Abstract
Intraductal carcinoma (IC) is the WHO designation for lesions previously called low-grade cribriform cystadenocarcinoma. The relationship of IC to salivary duct carcinoma (SDC) is controversial, but currently these are considered distinct entities. It is hypothesized that IC and SDC should have different genomic signatures that may be identifiable by next-generation-sequencing. A total of 23 ICs were identified: 14 pure IC and 9 invasive carcinomas with an intraductal component. Five invasive carcinomas were subjected to next-generation paired-end RNA-sequencing (RNAseq). Data analysis was performed using FusionSeq and Mutation detection algorithms (MuTect & VarScan) for variant callers. Gene fusion candidates were validated by FISH and RT-PCR, and mutations by Sanger sequencing. Among the 9 invasive carcinomas, all except one were apocrine SDCs with an intraductal component. The remaining case showed typical intercalated duct type IC with invasive adenocarcinoma. The 14 pure ICs had typical intercalated duct features (2 showed hybrid intercalated/apocrine features). RNAseq predicted a NCOA4-RET fusion, confirmed by RT-PCR, in the intercalated duct type IC invasive component. Six additional cases of pure IC showed RET rearrangement by FISH (7/15=47%). No apocrine carcinomas showed RET rearrangement. RNAseq and Sanger sequencing identified PIK3CA (p.E545K/p.H1047R) and/or HRAS (p.Q61R) hotspot mutations in 6/8 (75%) apocrine carcinomas. In conclusion, two distinctive types of intraductal lesions are emerging based on molecular analysis. Classic intercalated type ICs commonly harbour fusions involving RET and rarely show widespread invasion. Apocrine intraductal lesions are typically associated with widespread invasion with no pure examples and show similar PIK3CA and HRAS mutations to SDC.
INTRODUCTION
In recent years most of the common low to intermediate-grade salivary gland cancers have been found to have pathognomonic mutations and translocations. These alterations are generally present in the majority of cases and are specific within salivary gland for the entity in question. This includes CRTC1-MAML2 in mucoepidermoid carcinoma, MYB-NFIB in adenoid cystic carcinoma, EWSR1-ATF1 in hyalinizing clear cell carcinoma, ETV6-NTRK3 in mammary analogue secretory carcinoma, PRKD1 E.710D hotspot mutation in polymorphous low-grade adenocarcinoma and PRKD1-3 rearrangements in the related cribriform adenocarcinoma of salivary gland (1–6). On the other hand, high-grade salivary gland carcinomas, such as salivary duct carcinoma and adenocarcinoma, not otherwise specified (NOS) generally are more heterogeneous with no single genetic alteration accounting for a majority of cases (7–9). Common mutations in salivary duct carcinoma (SDC) include TP53 mutation, Her-2-Neu amplification, PIK3CA mutation and HRAS mutation (7–9).
One enigmatic low-grade salivary gland cancer that has yet to be genomically characterized is the so-called “low-grade cribriform cystadenocarcinoma”, recently classified by the World Health Organization (WHO) 2017 as “intraductal carcinoma” (10). This tumour has been surrounded by controversy since its discovery as it has gone by a number of terminologies, including “intraductal carcinoma”, “low-grade salivary duct carcinoma”, “salivary duct carcinoma in-situ” and “low-grade cribriform cystadenocarcinoma”, and because its relationship to SDC has remained uncertain. The term “low-grade salivary duct carcinoma” may be confusing to clinicians and this confusion may lead to unnecessary treatment. On the other hand, the term “low-grade cribriform cystadenocarcinoma” has been equally criticized for implying an invasive carcinoma and thus the term “intraductal carcinoma” (IC) was adopted by the WHO 2017 to clarify its generally non-invasive nature. The classic example of IC shows features similar to mammary atypical ductal hyperplasia or ductal carcinoma in-situ, is considered histologically low-grade and shows diffuse S100 positivity. These features are in sharp contrast to conventional SDC and therefore these lesions are generally accepted as distinct. This paradigm does not however fully answer the question of whether these lesions are at all related to salivary duct carcinoma, nor does it account for occasional well-documented IC with subsequent widespread invasion (11).
There have been no genomic studies to investigate the relationship of these tumours with an intraductal growth pattern and specifically no genomic studies of pure S100 positive IC. A group of tumours on the morphologic spectrum of pure IC, intraductal and invasive carcinomas with S100 positivity, and apocrine salivary duct carcinoma with an apocrine appearing intraductal component, were collected and subjected to various molecular methods. This included whole transcriptome RNA sequencing for novel fusion discovery and mutation analysis. ETV6 FISH was also investigated on ICs to explore the relationship to MASC, which shares architectural features and S100 positivity.
MATERIAL AND METHODS
The institutional pathology files of the authors were searched for the diagnosis of “Intraductal Carcinoma”, “Low-grade cribriform cystadenocarcinoma”, “Low-grade salivary duct carcinoma”, “Ductal Carcinoma In-Situ” and “Salivary Duct Carcinoma”, the latter with any additional reference to an intraductal component. Hematoxylin and Eosin (H&E) stained slides were available for review in all cases. Tumours were classified as being purely intraductal when they were composed of any combination of solid, cribriform and papillary-cystic nests surrounded by non-neoplastic myoepithelial cells and showing a lobular growth pattern. These lesions had the appearance of mammary type atypical ductal hyperplasia or ductal carcinoma in-situ (DCIS) of any grade. Tumours were classified as being both intraductal and invasive when a clearly defined infiltrative high-grade carcinoma showed a background component similar to a pure intraductal lesion. Purely invasive tumours were excluded. Invasive tumours with debatable focal intraductal components were excluded as well as they were deemed to potentially represent “cancerization of ducts” by the invasive component as opposed to having a true precursor lesion, as defined by Bahrami et al (12). The tumours were further classified into intercalated duct type intraductal carcinomas when they showed S100 positivity and small eosinophilic to amphophilic cuboidal cells, and apocrine type intraductal carcinomas when they showed AR positivity and/or abundant granular eosinophilic cytoplasm, large nucleoli and apocrine snouts and secretions (see results section for further details).
Fluorescence In-Situ Hybridization (FISH)
FISH was performed on 4 micron thick sections of formalin fixed paraffin embedded tissue on all cases. Custom bacterial artificial chromosome (BAC) probes, flanking the RET, KIF13B, KIF5B, ARID1A, NCOA4, ALK and ROS1 genes, were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org) and were chosen according to the UCSC genome browser (http://genome.ucsc.edu) (Supplementary Table 1). FISH was performed as previously described (13). Briefly, DNA from individual BACs was isolated according to the manufacturer’s instructions and labeled with different fluorochromes in a nick translation reaction (13). They were then denatured, hybridized to pretreated unstained coated slides, incubated, washed, and mounted with DAPI in an antifade solution (13). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred non-overlapping nuclei were scored using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Watertown, MA, USA). A case was confirmed as positive for rearrangement when ≥ 20% of the nuclei examined showed a break-apart signal pattern. In addition, FISH for ETV6 was previously performed on all pure intraductal lesions included in the study using a commercially available probe (Abbot Molecular, Des Plains, IL, USA) and analyzed similarly to those tested by BAC probes.
