Abstract
Bile acids are an important signal for germination of Clostridioides difficile spores; however, the bile acid signal alone is not sufficient. Amino acids, such as glycine, are another signal necessary for germination by C. difficile spores. Prior studies on the amino acid signal required for germination have shown that there is a preference for the amino acid used as a signal for germination. Previously we found that D-alanine can function as a co-germinant for C. difficile spores at 37 °C but not at 25 °C. Here, we tested the ability of other amino acids to act as co-germinants with taurocholate (TA) at 37 °C and found that many amino acids previously categorized as non-co-germinants are co-germinants at 37 °C. Based on the EC50 values calculated for two different strains, we found that C. difficile spores recognize different amino acids with varying efficiencies. Using this data, we ranked the amino acids based on their effect on germination and found that in addition to D-alanine, other D-forms of amino acids are also used by C. difficile spores as co-germinants. Among the different types of amino acids, ones with branched chains such as valine, leucine, and isoleucine are the poorest co-germinants. However, glycine is still the most effective amino acid signal for both strains. Our results suggest that the yet-to-be-identified amino acid germinant receptor is highly promiscuous.
Keywords: C. difficile, spore, germination, co-germinant, amino acid
Introduction
Clostridioides difficile (recently renamed from Clostridium difficile [1, 2]) is a Gram-positive, spore-forming, obligately anaerobic human pathogen that causes symptoms ranging from mild diarrhea to life threatening colonic inflammation [3, 4]. Antibiotic use is common in the healthcare environment, and prior antibiotic treatment, either related or unrelated to C. difficile infection (CDI), is the greatest risk factor for CDI. A healthy gut microbiota protects against CDI and the use of broad spectrum antibiotics disrupts the gut microbiota so that the gut becomes a suitable environment for C. difficile colonization [3–5]. During colonization and disease, C. difficile forms metabolically dormant spores that are resistant to adverse environmental factors including most disinfectants and it is the spore-form that can transmit disease or cause re-infection [5–7].
Bacterial endospores are metabolically dormant structures that some bacteria produce when the cells are under nutrient deprivation or other environmental stresses. Similar to what is observed in Bacillus subtilis, a model organism for studying spore germination, the C. difficile spore core contains DNA, RNA, and the proteins necessary for the spores to outgrow after germination initiates [6, 7]. The core is rich in calcium dipicolinic acid (DPA) which contributes to the spores’ significant resistance to heat [8–11]. The core is surrounded by an inner membrane, a germ cell wall layer, a cortex layer, an outer membrane, a spore coat layer and an exosporium layer [6]. To initiate germination, small molecule germinants must penetrate these layers to interact with their receptors. Because C. difficile is found in the gut, the key germination signals that its spores respond to are bile salts, such as taurocholate (TA), and amino acids, such as glycine [12]. Spore germination is a crucial step in causing CDI, therefore it is an attractive therapeutic target for treatment [13, 14].
Though bile acids are essential signals for C. difficile spore germination, they are not sufficient [12, 13]. C. difficile spores also require an amino acid signal (e.g., glycine) to trigger the germination process [12, 15, 16]. Even though some of the amino acids that trigger germination in other organisms can activate germination in C. difficile, C. difficile does not encode orthologues of the well-studied ger receptors [13, 17]. In fact, C. difficile spores germinate through novel spore germination pathway involving activation of a pseudoprotease, CspC, present within the cortex layers [6, 18]. In our working model, when taurocholate (TA) binds to CspC, the protein becomes activated such that it transmits its signal to the CspB protease, by an unknown mechanism [6, 18, 19]. Activated CspB cleaves pro-SleC into its active, cortex-degrading, form [20, 21]. DPA is then released from the core, the core becomes hydrated, and the germinated spore prepares for outgrowth [18, 19].
C. difficile spore germination cannot occur without a co-germinant (historically an amino acid, but calcium may play an unknown role [22]) and the amino acid germinant receptor is still unknown. Amino acids are frequently used as germinants. For example, in B. subtilis, spores germinate in response to L-alanine or to the combination of L-asparagine, D-glucose, D-fructose and potassium ions (AGFK) [23]. The GerA germinant receptor (a combination of GerAA, GerAB and GerAC proteins) of B. subtilis recognizes L-alanine as a germinant. Prior work has suggested two possibilities regarding the identity of the amino acid germinant receptor in C. difficile: (i) there are multiple amino acid-recognizing germinant receptors; or (ii) the receptor recognizes multiple amino acids as germinants [15].
