Abstract
Despite substantial advances in the treatment of various cancers, many patients still receive anti-cancer therapies that hardly eradicate tumor cells but inflict considerable side effects. To provide the best treatment regimen for an individual patient, a major goal in molecular oncology is to identify predictive markers for a personalized therapeutic strategy. Regarding novel targeted anti-cancer therapies, there are usually good markers available. Unfortunately, however, targeted therapies alone often result in rather short remissions and little cytotoxic effect on the cancer cells. Therefore, classical chemotherapy with frequent long remissions, cures, and a clear effect on cancer cell eradication remains a corner stone in current anti-cancer therapy. Reliable biomarkers which predict the response of tumors to classical chemotherapy are rare, in contrast to the situation for targeted therapy. For the bulk of cytotoxic therapeutic agents, including DNA-damaging drugs, drugs targeting microtubules or antimetabolites, there are still no reliable biomarkers used in the clinic to predict tumor response. To make progress in this direction, meticulous studies of classical chemotherapeutic drug action and resistance mechanisms are required. For this purpose, novel functional screening technologies have emerged as successful technologies to study chemotherapeutic drug response in a variety of models. They allow a systematic analysis of genetic contributions to a drug-responsive or −sensitive phenotype and facilitate a better understanding of the mode of action of these drugs. These functional genomic approaches are not only useful for the development of novel targeted anti-cancer drugs but may also guide the use of classical chemotherapeutic drugs by deciphering novel mechanisms influencing a tumor’s drug response. Moreover, due to the advances of 3D organoid cultures from patient tumors and in vivo screens in mice, these genetic screens can be applied using conditions that are more representative of the clinical setting. Patient-derived 3D organoid lines furthermore allow the characterization of the “essentialome”, the specific set of genes required for survival of these cells, of an individual tumor, which could be monitored over the course of treatment and help understanding how drug resistance evolves in clinical tumors. Thus, we expect that these functional screens will enable the discovery of novel cancer-specific vulnerabilities, and through clinical validation, move the field of predictive biomarkers forward. This review focuses on novel advanced techniques to decipher the interplay between genetic alterations and drug response.
Keywords: Functional genetic screens, Chemotherapy, CRISPR/Cas9, Haploid cells, Insertional mutagenesis, 3D organoids, Gene essentiality, DNA damage, Predictive markers
1. Introduction
Anti-cancer drug resistance is the major cause of death of cancer patients with disseminated tumors (Borst, 2012). In some patients intrinsic (or primary) drug resistance is already observed from the start (i.e. prior to chemotherapy) and tumors grow in the presence of chemotherapy (Holohan et al., 2013). Such intrinsic drug resistance can be a cancer-type specific or caused by individual cancer features (Gottesman, 2002). Frequently however, resistance arises in two steps. The tumor initially responds, but not all tumor cells are eradicated. From the residual disease the tumor regrows and eventually becomes resistant to all available chemotherapeutic drugs (Borst, 2012). We have recently reviewed various mechanisms that may cause minimal residual disease (Blatter and Rottenberg, 2015). Although residual disease may already contain selected drug-refractory tumor cells, it is also possible that the residual tumors are only transiently resistant due to cell cycle characteristics (Pajic et al., 2017). Then, drug resistance is acquired during the course of treatment (Housman et al., 2014). This secondary resistance is often due to (epi-)genetic alterations arising during the treatment that lead to, for instance, the activation of alternative signaling pathways, increased drug efflux, altered drug target availability, or rewiring of the DNA damage response (Holohan et al., 2013; Borst, 2012; Bouwman and Jonkers, 2012). To attenuate the development of drug resistance, combinational therapies of several drugs with different molecular mechanisms are frequently given to cancer patients (Al-Lazikani et al., 2012). Another approach is to re-sensitize resistant tumor cells by drugs targeting the resistance mechanism or the tumor microenvironment (De Henau et al., 2016; Callaghan et al., 2014). Unfortunately, we often lack knowledge about the mechanisms underlying resistance and therefore we usually lack a personalized strategy how to treat patients with (relapsing) tumors.
In the past decades, progress in the treatment of disseminated cancers has reduced cancer-related mortality (Kort et al., 2009). In addition to classical chemotherapy, also targeted anti-cancer drugs further improved cancer remission (Motzer et al., 2015; Zhou et al., 2011). Despite these advances, treatment failure due to drug resistance remains a substantial challenge in the clinical management of cancer. Treating a non-responsive tumor causes side effects without providing a benefit for the patient. Moreover, it incurs unnecessary costs and may even decrease the likelihood of success of subsequent treatments with other regimens (Siddiqui and Rajkumar, 2012).
To improve cancer therapy outcome, precision oncology is a promising strategy. Through the assessment of a tumor’s specific genetic or proteomic changes, i.e. its biomarkers (Mehta et al., 2010), an individualized best treatment regimen can be chosen. Prognostic gene expression signatures are clinically well established, because prognosis of tumor recurrence directly depends on the altered expression of a number of genes involved in tumor progression and metastasis (Reyal et al., 2008; Wirapati et al., 2008; Cardoso et al., 2016). Conversely, a tumor’s response to a particular treatment can fail due to the alteration of a single gene, such as the drug target or drug entry transporter (Borst and Wessels, 2010). Such alterations may not reliably be picked up by standard gene expression profiling. Thus, it is not surprising that only few predictive biomarkers are established, and even those remain imperfect in predicting therapy success. Currently, biomarkers are only available for targeted therapies, which block or stimulate specific pathways of tumor cells (Twomey et al., 2017) and usually yield good initial response with a modest effect on overall survival (Fojo and Parkinson, 2010). In contrast, classical cytotoxic chemotherapy interferes with all rapidly dividing cells, does not rely on oncogenic protein or pathway alterations, but often results in long-term remission and even cures some cancer types, and reduces cancer-related mortality. Unfortunately, not all patients benefit from the treatment and many eventually become resistant to all drugs available. Hence, there is a lack of clinically validated predictive biomarkers for classical chemotherapy.
Regarding targeted therapy, an early example of a predictive biomarker is HER2 expression status for trastuzumab treatment in metastatic breast cancer, an anti-cancer drug approved by the FDA in 1998. In combination with classical chemotherapy, trastuzumab efficiently decreases disease progression in HER2-amplified metastatic breast cancer (Cobleigh et al., 1999). In contrast, trastuzumab provides no benefit in breast cancer patients lacking HER2. Unfortunately, only about 30% of all HER2-positive breast cancer patients respond to trastuzumab-containing chemotherapy (De Palma and Hanahan, 2012). Thus, there are additional factors that influence therapy response, such as the intertumoral heterogeneity among a cancer (sub-)type, reflecting variations in molecular profiles of cancers in different patients. Additionally, the intratumoral heterogeneity complicates predictions of drug response (Ng et al., 2014). Molecular and genetic profiling of tumors has become cheaper and is often readily available. For mutations in specific genes, for instance BRAF, the effect on therapy response has been well characterized, so that sequencing of the corresponding genomic region will directly yield a predictive marker for therapy response. Unfortunately, the number of such well-defined biomarkers is limited, and to date only a small fraction of cancer patients directly benefit from established biomarkers. This is aggravated by the fact that not all patients bearing BRAF mutations do respond equally well to targeted BRAF inhibitors (Corcoran et al., 2015; Long et al., 2014; Prahallad et al., 2012). Thus, even such well-defined biomarkers are not sufficient, and additional characterization of the tumor is needed.
Several approaches have successfully identified novel molecular peculiarities which serve as predictive biomarkers. Hypothesis-driven approaches have, for instance, resulted in the establishment of BRCA1/2 mutational status in predicting a positive response upon PARP inhibitor treatment in breast and ovarian cancer (Farmer et al., 2005; Tutt et al., 2010; Bryant et al., 2005). Analyses of large, population-based clinical trials have also identified subgroups of responsive patients (Uryniak et al., 2011), e.g. leukemia patients with the Philadelphia chromosome responded better to imatinib treatment (Druker et al., 2001). Predictive markers based on clinical data have also been suggested for classical chemotherapeutics, including high HER2 or low tau expression as markers for paclitaxel sensitivity (Pusztai, 2007). Besides BRCA1/2 status, these markers have not entered the clinic, however, and still require additional validating clinical studies (Schork, 2015).
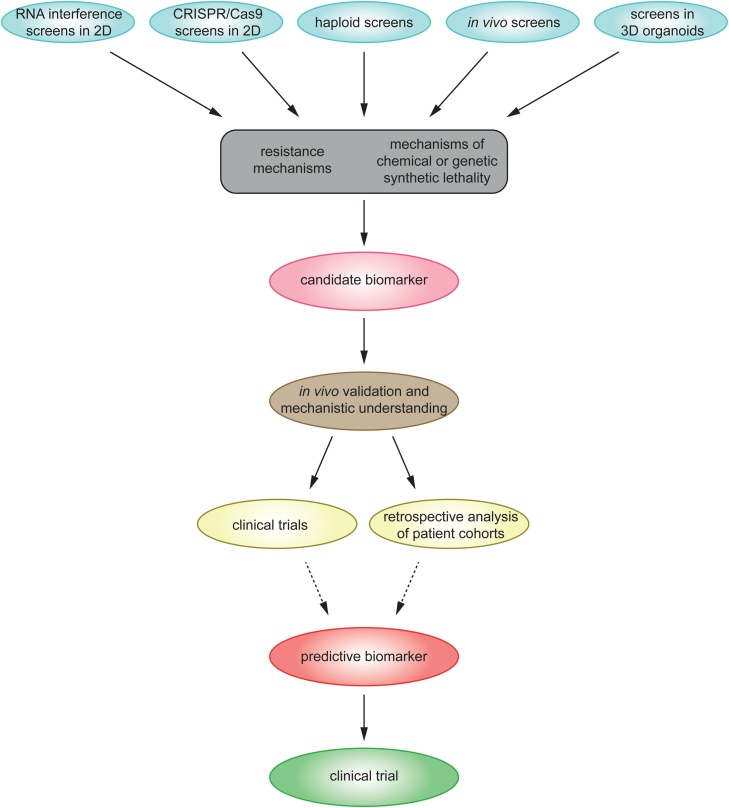
In recent years, advances in experimental genetic screening techniques have linked many genotypes to novel phenotypes in mammalian cells (Chen et al., 2015; Brockmann et al., 2017; Zhou et al., 2014b; Blomen et al., 2015; Wang et al., 2015b; Hart et al., 2015). Furthermore, genome-wide screens have broadened our understanding of molecular mechanisms responsible for therapy response (Ruiz et al., 2016; Berns et al., 2016; Planells-Cases et al., 2015; Wijdeven et al., 2015, for instance; and Table 1). Thus, these screens are valuable tools which can reveal novel mechanisms of resistance or hypersensitivity towards drugs, and facilitate a better understanding of drug response which might ultimately result in novel predictive biomarkers (Fig. 1). While most targeted anti-cancer therapeutics exploit gain-of-function alterations, e.g. in terms of oncogene addiction (Pagliarini et al., 2015), not all tumors bear targetable gain-of-function mutations. Inactivation of tumor suppressor genes is frequent, and cannot be directly targeted with a drug. However, as shown by the example of PARP inhibitor treatment in BRCA1/2 mutated tumors, loss of a tumor suppressor can offer a treatment option with low side effects for healthy tissue. The study of synthetic lethality and context-dependent gene essentiality has been challenging in mammalian cells and was for long time limited to few model organisms. With the development of CRISPR/Cas9 genome editing and insertional mutagenesis in haploid human cells, it is now possible to efficiently study genetic interactions as well as the functional consequence of genetic mutations and possibly reveal new predictive biomarkers by linking drug-responsive phenotypes to genotypes.
Table 1.
Examples of screens performed to identify unknown factors of drug response, to suggest potential therapeutic strategies or to exploit novel screening concepts.
