Abstract
BACKGROUND AND PURPOSE
Diffusion magnetic resonance imaging (dMRI) provides preoperative maps of neurosurgical patients’ white matter tracts, but these maps suffer from echo-planar imaging (EPI) distortions caused by magnetic field inhomogeneities. In clinical neurosurgical planning, these distortions are generally not corrected and thus contribute to the uncertainty of fiber tracking. Multiple image processing pipelines have been proposed for image-registration-based EPI distortion correction in healthy subjects. In this article, we perform the first comparison of such pipelines in neurosurgical patient data.
METHODS
Five pipelines were tested in a retrospective clinical dMRI dataset of 9 patients with brain tumors. Pipelines differed in the choice of fixed and moving images and the similarity metric for image registration. Distortions were measured in two important tracts for neurosurgery, the arcuate fasciculus and corticospinal tracts.
RESULTS
Significant differences in distortion estimates were found across processing pipelines. The most successful pipeline used dMRI baseline and T2-weighted images as inputs for distortion correction. This pipeline gave the most consistent distortion estimates across image resolutions and brain hemispheres.
CONCLUSIONS
Quantitative results of mean tract distortions on the order of 1–2 mm are in line with other recent studies, supporting the potential need for distortion correction in neurosurgical planning. Novel results include significantly higher distortion estimates in the tumor hemisphere and greater effect of image resolution choice on results in the tumor hemisphere. Overall, this study demonstrates possible pitfalls and indicates that care should be taken when implementing EPI distortion correction in clinical settings.
Keywords: Diffusion tensor imaging, EPI distortion correction, image registration, neurosurgical planning, tractography
Introduction
Diffusion magnetic resonance imaging (dMRI) can provide preoperative maps of patients’ white matter fiber tracts1 for neurosurgical planning, and the use of dMRI has been shown to increase the likelihood of complete tumor resection and time of survival.2 In dMRI, the acquisition of diffusion-weighted images (DWIs) is usually performed using echo-planar imaging (EPI), which allows rapid scanning at the expense of image distortions due to eddy currents and static magnetic field inhomogeneities.3,4 The sensitivity of EPI to magnetic field inhomogeneities produces nonlinear geometric distortions, mainly along the phase encoding direction,4 with consequent displacement of anatomical structures. Ignoring these artifacts can result in inaccuracies in diffusion tensor image (DTI) estimation and reduce accuracy in fiber tract tracing.5–8 Local magnetic susceptibility artifacts have been reported in or near brain tumors,9–11 but because EPI distortions caused by magnetic field inhomogeneities are generally expected to be localized to the anterior part of frontal and temporal lobes12–14 and may in many cases be distant from the lesion, in general distortion correction is not yet addressed in the clinic.
One technique widely employed for EPI distortion correction is based on nonrigid image registration, where a distorted EPI image is aligned to a T1- or T2-weighted structural target image.5,15,16 This technique has high potential for future implementation in neuronavigation systems, as it requires only MRI data that are standardly acquired for presurgical planning. The literature includes many different pipelines for image-registration-based EPI distortion correction.5,8,12,15,17–20 Nonetheless, none of the proposed pipelines is specifically addressed to neurosurgical planning, and despite the multitude of the options available, their implementation is not widespread for clinical use. To our knowledge, only one group has investigated an image-registration-based EPI distortion correction pipeline in neurosurgical patient data.5 Therefore, in this article, we perform the first comparison of multiple image registration pipelines for EPI distortion correction in neurosurgical patient data. We evaluate the effects of image-registration-based EPI distortion correction pipelines on two clinically relevant anatomical pathways, the corticospinal tract (CST)21 and the arcuate fasciculus (AF),22,23 as well as the patient-specific tumor regions.
Methods
Data Acquisition and Patient Selection
For this retrospective study, we selected 9 consecutive patients (Table 1) with brain tumors who had dMRI, T2-weighted, and contrast-enhanced T1-weighted MRI images acquired presurgically. All of our clinical high-resolution T1 images are acquired with gadolinium contrast injection, an acquisition modality that is widely used for monitoring brain tumors. All imaging was acquired with a Siemens 3T scanners (Siemens Trio and Verio, Siemens Healthcare, Erlangen, Germany) equipped with a 12-channel head coil. dMRI was acquired using an EPI sequence (30 gradient directions, 1 baseline (b = 0) image, b = 2,000 s/mm2, TR = 12,700, TE = 98, flip angle = 90, matrix = 100 × 90, FOV = 22 cm, 59 axial slices, voxel size = 2.3 mm3). High-resolution anatomical T1 (with gadolinium contrast) and T2-weighted scans were acquired as clinically indicated for each patient (Fig 1A and B). The study was approved by the hospital Institutional Review Board, and informed consent was obtained from all participants prior to scanning.
Table 1.
Patient Demographic Data and Pathology
| Subjects | Age | Gender | Tumor Type | Tumor Location | Tumor Volume (mm3) |
|---|---|---|---|---|---|
| 1 | 28 | F | Oligodendroglioma W.H.O. Grade II | Right frontal | 7,525 |
| 2 | 34 | F | Recurrent metastatic carcinoma, lung primary | Left frontal | 5,109 |
| 3 | 57 | M | Glioblastoma, W.H.O. Grade IV | Right parietal | 91,683 |
| 4 | 66 | F | Glioblastoma, W.H.O. Grade IV | Left temporal | 25,091 |
| 5 | 63 | M | Metastatic melanoma | Right frontal/right temporal/left temporal | 28,497 |
| 6 | 52 | F | Metastatic carcinoma, breast primary | Right frontal/right temporo-occipital | 3,261 |
| 7 | 70 | M | Anaplastic astrocytoma, W.H.O. Grade III | Left frontal | 78,142 |
| 8 | 26 | F | Anaplastic astrocytoma, W.H.O. Grade III | Left frontal | 51,825 |
| 9 | 57 | F | Diffuse astrocytoma W.H.O. Grade II | Right frontal | 41,062 |
A total of 9 consecutive patients with brain tumors who had diffusion MRI, T2-weighted, and contrast-enhanced T1-weighted MRI images acquired presurgically were selected. W.H.O. = World Health Organization.
Fig 1.
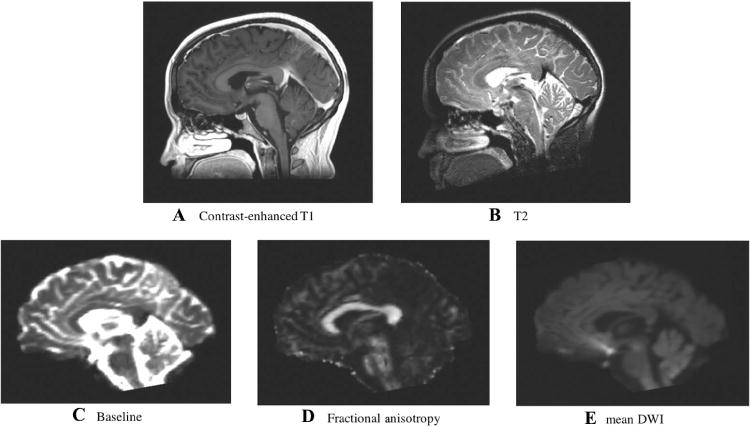
Target and moving images.
