Abstract
Objective
To investigate whether altered DNA methylation contributes to the inappropriate expression of LINE-1 (L1) retroelements in primary Sjogren’s syndrome (SS) and systemic lupus erythematosus (SLE).
Methods
Minor salivary glands (MSG) were obtained from 42 patients with primary SS [23 without adverse predictors for lymphoma development (SS-low risk), 7 SS-high risk and 12 complicated by B-cell lymphoma (SS-lymphoma)] and 17 sicca controls (SC). Additionally, kidney biopsy specimens and PBMCs were obtained from 23 and 73 lupus patients, respectively. Relative mRNA expression was quantified for full-length L1 transcripts, along with mediators of methylation. In an independent set of 44 MSG samples (11 SS-low risk, 10 SS-high risk, 15 SS-lymphoma and 8 SC), methylation levels of the L1 promoter were determined by bisulphite pyrosequencing.
Results
A strong positive correlation was demonstrated between L1 transcripts and gene products that mediate de novo and constitutive DNA methylation, DNA methyltransferase (DNMT)3B, DNMT1, and methyl CpG binding protein 2 (MeCP2), in both SS MSG and lupus renal tissues. A significant negative correlation was observed between expression of L1 and lymphoid-specific helicase (LSH, encoded by HELLS) in both SS MSG and SLE kidney tissues, as well as between DNMT3A transcripts and L1 expression in SLE kidney tissues and PBMCs. Reduced levels of L1 promoter methylation along with increased DNMT3B, DNMT1, and MeCP2, but reduced LSH levels were detected in SS-low risk patients compared to both SS-lymphoma and SC. The SS-lymphoma group was also characterized by a profound decrease of MeCP2 and DNMT3B compared to SC.
Conclusion
Our data support a contributory role of altered methylation mechanisms in the pathogenesis of systemic autoimmune disorders and related lymphoproliferative processes and suggest that LSH and DNMT3A should be investigated as candidate upstream mediators of decreased L1 promoter methylation and increased L1 expression.
Keywords: Systemic lupus erythematosus (SLE), Sjogren’s syndrome (SS), methylation, LINE-1 expression endogenous retroelements
1. Introduction
The abundant representation in the human genome of virus-like repetitive DNA sequences derived from transposable elements and the documented capacity of some elements to be transcribed and in some cases reinserted into new genomic locations supports those elements as important contributors to gene regulation and the evolution of eukaryotic genomes. Inadequate control of these elements may impose a threat to genome integrity and contribute to disease pathogenesis [1], [2]. We have previously demonstrated that long interspersed nuclear element 1 (LINE-1; L1), among the most abundant members of the transposon family[3], is overexpressed in target tissues from two systemic autoimmune disorders, minor salivary gland (MSG) biopsies from patients with Sjogren’s syndrome (SS) and renal tissues from patients with lupus nephritis, and is mechanistically linked to type I interferon (IFN) production[4], an important contributor to the pathogenesis of those diseases [5, 6]. Additionally, we have also shown an inverse correlation between L1 expression and methylation of CpG sites in the L1 promoter in MSG tissues derived from patients with SS, suggesting methylation alterations as significant contributors to the L1 derepression seen in those tissues[4].
L1 retroelements are retroviral-like, endogenous DNA sequences that are able to amplify and transpose to new locations in the genome through an RNA intermediate. Approximately 80–100 full length active copies have been identified, and though their expression was once thought to be restricted to the germline, it has been increasingly appreciated that L1 expression can occur in several somatic tissues and human neoplasms [3, 7, 8].
Given the potential threat of L1 transposition to the integrity of the human genome, a number of host strategies have evolved to maintain control of L1 expression under physiological conditions. Methylation of the L1 promoter [9], destruction of transcripts mediated by members of the APOBEC family [10], small inhibitory RNA interference mechanisms [11, 12], transcriptional and posttranscriptional regulation by members of the SOX family and the MOV10 helicase [13, 14], interactions with DNA repair enzymes [15], histone modification through retinoblastoma proteins [16], SAMHD1 mediated formation of stress granules[17] and type I IFNs are among the mechanisms that contribute to regulation of L1 transcription and activity[18].
Methylation has been suggested as a major repression mechanism for these elements [9, 19], mediated by a complex interaction of methylating enzymes. Previous observations supported the presence of inter-individual differences in whole blood global L1 methylation, with male sex associated with increased L1 methylation levels [20, 21]. In contrast, age and hormonal factors did not seem to affect L1 methylation in peripheral leucocytes [21]. The methylation machinery includes at least three independent DNA methyltransferases: DNA methyltransferase 1 (DNMT1), DNA methyltransferase 3A (DNMT3A) and DNA methyltransferase 3B (DNMT3B). DNMT1 is the enzyme responsible for copying methylation patterns after DNA replication, and therefore is often referred to as the ‘maintenance’ methyltransferase. DNMT3A and B are implicated in establishing methylation patterns at specific genome locations early in development and are termed de novo methyltransferases [22]. Overlapping functions between the various DNMTs are essential for maintenance of the methylation status of repetitive L1 elements [23]. DNMT3B has been also shown to be essential for methylation of L1 CpG islands on the inactive X chromosome [24], while recruitment of methyl-CpG binding protein 2 (MeCP2) by methylated L1 sequences has the potential to repress L1 expression in somatic tissues [25, 26]. Finally, another chromatin remodeling protein, namely lymphocytic specific helicase (LSH, encoded by the HELLS gene) has been shown to be essential for completion of meiosis and transcriptional repression of repetitive elements in the female gonad [27–30].