RNA Sequencing
Total RNA was prepared for RNA sequencing in accordance with the standard Illumina mRNA sample preparation protocol (Illumina), as previously described (5). Briefly, mRNA was isolated with oligo(dT) magnetic beads from total RNA (2 μg) extracted from the index cases. The mRNA was fragmented by incubation at 94°C for 2.5 min in fragmentation buffer. Artifactual chimeric transcripts due to random priming of transcript fragments into the sequencing library (because of inefficient A-tailing reactions) were excluded with an additional gel size-selection step (capturing 350–400 bp) prior to the adapter ligation step (14). The adaptor-ligated library was then enriched by PCR for 15 cycles and purified. The library was sized and quantified using DNA1000 kit (Agilent) on an Agilent 2100 Bioanalyzer according to the manufacturer’s instructions. Paired-end RNA-sequencing at read lengths of 51 bp was performed with the HiSeq 2000 (Illumina).
Analysis of RNA Sequencing Results with FusionSeq
The RNA sequencing and FusionSeq analysis was performed as previously described (5) on 5 cases of invasive carcinoma with a prominent intraductal component. All reads were independently aligned with STAR alignment software against the human genome reference sequence (hg19) and a splice junction library, simultaneously. The mapped reads were converted into Mapped Read Format (15) and analyzed with FusionSeq (16) to identify potential fusion transcripts. The FusionSeq algorithm is a computational method successfully applied to paired-end RNA-seq data for the identification of chimeric fusion transcripts (17). Briefly, paired-end reads mapped to different genes are first used to identify candidate chimeric transcripts. A cascade of filters, each taking into account different artifacts in RNA-sequencing results was then applied to remove spurious fusion transcript candidates, such as “read throughs”. A list of fusion candidates was generated which were statistically ranked according to their probability of being a significant result. This was to prioritize multiple fusion candidates for experimental validation. Usually, only one significant candidate is produced from this data in any given case. In this study, the DASPER score was used to assign a confidence weight to the potential candidates as previously described (5, 17). The DASPER is the difference between the observed and analytically calculated expected SPER with a higher DASPER score indicating a greater likelihood that the potential fusion candidate is authentic. See Sboner A, et al. (16) for further details about FusionSeq. In addition, RNA sequencing data from all 5 cases was also used for gene mutation calls, as previously described (18). Briefly, BAM files were generated by STAT alignment, followed by PicardTools (ver. 1.130) standard preprocessing. MuTect (ver. 1.15) and VarScan (ver. 2.3.8) variant callers were used for mutation detection, followed by vcf2maf for converting VCF into MAF files, with the annotation added by the Variant Effect Predictor tool provided by Ensembl. Sanger sequencing validation was performed subsequently on the cases and was also performed on all invasive carcinomas in the study.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
An RNA aliquot of the index invasive carcinoma with a prominent intraductal carcinoma in the background was used for RT-PCR to confirm the potential novel fusion transcripts. The RNA quality was determined by Eukaryote Total RNA Nano Assay and cDNA quality was tested for PGK housekeeping gene (247 bp amplified product). RT-PCR was performed using the advantage 2 PCR kit (Clonetech, Mountain View, CA). RT-PCR was run using the following primers: NCOA4 exon 6 fwd: 5′-CACTTGATGGCTCATGCTAGTTCAG-3′ and RET exon 12 rev: 5′-GTGTACCCTGCTCTGCCTTTCAG-3′). The PCR product was confirmed by Sanger sequencing in this case.
RESULTS
Histologic and Clinical Findings
The study cohort consisted of 9 invasive carcinomas with an intraductal component and 14 additional pure intraductal carcinomas with no invasion identified (see Table 1). The patients with available clinical information included 16 males and 5 females, with a wide age-range at diagnosis of 25–80 years (mean 61.3 years). The invasive carcinomas occurred in patients ranging from 38–82 years (mean 60.4 years) and the pure intraductal carcinomas ranged similarly from 25–80 years old (mean 62.2). The invasive carcinomas were mostly interpreted as salivary duct carcinoma (SDC) at the time of diagnosis. The tumours occurred exclusively in the parotid gland for both groups. The tumours ranged from 0.5 to 4.1 cm (mean 1.9 cm), although the mean size for the invasive tumours was 2.4 cm while the mean size for the pure intraductal carcinomas was 1.4 cm.
Table 1.
Clinicopathologic and Molecular Features
| Case# | Age/Sex | Diagnosis | Next Generation Sequencing Results | S100/AR | RET FISH | Other FISH | ETV6 FISH | Direct Sequencing |
|---|---|---|---|---|---|---|---|---|
| 1* | 82M | Intraductal and invasive adenocarcinoma of intercalated duct type. | NCOA4-RET | +/− | − | − | ND | NCOA4-RET |
| 2 | NA | Intraductal carcinoma with hybrid intercalated duct and apocrine features. | ND | +/F+ | Pos** | − | ND | ND |
| 3 | 25M | Intraductal carcinoma of intercalated duct type. | ND | +/ND | − | − | − | ND |
| 4 | 56M | Intraductal carcinoma of intercalated duct type. | ND | +/ND | − | − | − | ND |
| 5 | 80M | Intraductal carcinoma of intercalated duct type. | ND | +/ND | Pos** | − | − | ND |
| 6 | 72M | Intraductal carcinoma of intercalated duct type. | ND | +/ND | − | − | − | ND |
| 7 | 80M | Intraductal carcinoma of intercalated duct type. | ND | +/ND | − | − | − | ND |
| 8 | 53M | Intraductal carcinoma of intercalated duct type. | ND | +/ND | − | − | − | ND |
| 9 | 62F | Intraductal carcinoma of intercalated duct type. | ND | +/ND | − | − | − | ND |
| 10 | NA | Intraductal carcinoma of intercalated duct type. | ND | +/− | − | − | − | ND |
| 11 | 66F | Intraductal carcinoma of intercalated duct type. | ND | +/ND | − | − | − | ND |
| 12 | 73F | Intraductal carcinoma of intercalated duct type. | ND | +/F+ | Pos** | ND | − | ND |
| 13 | 62F | Intraductal carcinoma of intercalated duct type. | ND | +/− | Pos** | ND | − | ND |
| 14 | 50M | Intraductal carcinoma with macrocystic architecture and papillary changes. | ND | +/− | Pos** | ND | − | ND |
| 15 | 66M | Intraductal carcinoma with hybrid intercalated duct and apocrine areas. | ND | +/F+ | Posˆ | ND | ND | ND |
| 16* | 67M | Apocrine invasive and focally intraductal carcinoma. | PIK3CA p.E545K | ND/+ | − | − | ND | PIK3CA p.E545K |
| 17* | 58M | Apocrine invasive and focally intraductal carcinoma. | HRAS p.Q61R PIK3CA p.H1047R | ND/+ | − | − | ND | HRAS p.Q61R PIK3CA p.H1047R |
| 18* | 74M | Apocrine invasive and focally intraductal carcinoma. | DFFA-ARID1AHRAS p.Q61R PIK3CA p.1047R | ND/+ | − | ARID1A Positive | ND | HRAS p.Q61R PIK3CA p.1047R |
| 19* | 38M | Focally invasive apocrine carcinoma with predominant intraductal architecture. | KIF13B-EPB41L4B HRAS p.Q61R | −/+ | − | KIF13B Positive | ND | HRAS p.Q61R |
| 20 | 70M | Apocrine invasive and focally intraductal carcinoma. | ND | −/+ | − | − | ND | PIK3CA p.E545K |
| 21 | 42M | Apocrine invasive and focally intraductal carcinoma. | ND | ND/+ | − | − | ND | PIK3CA p.H1047R |
| 22 | 46M | Apocrine invasive and focally intraductal carcinoma. | ND | ND/+ | − | − | ND | − |
| 23 | 67F | Apocrine invasive and focally intraductal carcinoma. | ND | −/+ | − | − | ND | − |
These cases represented the index cases sequenced by RNA sequencing and analyzed by FusionSeq.
