Abstract
In order to better understand the process of breast cancer metastasis, we have generated a mammary epithelial progression series of increasingly aggressive cell lines that metastasize to lung. Here, we demonstrate that up-regulation of an Endoplasmic Reticulum (ER) to Golgi trafficking gene signature in metastatic cells enhances transport kinetics, which promotes malignant progression. We observe increased ER-Golgi trafficking, an altered secretome and sensitivity to the retrograde transport inhibitor brefeldin A (BFA) in cells that metastasize to lung. CREB3 was identified as a transcriptional regulator of up-regulated ER-Golgi trafficking genes ARF4, COPB1 and USO1, and silencing of these genes attenuated the metastatic phenotype in vitro and lung colonization in vivo. Furthermore, high trafficking gene expression significantly correlated with increased risk of distant metastasis and reduced relapse-free and overall survival in breast cancer patients, suggesting that modulation of ER-Golgi trafficking plays an important role in metastatic progression.
Keywords: breast cancer, metastasis, ER-Golgi trafficking
Introduction
As the majority of cancer-associated deaths are caused by metastasis, there is significant interest in identifying the mechanisms through which cancer cells acquire the ability to metastasize, in the hope that such mechanisms may be targeted therapeutically. Our laboratory has previously demonstrated that the RNA binding protein hnRNP E1, a tumor suppressor, is an important regulator of the epithelial to mesenchymal transition (EMT)1–4. This transition is a normal cellular process involved in wound healing and embryonic development, and thought to be aberrantly activated in the early stages of metastasis. The transition to a mesenchymal phenotype is associated with a loss of apical/basolateral polarity and the acquisition of migratory, invasive and stem cell-like properties5. Silencing of hnRNP E1 in non-tumorigenic mammary epithelial cells induces this transition resulting in a mesenchymal, invasive phenotype1. The in vivo selection of these tumorigenic hnRNP E1 silenced (E1KD) cells provides a unique system to interrogate gene signatures in mammary epithelial cells associated with cancer initiation, tumorigenesis and metastatic progression.
Here, we describe the co-regulation of several ER-Golgi trafficking genes in our mammary epithelial cell series that alter traffic kinetics and in turn metastatic progression. The ER and Golgi are essential for processing and trafficking of a large portion of the proteome. ER-processed proteins are transported to the Golgi through COPII vesicles regulated by RAB GTPases6. N-glycan modification and O-linked glycosylation of proteins occur within the linked cisternae that comprise the Golgi ribbon, before transport to the Trans-Golgi Network (TGN) for sorting6,7. Retrograde transport of ER-resident proteins from the Golgi to the ER occurs through the regulation of ADP-ribosylation factors (ARFs), and their guanine nucleotide exchange factors (GEFs) which control COPI vesicle budding6,8. Golgi mediated regulation of multiple processes including mitosis, apoptosis and migration is described9–12. Recent studies have also demonstrated a role for ER-Golgi trafficking genes in promoting cancer progression through alteration of the secretome13,14.
ER stress sensing and the downstream induction of the unfolded protein response (UPR) have been well characterized in the literature15. Stress, such as the accumulation of unfolded or misfolded proteins, activates the three branches of this response mediated by ER-resident protein kinase R-like kinase (PERK), activating transcription factor-6 (ATF6) and inositol-requiring enzyme 1α (IRE1α)16. The UPR acts to alleviate stress and restore ER function by blocking translation and promoting degradation of misfolded proteins through downstream effectors such as PERK-activated eIF2α and IRE1α-induced splicing of the X-box Binding Protein (XBP1)17. Acute or prolonged ER stress, in which ER homeostasis cannot be restored, induces apoptosis through effectors including CCAAT/enhancer-binding protein homologous protein (CHOP)18. Activation of a Golgi stress response has been reported in several studies12,19–22; this response is hypothesized to restore Golgi function following stressors such as increased protein load and viral infection23. However, the interdependence between anterograde and retrograde ER-Golgi trafficking confounds analyses of stress responses originating from the Golgi. Consequently, the stimuli and downstream effectors that regulate Golgi homeostasis are poorly understood.
The cAMP responsive element binding protein 3 (CREB3) subfamily of basic leucine zipper transcription factors (TFs) consists of CREB3, CREB3L1, CREB3L2, CREB3L3 and CREB3L4. These ER-localized TFs function in numerous processes including secretion, UPR, osteogenesis and chondrogenesis24–28. Activation of CREB3 TFs occurs through regulated intramembrane proteolysis (RIP), similar to ATF6 and sterol-regulatory element-binding protein (SREBP) activation, where C-terminal transmembrane domains are cleaved by site 1 protease (S1P) and site 2 protease (S2P) localized in the Golgi24,29,30. We hypothesize the CREB3 activation in our model up-regulates ER-Golgi trafficking gene expression in metastatic cells driving malignant progression. Here, we demonstrate CREB3 regulation of ER-Golgi trafficking genes in cells derived from our mouse metastatic progression model. Increased ER-Golgi trafficking and secretion in these cell lines associated with an invasive phenotype, which was attenuated by silencing of ARF4, COPB1 and USO1.
Results
Isolation of tumorigenic and metastatic cell lines through the in vivo selection of mammary epithelial cells
We have developed a mouse model of metastasis utilizing the non-transformed normal murine mammary gland (NMuMG) cell line. NMuMG cells exhibit an non-invasive, epithelial phenotype and transition to an invasive mesenchymal phenotype upon silencing of the RNA binding protein hnRNP E1, which regulates the epithelial-to-mesenchymal transition1. In the absence of hnRNP E1 expression, NMuMG cells become both tumorigenic and metastatic, with metastases identified in the lungs of mammary fat pad injected NOD/SCID mice. In vivo passaging of hnRNP E1 knockdown (E1KD) cells via mammary fat pad xenograft led to the isolation of L1P and L2P cells that metastasize from the mammary fat pad to lung. In addition, the M1P and M2P cell lines were isolated from primary mammary tumors (Supplementary Fig S1A).
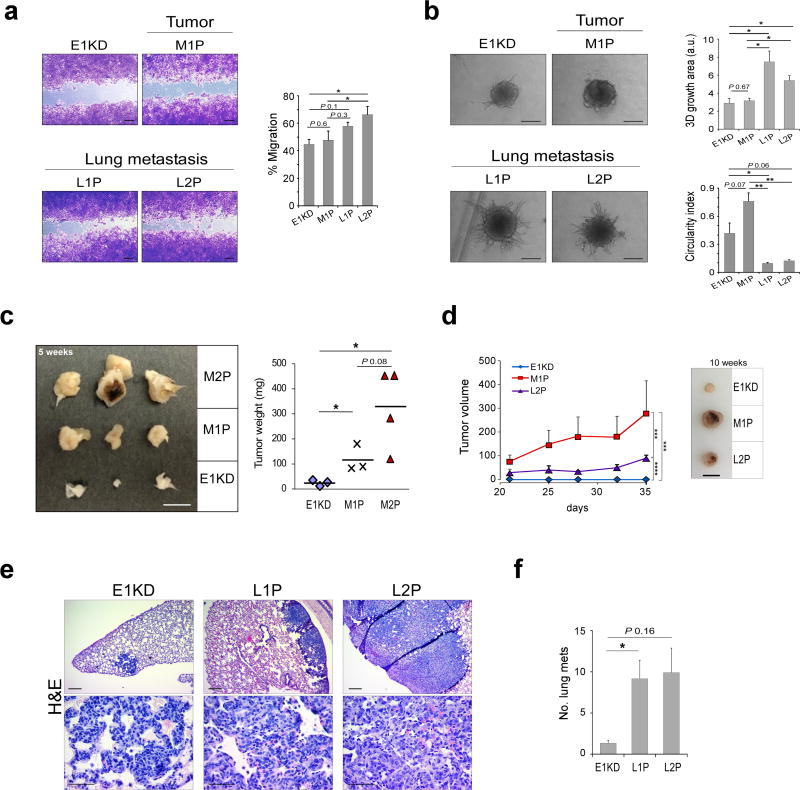
When cultured ex vivo the passaged cell lines retain hnRNP E1 knockdown and a mesenchymal phenotype, as assessed by reduced E-cadherin cell surface expression and actin cytokeleton organization compared to NMuMG cells (Supplementary Fig S1B and G). In addition, in vivo passaged cells and E1KD cells express the pSilencer shRNA used to silence hnRNP E1, as determined by semi-quantitative PCR (Supplementary Fig S1C). We observed significantly enhanced migration of L2P cells, compared to E1KD and M1P cells (Fig 1A). In addition, increased invasiveness of L1P and L2P cells was observed compared to E1KD and M1P cells, as determined by increased sphere area and decreased circularity index (Fig 1B). Flow cytometric analysis of CD29/CD24 levels demonstrated an increase in CD29 levels and a decrease in CD24 levels in the L series of cells, indicative of a cancer stem cell phenotype (Supplementary Fig S1E). In agreement with these findings, an increase in mammosphere formation, a marker of cancer cell stemness, was observed in the metastatic L1P and L2P cells but not in the tumor-isolated M1P cell line (Supplementary Fig S1F). Next, to determine whether our passaged cells retain tumor forming (M series) or metastases forming (L series) ability following culture ex vivo, xenografts were performed. E1KD cells along with the M series (M1P and M2P) and L2P cells were injected into the mammary fat pad of NOD/SCID mice. A progressive increase in tumor size was observed following passaging of the M series, compared to E1KD and L2P cells (Fig 1C and D). Similarly, an increase in lung metastases was observed following tail vein injection of L1P and L2P cells compared to E1KD cells (Fig 1E and F).
Figure 1. Characterization of in vivo selected mammary epithelial cells.
(a) Representative images and quantitation of wound healing assay assessing migration of E1KD, M1P, L1P and L2P cells (n ≥ 4, paired t-test *P<0.05, scale bar: 200 µm). (b) Representative images and quantitation of 3D invasion assay assessing invasion of E1KD, M1P, L1P and L2P cells (n ≥ 3, unpaired t-test *P<0.05, scale bar: 200 µm). (c) Image of mammary fat pad tumors excised from NOD/SCID mice injected with E1KD, M1P or M2P cells (left panel). Graph of E1KD, M1P and M2P tumor weight (n ≥ 3, unpaired t-test *P<0.05, scale bar: 1 cm). (d) Graph of E1KD, M1P and L2P tumor volume (n ≥ 3, ***P < 0.001, **** P < 0.0001; two-way ANOVA with Bonferroni's post-hoc test, scale bar: 1 cm). (e) Representative images of H&E stained lung tissue sections from NOD/SCID mice tail vein injected with E1KD, L1P and L2P cells (n ≥ 7 mice per group). Scale bar: 200 µm (top panel) and 50 µm (bottom panel). (f) Lung metastases number in mice tail vein injected with E1KD, L1P and L2P cells (unpaired t-test, * P<0.05).