| Selection | Screening method | Model | Identified genes – proof of concept | Identified genes – novel findings | Proposed mechanism | Remarks | Reference |
|---|---|---|---|---|---|---|---|
| Trastuzumab (HER2-targeting antibody) | shRNA screen (7,914 genes), positive selection | HER2-amplified breast cancer cell line BT474 | PTEN | Loss of PTEN activates PI3K/AKT signaling | PI3K pathway activation as predictive marker | Berns et al. (2007) | |
| Vemurafenib (PLX4032, BRAF inhibitor) | Kinome shRNA screen (535 genes), positive selection | Colorectal cancer cell line WiDr | EGFR | BRAF(V600E) inhibition activates EGFR and stimulates proliferation (feedback activation) | Melanoma cells express low levels of EGFR and are thus sensitive to BRAF inhibition; BRAF-mutant colon cancer might benefit from combination of BRAF and EGFR inhibitors | Prahallad et al. (2012) | |
| Trastuzumab (HER2-targeting antibody) | shRNA screen (7,914 genes), positive selection | HER2-amplified breast cancer cell lines BT474, SKBR3 and HCC1954 | PTEN | ARID1A | ARID1A loss activates ANXA1 which in turn activates AKT and causes resistance | High ANXA1 expression suggested as predictive marker | Berns et al. (2016) |
| Vemurafenib (BRAF inhibitor) | Genome-wide CRISPR/Cas9 screen (18,080 genes), positive selection | BRAF(V600E)-mutated melanoma cell line A375 | NF1, MED12 | NF2, CUL3, TADA1, TADA2B | TADA1 and TADA2 B (member of STAGA complex) recruit MED12 to c-myc to activate proliferation; MED12 activates TGF-βR signaling and MEK/ERK | Shalem et al. (2014) | |
| Cytosine arabinoside (antimetabolite) | Genome-wide CRISPR/Cas9 screen (18,080 genes), positive selection | Acute myeloid leukemia cell line U937 | DCK | SLC29A | Associated with nucleotide salvage pathway and required for the uptake and activation of Ara-C | Kurata et al. (2016) | |
| 6-thioguanine (antimetabolite) | Genome-wide CRISPR/Cas9 screen (19,150 genes), positive selection | Male mouse ES (JM8) cells | Mismatch repair genes (Mlh1, Msh2, Msh6, Pms2) | Hprt, GM15293, Letmd1, Olfr815, Prkg1, Tmem8c | Unknown candidate genes did not validate in subsequent in vitro experiments | Koike-Yusa et al. (2014) | |
| 6-thioguanine (antimetabolite) | Genome-wide CRISPR/Cas9 screen (7114 genes), positive selection | Human near-haploid chronic myeloid leukemia cell line KBM7 | Mismatch repair genes (MSH2, MSH6, MLH1, PMS2) | Wang et al. (2014b) | |||
| Etoposide (DNA topoisomerase II inhibitor) | Genome-wide CRISPR/Cas9 screen (7114 genes), positive selection | Human pseudo-diploid leukemic HL60 and near-haploid KBM7 cell lines | TOP2A | CDK6 | G1-cyclin dependent kinase involved in etoposide cytotoxicity | Wang et al. (2014b) | |
| ATR inhibitor | Genome-wide CRISPR/Cas9 screen (19,150 genes), positive selection | Mouse ES cells (KH2) | CDC25A | CDC25A prevents cells from premature entry into mitosis | CDC25A levels could serve as criterion for patients more likely to respond; rationale to combine ATR and WEE1 inhibitor treatment | Ruiz et al. (2016) | |
| Phenotypic selection | Genome-wide (18,543 human and 18,986 mouse genes) and focused (132 Ras-associated genes) CRISPR/Cas9 screen, negative selection | 12 acute myeloid leukemia cell lines and NRAS-engineered mouse CGN Ba/F3 cell line | Several genes involved in Ras maturation or downstream of MAPK signaling pathway | Cancers driven by oncogenic Ras require Rac/PAK signaling to activate MAPK signaling | PAK inhibition as potential synthetic lethal therapeutic strategy in Ras-driven cancers | Wang et al. (2017) | |
| Phenotypic selection | Genome-wide CRISPR/Cas9 screen (18,080 genes), negative selection | Glioblastoma stem-like and neural stem/progenitor cell lines | PKMYT1 | PKMYT1, essential to inhibit cyclin B-CDK1 activity, is lost in glioblastoma | PKMYT1 inhibition as potential synthetic lethal therapeutic strategy in glioblastoma | Toledo et al. (2015) | |
| Phenotypic selection | Genome-wide CRISPR/Cas9 screen (17,232 genes), negative selection | RNF43-mutant pancreatic ductal adenocarcinoma cell line | Components of Wnt pathway | FZD5 | FZD5 encodes the main receptor for Wnt-β-catenin signaling in this context | FDZ5 inhibiton as a potential synthetic lethal therapeutic strategy in RNF43-mutated pancreatic cancer | Steinhart et al. (2017) |
| Phenotypic selection | Genome-wide (18,360 genes) and mini-pool (300 genes) CRISPR/Cas9 screen, quantitiative protein measurement of SQSTM1 modulators | Human neuroglioma H4 cell line | MTOR complex 1 and canonical macroautophagy components | Ufmylation components | Ufmylation induces SQSTM1 expression | Dejesus et al. (2016) | |
| Lipopolysaccharide | Genome-wide CRISPR/Cas9 screen (21,786 genes), quantitative measurement of Tnf expression | Mouse bone-marrow derived dendritic cells | Tlr4, Myd88 (signal high), Zfp36 (signal low) | Components of OST complex, ER translocation pathway, PAF complex | Parnas et al. (2015) | ||
| Doxorubicin (DNA topoisomerase II inhibitor) | Viral gene-trap haploid screen, positive selection | Human haploid cell line HAP1 | ABCB1, Keap1 | SWI/SNF subunits, C9orf82, Eif4a1 | SWI/SNF regulates Topoisomerase II activity, C9orf82 negatively regulates DNA repair | Patients with low SWI/SNF expression should not be treated with doxorubicin but rather aclarubicin or topotecan | Wijdeven et al. (2015) |
| Carboplatin (platinum drug) | Viral gene-trap haploid screen, positive selection | Human near-haploid chronic myeloid leukemia cell line KBM7 | Components of volume-regulated anion channel (LRRC8D/LRRC8A) | 50% of cellular platinum drug uptake mediated via LRRC8A/D channels | Downregulation of LRRC8 subunits could have an impact on platinum resistance | Planells-Cases et al. (2015) | |
| 6-thioguanine (antimetabolite) | piggyBac transposon haploid screen, positive selection | Mouse haploid ES cells | Mismatch repair genes (Msh2, Msh6, Mlh1) | Validation of loss-of-function screen | Pettitt et al. (2013) | ||
| Olaparib (PARP inhibitor) | piggyBac transposon haploid screen, positive selection | Mouse haploid ES cells | Parp1 | Parp1 is a drug target and required for drug toxicity | Inhibited PAPR1 enzyme forms a toxic DNA lesion | Pettitt et al. (2013) | |
| 6-thioguanine (antimetabolite) | Viral gene-trap haploid screen, positive selection | Human near-haploid chronic myeloid leukemia cell line KBM7 | HPRT | Enzyme converting 6-thioguanine to a toxic metabolite | Carette et al. (2009) | ||
| Imatinib (tyrosine-kinase inhibitor) | Viral gene-trap haploid screen, positive selection | Human near-haploid chronic myeloid leukemia cell line KBM7 | NF1, PTPN1 | PTPN12 | Tyrosine phosphatase negatively regulates c-abl | Carette et al. (2009) | |
| Formaldehyde | Viral gene-trap haploid screen, positive selection | Human near-haploid chronic myeloid leukemia cell line KBM7 | 12 candidate genes | 6 out of 12 candidates validated | Shen et al. (2016) | ||
| Imatinib (tyrosine-kinase inhibitor) | Viral gene-trap haploid screen, positive selection | Human near-haploid chronic myeloid leukemia cell line KBM7 | CASP10, CUX1, NF1, LYRM9, ZPBP, CEBPG | Only LYRM9 validated; only NF1 was also identified by Carette et al. (2009) | Shen et al. (2016) | ||
| MK-1775 (Wee1 inhibitor) | Viral gene-trap haploid screen, positive selection | Human near-haploid chronic myeloid leukemia cell line KBM7 | SKP2,CUL1, CDK2 (among others) | Inactivation of S-phase can overcome Wee1 inhibitor resistance | Activity of DNA replication machinery could serve as selection criterion for Wee1 inhibitor treatment | Heijink et al. (2015) | |
| Talazoparib (PARP inhibitor) | piggyBac transposon haploid screen, negative selection | Brca2-mutated mouse haploid ES cells (H129.2) | Ewsr1 | Pettitt et al. (2017) | |||
| Phenotypic selection | Viral gene-trap haploid screen, quantitative protein measurement of AKT signaling | Human haploid cell line HAP1 | KCTD5; GNB1 and other genes encoding Gβγ subunits | KCTD5 negatively regulates GPCR signaling by triggering proteolysis of dissociated Gβγ subunits | Brockmann et al. (2017) | ||
| Phenotypic selection | Viral gene-trap haploid screen, quantitative protein measurement of WNT signaling | Human haploid cell line HAP1 with 7TG-WNT reporter | Several known regulators | Genes linked to WNT receptor complex, CTNNB1 destruction complex and others | Other processes than CTNNB1 protein levels, e.g. truncation of domains, might regulate WNT signaling | Lebensohn et al. (2016) | |
| Interferon-γ | Viral gene-trap haploid screen, quantitative protein measurement of PD-L1 abundance | Human haploid cell line HAP1 | IFNγR-pathway, IRF1, CD274 | CMTM6 | CMTM6/4 enhances PD-L1-mediated T-cell inhibition, stabilizes PD-L1 protein level | Novel potential target for immune-suppressive cancer therapy | Mezzadra et al. (2017) |
| Phenotypic selection | Comparison of genome-wide CRISPR/Cas9 (19,050 genes) and viral gene-trap haploid screen, quantitative protein measurement of ER-associated degradation of MHC class I molecules | Human near-haploid chronic myeloid leukemia cell line KBM7 with MHC-I-ERAD reporter | TXNDC11 | TXNDC11 encodes an EDEM2/3-associated disulphide reductase | Timms et al. (2016) | ||
| Phenotypic selection | Genome-wide CRISPR/Cas9 screen (20,611 genes), positive selection | Mouse non-small-cell lung cancer cell line transplanted into immunocompromised mice | Several candidate genes enriched in late primary tumors, high overlap of candidate genes in metastases | Enrichment of mutations in anti-apoptotic or other tumor suppressive pathways | Chen et al. (2015) | ||
| Monoclonal PD-1 antibody | Focused CRISPR/Cas9 screen (2,368 genes), positive selection | Mouse B16 melanoma cell line | CD47 | Ptpn2, and several genes involved in four distinct biological processes | Loss of Ptpn2 sensitize tumors to immunotherapy through increased antigen presentation and T-cell stimulation | Inhibition of Ptpn2 as a therapeutic strategy to increase the effect of anti-PD-1 immunotherapy | Manguso et al. (2017) |
| Phenotypic selection | Mini CRISPR/Cas9 screen (10 genes), positive selection | 3D mucociliary epithelial organoids from primary human basal cells | GRHL2 | GRHL2 plays a key role in apical-basal cell polarity and epithelial morphogenesis | Gao et al. (2015) |
Fig. 1.
Exemplary workflow to discover novel predictive biomarkers based on forward genetic screens.
Although some novel anti-cancer drugs have been successful and have yielded improvements for cancer patients, they remain imperfect (Fojo and Parkinson, 2010; Groenendijk and Bernards, 2014), and have also become a financial burden for the health system (Kantarjian et al., 2013; Aggarwal, 2010; Prasad and Mailankody, 2017; Fojo and Parkinson, 2010). During the course of treatment, most patients sooner or later also receive classical chemotherapy including platinum drugs, topoisomerase inhibitors, microtubule-targeting agents or antimetabolites as part of standard care (Gonen and Assaraf, 2012; Giovannetti et al., 2017). Their clinical use is based on empirical experience. However, these drugs are relatively cheap, effective and widely used. If clinical oncologists could be supported in their choice of classical chemotherapy based on molecular characteristics of a tumor, the therapeutic benefit of a standard treatment may increase and drugs to which the tumor is unlikely to respond would be avoided. To improve the proper selection of the treatment of choice and to expand our repertoire of drug response predictions, one needs to identify more molecular peculiarities of tumors which impact therapy response.
This review therefore elaborates on genome-wide screening techniques in mammalian cells with special emphasis on the response against classical cytotoxic drugs.
2. Forward genetic screens to improve our understanding of drug resistance and synthetic lethality
Using forward genetic screens, genetic mutations can efficiently be linked to phenotypes of interest and identify the crucial genes for the selected phenotype. The success of a screen depends on the choice of the best model as well as efficient gene suppression. Genome-wide screens were made possible in mammalian cells with the discovery of RNA interference, which introduces small specific RNA molecules into cells and targets the corresponding host mRNA for degradation (Carthew and Sontheimer, 2009; Brummelkamp et al., 2002). RNA interference screens have led to the discovery of novel predictive biomarkers for chemotherapy response which could be translated into the clinic. For instance, large-scale RNA interference screens identified loss of PTEN as a determinant of trastuzumab resistance in breast cancer (Berns et al., 2007) and feedback activation of EGFR as a resistance mechanism in BRAF-mutated colorectal cancer (Prahallad et al., 2012). However, for many applications RNA interference screens are imperfect, because this technique usually fails to inactivate gene function completely and remaining gene activity may mask the phenotype (Echeverri et al., 2006; Booker et al., 2011; Kaelin, 2012), although in some cases, such as the knock-down effect of essential genes on drug response, an incomplete gene inactivation might be required to detect an effect of the knock-down on drug response. Nevertheless, off-target effects complicate RNA interference screens through a high proportion of false positive hits. As RNA interference screens for biomarker discovery have been reviewed elsewhere (Mullenders and Bernards, 2009; Iorns et al., 2007; Swanton et al., 2008), we will focus here on more recent developments in screening technologies (Table 2 and Fig. 2).
Table 2.
Comparison of recent functional screening technologies.
| CRISPR/Cas9 in 2D cell lines | |
|---|---|
| Advantages | Disadvantages |
|
|
|
|
|
|
|
|
|
|
|
|
| Haploid insertional mutagenesis | |
|---|---|
| Advantages | Disadvantages |
|
|
|
|
|
|
|
|
| in vivo screens | |
|---|---|
| Advantages | Disadvantages |
|
|
|
|
|
|
|
|
|
|
| 3D cancer organoids | |
|---|---|
| Advantages | Disadvantages |
|
|
|
|
|
|
|
|
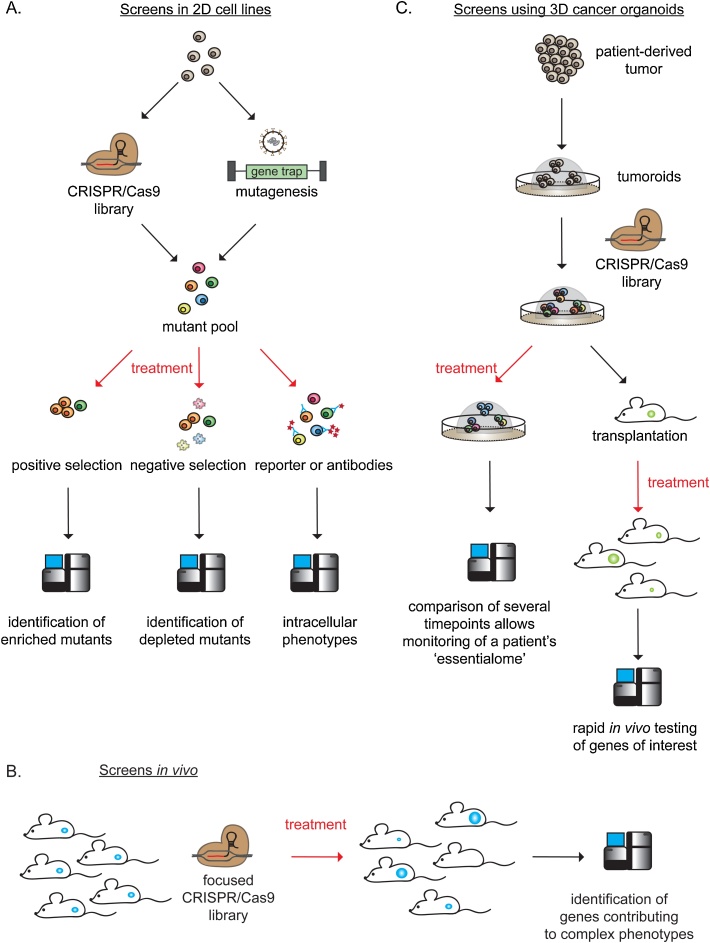
Fig. 2.
