Abstract
FOXC1 is a vital member of FOX families which play important roles in biological processes including proliferation, differentiation, apoptosis, migration, invasion, metabolism, and longevity. Here we are focusing on roles of FOXC1 and their mechanisms in cancers. FOXC1 promoted progress of many cancers, such as breast cancer (especially basal-like breast cancer), hepatocellular carcinoma, gastric cancer and so on. FOXC1 was also found to be associated with drug resistance of cancers. FOXC1 promoted metastasis of cancers by increasing expression of MMP7, NEDD9 and Snail. Proliferation and invasion of cancers were increased by FOXC1 by mediating NF-κB, MST1R and KLF4 expression. FOXC1 was associated with development by regulating expression of FGF19 and MSX1. Recently, FOXC1 was found to be required for niche of stem cells or development of stem cells by mediating expression of Gli2, CXCL12, SCF, NFATC1, BMP and Myh7. Overall, FOXC1 exerts its functions by many mechanisms and may be used as a potential biomarker for diseases.
Keywords: FOX family, FOXC1, cancer, drug resistance, stem cell
INTRODUCTION
FOX family
Forkhead box (FOX) proteins are describled with a common DNA-binding forkhead domain. FOX domain is a ∼100-amino-acid conserved forkhead domain which can bind DNA. FOX proteins which are a superfamily of evolutionarily conserved transcription factors play many important roles in biological processes including development, differentiation, proliferation, apoptosis, metabolism, migration, invasion and longevity [1–3]. Currently, human FOX gene family consists of 49 members including FOXA1-FOXS1. These forty-nine FOX members are categorized into 19 subfamilies (FOXA-FOXS) [4].
History of FOXC1 and axenfeld-rieger syndrome
Axenfeld-Rieger syndrome (ARS) is an eye disease which has an autosomal dominant genetic changes including systemic abnormalities and anterior segment dysgenesis. The eye disease was identified by Axenfeld and named as Axenfeld anomaly in 1920. Patients with congenital iris abnormalities including polycoria, iris hypoplasia and correctopia were described and named by Rieger in 1935. The combination of Reiger syndrome and Axenfeld anomaly is named as ARS.
FOXC1 (Forkhead Box C1) is also named as FKHL7. FOXC1 codes 553 amino acids and its molecular weight is 57KD. FOXC1 was firstly found to be associated with Axenfeld-Rieger syndrome and harbor mutations in patients which were diagnosed as Rieger anomaly (RA), Iris Hypoplasia (IH) and Axenfeld anomaly (AA) [5]. Mutations in FOXC1 were demonstrated to lead to many glaucoma phenotypes [6, 7]. More and more studies showed that there are new mutations in FOXC1, PITX2 and PAX6 in families which have anterior segment dysgenesis disorders [8, 9], that there were mutations in the wing region of the FOXC1 gene forkhead domain [10–15].
FOXC1 AND CANCERS
Functions of FOXC1 in cancers
FOXC1 and breast cancer
FOXC1 was firstly reported to be overexpressed in basal-like breast cancer (BLBC) by our group. Elevated FOXC1 expression was associated with poor prognosis of BLBC patients. Ectopic FOXC1 expression promoted cell migration, invasion and proliferation, whereas knockdown of FOXC1 expression induced opposite effects in breast cancer cells [16]. We further confirmed that FOXC1 exerted its functions in BLBC cells by activating NF-κB signaling [17]. FOXC1 expression by immunohistochemistry detection in triple negative invasive breast cancer is an independent biomarker of survival rate in BLBC patients [18]. Expression of FOXC1 was dramatically correlated with expression of EGFR in human breast cancers. FOXC1 expression was induced by EGFR in BLBC while inhibition of EGFR led to decrease of FOXC1 expression in xenograft tumors. Knockdown of FOXC1 inhibited the effects of EGF on migration, invasion and proliferation of BLBC cell [19]. Expression of FOXC1 was correlated with Gli2 expression and its downstream targets in breast cancers. In addition, overexpression of FOXC1 significantly decreases sensitivity to Hedgehog (Hh) inhibitors in BLBC cells as well as xenograft tumors. FOXC1 activates smoothened-independent Hh signal pathway in BLBC [20]. Combination of FOXC1 expression with matrix metalloprotease 7 (MMP7) expression can be used as an independent predictor of patient outcome in multivariate analyses of two breast cancer patient cohorts. FOXC1 promotes metastasis and invasion by inducing MMP7 expression in breast cancer [21]. GATA3 and BRCA1 were found to inhibit the development of BLBCs by inhibiting expression of FOXC1. Abnormal FOXC1 expression promotes the epithelial-to-mesenchymal transition (EMT)-like phenotypes and drug resistance which contributed to aggressive BLBCs [22]. Recently, we found that FOXC1 expression was closely associated with ERα expression. FOXC1 overexpression reduced ERα expression while knockdown of FOXC1 increase ERα expression. FOXC1 expression inhibited responses to estradiol (E2) and Tamoxifen in the MCF-7. In addition, FOXC1 expression reduced expression of ERα downstream targets including progesterone receptor and XBP1, and dramaticlly inhibited ERE luciferase activity caused by E2 [23].