Anatomical MRI images used as registration targets included (A) T2-weighted and (B) contrast-enhanced T1-weighted. Images derived from diffusion MRI including (C) baseline, (D) fractional anisotropy, and (E) mean diffusion-weighted images were used as moving images in the EPI distortion correction experiments. Displayed images are from the first subject in the study. Note that a skull-stripping mask was applied to all images prior to deformable registration.
Image Processing
DWIs were corrected for motion and eddy current distortions using DTIPrep (www.nitrc.org/projects/dtiprep).24 The 3D Slicer (www.slicer.org version 4.4) SlicerDMRI extension (www.dmri.slicer.org) was used to extract dMRI baseline (b0) images and compute fractional anisotropy (FA) maps (Fig 1C and D, respectively) and DTI directionally encoded color maps. Binary brain masks were created using the Brain Extraction Tool.25 Each mask was visually inspected and edited as needed in 3D Slicer.
Image Registration for EPI Distortion Correction
We tested five pipelines previously proposed in the literature (Table 2). The target image was an anatomically undistorted image, either T1- or T2-weighted (Fig 1A and B). Moving images to be registered to the target were selected from the DWI dataset: baseline, FA, and mean diffusion weighted images (Fig 1C–E). Cross-correlation (CC) or mutual information (MI) image similarity metrics were used as appropriate for contrast differences between the images.
Table 2.
Registration Pipelines Tested for EPI Distortion Correction
| Fixed Image | Moving Image | References | Abbreviations | Similarity Metric |
|---|---|---|---|---|
| T1 | Baseline | [5] | B0T1 | Mutual information |
| T2 | Baseline | [8, 12, 15, 18, 19, 35] | B0T2 | Cross-correlation |
| T1 | FA | [20] | FAT1CC | Cross-correlation |
| T1 | FA | n/a | FAT1MI | Mutual information |
| T1 | mDWI | [36, 50] | mDWIT1 | Mutual information |
Registration pipelines tested for echo-planar imaging (EPI) distortion correction in diffusion MRI, with corresponding references. Note the references relate to the fixed and moving images but not necessarily to the similarity metric. Similarity metrics in the literature include but are not limited to mutual information (MI), which is insensitive to intermodality contrast differences,28,51,52 and cross-correlation (CC), which is more suited to intramodality registration. For pipeline B0T1, the metric used in the original work was normalized MI,5 so in the current study we applied the most similar available metric (MI) provided by Advanced Normalization Tools (ANTs). In B0T2, the similarity of contrast between the two images allowed the use of the CC metric. For FA to T1 registration, the metric used in the original work was normalized CC,20 so we applied the most similar available metric (CC) provided by ANTs and also investigated MI. Pipeline mDWIT1 used MI for intermodality registration. FA = fractional anisotropy; mDWI = mean diffusion-weighted images.
The registration process was initialized with a rigid registration of T1 and T2 images into the DWI space to create fixed structural targets. (In this way, the T1 and T2 images were aligned with the phase encode direction of the DWIs.) Next, each moving image was registered to the corresponding structural target using a nonrigid diffeomorphic deformable registration. Deformations were restricted to the phase-encoding axis following the standard procedure for EPI distortion correction.8,12,15,18,19 Finally, the resulting deformation field was applied to the DWIs to shift the voxels in the areas affected by distortion. Diffeomorphic registration was performed with the nonlinear SyN algorithm,13 one of the highest ranking brain registration methods.26 We used Advanced Normalization Tools (ANTs) version 2.1.0 for all image registration steps (http://stnava.github.io/ANTs/). The ANTs method has approximately 28M degrees of freedom (for nonlinear rigid registrations).26
Because anatomical T1 and T2 MRI data generally have higher voxel resolution than dMRI data, the nonrigid registration reference voxel size can be chosen at the higher19,27 or lower28,29 resolution. To compare these two options, data were registered at both low resolution (where the target image was downsampled to the voxel size of the moving image) and high resolution (where the moving image was upsampled to the voxel size of the target image). For this second option, for consistency, the reference voxel dimension was that of the T2-weighted image even when we used the T1-weighted image as the fixed image.
All results were visualized for quality control by overlaying each EPI distortion corrected volume on the corresponding subject’s T1-weighted and the T2-weighted images to assess the presence of local mismatch.
Fiber Tract Reconstruction
Whole-brain tractography was performed within the entire brain mask using two-tensor unscented Kalman filter tractography (UKFt),30 which can model crossing fibers and is relatively insensitive to edema.31 UKFt was performed in the following datasets: uncorrected dMRI data, dMRI data corrected for motion and eddy currents only, and dMRI data corrected for motion, eddy currents, and EPI distortions using all five pipelines. Tractography was seeded 20 times in all voxels within the binary brain mask. Tracking stopped where FA fell below .15 (default value) or generalized anisotropy (used to assess the suitability of fitting a multifiber model) fell below .075 (a slight decrease below the default value to increase sensitivity to tracts passing through edema). Tractography visualizations were rendered with 3D Slicer.
Anatomical White Matter Pathways and Tumor Segmentation
Expert selection of CST and AF was performed using regions of interest (ROIs) based on the T2-weighted image. Additional visual reference was provided by both the T1-weighted image and the directionally encoded color map of an EPI distortion corrected image obtained with the B0T2 registration method. Selection of CST was achieved with four inclusion ROIs at the level of the pyramid, the middle three fifths of the midbrain, the posterior limb of the internal capsule, and the cortical areas including primary motor/M1 (Brodmann’s area 4), premotor, supplementary motor (Brodmann’s area 6), and the somatosensory cortex (Brodmann’s areas 3, 1, and 2). AF was selected with three ROIs. On a coronal plane passing through the precentral gyrus, the first ROI encompassed the anteroposteriorly oriented fibers adjacent to the lateral aspect of the CST, at the same level of the corpus callosum on a cranio-caudal axis. The second ROI was created on an axial plane immediately above the level of the anterior commissure and included only the vertically oriented fibers lateral to the atrium of the lateral ventricle. The third ROI was created on an axial plane, encompassing the anterolaterally oriented fibers lateral to the lateral ventricle at the junction between the atrium and the temporal horn. All ROIs were placed bilaterally. Exclusion masks were used to avoid spurious fibers; each exclusion ROI was specific for each tract and hemisphere. Tumor segmentation was performed with reference to the T1, T2, and b0 images.
