Abstract
Autophagy is an evolutionarily conserved cellular pathway that culminates in lysosomal degradation of selected substrates. Autophagy can serve dual roles in virus infection with either pro- or antiviral functions depending on the virus and the stage of the viral replication cycle. Recent studies have suggested a role for autophagy in Zika virus (ZIKV) replication by demonstrating the accumulation of autophagic vesicles following ZIKV infection in both in vitro and in vivo models. In human fetal neural stem cells, ZIKV inhibits Akt-mTOR signaling to induce autophagy, increase virus replication and impede neurogenesis. However, autophagy also has the potential to limit ZIKV replication, with separate studies demonstrating antiviral roles for autophagy at the maternal-placental-fetal interface, and more specifically, at the endoplasmic reticulum where virus replication is established in an infected cell. Interestingly, ZIKV (and related flaviviruses) has evolved specific mechanisms to overcome autophagy at the ER, thus demonstrating important roles for these autophagic pathways in virus replication and host response. This review summarizes the known roles of autophagy in ZIKV replication and how they might influence virus tissue tropism and disease.
1) Introduction
Zika virus (ZIKV), a mosquito-borne flavivirus of the Flaviviridae family of positive-strand RNA viruses, is closely related to other medically relavent global pathogens including dengue (DENV), yellow fever (YFV), West Nile (WNV) and tick-borne encephalitis (TBEV) viruses [1]. Typically, the clinical features caused by historic ZIKV infections consisted of a mild self-limiting flu-like fibrile illness [2]. However, the ZIKV epidemic first detected in 2015 in the Americas was associated with severe disease, including multi-organ failure, Guillain-Barre syndrome, and in expectant mothers, abnormal fetal brain development resulting in a spectrum of neurological disorders including mirocephaly [3]. Virus transmission occurs directly between Aedes mosquitoes and humans, although, evidence for sexual transmission indicates human-to-human transmission is also possible [4]. The target cells for ZIKV include peripheral dendritic cells (DCs) and macrophages, placental cells (Hofbauer cells, trophoblasts and endothelial cells) and neuronal cell types such as neuronal progenitor cells and less frequently mature neurons and astrocytes [reviewed in [5]].
The ZIKV genome of ~11 kb encodes three structural and seven non-structural proteins. The virus particle (~50nm in diameter) is composed of a nucleocapsid surrounded by a host-cell derived lipid bilayer decorated with the structural proteins (prM/M and E) on the surface [6]. The virus enters target cells via endocytosis and is thought to utilize receptors TYRO3, AXL and MER (receptor tyrosine kinases) to promote virus binding [7]. These receptors are highly expressed on the surface of T cells, DCs, macrophages, immature NKs, sertoli cells (testis) and retinal pigment epithelial cells and endothelial cells [8]. However, it is important to note that despite T cells being enriched for these receptors on their surface are still resistant to ZIKV infection suggesting that there could be other receptors or post-entry menchanisms that dictate viral tropism [9]. The relevance of these candidate receptors to ZIKV is likely to be specific to humans or non-human primates, as these receptors are nonessential in mouse models, in vivo [10]. Upon uncoating and release of the viral genome into the cytoplasm, the non-structural module of the viral genome (NS1-NS5) is sufficient to establish viral RNA replication in specialized sub-cellular structures that appear to be invaginations of the endoplamic reticulum (ER). Virus assembly occurs in close proximity to sites of viral RNA replication and may utilize ESCRT components for virus budding and release [11]. Noninfectious immature virus particles assemble in the ER which are then transported to the trans- golgi for virion maturation that requires furin-dependent cleavage of prM to M. Mature virion release into the extracellular space is thought to utilize the conventional protein secretory pathway, although the precise mechanism for viral exit remains to be determined. Given the intimate dependence on the host to successfully infect and establish viral replication, the cellular environment offers multiple host-pathogen interfaces that can serve as pro– or anti- viral platforms at each stage of the viral replication cycle. One important pathway is the lysosomal-dependent degradative pathway of autophagy [reviewed in [12–14]]. This review focuses on highlighting the function of autophagy in ZIKV replication and disease, including the viral determinants that may antagonize or manipulate different aspects of the pathway.