In order to identify alterations of host defense mechanisms against inappropriate expression of endogenous retroelements that might contribute to the increased expression of L1 and type I IFNs that we have observed in SS and SLE[4], we performed quantitative gene expression analysis of L1 and members of the methylation machinery in MSG tissue from SS patients and affected renal tissues and PBMCs from SLE patients. As we have previously observed reduced IFNα transcript levels in salivary gland tissues from SS patients complicated by lymphoma[31], we were particularly interested in comparing the relative expression and the methylation status of L1 retroelements, as well as expression of members of the methylation machinery, between SS-low risk (characterized by the presence of ≤2 risk factors for lymphoma development) [32] and SS-lymphoma patients.
2. Patients and Methods
2.1 Patients
The study population included 42 patients with primary SS according to the revised American-European Consensus Group Criteria [33] and 17 sicca controls (SC). SC had subjective and/or objective features of oral/ocular dryness but did not fulfill the above criteria [33]. Primary SS patients and controls had undertaken labial MSG biopsies at the Department of Pathophysiology of the School of Medicine at the University of Athens, Athens, Greece, as a routine part of the diagnostic evaluation for SS. A focus score was determined for each MSG biopsy sample, as previously described [34]. Forty one out of 42 SS patients and all SC were females, and they provided informed consent at the time of MSG biopsy. Primary SS cases were further classified into two groups: a. a low-risk SS group characterized by the presence of ≤2 risk factors previously described as adverse predictors for lymphoma development (Salivary gland enlargement, lymphadenopathy, Raynaud’s phenomenon, Anti-Ro/SSA and/or anti-La/S SB positivity, Rheumatoid factor positivity, Monoclonal gammopathy, and C4 hypocomplementemia) (n=23) [32], b. a high risk SS group characterized by the presence of more than 2 aforementioned adverse predictors (n=7) and c. an SS-lymphoma group, in which B-cell lymphoma had developed (n=12). The mean age±SD of patients with SS-low risk, SS-high risk, SS-lymphoma and SC was 51.3±14.4, 49.0±13.7, 52.6±14.0 and 48±12.1, respectively.
The methylation levels of the L1 promoter were determined in an independent set of 44 samples derived from 11 SS-low risk (11 females), 10 SS-high risk (9 females), 15 SS-lymphoma (15 females) and 8 SC (8 females).
A commercial preparation of total RNA from normal salivary glands pooled from 24 male/female Caucasians (ages 15–60; cause of death: sudden death) was used as a source of healthy donor RNA (Clontech Laboratories, Inc.).
The study was approved by the Laikon Hospital ethics committee and followed the Declaration of Helsinki guidelines.
2.2 Kidney biopsies
Kidney biopsies from 23 patients with lupus nephritis were obtained from the Department of Pathology at the New York Presbyterian Hospital, New York, NY and were classified according to the International Society of Nephrology ISN/RPS 2003 classification criteria (6 class III, 14 class IV, 4 class V) [35]. All but two patients were females between the ages of 11 and 55 at the time of biopsy, fulfilled the American College of Rheumatology classification criteria for SLE [36] and provided informed consent. A commercial preparation of total RNA from renal tissue derived from a single donor was used as a source of healthy donor RNA (#7976, Ambion, USA).
2.3 Peripheral blood mononuclear cells (PBMCs) collection
Seventy-three SLE patients, followed at the Hospital for Special Surgery (HSS) and fulfilling the American College of Rheumatology criteria [36], provided blood samples. These samples were part of the same SLE patient cohort previously studied for interferon-inducible gene expression [37]. Similarly, blood samples were collected from 20 healthy volunteers after they signed informed consent forms.
Additionally, PBMC-derived genomic DNA from 53 pediatric lupus patients obtained from the HSS Lupus Family Registry and 10 healthy controls was analyzed for methylation levels of the L1 promoter.
Twenty milliliters of heparinized blood was centrifuged and the plasma was removed and stored at −70°C. The blood was then further centrifuged over Ficoll-Hypaque to obtain PBMCs.
2.4 RNA isolation
Total RNA was isolated from MSG biopsies, kidney tissues and PBMCs according to standard procedures with the RNeasy Mini Kit (Qiagen, Chatsworth, CA). The quantity of the starting total RNA was 0.1 micrograms as calculated by spectrophotometry.
2.5 Preparation of cDNA
Total mRNA was reverse-transcribed using the Superscript III reverse transcriptase system from Invitrogen (Carlsbad, CA). Oligo-dT primer was used to amplify mRNA specifically and an RNAse inhibitor was included to prevent degradation.