Identification of a metastasis-associated ER-Golgi trafficking gene signature
As the accumulation of genetic and epigenetic alterations in cancer cells is thought to drive cancer progression, we were interested in utilizing Affymetrix array as an unbiased approach to characterize differential gene expression in our model. We anticipated the identification of a cohort of genes associated with tumor initiation and growth, as determined by comparison of the non-tumorigenic NMuMG with tumor-forming E1KD and M1P cell lines. In addition, we expected to observe gene expression changes linked to metastatic potential, whereby a change in metastasis-associated genes would be observed in L1P and L2P cells, compared to E1KD cells.
A 1.5 fold change in gene expression and P<0.05 was used to filter array data (Fig 2A and Supplementary Fig S2). To identify expression changes associated with tumorigenic versus metastatic cell traits, a 3-way comparison of M1P/E1KD, L1P/E1KD and L2P/E1KD gene sets was utilized, using Venny 2.1 software31 (Fig 2B and Supplementary Fig S2A and C). Gene set enrichment analysis was then performed using DAVID 6.8 software32,33, to identify processes enriched in L1P and L2P cells. Interestingly, Golgi-related pathways, such as ER to Golgi vesicle mediated transport and Golgi organization, were significantly enriched in L1P and L2P up-regulated gene sets (Fig 2C).
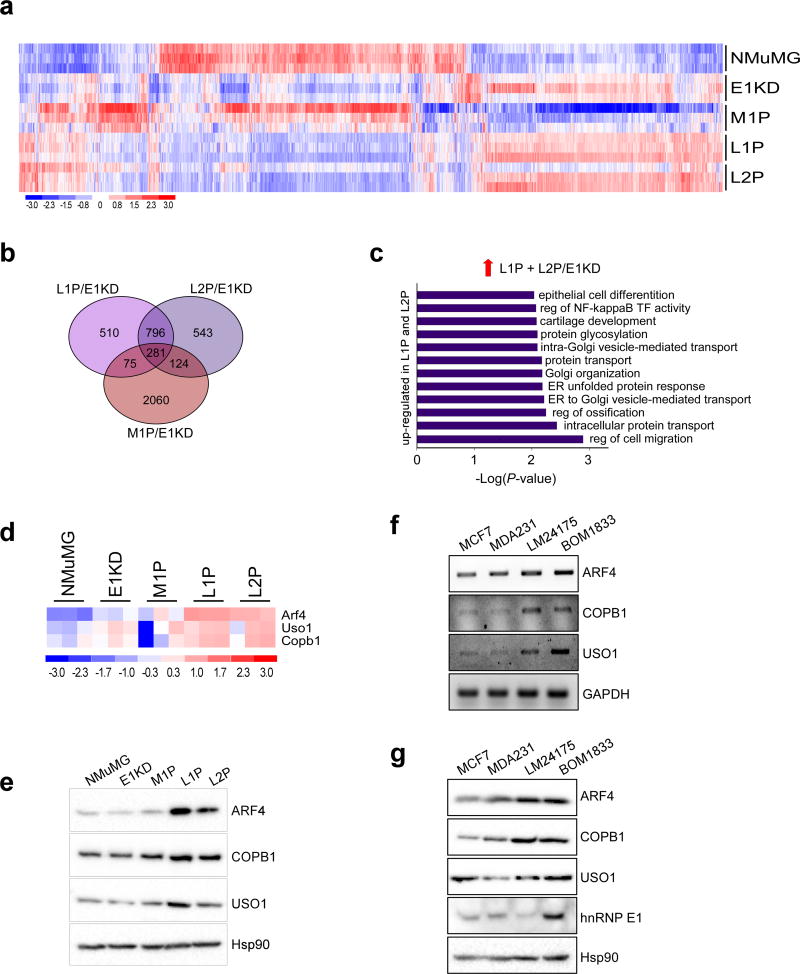
Figure 2. Identification of a metastasis-associated gene signature through the in vivo selection of mammary epithelial cells.
(a) Heat-map of differentially regulated genes in triplicate samples generated using dChip software; data was filtered using a P-value of 0.05 and a 1.5 fold cut off. (b) Venn diagram showing overlap between M1P/E1KD, L1P/E1KD and L2P/E1KD data sets. (c) Graph of enriched GO processes in up-regulated genes unique to L1P and L2P cells, filtered using a P-value of 0.01. (d) Heat-map of ARF4, COPB1 and USO1 trafficking genes up-regulated in L1P and L2P cells compared to E1KD cells. (e) Immunoblot analysis of ARF4, COPB1 and USO1 levels in the NMuMG progression series. Hsp90 was used as a loading control. (f) Transcript levels of ARF4, COPB1, USO1 and GAPDH in MCF-7, MDA-231, LM2-4175 and BOM-1833 cells. (g) Protein levels of ARF4, COPB1, USO1 and hnRNP E1 in MCF-7, MDA-231, LM2-4175 and BOM-1833 cells. Hsp90 was used as a loading control.
From the list of enriched ER-Golgi trafficking genes in the metastatic L series, we focused on validating the up-regulation of ARF4, COPB1 and USO1 (Fig 2D and E). ARF4 and COPB1 act in retrograde ER-Golgi trafficking; the GTPase ARF4, regulates COPI vesicle formation required for retrograde transport and COPB1 is a component of these COPI vesicles34–37. The anterograde trafficking gene USO1/p115 acts as a tethering factor for COPII vesicles38,39. A significant up-regulation of these trafficking genes at the transcript level was observed in L1P and L2P cells (Fig 2D) and increased expression at the protein level was also observed (Fig 2E). To validate our findings in a human progression series, we utilized the MDA-MB-231 (MDA-231) series of cell lines isolated from lung (LM24175) and bone (BOM1833) metastases40. An increase in ARF4, COPB1 and USO1 transcript (Fig 2F) and protein (Fig 2G) expression was observed in MDA-231 cells isolated from lung or bone metastases compared to the parental cell line and the less aggressive MCF-7 cell line. Expression of hnRNP E1 protein did not correlate with ARF4, COPB1 and USO1 expression in these cell lines, suggesting that up-regulation of this trafficking signature is associated with the development of distant metastases and not hnRNP E1 silencing alone.
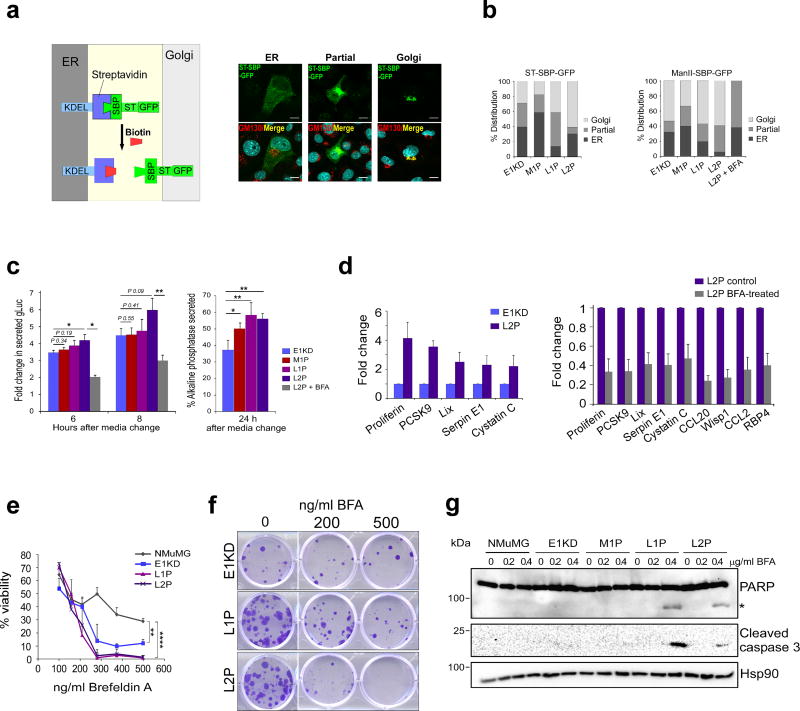
We hypothesized that up-regulation of trafficking genes in our model may alter ER-Golgi trafficking kinetics and secretion. To test this, we initially assessed trafficking kinetics using the RUSH reporters ST-SBP-GFP and ManII-SBP-GFP41 which traffic from the ER to the Golgi upon biotin addition (Fig 3A). Increased ER-Golgi trafficking kinetics of ST-SBP-GFP and ManII-SBP-GFP reporters were observed in L1P and L2P cells compared to E1KD cells, as determined by the decrease in ER retention of reporters 15 min following biotin addition (Fig 3B). As a control in this experiment, ER-Golgi trafficking in L2P cells was blocked using the ARF-GEF inhibitor brefeldin A, which resulted in attenuation of ER-Golgi trafficking kinetics. Secretion of transiently transfected Gaussia luciferase and alkaline phosphatase was also assessed in our model. Consistent with RUSH reporter data, increased secretion of Gaussia luciferase and alkaline phosphatase was observed in L1P and L2P cells compared to E1KD cells, and Gaussia luciferase secretion was attenuated by brefeldin A treatment (Fig 3C). Analysis of secreted cytokines and growth factors from E1KD and L2P cells revealed an increase in secretion of multiple pro-metastatic factors including serpin E1 and LIX/CXCL5 (Fig 3D and Supplementary Fig S3A). Secretion of these up-regulated factors was sensitive to brefeldin A, suggesting that increased ER-Golgi transport kinetics in the L series contributes to increased levels of these pro-metastatic secreted factors (Fig 3D).
Figure 3. Up-regulation of ER-Golgi trafficking and sensitization to brefeldin A in L1P and L2P cells.
