Abstract
Mice deficient for microRNA (miRNA) cluster mirn23a exhibit increased B lymphopoiesis at the expense of myelopoiesis while hematopoietic stem and progenitor (HSPC) populations are unchanged. Mammals possess a paralogous mirn23b gene that can give rise to 3 mature miRNAs (miRs -23b, 24-1, and -27b) that have identical seed/mRNA targeting sequences to their mirn23a counterparts. To assess whether compound deletion of mirn23a and mirn23b exacerbates the hematopoietic phenotype observed in mirn23a−/− mice, we generated a compound mirn23a−/−mirn23bfl/fl: Mx1-Cre conditional knockout mouse and assayed hematopoietic development after excision of mirn23b. Loss of both genes in adult bone marrow further skewed HSPC differentiation towards B cells at the expense of myeloid cells demonstrating a dosage dependent effect on regulating cell differentiation. Strikingly, double knockout mice had decreased bone marrow cellularity with significantly decreased HSC and progenitor populations, a phenotype not observed in mice deficient for mirn23a alone. Competitive transplant assays showed decreased contribution of mirn23a−/−mirn23b−/− HSPCs to hematopoietic lineages at 6 and 12 weeks post transplant. Defects in the proliferation of mirn23a−/−b−/− HSPCs was not observed, however double knockout cells were more apoptotic compared to both wildtype and mirn23a−/− cells. Together, our data shows that complete loss of mirn23a/mirn23b miRNAs results in decreased blood production and affects lineage output in a concentration dependent manner.
Introduction
Effector cells of the hematopoietic system are typically short lived and are constantly being replenished by a hierarchy of hematopoietic stem and progenitor cells. The long-term hematopoietic stem cell (LT-HSC) resides atop this hierarchy and can give rise to all mature blood lineages. While the LT-HSC has been extensively studied since its discovery, the genetic mechanisms governing its balance of quiescence, self-renewal, differentiation, and interactions within the hematopoietic niche remain largely unknown [1, 2]. Initial studies of the HSC were predominantly aimed at trying to identify or elucidate the role of hematopoietic cytokine/receptor interactions (notably c-Kit/SCF and c-Mpl/TPO), cell signaling pathways (Wnt, PI3K/AKT, TGF-β/Smad, Notch), and hematopoietic transcription factors (such as FoxO3a, Gata2, Gfi1, and HoxB4) in regards to HSC maintenance and function[3–5] More recently, alternative mechanisms of gene regulation have been explored. Epigenetic factors have emerged as important factors in HSC maintenance and function, with the best examples being DNA methyltransferase Dnmt3a and polycomb-group protein Bmi-1, whose functions are critical in the HSC[6–8]. More recently, the discovery of noncoding RNA molecules as regulators of gene expression has implicated them as potential regulators of HSC maintenance.
MicroRNAs (miRNAs) are a class of short (~22 nt) non-coding RNA molecules that directly bind to the 3′ UTR of target mRNAs and inhibit their translation [9, 10]. Several groups, including our own, have shown that microRNAs influence hematopoietic cell fate by targeting essential lineage specific transcription factors in overexpression and knockdown studies [11–20]. Several miRNAs have also been implicated in HSC maintenance under steady state conditions. MiR-125a has recently emerged as an important regulator of HSC maintenance, as it is enriched in LT-HSCs and forced expression results in decreased apoptosis and a significant increase in the LT-HSC pool [21]. MiR-29a appears to be important in the HSC as both overexpression and knockout of the miRNA results in increased HSC cell cycling [22, 23]. Loss of miR-126 results in increased HSC proliferation, suggesting it may have an endogenous role of maintaining HSC pool size [24]. Lastly, miR-22 expression has also been shown to be critical for HSC self-renewal, as forced expression results in increased HSC self-renewal and impaired differentiation, while inhibition exerts the opposite phenotype [25].
We previously described that the miRNA cluster transcribed from the mirn23a gene (referred to hereafter as the mirn23a miRNA cluster), which codes for three mature miRNAs (miRs -23a, -24-2, and -27a), has the ability to promote myeloid development at the expense of B cell development as demonstrated by both overexpression and genetic knockout studies [18, 19]. When characterizing the mirn23a−/− mouse, our results showed no apparent HSC defects at steady state conditions since HSC populations were unchanged and overall cellularity in the mouse bone marrow was unperturbed. These mice developed no apparent hematological disease and could breed normally. Despite not observing an HSC defect, we did observe that the number of bone marrow and splenic B cells was significantly increased in mirn23a−/− mice compared to their wildtype counterparts, and that this was accompanied by a concomitant decrease in myeloid cell populations, suggesting a role for the mirn23a cluster in a lymphoid vs myeloid cell fate decision [18].