2) Basics of autophagy
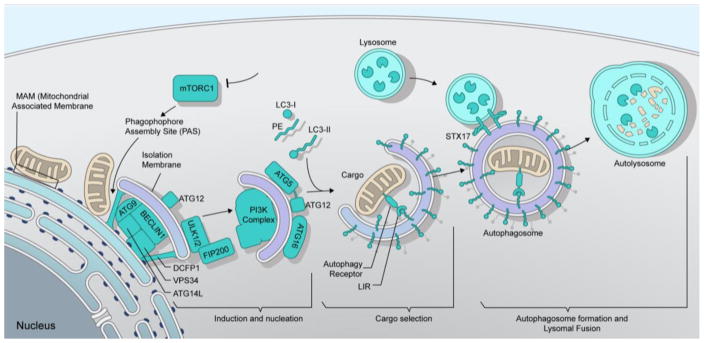
Autophagy is a complex and tightly regulated cellular pathway that involves non-selective and selective mechanisms to target a plethora of substrates ranging from protein aggregates to cellular organelles and pathogenic components for lysosomal degradation. Although autophagy is degradative in nature, an additional role for autophagy in unconventional secretion of leaderless cyctosolic proteins is well established [15]. Importantly, autophagy is always “on” and occurs at a basal level in most cells of an organism. The three types of autophagy described so far include microautophagy, macroautophagy and chaperon-mediated autophagy. Macroautopahgy (henceforth referred to as autophagy) is the most well characterized of the three and is highly conserved from yeast to mammalian cells. In general, the pathway of autophagy can be divided into distinct stages including induction, autophagosome formation, cargo selection, lysosomal fusion and degradation (Figure 1). The induction step requires the formation of double membrane structures at organelle interfaces known as mitochondrial associated membranes (MAMs) that are points of contact between the ER and mitochondria [16]. This membrane structure, also referred to as the omegasome, expands around the selected autophagy substrate and matures into a double membrane vesicle known as the autophagsosome [17]. As a part of the maturation process, autophagosomes can associate with vesicles or cargo from the endomembrane system such as early and late endosomes to form intermediate compartments called amphisomes. Subsequently, autophagosomes/amphisomes can fuse with lysosomes to form autolysosomes that results in degradation of cargo. The measure of autophagy is referred to as “autophagic flux” that represents the total number of autophagic vesicles formed and degraded [18]. At basal levels, autophagic flux is constant. However, upon different stimuli including starvation, stress or pathogen infection, the rate of autophagic flux can either be enhanced or inhibited. Furthermore, the autophagic capacity of a cell is directly proportional to lysosomal function and the efficiency of autophagosomes to fuse with lysosomes [18].
Figure 1. Autophagy pathway.
Autophagy is initiated through inhibition of mTORC1 signaling. This results in the recruitment and coordinated function of induction and nucleation factors (DFCP1, ATG9, ULK1/2, FIP200 Beclin1, VPS34, ATG14L, classIIIPI3K complex, ATG5, ATG12 and ATG16) at the phagophore assembly site (PAS) that assembles at the MAM (mitochondrial associated membrane). Induction and nucleation is followed by the conjugation of phosphatidylethanolamine (PE) onto LC3-I through a reaction involving the ATG12-ATG5-ATG16 complex to form LC3-II bound onto the autophagosome membrane. LC3-II directly interacts with selective autophagy receptors via their LC3-interacting region [18] to specifically sequester cargo for autophagic degradation. Upon selection of cargo and closure of the nascent autophagosomal membrane, mature autophagosomes fuse with lysosomes utilizing a SNARE protein, syntaxin 17 (STX17), to form the autolysosome which facilitates degradation of cargo.
The molecular basis of autophagy, spanning from induction to cargo selection and autolysosomal degradation, commands the intricate co-ordination of multiple proteins including post-translation modifications to engage a functional autophagic response [19, 20]. Autophagy – related (Atg) proteins constitute the main components of the core autophagic machinery, of which there are more than 30 identified in yeast with most of them conserved in mammals.
Induction and nucleation
Autophagy (autophagic flux) occurs at a low level under normal conditions. Pathogen infection, cellular stress and extracellular cues can all efficiently induce autophagy. The target of rapamycin complex 1 (TORC1) is a nutrient-sensing pathway and is essential for monitoring changes in nutrient availability. In mammalian cells, inhibition of TORC1 function following nutrient deprivation or rapamycin treatment leads to autophagy induction. Mechanistically, the induction of autophagy requires the autophosphorylation of Unc-15 like kinase 1 (ULK1) and -2 (ULK2) that in turn phosphorylate FIP200 (focal adhesion kinase family-interacting protein of 200kD) and mammalian Atg13 (mAtg13). The assembly of this multi-protein complex is crucial for autophagy since it serves as a scaffold in recruiting other Atg proteins and molecules to the phagophore assembly site (PAS) [19, 21, 22](Figure 1). PAS formation is followed by the mobilization of a small group of molecules to this site to convert it to the active sequestering platform for autophagy. This process is referred to as nucleation and acts as an amplification signal that results in the recruitment of other proteins and phagophore expansion. Of these, the class III Ptdlns3K complex I (VPS34 and Beclin-1) is one of the key components responsible for the production of phosphatidylinositol-3-phosphate (PIP3) from phosphatidylinositol [23]. PIP3, the major lipid species in the autophagy pathway, is produced on precursor autophagic membranes and is indispensable for proper localization of subsequent factors that help in expansion and closure of nascent autophagosomes.