(a) Schematic of RUSH trafficking reporter system (left panel). Immunofluorescence images show localization of ST-SBP-GFP reporter at the ER or Golgi 15 min following biotin treatment in L1P cells. GM130 was used as a Golgi marker in this experiment, scale bar: 10µm. (b) Graphs of ST-SBP-GFP and ManII-SBP-GFP reporter localization 15 min following biotin treatment (n ≥ 15 cells per sample). (c) Secreted levels of Gaussia luciferase (n=6) and alkaline phosphatase (n=5) in E1KD, M1P, L1P and L2P cells and L2P cells treated with brefeldin A (*P < 0.05, ** P < 0.01; paired t-test). (d) Cytokine array data of secreted factors released from E1KD and L2P cells, in addition to control and BFA-treated (200 ng/ml) L2P cells. Densitometry was performed on three exposures per array using ImageJ software. A two fold cut-off was used to filter data. (e) MTT assay assessing cell viability in 72h BFA-treated NMuMG, E1KD, L1P and L2P cells (n = 3, **P < 0.01, **** P < 0.0001; two-way ANOVA with Bonferroni's post-hoc test). (f) Colony formation assay assessing clonogenic growth in BFA-treated cells. (g) Immunoblot analysis of PARP and caspase 3 cleavage, indicative of apoptosis, in BFA-treated cells (*denotes cleaved PARP). Hsp90 was used as a loading control.
Increased sensitivity of metastatic cells to the retrograde ER-Golgi inhibitor Brefeldin A
Due to the increased trafficking kinetics of metastatic cell lines, we next tested whether these cells exhibited altered sensitivity to long-term ER-Golgi trafficking inhibition by brefeldin A. Golgi fragmentation, as assessed by GM130 immunofluorescence, was observed in all cell lines following BFA treatment (Supplementary Fig S3B). Interestingly, L1P and L2P cells demonstrated increased sensitivity to BFA treatment as determined by MTT assay and colony formation assay (Fig 3E and F). In addition, apoptosis induction was detected only in L1P and L2P cells following BFA as assessed by cell morphology, and caspase 3 and PARP cleavage (Fig 3G and Supplementary Fig S3C). Sensitization to brefeldin A in our model was partially due to an up-regulation of retrograde ER-Golgi trafficking genes, as knockdown of ARF4 and to a lesser extent COPB1 resulted in increased resistance to BFA cytotoxicity (Supplementary Fig S6E). These findings are in agreement with previous studies, demonstrating ARF4-dependent BFA sensitivity19. A similar sensitivity to ER-Golgi blockade was observed in MDA-231 cells isolated from lung or bone metastases compared to the less aggressive MCF-7 cell line and parental MDA-231 cells (Supplementary Fig S3D and E).
To determine if this effect was a general sensitization to apoptosis inducers or ER stress, we assessed UPR-regulated transcript expression and response to the chemotherapeutics cisplatin and etoposide and the ER stress inducers, thapsigargin and tunicamycin. Interestingly, the metastatic L series demonstrated similar sensitivity to cisplatin, etoposide, thapsigargin and tunicamycin as NMuMG, E1KD and M1P cells (Supplementary Fig S4A and B). In addition, no significant difference in BiP and Chop expression was observed between E1KD, L1P and L2P cells, whereas, an increase in these transcripts was observed in M1P cells (Supplementary Fig S4C). Analyses of Chop and PERK expression by immunoblot revealed a decrease in Chop protein induction, in response to thapsigargin and tunicamycin, in M1P, L1P and L2P cells compared to NMuMG and E1KD, whereas, levels of phosphorylated PERK and ER-stress induced XBP1 splicing were similar across cell lines in the series (Supplementary Fig S4D and F).
CREB3-mediated regulation of ER-Golgi trafficking genes
To assess the upstream regulators of ER-Golgi trafficking genes in our progression model, we performed meta-analysis of breast cancer patient array data which revealed co-regulation of candidate genes (Supplementary Fig S5A and B). We looked for common transcription factor motifs present in the promoters of our genes of interest, which might explain the co-regulation of these genes. This analysis revealed CREB3 and CREB3-like motifs, which are conserved across higher eukaryotes, present in the promoters of up-regulated genes, including ARF4, COPB1 and USO1 (Supplementary Fig S5C). Consistent with a role of CREB3 and CREB3-like TFs in regulating these genes, expression of CREB3 in breast cancer correlated with ARF4, COPB1 and USO1 but not GGA2, a TGN-Endosome trafficking adaptor protein which does not contain a conserved CREB3 site (Fig 4A).
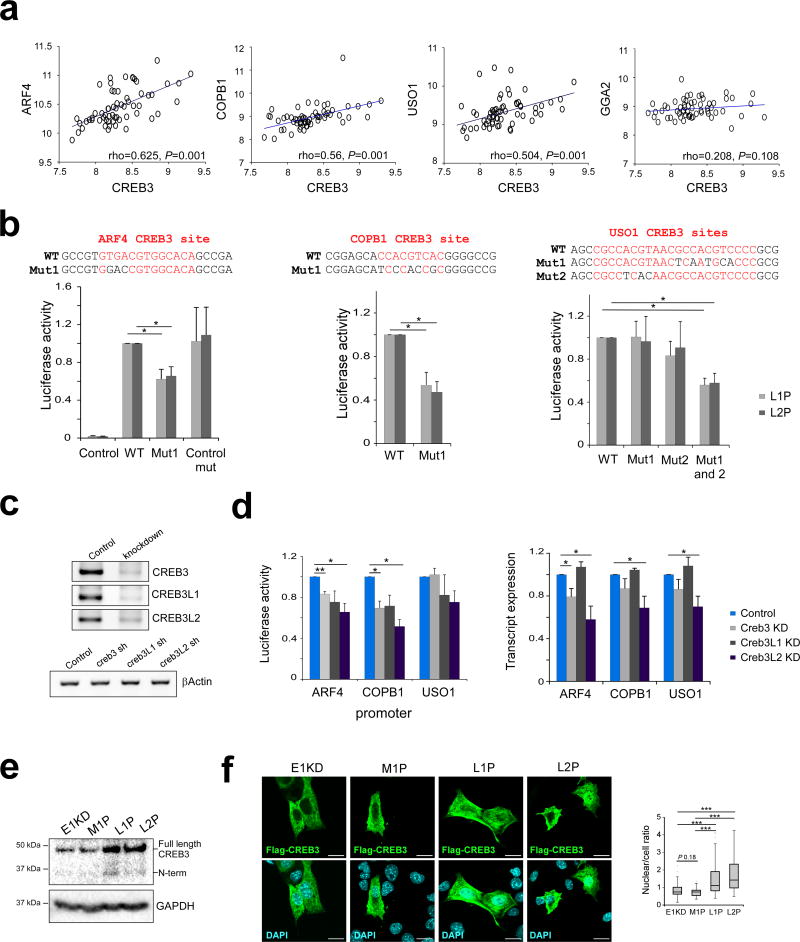
Figure 4. CREB3-mediated regulation of ER-Golgi trafficking genes.
(a) Correlation between CREB3 gene expression and expression of ARF4, COPB1, USO1 and GGA2. Spearman's rho and significance determined using SPSS software (n = 61). (b) CREB3 sites (in red) were mutated in ARF4, COPB1 and USO1 luciferase reporters by site directed mutagenesis and luciferase activity was tested in L1P and L2P cells (n ≥ 3, *P<0.05; paired t-test). (c) CREB3, CREB3L1 and CREB3L2 were silenced by shRNA in L2P cells. Levels of transcription factor knockdown were assessed by semi-quantitative PCR. (d) Luciferase activity (left panel) and transcript levels of ARF4, COPB1 and USO1 (right panel) was tested in scrambled control, CREB3 KD, CREB3L1 KD and CREB3L2 KD L2P cells (n ≥ 3, *P<0.05, **P < 0.01; paired t-test). Transcript levels were assessed by semi-quantitative PCR using βActin as a normalizer. (e) CREB3 protein expression in MG132-treated E1KD, M1P, L1P and L2P cells; GAPDH was used as a loading control. (f) Flag-CREB3 localization in E1KD, M1P, L1P and L2P cells transiently transfected with an N-terminal Flag-tagged CREB3 construct, as assessed by immunofluorescence using an anti-Flag antibody. Mean whole cell and nuclear signal intensities were assessed by ImageJ and nuclear/cell ratios were calculated using the formula: mean intensity nucleus/ mean intensity cell (n ≥ 27 cells per sample, ***P < 0.001; unpaired t-test, scale bar: 20 µm).
To assess the role of CREB3 and CREB3-like TF regulation of these genes in our progression model, the promoter sequences of ARF4, COPB1 and USO1 containing CREB3 sites were cloned upstream of a luciferase reporter. Mutation of the CREB3 sites present in these promoter constructs resulted in significantly reduced luciferase activity in both L1P and L2P cells (Fig 4B). The effect of silencing CREB3, CREB3L1 and CREB3L2, which are expressed in our cell lines, was next tested in L2P cells. Luciferase activity of ARF4 and COPB1 reporters was significantly reduced upon CREB3 and CREB3L2 silencing in L2P cells. Consistent with this data, a significant reduction of ARF4, COPB1 and USO1 transcript levels was also observed upon CREB3L2 silencing, with CREB3 silencing significantly reducing ARF4 transcript levels (Fig 4C and D).
Levels of active CREB3 and CREBL2 in our progression series were subsequently characterized. If these TFs are regulators of ER-Golgi trafficking genes in our model, we would expect to see higher TF activity in L1P and L2P cells. To test this hypothesis, levels of the full-length and N-terminal active form of CREB3 and CREB3L2 were assessed by immunoblot in cells treated with the proteasome inhibitor MG132 to prevent degradation (Supplementary Fig S5D). Higher expression of the full-length and N-terminal active forms of CREB3 was observed in L1P and L2P compared to E1KD and M1P cells (Fig 4E). An increase in full-length CREB3L2 was not observed in L1P and L2P cells; however, a slight increase in the N-terminal form was detected, compared to E1KD cells (Supplementary Fig S5D).
Next, localization of a transiently transfected N-terminal Flag-tagged CREB3 was determined by immunofluorescence in E1KD, M1P, L1P and L2P cells. Whole cell and nuclear signal intensities of Flag-CREB3 were measured using ImageJ software and nuclear/cell ratios were calculated using the formula (mean intensity nucleus/ mean intensity cell). A significant increase in CREB3 nuclear localization was observed in L1P and L2P cells compared to both E1KD and M1P cells (Fig 4F and Supplementary Fig S5G). Consistent with these findings, increased nuclear localization of endogenous CREB3L2 was observed in L1P and L2P compared to E1KD and M1P cells, nuclear localization was also induced following BFA treatment (Supplementary Fig S5E and F).