Case 1 showed an NCOA4-RET fusion gene which was confirmed by RT-PCR. RET FISH failed to detect this inversion/rearrangement.
Case #s 2, 5, and 12–14 showed a RET FISH signal pattern indicative of an inversion/rearrangement.
Case #15 showed a classic break-apart signal pattern. NA=not available. ND=not done F+=focally positive.
The tumours were classified further into 2 groups: those that showed an intercalated duct phenotype and those with an apocrine phenotype. This was determined both histologically and immunohistochemically. A summary of the clinicopathologic findings is found in Table 1. Intercalated duct-like neoplasms morphologically showed smaller cells with amphophilic to eosinophilic cytoplasm and small nuclei, similar to normal intercalated ducts. They sometimes showed apocrine differentiation with two cases showing hybrid features with marked apocrine areas. They were typically S100 positive and AR negative, similar to other intercalated duct lesions and carcinomas (eg. intercalated duct hyperplasia and epithelial-myoepithelial carcinomas). All 14 pure intraductal carcinomas were S100 positive and three variably co-expressed AR, with two of these being the cases showing hybrid intercalated and apocrine features. A single case (case #14) showed a macrocystic architecture lined by papillary tufts and having vacuolated cells with pigment, superficially resembling MASC. These lesions were all considered “intercalated duct type” for the purposes of this study and are labeled as group 1. A single invasive carcinoma (case #1) was also S100 positive and AR negative, and showed the same immunophenotype in the intraductal carcinoma component, which showed typical features similar to the other pure intraductal neoplasms. This accounted for a total of 15 cases harboring an intercalated duct type intraductal carcinoma. The invasive carcinoma in this case showed an unusual combination of cystic change, micropapillary tufts and detached papillary elements floating in the cysts, reminiscent of “spread through alveolar spaces” in pulmonary adenocarcinomas (Fig. 1a–d). The remaining cases were all widely invasive and showed morphologic evidence of pure apocrine differentiation and higher-grade histology and represented group 2 lesions. This included apocrine snouts, decapitation secretions and large nuclei with prominent nucleoli. These tumours were best defined as salivary duct carcinoma (SDC) with varying morphologies (Fig. 2a–d). Some were typical SDC with high-grade apocrine carcinomas showing comedonecrosis, while others were more tubular appearing apocrine carcinomas. The intraductal lesions in this group showed similar apocrine features and lacked any evidence of the intercalated duct morphology seen in group 1 lesions. These tumours were always AR positive, although one case showed AR positivity only in the intraductal component (case #23) with complete AR loss in the invasive carcinoma. This case has been described previously (11). All 3 apocrine cases previously tested were S100 negative.
Fig. 1.
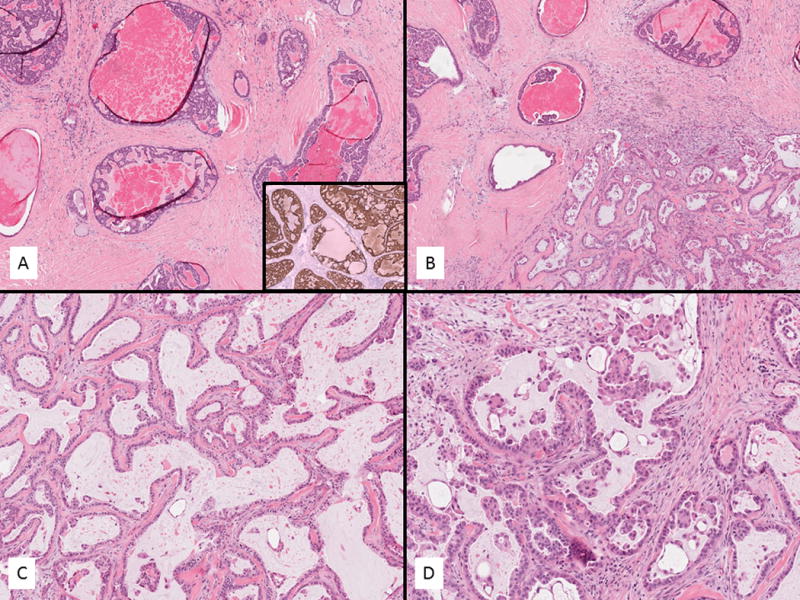
Index case for sequencing of intraductal and invasive adenocarcinoma of intercalated duct type (case 1) (A–D). The intraductal component showed small bland cells forming solid and cribriform type nests with amphophilic cytoplasm (A). This component and all other intraductal carcinomas were S100 positive (Inset A). The invasive component showed cystic change with a fibrotic stroma and displayed prominent papillary tufts (B–C). The transition zone is seen in B. The papillary tufts floated in the spaces mimicking “spread through alveolar spaces” in pulmonary adenocarcinoma (D).
Fig. 2.