FOXC1 and liver cancer
Expression of FOXC1 was significantly higher in Hepatocellular carcinoma (HCC) tissues than that in corresponding normal tissues. HCC patients with low FOXC1 expression had longer overall survival times and lower recurrence rates than those with high FOXC1 expression. FOXC1 expression was an independent, important factor for survival and recurrence after operation. FOXC1 overexpression led to significant changes of EMT and improvement of invasion in HCC cell and lung metastasis while knockdown of FOXC1 reduced these processes. FOXC1 induced expression of Snai1, but inversely reduced E-cadherin expression in human HCC tissues [24]. FOXC1 was showed to increase microvascular invasion of primary HCC via regulating EMT [25]. It was reported that IL-8 activates expression of FOXC1 via PI3K/AKT pathway and hypoxia-inducible factor 1α, and that FOXC1 expression induced transactivation of CXCR1 and CCL2, and promoted inflammation, migration and invasion of HCC cells [26].
FOXC1 and lung cancer
Both FOXC1 mRNA expression and protein expression in the tissues of NSCLC were dramatically higher than those in adjacent normal tissues. Overexpression of FOXC1 was related to poor prognosis of tumor, stage of tumor node metastasis, and lymph node metastasis. Patients with low FOXC1 expression had higher overall survival rate than those with high expression. According to clinical statistics, FOXC1 expression could be used as an independent prognostic biomarker in NSCLC patients [27]. Silencing FOXC1 may significantly inhibit the migration of lung cancer cells by changing the EMT process through inhibiting expression of cadherin, being associated with the expressions of extracellular MMPs [28].
FOXC1 and leukemia
FOXC1 was expressed in more than 20% of human Acute Myeloid Leukemia (AML) patients while there was no expression of FOXC1 in normal hematopoietic populations. FOXC1 expression was significantly correlated with expression of the HOXA/B locus in AML. FOXC1 inhibits differentiation of monocyte/macrophage and increases clonogenic potential. In vivo analyses, FOXC1 dramatically promoted the onset of symptomatic leukemia with HOXA9. Human AML cases who have high FOXC1 expression showed little morphologic monocytic differentiation and inferior survival [29].
FOXC1 and other cancers
FOXC1 overexpression caused by hypomethylation of FOXC1 gene promoted proliferation, migration and invasion of melanoma cells. FOXC1 exerted functional roles by activating the MST1R/PI3K/AKT pathway in melanoma. FOXC1 expression can be used as a biomarker in melanoma patient [30].
mRNA and protein expression of FOXC1 in gastric cancer (GC) tissues were dramatically higher than those in adjacent normal tissues. Elevated FOXC1 expression was correlated with the degree of histological differentiation, lymph node metastasis, distant metastasis, TNM stage and invasive depth. Survival analysis showed that patients with low FOXC1 expression had higher overall survival rate than those with high expression. Clinical statistical analysis demonstrated that overexpression of FOXC1 could be used as an independent prognostic biomarker in GC patients [31].
FOXC1 mRNA and protein was highly expressed in pancreatic ductal adenocarcinoma (PDA) tissues compared with corresponding normal tissues. Elevated FOXC1 expression was correlated with the degree of histological differentiation, lymph node metastasis, distant metastasis, TNM stage and invasive depth. Survival analysis demonstrated that overexpression of FOXC1 is closely correlated with a poor prognosis [32].
Esophageal squamous cell carcinoma (ESCC) patients with low FOXC1 expression have a significantly better prognosis than those with high FOXC1 expression. FOXC1 significantly increased cell proliferation, colony formation, migration and invasion in ESCC cells [33].
Upregulation of FOXC1 was associated with progression of prostate cancer [34].