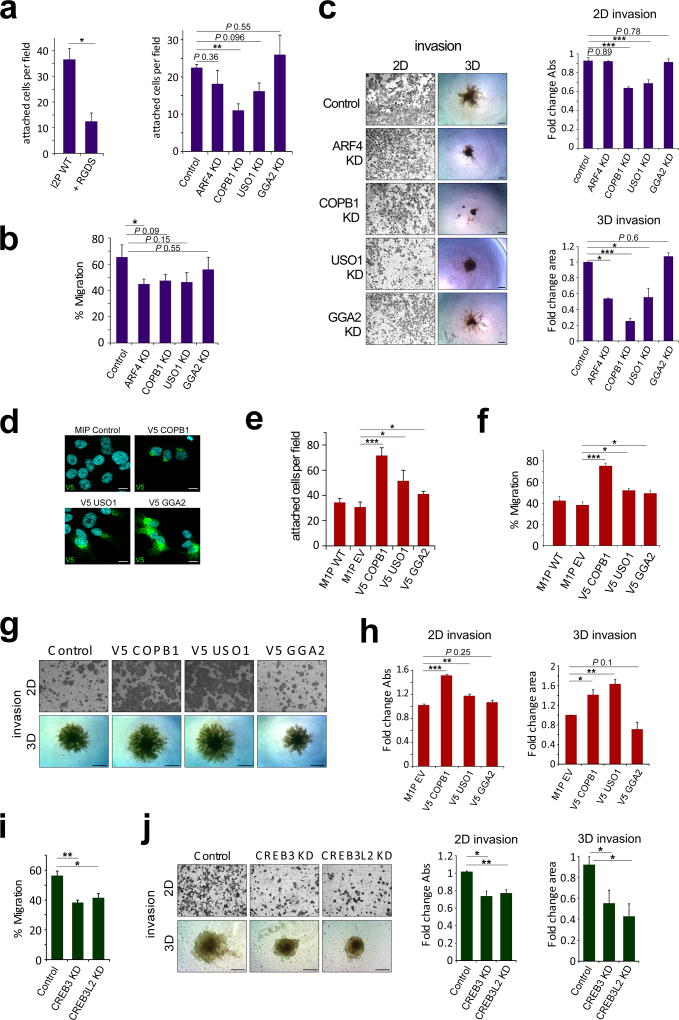
Up-regulation of ER-Golgi trafficking genes enhances trafficking kinetics and promotes cell adhesion, migration and invasion
In order to determine the functional significance of ER-Golgi trafficking up-regulation in the metastatic L series, ARF4, COPB1, and USO1 were silenced by shRNA in L2P cells (Supplementary Fig S6A). In addition, we included the TGN-Endosome trafficking adaptor protein GGA2 as a control42. Stable knockdown (KD) of ARF4, COPB1 and USO1 did not notably alter Golgi morphology as assessed by GM130 immunofluorescence (Supplementary Fig S7B), but did result in reduced ER-Golgi transport kinetics as assessed by the RUSH reporter ST-SBP-GFP (Supplementary Fig S6B). Moreover, a reduction in several pro-metastatic secreted cytokines was observed upon ARF4, COPB1 and USO1 silencing (Supplementary Fig S6D). Next, we assessed factors associated with metastatic progression including proliferation rate, adhesion, migration and invasion. We observed no difference in proliferation rate between scrambled control and KD cells (Supplementary Fig S6F); however, we did observe significantly reduced adhesion onto fibronectin upon COPB1 KD. A trend towards ARF4 and USO1 silencing affecting adhesion was also observed; whereas, silencing GGA2 did not alter adhesion in L2P cells (Fig 5A). Significantly reduced migration in ARF4 KD cells was observed as assessed by wound healing assay (Fig 5B). In addition, decreased 2D invasion of COPB1 and USO KD cells, as well as decreased 3D spheroid area and increased circularity index in ARF4, COPB1 and USO1 KD cells were observed (Fig 5C, Supplementary Fig S6C). These data indicate that reduced expression of the ER-Golgi trafficking genes, ARF4, COPB1 and USO1, but not the TGN-endosome gene GGA2, attenuates the metastatic phenotype of L2P cells. Consistent with these findings, overexpression of V5-tagged COPB1 and USO1 in M1P cells resulted in increased ER-Golgi trafficking (Supplementary Fig S6E and F) and an increase in cell adhesion, migration and invasion, with no change in proliferation rate (Fig 5D–H, Supplementary Fig S6H). Inhibition of ARF4, COPB1 and USO1 also attenuated the invasive phenotype of LM2-4175 cells, derived from lung metastasis of MDA-231 cells (Supplementary Fig S6I–K). These data indicate that enhanced ER-Golgi trafficking may be an important mediator of metastatic disease.
Figure 5. ER-Golgi trafficking genes promote cell adhesion, migration and invasion.
(a) Adhesion onto fibronectin-coated plates in L2P KD cells, an RGDS peptide control was included to inhibit cell surface integrin binding to fibronectin coated wells (n ≥ 3, *P < 0.05, ** P < 0.01; unpaired t-test). (b) Quantitation of wound healing assay assessing migration in L2P KD cells (n ≥ 3, *P < 0.05; unpaired t-test). (c) Representative images of 2D and 3D invasion assays demonstrating invasive capability of ARF4, COPB1, USO1 and GGA2 KD cells compared to control L2P cells (left panel), Quantitation of 2D and 3D invasion assays (n ≥ 3, *P < 0.05, *** P < 0.001; unpaired t-test, scale bar: 200 µm). (d) Representative images of V5 tagged COPB1, USO1 and GGA2 expression and localization in M1P cells as assessed by immunofluorescence analysis; scale bar: 10µm. (e) Adhesion onto fibronectin-coated plates (n = 11 fields) in M1P overexpressing cells (*P < 0.05, *** P < 0.001; unpaired t-test) (f) Quantitation of wound healing assay assessing migration (n = 3) in M1P overexpressing cells (*P < 0.05, *** P < 0.001; unpaired t-test). (g) Representative images of 2D and 3D invasion assays demonstrating invasive capability of M1P overexpressing cells. (h) Quantitation of 2D and 3D invasion assays (n = 3, *P < 0.05, **P < 0.01, *** P < 0.001; unpaired t-test, scale bar: 200 µm). (i) Quantitation of wound healing assay assessing migration (n = 3) in L2P CREB3 and CREB3L2 KD cells (*P < 0.05, ** P < 0.01; unpaired t-test). (j) Representative images of 2D and 3D invasion assays demonstrating invasive capability of L2P CREB3 and CREB3L2 KD cells compared to scrambled control L2P cells (left panel). Quantitation of 2D and 3D invasion assays (n ≥ 3, *P < 0.05, **P < 0.01; unpaired t-test, scale bar: 200 µm).
As we observe reduced trafficking gene expression upon CREB3 and CREB3L2 silencing, we wanted to test whether CREB3 modulation impacted metastatic progression in our model. Cell migration and invasion rates in L2P cells silenced for CREB3 or CREB3L2 were compared to scrambled control. A significant reduction in cell migration and invasion was observed when these transcription factors were silenced (Fig 5I and J), consistent with previous data reporting CREB3 regulation of cell adhesion, migration and invasion43.
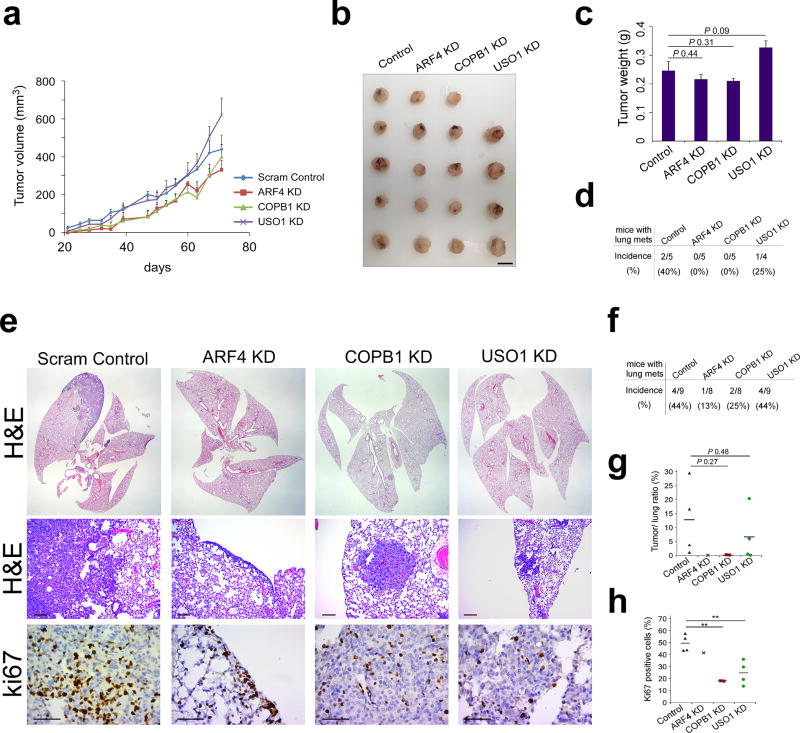
Next, we assessed the effect of ARF4, COPB1 and USO1 KD in L2P cells in vivo. Mammary fat pad injection of scrambled control, ARF4, COPB1 and USO1 KD L2P cells in NOD/SCID mice resulted in no significant difference in tumor formation (Fig 6A–C); however, reduced incidence in lung metastasis was noted (Fig 6D). Lung colonization was further investigated by tail vein injection of control and KD L2P cells. A trend towards reduced lung metastasis incidence and volume upon silencing of ARF4 and COPB1 was observed, whereas, a slight reduction in tumor/lung ratio was observed upon USO1 silencing (Fig 6E–G). In accordance with these findings, Ki67 positivity, an indicator of proliferation, was significantly reduced in COPB1 and USO1 KD metastases compared to scrambled control (Fig 6H).
Figure 6. ER-Golgi trafficking genes promote metastatic progression.