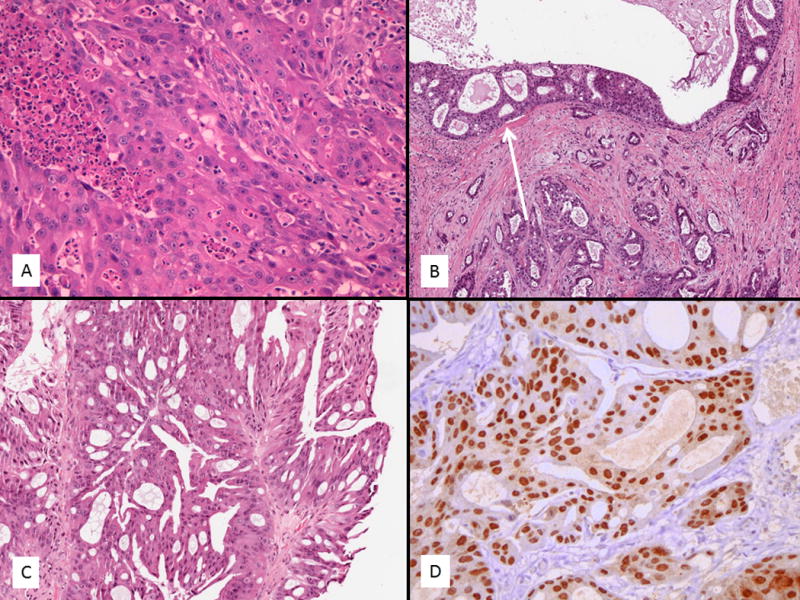
Apocrine/salivary duct adenocarcinomas were always invasive with varying morphologies and showed focal intraductal components (A–D). Some cases showed typical features of salivary duct carcinoma with high-grade solid nests and comedonecrosis (A). A number of cases showed a prominent tubular morphology (B) and all cases had at least a focal intraductal component (arrow). A single case in a 38 year old showed a low to intermediate grade apocrine carcinoma not typical of conventional salivary duct carcinoma (C). However, all cases were androgen receptor (AR) positive (D) and considered in the salivary duct carcinoma spectrum.
Mitotic activity was absent in all pure intraductal lesions and was very brisk in all invasive carcinomas. This did not contribute to the diagnostic grouping. The invasive carcinomas generally showed extensive atypia and necrosis and the intraductal lesions associated with the invasive apocrine carcinomas generally showed a range of atypia from low to high-grade atypia/dysplasia. The 2 exceptions to this were the invasive intercalated duct type carcinoma (case #1) that showed only low-grade atypia in the intraductal component, as well as a 38 year old patient with an apocrine carcinoma with only low to intermediate grade atypia/dysplasia in both the intraductal and invasive components (case #19). This latter lesion was not considered a typical SDC but was grouped in the apocrine subtype (group 2).
Follow up and treatment was not available on the pure intraductal group as these were often incidental lesions or lost to follow up without treatment beyond surgery. Based on previous literature for pure intraductal lesions, they are deemed likely cured of disease, as no recurrences or distant metastases have been reported to date. In the invasive group, there were 8 invasive apocrine carcinomas (SDC) with 5 presenting with lymph node metastases. A total of 7 had follow up ranging from 17–96 months (mean 43 mths). Four patients showed no evidence of disease with follow up ranging from 17–96 months (mean 37 mths). One patient showed a skull base recurrence at 43 months, a skin recurrence at 72 months and was deemed palliative at last follow up at 111 months. Two patients had distant metastases: one to bone and pleura at 16 and 18 months, and the other to bone, brain and lung at 61 months. These patients were palliative at last follow up at 20 and 61 months, respectively. The single patient with the intercalated duct type adenocarcinoma and intraductal component (case #1) showed no evidence of lymph node metastases.
Molecular Findings
A novel NCOA4-RET fusion was identified in the index case of Intraductal and Invasive Carcinoma of Intercalated Duct type
The samples with frozen material (cases #1 and #16–19) were subjected to RNA-sequencing to identify potential fusion candidates. An NCOA4-RET fusion transcript was identified by FusionSeq as the top candidate in case 1. Read alignments suggested a fusion of NCOA4 exon 6 on chromosome 10 with RET exon 12 on chromosome 10 (Fig. 3). NCOA4 and RET genes are located nearby on the 10q11.2 locus in opposite directions of transcription. Thus a functional intrachromosomal fusion would require a more complex, typically unbalanced, break and inversion of the RET gene. Indeed, FISH testing using break-apart probes for these closely located genes (RET and NCOA4) could not identify any structural abnormalities. However, RT-PCR confirmed the presence of a fusion transcript of NCOA4 exon 6 to exon 12 of the RET gene (Fig 3). The morphologic appearance of this tumour was that of an invasive adenocarcinoma with micropapillary tufts mimicking “spread through alveolar spaces” in pulmonary adenocarcinomas.
Fig. 3.
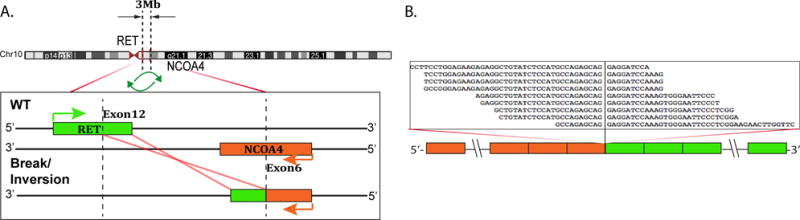
NCOA4-RET gene fusion in an invasive and intraductal adenocarcinoma with an intercalated duct phenotype (case #1). (A) Schematic representation of the NCOA4-RET fusion indicating the loci that are joined together; NCOA4 exon 6 being fused to RET exon 12 (left side of image). (B) RNA reads covering the fusion junction were isolated independent to FusionSeq analysis work flow, supporting the NCOA4-RET fusion candidate and confirmed by RT-PCR.
FISH screening identified RET gene rearrangements in 6 additional cases of pure Intraductal Carcinoma of the Intercalated Duct type
Despite a negative FISH result for RET rearrangement/inversion in case #1, FISH detected RET gene rearrangements in six of fourteen additional cases of pure intraductal carcinoma of the intercalated duct type (Fig. 4). Of the fifteen cases that showed an intercalated duct type intraductal carcinoma (14 pure intraductal and 1 intraductal and invasive), a total of 7/15 cases showed evidence of a fusion involving RET (47%). All seven cases represented S100 positive tumours that would also be classified as “low-grade cribriform cystadenocarcinoma” in the old WHO terminology (19). Two of these pure intraductal tumours positive for RET showed mixed intercalated and apocrine features and dual S100 and AR positivity (cases #2 and 15). In case #15 the apocrine areas lost S100 staining and gained AR positivity. A third case showed focal isolated AR positive cells (case #12). All cases with a pure apocrine morphology showed negativity for RET rearrangement by FISH. Because of the common occurrence of RET, ALK and ROS1 in lung adenocarcinomas and shared kinase activity for these genes, the RET-negative cases were also tested by FISH for ALK and ROS1 gene abnormalities, but none were positive.
Fig. 4.