FOXC1 and resistance of cancer drug
FOXC1 plays important roles in and is involved in development of many kinds of stem cells. FOXC1 promoted progress of cancers by activating many signal pathways. More and more study showed that cancer stem cells (CSCs) which were associated with metastasis, relapse of cancers and drug resistance could dramatically influence treatment of tumor. Overexpression of FOXC1 led to resistance of melanoma cells to BRAF inhibitor PLX4032 [30]. MCF-7-FOXC1 cells with high FOXC1 expression are less sensitive to Tamoxifen than MCF-7 control cells without FOXC1 expression [23]. Overexpression FOXC1 led to resistance of Hh inhibitors in BLBC and xenograft tumors [20]. Recently, our data showed that overexpression of FOXC1 increased resistance to Doxorubicin or Paclitaxel in triple negative breast cancer cells and tumors (Data wasn’t published now).
FOXC1 AND OTHER DISEASES
FOXC1 is involved in many cellular processes including development, proliferation, differentiation, cell cycle, migration and invasion, metabolism and DNA damage response of cells.
Stem cell
FOXC1 is involved in development of many stem cells. In the embryo, it was well studied that progenitors of vascular smooth muscle cells (SMCs) which are originated from multiple types of progenitor cells are the cardiac neural crest stem cells. Differentiation of Smooth muscle in the neural crest lineage is mediated by FOXC1 [35]. The periocular mesenchyme developed from neural crest (NC) stem cell partly promoted development of eye. TGF-β2 from the lens promotes expression of Pitx2 and FOXC1 in NC-derived cornea [36]. Bone marrow provides special microenvironments for both haematopoietic stem and progenitor cells. FOXC1, known as a specific transcriptional regulator, play important roles in maintaining development of haematopoietic stem and progenitor cells [37]. Many BLBC cells which have CSC properties were activated through Smoothened (SMO)-independent Hh signal pathway by Gli2 [20]. FOXC1-deficient hair follicle stem cells (HFSCs) spend less time in quiescence, which led to significantly shorten resting periods between hair cycles. FOXC1 sustains niche of the HFSCs and regulated stem cell quiescence to preserve long-term tissue-regenerating potential [38]. In self-renewing hair follicle stem cells, both nuclear factor of activated T-cells 1(NFATC1) and bone morphogenetic protein (BMP) signal pathways were active by FOXC1. FOXC1 plays important role in strengthening quiescence in maintaining hair follicle stem cells [39]. FOXC1 regulated the early stage of cardiomyogenesis by many pathways such as calcium signaling, tight and gap junctions, cell adhesion, and actin cytoskeleton. Myh7 is expressed during cardiac development and is a downstream target of FOXC1. In addition, FOXC1 mediated the functional properties of cardiomyocytes which are derived from embryonic stem cell (ESC) [40]. Bingchen Han et al reported that expression of FOXC1 was correlated with Gli2 expression and its downstream targets in breast cancers, and that FOXC1 maintained CSC properties in BLBC cells in Vivo and in Vitro by mediating non-canonica Gli2-Hh signal pathway [20].
FOXC1 and other diseases
It was reported that mutations in the FOXC1 gene caused defects in the anterior segment of the eye, mitral valve defects and atrial septal defects [41]. FOXC1 was found to play an important role in the morphogenesis of the cardiovascular system [42]. Both FOXC1 and FOXC2 are need for the morphogenesis of the cardiac outflow tract [43]. FOXC1 overexpression increased expression of Notch1 and its corresponding ligand Delta-like 4 (Dll4), which are known as arterial markers. Both FOXC1 and FOXC2 can bind to Dll4 promoter via a Foxc-binding site. In addition, mutation of FOXC1 inhibited the initial development of lymphatic endothelial cells [44]. The early organogenesis of the kidney and urinary tract was promoted by FOXC1 [45, 46].
Many studies show that FOXC1 is an important component of the signal pathways which mediates embryonic heart development and cardiogenesis, playing key important roles in development of heart [47]. Both mutations in FOXC1 and the abnormal expression of FOXC1 significantly affect the congenital heart disease (CHD) of humans [48].