Autophagosome formation and lysosomal fusion
The hallmark of autophagy is the formation of double-membraned vesicles known as autophagosomes that represent the mature form of the phagophore (Figure 1). The expansion of phagophores to mature autophagosomes is carried out by the coordinated action of two ubiquitin-like (Ubl) conjugation systems that involve Ubl proteins Atg12 and Atg8 that share structural similarity to ubiquitin but are not genuine homologs [reviewed in [24]]. Atg12 is conjugated to Atg5 in an Atg7 and Atg10 (E1 and E2 enzymes)-dependent mechanism to bind Atg16 and form the Atg12-Atg5-Atg16 complex. Mammalian cells have seven Atg8 homologs: MAP1LC3 A/B/C (microtubule- associated protein 1A/1B light chain), GABARAP (gamma-aminobutyric acid receptor-associated protein) and GABARAPL1/2/3 (henceforth indicated as LC3). LC3 is a broadly used marker for autophagosome detection in microscopy. On synthesis, LC3 is C-terminally cleaved by Atg4 (cysteine protease) to expose a glycine residue to form LC3-I. Subsequently, LC3-I is lipidated with PE (phosphatidylethanolamine) to form phagophore/autophagsome bound, LC3-II. This reaction involves Atg7 (E1 enzyme), Atg3 (E2 enzyme) and the Atg12-Atg5-Atg16 complex (E3-like ligase) [25, 26]. The relative amount of LC3-II is used to quantify autophagic activity in cells via immunoblot analysis, although this is only partially informative as it does not discriminate between an induction or blockage of autophagy. The Atg-conjugation system (Atg3, Atg5 and Atg7) is crucial to ensure autophagosome formation, since defects in these genes results in accumulation of partially formed “unclosed” autophagosomes [27]. More recently, this conjugation system was shown to be necessary for efficient degradation of the inner autophagic membrane but is not required for autophagosome-lysosome fusion [28]. Another important aspect in autophagosome biogenesis is the origin of the autophagosomal membrane. As mentioned earlier, class III phosphatidylinositol 3 kinase complex I-dependent PIP3 production is essential for autophagosome formation, which also activates and recruits additional factors WIPI2, DFCP1 and Atg14 to MAMs [17, 29, 30]. Atg14 confers membrane curvature to the nascent autophagosome and also interacts with the SNARE (soluble NSF attachment protein receptor) protein, syntaxin 17, to promote autophagosome and autolysosome formation upon fusion with lysosomes [30, 31]. In addition, Rab7, a member of the Rab-GTPase family usually localized to late endosomes and needed for maturation of endosomes to endolysosomes, is important for autophagosome-lysosome fusion [32]. Importantly, autophagy is transcriptionally coordinated with lysosome biogenesis by transcription factor EB (TFEB) that drives expression of both autophagy and lysosomal genes [33]. Finally, autolysosomal degradation, which is the last step in autophagy, is regulated by the membrane concentration of V-ATPase proton pumps, lysosomal hydrolases and cysteine proteases known as cathepsins [34, 35].
Cargo Selection
Autophagy being a crucial degradation pathway can target a diverse range of intracellular and extracellular substrates. Under starvation conditions, autophagy exerts a nonselective catabolic function in degrading cytoplasmic constituents (cargo) including protein aggregates, endosomes, lysosomes, mitochondria, ER, peroxisomes and lipid droplets as a source of membranes or amino acids and lipids to restore cellular homeostasis. Autophagosomes can recognize multiple cargos simultaneously. Cargo selection usually begins with a specific signal (mostly ubiquitin or galectins), that tags cargo (proteins aggregate, organelle or pathogen) to be recognized by dedicated receptors through direct or indirect interaction with LC3 on nascent autophagosomal membranes [20, 36]. Autophagy receptors interact with LC3 on autophagasome membranes via LC3-interacting regions (LIRs) that have an evolutionary conserved consensus sequence Trp/Phe/Tyr-X-X-Leu/Ile/Val (W/F/Y-X-X-L/I/V) [37]. The ability to bind LC3 is not exclusive to autophagy receptors, since core autophagy proteins ULK1 and ULK 2 kinase, Atg13, FIP200 also interact with LC3 through their LIRs classifying them as autophagy adaptors [38]. Several autophagy pathways (mitophagy – mitochondria, ER – ERphagy or reticulophagy, protein aggregates – aggrephagy, liposomes – lipophagy, and pathogens – xenophagy) have been described and termed according to the type of selected cargo [reviewed in [20]].
3) Roles for autophagy in ZIKV replication and development of microcephaly
Functional autophagy is extremely important in the development of an organism [21]. Impaired autophagy can severely stunt embryonic development, organanogeneis, and the ability to confer a robust immune response to protect a developing embryo from pathogenic infection [39]. Therefore, understanding the role of autophagy in ZIKV infection may have implications for understanding pathogenesis and aid in identifying therapeutic targets.
Membrane re-arrangements and induction of autophagy following ZIKV infection
Recent studies have demonstrated that analogous to other flaviviruses (DENV, WNV and TBEV), ZIKV infection of skin fibroblast or human neural progenitor cells (hNPCs) induces rearrangements of the ER resulting in clusters of vesicles termed vesicle packets (VPs) and swollen ER-cisternae containing regular arrays of virus particles referred to as virus bags. These ultrastructural sites are considered the sites of viral RNA replication and assembly, respectively [40–42]. Analysis of infected primary fibroblasts revealed the presence of multi-membrane structures in ZIKV infected cells reminiscent of autophagic vesicles [9]. Importantly, immunolblotting of ZIKV-infected placentas (5 days postinfection) showed increased accumulation of LC3-II compared to uninfected controls, implying a role for autophagy in in vivo models of ZIKV infection [43]. The role for autophagy is proviral in function, as infection of pregnant mice hypomorphic (HM) for ATG16 (Atg16l1HM) exhibiting decreased autophagic activity showed significantly lower levels of ZIKV infection in placentas compared to wild-type controls. Importantly, ZIKV titers in Atg16l1-deficient fetuses was notably lower compared to control mice[43]. These data suggests that ATG16 (ATG16L1) plays a crucial role in influencing placental suseptibilty and vertical transmission of ZIKV infection.