In C. difficile, glycine is the most effective co-germinant and can be found in most rich media and in an antibiotic-treated host environment; indeed glycocholate, a host derived bile acid, can be deconjugated to glycine and cholate [24, 25]. Other L-forms of amino acids (e.g, L-alanine or L-cysteine) also trigger C. difficile spore germination when added with TA [15, 26]. Yet, not all amino acids (e.g., L-lysine or L-methionine) were identified as co-germinants [15].
Previously we showed that at a physiologically-relevant temperature, amino acids that were reported not to be co-germinants did trigger germination when added with TA (e.g., D-alanine or D-serine) [26]. These results led us to revisit germination of other amino acids which have not been previously shown to stimulate germination at room temperature. Since many enzymes have an optimal temperature of 37 °C, we tested the germination with TA and other amino acids at room temperature as well as at 37 °C. To quantify the interaction of the amino acids with the C. difficile spore, we calculated the EC50 (the concentration that achieves half-maximum germination rate) values for many amino acids from the data generated from the DPA release assay, which serves as a measure for spore germination. Based on the EC50 values, we were able to rank the amino acids from most effective to least effective as co-germinants. However, a few other amino acids such as L-isoleucine, L-leucine, and L-valine were still not able to stimulate germination at 37 °C. This leads us to two more hypotheses regarding the role of amino acids, (i) the amino acid receptor has specificity to certain amino acids and this specificity decreases as the temperature is increased to 37 °C (e.g., in a host), (ii) any amino acid can act as the second signal, but the affinity of the amino acid for its receptor varies.
Materials and Methods
Strains and their growth conditions
C. difficile strains UK1 (027 ribotype, [13, 27, 28]) and M68 (017 ribotype, [29, 30]) were grown on BHIS agar medium (37 g of brain heart infusion, 5 g of yeast extract and 1g of cysteine per liter) at 37 °C under anaerobic conditions (10% H2, 5% CO2, 85% N2) for 3–4 days before harvesting for spore purification.
Spore purification
Spores derived from the UK1 and M68 strains were purified as described previously [18, 19, 31]. Briefly, the strains were grown on BHIS agar medium (20–30 plates) before scraping into microcentrifuge tubes containing 1 mL sterile water and kept at 4 °C overnight. The suspension containing spores and vegetative cell debris was washed with water five times by centrifuging for 1 min at 14,000 x g per wash. Spores were combined into 2–4 mL of water and further purified with a sucrose gradient (added 2 mL spores on top of 8mL of 60% sucrose and centrifuged at 4,000 x g for 20 min). The supernatant containing cell debris was discarded and the remaining pellet was washed again five times with sterile water. The spores were stored at 4 °C until use and heat activated at 65 °C for 30 minutes before use.
Germination assay
To compare the germination of spores at two different temperatures, spore germination was analyzed at 25 °C and 37 °C by measuring both OD600 and DPA release. For the OD assay, the germination was carried out in clear Falcon 96-well pates in a final volume of 100 μL and final concentration of 10 mM TA, 30 mM amino acid, 50 mM HEPES, 100 mM NaCl pH 7.5. All amino acids stocks were dissolved in water, the pH was adjusted to 7.5 and filtered-sterilized before use.
Similarly, 96-well black opaque plates were used to measure DPA released from the spores when exposed to a final concentration of 10 mM TA, 30 mM amino acid, 250 μM TbCl3, 50 mM HEPES, 100 mM NaCl at pH 7.5. Spores were added to final OD600 of 0.5 and germination was analyzed for 1 hr using a plate reader (Spectramax M3 Plate Reader, Molecular Devices, Sunnyvale, CA). Tb fluorescence was monitored at 275 nm excitation and 545 nm emission.
Calculation of EC50 values
DPA release was measured using concentration of solutions described earlier with some changes [26]. Various amounts of amino acids ranging from 0 mM to 500 mM were used for the DPA release and this assay was run at 37 °C for 2 hours. The values acquired from the DPA release were normalized and any DPA release that occurred in the absence of amino acid was subtracted before calculating the EC50 values for each amino acid. EC50 values were calculated by deriving a rate curve from slopes of the germination plot against time. All germination experiments were done in triplicate and the average mean and standard deviation values were reported.