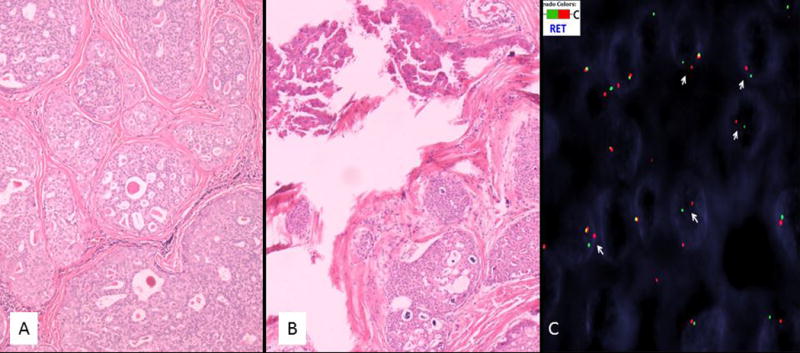
Six additional pure intraductal carcinoma of the intercalated duct type showed RET rearrangement by FISH (A–D). Four of these cases showed a pure intraductal carcinoma of the intercalated duct type (A) and the other two showed a mixed intercalated duct type intraductal carcinoma with apocrine areas (B). The apocrine areas in case #2 showed prominent papillary architecture (top left of image). S100 was positive in all pure intraductal cases (not shown). FISH for RET shows break apart signals with one normal fused yellow signal and separate 5′ green and 3′ red signals indicating rearrangement of the gene (arrows) (case #2). Note the consistent distance of the split signals owing to the putative inversion in this case, which was the case in all RET rearranged tumours other than case #15 that showed a classic break-apart signal (not shown).
Additional DFFA-ARID1A and KIF13B-EPB41L4B fusions were identified by RNA-seq in invasive apocrine/salivary duct carcinomas with an intraductal component
A DFFA-ARID1A fusion transcript was selected by FusionSeq as the top candidate in case #18. This was a high-grade apocrine carcinoma with a predominant tubular growth pattern and was largely invasive, typical of SDC. Read alignments suggested an intra-chromosomal fusion/inversion of DFFA exon 4 on chromosome 1 with ARID1A exon 8 on chromosome 1 (Supplementary Fig. 1). The ARID1A gene rearrangement was further validated by FISH (Supplementary Fig. 1).
Finally, a KIF13B-EPB41L4B fusion was selected by FusionSeq as the top candidate in case #19. This was the 38 year-old patient with an unusual low to intermediate grade apocrine carcinoma not typical of a salivary duct carcinoma. The tumour was located within the parotid and immediately beneath the skin, which showed a predominantly low-grade intraductal tumour containing Roman-bridge type architecture, cribriform spaces and focally higher-grade atypia (Fig. 2c). There was a myoepithelial layer demonstrated around most of the tumour with antibodies against calponin, actin and CK14, and this also demonstrated clear-cut invasion through the absence of a myoepithelial layer around parts of the tumour. Read alignments predicted a fusion of KIF13B exon 37 on chromosome 8 with EPB41L4B intron 1 on chromosome 9 (Supplementary Fig. 2). The KIF13B rearrangement was further confirmed by FISH break-apart assay (Supplementary Fig. 2). Neither of these fusions was recurrent in the study set.
As KIF13B has significant sequence homology with KIF5B gene, which has been previously reported as a fusion partner with both ALK and RET in lung adenocarcinoma (20–21), we have also investigated for KIF5B gene alterations by FISH but none were found in 9 cases tested. All 12 pure intraductal carcinomas tested for ETV6 by FISH were negative for rearrangement as well.
PIK3CA and HRAS hotspot mutations were identified only in invasive apocrine/salivary duct carcinomas with an intraductal component
There was no fusion candidate detected by FusionSeq in two of the five cases sequenced, however the mutation detection algorithms (MuTect and VarScan) detected recurrent hotspot mutations in four of the five cases initially sequenced. This included PIK3CA and HRAS mutations in 3 cases each with two cases having concurrent mutations in both genes. The mutations were PIK3CA p.H1047R (2 cases), PIK3CA p.E545K (1 case) and HRAS p.Q61R (3 cases) (Fig. 5). One of the cases with an HRAS p.Q61R mutation was the case also shown to have a KIF13B-EPB41L4B fusion and one of the two cases with both PIK3CA p.H1047R and HRAS p.Q61R mutations was the case harboring a DFFA-ARID1A fusion. These mutations were validated by direct sequencing and then tested for in the remaining apocrine group. The additional 4 cases tested revealed PIK3CA p.E545K and PIK3CA p.H1047R in one case each. A total of 6/8 (75%) invasive carcinomas had mutation in one or both of these genes, typical of conventional salivary duct carcinomas. No fusions or mutations were detected in the two remaining cases.
Fig. 5.
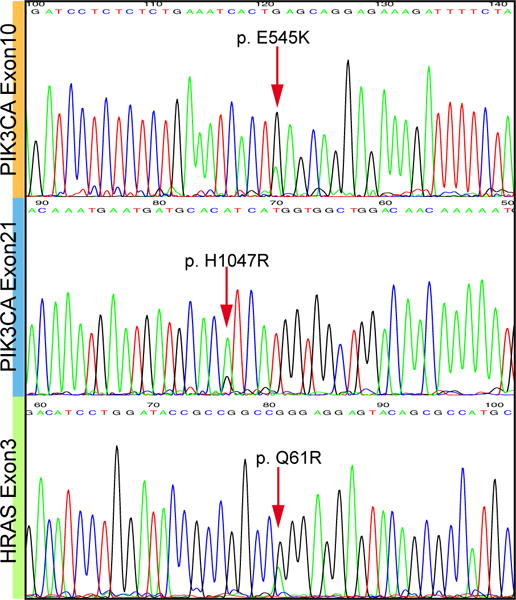
The invasive apocrine/salivary duct carcinoma group showed common mutations in PIK3CA and HRAS. These included PIK3CA p.E545K (2 cases), PIK3CA p.H1047R (3 cases) and HRAS p.Q61R (3 cases). A total of 6/8 (75%) of these invasive carcinomas harbored mutations in one or both genes, suggesting that they are best classified as salivary duct carcinoma even when they show unusual morphology.
DISCUSSION
The current World Health Organization (WHO) classification system of “intraductal carcinoma” of salivary glands is controversial and somewhat confusing as it incorporates lesions that have gone by a number of different names in the literature (10). This includes “intraductal carcinoma”, “low-grade salivary duct carcinoma”, “low-grade cribriform cystadenocarcinoma”, and “salivary duct carcinoma in-situ” (19; 22–24). The original description by Chen in 1983 used the term “intraductal carcinoma” to describe a single case arising in minor salivary glands of the oral cavity (22). Subsequently, Cheuk et al described a case report with higher-grade dysplasia arising in the oral cavity and suggested revised criteria to include any intraductal proliferation of salivary gland showing varying degrees of dysplasia from low to high-grade (25). In the meantime, Delgado et al had described the first case series of a new salivary gland entity under the appellation “low-grade salivary duct carcinoma” (LGSDC) in 1996, which presumably represented a low-grade counterpart of conventional salivary duct carcinoma (23). They deemed it to be a predominantly low-grade intraductal carcinoma, although a single case had focal high-grade atypia and a second case had microinvasion (23). All cases studied had diffuse co-expression of S100 and CK903 and all showed good behavior (23). A second case series by Brandwein-Gensler et al in 2004 expanded on this entity using the LGSDC name and similarly showed predominance of intraductal growth, although 25% of cases showed microinvasion and 12.5% showed areas of high-grade atypia (26). Similar to Delgado et al, this study found diffuse S100 expression and good outcome. The combined features, including S100 expression, good outcome and predominant intraductal growth suggested that this entity is unrelated to conventional salivary duct carcinoma. The WHO classification in 2005 reflected this by using a descriptive name for the entity, namely “low-grade cribriform cystadenocarcinoma” (19), despite the fact that this name had never appeared in the literature to that point.