MECHANISMS OF FOXC1
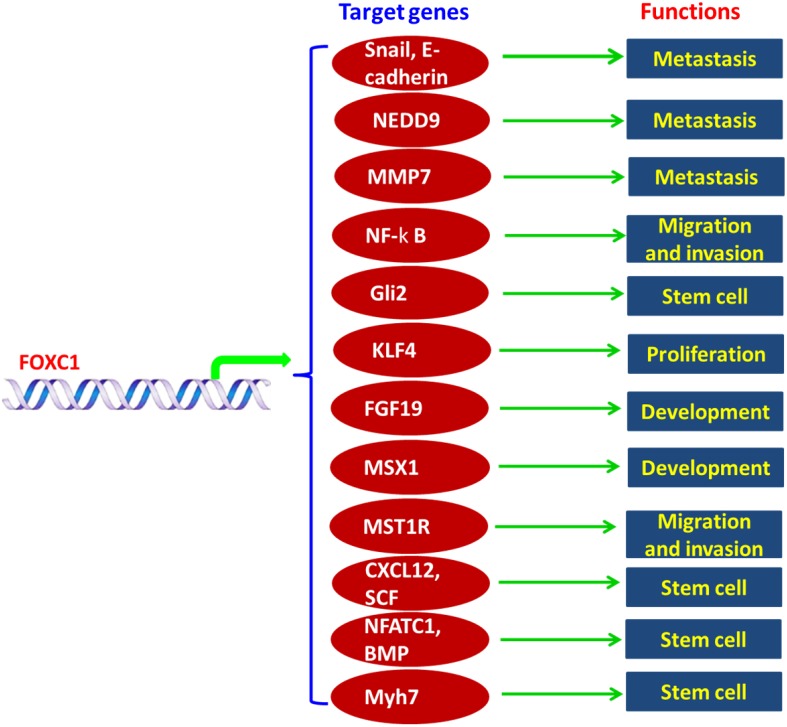
FOXC1 plays complex and important roles in biological processes from development and organogenesis. FOXC1 exerts many functions by different mechanisms (Figure 1).
Figure 1. Functions and mechanisms of FOXC1 in human.
FOXC1 induced snai1 expression and promoted epithelial-to-mesenchymal transition
FOXC1 induced expression of Snai1, but inversely reduced E-cadherin expression in human HCC tissues. FOXC1 increased expression of Snail by directly binding to its promoter, thereby inhibited expression of E-cadherin and promoted EMT [24, 25].
FOXC1 induced neural precursor cell expressed developmentally down-regulated 9 (NEDD9) expression and promoted FOXC1 invasion and metastasis
Compelling evidence, such as DNA microarray, site directed mutagenesis et al, confirmed that NEDD9 is a downstream target of FOXC1. FOXC1 upregulated the expression of NEDD9 and promoted invasion and metastasis of HCC [24].
FOXC1 induced MMP7 expression and promotes invasion of breast cancer
FOXC1 expression is significantly associated with expression of MMP7 in breast cancer samples and cell lines at both the mRNA and protein levels. Ectopic overexpression of FOXC1 in nontransformed mammary epithelial cell lines significantly increased expression of MMP7. On the contrary, knockdown of FOXC1 inhibited expression of MMP7 without affecting expression of other matrix metalloproteinases in BLBC cell lines [21].
FOXC1 activated NF-κB pathway
FOXC1 inhibited P65 degradation by increasing expression of PIN1 (a peptidyl-prolyl isomerase) and reducing expression of SOCS1 (suppressor of cytokine signaling 1) in triple negative breast cancer cells. FOXC1 overexpression caused more sensitive to pharmacological inhibition of NF-κB breast cancers. FOXC1 promoted cell proliferation, migration and invasion by activating NF-κB pathway [17].
FOXC1 activated smoothened-independent hedgehog signal pathway
Expression of FOXC1 was significantly correlated with that of Gli2 and its downstream targets in breast cancers. Many BLBC cells which were activated by FOXC1 maintained CSC properties via activation of Smoothened (SMO)-independent Hh signaling by Gli2. FOXC1 directly binds to the promoter of Gli2 and regulated Gli2 expression. Overexpression of FOXC1 caused more resistance to Hh inhibitors in BLBC cells and xenograft tumors. [20].
FOXC1 reduced expression of KLF4 and collaborated with HOXA9
FOXC1 inhibited differentiation of monocyte/macrophage and enhanced clonogenic potential by reducing expression of KLF4. In vivo analyses, FOXC1 significantly accelerated the onset of symptomatic leukemia by collaborating with HOXA9 [29].
FOXC1 activated the FGF19–FGFR4-MAPK pathway
FOXC1 positively mediates expression of FGF19 in both corneal and periocular mesenchymal cells which are from zebrafish embryos or cultured cells. FGF19 was confirmed as a downstream target of FOXC1 in the eye. FGF19 increases MAPK phosphorylation by the FGFR4 tyrosine kinase in the both developing and mature cornea. Knockout of FOXC1 expression or FGF19 expression caused complementary, but distinct, anterior segment dysgeneses. FOXC1 plays an important role in both the development and maintenance of anterior segment structures within the eye by activating FGF19–FGFR4-MAPK pathway [49].