Results
Effect of temperature on germination by C. difficile spores
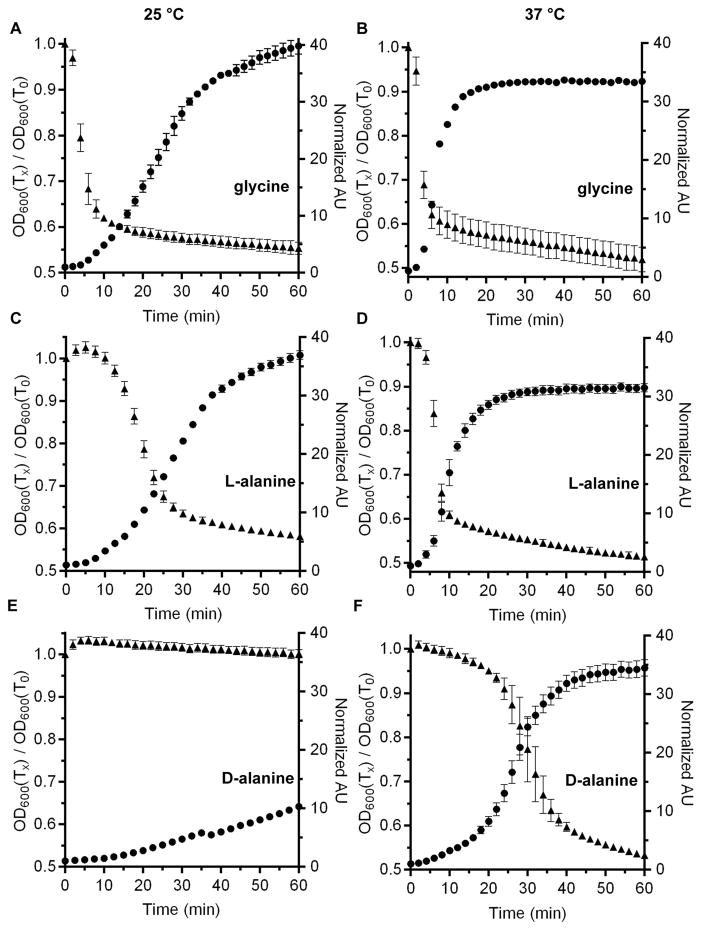
Bile acids are essential, but not sufficient, to stimulate C. difficile spore germination. Amino acids are a required second signal. The best amino acid co-germinant is glycine, but others can substitute (e.g., L-alanine) [6, 12, 15]. Previously, we demonstrated that temperature plays an important role in germination of C. difficile spores; other amino acids were revealed to be co-germinants only when C. difficile spore germination was tested at 37 °C [26]. To further investigate the effect of this physiologically-relevant temperature on C. difficile spore germination, we compared the ability of glycine, L-alanine and D-alanine to induce germination at 25 °C and at 37 °C (Figure 1). When C. difficile UK1 spores were germinated with TA and glycine at 25 °C, the OD of the spore suspension rapidly dropped and DPA was rapidly released from the spore core (Figure 1A). When germinated at 37 °C, the rate of OD drop and DPA release increased (Figure 1B). We also observed the same trend for L-alanine (Figure 1C and 1D). When we tested D-alanine at 25 °C, we observed little change in the OD of the spore suspension and a minor amount of DPA release (Figure 1E). However, when analyzed at 37 °C, D-alanine functioned as a strong co-germinant for C. difficile spores (Figure 1F). Although, the efficiency with which D-alanine functioned as a co-germinant was less than that of L-alanine and glycine. These results suggest that there may be a hierarchy for amino acids functioning as co-germinants at 37 °C.
Figure 1. Comparison of C. difficile spore germination at 25 °C and 37 °C.
Purified C. difficile UK1 spores were suspended in buffer supplemented with 10 mM taurocholate and 30 mM of the indicated amino acid. The change in OD600 during germination (▲) and the release of DPA (●) was measured over time at 25 °C (A, C, E) or 37 °C (B, D, F). Glycine (A & B), L-alanine (C & D) or D-alanine (E & F) were tested as co-germinants. The data represent the average of three independent measurements and error bars represent the standard deviation from the mean.
Identifying other co-germinants for C. difficile spore germination
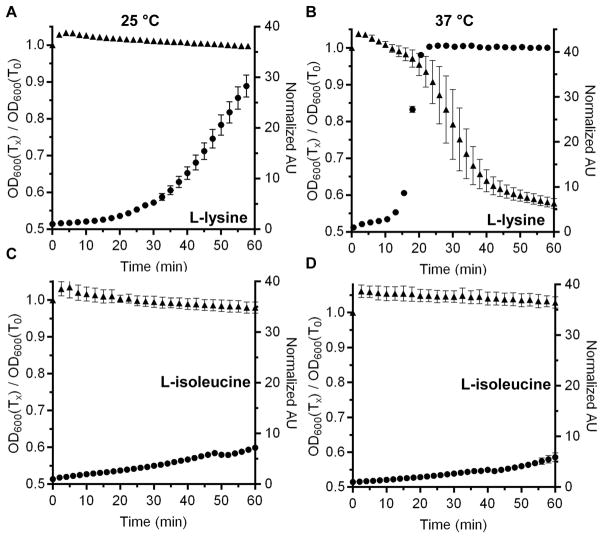
Because D-alanine functioned as a co-germinant at 37 °C but not at 25 °C, we reasoned that there may be other amino acids that can act with TA to stimulate C. difficile spore germination at 37 °C. Thus, we tested every other soluble amino acid for its ability to function as a co-germinant; L-tyrosine and L-tryptophan were insoluble. As we hypothesized, other amino acids were co-germinants with TA when incubated with spores at 37 °C. For example, L-lysine was a very poor germinant at 25 °C (Figure 2A) but led to a substantial decrease in the OD of the spores and an increase the release of DPA when tested at 37 °C (Figure 2B). Importantly, not every tested amino acid functioned as a co-germinant with TA. Incubation of C. difficile spores with TA and L-isoleucine at 25 °C (Figure 2C) or at 37 °C (Figure 2D) did not lead to an appreciable decrease in OD or DPA release. These results suggest that other amino acids function as co-germinants with TA but not all amino acids are recognized as co-germinants during C. difficile spore germination.