Several publications that followed showed features that suggested that an overlap with salivary duct carcinoma may sometimes occur (11–12, 24). Weinreb et al argued that this terminology was confusing and that the original description by Chen and supported by Cheuk et al of “intraductal carcinoma” was the most reflective of its growth pattern and would lessen the risk of confusion with a fully invasive carcinoma, presumably requiring drastically different treatment (11). The study also demonstrated three cases, which suggested at least some overlap with conventional salivary duct carcinoma. First, all three intraductal carcinomas in the study were of apocrine phenotype with AR positivity, similar to salivary duct carcinoma. Second, one showed a clinically long standing intraductal carcinoma with subsequent wide spread invasion and lymph node metastasis (11), which was the first evidence of metastatic behavior in one of these lesions. Simpson et al followed this up with a study of three cases of pure in situ high-grade carcinoma with features of salivary duct carcinoma (24). This demonstrated that salivary duct carcinoma could represent a pure intraductal lesion as well, albeit rarely. One of these cases even shared Her-2-neu gene amplification with conventional salivary duct carcinoma (24). Finally, in a study by Bahrami et al, it was shown that conventional salivary duct carcinomas, with a background intraductal lesion resembling “low-grade cribriform cystadenocarcinoma” have no PLAG1 or HMGA2 rearrangement, unlike a large proportion of salivary duct carcinoma arising in pleomorphic adenoma (12). This suggested that these cases did not simply represent “cancerization of ducts” by salivary duct carcinoma but were a putative low-grade precursor lesion. The current WHO classification of 2017 now combines all these lesions under the name “intraductal carcinoma” and allows for any grade, however it does not make a definitive statement on whether conventional salivary duct carcinoma is related to this group or not (10). It only states that these lesions must be distinguished from “variants of adenocarcinoma, NOS, including cystadenocarcinoma” (10). These lesions must also be separated from “mammary analogue secretory carcinoma” which shows ETV6-NTRK3 fusion (3). This distinction is important as MASC does not have a significant intraductal component, but may share diffuse S100 positivity and similar architectural features, such as cribriform-like secretory spaces, cystic architecture and papillary growth (3). One “low-grade cribriform cystadenocarcinoma” tested as part of the control group in the original description of MASC was ETV6 negative by FISH (3). A second recent study also showed ETV6 negativity by FISH in these tumours in 5 cases tested (27).
Our previous observations of widely invasive carcinomas with background intraductal low-grade lesions (11–12), coupled with a perception that these lesions anecdotally behave better than conventional salivary duct carcinoma even when widely invasive, lead to the ongoing question of whether conventional salivary duct carcinomas share any relationship to “intraductal carcinomas”. Previously, salivary duct carcinomas have shown a number of different genomic alterations including Her-2-neu gene amplification, and hotspot PIK3CA and HRAS mutations (7). We and others have also found multiple salivary gland carcinomas to have novel and tumour specific translocations, including CRTC1-MAML2 in mucoepidermoid carcinoma, MYB-NFIB in adenoid cystic carcinoma, EWSR1-ATF1 in hyalinizing clear cell carcinoma, ETV6-NTRK3 in mammary analogue secretory carcinomas and ARID1A-PRKD1 and variant PRKD1, PRKD2 and PRKD3 fusions in cribriform adenocarcinoma of minor salivary glands (1–5). There have been no previous genomic studies of pure “intraductal carcinomas” or of widely invasive carcinomas with a significant low-grade intraductal component. In this study we examined a group of pure intraductal carcinomas and invasive carcinomas with an intraductal component by next generation sequencing to identify novel fusion transcripts.
The tumors studied were separated into intercalated duct type lesions and apocrine type lesions. The former group showed features identical to previous reports of S100 positive intraductal carcinomas, under the names “low-grade salivary duct carcinoma” and “low-grade cribriform cystadenocarcinoma”. Delgado et al first proposed that these lesions were of intercalated duct type differentiation due to their small cells, eosinophilic to amphophilic cytoplasm and S100 positivity, similar to other intercalated duct type salivary lesions. Although previous microinvasion has been reported, the case in this study with widespread invasion (case #1) is the first bona fide S100 positive invasive tumour with a similar intraductal component. This represented the index case for sequencing in this study. In addition, 14 pure intraductal carcinomas with S100 positivity were used in this group, including one of the cases previously reported by Weinreb et al in 2006 to have apocrine morphology and AR positivity (11). Upon re-review of this case, it showed dual intercalated duct and apocrine differentiation with combined S100 and AR positivity, but the predominant morphology was the intercalated duct type of differentiation (see Fig. 4B and case 2 in Table 1). A second case in this study showed similar mixed intercalated and apocrine differentiation (case #15). The second group in this study represented invasive carcinomas with an intraductal component and with apocrine differentiation but no intercalated duct features. These showed variable morphology including typical high-grade salivary duct carcinoma, moderately differentiated tubular apocrine carcinomas and 1 case with a low to intermediate grade apocrine carcinoma (case #19 in Table 1 and Fig 2C). This group also included case 3 from the previous study by Weinreb et al in 2006, which showed the long standing intraductal apocrine lesion and widely invasive carcinoma with lymph node metastases (11) and is labeled case #23 in this study (see Table 1). This case showed S100 negativity and AR positivity in the intraductal component and was negative for both markers in the invasive component, which was initially labeled an adenosquamous carcinoma rather than salivary duct carcinoma due to its AR negativity.