2.6 Quantitative Real-Time Polymerase Chain Reaction
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) was used to quantify specific cDNAs in MSG and kidney tissues as well as in PBMCs samples using the Bio-Rad SYBR Green intercalating fluorophore system with a Bio-Rad I-Cycler thermocycler and fluorescence detector (Bio-Rad, Hercules, CA). Primers for the following genes were designed with Beacon Designer software (Supplementary Table 1): the 5′ UTR of L1 (reflecting full-length transcripts), DNMT3B, DNMT3A, DNMT1, MeCP2 (mediators of methylation) and LSH, - a chromatin remodeling protein. The reaction was carried out in a total volume of 25 μL per reaction. The reaction mixture included 2 μL of template cDNA, 0.4 μM of each primer, 12.5μl of 2x IQ SYBR Green SuperMix (Bio-Rad), and sterile water. The amplification protocol started with 95°C for 4 min followed by 42 cycles at 95°C for 10s, 60°C for 30s and 72°C for 30s. To assess product specificity, amplicons were checked by melting curve analysis. Melting curves were generated from 65°C to 95°C with increments of 0.5°C/cycle for 15s at each cycle and all inconsistent results were discarded. All reactions were performed in duplicate. The amount of Glyceraldehyde Phosphate Dehydrogenase (GAPDH) cDNA was quantified in the samples to control for background gene expression. The threshold values were recorded for each sample in the logarithmic portion of the amplification curve. Standard curves using known quantities of cDNA were created to control for differing efficiency of the PCR reaction at different substrate concentrations.
2.7 DNA Extraction
MSG tissues and PBMCs were immediately stored at −80°C upon collection. DNA extraction was performed with TRIzol Reagent (Ambion, Life Technologies, USA) in an independent set of 22 MSG tissues (8 SS-low risk, 11 SS-lymphoma and 3 SC). Genomic DNA derived from 53 lupus PBMCs was isolated using the Qiagen DNA isolation kit. The quantity and quality of DNA samples was measured spectrophotometrically (Biospec Nano, Japan).
2.8 Quantification of L1 promoter methylation in salivary gland tissues and lupus PBMC
A prevalidated pyrosequencing-based methylation assay was used to assess 4 CpG sites in the promoter region of L1. PCR and pyrosequencing for L1 methylation were performed as previously described [38, 39] using the Pyro-Mark kit (Qiagen, Valencia, CA) in minor salivary gland tissues.
Methylation analysis of the L1 promoter in PBMC genomic DNA derived from 53 lupus patients and 10 healthy controls was performed (GenBank accession number AH005269) by bisulfite-PCR Pyrosequencing [40]. DNA (1 μg) was bisulfite modified using the EZ DNA Methylation kitTM, according to the manufacturer’s instructions (Zymo Research, Orange, CA, USA). The bisulfite treated DNA was subsequently amplified. PCR cycling conditions were 95°C for 15min; (95°C 30 s; 63°C 30 s; 72°C 30 s) for 45 cycles, 72°C for 5 min. The PCR product, with one of the strands biotin labeled, was purified using streptavidin coated sepharose beads (GE Healthcare). The single stranded DNA was generated using Biotage’s Vacuum Prep Tool (Biotage, Uppsala, Sweden) as per the manufacturer’s recommendation. To quantify the methylation level of the L1 promoter, we performed sequencing of the PCR product by pyrosequencing using PyroMark MD System (The Biotage, Uppsala, Sweden). Two pyrosequencing primers (5′-AGTCGACTGAGTCGAGCTGGA-GGTCGTCGATA -3′) and 5′-ATCATGTCGATGTCGAT -3′) annealed to the ssDNA were used to sequence the target regions and quantify the methylation level of 6 CpG sites (labeled CpG pos#1 through CpG pos#6) of the L1 promoter (CpG island: 49-427).
The degree of methylation was expressed as percentage of methylation based on the allele quantification of artificial “C/T” SNPs. The completion of bisulfite modification was verified by a built-in non-CpG cytosine control. The high and low methylated DNA controls (EpigenDx, MA, USA) were used to ensure the quality of the quantification results. The methylation assays were performed at EpigenDX (Worcester, MA, USA).
2.9 Statistics
Differences were considered statistically significant for p<0.05. Comparison between groups was performed by unpaired t-test or Mann Whitney test for Gaussian and non-Gaussian distributions, respectively. Correlation between gene expression data was determined using non-parametric Spearman’s test.
3. Results
3.1 Methylation levels of the L1 promoter in MSG tissues and lupus PBMC
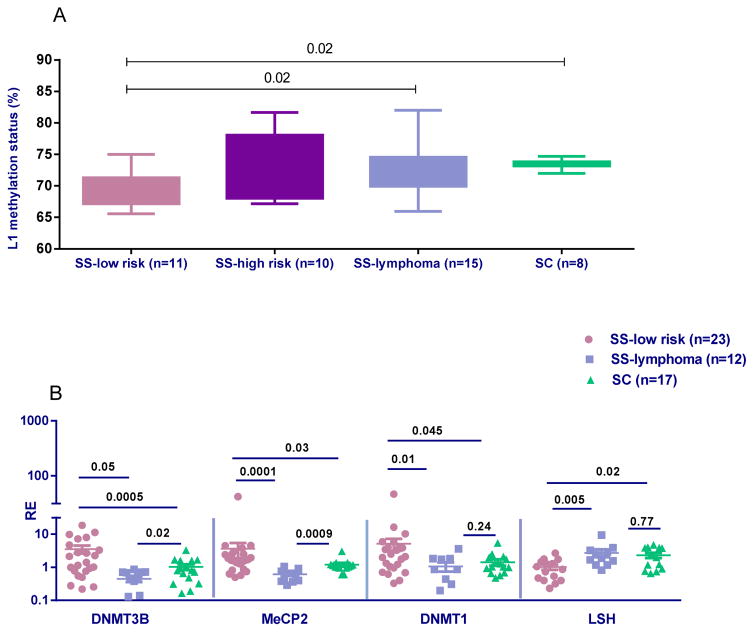
In view of our previous data showing that L1 expression in MSG SS tissues is negatively correlated with the methylation status of the L1 promoter and positively correlated with mRNA IFNα levels [4], together with the low mRNA IFNα levels detected in MSG tissues derived from SS-lymphoma patients [31], we wished to investigate the L1 promoter methylation status in our SS patient groups. In accord with our initial hypothesis, the mean methylation levels of the L1 promoter in DNA extracted from MSG tissues were significantly reduced in SS-low risk patients compared to both SS-lymphoma and SC samples (69.2±3.2 vs 72.7±4.0 vs 73.6±0.8, p-values: 0.02, for both comparisons) (Figure 1A). While no statistically significant difference was detected between SS-lymphoma, SS-high risk and SC samples, a borderline significance between high- and low-risk SS was observed (p-value: 0.08).