Mean Absolute Tract Displacement
For each patient dataset and registration pipeline, we calculated the mean absolute displacement of CST and AF estimated by nonrigid image registration for EPI distortion correction. For each DWI that underwent image distortion correction, we obtained the estimated displacement in the phase encoding direction (in mm) (Figs 2A, B, and 3). Next, on the corrected DWIs, we performed whole-brain tractography, from which the two anatomical tracts of AF and CST were selected (Fig 2C and D). At this point, we computed the magnitude of the deformation (equal to the absolute value of the y component of the deformation field). Next, we created a binary mask of all voxels intersected by the fiber tract. Finally, the mean deformation was computed within this binary mask (Fig 2E). The total tract volume was also computed. In 3D Slicer, this process used the modules Tractography to Mask Image and Label Statistics. This procedure allowed the quantification of mean tract displacement and tract volume.
Fig 2.
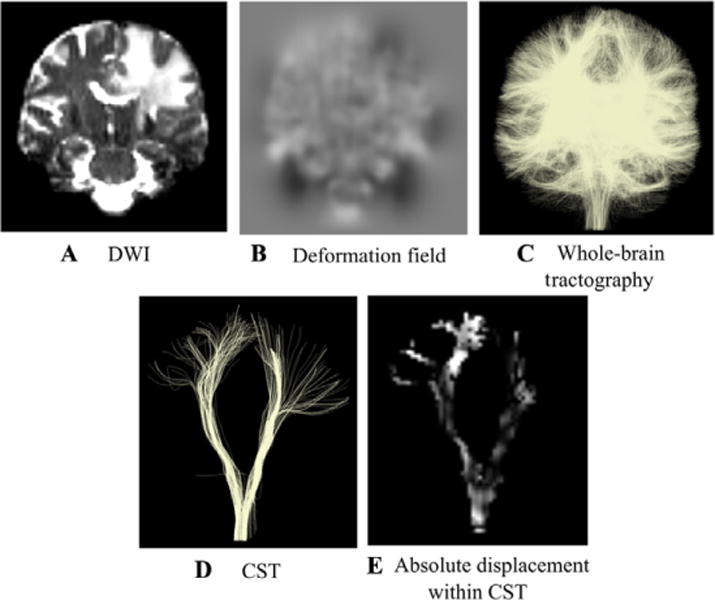
Mean absolute displacement measurement within anatomical tracts.
Steps used to obtain measurements of the absolute displacement within the corticospinal tract (CST) and the arcuate fasciculus (AF, not shown). Each diffusion-weighted image (DWI) (A) underwent image distortion correction. From the distortion correction along the phase encoding direction, a deformation field (B) was obtained. The grayscale value indicates the amount of displacement with −15 mm in black and +15 mm in white. (C) Whole-brain tractography was performed on every distortion corrected volume and anatomical tracts such as CST (D) and AF were obtained. Within each of the tracts, we quantified the mean absolute displacement derived from the deformation field (E).
Fig 3.
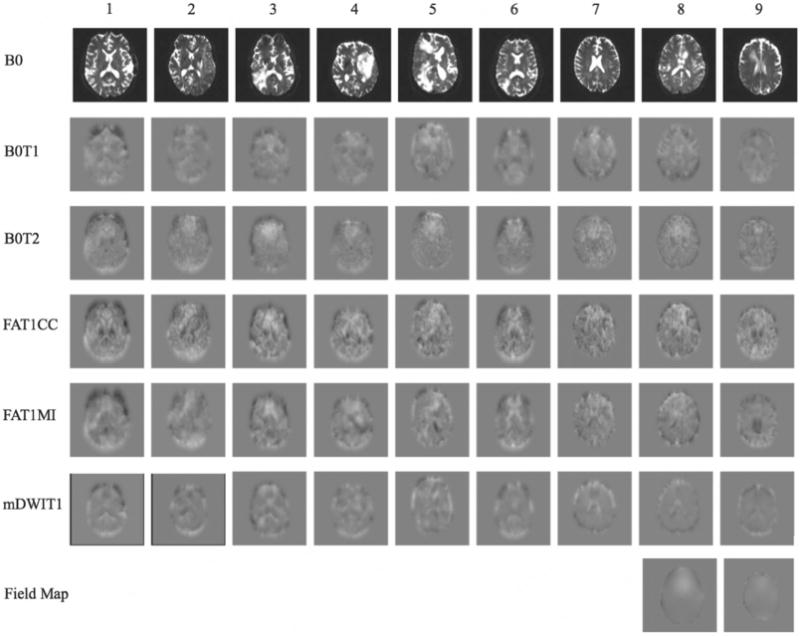
Estimated displacement in the phase encode direction.
Estimated distortion using high-resolution data, where each column corresponds to one subject and each row corresponds to a pipeline. Deformation fields visualization is in the axial plane and indicates translation in the anterior-posterior direction (phase encode direction), as estimated by the registration pipelines. The grayscale value indicates the amount of displacement with −15 mm in black and +15 mm in white. Field maps: maps representing the field inhomogeneity across the images (available only for 2 subjects). B0 = baseline; B0T1 = baseline toT1- weighted anatomical image (T1) registration pipeline; B0T2 = baseline to T2-weighted anatomical image (T2) registration pipeline; FAT1CC = fractional anisotropy to T1 registration pipeline; FAT1MI = fractional anisotropy to T1 registration pipeline; mDWIT1 = mean diffusion-weighted image to T1 registration pipeline.
Mean Absolute Tumor Displacement
For each patient and registration pipeline, we calculated the mean absolute displacement within the tumor region as estimated by the nonrigid image registration. The magnitude of the deformation was computed within the binary mask of the tumor segmentation.
Statistical Analysis
Data were analyzed using Matlab (The MathWorks Inc., Natick, MA, USA) version 2015b. To test for differences in tract displacement between pipelines, we used a one-way ANOVA test. To test for differences in tract displacement across hemispheres, we used a paired t-test, which was also used for testing differences in tract displacement between low-resolution and high-resolution data. For all the tests, the significance threshold for the reported two-tailed P values was .05.
Computational Time
Typical run times of the complete registration process on a large multiprocessor machine were 2.3 minutes (low resolution) and 13.3 minutes (high resolution), where the maximum numbers of concurrent processing threads were 17 and 26. Both run times were feasible for neurosurgical planning; however, the total CPU time for the high-resolution data registration (the time it would take a single processor system) was 2 hours and 38 minutes.
Results
Visualization of Computed Deformations
Visualization of the amount of displacement estimated in the phase encode direction (Fig 3), demonstrates the apparent high variability across pipelines. For 2 subjects, a field map was acquired, which is known to be proportional to the EPI distortion.32,33 We note that, when compared to the image registration results, the two field maps appear visually more consistent across subjects in terms of the spatial locations of the distortions.