Simplified layouts of the screening technologies presented in the current review. A.: Screens in 2D cell lines using CRISPR/Cas9 or insertional mutagenesis are useful to study genetic contributions to a phenotype of interest upon drug treatment. 2D cell lines can be modified with either CRISPR/Cas9 or insertional mutagenesis before drug exposure. Depending on the research question, the screens can be analyzed for enrichment (positive selection, potential drug resistance genes) or depletion of mutants (negative selection, potential drug hypersensitivity genes) or intracellular phenotypes by employing antibody- or reporter-based assays. B.: Mice bearing CRISPR/Cas9-modified tumors can be treated and analyzed efficiently to study complex phenotypes such as metastasis formation. C.: Patient-derived organoids can be modified with CRSIPR/Cas9 and used for both rapid in vivo testing of a gene panel of interest and monitoring of the tumor’s ‘essentialome’.
2.1. CRISPR/Cas9 screens in 2D cell lines
The discovery of the bacterial Cas9 endonuclease has revolutionized genome engineering. A 20-basepairs site-specific single guide RNA (sgRNA) directs the endonuclease to its corresponding target site and introduces a DNA break. This break stimulates repair by non-homologous end-joining (NHEJ) or homology-directed repair (HDR) by frequently introducing frameshifts by indel mutations, resulting in a premature stop codon or nonsense-mediated decay of the transcript. Consequently, gene inactivation is achieved at the genomic level by CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 by creating a gene knock-out which, for many applications, outperforms gene knock-down by RNA interference (Evers et al., 2016; Hart et al., 2015; Wang et al., 2015b). CRISPR/Cas9 also allows the study of inactivation of non-transcribed elements (Wang et al., 2014b).
2.1.1. Choice of cellular model
The universal applicability of CRISPR/Cas9 screens in a broad repertoire of models (Wang et al., 2015b) makes this technique a powerful tool to study cancer-type-specific genetic requirements. The most outstanding advantage of CRISPR/Cas9 is that genetic mutations can be introduced into versatile cellular models. Some mechanisms of drug resistance or hypersensitivity might only be present in a certain lineage or cancer type. Melanoma patients bearing the valine-to-glutamate change at residue 600 (V600E) of BRAF, for instance, respond well to a BRAF inhibitor in combination with a mitogen/extracellular signal-regulated kinase (MEK) inhibitor (Long et al., 2014), whereas the same treatment in colorectal patients bearing the same BRAF mutation is less efficient (Corcoran et al., 2015). It may therefore be necessary to study specific genetic requirements in several models to identify novel genetic dependencies originating from pathway rewiring following genetic perturbations. Exploiting such lineage-specific vulnerabilities could identify targets for use in personalized medicine (Muller et al., 2012).
Despite this major advantage, the large genetic variation between tumor cell lines may also hinder the successful identification of genetic contributions to a phenotype. The efficacy of genetic modification of CRISPR/Cas9 depends on the number of target loci in the genome. Whereas most human cells are diploid, cancer cell lines often have increased numbers of chromosomes or individual gene amplifications due to genomic instability (Stratton et al., 2009). To inactivate genes in HeLa cells, for instance, which have up to five copies of a chromosome (Landry et al., 2013), Cas9 needs to cleave the locus with higher efficiency compared to a cell line with a diploid gene set. Screens performed in diploid cells will therefore likely yield more robust data with a higher signal-to-noise ratio compared to polyploid cells. Furthermore, cells adapt to genetic mutations with secondary genetic changes or other forms of compensation (Teng et al., 2013). This needs to be particularly considered when comparing studies in long-term cultured isogeneic cells (Housden et al., 2017). Due to the individual variation of genes affecting their genetic interactions and fitness requirements, every cell line will have a different “essentialome”. Combining several screens from multiple cell lines can therefore identify the “core essentialome” of a lineage, while at the same time some information will be missed as a result of the variation in genetic context between cell lines. Hence, the properties of the chosen cell line directly impact screening results.
Moreover, the activity of DNA repair pathways in a cell line influences mutation efficiency by CRISPR/Cas9. The introduced DNA break can be repaired by HDR, which uses a homologous DNA template such as a sister chromatid. HDR is usually error-free, resulting in an unchanged genomic locus. The success of genetic inactivation using CRISPR/Cas9 therefore depends on the error-prone NHEJ repair pathway. The DNA cleavage site gets trimmed before re-ligation of the two DNA ends. This causes insertions or deletions of base pairs, resulting in a frameshift. If both HDR and NHEJ are active in a chosen cell line, one can expect a lower probability for complete gene inactivation in every locus (Miles et al., 2016). However, in cell lines lacking a functional HDR, CRISPR/Cas9-mediated gene knock-out has a high success rate (Miles et al., 2016).
2.1.2. Impact of the sgRNA library on screening success
To date, three major classes of CRISPR/Cas9-mediated genomic modifiers are available. The most common CRISPR libraries are knock-out libraries, but gene repression (CRISPRi) and activation (CRISPRa) libraries are also employed for specific research questions (Lopes et al., 2016; Miles et al., 2016; Gilbert et al., 2014; Dominguez et al., 2016). Libraries can either be genome-wide, targeting for example about 20,000 genes (Sanjana et al., 2014), or specifically designed to study one particular pathway or a selection of genes of interest (Zhou et al., 2014b). For instance, within a genetic region of interest, CRISPR/Cas9 tiling screens are able to dissect which genetic segments encode functional domains relevant for a phenotype (Korkmaz et al., 2016). Recent developments of the CRISPR system for RNA targeting or base editing will further broaden the investigable research fields (Cox et al., 2017; Gaudelli et al., 2017).
The selection of the individual sgRNAs in a library directly impacts screening results. For loss-of-function screens, sgRNAs should preferably target an early, constitutively expressed exon of a gene of interest. Ideally, this leads to nonfunctional transcripts of all gene variants with no off-target activity on other genes. Most commercially available libraries contain 5–10 sgRNAs per gene designed to achieve high cleavage efficacy and low off-target activity. However, these libraries do not take potential genomic pleomorphisms of different cell lines into account, and for some genes, shorter sequence variants might not be targeted and retain some gene functionality.
Off-target effects arise from partial complementarity of the sgRNA target site with additional unintended target sites. Off-target effects can be reduced by the use of improved Cas9 nuclease variants, e.g. by a decreased binding efficacy and increased site specificity, by the use of alternative endonucleases or improved bioinformatics algorithms to design specific sgRNAs (Kleinstiver et al., 2016; Slaymaker et al., 2016; Kim et al., 2016; Zetsche et al., 2015; Doench et al., 2014). This, in turn, increases the need for a perfect alignment of the sgRNA sequence with the target site. A single nucleotide mismatch as a cell line-specific genetic variation or the introduction of a terminal G during sgRNA design could by itself prevent sgRNA alignment with the target site. Moreover, optimized PAM sequence variants may also improve site specificity (Kleinstiver et al., 2015).
Another crucial step in CRISPR/Cas9 screens is the efficacy of library transduction in a cell line. Typically, sgRNA libraries are transduced at a MOI of 0.4–0.6 to ensure that every cell contains only a single sgRNA (Shalem et al., 2014; Wang et al., 2014b). For successful screens, a single copy of each sgRNA needs to cleave both (or even more) copies of the target locus (Wang et al., 2014b). If more than one sgRNA is introduced into a single cell (as usually achieved through direct transfection), it is challenging to determine which sgRNA caused the phenotype due to the relatively low number of distinct sgRNAs. In contrast, less than one sgRNA per cell can lead to insufficient library coverage at the end of a screen.
Depending on the plasmid system used to deliver the library, a stable expression of both sgRNA as well as Cas9 increases the genomic cleavage over time (Wang et al., 2014b). This may be on purpose, but it might also be problematic when screens are performed over a long time period, as not only on-target sites but also off-target sites will be cleaved repeatedly. DNA double strand breaks also result in genotoxic stress, causing a non-specific anti-proliferative effect which limits the use of CRISPR/Cas9 for highly amplified genomic regions (Aguirre et al., 2016; Munoz et al., 2016; Housden et al., 2017).
2.1.3. Data analysis
Owing to the popularity of CRISPR/Cas9 screens, a broad spectrum of resources for data analysis is available. To analyze a screen, sgRNA sequences are amplified by PCR before deep sequencing. The library sequencing reads may subsequently be analyzed using publically available algorithms, including HitSelect (Diaz et al., 2015) or MaGeCK (Li et al., 2014b). Although they require some programming experience, these algorithms are widely used, relatively user-friendly and provide a certain consistency in the analysis of many CRISPR/Cas9 screens.
When analyzing CRISPR/Cas9 screens, it is crucial to remember that sgRNA copy abundance is counted as an indirect measurement of mutations. Not every sgRNA necessarily creates a frameshift mutation, since the DNA break can also be repaired error-free by HDR or with an in-frame mutation retaining functionality of the transcript. When a sgRNA introduces both frame-shift and in-frame mutations, resulting in a mixed phenotype of this sgRNA, it is possible that this sgRNA will not score as being significant in the analysis (Shalem et al., 2015).
2.1.4. Applications to study drug response
CRISPR/Cas9 screening technology has enabled a systematic analysis of gene function in mammalian cells in terms of both positive (enrichment of mutants) and negative selection (depletion of mutants) (Wang et al., 2014b; Shalem et al., 2014; Zhou et al., 2014b; Wang et al., 2015b). Already at an early stage of development of the CRISPR/Cas9 technology, it has been shown that multiplexing with several sgRNAs can be achieved in vitro as well as in vivo (Horii et al., 2013; Wang et al., 2013; Cong et al., 2013; Mali et al., 2013).
In general, positive selection screens are less prone to alterations in library representation than negative selection screens which require a quantitative evaluation of sgRNA abundance. A good coverage and few false positives made CRISPR/Cas9 screens a valuable tool to study resistance phenotypes. In terms of anti-cancer drug resistance, for instance, genes were identified whose loss-of-function-mutations cause resistance to vemurafenib, cytosine arabinoside, 6-thioguanine and DNA topoisomerase II inhibitors (Shalem et al., 2014; Kurata et al., 2016; Koike-Yusa et al., 2014; Wang et al., 2014b). As an example, Shalem et al. identified two known mediators of resistance against the BRAF inhibitor vemurafenib, NF1 and MED12 (Huang et al., 2012; Whittaker et al., 2013) in a genome-wide CRISPR/Cas9 library screen targeting 18,080 genes with 3–4 sgRNAs/gene (Shalem et al., 2014). Additionally, the authors found novel genes which have not been linked to BRAF inhibitor response so far, and validated their inactivation as resistance factors towards vemurafenib in vitro. Among those genes were members of the STAGA complex which recruit Mediator complex proteins including MED12, which negatively regulates TGF-β, to c-myc activating cell proliferation (Liu et al., 2008). Thus, CRISPR/Cas9 screens are able to identify novel mechanisms of anti-cancer drug resistance.
For the analysis of negative selection screens, the abundance of all sgRNAs needs to be quantified to identify the sgRNAs that were depleted from the population. As a consequence, gene knock-out efficacy needs to be close to 100% because every remaining non-modified cell or in-frame modification may mask the phenotype. These screens are less robust compared to positive selection screens and thus depend on more extensive statistical analysis, which in turn could be complicated by poor sgRNA-mediated cleavage efficacy. Despite these technical hurdles, CRISPR/Cas9 screens have successfully identified essential genes in mammalian cells (Wang et al., 2015b; Hart et al., 2015), as well as synthetic lethal genetic interactions which could potentially serve as targets for cancer therapy (Toledo et al., 2015; Steinhart et al., 2017). For instance, a study focusing on synthetic lethal interactions with oncogenic Ras identified critical regulators of the MAPK pathway in acute myeloid leukemia cell lines (Wang et al., 2017).
Moreover, CRISPR/Cas9 screens have proven useful to study intracellular phenotypes by employing reporter- or antibody-labelled quantitative protein measurements (Dejesus et al., 2016; Parnas et al., 2015). This type of screen compares cell populations with high and low signal measurements, and couples genes to an intracellular phenotype which does not necessarily result in altered cell viability. Studying such intracellular phenotypes broadens the understanding of key biological processes, and may identify crucial genetic dependencies of a phenotype of interest.
2.2. Haploid insertional mutagenesis screens
Insertional mutagenesis screens are a powerful alternative to CRISPR/Cas9 screens. They provide the possibility of applying principles of classical genetics in mammalian cells to uncover fundamental biological processes in a highly comparative manner. In particular, recessive genetic screens in haploid yeast have substantially contributed to gene discovery and our understanding of development, basic physiology, and various diseases (Giaever and Nislow, 2014). It was therefore a great achievement of Thijn Brummelkamp and coworkers to establish insertional mutagenesis screens in haploid human cell lines and thereby increase the power of insertional mutagenesis in a mammalian system (Carette et al., 2009).
2.2.1. Haploid cell lines
Insertional mutagenesis screens crucially depend on haploid (or near-haploid) cell lines, because no second copy of the gene exists which could phenotypically mask the effect of gene inactivation. Thus, genetic manipulation is highly efficient in haploid cells. To date, only few haploid cell lines exist. The KBM7 cell line, derived from a chronic myeloid leukemia patient, is haploid except for chromosome 8 (Kotecki et al., 1999). Its non-hematopoietic derivate HAP1 is haploid for all chromosomes, except of a duplicated 30-megabase fragment of Chromosome 15 fused to Chromosome 19, which was excised to obtain the fully haploid eHAP cell line (Essletzbichler et al., 2014). With the isolation of haploid human, murine, rat and monkey embryonic stem cells, a broader repertoire of haploid cell lines is currently being established, also by further differentiating these cells into several lineages (Elling et al., 2011; Leeb and Wutz, 2011; Sagi et al., 2016; Yang et al., 2013; Li et al., 2014a).