Figure 1. Methylation levels of the L1 promoter and mRNA expression of mediators of methylation in MSG tissues from SS patients and controls.
A. Statistically significant decreased mean levels of % methylation at 4 CpG sites of the L1 promoter detected by bisulphite pyrosequencing in SS-low risk patients (n=11) compared to SS-lymphoma patients (n=15) and SC (n=8) (mean±SD: 69.2±3.2 vs 72.7±4.0 vs 73.6±0.8, p-values: 0.02 for both comparisons). No other statistically significant differences were detected in L1 methylation status between 10 high risk SS patients (73.0±5.1) compared to all groups studied. Of note, a borderline significance between high- and low-risk SS was observed (p-value: 0.08). B. Relative gene expression (RE) of methylating enzymes in MSG tissues from SS-low risk patients (n=23), SS-lymphoma patients (n=12) and SC (n=17). All transcripts were measured by real-time PCR. Transcript levels of DNMT1, DNMT3B and MeCP2 were higher in SS compared to both SS-lymphoma (p-values: 0.01, 0.0005 and 0.0001, respectively) and SC group (p-values: 0.045, 0.05 and 0.03, respectively). Significantly decreased LSH levels were observed in the SS-low risk compared to both SS-lymphoma (p-value: 0.005) and SC patients (p-value: 0.02). A profound decrease of MeCP2 and DNMT3B transcripts compared to SC was observed in the SS-lymphoma group (p=0.0009 and p=0.02, respectively).
Additionally, whole blood genomic DNA derived from 53 lupus patients exhibited significantly lower methylation levels at several L1 CpG sites compared to healthy controls (Table 1).
Table 1.
Reduced L1 promoter methylation levels in lupus PBMC compared to healthy controls (HC).
| % mean methylation ± SD | |||||||
|---|---|---|---|---|---|---|---|
| CpG pos #1 | CpG pos #2 | CpG pos #3 | CpG pos #4 | CpG pos #5 | CpG pos #6 | Mean | |
| SLE (n=53) | 45.3±1.05 | 44±0.9 | 35.4±0.9 | 54±1.3 | 31.7±1.0 | 49.1±1.9 | 43.3±1.2 |
| HC (n=10) | 46.1±0.9 | 45.32±1.3 | 36.56±0.7 | 55.23±1.4 | 32.88±1.2 | 50.04±2.89 | 44.4±1.6 |
| p-value | 0.06 | 0.005 | 0.004 | 0.02 | 0.09 | 0.52 | 0.01 |
3.2 Mediators of methylation in MSG tissues
In order to investigate whether differential methylation of the L1 promoter between the SS groups is related to alterations in enzymes involved in the methylation machinery, transcript levels of DNMT1, DNMT3A, DNMTB, LSH and MeCP2 were measured in MSG biopsies from SS-low risk, SS-lymphoma and SC using quantitative real-time PCR. Transcript levels of DNMT1, DNMT3B and MeCP2 were significantly increased in SS-low risk compared to both SS-lymphoma (p-values: 0.01, 0.0005 and 0.0001, respectively) and SC group (p-values: 0.045, 0.05 and 0.03, respectively). On the other hand, significantly decreased LSH levels were observed in the SS-low risk group compared to both SS-lymphoma (p-value: 0.005) and SC (p-value: 0.02) groups (Figure 1B). Furthermore, SS-lymphoma patients were characterized by a profound decrease of MeCP2 and DNMT3B transcripts compared to SC (p-values: 0.0009 and 0.02, respectively). No differences were noted between patient groups for DNMT3A transcripts (data not shown).
In regard to the SS-high risk group, DNMT3b gene expression was found to be significantly lower compared to SS-low risk (0.8±0.9 vs 3.6±4.6, p-value: 0.04) and MeCP2 transcripts were significantly higher compared to SS-lymphoma (1.5±1.1 vs 0.6 ±0.2, p-value: 0.007). All other comparisons between groups and other methylating enzymes were non-significant (data not shown).