Visualization of Distortion-Corrected Tracts
CST and AF tractography results using original uncorrected data, data corrected for eddy currents and head motion, and data obtained from all five registration pipelines were visualized. Overall, the location of all tracts was similar when viewed in 3D. However, a closer examination of the tracts’ intersection with T1 anatomical images indicated some variability across pipelines, which could affect the clinically important spatial relationship of the tracts to the tumor (Fig 4).
Fig 4.
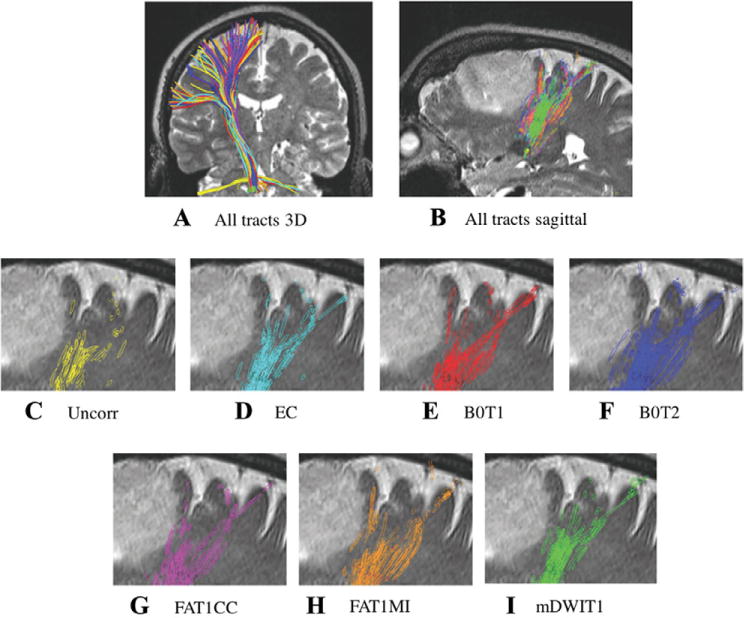
Spatial relationship of the tracts to the tumor.
Spatial relationship of the tracts with the tumor: the distance from the tracts to the lesion can vary with the registration method. (A) 3D representation of tractography results in corticospinal tract from patient 9 with diffuse astrocytoma. After application of registration pipelines, the location of the seeded tracts appears visually similar in 3D. (B) Tracts from all pipelines, showing their intersection with a T2 sagittal slice. (C-I) Tracts’ intersection with a T2-weighted anatomical image shows the variability across pipelines. EC = Eddy current and movement corrected data; Uncorr = uncorrected data; B0T1 = baseline to T1-weighted anatomical image (T1) registration pipeline; B0T2 = baseline to T2-weighted anatomical image (T2) registration pipeline; FAT1CC = fractional anisotropy to T1 registration pipeline; FAT1MI = fractional anisotropy to T1 registration pipeline; mDWIT1 = mean diffusion-weighted image to T1 registration pipeline.
Analysis of Mean Tract Displacement
Expert tractography selection identified CST bilaterally in all 9 patients and AF bilaterally in 6 patients. Due to peritumoral edema and mass effect, it can be a challenge to trace AF and to select AF with standard anatomical ROIs, especially in the right hemisphere where AF may be smaller. The following statistical analyses were performed on N= 9 patients (CST) and N= 6 patients (AF).
Mean absolute displacement of AF and CST was significantly different across the distortion correction pipelines, in both low-resolution (AF, P .012; CST, P .014) and high-resolution (AF, P < .001; CST, P < .001) data (Fig 5), after averaging across hemisphere within each subject. The FAT1CC pipeline was inconsistent with the other pipelines in that it estimated the largest distortions, which are likely to be incorrect overestimates (see also Fig 6). We found greater displacement in the tumor hemisphere for CST, but not AF, for both low-resolution (P < .002) and high-resolution (P .014) data (Table 3). Finally, we found greater displacement when using low-resolution data relative to high-resolution data (comparison of data in Table 3), in both the tumor and contralateral hemispheres (AF-ipsilateral, P .011; AF-contralateral, P .047; CST-ipsilateral, P .011; CST-contralateral, P .009).
Fig 5.
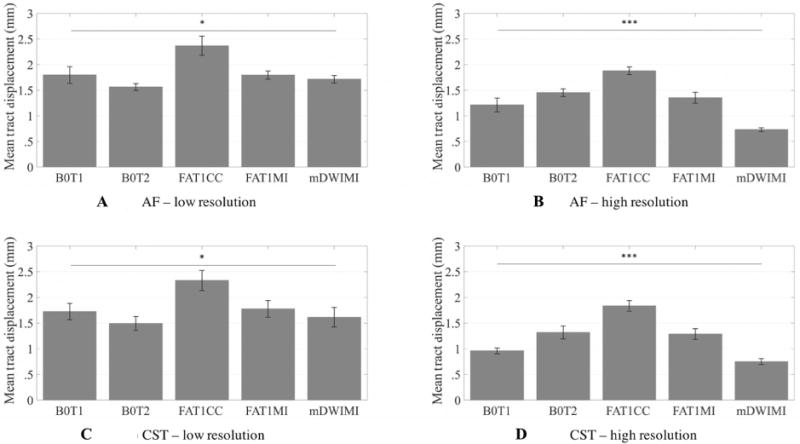
Estimated mean tract displacement.
Comparison across registration pipelines shows significant differences in estimated displacement of critical white matter structures. Mean absolute displacement of arcuate fasciculus (low-resolution (A) and high-resolution (B) data) and corticospinal tract (low-resolution (C) and high-resolution (D) data), after averaging over hemisphere. Statistics were performed on mean displacement values (in mm) on N= 6 subjects for arcuate fasciculus (AF) and on N= 9 subjects for corticospinal tract (CST) using a one-way ANOVA (low-resolution data: AF, P= .012; CST, P= .014), * = P < .05; (high-resolution data: AF, P < .001; CST, P < .001), *** = P < .001. Significant comparisons are indicated by asterisks (*P < .05, **P < .001, ***P < .0001). B0T1 = baseline to T1-weighted anatomical image (T1) registration pipeline; B0T2 = baseline to T2-weighted anatomical image (T2) registration pipeline; FAT1CC = fractional anisotropy to T1 registration pipeline; FAT1MI = fractional anisotropy to T1 registration pipeline; mDWIT1 = mean diffusion-weighted image to T1 registration pipeline.
Fig 6.