Moreover, treatment of primary fibroblasts and a human cytotrophoblast cell line (JEG-3) with torin1 or rapamycin (autophagy inducers) infected with ZIKV infection resulted in a modest increase of viral RNA copy number and release of infectious virus particles from cells [9, 43, 44]. Conversely, the addition of a class III PIK3 kinase inhibitor 3-methyladinine (3-MA), resulted in a slight reduction in copy number of viral RNA and virus production [9, 43]. 3-MA has been broadly used as an autophagy inhibitor. However, the effects of 3-MA have been extended to endocytic trafficking, which is important for virus entry and replication [45] suggesting that effects of 3-MA on virus replication might not be a direct consequence of autophagy inhibition. Importantly, hydroxychloroquine (HCO) treatment [inhibitor of autophagy in vivo [46]] of human trophoblast cells resulted in decreased viral replication, further supporting a proviral function of autophagy in ZIKV infection [43]. In further support of autophagy induction by ZIKV, infection of a human cytotrophoblast cell line (JEG-3) and human fetal neural stem cells (fNSCs) at a low multiplicity of infection with ZIKV strains Paraiba (2015), MR766 and ibH30656 increased conversion of cytosolic LC3-I to autophagosome bound LC3-II in the presence and absence of lysosomal inhibitors [43, 44]. This is not dissimilar to infection of Huh7 cells with the closely related DENV, that caused an induction of autophagic flux early in infection. However, synchronized infection (high MOI) with DENV resulted in a subsequent inhibition in autophagic degradation later in infection prior to the onset of apoptosis [47]. This block in autophagy may be difficult to observe at low MOIs during a spreading infection and thus it would be useful to compare DENV and ZIKV effects on autophagy during one-step growth curves to fully understand the impact of these viruses on general induction autophagy pathways.
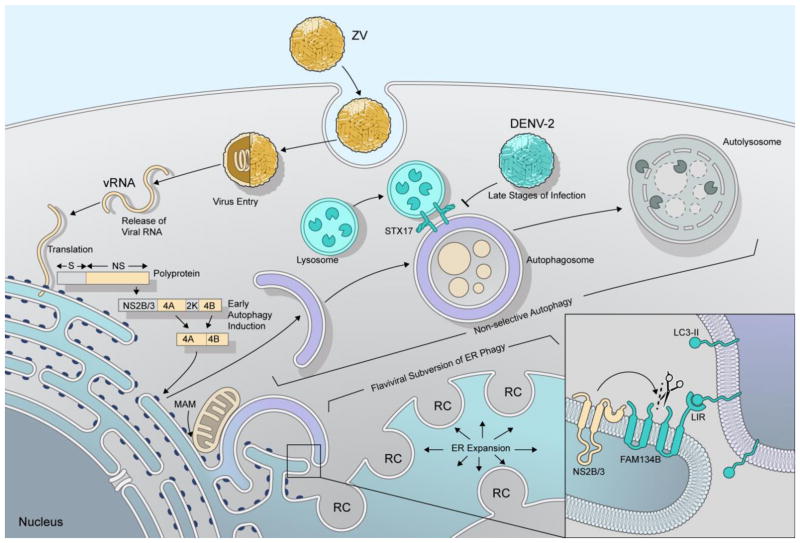
In the context of flavivirus infections, autophagy induction is thought to be triggered mainly by ER stress. This cellular stress pathway is induced in part through co-expression of the viral proteins NS4A and NS4B that function to facilitate membrane curvature of the ER to form flaviviral replication complexes [48–50]. Intriguingly, co-expression of NS4A and NS4B from ZIKV in Hela or fNSCs resulted in a significant increase in GFP-LC3 punctate numbers per cell and also showed enhanced conversion of LC3-I to LC3-II in the presence or absence of lysosomal degradation. This work demonstrates the ability of NS4A and NS4B specifically to induce autophagy in these cells and recapitulate observations following virus infection [44] (Figure 2). Importantly, individual expression of all 10 ZIKV proteins revealed that expression of NS4A or NS4B alone or together decreased Akt phosphorylation and subsequently inhibited mTOR activity to promote autophagy induction. In addition, expression of ZIKV NS4A and NS4B individually or in combination resulted in a dramatic effect on the ability of fNSCs to form neuroshperes. This effect was not observed following expression of DENV NS4A and NS4B, suggesting a link between the function of NS4A/4B and neurological disease specifically in ZIKV-infected neonates [44]. Thus, as a consequence of ZIKV infection, inhibition of mTOR activity to trigger pro-viral autophagy early in infection which is essential for vertical tramsmission but may also negatively impact neurogenesis in the fetal brain and thereby contribute to the unique pathogenesis observed in the context of ZIKV.
Figure 2. Autophagy and flavivirus infection.