3.3 Strong correlation of several mediators of methylation with L1 expression in SS MSG biopsies
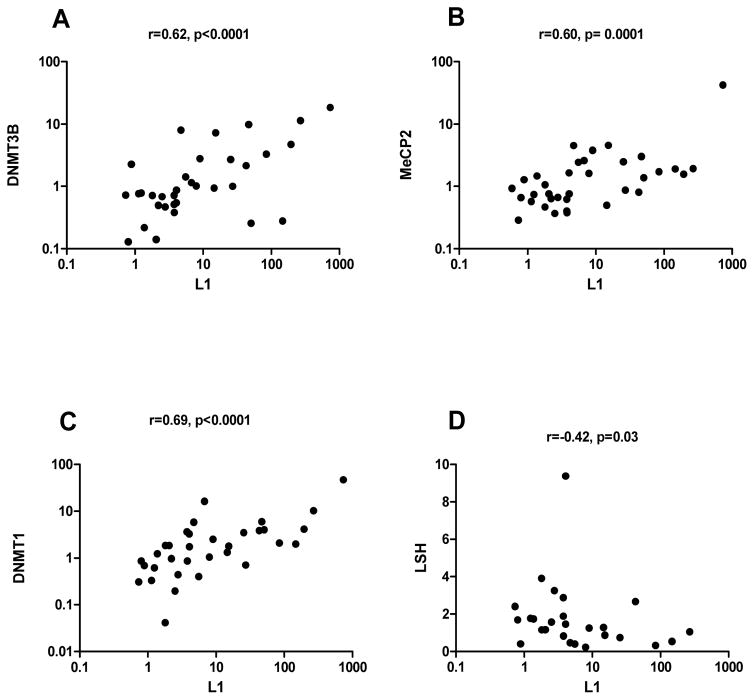
To explore potential alterations of epigenetic mechanisms of L1 regulation in SS-low risk and SS-lymphoma patients, we investigated whether expression of full-length L1 transcripts correlates with expression of methylating enzymes in MSG tissue. Figure 2 illustrates a strong positive correlation of L1 full length mRNA levels with DNMT3B, MeCP2 and DNMT1 transcripts, suggesting their potential compensatory role in controlling inappropriate expression of L1 retroelements (r=0.62, p<0.0001, r=0.60, p=0.0001, r=0.69, p<0.0001, panels A, B, C respectively). Of interest, a negative correlation between L1 and LSH transcript levels was also observed, implying a potential role of this regulator in the defective L1 methylation noted in SS MSG tissues (r=−0.42, p=0.03, panel D). A trend toward a negative relationship between DNMT3A transcripts and L1 was found (r=−0.3, p=0.09) (data not shown). In MSG derived from SC, no associations were detected between L1 and methylating enzyme mRNA levels (Supplementary table 2).
Figure 2. Association between L1 retroelements and members of the methylation machinery.
Relative expression of L1 retroelements and DNMT3B, MeCP2, DNMT1 and LSH was measured by real-time PCR in MSG biopsy specimens obtained from SS-low risk and SS-lymphoma patients. Association between L1 mRNA levels and members of the methylation machinery were tested using Spearman’s nonparametric correlation test. L1 expression was positively correlated with DNMT3B, MeCP2, DNMT1 transcripts (r=0.62, p<0.0001, r= 0.60, p=0.0001, r=0.69, p<0.0001 respectively, panels A–C) and negatively correlated with LSH mRNA levels (r=−0.42, p=0.03, panel D).
3.4 Mediators of methylation in renal biopsies and PBMCs from patients with SLE
We have previously demonstrated higher L1 expression in affected renal tissues and PBMCs from SLE patients compared to HC [4]. To explore a potential regulatory role of members of the methylation machinery in the inappropriate L1 expression in SLE, we measured mRNA expression of DNMT1, DNMT3A, DNMT3B, LSH and MeCP2 in renal biopsies and PBMCs from patients with SLE. In a similar pattern to that observed in SS MSG tissue, a coordinate expression of MeCP2, DNMT1 and DNMT3B, was found at the level of renal tissues from SLE patients (r=0.48, p=0.02; r=0.52, p=0.01; and r=0.63, p=0.001, respectively). On the other hand, significant negative correlations were noted between DNMT3A, LSH and L1 expression, suggesting that primary defects of these enzymes could account for the increased L1 expression, at least in some of the SLE patients. Similar observations were noted when L1 expression was related to a panel of methylating gene transcripts in the PBMC from these patients (Supplementary Table 3).
4. Discussion
The burden of transposable elements in the mammalian genome and their potential for genome disruption demand redundant and strict control mechanisms to suppress the activation and mobility of those parasitic repeat elements. Although transcriptional regulation of L1 elements is not fully elucidated, methylation of the L1 promoter is among the major mechanisms of L1 suppression [41]. In the current study, we present evidence of defective L1 promoter methylation as well as reduced expression of the chromatin remodeling enzyme LSH in low risk SS compared to both SS-lymphoma patients and SC, as well as an inverse correlation between LSH transcripts and L1 expression in SS and SLE-derived target tissues. Low risk SS patients – with a probability for lymphoma development being less than 5%, are those characterized by the presence of ≤2 independent risk factors recently designated as adverse predictors for lymphoma development including salivary gland enlargement, lymphadenopathy, Raynaud’s phenomenon, anti-Ro/SSA and/or anti-La/S SB positivity, rheumatoid factor positivity, monoclonal gammopathy, and C4 hypocomplementemia [32]. A significantly negative correlation between L1 and DNMT3A was also detected in both lupus kidneys and PBMCs, providing a potential mechanism for the inadequate L1 control observed in these patients. Furthermore, the observed upregulation of the methylation mediators DNMT1, MeCP2 and DNMT3B levels along with the strong correlation with L1 expression in target tissues from both low risk SS and SLE implies the mobilization of epigenetic host defense responses against inappropriate expression of L1 endogenous retroelements in these disorders. An opposite pattern was observed in SS patients complicated by lymphoma, in which a profound decrease of DNMT1, MeCP2 and DNMT3B along with higher L1 methylation and LSH levels (though comparable to that observed in SC) was detected compared to low risk SS. The observed differential methylation patterns of the L1 promoter provide a potential explanation for the difference in L1 expression levels between the two SS groups.