FOXC1 activated MSX1- NKX3-1 pathway
FOXC1 mediated activation of MSX1 transcription. Overexpression of FOXC1 induced upregulation of MSX1. Overexpression of the both transcriptional factor GATA3 and TAL1 significantly decreased expression level of MSX1 in PER-117 cells while knockdown of GATA3 and TAL1 markedly induced expression of MSX1 in JURKAT cells. Knockdown of MSX1 reduced expression of NKX3-1, which demonstrated that MSX1 activated NKX3-1 as well. MSX1 regulated the NKL homeobox gene NKX3-1 which may play an important role in T-cell acute lymphoblastic leukemia(T-ALL)[50].
MST1R/PI3K/AKT pathway was activated by FOXC1
FOXC1 play an important role in melanoma cells, such as invasion, migration and proliferation, colony formation and growth in 3D Matrigel. Ectopic FOXC1 expression induced MST1R expression and promoted migration and invasion of melanoma cells by activating the MST1R/PI3K/AKT pathway. Inhibition of PI3K/AKT significantly reduced migration and invasion mediated by FOXC1 [30].
FOXC1 regulated SCF and CXCL12 expression
FOXC1 is found to be expressed in the CXC chemokine ligand (CXCL) 12-abundant reticular (CAR) cells which are necessary for maintenance of haematopoietic stem and progenitor cell in vivo in the developing and adult bone marrow. Haematopoietic stem and progenitor cells were significantly reduced if FOXC1 was knock out in CAR cells or marrow mesenchymal cells. FOXC1 upregulates expression of CXCL12 and stem cell factor (SCF) and promotes development of CAR cells. FOXC1 plays an important role in maintaining the mesenchymal niches for haematopoietic stem and progenitor cells [37].
FOXC1 activated Nfatc1 and bone morphogenetic protein (BMP) signaling
Expression of FOXC1 is increased when murine hair follicle stem cells (SCs) is activated. SCs can’t reestablish quiescence and allow premature activation if FOXC1 is deleted in activated SCs. The loss of old hair was caused by knockdown of FOXC1 in the SC. FOXC1 mediates BMP and Nfatc1 signal pathways to govern quiescence in self-renewing SCs [39].
FOXC1 regulated expression of Myh7 during cardiac development
FOXC1 regulated many pathways which are implicated in cardiac functions, such as calcium signaling, actin cytoskeleton, gap and tight junctions, and cell adhesion. Expression of Myh7 is regulated by FOXC1 during cardiac development. FOXC1 mediated the functional properties of cardiomyocytes which are derived from ESC [40].
CONCLUSIONS AND FUTURE PERSPECTIVES
Up to now, it is unclear how FOXC1 expression is regulated. Some studies showed that epigenetic changes of FOXC1 were involved in FOXC1 expression. It is very important to study FOXC1 expression regulated by epigenetics and elucidate the mechanism of FOXC1 expression in the future. Study and RNA-Seq data showed that expression FOXC1 was involved with inflammation. More studies will be focused on the effects of FOXC1 on inflammation in cancer microenvironment. More importantly, there is no inhibitor of FOXC1. It is necessary to establish high throughput screen model for screening FOXC1 inhibitor and develop FOXC1 inhibitor as an anti-cancer drug.
FOXC1, an important member of FOX family, is highly expressed and plays critical roles in many cancers and stem cells. FOXC1 can be used as a potential biomarker of diseases.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81573454) and supported by Beijing Natural Science Foundation (7172142). This work was also supported by CAMS Innovation Fund for Medical Sciences (2016-I2M-3-007).
