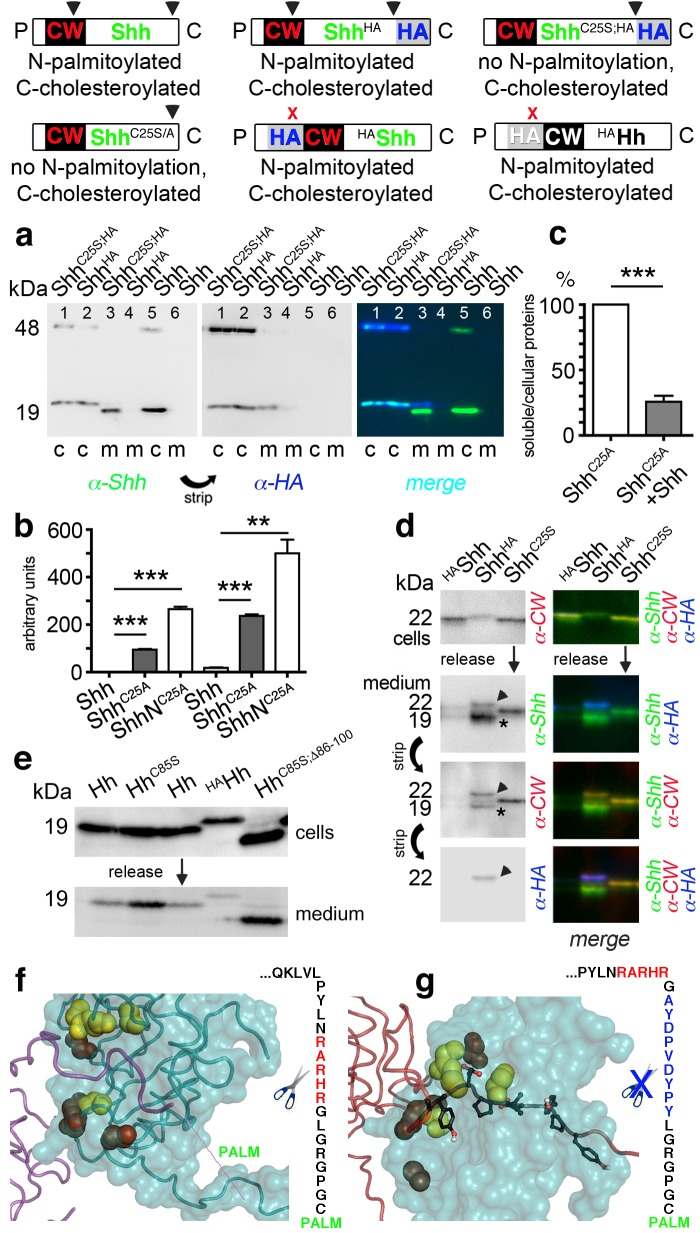
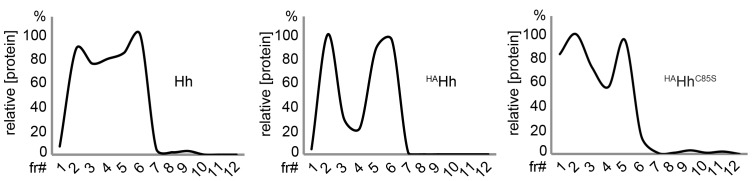
Figure 6. N-palmitoylation controls Hh morphogen release in vitro.
Top: Schematics of transgenes. Arrowheads indicate cleavage sites, the x denotes blocked cleavage. (a) Palmitoylated vertebrate Hh orthologs Shh and ShhHA and non-palmitoylated ShhC25A;HA were expressed in Bosc23 cells, and the proteins in the cellular (c) and corresponding soluble fractions (m) were analyzed by immunoblotting. To better demonstrate protein processing during release, we inverted grayscale blots and colored them (right: green: α-Shh, blue: α-HA). Green signals label untagged or processed Shh, and cyan signals label unprocessed HA-tagged proteins. Higher electrophoretic mobility confirms terminal processing during release. Tagged and untagged palmitoylated proteins are not efficiently released. (b) Compared dual-lipidated, monolipidated and non-lipidated Shh release after 1 hr (left) and 4 hr (right). ***p≤0.0001, **p≤0.001, n = 3. (c) Release of ShhC25A is downregulated 4-fold upon dual-lipidated Shh coexpression (27.7 ± 4.6% if compared to ShhC25A release alone, which was set to 100%, n = 7, ***p≤0.0001). (d) Impaired release of N-terminally HA-tagged HAShh. α-CW antibodies detect the N-terminal CW motif (KRRHPKK). Bright cellular signals in merged blots denote unprocessed proteins (arrowhead), orange signals denote C-processed/N-unprocessed proteins, and green signals confirm the removal of N- and C-terminal peptides (asterisk). Note ShhHA processing at the CW site during release. By contrast, N-terminal processing of ShhC25S is impaired. (e) Immunoblot analysis of recombinant Hh proteins released into media of transfected Drosophila S2 cells (left). Top row: S2 cells express palmitoylated and non-palmitoylated proteins to comparable levels. Bottom row: S2 cells released high levels of unpalmitoylated HhC85S and HhC85S;Δ86-100, and lower levels of palmitoylated Hh, into the media. Only very low levels of HAHh were released in unprocessed form (top band). (f) Intermolecular interactions of Drosophila Hh N-terminal peptides. Right: schematic of the palmitoylated N-terminal ‘stem’ peptide, including basic CW residues (red) serving as the predicted membrane-proximal cleavage site. (g) Modeled insertion of the HA tag upstream of the N-terminal CW motif (located between Hh residues L91 and G92) of Drosophila Hh. This moves the CW motif nine amino acids more distal to the membrane and replaces its previous position with the HA tag (blue, right). Modeled N-terminal palmitate is shown as a zigzag line (pointing to the right). Yellow spheres denote Drosophila Hh residues corresponding to Shh residues that interact with Ptc (Drosophila Hh H193, H194, H200, H240) (Bosanac et al., 2009). Red spheres denote residues corresponding to Shh amino acids bound by the Shh inhibitory antibody 5E1 (K105, R147, R213, R238, R239 in Drosophila Hh) (Maun et al., 2010).