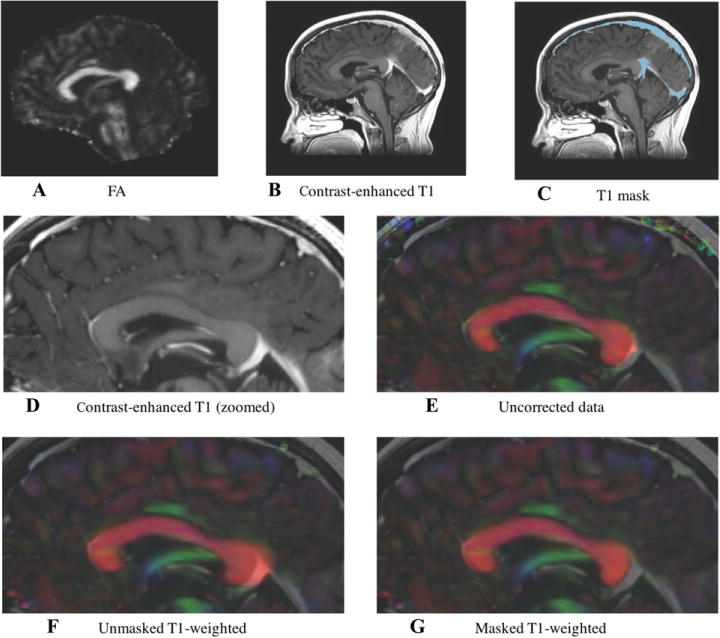
Effect of contrast enhancement on T1-based distortion correction method.
Directionally encoded color images of the corpus callosum for one sample subject, showing the effect of contrast enhancement on the T1- based distortion correction method. (A) Fractional anisotropy map; (B-D) original contrast-enhanced T1-weighted image; (C) contrast-enhanced T1-weighted image with mask overlaid; (E) uncorrected DTI volume; (F) echo-planar imaging (EPI) distortion corrected data obtained with FAT1CC registration demonstrates “overstretching” of the corpus callosum; and (G) EPI distortion corrected data obtained with FAT1CC after masking brighter venous drainage resulting from the contrast enhancement in the T1-weighted volume.
Table 3.
Mean Absolute Tract Displacement
| Mean Absolute Displacement AF (mm)
| ||||
|---|---|---|---|---|
| Low Resolution
|
High Resolution
|
|||
| Pipeline | Tumor | Contralateral (Healthy) | Tumor | Contralateral (Healthy) |
| B0T1 | 2.0 ± .8 | 1.6 ± .3 | 1.3 ± .5 | 1.1 ± .3 |
| B0T2 | 1.7 ± .3 | 1.5 ± .1 | 1.5 ± .3 | 1.4 ± .1 |
| FAT1CC | 2.6 ± 1.0 | 2.1 ± .3 | 1.9 ± .3 | 1.9 ± .2 |
| FAT1MI | 1.8 ± .4 | 1.8 ± .2 | 1.4 ± .5 | 1.3 ± .3 |
| mDWIT1 | 1.6 ± .4 | 1.8 ± .4 | .6 ± .2 | .8 ± .1 |
| Average | 1.9 ± .6† | 1.8 ± .3† | 1.3 ± .3† | 1.3 ± .2† |
|
Mean Absolute Displacement CST (mm)
| ||||
|
Low Resolution
|
High Resolution
|
|||
| Pipeline | Tumor | Contralateral (Healthy) | Tumor | Contralateral (Healthy) |
|
| ||||
| B0T1 | 2.0 ± .6 | 1.5 ± .5 | 1.1 ± .3 | .9 ± .1 |
| B0T2 | 1.6 ± .6 | 1.3 ± .3 | 1.5 ± .6 | 1.2 ± .3 |
| FAT1CC | 2.5 ± .8 | 2.1 ± .5 | 1.9 ± .3 | 1.8 ± .4 |
| FAT1MI | 1.9 ± .8 | 1.7 ± .7 | 1.3 ± .3 | 1.2 ± .4 |
| mDWIT1 | 1.8 ± .8 | 1.4 ± .5 | .8 ± .2 | .7 ± .2 |
| Average | 2.0 ± .7†* | 1.6 ± .5†* | 1.3 ± .3†* | 1.1 ± .3†* |
Mean absolute displacement ± standard deviation of the arcuate fasciculus (AF) and the corticospinal tract (CST), as estimated by the five registration pipelines. The mean absolute displacement of the CST was significantly higher in the tumor hemisphere for both low-resolution and high-resolution data (paired t-test, P= .002 and P= .014, respectively). Higher displacement was found when using low-resolution data relative to high-resolution data in both the ipsilateral (tumor) and contralateral (healthy) hemispheres (AF-ipsilateral, P: .011; AF-contralateral, P: .047; CST-ipsilateral, P: .011; CST-contralateral, P: .009).
= P < .05, comparison of tumor versus contralateral hemisphere;
= P < .05, comparison of low-resolution versus high-resolution data.
In order to assess which of the five registration pipelines might be most suitable for clinical use, we evaluated consistency of the estimated tract displacement across hemispheres and resolutions. To quantify consistency, we computed the differences in mean absolute tract displacement between resolutions employed for the registration (high and low) (Table 5) and between hemispheres (tumor and contralateral) (Table 6). The B0T2 pipeline was the most consistent: it was the least affected by the resolution of the image used, and it was not highly affected by the presence of the tumor (Tables 5 and 6). Across all methods, the choice of resolution had the most impact in the tumor hemisphere: the difference between displacements estimated at high and low resolution was greater in the tumor hemisphere (in CST: paired t-test, P= .036).
Table 5.
Differences in Mean Absolute Tract Displacement across Resolutions
| Low Resolution – High Resolution (AF)
|
Low Resolution – High Resolution (CST)
|
|||
|---|---|---|---|---|
| Pipeline | Tumor | Contralateral (Healthy) | Tumor | Contralateral (Healthy) |
| B0T1 | .7 ± .4 | .5 ± .2 | .9 ± .5 | .6 ± .5 |
| B0T2 | .2 ± .2 | .04 ± .3 | .2 ± .3 | .2 ± .2 |
| FAT1CC | .7 ± 1.0 | .3 ± .2 | .6 ± .7 | .4 ± .3 |
| FAT1MI | .4 ± .3 | .5 ± .3 | .5 ± .4 | .4 ± .4 |
| mDWIT1 | 1.0 ± .3 | 1.0 ± .4 | 1.0 ± .6 | .7 ± .4 |
| Average | .6 ± .4 | .5 ± .3 | .7 ± .5* | .5 ± .4* |
Low difference across resolutions indicates high pipeline consistency or robustness, and by this measure the B0T2 pipeline is superior, with the lowest differences in all experiments (second row). The mDWIT1 and B0T1 methods perform worst by this metric, with the highest differences. Note that in the corticospinal tract, results in the tumor hemisphere are more affected by the choice of resolution than results in the contralateral hemisphere (paired t-test, P= .036).
= P < .05, comparison of tumor versus contralateral hemisphere. AF = arcuate fasciculus; CST = corticospinal tract.
Table 6.