Footnotes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Yamashita H, Amponsa VO, Warrick JI, Zheng Z, Clark PE, Raman JD, Wu XR, Mendelsohn C, DeGraff DJ. On a FOX hunt: functions of FOX transcriptional regulators in bladder cancer. Nat Rev Urol. 2017;14:98–106. doi: 10.1038/nrurol.2016.239. [DOI] [PubMed] [Google Scholar]
- 2.Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Benayoun BA, Caburet S, Veitia RA. Forkhead transcription factors: key players in health and disease. Trends Genet. 2011;27:224–32. doi: 10.1016/j.tig.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. Hum Genomics. 2010;4:345–52. doi: 10.1186/1479-7364-4-5-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet. 1998;19:140–7. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 6.Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, Marshall J, Gould DB, Pearce W, et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet. 1998;63:1316–28. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komatireddy S, Chakrabarti S, Mandal AK, Reddy AB, Sampath S, Panicker SG, Balasubramanian D. Mutation spectrum of FOXC1 and clinical genetic heterogeneity of Axenfeld-Rieger anomaly in India. Mol Vis. 2003;9:43–8. [PubMed] [Google Scholar]
- 8.Cella W, de Vasconcellos JP, de Melo MB, Kneipp B, Costa FF, Longui CA, Costa VP. Structural assessment of PITX2, FOXC1, CYP1B1, and GJA1 genes in patients with Axenfeld-Rieger syndrome with developmental glaucoma. Invest Ophthalmol Vis Sci. 2006;47:1803–9. doi: 10.1167/iovs.05-0979. [DOI] [PubMed] [Google Scholar]
- 9.Berry FB, Lines MA, Oas JM, Footz T, Underhill DA, Gage PJ, Walter MA. Functional interactions between FOXC1 and PITX2 underlie the sensitivity to FOXC1 gene dose in Axenfeld-Rieger syndrome and anterior segment dysgenesis. Hum Mol Genet. 2006;15:905–19. doi: 10.1093/hmg/ddl008. [DOI] [PubMed] [Google Scholar]
- 10.Panicker SG, Sampath S, Mandal AK, Reddy AB, Ahmed N, Hasnain SE. Novel mutation in FOXC1 wing region causing Axenfeld-Rieger anomaly. Invest Ophthalmol Vis Sci. 2002;43:3613–6. [PubMed] [Google Scholar]
- 11.Kawase C, Kawase K, Taniguchi T, Sugiyama K, Yamamoto T, Kitazawa Y, Alward WL, Stone EM, Nishimura DY, Sheffield VC. Screening for mutations of Axenfeld-Rieger syndrome caused by FOXC1 gene in Japanese patients. J Glaucoma. 2001;10:477–82. doi: 10.1097/00061198-200112000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Micheal S, Siddiqui SN, Zafar SN, Villanueva-Mendoza C, Cortes-Gonzalez V, Khan MI, den Hollander AI. A novel homozygous mutation in FOXC1 causes Axenfeld Rieger syndrome with congenital glaucoma. PLoS One. 2016;11:e0160016. doi: 10.1371/journal.pone.0160016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar M, Chambers C, Dhamija R. Axenfeld-Rieger syndrome and leukoencephalopathy caused by a mutation in FOXC1. Pediatr Neurol. 2017;66:113–4. doi: 10.1016/j.pediatrneurol.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Pasutto F, Mauri L, Popp B, Sticht H, Ekici A, Piozzi E, Bonfante A, Penco S, Schlotzer-Schrehardt U, Reis A. Whole exome sequencing reveals a novel de novo FOXC1 mutation in a patient with unrecognized Axenfeld-Rieger syndrome and glaucoma. Gene. 2015;568:76–80. doi: 10.1016/j.gene.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Kaur K, Ragge NK, Ragoussis J. Molecular analysis of FOXC1 in subjects presenting with severe developmental eye anomalies. Mol Vis. 2009;15:1366–73. [PMC free article] [PubMed] [Google Scholar]
- 16.Ray PS, Wang J, Qu Y, Sim MS, Shamonki J, Bagaria SP, Ye X, Liu B, Elashoff D, Hoon DS, Walter MA, Martens JW, Richardson AL, et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010;70:3870–6. doi: 10.1158/0008-5472.CAN-09-4120. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Ray PS, Sim MS, Zhou XZ, Lu KP, Lee AV, Lin X, Bagaria SP, Giuliano AE, Cui X. FOXC1 regulates the functions of human basal-like breast cancer cells by activating NF-kappaB signaling. Oncogene. 2012;31:4798–802. doi: 10.1038/onc.2011.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray PS, Bagaria SP, Wang J, Shamonki JM, Ye X, Sim MS, Steen S, Qu Y, Cui X, Giuliano AE. Basal-like breast cancer defined by FOXC1 expression offers superior prognostic value: a retrospective immunohistochemical study. Ann Surg Oncol. 2011;18:3839–47. doi: 10.1245/s10434-011-1657-8. [DOI] [PubMed] [Google Scholar]