Reduced levels of LSH in MSG tissues derived from patients with low risk SS along with the observed negative correlation between LSH and L1 expression in SS MSG and lupus kidney tissues might suggest LSH as an important L1 suppressor which is deranged in systemic autoimmune diseases leading to L1 hypomethylation, L1 overexpression and induction of the type I IFN pathway. LSH is a member of the SNF2 subfamily of helicases. Limited data so far indirectly link LSH to the pathogenesis of autoimmune disease. E2F1 transcription factors, deficiency of which leads to type I diabetes and SS in non-obese diabetic mice [42], have been shown to physically interact with the LSH promoter resulting in increased expression of LSH at both transcriptional and translational levels [43]. LSH has been also shown to be essential for the proliferation of T lymphocytes, is expressed in the lymphoid tissue of the adult mice, and is involved in the maintenance of methylation in female gonads, providing a potential clue for the gender bias towards females in SLE and SS [27, 44].
Additionally, our data show that DNMT3A levels negatively correlate with L1 expression at the level of renal tissues and PBMCs from lupus patients. Together, our data identify LSH and DNMT3A as candidate enzymes that might be deficient in systemic autoimmune disease and contribute to impaired control of L1 expression. In contrast to our results, a recent study reported increased DNMT3A transcripts in PMBCs from lupus patients compared to controls, especially among patients of African American origin [45] and another showed decreased DNMT1 expression and activity in lupus T cells [46]. A study of salivary gland epithelial cells derived from SS patients showed decreased DNMT1 levels along with decreased global methylation levels [47]. Methylation enzyme expression may differ depending on cell type studied as well as the activation status of those cells.
Next, a positive correlation of several methylating enzymes, including DNMT3B, MeCP2 and DNMT-1, along with L1 expression was detected in both SLE kidney tissue and SS MSG tissues. Similar observations were also made in lupus PBMCs in regard to MeCP2 and DNMT3B but not DNMT1 levels. Of interest, the above methylation mediators have been previously implicated in L1 control under physiological conditions, and MeCP2 polymorphisms were found to confer increased SLE and SS risk [23–25, 48, 49]. In addition, the risk MeCP2 variant was found to be associated with increased IFN-inducible gene expression in patients with lupus, a hallmark of disease pathogenesis [37, 50]. Furthermore, increased levels of DNMT3B were previously reported to be higher in PBMC from Caucasian but not African American SLE patients compared to healthy controls, along with a global decrease in 5-methylcytosine [45].
While SS-low risk and SLE patients seem to share common patterns in the expression of various methylation mediators, to our surprise we noted a distinct epigenetic profile in MSG tissues from SS patients complicated by lymphoma. In accord with our previous findings showing reduced L1 expression levels in SS-lymphoma patients compared to low risk SS, methylation levels of the L1 promoter along with LSH transcripts were higher in SS patients complicated by lymphoma compared to the benign SS subset. Of interest, differences in methylation status between SS and SS lymphoma groups are revealed only when SS group is divided into low and high risk since L1 promoter methylation in the high risk-SS group was comparable to the SS-lymphoma group and higher (borderline significance) than the low SS risk group. This finding could provide a potential explanation for the lack of association between methylation status and lymphoma development in a previous report, including a mixture of low risk and high risk SS patients [39]. On the other hand, decreased expression of several methylating enzymes, including DNMT3B, MeCP2 and DNMT1, were observed in SS patients complicated by lymphoma compared to SS alone and SC, implying a possible role of these methylating enzymes in SS-related lymphomagenesis. Though L1 hypomethylation and increased L1 expression has been reported for numerous solid cancers and may represent a mechanism that contributes to genomic instability, there is little evidence implicating L1 hypomethylation in the pathogenesis of lymphomas [51]. Methylation alterations have been implicated in tumorigenesis either by suppressing tumor suppressor genes or by re-depressing normally silenced oncogenes. Several reports viewed de novo methyltransferases mainly as oncogenes, since they were found to be increased in several cancers and lymphomas and connoted adverse prognosis [52, 53]. However, and in line with our current findings in SS related lymphomas, a growing body of data suggest rather a tumor suppressive role with inhibition of de novo methylation arising as a central oncogenic event in the pathogenesis of hematological malignancies [54, 55]. Of interest, inactivating DNMT3A mutations have been also linked to both acute myelogenous leukemia and myelodysplastic syndrome [56, 57].
The coordinate expression of endogenous host defense mechanisms that regulate endogenous retroelements, including mediators of methylation in association with increased expression of L1 mRNA in SS MSG and SLE renal tissue, supports the in vivo relevance of these retrotransposon transcripts in patients with autoimmune disorders and may reflect a compensatory mechanism aimed at controlling expression of potentially pathogenic L1 elements. Defective LSH and/or DNMT3A expression, based on genetic variation or environmental influences, might underlie the impaired methylation patterns seen in SS and SLE patients, identifying LSH as a significant gatekeeper against aberrant autoimmune responses through methylation and silencing of endogenous retroelements. Finally, distinct epigenetic signatures between SS patients with or without complication with lymphoma suggest a potential contribution of these altered epigenetic mechanisms to SS-related lymphomagenesis and need to be further explored. Environmental triggers in association with genetic variations might precede the epigenetic alterations observed.