Figure 2. Analysis of the effects of different co-germinants on C. difficile spore germination.
Purified C. difficile UK1 spores were suspended in buffer supplemented with 10 mM taurocholate and 30 mM of the indicated amino acid. The change in OD600 during germination (▲) and the release of DPA (●) was measured over time at 25 °C (A & C) or 37 °C (B & D). L-lysine (A & B) or L-isoleucine (C & D) were tested as co-germinants. The data represent the average of three independent measurements and error bars represent the standard deviation from the mean.
Determining the effectiveness of various amino acids as a co-germinant
To test if there indeed is a hierarchical rank of amino acids that function as co-germinants, we determined the EC50 values for each amino acid. These values have been used to provide information about how germinants interact with the C. difficile spore [15, 16, 26, 27, 31–38]. To calculate the EC50 values we incubated C. difficile UK1 spores in buffer supplemented with 10 mM TA and increasing concentrations of tested amino acid; the pH of the germination solution was adjusted to pH 7.5 before spores were added. The release of DPA during C. difficile spore germination was monitored for up to 2 hours at 37 °C. The values were normalized and plotted against time. From this data, the maximum rates of DPA release were determined and used to calculate the EC50 using the Michaelis-Menten equation, as described previously [15, 16, 26, 27, 31–38]. Indeed, we observed a hierarchical order of amino acids that can stimulate germination in combination with TA (Table 1). As expected, glycine functioned as the best co-germinant and yielded an EC50 of ~190 μM. This was followed by L-alanine (1.1 mM), taurine (2.5 mM), L-glutamine (3.1 mM) and L-histidine (4.1 mM). For the C. difficile UK1 strain the order was glycine > L-alanine > taurine > L-glutamine > L-histidine > L-serine > L-arginine > L-lysine > L-threonine > D-alanine > D-serine > L-proline > L-methionine > D-lysine > L-glutamate > L-aspartic acid.
Table 1.
EC50 values for amino acids and C. difficile UK1 spores.
| Amino Acids | C. difficile UK1 |
|---|---|
| EC50 (mM) | |
| glycine | 0.19 ± 0.01 |
| L-alanine | 1.1 ± 0.2 |
| taurine | 2.5 ± 1.6 |
| L-glutamine | 3.1 ± 2.1 |
| L-histidine | 4.1 ± 1.6 |
| L-serine | 5.6 ± 0.5 |
| L-arginine | 6.1 ± 0.6 |
| L-lysine | 13.8 ± 1.3 |
| L-threonine | 14.4 ± 3.2 |
| D-alanine | 15.1 ± 9.1 |
| D-serine | 24.3 ± 1.6 |
| L-proline | 28.5 ± 5.2 |
| L-methionine | 35.2 ± 10.5 |
| D-lysine | 72.8 ± 36.1 |
| L-glutamate | 98.7 ± 45.6 |
| L-aspartic acid | 115 ± 17 |
| L-leucine | >500 mM |
| L-isoleucine | >500 mM |
| L-valine | >500 mM |
| L-phenylalanine | >500 mM |
| L-tyrosine | Not soluble |
| L-tryptophan | Not soluble |
| L-cysteine | Interfered with DPA measurements |
EC50 values were calcuated from the germination rate curves as described in the materials and methods. The values reported are the averages from three indepented experiments with the standard deviation from the mean.
In order to determine if spores from a different C. difficile isolate germinate with the same rank order of amino acids, or if the observed effects on spore germination are specific to the UK1 strain, we determined EC50 values for each amino acid and C. difficile M68 spores (Table 2). There was a slight difference in the rank order for the two strains of C. difficile that we tested. C. difficile M68 yielded a rank order of: glycine > L-arginine > L-alanine > taurine > L-glutamine > L-histidine > L-lysine > L-serine > D-alanine > D-serine > D-lysine > L-methionine > L-threonine > L-proline > L-aspartic acid.