- 19.Jin Y, Han B, Chen J, Wiedemeyer R, Orsulic S, Bose S, Zhang X, Karlan BY, Giuliano AE, Cui Y, Cui X. FOXC1 is a critical mediator of EGFR function in human basal-like breast cancer. Ann Surg Oncol. 2014;21:S758–66. doi: 10.1245/s10434-014-3980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han B, Qu Y, Jin Y, Yu Y, Deng N, Wawrowsky K, Zhang X, Li N, Bose S, Wang Q, Sakkiah S, Abrol R, Jensen TW, et al. FOXC1 activates smoothened-independent hedgehog signaling in basal-like breast cancer. Cell Rep. 2015;13:1046–58. doi: 10.1016/j.celrep.2015.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sizemore ST, Keri RA. The forkhead box transcription factor FOXC1 promotes breast cancer invasion by inducing matrix metalloprotease 7 (MMP7) expression. J Biol Chem. 2012;287:24631–40. doi: 10.1074/jbc.M112.375865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tkocz D, Crawford NT, Buckley NE, Berry FB, Kennedy RD, Gorski JJ, Harkin DP, Mullan PB. BRCA1 and GATA3 corepress FOXC1 to inhibit the pathogenesis of basal-like breast cancers. Oncogene. 2012;31:3667–78. doi: 10.1038/onc.2011.531. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Xu Y, Li L, Wang L, Yao R, Sun Q, Du G. FOXC1 is associated with estrogen receptor alpha and affects sensitivity of tamoxifen treatment in breast cancer. Cancer Med. 2017;6:275–87. doi: 10.1002/cam4.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia L, Huang W, Tian D, Zhu H, Qi X, Chen Z, Zhang Y, Hu H, Fan D, Nie Y, Wu K. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2013;57:610–24. doi: 10.1002/hep.26029. [DOI] [PubMed] [Google Scholar]
- 25.Xu ZY, Ding SM, Zhou L, Xie HY, Chen KJ, Zhang W, Xing CY, Guo HJ, Zheng SS. FOXC1 contributes to microvascular invasion in primary hepatocellular carcinoma via regulating epithelial-mesenchymal transition. Int J Biol Sci. 2012;8:1130–41. doi: 10.7150/ijbs.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W, Chen Z, Zhang L, Tian D, Wang D, Fan D, Wu K, Xia L. Interleukin-8 induces expression of FOXC1 to promote transactivation of CXCR1 and CCL2 in hepatocellular carcinoma cell lines and formation of metastases in mice. Gastroenterology. 2015;149:1053–67 e14. doi: 10.1053/j.gastro.2015.05.058. [DOI] [PubMed] [Google Scholar]
- 27.Wei LX, Zhou RS, Xu HF, Wang JY, Yuan MH. High expression of FOXC1 is associated with poor clinical outcome in non-small cell lung cancer patients. Tumour Biol. 2013;34:941–6. doi: 10.1007/s13277-012-0629-3. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Jiao S, Jia Y, Li Y. Effects of targeted silencing of FOXC1 gene on proliferation and in vitro migration of human non-small-cell lung carcinoma cells. Am J Transl Res. 2016;8:3309–18. [PMC free article] [PubMed] [Google Scholar]
- 29.Somerville TD, Wiseman DH, Spencer GJ, Huang X, Lynch JT, Leong HS, Williams EL, Cheesman E, Somervaille TC. Frequent derepression of the mesenchymal transcription factor gene FOXC1 in acute myeloid leukemia. Cancer Cell. 2015;28:329–42. doi: 10.1016/j.ccell.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Li L, Liu S, Zhao Y, Wang L, Du G. FOXC1 promotes melanoma by activating MST1R/PI3K/AKT. Oncotarget. 2016;7:84375–87. doi: 10.18632/oncotarget.11224. https://doi.org/10.18632/oncotarget.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Shao QS, Yao HB, Jin Y, Ma YY, Jia LH. Overexpression of FOXC1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2014;64:963–70. doi: 10.1111/his.12347. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Gu F, Liu CY, Wang RJ, Li J, Xu JY. High level of FOXC1 expression is associated with poor prognosis in pancreatic ductal adenocarcinoma. Tumour Biol. 2013;34:853–8. doi: 10.1007/s13277-012-0617-7. [DOI] [PubMed] [Google Scholar]