(a) Tumor volume of mammary fat pad injected L2P cells stably expressing scrambled control, ARF4, COPB1 and USO1 shRNA (n = 5 animals for control, ARF4 KD and COPB1 KD groups, n = 4 animals for USO1 KD group). (b) Image of excised tumors and (c) quantitation of tumor weight from mice that were mammary fat pad injected with L2P cells stably expressing scrambled control, ARF4, COPB1 and USO1 shRNA (data not significant; unpaired t-test, scale bar: 1 cm). (d) Incidence of lung metastasis in mice following mammary fat pad injection of control, ARF4, COPB1 and USO1 KD L2P cells. (e) Representative images of H&E and Ki67 stained lung tissue sections following tail vein injection of control, ARF4, COPB1 and USO1 KD L2P cells. Scale bar: 100 µm (H&E) and 50 µm (Ki67). (f) Incidence of lung metastasis, (g) Tumor/lung ratio and (h) Ki67 positivity in lungs of mice tail vein injected with control, ARF4, COPB1 and USO1 KD L2P cells (n = 9 animals for control and USO1 KD groups, n = 8 animals for ARF4 KD and COPB1 KD groups, **P < 0.01; unpaired t-test).
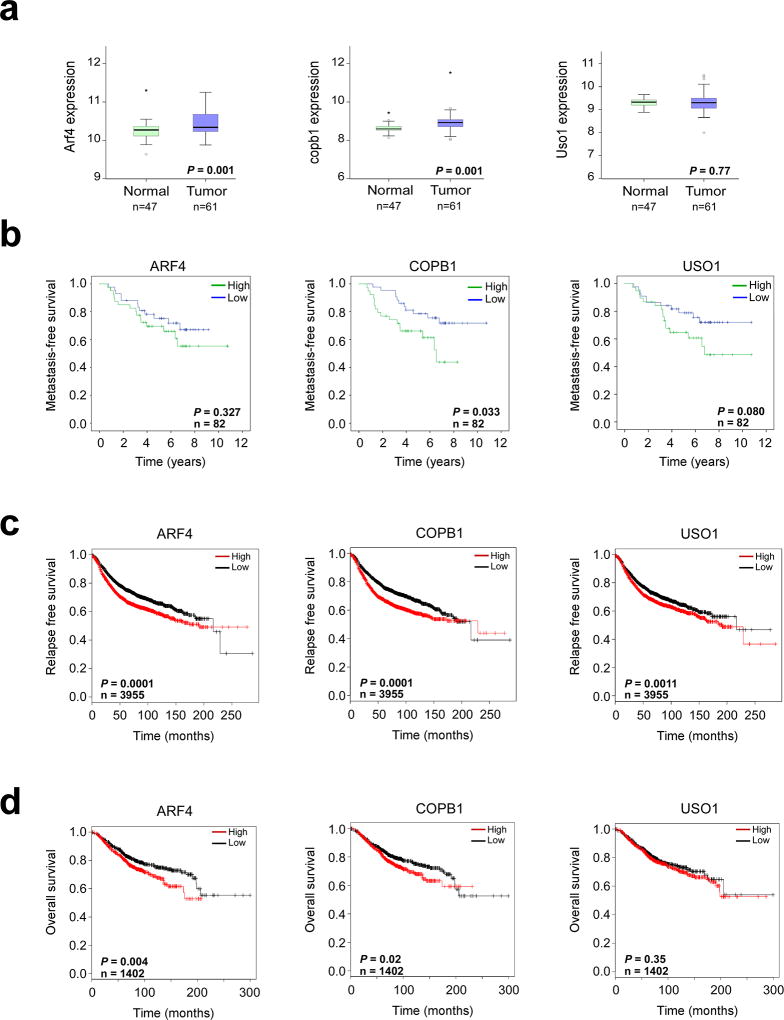
Up-regulation of ER-Golgi trafficking genes associated with increased risk of distant metastasis and reduced survival in breast cancer
Initially, the significance of ER-Golgi trafficking gene up-regulation in human breast cancer was assessed by meta-analysis of normal and breast cancer microarray data from the NCBI Gene Expression Omnibus (GEO). Comparison of ARF4, COPB1 and USO1 expression levels in normal and breast cancer tissue revealed a statistically significant increase in ARF4 and COPB1 transcript in cancer tissue (Fig 7A). Further analyses of breast cancer patient data from GEO revealed a significant association between high expression of COPB1 and increased risk of metastasis to lung or bone; whereas, a trend between high USO1 expression and increased risk of metastatic event were observed (Fig 7B). Of interest, a significant association between COPB1, ARF4 and USO1 expression and either relapse-free survival or overall survival was also observed when breast cancer patient data was analyzed using KM plotter (Fig 7C and D). The association between high ARF4 expression and survival in this dataset was stronger in ER negative tumors that typically have a less favorable prognosis than ER positive tumors (Supplementary Fig S8). Overall, these data indicate a pro-metastatic role of the ER-Golgi trafficking pathway in breast cancer, consistent with the findings of our study.
Figure 7. ER-Golgi trafficking genes associated with metastatic risk and survival in human breast cancer.
(a) Meta-analysis of ARF4, COPB1 and USO1 mRNA expression levels in 47 normal and 61 breast tumor tissue (GEO series GSE37751, P<0.05 considered significant; unpaired t-test). (b) Kaplan-Meier analysis of metastasis-free survival in 82 breast cancer patients. High and low transcript expression determined based on median expression value across all arrays. (GEO series GSE2603, P<0.05 considered significant, log rank test). (c) Kaplan-Meier analysis of relapse-free survival (n = 3955) and (d) overall survival (n = 1402) in breast cancer patients stratified by median trafficking gene expression values (KM-plotter, P<0.05 considered significant, log rank test).
Discussion
We have developed a progression series of increasingly metastatic cell lines generated from the normal murine mammary epithelial cell line, NMuMG, to distinguish between early changes in cancer initiation and metastasis-promoting alterations. Enriched pathways associated with cancer initiation in our series (deregulated genes compared to NMuMG cells) included epithelial cell differentiation, changes to cell metabolism, blood vessel morphogenesis and inflammatory response, which have been linked to tumorigenesis previously44. Due to the impact of metastatic disease on cancer patient mortality, we focused on pathways enriched in the metastatic L1P and L2P cells. In this dataset, enriched pathways including migration, adhesion and steroid/cholesterol metabolism have been described previously as metastasis-promoting45–47. The role of the enriched ER-Golgi trafficking pathway in metastatic progression was further characterized due to the increased trafficking kinetics and secretion observed in L1P and L2P cells.
We hypothesize that the demand for increased secretory capacity in metastatic cells leads to a Golgi stress response and an up-regulation of ER-Golgi genes through mediators including CREB3. Indeed, increased Golgi capacity has been described in cells with higher secretion demands, such as acinar cells of mammary glands during lactation48. Moreover, a recent study demonstrates an extended Golgi morphology in metastatic cells, consistent with our hypothesis13. Data obtained from knockdown of trafficking genes in L2P cells and their overexpression in M1P cells indicates that modulation of ER-Golgi trafficking does not alter cell proliferation but does effect cell adhesion, migration and invasion which contribute to metastatic progression (Fig 5A–C, Supplementary Fig S6F).. These data are consistent with our in vivo findings, where gene silencing in L2P cells did not significantly alter tumor growth but did reduce lung metastasis incidence and metastases growth (Fig 6). We propose that the pro-metastatic role of increased Golgi trafficking is due to a combination of an altered secretome, where factor such as serpin E1 and LIX are up-regulated promoting metastatic progression, and changes in receptor cycling which alter cell signaling, adhesion and migration.
Analysis of potential transcriptional regulators of ER-Golgi trafficking genes led to the identification of CREB3 and CREB3-like transcription factors. The regulation of ARF4, COPB1 and USO1 by CREB3 and CREB3-like TFs in our model is consistent with previous studies in Drosophila, where the CREB3L1/L2 orthologue, CREBA, regulates multiple genes in the secretory pathway, including COPB125. Moreover, CREB3 regulation of ARF4 in human cancer cells has been described19. In addition to CREB3 regulation of ER-Golgi genes, a Golgi apparatus stress response element (GASE) has been described in the promoter of several Golgi-associated genes up-regulated by the Golgi stressor, Monensin20. The transcription factors TFE3 and MLX are reported to bind to the GASE motif sequence, ACGTGGC, which shows similarity to the CREB3 motif21,49. How these Golgi stress sensing TFs are activated is largely unknown. CREB3 and CREB3-like TFs localize to the ER through transmembrane domains and upon activation, these factors are transported to the Golgi where they are processed by S1P and S2P prior to translocation to the nucleus24. The stimuli and escort proteins required for CREB3 translocation and processing have yet to be identified. ER stress induction is reported to induce these TFs, however, these findings are inconclusive with several studies showing no or low level activation upon ER stress induction by tunicamycin and thapsigargin compared to brefeldin A26,50–52.
Several stimuli of Golgi stress response have been proposed including differentiation of secretory cells, changes in lipid content at Golgi vesicles and viral infection. Cholesterol levels have been shown to regulate a subset of SNARE proteins required for proper Golgi transport with cholesterol depletion leading to impaired trafficking, reduced levels of cell surface α5β1 Integrin and attenuation of migration53. In our series, we observed a significant reduction in cholesterol biosynthesis gene expression in L1P and L2P cells. Moreover, secreted levels of the serine protease PCSK9 which regulates low-density lipoprotein receptor (LDLR) and in turn cholesterol54 were up-regulated in L2P cells and were sensitive to BFA treatment (Fig 3D). These data suggest that cholesterol levels are altered in the metastatic L1P and L2P cells which may contribute to the induction of ER-Golgi trafficking genes via CREB3. Further research is warranted to characterize stimuli mediating the Golgi stress response in our model.
Gene set enrichment analyses did demonstrate an enrichment of “ER unfolded protein response” genes in L1P and L2P cells, namely Ero1l, Cdk5rap3, Dnajc3, Edem2, Uggt2, Cyclin D1, and Nfe2l2. However, our data indicates that metastatic L1P and L2P cells are uniquely sensitive to Golgi stress as opposed to ER stress, as sensitization to the ER stress inducers, thapsigargin and tunicamycin was not observed (Supplementary Fig S4B). Previous research has linked EMT induction to enhanced secretory capacity associated with increased PERK activation and sensitivity to thapsigargin and tunicamycin in human mammary epithelial cells55. In our model, PERK expression was higher in mesenchymal E1KD cells compared to epithelial NMuMG cells; however, this increase in PERK did not correlate with thapsigargin and tunicamycin sensitivity (Supplementary Fig S4B and D). In M1P, L1P and L2P cells derived from E1KD cells, increased thapsigargin and tunicamycin sensitivity or enhanced PERK activation was not observed. In addition, ER-stress induced XBP1 splicing was similar across all cell lines in the series (Supplementary Fig S4F). We also characterized UPR induction following brefeldin A treatment, as blockade of retrograde trafficking can result in ER stress and the induction of UPR, which may contribute to apoptosis induction56. Upon BFA treatment at a concentration that induces cell death (400 ng/ml), pro-apoptotic CHOP was not detected and minimal PERK activation was observed 6 h following treatment, with a slight increase in activation in L1P and L2P cells observed at 24 h (Supplementary Fig S4E). Increased induction of pro-survival XBP1s was observed in L1P and L2P cells following 6 h BFA treatment, compared to E1KD cells, indicating a potential role for IRE1α in adaptation to stress in BFA-treated cells (Supplementary Fig S4F). These findings highlight the need for further characterization of the interaction between ER and Golgi stress responses, in particular, the stimuli required to induce such responses and the degree of cross-talk that occurs between pathways.