Table 2.
EC50 values for amino acids and C. difficile M68 spores.
| Amino Acids | C. difficile M68 |
|---|---|
| EC50 (mM) | |
| glycine | 0.48 ± 0.2 |
| L-arginine | 3.6 ± 0.7 |
| L-alanine | 5.1 ± 0.7 |
| taurine | 7.7 ± 1.9 |
| L-glutamine | 10.1 ± 0.6 |
| L-histidine | 12.4 ± 5.8 |
| L-lysine | 18.5 ± 4.6 |
| L-serine | 19.4 ± 4.2 |
| D-alanine | 25.0 ± 6.6 |
| D-serine | 54.4 ± 2.6 |
| D-lysine | 56.9 ± 19.4 |
| L-methionine | 58.7 ± 4.6 |
| L-threonine | 59.8 ± 25.3 |
| L-proline | 73.2 ± 18.0 |
| L-aspartic acid | 425 ± 170 |
| L-glutamate | CND |
| L-leucine | >500 mM |
| L-isoleucine | >500 mM |
| L-valine | >500 mM |
| L-phenylalanine | >500 mM |
| L-tyrosine | Not soluble |
| L-tryptophan | Not soluble |
| L-cysteine | Interfered with DPA measurements |
EC50 values were calcuated from the germination rate curves as described in the materials and methods. The values reported are the averages from three indepented experiments with the standard deviation from the mean.
The most effective amino acid co-germinant was glycine and the least effective co-germinant was L-aspartic acid for both the strains. Unfortunately, we could not determine the EC50 values for the remaining amino acids. Even though L-cysteine was shown to be a germinant in a prior report [15], L-cysteine interfered with the DPA release assay. L-tyrosine and L-tryptophan were not soluble. L-phenylalanine, L-valine, L-leucine, L-isoleucine, L-asparagine were very poor germinants at the concentrations that were soluble.
Discussion
Apart from the bile acid signal, amino acids are also an important signal for C. difficile spore germination [6]. Glycine was first identified as the co-germinant required to initiate C. difficile spore gerimination, though other amino acids could substitute for glycine [12, 15]. Later, the requirements for the amino acid structure in functioning as a co-germinant were tested by Howerton & Abel-Santos [15]. Here, the authors found that the EC50 for glycine was approximately 8.7 mM2 for the C. difficile CD630 strain, using the OD assay as a readout [15]. Importantly, these two studies analyzed germination at either room temperature or 30 °C [12, 15]. Recently, we found that when C. difficile spore germination occurs at 37 °C, the spores recognize amino acids previously reported to not activate germination (e.g., D-alanine) [26]. Therefore, we hypothesized that other amino acids may be recognized by the spore when tested at physiologically relevant concentrations.
When tested at 37 °C, we observed that C. difficile spores recognized a diverse set of amino acids as co-germinants. Using two different C. difficile strains (UK1 and M68, ribotypes 027 and 017, respectively), we found that glycine is the most preferred amino acid co-germinant (Table 1). By using kinetic analysis of spore germination, we calculated the EC50 of glycine to be 190 μM and 420 μM for C. difficile UK1 and C. difficile M68, respectively (Table 1). These values are below the previously-reported value for glycine [15]. There are several factors that might have affected germination in response to these amino acid co-germinants. For example, the temperature previously used to test germination, strain considerations and the assay conditions. All prior studies that determined the EC50 values for germinants/co-germinants and the C. difficile spore used the OD assay for the measurements [15, 16, 27, 31, 34–36]. Here, we used a DPA release assay to calculate the EC50 in order to test germination at 37 °C. The SpectroMax plate reader we routinely use for these assays will also accommodate OD measurements at 37 °C. Importantly, though, due to the small light path of the 96-well plate, much higher spore amounts are needed to achieve an OD600 ~0.5 so that we can monitor sufficient OD change during germination. These higher spore concentrations lead to a concominant increase in the unidentified amino acid co-germinant receptor concentration(s) which would influence EC50 calculations (an increased receptor concentration would lead to an increase in the EC50 value).
One of the reasons why glycine might be more effective as a co-germinant is because of its size. Glycine is the simplest amino acid, which may help with diffusion through the spore layers. Importantly, though, size alone cannot explain why glycine functions as the best amino acid co-germinant. Since, the next best amino acid co-germinants were L-alanine, taurine and L-glutamine (for C. difficile UK1) and some of these amino acids are larger than L-serine, which functioned as a poorer co-germinant (Table 1). These observations also held true for C. difficile M68 spore germination (Table 2). Though the hierarchical order of co-germinant efficacy was slightly different, L-serine was a poorer spore germinant than L-arginine and L-alanine (Table 1).