- 33.Pan F, Yao J, Chen Y, Zhou C, Geng P, Mao H, Fang X. A novel long non-coding RNA FOXCUT and mRNA FOXC1 pair promote progression and predict poor prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:2838–49. [PMC free article] [PubMed] [Google Scholar]
- 34.van der Heul-Nieuwenhuijsen L, Dits NF, Jenster G. Gene expression of forkhead transcription factors in the normal and diseased human prostate. BJU Int. 2009;103:1574–80. doi: 10.1111/j.1464-410X.2009.08351.x. [DOI] [PubMed] [Google Scholar]
- 35.Hirschi KK, Majesky MW. Smooth muscle stem cells. Anat Rec A Discov Mol Cell Evol Biol. 2004;276:22–33. doi: 10.1002/ar.a.10128. [DOI] [PubMed] [Google Scholar]
- 36.Ittner LM, Wurdak H, Schwerdtfeger K, Kunz T, Ille F, Leveen P, Hjalt TA, Suter U, Karlsson S, Hafezi F, Born W, Sommer L. Compound developmental eye disorders following inactivation of TGFbeta signaling in neural-crest stem cells. J Biol. 2005;4:11. doi: 10.1186/jbiol29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Omatsu Y, Seike M, Sugiyama T, Kume T, Nagasawa T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature. 2014;508:536–40. doi: 10.1038/nature13071. [DOI] [PubMed] [Google Scholar]
- 38.Lay K, Kume T, Fuchs E. FOXC1 maintains the hair follicle stem cell niche and governs stem cell quiescence to preserve long-term tissue-regenerating potential. Proc Natl Acad Sci U S A. 2016;113:E1506–15. doi: 10.1073/pnas.1601569113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Siegenthaler JA, Dowell RD, Yi R. Foxc1 reinforces quiescence in self-renewing hair follicle stem cells. Science. 2016;351:613–7. doi: 10.1126/science.aad5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambers E, Arnone B, Fatima A, Qin G, Wasserstrom JA, Kume T. Foxc1 regulates early cardiomyogenesis and functional properties of embryonic stem cell derived cardiomyocytes. Stem Cells. 2016;34:1487–500. doi: 10.1002/stem.2301. [DOI] [PubMed] [Google Scholar]
- 41.Gripp KW, Hopkins E, Jenny K, Thacker D, Salvin J. Cardiac anomalies in Axenfeld-Rieger syndrome due to a novel FOXC1 mutation. Am J Med Genet A. 2013;161A:114–9. doi: 10.1002/ajmg.a.35697. [DOI] [PubMed] [Google Scholar]
- 42.Swiderski RE, Reiter RS, Nishimura DY, Alward WL, Kalenak JW, Searby CS, Stone EM, Sheffield VC, Lin JJ. Expression of the Mf1 gene in developing mouse hearts: implication in the development of human congenital heart defects. Dev Dyn. 1999;216:16–27. doi: 10.1002/(SICI)1097-0177(199909)216:1<16::AID-DVDY4>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 43.Seo S, Kume T. Forkhead transcription factors, Foxc1 and Foxc2, are required for the morphogenesis of the cardiac outflow tract. Dev Biol. 2006;296:421–36. doi: 10.1016/j.ydbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Seo S, Fujita H, Nakano A, Kang M, Duarte A, Kume T. The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev Biol. 2006;294:458–70. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 45.Kume T, Deng K, Hogan BL. Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development. 2000;127:1387–95. doi: 10.1242/dev.127.7.1387. [DOI] [PubMed] [Google Scholar]
- 46.Nakano T, Niimura F, Hohenfellner K, Miyakita E, Ichikawa I. Screening for mutations in BMP4 and FOXC1 genes in congenital anomalies of the kidney and urinary tract in humans. Tokai J Exp Clin Med. 2003;28:121–6. [PubMed] [Google Scholar]
- 47.Kume T, Jiang H, Topczewska JM, Hogan BL. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001;15:2470–82. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouvagnet P. Unraveling de novo copy number variants in congenital heart defects: the bottom of the iceberg is under attack. Circ Cardiovasc Genet. 2016;9:4–5. doi: 10.1161/CIRCGENETICS.116.001360. [DOI] [PubMed] [Google Scholar]
- 49.Tamimi Y, Skarie JM, Footz T, Berry FB, Link BA, Walter MA. FGF19 is a target for FOXC1 regulation in ciliary body-derived cells. Hum Mol Genet. 2006;15:3229–40. doi: 10.1093/hmg/ddl400. [DOI] [PubMed] [Google Scholar]
- 50.Nagel S, Ehrentraut S, Meyer C, Kaufmann M, Drexler HG, MacLeod RA. Repressed BMP signaling reactivates NKL homeobox gene MSX1 in a T-ALL subset. Leuk Lymphoma. 2015;56:480–91. doi: 10.3109/10428194.2014.924119. [DOI] [PubMed] [Google Scholar]