The index tumour sequenced (case #1) with intraductal and invasive intercalated duct type adenocarcinoma demonstrated NCOA4-RET fusion by next generation RNA sequencing and FusionSeq analysis and this was confirmed by direct sequencing. RET rearrangement by FISH was found in six additional pure intraductal carcinomas with intercalated duct type differentiation and S100 positivity. A total of 47% of intercalated duct type intraductal carcinomas showed evidence of a rearrangement involving RET. This was not found in any of the pure apocrine S100 negative lesions. ROS-1 and ALK-1 rearrangement was not found in any of these tumours. In addition, ETV6 FISH was negative in all pure intraductal carcinomas in this group, separating these lesions from mammary analogue secretory carcinoma. This includes a single case (case #14) with macrocystic architecture, papillary growth and vacuolated cells simulating MASC. The index case with NCOA4-RET showed an unusual cystic and sclerotic pattern with prominent papillary tufts and islands of cells floating in the cystic spaces, mimicking “spread through alveolar spaces” in lung adenocarcinomas. This fusion is not specifically associated with this morphology in lung adenocarcinomas, despite being seen in a small percentage of lung tumours overall (28). NCOA4-RET has recently been described in two salivary duct carcinoma cases by Wang et al (8) and a third case showed CCDC6-RET in an adenocarcinoma, NOS. Treatment responses were seen with anti-RET therapies as well (8). These cases were not well illustrated for comparison and no reference to an intraductal component or similar features to our own case were presented. No reference to S100 staining was made either and the image of AR positivity appeared to show focal staining. It is not clear whether these cases represent true conventional salivary duct carcinomas based on the information currently available. The finding of NCOA4-RET in an S100 positive adenocarcinoma with unique features and an intraductal component has not been described previously and six pure intraductal low-grade carcinomas with RET rearrangement represent the first report of RET being an early driver of carcinogenesis. The partner gene(s) for these six cases are not known yet and it could be that the specific RET fusion determines the final morphology and potential for invasion of a ductal derived salivary tumour. It is also possible that the location of the cell of origin determines the line of differentiation. For instance, salivary duct carcinoma has been thought of as differentiating towards the large excretory duct phenotype, whereas intercalated duct tumors are believed to differentiate towards a smaller intercalated duct phenotype. In theory, the location of the cell of origin acquiring the genomic alteration could determine the final morphology rather than the partner gene. However, it is more likely that other molecular or epigenetic mechanisms are responsible for the final phenotype of the tumor rather than the cell of origin or even RET or its partners. Finally, the presence of RET in early tumors may suggest that this genomic alteration could be insufficient for a widely invasive adenocarcinoma and that additional, as yet undiscovered mutations determine the final tumor status and behavior. More cases of RET rearranged tumors with more comprehensive morphological evaluation are required to answer these questions.
The apocrine group showed two non-recurrent fusions involving one tubular appearing adenocarcinoma with a DFFA-ARID1A fusion, and the unusual low to intermediate-grade carcinoma, which showed a KIF13B-EPB41L4B fusion. However, recurrent hotspot mutations in PIK3CA and HRAS were also found in these two tumors and found in 75% of the apocrine group as a whole. This suggests that these cases all belong in the conventional salivary duct carcinoma category despite the low-grade intraductal component and the varying and occasionally unusual morphology in some of the cases. This is contrary to our initial hypothesis that these varied tumors may represent a different pathway to conventional SDC. PIK3CA and HRAS mutations found in the two non-recurrent fusion cases suggests either that these fusions were early events in the intraductal component with subsequent invasion associated with the hotspot mutations, or that they are non-driver events. As a group, the behavior was mixed with some showing no evidence of disease at last presentation and others showing aggressive features, suggesting that the perception of better behavior in this group as compared to conventional salivary duct carcinoma as a whole could not be confirmed. More cases with these findings would need to be studied to examine the relationship between outcome and varying features in this group and for now it is reasonable to conclude that all pure widely invasive apocrine carcinomas of salivary gland are best classified as salivary duct carcinoma, similar to the conclusions by Williams et al (29).
In summary, recurrent rearrangements involving RET have been identified in 47% of cases with a low-grade intraductal carcinoma with S100 positivity and intercalated duct phenotype. NCOA4-RET specifically was found in an intraductal and invasive intercalated duct type adenocarcinoma, NOS for the first time, although this rearrangement has rarely been reported in possible fully invasive salivary duct carcinomas. This is the first description of RET as an early driver in a non-invasive salivary gland cancer. On the other hand, apocrine invasive carcinomas with possible low-grade intraductal precursors appear not to have RET rearrangement and have common mutations in PIK3CA and HRAS suggesting they best fit in the conventional salivary duct carcinoma category. They have a worse prognosis in this small series than appreciated previously and are best classified as salivary duct carcinoma even when they have unusual morphology. This dichotomy between intercalated predominant and pure apocrine carcinomas and their differing genomic alterations suggest that the current term “intraductal carcinoma” in the WHO 2017 does not completely account for the differing clinicopathologic characteristics of these tumors (10). However, the potential shared NCOA4-RET between this series and occasional SDCs in the Wang et al series suggests significant work needs to be done to fully explore this classification and role of RET in salivary gland cancer. This is particularly true if it is to be used as a therapeutic target or tested routinely in high-grade adenocarcinomas of salivary gland. There would be no rationale for treating indolent pure intraductal carcinomas with RET rearrangement with an anti-RET therapy for instance. Given the specific nature of fusions in salivary gland cancers described thus far (1–5), it would be somewhat surprising to have NCOA4-RET and RET rearrangement in general in multiple different tumor types. However, the precedent for RET, ALK-1 and ROS-1 in lung adenocarcinomas is that the fusions are not reliably predicted by morphology and that a large number of tumors require testing for rare cases with gene rearrangement to be identified and benefit from targeted therapies. Therefore, there may be a role for a similar screening program in salivary gland cancers in the future.
Supplementary Material
Supplementary Fig. 1. DFFA-ARID1A gene fusion in an apocrine invasive adenocarcinoma (case #15). (A) Schematic representation of the DFFA-ARID1A fusion indicating the loci that are joined together; DFFA exon 4 being fused to ARID1A exon 8 (left side of image). (B) RNA reads covering the fusion junction were isolated independent to FusionSeq analysis work flow, supporting the DFFA-ARID1A fusion candidate (top image). FISH for ARID1A shows break apart signals with one normal fused yellow signal and separate 5′ green and 3′ red signals indicating rearrangement of the gene (arrows). Note the consistent distance of the split signals owing to the putative inversion in this case.
Supplementary Fig. 2. KIF13B-EPB41L4B gene fusion in a low to intermediate grade apocrine intraductal and focally invasive adenocarcinoma (case #16). (A) Schematic representation of the KIF13B-EPB41L4B fusion indicating the loci that are joined together; KIF13B exon 37 being fused to EPB41L4B intron 1 (left side of image). (B) RNA reads covering the fusion junction were isolated independent to FusionSeq analysis work flow, supporting the KIF13B-EPB41L4B fusion candidate (top image). FISH for KIF13B shows break apart signals with one normal fused yellow signal and multiple separate 3′ red signals indicating rearrangement of the gene (arrows).