Differences in Mean Absolute Tract Displacement across Hemispheres
| Tumor – Contralateral (AF)
|
Tumor – Contralateral (CST)
|
|||
|---|---|---|---|---|
| Pipeline | Low Resolution | High Resolution | Low Resolution | High Resolution |
| B0T1 | .3 ± .6 | .1 ± .3 | .5 ± .6 | .2 ± .3 |
| B0T2 | .2 ± .3 | .04 ± .2 | .3 ± .4 | .3 ± .5 |
| FAT1CC | .5 ± 1.0 | .1 ± .2 | .4 ± .6 | .1 ± .3 |
| FAT1MI | −.04 ± .4 | .01 ± .5 | .2 ± .7 | .1 ± .4 |
| mDWIT1 | −.3 ± .7 | −.2 ± .2 | .4 ± .7 | .1 ± .2 |
| Average | .1 ± .6 | .1 ± .3 | .4 ± .6 | .2 ± .3 |
Smaller magnitude differences across hemispheres (differences near zero) indicate a low impact of the tumor on the estimation of displacements. By this measure, the B0T2 pipeline also performs well, as it gives a relatively low difference across hemispheres that is consistent across the resolutions (second row). The mDWIT1 method apparently performs worst by this metric, as it disagrees with other methods in the arcuate fasciculus, attributing larger displacements to the healthy hemisphere. AF = arcuate fasciculus; CST = corticospinal tract.
We found that one important limitation for image-registration-based distortion correction may derive from the clinical acquisition of contrast-enhanced T1-weighted images. All of our clinical high-resolution T1 images are acquired with gadolinium contrast injection, an acquisition modality that is widely used for monitoring brain tumors. Upon visual inspection, we found that in some cases, in particular in the FAT1CC registration, the distortion correction was affected by the bright contrast in the brain’s venous system. This may have caused overstretching of the regions adjacent to brighter venous drainage resulting from the contrast enhancement (Fig 6). Others have also observed overstretching in EPI distortion correction due to high T1 signal for the skull and soft tissue areas,34 even without the image registration challenge of contrast injection.
Analysis of Mean Tumor Displacement and Average Tumor Volume
Tumor segmentation was performed for all patients. Mean absolute displacement of the tumor area was significantly different across the distortion correction pipelines for both the low-resolution data (P: .046) and high-resolution data (P: .043). Mean displacement was higher when using low-resolution data, as previously shown for calculation of mean absolute tract displacement (Table 4). Finally, we did not find any correlation between the tumor volume (Table 1) and the amount of tumor displacement.
Table 4.
Mean Absolute Tumor Displacement
| Mean Absolute Displacement Tumor Area (mm)
| ||
|---|---|---|
| Pipeline | Low Resolution | High Resolution |
| B0T1 | 2.1 ± 1.3 | 1.3 ± .9 |
| B0T2 | 1.6 ± .5 | 1.6 ± .5 |
| FAT1CC | 3.6 ± 1.5 | 2.0 ± .6 |
| FAT1MI | 2.2 ± 1.0 | 1.4 ± .6 |
| mDWIT1 | 2.1 ± 1.0 | 1.1 ± .3 |
| Average | 2.3 ± 1.0 | 1.5 ± .6 |
Mean absolute displacement ± standard deviation of the tumor area, as estimated by the five registration pipelines. Higher displacement was found when using low-resolution data relative to high-resolution data. In accordance with tract-based results, the B0T2 pipeline gave consistent results across image resolutions.
Average Tract Volume
To confirm that results were not confounded by volume variability of fiber tracts, we compared the average tract volume across pipelines for both AF and CST of the low- resolution data. Differences in average tract volume across pipelines did not reach statistical significance in the tumor hemisphere nor in the contralateral hemisphere (AF-ipsilateral, P: .998; AF-contralateral, P: .870; CST-ipsilateral, P: .545; CST-contralateral, P: .924).
Discussion
In this study, we evaluated five pipelines for image-registration-based EPI distortion correction (Table 2) that have previously been described and assessed.5,8,12,15,18–20,35,36 To our knowledge, this is the first time these pipelines have been evaluated in the context of neurosurgical patient data.
Our results indicate small mean displacements on the order of 1 or 2 mm within AF and CST (Table 3), and of 1–3 mm within the tumor volume, in line with the current clinical practice of neglecting EPI distortion correction. However, this is the mean value over a large volume (an entire AF or CST tract or tumor volume), and as Figure 3 shows, locally larger deformations are also computed. Overall, the mean displacement was found to be of similar magnitude both within the tumor and within the tracts (Tables 3 and 4). Plots of tumor size versus tumor distortion measurement (not shown) indicated no apparent relationship in our small study. Given the variety of tumor types and locations, a larger study would be needed to assess the influence of the tumor (Table 1). Furthermore, the computed deformations were sufficient to affect the spatial relationship between the tractography and the tumor (Fig 4B-G). We note that evaluation of tractography for neurosurgery is a challenging and open problem.37 However, these quantitative results demonstrate the existence of EPI distortions in major fiber tracts and support the potential need for distortion correction in neurosurgical planning.
To our knowledge, two other groups have quantified EPI distortions in dMRI data from patients with brain tumors. One group performed EPI distortion correction using a nonlinear registration method (baseline image to T1 MPRAGE) in 14 retrospective patients.5 Their approach did not restrict deformations to the phase encode direction or perform tractography after distortion correction to take advantage of expected improvements in local tract continuity.6–8 Distortion correction was instead applied directly to previously performed CST tractography, giving a mean displacement of 2.4 mm in the internal capsule and maximum displacements up to 15 mm. A second group evaluated the reversed gradient method for distortion correction in a retrospective fashion in a large cohort of patients with brain tumors, finding a median EPI distortion of 2.1 mm in the whole brain,38 similar to the distortion magnitudes we found. However, as the entire brain was evaluated, specific information about clinically relevant tracts was not available for comparison. Another possibility for reducing possible influence of the tumor on image registration is to mask the tumor from the registration. This approach has been proposed for when both images do not contain the tumor (eg, registration of atlas to patient)39,40 or for pre- to intraoperative image registration when changes due to surgery have taken place, such as partial or complete tumor resection.41 In our experiments, the tumor was present in all images, so we did not remove tumor information by masking, though this approach could have potential for enhancing tumors if T1 contrast-enhanced images are the only ones available for image registration.