The significance of trafficking gene deregulation was assessed in human breast cancer, through array meta-analysis, which revealed the association of high trafficking gene expression with reduced relapse-free and overall survival and increased risk of distant metastasis of breast cancer, with a stronger association observed between ARF4 expression and survival in ER negative tumors (Fig 7B–D, Supplementary Fig S8). These data point toward the potential use of Golgi trafficking genes as prognostic markers in cancer. To date, several Golgi-associated genes have been studied as prognostic markers in cancer, including Golgi phosphoprotein-3 (GOLPH3), Golgi protein 73 (GP73) and progestin and adipoQ receptor 3 (PAQR3)57–60. Further studies focused on elucidating the mechanisms regulating Golgi secretion capacity are warranted and may lead to therapeutic targeting of discrete components of this pathway in metastatic disease.
Materials and Methods
Cell lines and treatments
All cell lines were cultured in DMEM high glucose supplemented with 5% FBS/5% Calf Serum and 1% antibiotic/antimycotic solution (penicillin G, streptomycin, amphotericin B) at 37 °C, 5% CO2. NMuMG and MCF-7 cells were obtained from the American Type Culture Collection (ATCC). Lenti-X 293T cells were obtained from Clontech. LM2-4175 and BOM-1833 cells were kindly provided by Dr. Massague (Memorial Sloan-Kettering Cancer Center). Generation of hnRNP E1 silenced NMuMG (E1KD) cells and the isolation of M1P, M2P, L1P and L2P cell lines have been described previously61. Brefeldin A was purchased from Cell Signaling Technology. Cisplatin, etoposide, MG132, tunicamycin and D-biotin were purchased from Sigma-Aldrich. Exo1 was purchased from Abcam and thapsigargin was gifted by Dr. Alan Diehl (Medical University of South Carolina).
Animal studies
All animal procedures were approved by the Animal Care and Use Committees of the Medical University of South Carolina. Nonobese diabetic, severe combined immunodeficient (NOD/SCID) mice (NOD.CB17-PrkdcSCID/J) were supplied by Envigo. No randomization was used to select animals for each group and studies were not blinded. Mammary fat pad injection of 5 × 104 cells in 100µl PBS into the right inguinal mammary fat pad of NOD/SCID females was performed in order to in vivo passage E1KD cells and for mammary fat pad xenograft of E1KD, M1P, M2P and L2P KD cells. Tumor volumes (mm3) were measured biweekly using digital calipers and tumors were weighted at experimental endpoints. For lung colonization experiments, a cell suspension of 105 cells in 100 µl PBS was injected into the tail vein of NOD/SCID females. Formalin fixed, paraffin embedded lung sections were cut at 5 µm and stained with hematoxylin and eosin (H&E) for histopathological evaluation or ki67 immunohistochemical analysis by the Biorepository and Tissue Analysis Resource at MUSC. Micrographs of stained sections were taken using a Leica DMIL LED microscope with Amscope camera and acquisition software. Lung and metastases area was determined using ImageJ software in order to calculate tumor/lung ratios (%).
Migration assay
Migration was assessed using CytoSelect™ 24-Well Wound Healing inserts (Cell Biolabs) that were placed in wells upon cell seeding. Cells were fixed and stained at 0 and 17 h following removal of inserts from confluent monolayers. Wound area at 0h and 17 h was determined using ImageJ software in order to calculate % wound recovery using the formula: (1-(area 17h/area 0h))*100.
2D and 3D invasion assays
Invasion across a basement membrane was performed using BD BioCoat™ Matrigel™ Invasion Chambers (BD Biosciences) as per manufacturer’s instructions. After 22 h, invasive cells located on the underside of chambers were stained with crystal violet and imaged. Crystal violet stain was then extracted from chambers using 33 % acetic acid and extracted dye was quantified by measuring absorbance at 570nm using a Wallac Victor3 plate reader. 3D Invasion was assessed using a modified Trevigen spheroid invasion assay. Briefly, a volume of 50 µl single cell suspension in serum-free media was prepared at a density of 3000 cells per well of a round-bottomed, low attachment 96-well plate. Once seeded, plates were centrifuged at 200 g for 3 min, followed by incubation at 37°C/5% CO2 for 48 h. Plates were placed on ice for 15 min to cool prior to addition of 50 µl Trevigen spheroid invasion matrix per well. Plates were then centrifuged at 300 g for 3 min and incubated for 1 h at 37°C to allow matrix to solidify prior to addition of 100 µl chemoattractant (DMEM supplemented with 10% FBS and 20 ng/ml EGF). Invasive protrusions from spheroids were imaged 5–7 days following matrix addition for the NMuMG progression series and 3 days for the MDA-231 series. Area and perimeter of spheroids were calculated using imageJ software, a circularity index was generated using the formula: 4π*(area/perimeter2).
Flow Cytometry
Single-cell suspensions of E1KD, L1P and L2P cells were incubated in PBS/1% BSA containing PE-conjugated CD24 (M1/69) and FITC-conjugated CD29 (HMβ1-1) antibody (Biolegend) for 30 min on ice, as per manufacturer’s instructions. Cells were then washed three times in PBS/1% BSA and resuspended in 500 µl PBS. Samples were analyzed using the CyAn ADP Cellular Analyzer and Summit software (Beckman Coulter).
Microarray processing and Pathway analysis
Conversion of total RNA into labelled material, mouse genome 430 2.0 GeneChip hybridization, and post hybridization washing, staining, and scanning were performed in accordance with Affymetrix protocols by the MUSC Proteogenomics Core Facility. Hybridization data were processed with Affymetrix Expression Console software to obtain normalized hybridization data (RMA algorithm) and detection scores (MAS5 algorithm). This data was imported into dChip software for hierarchical clustering and comparative analysis where a combination of fold change and Student’s t-test (unpaired) was utilized to identify genes changing significantly for pairwise relationships. Pathway analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and Molecular Signature Database (MSigDB) platforms. Enriched biological processes from Gene ontology (GO) were identified in gene sets using a P-value cutoff of 0.05. Raw data files were deposited in the NCBI Gene Expression Omnibus (GEO) repository as series GSE94637.
ER-Golgi trafficking reporters
The RUSH reporter plasmids ST-SBP-GFP and ManII-SBP-GFP obtained from Addgene were transiently transfected into cells using Lipofectamine LTX as per manufacturer’s instructions. Transfected cells were cultured for 48 h prior to addition of 40 µM D-Biotin to induce ST-SBP-GFP and ManII-SBP-GFP transport to the Golgi. Trafficking of reporters to the Golgi was monitored 0 min, 15min and 60 min at which time samples were fixed in 4% paraformaldehyde and prepared for immunofluorescence microscopy. In BFA treated samples, brefeldin A (500 ng/ml) was added 30 minutes prior to biotin addition.
Gaussia Luciferase assay
pCMV-GLuc 2 control plasmid obtained from NEB was transiently transfected into cells seeded in 24-well plates using Lipofectamine LTX as per manufacturer’s instructions. Transfected cells were cultured for 24 h prior to PBS wash and addition of 1 ml fresh media. Media (25 µl) was collected at 0, 2, 4, 6 and 8 h following media change to monitor secretion of Gaussia luciferase. Levels of secreted Luciferase in media were determined by addition of Coelentrazine (Sigma-Aldrich) to samples and subsequent luminescence detection using a Wallac Victor3 plate reader (Perkin Elmer). In BFA treated samples, brefeldin A (500 ng/ml) was added immediately following media change.
Alkaline Phosphatase assay
A placental alkaline phosphatase PLX304 lentiviral vector obtained from the DNASU Plasmid Repository was transduced into cells seeded in 24-well plates. A media change was performed 4 h after transduction and conditional media was collected after 24 h. Levels of secreted and intracellular alkaline phosphatase were determined using the Great EscAPe SEAP Chemiluminescence Kit (Clontech) and Wallac Victor3 plate reader. Secreted alkaline phosphatase levels were normalized to intracellular levels of the phosphatase.
Cytokine array
Conditioned media from E1KD and L2P cells, harvested 24 h following media change, were assayed using the R&D systems Proteome Profiler Mouse XL Cytokine Array according to manufacturer’s instructions. In BFA treated samples, brefeldin A (200 ng/ml) was added immediately following media change. Chemiluminescence was detected by CCD camera (Biorad Chemidoc MP). Signal intensities from three exposures were determined by densitometry using ImageJ software, values were corrected by median normalization and values for duplicate spots were averaged. A two fold cut-off in cytokine levels was used in pair-wise comparisons.
MTT assay
Cell lines were seeded at 103 cells per well of a 96 well plate and cultured overnight prior to any drug treatments. MTT assay was performed by addition of 20 µl of a 5 mg/ml MTT solution and incubation for 3 h at 37°C. To stop the reaction, 100 µl MTT stop mix (40% dimethyl formamide, 20% SDS) was added and plates were incubated for 1 h at RT with shaking. The reduction of yellow MTT to purple formazan by viable cells was detected by reading absorbance at 590 nm using a Wallac Victor3 plate reader.
Colony formation assay
Cell lines were seeded at 500 cells per well of a 6 well plate and cultured until visible colonies were detected. Colonies were fixed with 70% Ethanol and stained with crystal violet stain.