Acknowledgments
Supported in part by: P01CA47179 (CRA), P50 CA 140146-01 (CRA)
Footnotes
Conflict of interest: none
References
- 1.Antonescu CR, Katabi N, Zhang L, et al. EWSR1-ATF1 fusion is a novel and consistent finding in hyalinizing clear-cell carcinoma of salivary gland. Genes Chromosomes Cancer. 2011;50(7):559–70. doi: 10.1002/gcc.20881. [DOI] [PubMed] [Google Scholar]
- 2.Persson M, Andrén Y, Mark J, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106:18740–4. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skalova A, Vanecek T, Sima R, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- 4.Tonon G, Modi S, Wu L, et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33:208–13. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]
- 5.Weinreb I, Zhang L, Tirunagari LM, et al. Novel PRKD gene rearrangements and variant fusions in cribriform adenocarcinoma of salivary gland origin. Genes Chromosomes Cancer. 2014;53:845–56. doi: 10.1002/gcc.22195. [DOI] [PubMed] [Google Scholar]
- 6.Weinreb I, Piscuoglio S, Martelotto LG, et al. Hotspot activating PRKD1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–9. doi: 10.1038/ng.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiosea SI, Williams L, Griffith CC, et al. Molecular characterization of apocrine salivary duct carcinoma. Am J Surg Pathol. 2015;39(6):744–52. doi: 10.1097/PAS.0000000000000410. [DOI] [PubMed] [Google Scholar]
- 8.Wang K, Russell JS, McDermott JD, et al. Profiling of 149 Salivary Duct Carcinomas, Carcinoma Ex Pleomorphic Adenomas, and Adenocarcinomas, Not Otherwise Specified Reveals Actionable Genomic Alterations. Clin Cancer Res. 2016;22:6061–6068. doi: 10.1158/1078-0432.CCR-15-2568. [DOI] [PubMed] [Google Scholar]
- 9.Chiosea SI, Thompson LD, Weinreb I, et al. Subsets of salivary duct carcinoma defined by morphologic evidence of pleomorphic adenoma, PLAG1 or HMGA2 rearrangements, and common genetic alterations. Cancer. 2016;122(20):3136–3144. doi: 10.1002/cncr.30179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loening T, Leivo I, Simpson RHW, et al. In: WHO Classification of Head and Neck Tumours. El-Naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ, editors. Lyon: IARC Press; 2017. pp. 170–1. [Google Scholar]
- 11.Weinreb I, Tabanda-Lichauco R, Van der Kwast T, Perez-Ordoñez B. Low-grade intraductal carcinoma of salivary gland: report of 3 cases with marked apocrine ifferentiation. Am J Surg Pathol. 2006;30:1014–21. doi: 10.1097/00000478-200608000-00013. [DOI] [PubMed] [Google Scholar]
- 12.Bahrami A, Perez-Ordonez B, Dalton JD, Weinreb I. An analysis of PLAG1 and HMGA2 rearrangements in salivary duct carcinoma and examination of the role of precursor lesions. Histopathology. 2013;63(2):250–62. doi: 10.1111/his.12152. [DOI] [PubMed] [Google Scholar]
- 13.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1gene. Genes Chromosomes Cancer. 2010;49(12):1114–24. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quail MA, Kozarewa I, Smith F, et al. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habegger L, Sboner A, Gianoulis TA, et al. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–283. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sboner A, Habegger L, Pflueger D, et al. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosquera JM, Sboner A, Zhang L, et al. Novel MIR143-NOTCH fusions in benign and malignant glomus tumors. Genes Chromosomes Cancer. 2013;52:1075–87. doi: 10.1002/gcc.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang SC, Alaggio R, Sung YS, et al. Frequent HRAS mutations in malignant ectomesenchymoma: overlapping genetic abnormalities with embryonal rhabdomyosarcoma. Am J Surg Pathol. 2016;40:876–85. doi: 10.1097/PAS.0000000000000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandwein-Gensler MS, Gnepp DR. In: WHO Classification of Tumours Pathology and Genetics of Head and Neck Tumours. Barnes L, Eveson JW, Reichart P, Sidransky D, editors. Lyon: IARC Press; 2005. p. 233. [Google Scholar]
- 20.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–4. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–9. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 22.Chen KT. Intraductal carcinoma of the minor salivary gland. J Laryngol Otol. 1983;97(2):189–91. doi: 10.1017/s002221510009397x. [DOI] [PubMed] [Google Scholar]
- 23.Delgado R, Klimstra D, Albores-Saavedra J. Low grade salivary duct carcinoma. A distinctive variant with a low grade histology and a predominant intraductal growth pattern. Cancer. 1996;78(5):958–67. doi: 10.1002/(SICI)1097-0142(19960901)78:5<958::AID-CNCR4>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 24.Simpson RH, Desai S, Di Palma S. Salivary duct carcinoma in situ of the parotid gland. Histopathology. 2008;53(4):416–25. doi: 10.1111/j.1365-2559.2008.03135.x. [DOI] [PubMed] [Google Scholar]
- 25.Cheuk W, Miliauskas JR, Chan JK. Intraductal carcinoma of the oral cavity: a case report and a reappraisal of the concept of pure ductal carcinoma in situ in salivary duct carcinoma. Am J Surg Pathol. 2004;28(2):266–70. doi: 10.1097/00000478-200402000-00017. [DOI] [PubMed] [Google Scholar]
- 26.Brandwein-Gensler M, Hille J, Wang BY, et al. Low-grade salivary duct carcinoma: description of 16 cases. Am J Surg Pathol. 2004;28(8):1040–4. doi: 10.1097/01.pas.0000128662.66321.be. [DOI] [PubMed] [Google Scholar]
- 27.Stevens TM, Kovalovsky AO, Velosa C, et al. Mammary analog secretory carcinoma, low-grade salivary duct carcinoma, and mimickers: a comparative study. Mod Pathol. 2015;28:1084–100. doi: 10.1038/modpathol.2015.64. [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30:4352–9. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- 29.Williams L, Thompson LD, Seethala RR, et al. Salivary duct carcinoma: the predominance of apocrine morphology, prevalence of histologic variants, and androgen receptor expression. Am J Surg Pathol. 2015;39:705–13. doi: 10.1097/PAS.0000000000000413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. DFFA-ARID1A gene fusion in an apocrine invasive adenocarcinoma (case #15). (A) Schematic representation of the DFFA-ARID1A fusion indicating the loci that are joined together; DFFA exon 4 being fused to ARID1A exon 8 (left side of image). (B) RNA reads covering the fusion junction were isolated independent to FusionSeq analysis work flow, supporting the DFFA-ARID1A fusion candidate (top image). FISH for ARID1A shows break apart signals with one normal fused yellow signal and separate 5′ green and 3′ red signals indicating rearrangement of the gene (arrows). Note the consistent distance of the split signals owing to the putative inversion in this case.
Supplementary Fig. 2. KIF13B-EPB41L4B gene fusion in a low to intermediate grade apocrine intraductal and focally invasive adenocarcinoma (case #16). (A) Schematic representation of the KIF13B-EPB41L4B fusion indicating the loci that are joined together; KIF13B exon 37 being fused to EPB41L4B intron 1 (left side of image). (B) RNA reads covering the fusion junction were isolated independent to FusionSeq analysis work flow, supporting the KIF13B-EPB41L4B fusion candidate (top image). FISH for KIF13B shows break apart signals with one normal fused yellow signal and multiple separate 3′ red signals indicating rearrangement of the gene (arrows).