There are several possible limitations of our study. In our experiments, we only evaluated image-registration-based pipelines for correction of EPI distortions due to their simple applicability to neurosurgical planning. Other EPI distortion correction methods that require additional MRI acquisitions have been proposed, including fieldmap techniques4,6,14,32,33,42 and the reversed gradient (“blip-up blip-down”) method.12,43–47 While our set of tested registration pipelines was extensive, other pipelines are possible. It is also possible that each pipeline may need separate optimization to further improve the output. The parameters chosen were kept as constant as possible across pipelines and were obtained with reference to the ANTs documentation and consulting the ANTs forum. The sampling rate was the ANTs default of 100% for both CC and MI experiments. The gradient step size, the smoothing sigmas, and the shrink factor parameters were kept constant for all pipelines, while parameters specific to similarity metrics were set as region radius of 2 (CC) and bin number of 32 (MI).48 Another limitation of this study is the lack of ground truth information about tract displacement; our results are limited to an evaluation of consistency which, while useful to highlight differences across methods, does not directly provide information about method accuracy. Another possible limitation of using T1 as the registration reference is the clinically observed postcontrast reduction in contrast between gray and white matter, which could be an added challenge for the intensity-driven registration, in addition to the discussed problems caused by enhancing vasculature.
Overall, we find that care must be taken when implementing an image-registration-based EPI distortion correction pipeline in the context of neurosurgical patient data. The choice of input images, the choice of image resolution, and the performance of the pipeline in the presence of a tumor are all important factors. Differential registration performance in tumor versus contralateral hemispheres may relate to technical factors affecting registration algorithms, such as differences in image contrast in the presence of the lesion, or to pathological factors, as previous studies have reported local susceptibility artifacts in or near brain tumors.9–11
Conclusion
In conclusion, on the basis of this small study, we can provide some recommendations for groups choosing to use image- registration-based EPI distortion correction methods during neurosurgical planning. Based on quantitative tract-specific measurements, our experiments provided evidence that the dMRI baseline to T2 registration pipeline (B0T2) gave the most consistent information about tract displacement. Similarly, the B0T2 registration pipeline was the most consistent in the quantification of the mean absolute displacement of the tumor volume. It is thus plausible that B0T2 is the most reliable image-registration-based EPI distortion correction pipeline for application to patient data. Moreover, the B0T2 registration appears to be the most popular pipeline, outnumbering the other pipelines in terms of literature citations (Table 2). In addition, it avoids the use of a clinical contrast-enhanced T1 image, which was shown to give problematic registration results (eg, Fig 6) and could lead to likely overestimates of displacement (Table 3). If computation time permits, robust registration using the high-resolution voxel space of the T2 image should be preferred, rather than downsampling the T2 image.
In the future, we expect that new methods that go beyond simple pairwise image registration to take advantage of additional information, such as distortion correction using structural MRI, DWI, and reversed gradient data,34 or eddy current correction using all DWI images,49 have high potential for distortion correction for neurosurgical planning.
Acknowledgments
Disclosure of funding: We gratefully acknowledge funding provided by the following National Institutes of Health (NIH) grants: U01 CA199459, R03 NS088301, P41 EB015898 National Center for Image-Guided Therapy, R01 MH074794, P41 EB015902 Neuroimage Analysis Center, R25 CA089017, R01 MH097979, R21 CA198740, U01 NS083223. A.A. is supported by the Armenise-Harvard Summer Fellowship. C.T. is supported by a grant (No. 612.001.104) from the Physical Sciences Division of the Netherlands Organization for Scientific Research (NWO).
References
- 1.Jellison BJ, Field AS, Medow J, et al. Diffusion tensor imaging of cerebral white matter: a pictorial review of physics, fiber tract anatomy, and tumor imaging patterns. Am J Neuroradiol. 2005;25:356–69. [PMC free article] [PubMed] [Google Scholar]
- 2.Wu JS, Zhou LF, Tang WJ, et al. Clinical evaluation and follow-up outcome of diffusion tensor imaging-based functional neuronavigation: a prospective, controlled study in patients with gliomas involving pyramidal tracts. Neurosurgery. 2007;61:935–49. doi: 10.1227/01.neu.0000303189.80049.ab. [DOI] [PubMed] [Google Scholar]
- 3.Jezzard P, Clare S. Sources of distortion in functional MRI data. Hum Brain Mapp. 1999;8:80–5. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<80::AID-HBM2>3.0.CO;2-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jezzard P, Balaban RS. Correction for geometric distortion in echo planar images from B0 field variations. Magn Reson Med. 1995;34:65–73. doi: 10.1002/mrm.1910340111. [DOI] [PubMed] [Google Scholar]
- 5.Merhof D, Soza G, Stadlbauer A, et al. Correction of susceptibility artifacts in diffusion tensor data using non-linear registration. Med Image Anal. 2007;11:588–603. doi: 10.1016/j.media.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Lazar M, Lee J, et al. Correction of B0 EPI distortions in diffusion tensor imaging and white matter tractography. Proc Int Soc Magn Reson Med. 2004;11:2172. [Google Scholar]
- 7.Andersson J, Richter M, Richter W, et al. Effects of susceptibility distortions on tractography. Proc Int Soc Magn Reson Med. 2004;11:87. [Google Scholar]
- 8.Irfanoglu MO, Walker L, Sarlls J, et al. Effects of image distortions originating from susceptibility variations and concomitant fields on diffusion MRI tractography results. Neuroimage. 2012;61:275–88. doi: 10.1016/j.neuroimage.2012.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagley LJ, Grossman RI, Judy KD, et al. Gliomas: correlation of magnetic susceptibility artifact with histologic grade. Radiology. 1997;202:511–6. doi: 10.1148/radiology.202.2.9015082. [DOI] [PubMed] [Google Scholar]
- 10.Krings T, Reinges M, Erberich S, et al. Functional MRI for presurgical planning: problems, artefacts, and solution strategies. J Neurol Neurosurg Psychiatry. 2001;70:749–60. doi: 10.1136/jnnp.70.6.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim MJ, Holodny AI, Hou BL, et al. The effect of prior surgery on blood oxygen level–dependent functional MR imaging in the preoperative assessment of brain tumors. Am J Neuroradiol. 2005;26:1980–5. [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M, Chang LC, Walker L, et al. Comparison of EPI distortion correction methods in diffusion tensor MRI using a novel framework. MICCAI. 2008:321–9. doi: 10.1007/978-3-540-85990-1_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avants BB, Epstein CL, Grossman M, et al. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gholipour A, Kehtarnavaz N, Scherrer B, et al. On the accuracy of unwarping techniques for the correction of susceptibility-induced geometric distortion in magnetic resonance echo-planar images. IEEE. 2011:6997–7000. doi: 10.1109/IEMBS.2011.6091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao R, Fletcher PT, Gerber S, et al. A variational image-based approach to the correction of susceptibility artifacts in the alignment of diffusion weighted and structural MRI. Inf Process Med Imaging. 2009:664–75. doi: 10.1007/978-3-642-02498-6_55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhushan C, Haldar JP, Choi S, et al. Co-registration and distortion correction of diffusion and anatomical images based on inverse contrast normalization. Neuroimage. 2015;115:269–80. doi: 10.1016/j.neuroimage.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao G, He R, Poonawalla AH, et al. The correction of EPI-induced geometric distortions and their evaluation. IEEE. 2007;6:225. [Google Scholar]