Immunoblotting
Cells were lysed in RIPA buffer and protein concentrations were determined prior to Laemmli sample buffer addition and heat denaturation at 95°C for 5 min. Samples were resolved on SDS–PAGE gels and transferred to PVDF membrane. Blots were probed with antibodies specific to COPB1 (ab24359) and USO1 (ab102470) from Abcam, E-cadherin (3195), DYKDDDDK Tag (14793), Chop (2895), cleaved caspase-3 (9664) and PARP (9542) from Cell Signaling Technology, LZIP/CREB3 (sc515434), GAPDH (sc32233) and Hsp90 (sc13119) from Santa Cruz Biotechnology, hnRNP E1 (M01; Abnova), ARF4 (11673-1-AP; Proteintech), PERK (100-401-962; Rockland), BBF2H7/CREB3L2 (MABE1018: Millipore) and V5 (R96025: ThermoFisher). Chemiluminescence was detected by CCD camera (BioRad ChemiDoc system).
Lentiviral transductions
Lentiviral stocks were prepared by transfecting Lenti-X 293T cells with pLKO.1 or pLX304 vector, psPAX2 and pMD2.G and collection of viral supernatant 24 h and 48 h following transfection. Stable knockdown pools were generated by lentiviral transduction of pLKO.1 Neo vectors (Addgene) containing the following shRNA sequences: human/mouse ARF4 (GTAGATAGCAATGATCGTGAA), mouse COPB1 (CCCTAAATCAAGAATGAGTTT), mouse USO1 (GCAGTTCATATAGCTGGGATT), mouse GGA2 (GCGGAGATTCTTCAAGCAAAT), human COPB1 (GCATTCCTGTTCTGTCCGATT), human USO1 (GCAGCTTTGTACTATCCTAAT), mouse CREB3 (CAGGAGATGTCTAGGCTGAT) (CAGCCCTTCTTTGGTCATCTT), mouse CREB3L1 (ATCCGTCCTCCTCCGATAAA), mouse CREB3L2 (CAGACGCTTATTCCTAAGATT and GTCTTGTTCAACTGAGAACTT) and a scrambled control sequence (CCTAAGGTTAAGTCGCCCTCG). Stable overexpression pools of V5-tagged COPB1 (HsCD00434332), USO1 (HsCD00434283) and GGA2 (HsCD00440576) were generated through lentiviral transduction of cells with pLX304 vectors obtained from DNASU. Empty vector pLX304 was obtained from Addgene. Stable pools were selected in media containing 2 mg/ml G418 (pLKO.1 Neo) or 10 µg/ml blasticidin (pLX304).
Semi-quantitative PCR
Total RNA isolation and semi-quantitative PCR was performed as described previously61. Primer sequences are described in Supplementary data (Supplementary Table S1).
CREB3 and CREB3L2 localization studies
For CREB3 localization studies, an N-Flag CREB3 plasmid obtained from Sino Biological was transiently transfected into cells using Lipofectamine 3000 as per manufacturer’s instructions. Transfected cells were cultured for 48 h prior to harvesting. In both CREB3 and CREB3L2 localization studies, to prevent TF degradation 10 µM MG132 was added to samples 4 h prior to harvesting samples by fixation in 4% paraformaldehyde. In BFA treated samples, brefeldin A (500 ng/ml) was added 60 minutes prior to harvesting.
Immunohistochemistry
Ki67 staining of mouse lung tissue was performed by the Biorepository and Tissue Analysis core at MUSC. Formalin-fixed, paraffin-embedded sections were deparaffinized in xylene, rehydrated in alcohol, and processed as follows: Sections were incubated with target retrieval solution (Dako) in a steamer for 45 min followed by 3% hydrogen peroxide solution for 10 min and protein block (Dako) for 20 min at room temperature. Sections were incubated overnight in a humid chamber at 4°C with antibodies against Ki-67 (12202; Cell Signaling) followed by biotinylated secondary antibody (Vector laboratories) for 30 min and ABC reagent for 30 min. Immunocomplexes of horseradish peroxidase were visualized by DAB reaction (Dako), and sections were counterstained with hematoxylin before mounting. Micrographs of stained sections were taken using a Leica DMIL LED microscope with Amscope camera and acquisition software. Ki67 positive nuclei were counted using ImageJ software and % positivity was assessed using the following equation: ((positive nuclei per field/all nuclei per field) ×100).
Immunofluorescence
Cells seeded onto cover slips in 6-well plates were fixed in 4% paraformaldehyde solution for 10 min with gentle rocking. Fixed cells were washed three times in PBS, incubated in blocking buffer (PBS/2% BSA/0.2% Triton X-100) for 20 min and incubated with primary antibody diluted in blocking buffer at 4°C overnight. The following primary antibodies were used: E-cadherin (3195: Cell Signaling), V5 (R96025; ThermoFisher), SBP tag (SB19-C4; Santa Cruz), GM130 (ab31561; Abcam), DYKDDDDK Tag (14793: Cell Signaling), BBF2H7/CREB3L2 (MABE1018: Millipore). Following primary antibody incubation, cells were washed three times in PBS and incubated with Alexa Fluor 488- or 568-conjugated secondary antibody (Life Technologies) diluted in blocking buffer for 1 h, followed by three PBS washes and mounting of slides using DAPI Fluoromount G (Southern Biotech). For Phallodin immunofluorescence, rhodamine phallodin was diluted in blocking buffer and incubated at 4°C overnight, cells were then washed three times in PBS and mounted. Images were taken using an Olympus FV10i LIV laser scanning confocal microscope.
Adhesion assay
Adhesion onto fibronectin was assessed in serum starved cells seeded at 5 × 104 cells onto fibronectin (10 µg/ml; Sigma Aldrich) coated 24-well plates for 30 min. As a control, Arg-Gly-Asp-Ser (RGDS) tetrapeptide (MP Biomedical) was used at a concentration of 40 µM to inhibit adhesion. Following the 30 min incubation, media was aspirated and wells were washed in PBS, attached cells were fixed with 70% ethanol and stained with crystal violet stain. Images of attached cells were taken using a Leica DMIL LED microscope and at least 15 images at 10× magnification were used to calculate number of attached cells per field in each experiment.
Mammosphere assay
Cells cultured in mammosphere media (DMEM:F12, 20 ng/ml EGF, 20 ng/ml FGF) were seeded at a density of 5000 cells/ml in 96-well or 6-well low attachment plates. Mammosphere formation (spheres ≥ 50 µm) was assessed 7 days after seeding.
Array meta-analysis
Breast cancer array data was obtained from GEO. Expression of ARF4, COPB1 and USO1 between 47 normal and 61 breast cancer samples, as well as correlation analyses of CREB3, ARF4, COPB1, USO1 and GGA2 levels in 61 breast cancer samples, was performed using GEO series GSE37751 and SPSS software. Correlation analyses of ARF4, COPB1 and USO1 levels by SPSS was also performed using 1904 breast cancer samples from the METABRIC database62. Kaplan-Meier analysis of distant metastases risk in breast cancer was performed using GEO series GSE2603. Expression of ARF4, COPB1 and USO1 in this dataset was divided into high and low levels based on median expression across all arrays, Kaplan-Meier plots were generated using SPSS. Relapse-free survival (n=3955) and overall survival (n=1402) breast cancer data was divided into high and low levels based on median expression, and Kaplan-Meier plots were generated using KM-plotter63.
Luciferase assay
ARF4, COPB1 and USO1 promoter sequences were cloned into BglII and HindIII sites of the pGL3-basic vector (Promega) and site directed mutagenesis was performed using the QuickChange Lightning site directed mutagenesis kit (Agilent Technologies) with primers listed in Supplementary data (Supplementary Table 1). Cells seeded at 104 cells in 96-well plates were transfected with 100 ng luciferase constructs and 25 ng Renilla plasmid, luciferase activity was measured 24 h following transfection using a dual luciferase assay (Promega).
Statistical analyses
All data are presented as the mean ± SEM. Representative experiments were repeated at least twice. Statistical analyses were performed by two-tailed Student’s t-test of pair-wise comparisons, two-way ANOVA with Bonferroni's post-hoc test for group comparisons, Spearman rank correlation coefficient for correlations and Log Rank (Mantel cox) test for Kaplan–Meier analyses. P-values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by Grants CA055536 and CA154663 from the National Cancer Institute to PHH. BVH was supported by funding from the Abney Foundation. This study used the services of the MUSC Proteogenomics Facility supported by NIH/NIGMS (GM103342 and GM103499), the Cell & Molecular Imaging Shared Resource of MUSC supported by P30 CA138313, the MUSC Center for Oral Health Research (COHR), which is partially supported by P30 GM103331, the MUSC Flow Cytometry Facility supported by NIH/NIGMS GM103342, the Biorepository & Tissue Analysis Shared Resource, Hollings Cancer Center, MUSC and the Gene Targeting and Knockout Shared Resource at MUSC. We would like to thank Annamarie Dalton for assisting us with xenograft experiments and thank members of the Howe lab for providing helpful feedback during the preparation of this manuscript.
Footnotes
Author contributions BH designed experiments, performed research and analyzed the data. LL generated in vivo selected cell lines, performed research and analyzed data. SG performed research and analyzed data. MES performed xenograft experiments and analyzed data. PHH directed the research and analyzed data. BVH and PHH wrote the manuscript.