Compared to CRISPR/Cas9 screens, the restriction to a few haploid cell lines limits the use of haploid screens in the study of lineage-specific biology. However, this limitation simultaneously provides an opportunity, exemplified by the achievements of yeast genetics where a well-characterized single-model organism unveiled numerous insights into diverse biological processes (Giaever and Nislow, 2014; Boone et al., 2007). By introducing genetic alterations into a clearly defined, controlled genetic background, effects of gene inactivation can be studied and compared with precision, facilitating the investigation of complex genetic effects and interactions (Costanzo et al., 2016; Brockmann et al., 2017). Subsequently, findings from haploid screens may be validated in a variety of cell lines to address lineage-specific variations.
2.2.2. Insertional mutagenesis
In contrast to CRISPR/Cas9 screens which depend on a sgRNA library, mutations are usually achieved by random mutagenesis in haploid screens. The choice of the random mutagenesis technique is commonly determined by the ease of retrieving genetic mutation sites for analysis from a pool of cells, with transposon- or retroviral gene-trapping being the most frequent mutagenesis strategies to date. Mutagenesis performed by gene-trapping is highly efficient and delivers a molecular tag to identify mutations in the genome by integrating an exogenous viral DNA section in the host genome which marks the mutation site (Carette et al., 2009). The gene-trap cassette introduces a splice acceptor site, followed by a polyadenylation signal, which prematurely terminates gene splicing and translation and thereby creates mutants resembling knock-outs (Carette et al., 2009). Transposon-mediated gene-trap mutagenesis is achieved by co-transfecting a gene-trap vector with a plasmid for transposase expression. The transposase stimulates insertion of the gene-trap in the genome, and by varying transposase expression, the gene-trap insertion frequency can be influenced (Mates et al., 2009; Pettitt et al., 2015).
In theory, gene-trapping mutation events occur all over the genome and this technique therefore provides a very high coverage. However, the gene-trap integration is not completely random, as preferred viral or transposon integration sites exist (Lee et al., 2007; Wang et al., 2014b; Carette et al., 2009; Blomen et al., 2015). Nevertheless, a genome-wide distribution of integration sites, for example integrations in more than 98% of expressed genes by retroviral gene trapping (Carette et al., 2011a), is achieved, and improved vectors can reduce integration bias (Schnuetgen et al., 2008).
Compared to the use of sgRNA libraries which employ a guiding RNA strand defining the target site and directly depend on sgRNA representation as well as cleavage success and error-prone DNA repair, insertional mutagenesis creates individual mutations at a higher frequency and efficiency (Blomen et al., 2015). The phenotypic effect of a gene is determined indirectly in CRISPR/Cas9 screens, usually by determining the abundance of DNA encoding the sgRNAs. In contrast, sensitive amplification of gene-trap insertion sites allows a direct count of hundreds of independent mutations in a gene, achieving a high resolution of the observed phenotype (Elling and Penninger, 2014).
However, similar to the need of a sgRNA disrupting all gene variants simultaneously to abolish gene function, alternative splicing as well as gene-trap integrations near the 3′ end of a gene can also maintain (partial) gene activity (Blomen et al., 2015).
2.2.3. Data analysis
Deep sequencing following PCR amplification retrieves millions of unique gene-trap integration sites which are mapped to the genome, allowing subsequent direct counting of individual knock-outs that each has contributed to the phenotype of interest. The large number of individual mutants permits powerful statistical analyses. This enables an improved distinction between hits and background noise compared to other screening methods (Elling and Penninger, 2014). Due to the high efficacy of gene disruption and the absence of second gene copies, haploid cells further improve the signal to noise ratio compared to screens performed in diploid cells, enabling the identification of subtle fitness defects or advantages (Wang et al., 2014b; Blomen et al., 2015).
To analyze insertional mutagenesis screens, basic statistical tests such as Fisher's exact and/or binomial tests are usually employed. However, for complex research questions, such as the study of a cell line’s essentialome, more complex algorithms are used (Blomen et al., 2015). Recently, bioinformatics pipelines have been proposed for the analysis of gene-trap insertional mutagenesis screens (Yu and Ciaudo, 2017; Mayor-Ruiz et al., 2017).
2.2.4. Applications to study drug response
Initially, haploid insertional mutagenesis screens were predominantly employed to study resistance factors of host cells towards pathogens (Carette et al., 2009; Carette et al., 2011b; Guimaraes et al., 2011; Jae et al., 2013). Furthermore, haploid screens yielded novel insight into resistance against chemical compounds including chemotherapeutic agents like platinum drugs, topoisomerase II, PARP, and other chemical inhibitors (Wijdeven et al., 2015; Planells-Cases et al., 2015; Pettitt et al., 2013; Shen et al., 2016; Heijink et al., 2015). Regarding platinum drugs, it has been unclear which transporters are responsible for reduced drug uptake that may explain clinical drug resistance (Borst et al., 2008). Using a haploid screen, we identified volume-regulated anion channels composed of LRRC8A and D as a cellular uptake mechanism of cisplatin and carboplatin (Planells-Cases et al., 2015), providing a new lead to understand clinical resistance. Similarly, proposed resistance mechanisms against topoisomerase II inhibitors, such as doxorubicin, cannot explain all cases of therapy failure in the clinic (Pommier, 2013). Here, a haploid screen identified novel resistance mechanisms which reduce DNA double strand break formation or stimulate DNA repair (Wijdeven et al., 2015). Although gene trap viruses should be inactivating, a gain-of-function mutation cannot be excluded. For example, we found a significant enrichment of integrations that induce the ABCG2 gene expression upon topotecan selection (Guyader, Blomen, Gerhards, Brummelkamp and Rottenberg, unpublished results). Despite the technical differences between haploid and CRISPR/Cas9-positive selection screens, direct comparisons of both approaches showed high concordance (Marceau et al., 2016; Timms et al., 2016; Wang et al., 2015b).
The inactivation of genes at high efficiency makes haploid screens very effective for negative selections, as most sense integrations in an intron will lead to reproducible and complete depletion of a gene (Elling and Penninger, 2014; Burckstummer et al., 2013). Since negative selection aims at identifying mutations that are depleted from a population of mutants, which is easily influenced by environmental factors, a robust identification of depleted mutations of the pool of cells can be challenging. A unidirectional design of the gene-trap cassette overcomes this problem and has facilitated the identification of essential genes in human cells as well as genotype-specific gene requirements by using the distribution of sense and antisense orientations as a readout for gene essentiality (Wang et al., 2015b; Blomen et al., 2015; Haarhuis et al., 2017). Haploid screens have furthermore been employed to study genotypes which sensitize cells to chemical compounds (Pettitt et al., 2017). For instance, we have recently found that loss of the tumor suppressor FBXW7 sensitize cells to Vinca alkaloids. This finding might aid the choice of microtubule-targeting chemotherapeutic drugs in patients (Gerhards et al., submitted).
Similar to CRISPR/Cas9, reporter- or antibody-based haploid screens have deciphered genetic modifiers of various intracellular phenotypes regardless of their effect on the viability of a cell (Brockmann et al., 2017; Lebensohn et al., 2016; Mezzadra et al., 2017; Timms et al., 2016; Lee et al., 2013). For instance, a reporter-based haploid screen for transgene silencing demonstrated the potential of this methodology to study epigenetic changes in human cells (Tchasovnikarova et al., 2015). Epigenetic mechanisms such as DNA methylation or histone modifications are considered to drive secondary drug resistance by altering the expression of genes involved in drug transport, DNA repair or apoptosis (Wilting and Dannenberg, 2012; Brown et al., 2014). As genetic reporters are not available for all intracellular processes, antibody-based screens in fixed mutagenized cells further expanded the scale of query phenotypes that can be studied (Brockmann et al., 2017). Hence, haploid screens can contribute to our understanding of the epigenetic drivers of drug resistance and potentially identify novel therapeutic strategies.
2.3. In vivo screens
Screens in 2D models are affected by assay conditions, the choice of cell lines as well as their growth on plastic in high oxygen. It has been shown that results vary between labs despite similar approaches (Scholl et al., 2009; Babij et al., 2011). Furthermore, it has been found that tumors exhibit contrasting drug responses ex vivo and in vivo (Teicher et al., 1990) and that a tumor’s microenvironment impacts drug response (Straussman et al., 2012). Additionally, comparisons of patient-derived and control cell lines revealed that the variation rather reflects the genetic background, culture conditions and cell line history than a disease-relevant phenotype (Soldner and Jaenisch, 2012; Gillet et al., 2011). Thus, 2D cancer models encounter limitations regarding their clinical relevance and their application to study complex phenotypes such as metastasis formation or a host’s immune response. In vivo screens can overcome these limitations and bridge the gap to clinically more relevant settings. They allow not only assessing tumor cell-intrinsic contributions to drug sensitivity or resistance, but also take a body’s microenvironment and immune response into account.
2.3.1. Choice of model
In vivo screens may be conducted by using a large cohort of knock-out animal models (Van Der Weyden et al., 2017). However, this approach is not feasible for all laboratories, as the majority does not have an extensive collection of animal models. With the development of CRISPR/Cas9, powerful and fast alternatives became available. CRISPR/Cas9 can be employed for ex vivo gene editing of cells which are subsequently transplanted into recipient mice or for non-germline manipulation of tumors in vivo. Somatic delivery of Cas9-expressing cells can cause an immune response clearing those cells (Wang et al., 2015a). Therefore, Cas9-transgenic mice are frequently employed, which only require local or systemic delivery of the sgRNAs, for instance through lentiviruses, into mice (Platt et al., 2014; Annunziato et al., 2016). Alternatively, lentiviruses encoding both Cas9 and sgRNAs or ex vivo CRISPR/Cas9-modified cells can be injected into immunodeficient or Cas9-tolerant mice for in vivo screening (Chen et al., 2015; Braun et al., 2016). A genome-wide screen, targeting for instance 20,000 genes with 6 sgRNAs per gene, would consist of 120,000 sgRNAs and therefore require millions of transplanted tumor cells. This is often not feasible, but a genome-wide screen is usually not necessary to study the phenotype of interest in vivo. For most in vivo screening approaches, a focused library targeting selected biological processes is sufficient to address the research question.
2.3.2. Applications to study drug response
The major benefit of in vivo over in vitro screens is the contribution of the animal organism to the phenotype. A solid tumor in vivo is a multicellular complex that interacts with its surrounding tissue, differing substantially from a clonal cancer cell line in a cell culture dish. The tumor microenvironment and tumor-stromal interactions gained increasing importance as modulators of drug response. For instance, stroma-induced drug resistance has been described in several preclinical models for chemotherapeutic drugs such as resistance to doxorubicin, vincristine or vemurafenib (Mcmillin et al., 2013).
Until today, most in vivo screens were performed using RNA interference editing (Gargiulo et al., 2013; Zhou et al., 2014a; Beronja et al., 2013; Meacham et al., 2015; Rudalska et al., 2014; Schramek et al., 2014). In vivo CRISPR/Cas9 screens have been employed to assess the metastatic potential upon gene inactivation (Chen et al., 2015). Furthermore, novel immune modulatory factors which could serve as immunotherapeutic targets were identified in a CRISPR/Cas9-edited melanoma screen in vivo (Manguso et al., 2017). Additional biological processes for in vivo screening approaches include the role of tumor angiogenesis and hypoxia in drug response, and basic research topics such as tumor development and tissue regeneration. Screens for these complex phenotypes are still in the process of being optimized.
Nevertheless, limitations of pooled in vivo screens are encountered due to the complexity of the library, poor efficiency of virus delivery, loss of sgRNA representation and of diversity in the outgrown tumor after transplantation or injection, as well as the complex interactions of tumors with the host body (Chen et al., 2015). Furthermore, ethical concerns of performing in vivo screens may rise, as they require large numbers of experimental animals.
2.4. Screens using 3D cancer organoids
A promising model to recapitulate in vivo tumor behavior in an ex vivo setting is the use of 3D organoid cultures (Clevers, 2016). These have several advantages over 2D cell lines. Traditional cell lines are usually clonal, immortalized cells, genetically adapted to cell culture conditions and lacking a tumor’s heterogeneity as well as its complex multilayered cell organization. In a solid tumor, a drug may not reach all cells at equal levels due to varying vascularization. This situation is mimicked to some extent in a 3D culture, as drugs do not freely penetrate the viscous biomaterial in which organoids are embedded. In contrast, all cells in a monolayer will receive equal concentrations of an administered drug. Furthermore, the composition and architecture of organoids correspond to the tissue they are derived from. Organoids can usually be expanded easily, cryopreserved and biobanked and efficiently genetically manipulated (Van De Wetering et al., 2015; Schwank et al., 2013; Drost et al., 2017).
2.4.1. Choice of model
3D organoids have been generated from the eye (Eiraku et al., 2011; Nakano et al., 2012), brain (Lancaster et al., 2013; Pasca et al., 2015), intestine (Sato et al., 2009; Spence et al., 2011; Forster et al., 2014), kidney (Takasato et al., 2014; Takasato et al., 2015), liver (Takebe et al., 2013; Huch et al., 2013), lung (Dye et al., 2015), inner ear (Koehler et al., 2013) and other organs. Organoids have furthermore been derived from cancer tissues (so-called tumoroids) (Baker et al., 2016; Van De Wetering et al., 2015; Schutte et al., 2017; Pauli et al., 2017), or were transformed into cancer organoids by genetic modification (Drost et al., 2015; Matano et al., 2015). 3D cultures allow an organ-like architecture, facilitate interactions with the extracellular matrix and provide rudiments of functionality (Fatehullah et al., 2016).