ZIKV engages exposed surface receptors on the host cell and virions are internalized via receptor-dependent endocytosis. The acidic pH of in the endosomal compartments initiates structural changes of the envelope glycoproteins, triggering the fusion of viral and endosomal membranes and release of viral RNA (vRNA) into the cytoplasm. The vRNA is translated into a polyprotein, which is co- and post-translationally processed by viral and host proteases into structural (S) and nonstructural proteins (NS). Of the NS proteins, NS4A, NS4B and NS2B/3 have important roles in manipulating the autophagy pathway. Early in infection, NS4A and NS4B induce membrane curvature and induce autophagy, to mobilize membranes and lipids required for biogenesis of the viral replication complex (induction). The endoplasmic reticulum (ER) is the critical membrane platform on which virus replication is established. Viral manipulation of the ER must induce ER-Phagy, which is actively antagonized (subversion) by the cleavage of the ER-Phagy receptor FAM134B by flavivirusprotease (NS2B/3) (inset). Furthermore, the closely related flavivirus, DENV-2, can block autolysosome formation by preventing fusion of autophagosomes with lysosomes.
Selective autophagy of the ER in virus infection
Autophgy of the ER, or ER-phagy, has been suggested to contribute in ER- quality control (ERQC) to remove overexpressed and misfolded proteins from the ER. The selective receptors for ER-phagy include Family with sequence similarity 134 (FAM134) proteins, FAM134A, B and C. Depletion of FAM134B in human or mouse cells results in ER expansion whereas FAM134B overexpression lead to fragmentation of the ER and subsequent lysosomal degradation [51]. Notably, ER expansion is a consequence of flavivirus infection and thought to be essential in the biogenesis of the viral replication complex. A recent report examining replication of ZIKV, DENV or WNV has revealed that flaviviruses can expand the ER by antagonizing autophagy through specific targeting of ER-phagy receptor FAM134B [52]. In support of this, RNAi-mediated downregulation of FAM134B modestly enhanced replication of both ZIKV and DENV in human cells. Importantly, expression of the flavivirus protease (NS3 together with its essential cofactor NS2B) NS2B/3 from ZIKV, DENV and WNV was sufficient to cleave FAM134B at a single site in the cytoplasmic loop of the reticulon homology domain (RHD) (Figure 2). Reticulons are ER resident proteins that influence vesicle formation and membrane morphogenesis via their RHDs [53]. Therefore, NS2B/3 cleavage of the FAM134B RHD may interfere with formation of ER-phagy specific autophagosomes (Figure 2). Expression of the cleavage-resistant FAM134B mutant (FAM134BR142A) showed enhanced accumulation of NS2B/3 compared with wild-type (WT) FAM134B, suggesting that FAM134B may be directly antiviral through autophagic clearance of the viral protease, an essential component of the viral replication complex. The NS2B/3-mediated cleavage of FAM134B would result in a FAM134B product with the potential to bind LC3B via its C-terminal LIR region, but unable to induce ER-phagy of viral proteins [52]. Viral cleavage of FAM134B expands the ER and simultaneously prevents the autophagic removal of viral proteins embedded in the ER (Figure 2). The antiviral nature of FAM134B may be independent of type I interferon as it exhibits some virus specificity, with no effect of FAM134B observed during VSV replication [52], although FAM134B expression limited replication of Ebola virus [54] which may have occurred through general effects on ER capacity. Thus, it will be important to determine the contribution of ER-phagy as an antiviral pathway following infection in vivo of FAM134B−/− mice [51] to fully understand the impact of this pathway on replication of ZIKV and other flaviviruses. Furthermore, ZIKV infection may upregulate non-selective autophagy early on but inhibit selective ER-phagy to expand the ER and generate the surface area essential for biogenesis of flaviviral replication complexes. Additional research is needed to determine the spatiotemporal relationship of nonselective autophagy in supporting viral assembly and release, and viral evasion of antiviral selective autophagy (ER-phagy).
Autophagy at the maternal-placental-fetal complex
The maternal immune status during pregnancy must tolerate and accommodate the forming placenta and developing fetus, while also protecting against infectious insults. The maternal immune system along with placental-fetal immune response combine to respond to infections and is important for immune regulation [55, 56]. Interestingly, a recent study revealed a role for autophagy at the interface between the feto-placental unit and maternal blood which is composed of trophoblasts [57]. The study demonstrated that human placental trophoblasts (PHTs) isolated from normal term deliveries were highly resistant to infection by DNA and RNA viruses. Importantly, resistance to infection was not a result of pre-existing interferon (IFN)-dependent antiviral signaling although treating non-placental cells usually permissive to virus infection with conditioned medium from naïve PHT cells conferred resistance to infection. In addition to growth factors, hormones and signaling molecules, trophoblasts release various lipid- encapsulated extracellular vesicles into the maternal blood such as apoptotic cell- derived vesicles, microvesicles and exosomes [58]. Importantly, exosomes and microvesicles can be recovered from the placenta during the first and second trimester of pregnancy and their numbers increase throughout pregnancy [59]. The authors demonstrated that exosomes secreted from placental trophoblasts [57], contained a primate specific chromosome 19 cluster (C19MC) of microRNAs (miRNAs) as antiviral agents that could be transferred onto nonplacental cells and restrict virus infection. Interestingly, C19MC miRNA cluster is exclusively expressed in the placenta and strongly induces autophagy in target cells, when delivered from trophoblast derived exosomes [57]. Of the 46 miRNAs in the C19MC miRNA cluster, at least three miRNAs (MIR517-3p, MIR16B-5p and MIR512-3p) exhibited potent antiviral activity [57]. Furthermore, conditioned medium from resting PHT cells, purified exosomes released from PHTs, and individual C19MC miRNA all induced autophagy when added onto target cells. Importantly, autophagic induction was abolished when target cells were exposed to conditioned PHT medium in combination with PI3Kinase inhibitor 3-MA. The induced autophagy observed in target cells was a result of enhanced autophagic flux (induction) rather than a block in autolysosomal degradation. Furthermore, the authors could show that PHT cells exhibited high levels of basal autophagy suggesting that these cells are primed to mount an autophagic response in response to viral infections [57, 59]. Although autophagy is an important response by the cells, it is likely that additional antiviral factors are produced by PHT cells. A subsequent study revealed that the antiviral effects of C19MC miRNAs were significantly weaker than some unknown components in conditioned PHT medium [60]. The unknown factor within the PHT conditioned medium included type III interferons (IFNλ1) as an additional factor that was dominantly responsible for resistance to virus infection, including ZIKV and DENV [60]. Nevertheless, a role for IFNλ in autophagy induction in these cells, still remains to be determined.