Disclosure of potential conflicts of interest: No potential conflicts of interest were disclosed.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Hussey George S, Chaudhury A, Dawson Andrea E, Lindner Daniel J, Knudsen Charlotte R, Wilce Matthew CJ, et al. Identification of an mRNP Complex Regulating Tumorigenesis at the Translational Elongation Step. Mol Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-[beta]-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussey GS, Link LA, Brown AS, Howley BV, Chaudhury A, Howe PH. Establishment of a TGFβ-Induced Post-Transcriptional EMT Gene Signature. PLOS ONE. 2012;7:e52624. doi: 10.1371/journal.pone.0052624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, et al. PCBP1 Suppresses the Translation of Metastasis-Associated PRL-3 Phosphatase. Cancer Cell. 2010;18:52–62. doi: 10.1016/j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 5.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 6.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bard F, Chia J. Cracking the Glycome Encoder: Signaling, Trafficking, and Glycosylation. Trends Cell Biol. 2016;26:379–388. doi: 10.1016/j.tcb.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Spang A. Retrograde Traffic from the Golgi to the Endoplasmic Reticulum. Cold Spring Harb Perspect Biol. 2013;5:a013391. doi: 10.1101/cshperspect.a013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guizzunti G, Seemann J. Mitotic Golgi disassembly is required for bipolar spindle formation and mitotic progression. Proc Natl Acad Sci USA. 2016;113:E6590–E6599. doi: 10.1073/pnas.1610844113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadav S, Puri S, Linstedt AD. A Primary Role for Golgi Positioning in Directed Secretion, Cell Polarity, and Wound Healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bershadsky AD, Futerman AH. Disruption of the Golgi apparatus by brefeldin A blocks cell polarization and inhibits directed cell migration. Proc Natl Acad Sci U S A. 1994;91:5686–5689. doi: 10.1073/pnas.91.12.5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, You H, Mo X, He W, Tang X, Jiang Z, et al. GOLPH3 Mediated Golgi Stress Response in Modulating N2A Cell Death upon Oxygen-Glucose Deprivation and Reoxygenation Injury. Mol Neurobiol. 2016;53:1377–1385. doi: 10.1007/s12035-014-9083-0. [DOI] [PubMed] [Google Scholar]
- 13.Halberg N, Sengelaub Caitlin A, Navrazhina K, Molina H, Uryu K, Tavazoie Sohail F. PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion. Cancer Cell. 2016;29:339–353. doi: 10.1016/j.ccell.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 16.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 17.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 18.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiling JH, Olive AJ, Sanyal S, Carette JE, Brummelkamp TR, Ploegh HL et al. A CREB3–ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat Cell Biol. 2013;15:1473–1485. doi: 10.1038/ncb2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oku M, Tanakura S, Uemura A, Sohda M, Misumi Y, Taniguchi M, et al. Novel Cis-acting Element GASE Regulates Transcriptional Induction by the Golgi Stress Response. Cell Struct Funct. 2011;36:1–12. doi: 10.1247/csf.10014. [DOI] [PubMed] [Google Scholar]
- 21.Taniguchi M, Nadanaka S, Tanakura S, Sawaguchi S, Midori S, Kawai Y, et al. TFE3 Is a bHLH-ZIP-type Transcription Factor that Regulates the Mammalian Golgi Stress Response. Cell Struct Funct. 2015;40:13–30. doi: 10.1247/csf.14015. [DOI] [PubMed] [Google Scholar]
- 22.Serebrenik Y, Crews C. Specific Induction of Golgi Stress by Targeted Protein Destabilization. FASEB J. 2015;29:723–725. [Google Scholar]
- 23.Maag RS, Hicks SW, Machamer CE. Death from within: apoptosis and the secretory pathway. Curr Opin Cell Biol. 2003;15:456–461. doi: 10.1016/s0955-0674(03)00075-9. [DOI] [PubMed] [Google Scholar]
- 24.Chan C-P, Kok K-H, Jin D-Y. CREB3 subfamily transcription factors are not created equal: Recent insights from global analyses and animal models. Cell Biosci. 2011;1:6. doi: 10.1186/2045-3701-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox RM, Hanlon CD, Andrew DJ. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J Cell Biol. 2010;191:479–492. doi: 10.1083/jcb.201004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang G, Audas TE, Li Y, Cockram GP, Dean JD, Martyn AC et al. Luman/CREB3 Induces Transcription of the Endoplasmic Reticulum (ER) Stress Response Protein Herp through an ER Stress Response Element. Mol Cell Biol. 2006;26:7999–8010. doi: 10.1128/MCB.01046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol. 2009;11:1205–1211. doi: 10.1038/ncb1963. [DOI] [PubMed] [Google Scholar]
- 28.Saito A, Hino S, Murakami T, Kanemoto S, Kondo S, Saitoh M, et al. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol. 2009;11:1197–1204. doi: 10.1038/ncb1962. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Shen J, Prywes R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem. 2002;277:13045–13052. doi: 10.1074/jbc.M110636200. [DOI] [PubMed] [Google Scholar]
- 30.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 31.Oliveros JC. Venny. An interactive tool for comparing lists with Venn's diagrams. 2007–2015 http://bioinfogpcnbcsices/tools/venny/indexhtml.
- 32.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Tekaya H, Kahn RA, Hauri H-P. ADP Ribosylation Factors 1 and 4 and Group VIA Phospholipase A2 Regulate Morphology and Intraorganellar Traffic in the Endoplasmic Reticulum–Golgi Intermediate Compartment. Mol Biol Cell. 2010;21:4130–4140. doi: 10.1091/mbc.E10-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodonova SO, Diestelkoetter-Bachert P, von Appen A, Hagen WJH, Beck R, Beck M, et al. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science. 2015;349:195–198. doi: 10.1126/science.aab1121. [DOI] [PubMed] [Google Scholar]
- 36.Popoff V, Langer JD, Reckmann I, Hellwig A, Kahn RA, Brügger B, et al. Several ADP-ribosylation Factor (Arf) Isoforms Support COPI Vesicle Formation. J Bio Chem. 2011;286:35634–35642. doi: 10.1074/jbc.M111.261800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volpicelli-Daley LA, Li Y, Zhang C-J, Kahn RA. Isoform-selective Effects of the Depletion of ADP-Ribosylation Factors 1–5 on Membrane Traffic. Mol Biol Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez C, Garcia-Mata R, Hauri H-P, Sztul E. The p115-interactive Proteins GM130 and Giantin Participate in Endoplasmic Reticulum-Golgi Traffic. J Biol Chem. 2001;276:2693–2700. doi: 10.1074/jbc.M007957200. [DOI] [PubMed] [Google Scholar]
- 39.Waters MG, Clary DO, Rothman JE. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boncompain G, Divoux S, Gareil N, de Forges H, Lescure A, Latreche L, et al. Synchronization of secretory protein traffic in populations of cells. Nat Meth. 2012;9:493–498. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- 42.Puertollano R, Randazzo PA, Presley JF, Hartnell LM, Bonifacino JS. The GGAs Promote ARF-Dependent Recruitment of Clathrin to the TGN. Cell. 2001;105:93–102. doi: 10.1016/s0092-8674(01)00299-9. [DOI] [PubMed] [Google Scholar]
- 43.Kim HC, Choi KC, Choi HK, Kang HB, Kim MJ, Lee YH, et al. HDAC3 selectively represses CREB3-mediated transcription and migration of metastatic breast cancer cells. Cell Mol Life Sci. 2010;67:3499–3510. doi: 10.1007/s00018-010-0388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2008;26:273–287. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- 46.Kuzu OF, Noory MA, Robertson GP. The Role of Cholesterol in Cancer. Cancer Res. 2016;76:2063–2070. doi: 10.1158/0008-5472.CAN-15-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danilov AV, Danilova OV, Huber BT. Cell cycle control and adhesion signaling pathways in the development of metastatic melanoma. Cancer Metastasis Rev. 2008;27:707–714. doi: 10.1007/s10555-008-9159-2. [DOI] [PubMed] [Google Scholar]
- 48.Clermont Y, Xia L, Rambourg A, Turner JD, Hermo L. Structure of the Golgi apparatus in stimulated and nonstimulated acinar cells of mammary glands of the rat. Anat Rec. 1993;237:308–317. doi: 10.1002/ar.1092370303. [DOI] [PubMed] [Google Scholar]
- 49.Taniguchi M, Sasaki-Osugi K, Oku M, Sawaguchi S, Tanakura S, Kawai Y, et al. MLX Is a Transcriptional Repressor of the Mammalian Golgi Stress Response. Cell Struct Funct. 2016;41:93–104. doi: 10.1247/csf.16005. [DOI] [PubMed] [Google Scholar]
- 50.Kondo S, Saito A, Hino S, Murakami T, Ogata M, Kanemoto S, et al. BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol Cell Biol. 2007;27:1716–1729. doi: 10.1128/MCB.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nadanaka S, Okada T, Yoshida H, Mori K. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol. 2007;27:1027–1043. doi: 10.1128/MCB.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vellanki RN, Zhang L, Guney MA, Rocheleau JV, Gannon M, Volchuk A. OASIS/CREB3L1 Induces Expression of Genes Involved in Extracellular Matrix Production But Not Classical Endoplasmic Reticulum Stress Response Genes in Pancreatic β-Cells. Endocrinol. 2010;151:4146–4157. doi: 10.1210/en.2010-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reverter M, Rentero C, Garcia-Melero A, Hoque M, Vilà de Muga S, Álvarez-Guaita A, et al. Cholesterol Regulates Syntaxin 6 Trafficking at trans-Golgi Network Endosomal Boundaries. Cell Rep. 2014;7:883–897. doi: 10.1016/j.celrep.2014.03.043. [DOI] [PubMed] [Google Scholar]
- 54.Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, et al. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc Natl Acad Sci U S A. 2005;102:5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng Y-x, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JHL, Proia TA, et al. Epithelial-to-Mesenchymal Transition Activates PERK–eIF2α and Sensitizes Cells to Endoplasmic Reticulum Stress. Cancer Discov. 2014;4:702–715. doi: 10.1158/2159-8290.CD-13-0945. [DOI] [PubMed] [Google Scholar]
- 56.Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, et al. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donizy P, Kaczorowski M, Biecek P, Halon A, Szkudlarek T, Matkowski R. Golgi-Related Proteins GOLPH2 (GP73/GOLM1) and GOLPH3 (GOPP1/MIDAS) in Cutaneous Melanoma: Patterns of Expression and Prognostic Significance. Int J Mol Sci. 2016;17:1619. doi: 10.3390/ijms17101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Guo L, Chen S-W, Zhao X-H, Zhuang S-M, Wang L-P, et al. GOLPH3 overexpression correlates with tumor progression and poor prognosis in patients with clinically N0 oral tongue cancer. J Transl Med. 2012;10:168. doi: 10.1186/1479-5876-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ling ZQ, Guo W, Lu XX, Zhu X, Hong LL, Wang Z, et al. A Golgi-specific protein PAQR3 is closely associated with the progression, metastasis and prognosis of human gastric cancers. Ann Oncol. 2014;25:1363–1372. doi: 10.1093/annonc/mdu168. [DOI] [PubMed] [Google Scholar]
- 60.Zhou J, Xu T, Qin R, Yan Y, Chen C, Chen Y, et al. Overexpression of Golgi phosphoprotein-3 (GOLPH3) in glioblastoma multiforme is associated with worse prognosis. J NeuroOncol. 2012;110:195–203. doi: 10.1007/s11060-012-0970-9. [DOI] [PubMed] [Google Scholar]
- 61.Howley BV, Hussey GS, Link LA, Howe PH. Translational regulation of inhibin [beta]A by TGF[beta] via the RNA-binding protein hnRNP E1 enhances the invasiveness of epithelial-to-mesenchymal transitioned cells. Oncogene. 2016;35:1725–1735. doi: 10.1038/onc.2015.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.