Considering that organoids consist of primary cells, the passage number influences the organoid’s geno- and phenotype. During culturing, some cells of the heterogeneous cell pool will be lost, leading to a larger discrepancy between original tumor and the organoid line. When larger amounts of starting material is available, e.g. from large excised patient or mouse tumors, a higher complexity of the heterogeneous cell pool can be maintained for longer time than with little starting material such as a biopsy. Additionally, organoid culturing medium is frequently supplemented with various factors such as Noggin or R-spondin which further select cells or alter gene expression in the organoid culture. Thus, organoids are less robust and less well-characterized than cell lines, but the heterogeneity resembles more the tumors observed in patients.
Gene editing components to modify organoids can be delivered by adenoviruses, retroviruses or lentiviruses (Wang et al., 2014a; Koo et al., 2012; Duarte et al., in press). The subsequently achieved modifications create a polyclonal pool of cells with mixed genotypes. To efficiently select modified cells, reporter genes can be added to the viruses. However, the introduction of genetic modifications will also cause additional selective pressure so that one should evaluate to what extent the original tumor is still fully represented in the modified organoid culture after some passages.
2.4.2. Applications to study drug response
3D organoid cultures are useful to study drug response of patient-derived material in vitro. They can be archived easily and used for high-throughput analyses of drug response linked to their omics profiles (Van De Wetering et al., 2015). Using matched samples of a PARP-inhibitor-sensitive and –resistant BRCA2-mutated mammary tumor, we have recently shown that more sophisticated functional assays to measure drug response (e.g. replication fork stability) can be performed in tumoroids (Ray Chaudhuri et al., 2016). How reliable the in vitro drug responses are in predicting the drug response in the patient from whom the tumoroids were derived remains to be seen. When we tested several matched PARP-inhibitor-sensitive and –resistant BRCA1-mutated organoids in vitro, we found examples where the resistant tumors regained drug sensitivity in vitro, despite the stable in vivo resistance of tumors derived from these tumoroids (Duarte et al., in press). This illustrates that further optimization of the in vitro conditions is required to mimic the response of real tumors.
The CRISPR/Cas9 system facilitates rapid genome engineering as well as forward screening approaches in 3D organoids (Nie and Hashino, 2017). Given the heterogeneous nature of organoids, these screens are more complex, their statistical power is reduced compared to screens in well-defined genetic backgrounds and the variation of size and shape of organoids further affect phenotypic selections. However, the sampling error is low due to the cellular complexity of the drawn test sample from the given pool of cells in the tumoroid and thus 3D screening results are expected to validate in vivo. So far, a CRISPR screen in lung organoids with a miniature focused library discovered genes involved in ciliogenesis and barrier function in the airway epithelium (Gao et al., 2015). The high number of cells, required to ensure an appropriate coverage, needs to be taken into consideration when organoid screens will be performed. These numbers of cells are usually only obtained after several passages which might conflict with the ambition of high similarity between the organoids and the original tumor. Additionally, the success of organoid screens strongly depends on the control of organoid size with clear pathophysiological gradients, as seen in a solid tumor, while preventing a too large area of central necrosis. Due to a batch-to-batch variation of the extracellular biomaterial, reproducibility and scalability of experiments is thus far limited. Thus, large-scale screens remain technically challenging in tissue-derived organoids.
However, we found that a major advantage of tumoroids is that they can serve as a rapid intermediate step to introduce genetic modifications, and subsequently transplant the tumors into mice to test drug response in vivo (Duarte et al., in press). Using tumoroids from Brca1-mutated mouse mammary tumors we introduced Trp53bp1 mutations by CRISPR/Cas9 and show that the tumors derived from the Trp53bp1-depleted tumoroids are resistant to PARP inhibition. Importantly, we also observed that tumoroids exhibit high clonal heterogeneity in vivo and give rise to tumors that preserve the cellular complexity of the parental organoid population (Duarte et al., in press). We therefore conclude that these models are suitable to study the effects of intratumoral heterogeneity in vivo and are useful for in vivo screening approaches to identify mechanisms of drug resistance.
Since tumoroids are more readily cultured from patient tumors than patient-derived 2D cell lines (Boj et al., 2015; Van De Wetering et al., 2015; Duarte et al., in press), they also provide an opportunity to better follow a patient’s disease development. Using genome-wide CRISPR/Cas9 mutagenesis, the essentialome of an individual tumor may be determined to provide information about specific vulnerabilities of this particular tumor. The patient could subsequently receive a treatment accordingly, and if the tumor relapses or stops responding, a new organoid line could be obtained and the essentialome re-analyzed by another functional genome-wide screen. This “sensitive-to-resistant-essentialome”-comparison approach may yield new insights into drug resistance mechanisms, directly received from clinical patients, and provide useful information for new vulnerabilities and treatment options.
In summary, organoid screens are still in the process of optimization and technical hurdles need to be resolved. Nevertheless, they provide a very promising approach to study complex contributions to a phenotype of interest in vitro that resembles treatment responses in vivo. It is very likely that novel factors influencing drug response will be discovered soon in tumoroid models, and they will further broaden our repertoire to study cancer specimens directly derived from patients.
3. Conclusions
In contrast to targeted anti-cancer treatment, predictive biomarkers for classical chemotherapy have been much more difficult to identify. This might be explained by the complex heterogeneity between tumors due to secondary mutations, epigenetic alterations or other mechanisms of drug resistance which are still poorly understood (Borst, 2012). Even in a well-characterized genetically homogeneous tumor system, such as syngeneic transplantable tumors from genetically engineered mouse models, the identification of genes contributing to drug response is limited (Rottenberg et al., 2012). Only if a gene is altered in most of the analyzed tumors, gene expression analysis will detect it as a predictive candidate, whereas drug response mechanisms present only in a subset of tumors will be missed. Given the fact that a variety of mechanisms, either single- or multi-genetic in nature, influence drug response, together with various mechanisms of action of classical chemotherapeutics, it is not surprising that we have not been very successful to establish predictive biomarkers for these drugs.
The advances in screening technologies however might provide the necessary starting point to develop novel hypotheses which might eventually translate into clinical predictive biomarkers. Forward genetic screens in mammalian cells have substantially broadened our understanding of cell biology, the mode of action of chemical compounds and anti-cancer drug resistance or hypersensitivity. These approaches offer crucial tools to discover novel tumor vulnerabilities, while at the same time highlighting context-specific dependencies and mechanistic complexities of cancer. A variety of cellular features determine drug sensitivity of a cell, and drug response varies between individual tumors and patients, which represents a major hurdle for clinical oncologists. Frequently, cancer patients do not benefit from a chosen treatment but predominantly experience the side effects, and better predictive biomarkers are urgently needed. Forward genetic screens allow modeling of the complex mutational landscape of human cancer and identifying similarities or differences between drug effects. For instance, studies performed on cellular platinum uptake showed that carboplatin and cisplatin, but not oxaliplatin, are entering cells through LRRC8A/D-containing anion channels (Planells-Cases et al., 2015). Thus, for patients with low LRRC8A/D expression, cisplatin and carboplatin might not be an effective treatment regimen. Such findings may be clinically useful and, thus, more screens using well-established, classical chemotherapeutic drugs may further contribute to improved precision oncology.
We expect that further technical improvements of existing screening technologies and the comparison of screens for various drugs will aid in the development of novel treatment regimens. The potential use of patient-derived tumoroids for drug response screens can additionally be exploited for personalized medicine. Future treatment concepts will increasingly be based on personalized tumor features and thus knowledge about specific tumor vulnerabilities is essential to move the field forward. Since classical chemotherapeutic agents are widely available, inexpensive and well-characterized in terms of dosage and toxicity, more effort should be made to unravel the full potential of these drugs and assess their benefit in clinical trials for patient subgroups characterized by particular genetic mutations. Furthermore, new drug combinations could be suggested when the impact of a genotype on drug response is better understood.
Given the advantages and limitations of the screening techniques, the chosen approach should be carefully evaluated. Screening output might depend more on experimental design than on the technology itself, as shown by a study comparing CRISPR/Cas9 with optimized RNAi screens concluding that both techniques perform equally well (Morgens et al., 2016). In general, the most comprehensive picture will be achieved by combining several approaches. Clinical data from patients remain essential to validate the findings and assess the predictive potential of identified gene-drug dependencies. Thus, studies would benefit from more than one approach to answer the same question and their combination into a therapy response map might promote the translation into clinically predictive markers and effective treatments with improved patient life quality and overall survival.
Conflict of interest
The authors declare no potential conflicts of interest.
Acknowledgements
We are grateful to Piet Borst, Vincent Blomen (The Netherlands Cancer Institute, Amsterdam, The Netherlands), Sohvi Blatter and Kerstin Hahn (Institute of Animal Pathology, Vetsuisse Faculty, University of Bern, Switzerland) for critical feedback on the manuscript. Our research is supported by the Swiss National Science Foundation (P1BEP3_155461 and 310030_156869), the Dutch Cancer Society (2014-6532) and the European Union (ERC CoG-681572).
References
- Aggarwal S. Targeted cancer therapies. Nat. Rev. Drug Discov. 2010;9:427–428. doi: 10.1038/nrd3186. [DOI] [PubMed] [Google Scholar]
- Aguirre A.J., Meyers R.M., Weir B.A., Vazquez F., Zhang C.Z., Ben-David U., Cook A., Ha G., Harrington W.F., Doshi M.B., Kost-Alimova M., Gill S., Xu H., Ali L.D., Jiang G.Z., Pantel S., Lee Y., Goodale A., Cherniack A.D., Oh C., Kryukov G., Cowley G.S., Garraway L.A., Stegmaier K., Roberts C.W., Golub T.R., Meyerson M., Root D.E., Tsherniak A., Hahn W.C. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Lazikani B., Banerji U., Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 2012;30:679. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- Annunziato S., Kas S.M., Nethe M., Yucel H., Del Bravo J., Pritchard C., Bin Ali R., Van Gerwen B., Siteur B., Drenth A.P., Schut E., Van De Ven M., Boelens M.C., Klarenbeek S., Huijbers I.J., Van Miltenburg M.H., Jonkers J. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes. Dev. 2016;30:1470–1480. doi: 10.1101/gad.279190.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babij C., Zhang Y.H., Kurzeja R.J., Munzli A., Shehabeldin A., Fernando M., Quon K., Kassner P.D., Ruefli-Brasse A.A., Watson V.J., Fajardo F., Jackson A., Zondlo J., Sun Y., Ellison A.R., Plewa C.A., San Miguel T., Robinson J., Mccarter J., Schwandner R., Judd T., Carnahan J., Dussault I. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 2011;71:5818–5826. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- Baker L.A., Tiriac H., Clevers H., Tuveson D.A. Modeling pancreatic cancer with organoids. Trends Cancer. 2016;2:176–190. doi: 10.1016/j.trecan.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K., Horlings H.M., Hennessy B.T., Madiredjo M., Hijmans E.M., Beelen K., Linn S.C., Gonzalez-Angulo A.M., Stemke-Hale K., Hauptmann M., Beijersbergen R.L., Mills G.B., De Vijver M.J.V., Bernards R. A functional genetic approach identifies the PI3 K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Berns K., Sonnenblick A., Gennissen A., Brohee S., Hijmans E.M., Evers B., Fumagalli D., Desmedt C., Loibl S., Denkert C., Neven P., Guo W., Zhang F., Knijnenburg T.A., Bosse T., Van Der Heijden M.S., Hindriksen S., Nijkamp W., Wessels L.F.A., Joensuu H., Mills G.B., Beijersbergen R.L., Sotiriou C., Bernards R. Loss of ARID1A activates ANXA1, which serves as a predictive biomarker for Trastuzumab resistance. Clin. Cancer Res. 2016;22:5238–5248. doi: 10.1158/1078-0432.CCR-15-2996. [DOI] [PubMed] [Google Scholar]