Glycine is also important for C. difficile growth because it can be used in Stickland reactions to produce energy [39, 40]. During Stickland-based metabolism, amino acids are oxidatively decarboxlyated or deaminated to produce ATP and NADH [39]. In the reductive branch, D-proline or glycine are reduced by proline reductase or glycine reductase, respectively, to regenerate NAD+ [39]. C. difficile spores can use both glycine and D-proline as amino acid co-germinants (Table 1). However, in the two C. difficile strains tested, glycine was the best co-germinant while D-proline was a poor co-germinant (Table 1). An interesting hypothesis is that germinating C. difficile spores may preferentially utilize glycine as an energy source during outgrowth of a vegetative cell. In a prior publication, D-proline was found to be the preferred amino acid for vegetative growth while glycine was less important [40]. It would be interesting to test the effects of glycine and proline during outgrowth of C. difficile spores.
C. difficle UK1 and M68 strains are different ribotypes (UK1 is 027 while M68 is 017). In these two ribotypes, it was previously reported that the EC50 for TA was similar (~3 mM) [31, 34]. However, we found that the EC50 values of amino acid co-germinants varied between the two strains. We found that the hierarchy of amino acids that functioned as co-germinants differed except for glycine (the most efficient), and aspartic acid (the least efficient). Interestingly, germination of C. difficile UK1 was more efficient with most amino acids than was observed for C. difficile M68 strain with some exceptions (e.g., L-arginine and D-lysine had EC50 values lower for M68).
In a previous report, L-methionine, L-serine, L-lysine, L-histidine, and L-aspartic acid were not co-germinants for C. difficile spores [15]. However, we found that at 37 °C, these amino acids functioned as co-germinants to trigger germination. Previously, C. difficile CD630 (ribotype 012) was used to characterize germination in response to different amino acid co-germinants. In that study, 12 mM of amino acids was used to test amino acids as co-germinants. We found that the EC50 values for some of these amino acids to be below the 12 mM concentration used in the prior work. Thus, it is likely that 12 mM was not sufficient to trigger germination in this study at the lower temperature [15]. These results confirm the hypothesis that temperature at which germination is tested/occurs is an important consideration for C. difficile spore germination.
In our study, there were several amino acids whose EC50 values could not be calculated. L-phenylalanine and L-cysteine were previously shown to function as co-germinants [15]. However, L-cysteine interfered with DPA release assay and L-phenylalanine did not dissolve at a concentration necessary for calculating the EC50 value, and, thus, we could not determine their EC50 values. Also, we found that L-glutamate is a slow co-germinant, but we could not determine the EC50 value for the M68 strain (for unknown reasons, the kinetic analysis of spore germination with L-glutamate as a co-germinant yielded negative EC50 values).
Consistent with a prior study that analyzed germination at a 12 mM concentration, we found L-valine, L-isoleucine and L-leucine were poor germinants, even at the highest tested concentration (500 mM) [Table 1, [15]]. Although nonpolar, like glycine or alanine, these amino acids have branched methyl groups attached to their side chains closer to the alpha carbon. We also noticed that negatively charged amino acids, such as glutamic acid and aspartic acid, have higher EC50 values, making them poor co-germinants. However, positively charged amino acids were better co-germinants even though they have longer side chains.
Even though the D-form of amino acids are not normally recognized as activators of germination, we observed that the D-forms of amino acids could function as co-germinants with TA. However, these forms were weaker germinants compared to the L-form. We know from a previous report that the C. difficile alanine racemase, Alr2, has affinity for and can interconvert both L/D-alanine and L/D-serine [26]. Thus, it is possible that other racemases could be part of the spore coat and influence the ability of D-form amino acids to function as co-germinants similar to our prior observations for L/D alanine and L/D serine [26]. In fact, a prior study by Lawley and colleagues that characterized the spore proteome found that a putative proline racemase (CD3237) was present [41].