- 18.Kybic J, Thévenaz P, Nirkko A, et al. Unwarping of unidirectionally distorted EPI images. IEEE Trans Med Imaging. 2000;19:80–93. doi: 10.1109/42.836368. [DOI] [PubMed] [Google Scholar]
- 19.Ardekani S, Sinha U. Geometric distortion correction of high-resolution 3 T diffusion tensor brain images. Magn Reson Med. 2005;54:1163–71. doi: 10.1002/mrm.20651. [DOI] [PubMed] [Google Scholar]
- 20.Malinsky M, Peter R, Hodneland E, et al. Registration of FA and T1-weighted MRI data of healthy human brain based on template matching and normalized cross-correlation. J Digit Imaging. 2013;26:774–85. doi: 10.1007/s10278-012-9561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al Masri O. An essay on the human corticospinal tract: history, development, anatomy, and connections. Neuroanatomy. 2011;10:1–4. [Google Scholar]
- 22.Catani M, Mesulam M. The arcuate fasciculus and the disconnection theme in language and aphasia: history and current state. Cortex. 2008;44:953–61. doi: 10.1016/j.cortex.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dick AS, Tremblay P. Beyond the arcuate fasciculus: consensus and controversy in the connectional anatomy of language. Brain. 2012;135:3529–50. doi: 10.1093/brain/aws222. [DOI] [PubMed] [Google Scholar]
- 24.Oguz I, Farzinfar M, Matsui J, et al. DTIPrep: quality control of diffusion-weighted images. Front Neuroinform. 2014;8 doi: 10.3389/fninf.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–55. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein A, Andersson J, Ardekani BA, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Risholm P, Golby AJ, Wells W. Multimodal image registration for preoperative planning and image-guided neurosurgical procedures. Neurosurg Clin NAm. 2011;22:197–206. doi: 10.1016/j.nec.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pluim PJ, Maintz JA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imaging. 2003;22:986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 29.Klein S, Staring M, Murphy K, et al. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 30.Malcolm JG, Shenton ME, Rathi Y. Filtered multitensor tractography. IEEE Trans Med Imaging. 2010;29:1664–75. doi: 10.1109/TMI.2010.2048121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z, Tie Y, Olubiyi O, et al. Reconstruction of the arcuate fasciculus for surgical planning in the setting of peritumoral edema using two-tensor unscented Kalman filter tractography. Neuroimage Clin. 2015;7:815–22. doi: 10.1016/j.nicl.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cusack R, Brett M, Osswald K. An evaluation of the use of magnetic field maps to undistort echo-planar images. Neuroimage. 2003;189:127–42. doi: 10.1006/nimg.2002.1281. [DOI] [PubMed] [Google Scholar]
- 33.Hutton A, Bork A, Josephs O, et al. Image distortion correction in fMRI: a quantitative evaluation. Neuroimage. 2002;16:217–40. doi: 10.1006/nimg.2001.1054. [DOI] [PubMed] [Google Scholar]
- 34.Irfanoglu MO, Modi P, Nayak A, et al. DR-BUDDI (diffeomorphic registration for blip-up blip-down diffusion imaging) method for correcting echo planar imaging distortions. Neuroimage. 2015;106:284–99. doi: 10.1016/j.neuroimage.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daducci A, Gerhard S, Griffa A, et al. The connectome mapper: an open-source processing pipeline to map connectomes with MRI. PLoS One. 2012;7:e48121. doi: 10.1371/journal.pone.0048121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leemans A, Jeurissen B, Sijbers J, et al. ExploreDTI: a graphical toolbox for processing, analyzing, and visualizing diffusion MR data. 17th Annual Meeting of Intl Soc Mag Reson Med. 2009;209:3537. [Google Scholar]
- 37.Pujol S, Wells W, Pierpaoli C, et al. The DTI challenge: toward standardized evaluation of diffusion tensor imaging tractography for neurosurgery. J Neuroimaging. 2015;25:875–82. doi: 10.1111/jon.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treiber JM, White NS, Steed TC, et al. Characterization and correction of geometric distortions in 814 diffusion weighted images. PLoS One. 2016;11:e0152492. doi: 10.1371/journal.pone.0152472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaus MR, Warfield SK, Nabavi A, et al. Automated segmentation of MR images of brain tumors. Radiology. 2001;218:586–91. doi: 10.1148/radiology.218.2.r01fe44586. [DOI] [PubMed] [Google Scholar]
- 40.Brett M, Leff AP, Rorden C, et al. Spatial normalization of brain images with focal lesions using cost function masking. Neuroimage. 2001;14:486–500. doi: 10.1006/nimg.2001.0845. [DOI] [PubMed] [Google Scholar]
- 41.Clatz O, Delingette H, Talos IF, et al. Robust nonrigid registration to capture brain shift from intraoperative MRI. IEEE Trans Med Imaging. 2005;24:1417–27. doi: 10.1109/TMI.2005.856734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersson JL, Hutton C, Ashburner J, et al. Modeling geometric deformations in EPI time series. Neuroimage. 2001;13:903–19. doi: 10.1006/nimg.2001.0746. [DOI] [PubMed] [Google Scholar]
- 43.Chang H, Fitzpatrick JM. A technique for accurate magnetic resonance imaging in the presence of field inhomogeneities. IEEE Trans Med Imaging. 1992;11:319–29. doi: 10.1109/42.158935. [DOI] [PubMed] [Google Scholar]
- 44.Embleton KV, Haroon HA, Morris DM, et al. Distortion correction for diffusion-weighted MRI tractography and fMRI in the temporal lobes. Hum Brain Mapp. 2010;31:1570–87. doi: 10.1002/hbm.20959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holland D, Kuperman JM, Dale AM. Efficient correction of inhomogeneous static magnetic field-induced distortion in echo planar imaging. Neuroimage. 2010;50:175–83. doi: 10.1016/j.neuroimage.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gelman N, Silavi A, Anazodo U. A hybrid strategy for correcting geometric distortion in echo-planar images. Magn Reson Imaging. 2014;32:590–3. doi: 10.1016/j.mri.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 47.Hong X, To XV, Teh I, et al. Evaluation of EPI distortion correction methods for quantitative MRI of the brain at high magnetic field. Magn Reson Imaging. 2015;33:1098–105. doi: 10.1016/j.mri.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Avants BB, Tustison NJ, Song G, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–44. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersson JL, Sotiropoulos SN. An integrated approach to correction for offresonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–78. doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duda JT. NeuroBattery pipeline. Available at: http://jeffduda.github.io/NeuroBattery. Accessed August 21, 2017.
- 51.Wells WM, Viola P, Atsumi H, et al. Multimodal volume registration by maximization of mutual information. Med Image Anal. 1996;1:35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- 52.Maes F, Collignon A, Vandermeulen D, et al. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–98. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]