Supplementary Material
Supplementary table 1: Primer sequences
Supplementary table 2: Non statistically significant associations between L1 mRNA expression with methylating enzymes transcripts in MSG tissues from sicca controls (SC).
Supplementary table 3: Correlations of L1 mRNA expression with expression of several enzymes of the methylation machinery in PBMC from SLE patients.
Table 2.
L1 mRNA expression levels were positively associated with DNMT3B, MeCP2 and DNMT1 whereas negatively correlated with DNMT3A and LSH mRNA expression at the level of kidney tissues derived from 23 SLE patients. Spearman’s rho correlation coefficients and corresponding p-values are shown.
| DNMT1 | DNMT3A | DNMT3B | LSH | MeCP2 | ||
|---|---|---|---|---|---|---|
| L1 | rho | 0.52 | −0.44 | 0.63 | −0.46 | 0.48 |
| p-value | 0.01 | 0.003 | 0.001 | 0.03 | 0.02 | |
Highlights.
Defective L1 promoter methylation in MSG tissues derived from low risk SS and SLE patients compared to controls
Inverse correlation between the chromatin modelling protein LSH expression and L1 expression in SS and SLE-derived target tissues.
Coordinate expression of L1 mRNA expression with mediators of methylation MeCP2, DNMT1 and DNMT3B in SS and SLE-derived target tissues.
Acknowledgments
Funding: This study was supported by a Stavros Niarchos Fellowship grant through the Arthritis Foundation, New York Chapter to CPM and a Stavros Niarchos Foundation Research Grant to Department of Physiology, University of Athens; and NIH R01AI059893, a Novel Research Grant from the Lupus Research Institute, a Target Identification in Lupus grant from the Alliance for Lupus Research, and the Mary Kirkland Center for Lupus Research to MKC.
The authors would like to thank Prof HM Moutsopoulos, MD for providing MSG tissues from patients with SS, Dr E Piperi, DDS for providing 3 MSG tissues from sicca controls for methylation studies and E Kapsogeorgou, PhD for excellent technical help.
Footnotes
Conflicts of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hancks DC, Kazazian HH., Jr Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science. 2016;351:aac7247. doi: 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crow MK. Long interspersed nuclear elements (LINE-1): potential triggers of systemic autoimmune disease. Autoimmunity. 2010;43:7–16. doi: 10.3109/08916930903374865. [DOI] [PubMed] [Google Scholar]
- 4.Mavragani CP, Sagalovskiy I, Guo Q, Nezos A, Kapsogeorgou EK, Lu P, et al. Long interspersed nuclear element-1 retroelements are expressed in patients with systemic autoimmune disease and induce type I interferon. Arthritis Rheumatol. 2016 doi: 10.1002/art.39795. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mavragani CP, Crow MK. Activation of the type I interferon pathway in primary Sjogren’s syndrome. J Autoimmun. 35:225–31. doi: 10.1016/j.jaut.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Crow MK, Wohlgemuth J. Microarray analysis of gene expression in lupus. Arthritis Res Ther. 2003;5:279–87. doi: 10.1186/ar1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010;38:3909–22. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–70. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thayer RE, Singer MF, Fanning TG. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene. 1993;133:273–7. doi: 10.1016/0378-1119(93)90651-i. [DOI] [PubMed] [Google Scholar]
- 10.Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol. 2013;4:28. doi: 10.3389/fmicb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 12.Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ, et al. miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat Struct Mol Biol. 2015;22:824–31. doi: 10.1038/nsmb.3090. [DOI] [PubMed] [Google Scholar]
- 13.Tchenio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–5. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, et al. The MOV10 helicase inhibits LINE-1 mobility. J Biol Chem. 2013;288:21148–60. doi: 10.1074/jbc.M113.465856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst) 2008;7:983–9. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montoya-Durango DE, Liu Y, Teneng I, Kalbfleisch T, Lacy ME, Steffen MC, et al. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat Res. 2009;665:20–8. doi: 10.1016/j.mrfmmm.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu S, Li J, Xu F, Mei S, Le Duff Y, Yin L, et al. SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genet. 2015;11:e1005367. doi: 10.1371/journal.pgen.1005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Q, Carbone CJ, Katlinskaya YV, Zheng H, Zheng K, Luo M, et al. Type I interferon controls propagation of long interspersed element-1. J Biol Chem. 2015;290:10191–9. doi: 10.1074/jbc.M114.612374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–33. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–14. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 21.El-Maarri O, Walier M, Behne F, van Uum J, Singer H, Diaz-Lacava A, et al. Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PLoS One. 2011;6:e16252. doi: 10.1371/journal.pone.0016252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 23.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–91. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen RS. X inactivation-specific methylation of LINE-1 elements by DNMT3B: implications for the Lyon repeat hypothesis. Hum Mol Genet. 2003;12:2559–67. doi: 10.1093/hmg/ddg268. [DOI] [PubMed] [Google Scholar]