- Beronja S., Janki P., Heller E., Lien W.H., Keyes B.E., Oshimori N., Fuchs E. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501:185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter S., Rottenberg S. Minimal residual disease in cancer therapy–Small things make all the difference. Drug Resist. Updat. 2015;21–22:1–10. doi: 10.1016/j.drup.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Blomen V.A., Majek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., Van Diemen F.R., Olk N., Stukalov A., Marceau C., Janssen H., Carette J.E., Bennett K.L., Colinge J., Superti-Furga G., Brummelkamp T.R. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- Boj S.F., Hwang C.I., Baker L.A., Chio Ii Engle D.D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M.S., Gracanin A., Oni T., Yu K.H., Van Boxtel R., Huch M., Rivera K.D., Wilson J.P., Feigin M.E., Ohlund D., Handly-Santana A., Ardito-Abraham C.M., Ludwig M., Elyada E., Alagesan B., Biffi G., Yordanov G.N., Delcuze B., Creighton B., Wright K., Park Y., Morsink F.H., Molenaar I.Q., Borel Rinkes I.H., Cuppen E., Hao Y., Jin Y., Nijman I.J., Iacobuzio-Donahue C., Leach S.D., Pappin D.J., Hammell M., Klimstra D.S., Basturk O., Hruban R.H., Offerhaus G.J., Vries R.G., Clevers H., Tuveson D.A. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker M., Samsonova A.A., Kwon Y., Flockhart I., Mohr S.E., Perrimon N. False negative rates in Drosophila cell-based RNAi screens: a case study. BMC Genom. 2011;12:50. doi: 10.1186/1471-2164-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C., Bussey H., Andrews B.J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Borst P., Wessels L. Do predictive signatures really predict response to cancer chemotherapy? ABBV Cell Cycle. 2010;9:4836–4840. doi: 10.4161/cc.9.24.14326. [DOI] [PubMed] [Google Scholar]
- Borst P., Rottenberg S., Jonkers J. How do real tumors become resistant to cisplatin? ABBV Cell Cycle. 2008;7:1353–1359. doi: 10.4161/cc.7.10.5930. [DOI] [PubMed] [Google Scholar]
- Borst P. Cancer drug pan-resistance: pumps, cancer stem cells, quiescence, epithelial to mesenchymal transition, blocked cell death pathways, persisters or what? Open Biol. 2012;2:120066. doi: 10.1098/rsob.120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P., Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer. 2012;12:587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- Braun C.J., Bruno P.M., Horlbeck M.A., Gilbert L.A., Weissman J.S., Hemann M.T. Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E3892–E3900. doi: 10.1073/pnas.1600582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann M., Blomen V.A., Nieuwenhuis J., Stickel E., Raaben M., Bleijerveld O.B., Altelaar A.F.M., Jae L.T., Brummelkamp T.R. Genetic wiring maps of single-cell protein states reveal an off-switch for GPCR signalling. Nature. 2017;546:307–311. doi: 10.1038/nature22376. [DOI] [PubMed] [Google Scholar]
- Brown R., Curry E., Magnani L., Wilhelm-Benartzi C.S., Borley J. Poised epigenetic states and acquired drug resistance in cancer. Nat. Rev. Cancer. 2014;14:747–753. doi: 10.1038/nrc3819. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Burckstummer T., Banning C., Hainzl P., Schobesberger R., Kerzendorfer C., Pauler F.M., Chen D., Them N., Schischlik F., Rebsamen M., Smida M., Fece De La Cruz F., Lapao A., Liszt M., Eizinger B., Guenzl P.M., Blomen V.A., Konopka T., Gapp B., Parapatics K., Maier B., Stockl J., Fischl W., Salic S., Taba Casari M.R., Knapp S., Bennett K.L., Bock C., Colinge J., Kralovics R., Ammerer G., Casari G., Brummelkamp T.R., Superti-Furga G., Nijman S.M. A reversible gene trap collection empowers haploid genetics in human cells. Nat. Methods. 2013;10:965–971. doi: 10.1038/nmeth.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan R., Luk F., Bebawy M. Inhibition of the multidrug resistance P-glycoprotein: time for a change of strategy? Drug Metab. Dispos. 2014;42:623–631. doi: 10.1124/dmd.113.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso F., Van’t Veer L.J., Bogaerts J., Slaets L., Viale G., Delaloge S., Pierga J.Y., Brain E., Causeret S., Delorenzi M., Glas A.M., Golfinopoulos V., Goulioti T., Knox S., Matos E., Meulemans B., Neijenhuis P.A., Nitz U., Passalacqua R., Ravdin P., Rubio I.T., Saghatchian M., Smilde T.J., Sotiriou C., Stork L., Straehle C., Thomas G., Thompson A.M., Van Der Hoeven J.M., Vuylsteke P., Bernards R., Tryfonidis K., Rutgers E., Piccart M., Investigators M. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. New Engl. J. Med. 2016;375:717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- Carette J.E., Guimaraes C.P., Varadarajan M., Park A.S., Wuethrich I., Godarova A., Kotecki M., Cochran B.H., Spooner E., Ploegh H.L., Brummelkamp T.R. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- Carette J.E., Guimaraes C.P., Wuethrich I., Blomen V.A., Varadarajan M., Sun C., Bell G., Yuan B.B., Muellner M.K., Nijman S.M., Ploegh H.L., Brummelkamp T.R. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol. 2011;29 doi: 10.1038/nbt.1857. 542-U108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., Dal Cin P., Dye J.M., Whelan S.P., Chandran K., Brummelkamp T.R. Ebola virus entry requires the cholesterol transporter niemann-pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.D., Sanjana N.E., Zheng K.J., Shalem O., Lee K., Shi X., Scott D.A., Song J., Pan J.Q., Weissleder R., Lee H., Zhang F., Sharp P.A. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Cobleigh M.A., Vogel C.L., Tripathy D., Robert N.J., Scholl S., Fehrenbacher L., Wolter J.M., Paton V., Shak S., Lieberman G., Slamon D.J. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J. Clin. Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S.L., Barretto R., Habib N., Hsu P.D., Wu X.B., Jiang W.Y., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran R.B., Atreya C.E., Falchook G.S., Kwak E.L., Ryan D.P., Bendell J.C., Hamid O., Messersmith W.A., Daud A., Kurzrock R., Pierobon M., Sun P., Cunningham E., Little S., Orford K., Motwani M., Bai Y.C., Patel K., Venook A.P., Kopetz S. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600-mutant colorectal cancer. J. Clin. Oncol. 2015;33:4023–4031. doi: 10.1200/JCO.2015.63.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Vandersluis B., Koch E.N., Baryshnikova A., Pons C., Tan G.H., Wang W., Usaj M., Hanchard J., Lee S.D., Pelechano V., Styles E.B., Billmann M., Van Leeuwen J., Van Dyk N., Lin Z.Y., Kuzmin E., Nelson J., Piotrowski J.S., Srikumar T., Bahr S., Chen Y.Q., Deshpande R., Kurat C.F., Li S.C., Li Z.J., Usaj M.M., Okada H., Pascoe N., San Luis B.J., Sharifpoor S., Shuteriqi E., Simpkins S.W., Snider J., Suresh H.G., Tan Y.Z., Zhu H.W., Malod-Dognin N., Janjic V., Przulj N., Troyanskaya O.G., Stagljar I., Xia T., Ohya Y., Gingras A.C., Raught B., Boutros M., Steinmetz L.M., Moore C.L., Rosebrock A.P., Caudy A.A., Myers C.L., Andrews B., Boone C. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016:353. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017 doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Henau O., Rausch M., Winkler D., Campesato L.F., Liu C., Cymerman D.H., Budhu S., Ghosh A., Pink M., Tchaicha J., Douglas M., Tibbitts T., Sharma S., Proctor J., Kosmider N., White K., Stern H., Soglia J., Adams J., Palombella V.J., Mcgovern K., Kutok J.L., Wolchok J.D., Merghoub T. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature. 2016;539:443–447. doi: 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma M., Hanahan D. The biology of personalized cancer medicine: facing individual complexities underlying hallmark capabilities. Mol. Oncol. 2012;6:111–127. doi: 10.1016/j.molonc.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejesus R., Moretti F., Mcallister G., Wang Z.C., Bergman P., Liu S.M., Frias E., Alford J., Reece-Hoyes J.S., Lindeman A., Kelliher J., Russ C., Knehr J., Carbone W., Beibel M., Roma G., Ng A., Tallarico J.A., Porter J.A., Xavier R.J., Mickanin C., Murphy L.O., Hoffman G.R., Nyfeler B. Functional CRISPR screening identifies the ufmylation pathway as a regulator of SQSTM1/p62. Elife. 2016:5. doi: 10.7554/eLife.17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A.A., Qin H., Ramalho-Santos M., Song J.S. HiTSelect: a comprehensive tool for high-complexity-pooled screen analysis. Nucleic Acids Res. 2015:43. doi: 10.1093/nar/gku1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez A.A., Lim W.A., Qi L.S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J., Van Jaarsveld R.H., Ponsioen B., Zimberlin C., Van Boxtel R., Buijs A., Sachs N., Overmeer R.M., Offerhaus G.J., Begthel H., Korving J., Van De Wetering M., Schwank G., Logtenberg M., Cuppen E., Snippert H.J., Medema J.P., Kops G.J.P.L., Clevers H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521 doi: 10.1038/nature14415. 43-U329. [DOI] [PubMed] [Google Scholar]
- Drost J., Van Boxtel R., Blokzijl F., Mizutani T., Sasaki N., Sasselli V., De Ligt J., Behjati S., Grolleman J.E., Van Wezel T., Nik-Zainal S., Kuiper R.P., Cuppen E., Clevers H. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358:234–238. doi: 10.1126/science.aao3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker B.J., Sawyers C.L., Kantarjian H., Resta D.J., Reese S.F., Ford J.M., Capdeville R., Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the philadelphia chromosome. New Engl. J. Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- Duarte A.A., Gogola E., Sachs N., Barazas M., Annunziato S., De Ruiter J.R., Velds A., Blatter S., Van De Ven M., Clevers H., Borst P., Jonkers J., Rottenberg S. BRCA-deficient mouse mammary tumor organoids to study cancer drug resistance. Nat. Methods. 2018 doi: 10.1038/nmeth.4535. in press. [DOI] [PubMed] [Google Scholar]
- Dye B.R., Hill D.R., Ferguson M.A., Tsai Y.H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D., White E.S., Deutsch G.H., Spence J.R. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 2015:4. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C.J., Beachy P.A., Baum B., Boutros M., Buchholz F., Chanda S.K., Downward J., Ellenberg J., Fraser A.G., Hacohen N., Hahn W.C., Jackson A.L., Kiger A., Linsley P.S., Lum L., Ma Y., Mathey-Prevot B., Root D.E., Sabatini D.M., Taipale J., Perrimon N., Bernards R. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T., Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–U73. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Elling U., Penninger J.M. Genome wide functional genetics in haploid cells. FEBS Lett. 2014;588:2415–2421. doi: 10.1016/j.febslet.2014.06.032. [DOI] [PubMed] [Google Scholar]
- Elling U., Taubenschmid J., Wirnsberger G., O'malley R., Demers S.P., Vanhaelen Q., Shukalyuk A.I., Schmauss G., Schramek D., Schnuetgen F., Von Melchner H., Ecker J.R., Stanford W.L., Zuber J., Stark A., Penninger J.M. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essletzbichler P., Konopka T., Santoro F., Chen D., Gapp B.V., Kralovics R., Brummelkamp T.R., Nijman S.M., Burckstummer T. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014;24:2059–2065. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers B., Jastrzebski K., Heijmans J.P., Grernrum W., Beijersbergen R.L., Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 2016;34:631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- Farmer H., Mccabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., Martin N.M., Jackson S.P., Smith G.C., Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- Fojo T., Parkinson D.R. Biologically targeted cancer therapy and marginal benefits: are we making too much of too little or are we achieving too little by giving too much? Clin. Cancer Res. 2010;16:5972–5980. doi: 10.1158/1078-0432.CCR-10-1277. [DOI] [PubMed] [Google Scholar]
- Forster R., Chiba K., Schaeffer L., Regalado S.G., Lai C.S., Gao Q., Kiani S., Farin H.F., Clevers H., Cost G.J., Chan A., Rebar E.J., Urnov F.D., Gregory P.D., Pachter L., Jaenisch R., Hockemeyer D. Human intestinal tissue with adult stem cell properties derived from pluripotent stem cells. Stem Cell Rep. 2014;2:838–852. doi: 10.1016/j.stemcr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Bali A.S., Randell S.H., Hogan B.L.M. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J. Cell Biol. 2015;211:669–682. doi: 10.1083/jcb.201506014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargiulo G., Cesaroni M., Serresi M., De Vries N., Hulsman D., Bruggeman S.W., Lancini C., Van Lohuizen M. In vivo RNAi screen for BMI1 targets identifies TGF-beta/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell. 2013;23:660–676. doi: 10.1016/j.ccr.2013.03.030. [DOI] [PubMed] [Google Scholar]
- Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017 doi: 10.1038/nature24644. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Nislow C. The yeast deletion collection: a decade of functional genomics. Genetics. 2014;197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., Qi L.S., Kampmann M., Weissman J.S. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet J.P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18708–18713. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti E., Zucali P.A., Assaraf Y.G., Funel N., Gemelli M., Stark M., Thunnissen E., Hou Z., Muller I.B., Struys E.A., Perrino M., Jansen G., Matherly L.H., Peters G.J. Role of proton-coupled folate transporter in pemetrexed resistance of mesothelioma: clinical evidence and new pharmacological tools. Ann. Oncol. 2017;28:2725–2732. doi: 10.1093/annonc/mdx499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen N., Assaraf Y.G. Antifolates in cancer therapy: structure: activity and mechanisms of drug resistance. Drug Resist. Updat. 2012;15:183–210. doi: 10.1016/j.drup.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Gottesman M.M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- Groenendijk F.H., Bernards R. Drug resistance to targeted therapies: deja vu all over again. Mol. Oncol. 2014;8:1067–1083. doi: 10.1016/j.molonc.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes C.P., Carette J.E., Varadarajan M., Antos J., Popp M.W., Spooner E., Brummelkamp T.R., Ploegh H.L. Identification of host cell factors required for intoxication through use of modified cholera toxin. J. Cell Biol. 2011;195:751–764. doi: 10.1083/jcb.201108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis J.H.I., Van Der Weide R.H., Blomen V.A., Yanez-Cuna J.O., Amendola M., Van Ruiten M.S., Krijger P.H.L., Teunissen H., Medema R.H., Van Steensel B., Brummelkamp T.R., De Wit E., Rowland B.D. The cohesin release factor WAPL restricts chromatin loop extension. Cell. 2017;169:693–707. doi: 10.1016/j.cell.2017.04.013. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., Macleod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S., Mero P., Dirks P., Sidhu S., Roth F.P., Rissland O.S., Durocher D., Angers S., Moffat J. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Heijink A.M., Blomen V.A., Bisteau X., Degener F., Matsushita F.Y., Kaldis P., Foijer F., Van Vugt M.a.T.M. A haploid genetic screen identifies the G(1)/S regulatory machinery as a determinant of Wee1 inhibitor sensitivity. Proc. Natl. Acad. Sci. U. S. A. 2015;112:15160–15165. doi: 10.1073/pnas.1505283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13:714. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- Horii T., Tamura D., Morita S., Kimura M., Hatada I. Generation of an ICF syndrome model by efficient genome editing of human induced pluripotent stem cells using the CRISPR system. Int. J. Mol. Sci. 2013;14:19774–19781. doi: 10.3390/ijms141019774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden B.E., Muhar M., Gemberling M., Gersbach C.A., Stainier D.Y.R., Seydoux G., Mohr S.E., Zuber J., Perrimon N. Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. Nat. Rev. Genet. 2017;18:24–40. doi: 10.1038/nrg.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housman G., Byler S., Heerboth S., Lapinska K., Longacre M., Snyder N., Sarkar S. Drug resistance in cancer: an overview. Cancers. 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.D., Holzel M., Knijnenburg T., Schlicker A., Roepman P., Mcdermott U., Garnett M., Grernrum W., Sun C., Prahallad A., Groenendijk F.H., Mittempergher L., Nijkamp W., Neefjes J., Salazar R., Ten Dijke P., Uramoto H., Tanaka F., Beijersbergen R.L., Wessels L.F.A., Bernards R. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Dorrell C., Boj S.F., Van Es J.H., Li V.S., Van De Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J., Haft A., Vries R.G., Grompe M., Clevers H. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorns E., Lord C.J., Turner N., Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat. Rev. Drug Discov. 2007;6:556–568. doi: 10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]