Mice and other mammals also possess a paralogous mirn23b miRNA cluster located in the intronic region of the aminopeptidase O gene that can give rise to 3 mature miRNAs (miRs -23b, 24-1, and -27b) that have identical seed sequences (mRNA targeting) to their mirn23a counterparts. Mature miR-24-2 (mirn23a) and miR-24-1 (mirn23b) are identical. The mature a and b forms of miRs-23 and miR-27 differ by one nucleotide outside of the seed sequence. While mirn23a is the predominant source of mirn23 cluster miRNAs in blood, mirn23a−/− mice retain approximately ~30% of wildtype miR-24 expression through the mirn23b cluster suggesting that the hematopoietic phenotype in mirn23a−/− mice is due to hypomorphic expression of mirn23a/b miRNAs [18, 19]. To examine hematopoiesis in the complete absence of mirn23a/b miRNAs, we generated an inducible mirn23a−/−mirn23bfl/fl: Mx1-Cre mouse. A conditional allele of mirn23b was used since previous work from our laboratory demonstrated that shRNA knockdown of miRs-24-2 and -24-1 in embryonic stem cells (ESCs) results in a defect in generating HSPCs from mesoderm[26]. Additionally an independent CRISPR mediated deletion of the mirn23a and mirn23b genes in ESC results in defects in the development of all three germ layers[27].
Germline loss of only mirn23b resulted in no discernible hematopoietic defects. However downstream lineage outputs of myeloid and lymphoid cells were further skewed in double knockout mice. DKO mice showed a further increase in their CLP (Common Lymphoid Progenitor) and B cell populations at the expense of myeloid cells compared to mirn23a and wildtype mice. Surprisingly, in contrast to wildtype and mirn23a−/− mice, mirn23a−/−b−/− mice showed decreased bone marrow cellularity 4 weeks post mirn23b deletion. All hematopoietic lineages were affected with decreases in LT-HSCs, ST-HSCs, and MPPs (Multipotent Progenitors) also observed. Additionally double knockout HSPCs cells showed decreased contribution to hematopoietic lineages in a competitive transplant setting compared to wildtype and single mirn23a knockout cells. Annexin V/7AAD analysis revealed that mirn23a/b deficient hematopoietic stem and progenitor cells (HPSCs) exhibit increased apoptosis, suggesting this may be the mechanism for the decreased cellularity. Gene expression analyses revealed upregulation of several pro-apoptotic genes in mirn23a−/−b−/− lineage negative cells. Together, our data shows that complete loss of mirn23a/mirn23b miRNAs results in decreased blood production and affects lineage output in a concentration dependent manner.
Methods
Generation of mirn23a−/−mirn23bf/fl Mx1-Cre Mouse
Mir23btm1MTm/Mmjax (36298-JAX) were obtained from Jackson Laboratory (Bar Harbor, ME). To convert this mirn23b allele to a loxP flanked conditional allele (mirn23bf/f), mir23btm1MTm mice were mated to B6;SJL-Tg(ACTFLPe)9205Dym/J (Jackson Laboratory 003800) to excise the FLP site flanked β galactosidase gene and neomycin resistance cassette. In addition we generated a null allele of mirn23b allele by mating Mir23btm1MTm to B6.FVB-Tg(EIIa-cre)C5379Lmgd/J (Jackson Laboratory 003724). This allele lacked coding sequence for mirn23b, but retained the inserted β galactosidase gene and neomycin resistance cassette. To generate mirn23a−/−mirn23bf/fl Mx1-Cre mice, our mirn23a germline knockout mouse[18] was first crossed to the mirn23bf/f mice. Mirn23a−/−mirn23bf/f were then mated to B6.Cg-Tg(Mx1-cre)1Cgn/J mice (Jackson Laboratory 003556). The use of mice in these experiments was approved by the Indiana University School of Medicine Institutional Review Board and University of Notre Dame Institutional Animal Care and Use Committee (Protocols 13-017 and 16-022).