- 25.Yu F, Zingler N, Schumann G, Stratling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 2001;29:4493–501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–6. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8:1448–54. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- 28.Termanis A, Torrea N, Culley J, Kerr A, Ramsahoye B, Stancheva I. The SNF2 family ATPase LSH promotes cell-autonomous de novo DNA methylation in somatic cells. Nucleic Acids Res. 2016;44:7592–604. doi: 10.1093/nar/gkw424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Y, Ren J, Lee E, Xu X, Yu W, Muegge K. Lsh/HELLS regulates self-renewal/proliferation of neural stem/progenitor cells. Sci Rep. 2017;7:1136. doi: 10.1038/s41598-017-00804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burrage J, Termanis A, Geissner A, Myant K, Gordon K, Stancheva I. The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J Cell Sci. 2012;125:5524–34. doi: 10.1242/jcs.111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nezos A, Gravani F, Tassidou A, Kapsogeorgou EK, Voulgarelis M, Koutsilieris M, et al. Type I and II interferon signatures in Sjogren’s syndrome pathogenesis: Contributions in distinct clinical phenotypes and Sjogren’s related lymphomagenesis. J Autoimmun. 2015;63:47–58. doi: 10.1016/j.jaut.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fragkioudaki S, Mavragani CP, Moutsopoulos HM. Predicting the risk for lymphoma development in Sjogren syndrome: An easy tool for clinical use. Medicine (Baltimore) 2016;95:e3766. doi: 10.1097/MD.0000000000003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, et al. Classification criteria for Sjogren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Annals of the rheumatic diseases. 2002;61:554–8. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chisholm DM, Mason DK. Labial salivary gland biopsy in Sjogren’s disease. Journal of clinical pathology. 1968;21:656–60. doi: 10.1136/jcp.21.5.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weening JJ, D’Agati VD, Schwartz MM, Seshan SV, Alpers CE, Appel GB, et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol. 2004;15:241–50. doi: 10.1097/01.asn.0000108969.21691.5d. [DOI] [PubMed] [Google Scholar]
- 36.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 37.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 38.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–73. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fragkioudaki S, Nezos A, Souliotis VL, Chatziandreou I, Saetta AA, Drakoulis N, et al. MTHFR gene variants and non-MALT lymphoma development in primary Sjogren’s syndrome. Scientific Reports. 2017;7:7354. doi: 10.1038/s41598-017-07347-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piyathilake CJ, Badiga S, Alvarez RD, Partridge EE, Johanning GL. A lower degree of PBMC L1 methylation is associated with excess body weight and higher HOMA-IR in the presence of lower concentrations of plasma folate. PLoS One. 2013;8:e54544. doi: 10.1371/journal.pone.0054544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28:527–39. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 42.Salam MA, Matin K, Matsumoto N, Tsuha Y, Hanada N, Senpuku H. E2f1 mutation induces early onset of diabetes and Sjogren’s syndrome in nonobese diabetic mice. J Immunol. 2004;173:4908–18. doi: 10.4049/jimmunol.173.8.4908. [DOI] [PubMed] [Google Scholar]
- 43.Niu J, Chen T, Han L, Wang P, Li N, Tong T. Transcriptional activation of the senescence regulator Lsh by E2F1. Mech Ageing Dev. 2011;132:180–6. doi: 10.1016/j.mad.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Geiman TM, Muegge K. Lsh, an SNF2/helicase family member, is required for proliferation of mature T lymphocytes. Proc Natl Acad Sci U S A. 2000;97:4772–7. doi: 10.1073/pnas.97.9.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiley KL, Treadwell E, Manigaba K, Word B, Lyn-Cook BD. Ethnic differences in DNA methyltransferases expression in patients with systemic lupus erythematosus. J Clin Immunol. 2013;33:342–8. doi: 10.1007/s10875-012-9803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, et al. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–56. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 47.Thabet Y, Le Dantec C, Ghedira I, Devauchelle V, Cornec D, Pers JO, et al. Epigenetic dysregulation in salivary glands from patients with primary Sjogren’s syndrome may be ascribed to infiltrating B cells. J Autoimmun. 2013;41:175–81. doi: 10.1016/j.jaut.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Sawalha AH, Webb R, Han S, Kelly JA, Kaufman KM, Kimberly RP, et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS One. 2008;3:e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cobb BL, Fei Y, Jonsson R, Bolstad AI, Brun JG, Rischmueller M, et al. Genetic association between methyl-CpG binding protein 2 (MECP2) and primary Sjogren’s syndrome. Ann Rheum Dis. 2010;69:1731–2. doi: 10.1136/ard.2009.122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb R, Wren JD, Jeffries M, Kelly JA, Kaufman KM, Tang Y, et al. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 2009;60:1076–84. doi: 10.1002/art.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodic N, Burns KH. Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms? PLoS Genet. 2013;9:e1003402. doi: 10.1371/journal.pgen.1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amara K, Ziadi S, Hachana M, Soltani N, Korbi S, Trimeche M. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci. 2010;101:1722–30. doi: 10.1111/j.1349-7006.2010.01569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasanthakumar A, Lepore JB, Zegarek MH, Kocherginsky M, Singh M, Davis EM, et al. Dnmt3b is a haploinsufficient tumor suppressor gene in Myc-induced lymphomagenesis. Blood. 2013;121:2059–63. doi: 10.1182/blood-2012-04-421065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hlady RA, Novakova S, Opavska J, Klinkebiel D, Peters SL, Bies J, et al. Loss of Dnmt3b function upregulates the tumor modifier Ment and accelerates mouse lymphomagenesis. J Clin Invest. 2012;122:163–77. doi: 10.1172/JCI57292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–8. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1: Primer sequences
Supplementary table 2: Non statistically significant associations between L1 mRNA expression with methylating enzymes transcripts in MSG tissues from sicca controls (SC).
Supplementary table 3: Correlations of L1 mRNA expression with expression of several enzymes of the methylation machinery in PBMC from SLE patients.