- Jae L.T., Raaben M., Riemersma M., Van Beusekom E., Blomen V.A., Velds A., Kerkhoven R.M., Carette J.E., Topaloglu H., Meinecke P., Wessels M.W., Lefeber D.J., Whelan S.P., Van Bokhoven H., Brummelkamp T.R. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science. 2013;340:479–483. doi: 10.1126/science.1233675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W.G., Jr. Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science. 2012;337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H.M., Fojo T., Mathisen M., Zwelling L.A. Cancer drugs in the United States: justum pretium-the just price. J. Clin. Oncol. 2013;31:3600–3604. doi: 10.1200/JCO.2013.49.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Kim J., Hur J.K., Been K.W., Yoon S.H., Kim J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016;34:863-. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z.L., Gonzales A.P.W., Li Z.Y., Peterson R.T., Yeh J.R.J., Aryee M.J., Joung J.K. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z.L., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler K.R., Mikosz A.M., Molosh A.I., Patel D., Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500:217–221. doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H., Li Y.L., Tan E.P., Velasco-Herrera M.D., Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Koo B.K., Stange D.E., Sato T., Karthaus W., Farin H.F., Huch M., Van Es J.H., Clevers H. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods. 2012;9:81–U197. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- Korkmaz G., Lopes R., Ugalde A.P., Nevedomskaya E., Han R., Myacheva K., Zwart W., Elkon R., Agami R. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016;34:192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- Kort E.J., Paneth N., Vande Woude G.F. The decline in U.S. cancer mortality in people born since 1925. Cancer Res. 2009;69:6500–6505. doi: 10.1158/0008-5472.CAN-09-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecki M., Reddy P.S., Cochran B.H. Isolation and characterization of a near-haploid human cell line. Exp. Cell Res. 1999;252:273–280. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- Kurata M., Rathe S.K., Bailey N.J., Aumann N.K., Jones J.M., Veldhuijzen G.W., Moriarity B.S., Largaespada D.A. Using genome-wide CRISPR library screening with library resistant DCK to find new sources of Ara-C drug resistance in AML. Sci. Rep. 2016:6. doi: 10.1038/srep36199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J.J.M., Pyl P.T., Rausch T., Zichner T., Tekkedil M.M., Stutz A.M., Jauch A., Aiyar R.S., Pau G., Delhomme N., Gagneur J., Korbel J.O., Huber W., Steinmetz L.M. The genomic and transcriptomic landscape of a HeLa cell line. G3-Genes Genomes Genet. 2013;3:1213–1224. doi: 10.1534/g3.113.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebensohn A.M., Dubey R., Neitzel L.R., Tacchelly-Benites O., Yang E., Marceau C.D., Davis E.M., Patel B.B., Bahrami-Nejad Z., Travaglini K.J., Ahmed Y., Lee E., Carette J.E., Rohatgi R. Comparative genetic screens in human cells reveal new regulatory mechanisms in WNT signaling. Elife. 2016:5. doi: 10.7554/eLife.21459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Shah C., Xu E.Y. Gene trap mutagenesis: a functional genomics approach towards reproductive research. Mol. Human Reprod. 2007;13:771–779. doi: 10.1093/molehr/gam069. [DOI] [PubMed] [Google Scholar]
- Lee C.C., Carette J.E., Brummelkamp T.R., Ploegh H.L. A reporter screen in a human haploid cell line identifies CYLD as a constitutive inhibitor of NF-kappaB. PLoS One. 2013;8:e70339. doi: 10.1371/journal.pone.0070339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M., Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131–U164. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li X., Li T., Jiang M.G., Wan H., Luo G.Z., Feng C., Cui X., Teng F., Yuan Y., Zhou Q., Gu Q., Shuai L., Sha J., Xiao Y., Wang L., Liu Z., Wang X.J., Zhao X.Y. Genetic modification and screening in rat using haploid embryonic stem cells. Cell Stem Cell. 2014;14:404–414. doi: 10.1016/j.stem.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Li W., Xu H., Xiao T.F., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014:15. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.H., Vorontchikhina M., Wang Y.L., Faiola F., Martinez E. STAGA recruits mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol. Cell. Biol. 2008;28:108–121. doi: 10.1128/MCB.01402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G.V., Stroyakovskiy D., Gogas H., Levchenko E., De Braud F., Larkin J., Garbe C., Jouary T., Hauschild A., Grob J.J., Sileni V.C., Lebbe C., Mandala M., Millward M., Arance A., Bondarenko I., Haanen J. B. a. G., Hansson J., Utikal J., Ferraresi V., Kovalenko N., Mohr P., Probachai V., Schadendorf D., Nathan P., Robert C., Ribas A., Demarini D.J., Irani J.G., Casey M., Ouellet D., Martin A.M., Le N., Patel K., Flaherty K. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. New Engl. J. Med. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- Lopes R., Korkmaz G., Agami R. Applying CRISPR-Cas9 tools to identify and characterize transcriptional enhancers. Nat. Rev. Mol. Cell Biol. 2016;17:597–604. doi: 10.1038/nrm.2016.79. [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L.H., Esvelt K.M., Aach J., Guell M., Dicarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manguso R.T., Pope H.W., Zimmer M.D., Brown F.D., Yates K.B., Miller B.C., Collins N.B., Bi K., Lafleur M.W., Juneja V.R., Weiss S.A., Lo J., Fisher D.E., Miao D., Van Allen E., Root D.E., Sharpe A.H., Doench J.G., Haining W.N. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau C.D., Puschnik A.S., Majzoub K., Ooi Y.S., Brewer S.M., Fuchs G., Swaminathan K., Mata M.A., Elias J.E., Sarnow P., Carette J.E. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- Mates L., Chuah M.K.L., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J., Ma L., Samara-Kuko E., Gysemans C., Pryputniewicz D., Miskey C., Fletcher B., Vandendriessche T., Ivics Z., Izsvak Z. Molecular evolution of a novel hyperactive sleeping beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- Mayor-Ruiz C., Dominguez O., Fernandez-Capetillo O. Trap(Seq): an RNA sequencing-based pipeline for the identification of gene-trap insertions in mammalian cells. J. Mol. Biol. 2017;429:2780–2789. doi: 10.1016/j.jmb.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcmillin D.W., Negri J.M., Mitsiades C.S. The role of tumour-stromal interactions in modifying drug response: challenges and opportunities. Nat. Rev. Drug Discov. 2013;12:217–228. doi: 10.1038/nrd3870. [DOI] [PubMed] [Google Scholar]
- Meacham C.E., Lawton L.N., Soto-Feliciano Y.M., Pritchard J.R., Joughin B.A., Ehrenberger T., Fenouille N., Zuber J., Williams R.T., Young R.A., Hemann M.T. A genome-scale in vivo loss-of-function screen identifies Phf6 as a lineage-specific regulator of leukemia cell growth. Genes. Dev. 2015;29:483–488. doi: 10.1101/gad.254151.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Shelling A., Muthukaruppan A., Lasham A., Blenkiron C., Laking G., Print C. Predictive and prognostic molecular markers for cancer medicine. Ther. Adv. Med. Oncol. 2010;2:125–148. doi: 10.1177/1758834009360519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzadra R., Sun C., Jae L.T., Gomez-Eerland R., De Vries E., Wu W., Logtenberg M.E.W., Slagter M., Rozeman E.A., Hofland I., Broeks A., Horlings H.M., Wessels L.F.A., Blank C.U., Xiao Y.L., Heck A.J.R., Borst J., Brummelkamp T.R., Schumacher T.N.M. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles L.A., Garippa R.J., Poirier J.T. Design, execution, and analysis of pooled in vitro CRISPR/Cas9 screens. FEBS J. 2016;283:3170–3180. doi: 10.1111/febs.13770. [DOI] [PubMed] [Google Scholar]
- Morgens D.W., Deans R.M., Li A., Bassik M.C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016;34:634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer R.J., Hutson T.E., Glen H., Michaelson M.D., Molina A., Eisen T., Jassem J., Zolnierek J., Maroto J.P., Mellado B., Melichar B., Tomasek J., Kremer A., Kim H.J., Wood K., Dutcus C., Larkin J. Lenvatinib, everolimus, and the combination in patients with metastatic renal cell carcinoma: a randomised, phase 2, open-label, multicentre trial. Lancet Oncol. 2015;16:1473–1482. doi: 10.1016/S1470-2045(15)00290-9. [DOI] [PubMed] [Google Scholar]
- Mullenders J., Bernards R. Loss-of-function genetic screens as a tool to improve the diagnosis and treatment of cancer. Oncogene. 2009;28:4409–4420. doi: 10.1038/onc.2009.295. [DOI] [PubMed] [Google Scholar]
- Muller F.L., Colla S., Aquilanti E., Manzo V.E., Genovese G., Lee J., Eisenson D., Narurkar R., Deng P., Nezi L., Lee M.A., Hu B., Hu J., Sahin E., Ong D., Fletcher-Sananikone E., Ho D., Kwong L., Brennan C., Wang Y.A., Chin L., Depinho R.A. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–342. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz D.M., Cassiani P.J., Li L., Billy E., Korn J.M., Jones M.D., Golji J., Ruddy D.A., Yu K., Mcallister G., Deweck A., Abramowski D., Wan J., Shirley M.D., Neshat S.Y., Rakiec D., De Beaumont R., Weber O., Kauffmann A., Mcdonald E.R., Keen N., Hofmann F., Sellers W.R., Schmelzle T., Stegmeier F., Schlabach M.R. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov. 2016;6:900–913. doi: 10.1158/2159-8290.CD-16-0178. [DOI] [PubMed] [Google Scholar]
- Nakano T., Ando S., Takata N., Kawada M., Muguruma K., Sekiguchi K., Saito K., Yonemura S., Eiraku M., Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Ng C.K.Y., Weigelt B., A'hern R., Bidard F.C., Lemetre C., Swanton C., Shen R., Reis-Filho J.S. Predictive performance of microarray gene signatures: impact of tumor heterogeneity and multiple mechanisms of drug resistance. Cancer Res. 2014;74:2946–2961. doi: 10.1158/0008-5472.CAN-13-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Hashino E. Organoid technologies meet genome engineering. EMBO Rep. 2017;18:367–376. doi: 10.15252/embr.201643732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini R., Shao W., Sellers W.R. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep. 2015;16:280–296. doi: 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic M., Blatter S., Guyader C., Gonggrijp M., Kersbergen A., Kucukosmanoglu A., Sol W., Drost R., Jonkers J., Borst P., Rottenberg S. Selected alkylating agents can overcome drug tolerance of G0-like tumor cells and eradicate BRCA1-deficient mammary tumors in mice. Clin. Cancer Res. 2017;23:7020–7033. doi: 10.1158/1078-0432.CCR-17-1279. [DOI] [PubMed] [Google Scholar]
- Parnas O., Jovanovic M., Eisenhaure T.M., Herbst R.H., Dixit A., Ye C.J., Przybylski D., Platt R.J., Tirosh I., Sanjana N.E., Shalem O., Satija R., Raychowdhury R., Mertins P., Carr S.A., Zhang F., Hacohen N., Regev A. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O'rourke N.A., Nguyen K.D., Smith S.J., Huguenard J.R., Geschwind D.H., Barres B.A., Pasca S.P. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli C., Hopkins B.D., Prandi D., Shaw R., Fedrizzi T., Sboner A., Sailer V., Augello M., Puca L., Rosati R., Mcnary T.J., Churakova Y., Cheung C., Triscott J., Pisapia D., Rao R., Mosquera J.M., Robinson B., Faltas B.M., Emerling B.E., Gadi V.K., Bernard B., Elemento O., Beltran H., Demichelis F., Kemp C.J., Grandori C., Cantley L.C., Rubin M.A. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt S.J., Rehman F.L., Bajrami I., Brough R., Wallberg F., Kozarewa I., Fenwick K., Assiotis I., Chen L.N., Campbell J., Lord C.J., Ashworth A. A genetic screen using the piggyBac transposon in haploid cells identifies parp1 as a mediator of olaparib toxicity. PLoS One. 2013:8. doi: 10.1371/journal.pone.0061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt S.J., Tan E.P., Yusa K. PiggyBac transposon-based insertional mutagenesis in mouse haploid embryonic stem cells. Methods Mol. Biol. 2015;1239:15–28. doi: 10.1007/978-1-4939-1862-1_2. [DOI] [PubMed] [Google Scholar]