An important question is if our findings are physiologically (i.e., in vivo) relevant. The concentrations of most of the amino acids found in the distal colon of a healthy host are less then 3.5 mmol/kg (the effects of antibiotics on the amino acid content were not determined in this study) [42]. Physiologically, a prior report found that the concentration of alanine is ~0.89 mmol/kg and glycine is ~1.79 mmol/kg [42]. Because the EC50 for glycine is 190 μM for UK1, a concentration below the glycine concentration in the distal colon, this suggests that the concentration of glycine present in the distal colon is sufficient to for it to function as a co-germinant [42]. Interestingly, we found that taurine was a good co-germinant for C. difficile spores in both the UK1 (2.5 mM) and M68 isolates (7.7 mM). In the gut, taurine is normally found to be conjugated to cholic acid (taurocholic acid, a very effective bile acid germinant) [24, 25]. In healthy individuals, taurine was found to be at 3.53 mmol/kg in the descending colon and 2.89 mmol/kg in the rectum [42]. This concentration is well within the EC50 range and suggests that glycine and taurine could effectively be used as co-germinants in a healthy host. In contrast, though we find that L-methionine can function as a co-germinant (Tables 1 and 2), its concentration in the distal colon was only 0.03 mmol/kg [42]. Converesly, we found that L-glutamate was a poor germinant (Tables 1 and 2) but its concentration in the distal colon is relatively high 3.44 mmol/kg [42]. This would suggest that the co-germinants to which C. difficile spores respond are not merely due to being the most abundant amino acids in the gut, but likely are specific to metabolic processes required for growth.
Here, our findings suggest that C. difficile spores can respond to a variety of amino acids as co-germinants with taurocholic acid. Interestingly, there appears to be a hierarchy in recognition of these amino acids with glycine, alanine and taurine functioning as the most efficient co-germinants. To date, the germinant receptor that recognizes the amino acid co-germinants has not been identified. Because C. difficile is well-equipped to respond to a variety of amino acids as co-germinants, it either uses several different/redundant receptors to repond to this variety of germinants, or, as hypothesized previously, the yet-to-be-identied receptor is significantly promiscuous [15].
Highlights.
C. difficile spores recognize a diverse set of amino acids as co-germinants.
Amino acids previously determined to not be effective co-germinants (e.g., L-methionine) are co-germinants for C. difficile spores.
Temperature should be a consideration when determining factors that activate C. difficile spore germination.
Acknowledgments
We would like to thank other members of the Sorg laboratory for their helpful comments and edits during the preparation of this manuscript. We would also like to thank Dr. Leif Smith and members of the Smith laboratory at Texas A&M University for their helpful comments during the preparation of this manuscript.
This project was supported by awards 5R01AI116895 and 1U01AI124290 to J.A.S. from the National Institute of Allergy and Infectious Diseases to J.A.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prevot 1938. Anaerobe. 2016;40:95–9. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Oren A, Garrity GM. Notification that new names of prokaryotes, new combinations, and new taxonomic opinions have appeared in volume 66, part 9, of the IJSEM. Int J Syst Evol Microbiol. 2016;66:4921–3. doi: 10.1099/ijsem.0.001620. [DOI] [PubMed] [Google Scholar]
- 3.Elliott B, Androga GO, Knight DR, Riley TV. Clostridium difficile infection: Evolution, phylogeny and molecular epidemiology. Infect Genet Evol. 2017;49:1–11. doi: 10.1016/j.meegid.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 4.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primers. 2016;2:16020. doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen A. A gut odyssey: The impact of the microbiota on Clostridium difficile spore formation and germination. PLoS Pathog. 2015;11:e1005157. doi: 10.1371/journal.ppat.1005157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharjee D, McAllister KN, Sorg JA. Germinants and their receptors in Clostridia. J Bacteriol. 2016;198:2767–75. doi: 10.1128/JB.00405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 2014;22:406–16. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–25. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 9.Setlow P. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol. 2014;196:1297–305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Setlow P. Spore resistance properties. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.TBS-0003-2012. [DOI] [PubMed] [Google Scholar]
- 11.Berendsen EM, Boekhorst J, Kuipers OP, Wells-Bennik MH. A mobile genetic element profoundly increases heat resistance of bacterial spores. ISME J. 2016;10:2633–42. doi: 10.1038/ismej.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–12. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francis MB, Allen CA, Shrestha R, Sorg JA. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog. 2013;9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howerton A, Patra M, Abel-Santos E. A new strategy for the prevention of Clostridium difficile infection. J Infect Dis. 2013;207:1498–504. doi: 10.1093/infdis/jit068. [DOI] [PubMed] [Google Scholar]
