMT genotyping for monitoring patients in Jordan to be treated with doses of thiopurine drugs.
References
- Alves S, Rocha J, Amorim A, et al. Tracing the origin of the most common thiopurine methyltransferase (TPMT) variants:preliminary data from the patterns of haplotypic association with two CA repeats. Ann Hum Genet. 2004;68:313–23. doi: 10.1046/j.1529-8817.2004.00104.x. [DOI] [PubMed] [Google Scholar]
- Ameyaw MM, Collie-Duguid ES, Powrie RH, et al. Thiopurine methyltransferase alleles in British and Ghanaian populations. Hum Mol Genet. 1999;8:367–70. doi: 10.1093/hmg/8.2.367. [DOI] [PubMed] [Google Scholar]
- Ayesh BM, Harb WM, Abed AA. Thiopurine methyltransferase genotyping in Palestinian childhood acute lymphoblastic leukemia patients. BMC Hematol. 2013;13:3. doi: 10.1186/2052-1839-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad M, Kaviani S, Soleimani M, et al. Common polymorphism's analysis of thiopurine S-methyltransferase (TPMT) in Iranian population. Yakhteh Med J. 2009;11:311–6. [Google Scholar]
- Bahari A, Hashemi M, Bari Z, et al. Frequency of thiopurine S-methyltransferase (TPMT) alleles in southeast Iranian population. Nucleosides Nucleotides Nucleic Acids. 2010;29:237–44. doi: 10.1080/15257771003720418. [DOI] [PubMed] [Google Scholar]
- Balis FM, Adamson PC. Application of pharmacogenetics to optimization of mercaptopurine dosing. J Natl Cancer Inst. 1999;91:1983–5. doi: 10.1093/jnci/91.23.1983. [DOI] [PubMed] [Google Scholar]
- Barik S, Singh S, Gupta S, et al. Association of polymorphism of enzyme thiopurine methyl transferase in head and neck squamous cell cancer and treatment response to concurrent chemo radiotherapy. J of Med Sci and Cli Res. 2017:20832–41. [Google Scholar]
- Belen BF, Gursel T, Akyurek N, et al. Severe myelotoxicity associated with thiopurine S-Methyltransferase*3A/*3C polymorphisms in a patient with pediatric leukemia and the effect of steroid therapy. Turk J Haematol. 2014;31:276–85. doi: 10.4274/tjh.2013.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boson WL, Romano-Silva MA, Correa H, et al. Thiopurine methyltransferase polymorphisms in a Brazilian population. Pharmacogenomics J. 2003;3:178–82. doi: 10.1038/sj.tpj.6500175. [DOI] [PubMed] [Google Scholar]
- Bradford K, Shih DQ. Optimizing 6-mercaptopurine and azathioprine therapy in the management of inflammatory bowel disease. World J Gastroenterol. 2011;17:4166–73. doi: 10.3748/wjg.v17.i37.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer C, Marinaki AM, Lambooy LH, et al. Pitfalls in the determination of mutant alleles of the thiopurine methyltransferase gene. Leukemia. 2001;15:1792–3. doi: 10.1038/sj.leu.2402285. [DOI] [PubMed] [Google Scholar]
- Budhiraja P, Popovtzer M. Azathioprine-related myelosuppression in a patient homozygous for TPMT*3A. Nat Rev Nephrol. 2011;7:478–84. doi: 10.1038/nrneph.2011.74. [DOI] [PubMed] [Google Scholar]
- Corominas H, Domenech M, del Rio E, et al. Frequency of thiopurine S-methyltransferase alleles in different ethnic groups living in Spain. Med Clin (Barc) 2006;126:410–2. doi: 10.1157/13086124. [DOI] [PubMed] [Google Scholar]
- Derijks LJ, Gilissen LP, Hooymans PM, et al. Review article:thiopurines in inflammatory bowel disease. Aliment Pharmacol Ther. 2006;24:715–29. doi: 10.1111/j.1365-2036.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Dubinsky MC. Azathioprine, 6-mercaptopurine in inflammatory bowel disease:pharmacology, efficacy, and safety. Clin Gastroenterol Hepatol. 2004;2:731–43. doi: 10.1016/s1542-3565(04)00344-1. [DOI] [PubMed] [Google Scholar]
- Elawi AM, Irshaid YM, Ismail SI, et al. Thiopurine S-methytransferase gene polymorphism in rheumatoid arthritis. Arch Med Res. 2013;44:105–9. doi: 10.1016/j.arcmed.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Ford L, Kampanis P, Berg J. Thiopurine S-methyltransferase genotype-phenotype concordance:used as a quality assurance tool to help control the phenotype assay. Ann Clin Biochem. 2009;46:152–4. doi: 10.1258/acb.2008.008167. [DOI] [PubMed] [Google Scholar]
- Ganiere-Monteil C, Medard Y, Lejus C, et al. Phenotype and genotype for thiopurine methyltransferase activity in the French Caucasian population:impact of age. Eur J Clini Pharmacol. 2004;60:89–96. doi: 10.1007/s00228-004-0732-5. [DOI] [PubMed] [Google Scholar]