Conclusively, in vivo evidence for the role of autophagy in vertical transmission at the maternal – placental – fetal complex is well supported by experiments employing pregnant Atg16l1-deficient mice or systemic adminstraction of HCO [inhibitor of autophagy in vivo [46]] in pregnant mouse models of ZIKV infection [43]. This study showed that genetic or pharmacologically induced impairment of autophagy and resulted in reduced ZIKV infection in the placenta and fetus without influencing systemic infection of other maternal organs, demonstrating that autophagy inhibiton is favorable for the host to limit vertical transmission. These findings are in contrast to the previously reported role of autophagy as an antiviral pathway in infected cells within the placenta, and again suggests that ZIKV has specifically evolved strategies to utilize autophagy as a proviral mechanism [57, 59]. Moreover, if autophagy is proviral early in infection, potentially as a source of lipids to promote membrane biogenesis for virus replication, the heightened autophagy in PHTs may specifically support efficient viral infection at the maternal-fetal interface.
Conclusions & Perspectives
Viruses are known to subvert the autophagy pathway and utilize different components of autophagy to establish virus replication and evade antiviral innate immunity [12–14]. Studies thus far suggest a pro-viral role for autophagy in ZIKV replication and vertical transmission to the fetus that might involve upstream signaling events requiring the inhibition of mTOR activity specifically by ZIKV NS4A and NS4B. Interestingly, mTOR inhibition results in autophagy induction and also impacts neurogenesis and therefore may contribute to ZIKV- associated pathogenesis following infection of neonates. In the case of DENV, autophagy is induced early in replication, to mobilize membranes and lipids required for replication [40, 61] but inhibited late in infection before the onset of cell death [47]. Since, ZIKV infection also induces cell death in neural stem cells, it would be important to determine the status of autophagy late in infection and its direct contribution to cell death in a cell type- specific manner. Interestingly, autophagy is thought to be pro-viral in the cases of polio-, coxsackie- and rhinovirus infection. Cells infected with these viruses release large autophagosome derived vesicles, which when added onto naive cells, demonstrate high levels of infectivity [62, 63]. Interestingly, these autophagosome derived vesicles are highly enriched in phosphatidylserine. The masking of exposed PS on virus-containing, autophagosome-derived vesicles by Annexin V treatment results in blocking infection of naive cells [63]. This suggests that recognition of PS could serve as an additional factor to mediate virus entry and expand viral tropism. Notably, the molecules implicated as entry receptors for ZIKV (TYRO3, AXL and MER) function as receptor tyrosine kinases and are highly expressed on the surface of ZIKV target cells including DCs, macrophages, and endothelial and sertoli cells (testis). RTKs are involved in the sensing of and phagocytic clearance of apoptotic cells. Briefly, PS exposed on the surface of apoptotic cells is recognized by ligands (GAS6 and S protein) and interacts with RTKs on the surface phagocytic cells resulting in apoptotic cell clearance [64, 65]. Interestingly, among the TAM family RTKs, AXL alone is sufficient to induce autophagy in macrophages which might suggest a role for ZIKV entry and autophagy induction (Jihye Han, Autophagy 2016). The observed localization of ZIKV envelope protein (E) with LC3 positive structures [9] and the presence of ER-cisternae enclosing uniform arrays of virus particles raises the possibility that switching from degradative autophagy to secretory autophagy could facilitate release and spread of mature viral particles. How secretory autophagy bifurcates from degradative autophagy is not well understood. Studies have shown that E3 ubiquitin ligases belonging to the TRIM family of proteins, in particular TRIM16 can recognize IL-1β to interact with SNARE protein SEC22B to cooperatively target cargo to LC3B-II positive sequestration membranes and fecilitate secretion from cells [15]. How exactly viruses could usurp mechanisms of secretory autophagy to fecilitate viral assembly and release requires to be further investigated.