- Pettitt S.J., Krastev D.B., Pemberton H.N., Fontebasso Y., Frankum J., Rehman F.L., Brough R., Song F.F., Bajrami I., Rafiq R., Wallberg F., Kozarewa I., Fenwick K., Armisen-Garrido J., Swain A., Gulati A., Campbell J., Ashworth A., Lord C.J. Genome-wide barcoded transposon screen for cancer drug sensitivity in haploid mouse embryonic stem cells. Sci. Data. 2017:4. doi: 10.1038/sdata.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planells-Cases R., Lutter D., Guyader C., Gerhards N.M., Ullrich F., Elger D.A., Kucukosmanoglu A., Xu G., Voss F.K., Reincke S.M., Stauber T., Blomen V.A., Vis D.J., Wessels L.F., Brummelkamp T.R., Borst P., Rottenberg S., Jentsch T.J. Subunit composition of VRAC channels determines substrate specificity and cellular resistance to Pt-based anti-cancer drugs. EMBO J. 2015;34:2993–3008. doi: 10.15252/embj.201592409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt R.J., Chen S.D., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M., Graham D.B., Jhunjhunwala S., Heidenreich M., Xavier R.J., Langer R., Anderson D.G., Hacohen N., Regev A., Feng G.P., Sharp P.A., Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. Drugging Topoisomerases: lessons and challenges. ACS Chem. Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A., Sun C., Huang S.D., Di Nicolantonio F., Salazar R., Zecchin D., Beijersbergen R.L., Bardelli A., Bernards R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–U146. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Prasad V., Mailankody S. Research and development spending to bring a single cancer drug to market and revenues after approval. JAMA Intern. Med. 2017 doi: 10.1001/jamainternmed.2017.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusztai L. Markers predicting clinical benefit in breast cancer from microtubule-targeting agents. Ann. Oncol. 2007;18(Suppl. 12):xii15–20. doi: 10.1093/annonc/mdm534. [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J., John S., Day A., Crespo A.V., Shen B., Starnes L.M., De Ruiter J.R., Daniel J.A., Konstantinopoulos P.A., Cortez D., Cantor S.B., Fernandez-Capetillo O., Ge K., Jonkers J., Rottenberg S., Sharan S.K., Nussenzweig A. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyal F., Van Vliet M.H., Armstrong N.J., Horlings H.M., De Visser K.E., Kok M., Teschendorff A.E., Mook S., Van ‘T., Veer L., Caldas C., Salmon R.J., Van De Vijver M.J., Wessels L.F.A. A comprehensive analysis of prognostic signatures reveals the high predictive capacity of the proliferation, immune response and RNA splicing modules in breast cancer. Breast Cancer Res. 2008:10. doi: 10.1186/bcr2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S., Vollebergh M.A., De Hoon B., De Ronde J., Schouten P.C., Kersbergen A., Zander S.A., Pajic M., Jaspers J.E., Jonkers M., Loden M., Sol W., Van Der Burg E., Wesseling J., Gillet J.P., Gottesman M.M., Gribnau J., Wessels L., Linn S.C., Jonkers J., Borst P. Impact of intertumoral heterogeneity on predicting chemotherapy response of BRCA1-deficient mammary tumors. Cancer Res. 2012;72:2350–2361. doi: 10.1158/0008-5472.CAN-11-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudalska R., Dauch D., Longerich T., Mcjunkin K., Wuestefeld T., Kang T.W., Hohmeyer A., Pesic M., Leibold J., Von Thun A., Schirmacher P., Zuber J., Weiss K.H., Powers S., Malek N.P., Eilers M., Sipos B., Lowe S.W., Geffers R., Laufer S., Zender L. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat. Med. 2014;20:1138–1146. doi: 10.1038/nm.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Mayor-Ruiz C., Lafarga V., Murga M., Vega-Sendino M., Ortega S., Fernandez-Capetillo O. A genome-wide CRISPR screen identifies CDC25A as a determinant of sensitivity to ATR inhibitors. Mol. Cell. 2016;62:307–313. doi: 10.1016/j.molcel.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi I., Chia G., Golan-Lev T., Peretz M., Weissbein U., Sui L., Sauer M.V., Yanuka O., Egli D., Benvenisty N. Derivation and differentiation of haploid human embryonic stem cells. Nature. 2016;532:107–111. doi: 10.1038/nature17408. [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., Van De Wetering M., Barker N., Stange D.E., Van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459 doi: 10.1038/nature07935. 262-U147. [DOI] [PubMed] [Google Scholar]
- Schnuetgen F., Hansen J., De-Zolt S., Horn C., Lutz M., Floss T., Wurst W., Noppinger P.R., Von Melchner H. Enhanced gene trapping in mouse embryonic stem cells. Nucleic Acids Res. 2008:36. doi: 10.1093/nar/gkn603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C., Frohling S., Dunn I.F., Schinzel A.C., Barbie D.A., Kim S.Y., Silver S.J., Tamayo P., Wadlow R.C., Ramaswamy S., Dohner K., Bullinger L., Sandy P., Boehm J.S., Root D.E., Jacks T., Hahn W.C., Gilliland D.G. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Schork N.J. Personalized medicine: time for one-person trials. Nature. 2015;520:609–611. doi: 10.1038/520609a. [DOI] [PubMed] [Google Scholar]
- Schramek D., Sendoel A., Segal J.P., Beronja S., Heller E., Oristian D., Reva B., Fuchs E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343:309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte M., Risch T., Abdavi-Azar N., Boehnke K., Schumacher D., Keil M., Yildiriman R., Jandrasits C., Borodina T., Amstislavskiy V., Worth C.L., Schweiger C., Liebs S., Lange M., Warnatz H.J., Butcher L.M., Barrett J.E., Sultan M., Wierling C., Golob-Schwarzl N., Lax S., Uranitsch S., Becker M., Welte Y., Regan J.L., Silvestrov M., Kehler I., Fusi A., Kessler T., Herwig R., Landegren U., Wienke D., Nilsson M., Velasco J.A., Garin-Chesa P., Reinhard C., Beck S., Schafer R., Regenbrecht C.R., Henderson D., Lange B., Haybaeck J., Keilholz U., Hoffmann J., Lehrach H., Yaspo M.L. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat. Commun. 2017;8:14262. doi: 10.1038/ncomms14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., Van Der Ent C.K., Nieuwenhuis E.E., Beekman J.M., Clevers H. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Mchale C.M., Haider S.I., Jung C., Zhang S., Smith M.T., Zhang L.P. Identification of genes that modulate susceptibility to formaldehyde and imatinib by functional genomic screening in human haploid KBM7 cells. Toxicol. Sci. 2016;151:10–22. doi: 10.1093/toxsci/kfw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui M., Rajkumar S.V. The high cost of cancer drugs and what we can do about it. Mayo Clin. Proc. 2012;87:935–943. doi: 10.1016/j.mayocp.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F., Jaenisch R. iPSC disease modeling. Science. 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M., Shroyer N.F., Wells J.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhart Z., Pavlovic Z., Chandrashekhar M., Hart T., Wang X., Zhang X., Robitaille M., Brown K.R., Jaksani S., Overmeer R., Boj S.F., Adams J., Pan J., Clevers H., Sidhu S., Moffat J., Angers S. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat. Med. 2017;23:60–68. doi: 10.1038/nm.4219. [DOI] [PubMed] [Google Scholar]
- Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R., Morikawa T., Shee K., Barzily-Rokni M., Qian Z.R., Du J.Y., Davis A., Mongare M.M., Gould J., Frederick D.T., Cooper Z.A., Chapman P.B., Solit D.B., Ribas A., Lo R.S., Flaherty K.T., Ogino S., Wargo J.A., Golub T.R. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487 doi: 10.1038/nature11183. 500-U118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C., Szallasi Z., Brenton J.D., Downward J. Functional genomic analysis of drug sensitivity pathways to guide adjuvant strategies in breast cancer. Breast Cancer Res.: BCR. 2008;10:214. doi: 10.1186/bcr2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Becroft M., Vanslambrouck J.M., Stanley E.G., Elefanty A.G., Little M.H. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Chiu H.S., Maier B., Baillie G.J., Ferguson C., Parton R.G., Wolvetang E.J., Roost M.S., Chuva De Sousa Lopes S.M., Little M.H. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.R., Ueno Y., Zheng Y.W., Koike N., Aoyama S., Adachi Y., Taniguchi H. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Tchasovnikarova I.A., Timms R.T., Matheson N.J., Wals K., Antrobus R., Gottgens B., Dougan G., Dawson M.A., Lehner P.J. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348:1481–1485. doi: 10.1126/science.aaa7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher B.A., Herman T.S., Holden S.A., Wang Y.Y., Pfeffer M.R., Crawford J.W., Frei E. Tumor resistance to alkylating agents conferred by mechanisms operative only in vivo. Science. 1990;247:1457–1461. doi: 10.1126/science.247.4949.1457. [DOI] [PubMed] [Google Scholar]
- Teng X., Dayhoff-Brannigan M., Cheng W.C., Gilbert C.E., Sing C.N., Diny N.L., Wheelan S.J., Dunham M.J., Boeke J.D., Pineda F.J., Hardwick J.M. Genome-wide consequences of deleting any single gene. Mol. Cell. 2013;52:485–494. doi: 10.1016/j.molcel.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms R.T., Menzies S.A., Tchasovnikarova I.A., Christensen L.C., Williamson J.C., Antrobus R., Dougan G., Ellgaard L., Lehner P.J. Genetic dissection of mammalian ERAD through comparative haploid and CRISPR forward genetic screens. Nat. Commun. 2016:7. doi: 10.1038/ncomms11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo C.M., Ding Y., Hoellerbauer P., Davis R.J., Basom R., Girard E.J., Lee E., Corrin P., Hart T., Bolouri H., Davison J., Zhang Q., Hardcastle J., Aronow B.J., Plaisier C.L., Baliga N.S., Moffat J., Lin Q., Li X.N., Nam D.H., Lee J., Pollard S.M., Zhu J., Delrow J.J., Clurman B.E., Olson J.M., Paddison P.J. Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell Rep. 2015;13:2425–2439. doi: 10.1016/j.celrep.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A., Robson M., Garber J.E., Domchek S.M., Audeh M.W., Weitzel J.N., Friedlander M., Arun B., Loman N., Schmutzler R.K., Wardley A., Mitchell G., Earl H., Wickens M., Carmichael J. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- Twomey J.D., Brahme N.N., Zhang B. Drug-biomarker co-development in oncology – 20 years and counting. Drug Resist. Updat. 2017;30:48–62. doi: 10.1016/j.drup.2017.02.002. [DOI] [PubMed] [Google Scholar]
- Uryniak T., Chan I.S.F., Fedorov V.V., Jiang Q., Oppenheimer L., Snapinn S.M., Teng C.H., Zhang J. Responder analyses-a PhRMA position paper. Stat. Biopharm. Res. 2011;3:476–487. [Google Scholar]
- Van De Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., Van Houdt W., Van Gorp J., Taylor-Weiner A., Kester L., Mclaren-Douglas A., Blokker J., Jaksani S., Bartfeld S., Volckman R., Van Sluis P., Li V.S., Seepo S., Sekhar Pedamallu C., Cibulskis K., Carter S.L., Mckenna A., Lawrence M.S., Lichtenstein L., Stewart C., Koster J., Versteeg R., Van Oudenaarden A., Saez-Rodriguez J., Vries R.G., Getz G., Wessels L., Stratton M.R., Mcdermott U., Meyerson M., Garnett M.J., Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Weyden L., Arends M.J., Campbell A.D., Bald T., Wardle-Jones H., Griggs N., Velasco-Herrera M.D., Tuting T., Sansom O.J., Karp N.A., Clare S., Gleeson D., Ryder E., Galli A., Tuck E., Cambridge E.L., Voet T., Macaulay I.C., Wong K., Spiegel S., Speak A.O., Adams D.J., Project S.M.G. Genome-wide in vivo screen identifies novel host regulators of metastatic colonization. Nature. 2017;541:233–236. doi: 10.1038/nature20792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhang H.Y., Zhang B.Q., Liu W., Zhang Z.L., Qiao M., Zhang H.M., Deng F., Wu N.N., Chen X., Wen S., Zhang J.H., Liao Z., Zhang Q., Yan Z.J., Yin L.J., Ye J.X., Deng Y.L., Luu H.H., Haydon R.C., Liang H.J., He T.C. Adenovirus-Mediated efficient gene transfer into cultured three-dimensional organoids. PLoS One. 2014:9. doi: 10.1371/journal.pone.0093608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Mou H.W., Li S.Y., Li Y.X., Hough S., Tran K.R., Li J., Yin H., Anderson D.G., Sontheimer E.J., Weng Z.P., Gao G.P., Xue W. Adenovirus-mediated somatic genome editing of pten by CRISPR/Cas9 in mouse liver in spite of Cas9-Specific immune responses. Human Gene Ther. 2015;26:432–442. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., Sabatini D.M. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Yu H.Y., Hughes N.W., Liu B.X., Kendirli A., Klein K., Chen W.W., Lander E.S., Sabatini D.M. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic ras. Cell. 2017;168:890-. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker S.R., Theurillat J.P., Van Allen E., Wagle N., Hsiao J., Cowley G.S., Schadendorf D., Root D.E., Garraway L.A. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijdeven R.H., Pang B., Van Der Zanden S.Y., Qiao X., Blomen V., Hoogstraat M., Lips E.H., Janssen L., Wessels L., Brummelkamp T.R., Neefjes J. Genome-wide identification and characterization of novel factors conferring resistance to Topoisomerase II poisons in cancer. Cancer Res. 2015;75:4176–4187. doi: 10.1158/0008-5472.CAN-15-0380. [DOI] [PubMed] [Google Scholar]
- Wilting R.H., Dannenberg J.H. Epigenetic mechanisms in tumorigenesis: tumor cell heterogeneity and drug resistance. Drug Resist. Updat. 2012;15:21–38. doi: 10.1016/j.drup.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Wirapati P., Sotiriou C., Kunkel S., Farmer P., Pradervand S., Haibe-Kains B., Desmedt C., Ignatiadis M., Sengstag T., Schutz F., Goldstein D.R., Piccart M., Delorenzi M. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008:10. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liu Z., Ma Y., Zhong C., Yin Q., Zhou C., Shi L., Cai Y., Zhao H., Wang H., Tang F., Wang Y., Zhang C., Liu X.Y., Lai D., Jin Y., Sun Q., Li J. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Res. 2013;23:1187–1200. doi: 10.1038/cr.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Ciaudo C. Vector integration sites identification for gene-trap screening in mammalian haploid cells. Sci. Rep. 2017;7:44736. doi: 10.1038/srep44736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., Van Der Oost J., Regev A., Koonin E.V., Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C.C., Wu Y.L., Chen G.Y., Feng J.F., Liu X.Q., Wang C.L., Zhang S.C., Wang J., Zhou S.W., Ren S.X., Lu S., Zhang L., Hu C.P., Hu C.H., Luo Y., Chen L., Ye M., Huang J.N., Zhi X.Y., Zhang Y.P., Xiu Q.Y., Ma J., Zhang L., You C.X. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- Zhou P.H., Shaffer D.R., Arias D.a.A., Nakazaki Y., Pos W., Torres A.J., Cremasco V., Dougan S.K., Cowley G.S., Elpek K., Brogdon J., Lamb J., Turley S.J., Ploegh H.L., Root D.E., Love J.C., Dranoff G., Hacohen N., Cantor H., Wucherpfennig K.W. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature. 2014;506:52–57. doi: 10.1038/nature12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.X., Zhu S.Y., Cai C.Z., Yuan P.F., Li C.M., Huang Y.Y., Wei W.S. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]