- Garat A, Cauffiez C, Renault N, et al. Characterisation of novel defective thiopurine S-methyltransferase allelic variants. Biochem Pharmacol. 2008;76:404–15. doi: 10.1016/j.bcp.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Haglund S, Lindqvist M, Almer S, et al. Pyrosequencing of TPMT alleles in a general Swedish population and in patients with inflammatory bowel disease. Clin Chem. 2004;50:288–95. doi: 10.1373/clinchem.2003.023846. [DOI] [PubMed] [Google Scholar]
- Hakooz N, Arafat T, Payne D, et al. Genetic analysis of thiopurine methyltransferase polymorphism in the Jordanian population. Eur J Clin Pharmacol. 2010;66:999–1003. doi: 10.1007/s00228-010-0826-1. [DOI] [PubMed] [Google Scholar]
- Hamdy SI, Hiratsuka M, Narahara K, et al. Genotype and allele frequencies of TPMT, NAT2, GST, SULT1A1 and MDR-1 in the Egyptian population. Br J Clin Pharmacol. 2003;55:560–9. doi: 10.1046/j.1365-2125.2003.01786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai H, Iida T, Takeuchi K, et al. Thiopurine maintenance therapy for ulcerative colitis:the clinical significance of monitoring 6-thioguanine nucleotide. Inflamm Bowel Dis. 2010;16:1376–81. doi: 10.1002/ibd.21190. [DOI] [PubMed] [Google Scholar]
- Hon YY, Fessing MY, Pui CH, et al. Polymorphism of the thiopurine S-methyltransferase gene in African-Americans. Hum Mol Genet. 1999;8:371–6. doi: 10.1093/hmg/8.2.371. [DOI] [PubMed] [Google Scholar]
- Katsanos K, Tsianos E. Azathioprine/6-mercaptopurine toxicity:the role of the TPMT gene. Ann Gastroenterol. 2008;20:251–64. [Google Scholar]
- Kim HY, Lee SH, Lee MN, et al. Complete sequence-based screening of TPMT variants in the Korean population. Pharmacogenet Genomics. 2015;25:143–6. doi: 10.1097/FPC.0000000000000117. [DOI] [PubMed] [Google Scholar]
- Krynetski E, Evans WE. Drug methylation in cancer therapy:lessons from the TPMT polymorphism. Oncogene. 2003;22:7403–13. doi: 10.1038/sj.onc.1206944. [DOI] [PubMed] [Google Scholar]
- Kubota T, Chiba K. Frequencies of thiopurine S-methyltransferase mutant alleles (TPMT*2, *3A, *3B and *3C) in 151 healthy Japanese subjects and the inheritance of TPMT*3C in the family of a propositus. Br J Clin Pharmacol. 2001;51:475–7. doi: 10.1046/j.1365-2125.2001.01371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larovere LE, de Kremer RD, Lambooy LH, et al. Genetic polymorphism of thiopurine S-methyltransferase in Argentina. Ann Clin Biochem. 2003;40:388–93. doi: 10.1258/000456303766477039. [DOI] [PubMed] [Google Scholar]
- Lennard L, Cartwright CS, Wade R, et al. Thiopurine methyltransferase genotype-phenotype discordance and thiopurine active metabolite formation in childhood acute lymphoblastic leukaemia. Br J Clin Pharmacol. 2013;76:125–36. doi: 10.1111/bcp.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E, Belaiche J. Optimizing treatment with thioguanine derivatives in inflammatory bowel disease. Best Pract Res Clin Gastroenterol. 2003;17:37–46. doi: 10.1053/bega.2002.0346. [DOI] [PubMed] [Google Scholar]
- Lu HF, Shih MC, Hsueh SC, et al. Molecular analysis of the thiopurine S-methyltransferase alleles in Bolivians and Tibetans. J Clin Pharm Ther. 2005;30:491–6. doi: 10.1111/j.1365-2710.2005.00640_1.x. [DOI] [PubMed] [Google Scholar]
- McLeod HL, Pritchard SC, Githang'a J, et al. Ethnic differences in thiopurine methyltransferase pharmacogenetics:evidence for allele specificity in Caucasian and Kenyan individuals. Pharmacogenetics. 1999;9:773–6. doi: 10.1097/00008571-199912000-00012. [DOI] [PubMed] [Google Scholar]
- Moini M, Ghaderi F, Sagheb MM, et al. The frequency and distribution of thiopurine S-methyltransferase alleles in south Iranian population. Mol Biol Rep. 2012;39:4581–7. doi: 10.1007/s11033-011-1248-6. [DOI] [PubMed] [Google Scholar]
- Nguyen CM, Mendes MA, Ma JD. Thiopurine methyltransferase (TPMT) genotyping to predict myelosuppression risk. PLoS Curr. 2011;3:Rrn1236. doi: 10.1371/currents.RRN1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira E, Quental S, Alves S, et al. Do the distribution patterns of polymorphisms at the thiopurine S-methyltransferase locus in sub-Saharan populations need revision?Hints from Cabinda and Mozambique. Eur J Clin Pharmacol. 2007;63:703–6. doi: 10.1007/s00228-007-0310-8. [DOI] [PubMed] [Google Scholar]