In addition to the pro-viral role of autophagy in ZIKV infection, recent efforts have also highlighted the importance of viral antagonism of selective autophagy by targeting the ER-phagy specific receptor FAM134B. Indeed, the viral protease NS2B/3 cleaves FAM134B thereby inactivating the receptor to mediate ER-phagy to degrade sites of viral RNA replication [52]. Depletion or inactivation of FAM134B results in ER expansion which is conducive for providing membrane surfaces to build invaginations for viral replication. The relevance of ER expansion in ZIKV infection was recently described with the formation of large ER-derived vacuoles upon ZIKV infection of human epithelial cells, primary skin fibroblasts and astrocytes [66]. Importantly, the accumulation of ER-derived vacuoles upon ZIKV infection inevitably induces a process similar to paraptosis, caspase-independent, non-apoptotic form of cell death associated with the formation of massive cytoplasmic vacuoles [66, 67]. Notably, expression of interferon-induced transmembrane protein 3 (IFITM3) reduced cytoplasmic vacuole formation as well as reducing virus replication and cell death [66]. However, the study also showed that ZIKV-induced paraptosis-like cell death required the action of class 1 and not class 3 PI3K activity suggesting an important role for PI3K/Akt pathway in virus induced cell death and no major role of autophagy in this step. Thus, it appears unlikely that ZIKV requires autophagy later infection to induce cell death as a mechanism for virus spread.
Taken together, the role of autophagy is important to establish initial stages of replication, although it is quite possible that other functions of autophagy could be exploited by the virus depending on precise stage in the viral replication cycle. Finally, components of the core autophagy machinery have been shown to have autophagy-independent roles in virus replication and innate immunity [68, 69], but specific roles of autophagy in innate immunity against ZIKV have not been explored. Furthermore, applying recently developed assays to monitor different aspects of autophagy [18], can definitively reveal the mechanistic interplay between autophagy and ZIKV infection. Understanding the molecular details of how viruses engage autophagy will shed light on the roles of autophagy in immunity and may aid in the rational design effective therapeutics to control virus infection and potentially treat disease.
Highlights.
Autophagy is pro- and antiviral in ZIKV infection.
ZIKV evades ER-Phagy by cleaving autophagy receptor FAM134B.
Autophagy is a critical antiviral response at the maternal-placental-fetal complex.
Acknowledgments
We would like to thank Dr. Heinz Feldmann for critically reading the manuscript and for valuable suggestions in the preparation of the manuscript. Also, we would like to thank Mr. Ryan Kissinger for generating all illustrations used in this artile. This work was supported by the Division of Intramural Research (DIR), National Institutes of Allergy and Infectious Diseases (NIAID), Natonal Institutes of Health (NIH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gould EA, Solomon T. Pathogenic flaviviruses. Lancet. 2008;371(9611):500–9. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- 2.Duffy MR, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360(24):2536–43. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 3.Dos Santos T, et al. Zika Virus and the Guillain-Barre Syndrome - Case Series from Seven Countries. N Engl J Med. 2016;375(16):1598–1601. doi: 10.1056/NEJMc1609015. [DOI] [PubMed] [Google Scholar]
- 4.Foy BD, et al. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17(5):880–2. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miner JJ, Diamond MS. Zika Virus Pathogenesis and Tissue Tropism. Cell Host Microbe. 2017;21(2):134–142. doi: 10.1016/j.chom.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sirohi D, et al. The 3.8 A resolution cryo-EM structure of Zika virus. Science. 2016;352(6284):467–70. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard AS, et al. AXL-dependent infection of human fetal endothelial cells distinguishes Zika virus from other pathogenic flaviviruses. Proc Natl Acad Sci U S A. 2017;114(8):2024–2029. doi: 10.1073/pnas.1620558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pagani I, et al. Human Endometrial Stromal Cells Are Highly Permissive To Productive Infection by Zika Virus. Sci Rep. 2017;7:44286. doi: 10.1038/srep44286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamel R, et al. Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015;89(17):8880–96. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastings AK, et al. TAM Receptors Are Not Required for Zika Virus Infection in Mice. Cell Rep. 2017;19(3):558–568. doi: 10.1016/j.celrep.2017.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabata K, et al. Unique Requirement for ESCRT Factors in Flavivirus Particle Formation on the Endoplasmic Reticulum. Cell Rep. 2016;16(9):2339–47. doi: 10.1016/j.celrep.2016.07.068. [DOI] [PubMed] [Google Scholar]
- 12.Chiramel AI, Brady NR, Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2(1):83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan TX, Randall G. Manipulation or capitulation: virus interactions with autophagy. Microbes Infect. 2012;14(2):126–39. doi: 10.1016/j.micinf.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul P, Munz C. Autophagy and Mammalian Viruses: Roles in Immune Response, Viral Replication, and Beyond. Adv Virus Res. 2016;95:149–95. doi: 10.1016/bs.aivir.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Kimura T, et al. Cellular and molecular mechanism for secretory autophagy. Autophagy. 2017:1–2. doi: 10.1080/15548627.2017.1307486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–74. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 17.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015;25(6):354–63. doi: 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26(1):6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Kaizuka T, Mizushima N. Atg13 Is Essential for Autophagy and Cardiac Development in Mice. Mol Cell Biol. 2015;36(4):585–95. doi: 10.1128/MCB.01005-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto H, et al. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev Cell. 2016;38(1):86–99. doi: 10.1016/j.devcel.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010;584(7):1302–12. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2(3):211–6. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 25.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 26.Otomo C, et al. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013;20(1):59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyama-Honda I, Tsuboyama K, Mizushima N. ATG conjugation-dependent degradation of the inner autophagosomal membrane is a key step for autophagosome maturation. Autophagy. 2017:0. doi: 10.1080/15548627.2017.1319041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuboyama K, et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354(6315):1036–1041. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 29.Dooley HC, et al. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55(2):238–52. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495(7441):389–93. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 31.Diao J, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520(7548):563–6. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1(1):1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 33.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–33. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marshansky V, Rubinstein JL, Gruber G. Eukaryotic V-ATPase: novel structural findings and functional insights. Biochim Biophys Acta. 2014;1837(6):857–79. doi: 10.1016/j.bbabio.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Perera RM, Zoncu R. The Lysosome as a Regulatory Hub. Annu Rev Cell Dev Biol. 2016;32:223–253. doi: 10.1146/annurev-cellbio-111315-125125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S, et al. Galectins and TRIMs directly interact and orchestrate autophagic response to endomembrane damage. Autophagy. 2017:1–2. doi: 10.1080/15548627.2017.1307487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalvari I, et al. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy. 2014;10(5):913–25. doi: 10.4161/auto.28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci. 2013;126(Pt 15):3237–47. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 39.Lippai M, Szatmari Z. Autophagy-from molecular mechanisms to clinical relevance. Cell Biol Toxicol. 2017;33(2):145–168. doi: 10.1007/s10565-016-9374-5. [DOI] [PubMed] [Google Scholar]
- 40.Welsch S, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5(4):365–75. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Offerdahl DK, et al. Cytoarchitecture of Zika virus infection in human neuroblastoma and Aedes albopictus cell lines. Virology. 2017;501:54–62. doi: 10.1016/j.virol.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortese M, et al. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017;18(9):2113–2123. doi: 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao B, et al. Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. J Exp Med. 2017;214(8):2303–2313. doi: 10.1084/jem.20170957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang Q, et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell. 2016;19(5):663–671. doi: 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishnan MN, et al. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J Virol. 2007;81(9):4881–5. doi: 10.1128/JVI.02210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenfeld MR, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10(8):1359–68. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metz P, et al. Dengue Virus Inhibition of Autophagic Flux and Dependency of Viral Replication on Proteasomal Degradation of the Autophagy Receptor p62. J Virol. 2015;89(15):8026–41. doi: 10.1128/JVI.00787-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller S, et al. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem. 2007;282(12):8873–82. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 49.Miller S, Sparacio S, Bartenschlager R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J Biol Chem. 2006;281(13):8854–63. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- 50.Paul D, Bartenschlager R. Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annu Rev Virol. 2015;2(1):289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 51.Khaminets A, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522(7556):354–8. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 52.Lennemann NJ, Coyne CB. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy. 2017;13(2):322–332. doi: 10.1080/15548627.2016.1265192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J, Prinz WA, Rapoport TA. Weaving the web of ER tubules. Cell. 2011;147(6):1226–31. doi: 10.1016/j.cell.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiramel AI, et al. FAM134B, the Selective Autophagy Receptor for Endoplasmic Reticulum Turnover, Inhibits Replication of Ebola Virus Strains Makona and Mayinga. J Infect Dis. 2016;214(suppl 3):S319–S325. doi: 10.1093/infdis/jiw270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colucci F, et al. How does variability of immune system genes affect placentation? Placenta. 2011;32(8):539–45. doi: 10.1016/j.placenta.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaynor LM, Colucci F. Uterine Natural Killer Cells: Functional Distinctions and Influence on Pregnancy in Humans and Mice. Front Immunol. 2017;8:467. doi: 10.3389/fimmu.2017.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delorme-Axford E, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U S A. 2013;110(29):12048–53. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouyang Y, et al. Review: placenta-specific microRNAs in exosomes - good things come in nano-packages. Placenta. 2014;35(Suppl):S69–73. doi: 10.1016/j.placenta.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delorme-Axford E, Sadovsky Y, Coyne CB. The Placenta as a Barrier to Viral Infections. Annu Rev Virol. 2014;1(1):133–46. doi: 10.1146/annurev-virology-031413-085524. [DOI] [PubMed] [Google Scholar]
- 60.Bayer A, et al. Type III Interferons Produced by Human Placental Trophoblasts Confer Protection against Zika Virus Infection. Cell Host Microbe. 2016;19(5):705–12. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8(5):422–32. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng Z, et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496(7445):367–71. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen YH, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160(4):619–30. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Best SM. Viruses PLAY DEAD to TAMe interferon responses. Cell Host Microbe. 2013;14(2):117–8. doi: 10.1016/j.chom.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhattacharyya S, et al. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host Microbe. 2013;14(2):136–47. doi: 10.1016/j.chom.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monel B, et al. Zika virus induces massive cytoplasmic vacuolization and paraptosis-like death in infected cells. EMBO J. 2017 doi: 10.15252/embj.201695597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaattela M, Tschopp J. Caspase-independent cell death in T lymphocytes. Nat Immunol. 2003;4(5):416–23. doi: 10.1038/ni0503-416. [DOI] [PubMed] [Google Scholar]
- 68.Mauthe M, et al. An siRNA screen for ATG protein depletion reveals the extent of the unconventional functions of the autophagy proteome in virus replication. J Cell Biol. 2016;214(5):619–35. doi: 10.1083/jcb.201602046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu Q, et al. Homeostatic Control of Innate Lung Inflammation by Vici Syndrome Gene Epg5 and Additional Autophagy Genes Promotes Influenza Pathogenesis. Cell Host Microbe. 2016;19(1):102–13. doi: 10.1016/j.chom.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]