- 15.Howerton A, Ramirez N, Abel-Santos E. Mapping interactions between germinants and Clostridium difficile spores. J Bacteriol. 2011;193:274–82. doi: 10.1128/JB.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez N, Liggins M, Abel-Santos E. Kinetic evidence for the presence of putative germination receptors in Clostridium difficile spores. J Bacteriol. 2010;192:4215–22. doi: 10.1128/JB.00488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nature genetics. 2006;38:779–86. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 18.Francis MB, Allen CA, Sorg JA. Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J Bacteriol. 2015;197:2276–83. doi: 10.1128/JB.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis MB, Sorg JA. Dipicolinic acid release by germinating Clostridium difficile spores occurs through a mechanosensing mechanism. mSphere. 2016;1 doi: 10.1128/mSphere.00306-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams CM, Eckenroth BE, Putnam EE, Doublie S, Shen A. Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog. 2013;9:e1003165. doi: 10.1371/journal.ppat.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutelius D, Hokeness K, Logan SM, Reid CW. Functional analysis of SleC from Clostridium difficile: an essential lytic transglycosylase involved in spore germination. Microbiology. 2014;160:209–16. doi: 10.1099/mic.0.072454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kochan TJ, Somers MJ, Kaiser AM, Shoshiev MS, Hagan AK, Hastie JL, et al. Intestinal calcium and bile salts facilitate germination of Clostridium difficile spores. PLoS Pathog. 2017;13:e1006443. doi: 10.1371/journal.ppat.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Setlow P, Wang S, Li YQ. Germination of spores of the orders Bacillales and Clostridiales. Annu Rev Microbiol. 2017;71:459–77. doi: 10.1146/annurev-micro-090816-093558. [DOI] [PubMed] [Google Scholar]
- 24.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–8. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–59. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Shrestha R, Lockless SW, Sorg JA. A Clostridium difficile alanine racemase affects spore germination and accommodates serine as a substrate. J Biol Chem. 2017;292:10735–42. doi: 10.1074/jbc.M117.791749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol. 2010;192:4983–90. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, Pepin J, et al. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol. 2008;46:431–7. doi: 10.1128/JCM.01484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–9. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drudy D, Harnedy N, Fanning S, O’Mahony R, Kyne L. Isolation and characterisation of toxin A-negative, toxin B-positive Clostridium difficile in Dublin, Ireland. Clin Microbiol Infect. 2007;13:298–304. doi: 10.1111/j.1469-0691.2006.01634.x. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharjee D, Francis MB, Ding X, McAllister KN, Shrestha R, Sorg JA. Reexamining the germination phenotypes of several Clostridium difficile strains suggests another role for the CspC germinant receptor. J Bacteriol. 2015;198:777–86. doi: 10.1128/JB.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girinathan BP, Monot M, Boyle D, McAllister KN, Sorg JA, Dupuy B, et al. Effect of tcdR mutation on sporulation in the epidemic Clostridium difficile strain R20291. mSphere. 2017;2 doi: 10.1128/mSphere.00383-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen CA, Babakhani F, Sears P, Nguyen L, Sorg JA. Both fidaxomicin and vancomycin inhibit outgrowth of Clostridium difficile spores. Antimicrob Agents Chemother. 2013;57:664–7. doi: 10.1128/AAC.01611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Francis MB, Allen CA, Sorg JA. Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS One. 2013;8:e73653. doi: 10.1371/journal.pone.0073653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liggins M, Ramirez N, Magnuson N, Abel-Santos E. Progesterone analogs influence germination of Clostridium sordellii and Clostridium difficile spores in vitro. J Bacteriol. 2011;193:2776–83. doi: 10.1128/JB.00058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez N, Abel-Santos E. Requirements for germination of Clostridium sordellii spores in vitro. J Bacteriol. 2010;192:418–25. doi: 10.1128/JB.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akoachere M, Squires RC, Nour AM, Angelov L, Brojatsch J, Abel-Santos E. Indentification of an in vivo inhibitor of Bacillus anthracis spore germination. J Biol Chem. 2007;282:12112–8. doi: 10.1074/jbc.M611432200. [DOI] [PubMed] [Google Scholar]
- 38.Stoltz KL, Erickson R, Staley C, Weingarden AR, Romens E, Steer CJ, et al. Synthesis and biological evaluation of bile acid analogues inhibitory to Clostridium difficile spore germination. J Med Chem. 2017;60:3451–71. doi: 10.1021/acs.jmedchem.7b00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouillaut L, Dubois T, Sonenshein AL, Dupuy B. Integration of metabolism and virulence in Clostridium difficile. Res Microbiol. 2015;166:375–83. doi: 10.1016/j.resmic.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouillaut L, Self WT, Sonenshein AL. Proline-dependent regulation of Clostridium difficile Stickland metabolism. J Bacteriol. 2013;195:844–54. doi: 10.1128/JB.01492-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawley TD, Croucher NJ, Yu L, Clare S, Sebaihia M, Goulding D, et al. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol. 2009;191:5377–86. doi: 10.1128/JB.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahlman B, Ljungqvist O, Persson B, Bindslev L, Wernerman J. Intestinal amino acid content in critically ill patients. JPEN J Parenter Enteral Nutr. 1995;19:272–8. doi: 10.1177/0148607195019004272. [DOI] [PubMed] [Google Scholar]