- Peregud-Pogorzelski J, Tetera-Rudnicka E, Kurzawski M, et al. Thiopurine S-methyltransferase (TPMT) polymorphisms in children with acute lymphoblastic leukemia, and the need for reduction or cessation of 6-mercaptopurine doses during maintenance therapy:the Polish multicenter analysis. Pediatr Blood Cancer. 2011;57:578–82. doi: 10.1002/pbc.23013. [DOI] [PubMed] [Google Scholar]
- Phan VH, Moore MM, McLachlan AJ, et al. Ethnic differences in drug metabolism and toxicity from chemotherapy. Expert Opin Drug Metab Toxicol. 2009;5:243–57. doi: 10.1517/17425250902800153. [DOI] [PubMed] [Google Scholar]
- Rossino R, Vincis C, Alves S, et al. Frequency of the thiopurine S-methyltransferase alleles in the ancient genetic population isolate of Sardinia. J Clin Pharm Ther. 2006;31:283–7. doi: 10.1111/j.1365-2710.2006.00736.x. [DOI] [PubMed] [Google Scholar]
- Sahasranaman S, Howard D, Roy S. Clinical pharmacology and pharmacogenetics of thiopurines. Eur J Clin Pharmacol. 2008;64:753–67. doi: 10.1007/s00228-008-0478-6. [DOI] [PubMed] [Google Scholar]
- Samochatova EV, Chupova NV, Rudneva A, et al. TPMT genetic variations in populations of the Russian Federation. Pediatr Blood Cancer. 2009;52:203–8. doi: 10.1002/pbc.21837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffeler E, Fischer C, Brockmeier D, et al. Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German-Caucasians and identification of novel TPMT variants. Pharmacogenetics. 2004;14:407–17. doi: 10.1097/01.fpc.0000114745.08559.db. [DOI] [PubMed] [Google Scholar]
- Schaeffeler E, Zanger UM, Eichelbaum M, et al. Highly multiplexed genotyping of thiopurine s-methyltransferase variants using MALD-TOF mass spectrometry:reliable genotyping in different ethnic groups. Clin Chem. 2008;54:1637–47. doi: 10.1373/clinchem.2008.103457. [DOI] [PubMed] [Google Scholar]
- Serpe L, Calvo PL, Muntoni E, et al. Thiopurine S-methyltransferase pharmacogenetics in a large-scale healthy Italian-Caucasian population:differences in enzyme activity. Pharmacogenomics. 2009;10:1753–65. doi: 10.2217/pgs.09.103. [DOI] [PubMed] [Google Scholar]
- Szumlanski C, Otterness D, Her C, et al. Thiopurine methyltransferase pharmacogenetics:human gene cloning and characterization of a common polymorphism. DNA Cell Biol. 1996;15:17–30. doi: 10.1089/dna.1996.15.17. [DOI] [PubMed] [Google Scholar]
- Taja-Chayeb L, Vidal-Millan S, Gutierrez O, et al. Thiopurine S-methyltransferase gene (TMPT) polymorphisms in a Mexican population of healthy individuals and leukemic patients. Med Oncol. 2008;25:56–62. doi: 10.1007/s12032-007-9002-6. [DOI] [PubMed] [Google Scholar]
- Tumer TB, Ulusoy G, Adali O, et al. The low frequency of defective TPMT alleles in Turkish population:a study on pediatric patients with acute lymphoblastic leukemia. Am J Hematol. 2007;82:906–10. doi: 10.1002/ajh.20947. [DOI] [PubMed] [Google Scholar]
- Wang L, Pelleymounter L, Weinshilboum R, et al. Very important pharmacogene summary:thiopurine S-methyltransferase. Pharmacogenet Genomics. 2010;20:401–5. doi: 10.1097/FPC.0b013e3283352860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics:insights, challenges and future directions. Oncogene. 2006;25:1629–38. doi: 10.1038/sj.onc.1209372. [DOI] [PubMed] [Google Scholar]
- Yates CR, Krynetski EY, Loennechen T, et al. Molecular diagnosis of thiopurine S-methyltransferase deficiency:genetic basis for azathioprine and mercaptopurine intolerance. Annals of inter med. 1997;126:608–14. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- Zeglam HB, Benhamer A, Aboud A, et al. Polymorphisms of the thiopurine S-methyltransferase gene among the Libyan population. Libyan J Med. 2015;10:27053. doi: 10.3402/ljm.v10.27053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JP, Guan YY, Wu JH, et al. Phenotyping and genotyping study of thiopurine S-methyltransferase in healthy Chinese children:a comparison of Han and Yao ethnic groups. Br J Clin Pharmacol. 2004;58:163–8. doi: 10.1111/j.1365-2125.2004.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JP, Zhou SF, Chen X, et al. Determination of intra-ethnic differences in the polymorphisms of thiopurine S-methyltransferase in Chinese. Clin Chim Acta. 2006;365:337–41. doi: 10.1016/j.cca.2005.09.005. [DOI] [PubMed] [Google Scholar]