Genotyping of mice
Mice were genotyped by isolating DNA from ear punches and performing polymerase chain reaction (PCR) with gene specific primers. For genotyping mirn23b wildtype and floxed alleles we followed the protocol provided by Jackson Laboratory for stock number 36298-JAX, which was updated 11/2/2011. For detection of Mx1-CRE transgene we used the master protocol provided by Jackson Laboratory for detecting CRE alleles, which was updated 09/24/2009. Mirn23a locus was genotyped as previously described[18]. Lastly to determine if the floxed mirn23b allele was deleted after pIpC deletion DNA was isolated from nucleated bone marrow cells and subjected to PCR using the primers: 5′-GCA ATT GGA GAA CAG GGT GT -3′ and 5′-AGC CTC TGA CCT CCA CTT GA-3′.
Polyinosinic:polycytidylic acid (pIpC) Injections
Mice were given an intraperitoneal injection of 250ug pIpC in 200uL of sterile phosphate buffered saline (PBS) every other day for 5 days (3 total injections). DNA was harvested from the bone marrow and spleen of mice 4 or 8 weeks post final injection by DNAzol extraction (ThermoFisher Scientific, Waltham, MA). Confirmation of mirn23a/mirn23b deletion was done by genomic DNA PCR and quantitative real-time PCR (qRT-PCR).
Flow Cytometry
Bone marrow cells were collected from the femurs and tibias of 8–9 week old mice 4 weeks post treatment with pIpC. Splenic cells were isolated by passing cells through a 70μM cell strainer (Fisher, Pittsburgh, PA, USA). Mature erythroid cells were removed by ammonium chloride lysis for 7 minutes at room temperature. Bone marrow and spleen cells were stained to characterize B cell, myeloid, erythroid, CLP, CMP, HSC, and multipotent progenitor (MPP) cell populations. Unless stated otherwise, all antibodies were obtained from BioLegend (San Diego, CA, USA). Prior to staining, Fcγ receptors were blocked by incubating cells with anti-FcγRIII/II (CD16/32). Single cell suspensions were incubated with the following combinations of antibodies: B cell/myeloid- B220 (RA3-6B2)-allophycocyanin (APC), CD11b (M1-70)-allophycocyanin-Cy7 (APC/Cy7). Bone marrow B cell development- IgM (RMM-1)-FITC, CD19 (6D5)-PE, B220 (RA3-6B2)-APC, IgD (11-26c.2A)-APC/Cy7. Bone marrow pre-pro B cells – CD19 (6D5)-FITC, B220 (RA3-6B2)-PE, CD93 (AA4.1)-APC, cKit (2B8)-APC/cy7. Splenic B, T, and myeloid cell populations – CD4-FITC, CD8-PE, B220-APC, CD11b-APC/Cy7. CLP – Flt3 (A2F10)-PE, lineage cocktail - CD11b (RM 2801)-biotin, B220 (RA3-6B2)-biotin, CD19 (6D5)-biotin, GR1 (RB6-8C5)- biotin, Terr119 (Ter-119)-biotin, CD3e (145-2c11)-biotin), Texas red-streptavidin (TR), IL7R (A7R34)-APC, cKit (2B8)-APC/Cy7. CMP- CD34 (RAM34)-FITC, FcγRIII/II (93)-PE, cKit (ACK2)-APC, lineage cocktail-biotin, Sca-1 (D7)-biotin, IL7R (A7R34)-biotin, and APC/Cy7-streptavidin. HSC/MPP – Sca1 (D7)-FITC, CD48 (HM48-1)-PE, lineage cocktail-biotin, TR-streptavidin, CD150 (TC15-12F12.2)-APC, cKit (2B8)-APC/cy7. Apoptosis Panel – Annexin Stained cells were subsequently assessed using Beckman Coulter FC500 Flow Cytometer (Brea, CA, USA) and data was analyzed using Flowjo software (Tree Star, Ashland, OR, USA). Dead cells were removed from analysis by the use of FSC/SSC gating and/or exclusion of propidium iodide (Sigma-Aldrich, St. Louis, MO, USA). Basis of gates was determined with the use of fluorescence minus one (FMO) controls when necessary. Results are presented as standard error of the mean (SEM) for averages of each mouse genotype.
Competitive Transplant Assays
Three week old CD45.2+ donor mice (wildtype, mirn23a−/−mirn23bf/f, and mirn23a−/−mirn23bf/f:MX1-CRE) were injected with pIpC as described above. Donor mice received a total of 6 injections over 10 days. Donor mice were sacrificed 2 weeks after the last injection. Femurs and tibias were removed for isolation of bone marrow. Whole marrow was pooled according to genotype. Red cells were removed by Ammonium-Chloride-Potassium (ACK) cell lysis buffer. Nucleated cells were mixed at 1:1 ratios (WT: WT CD45.1; mirn23a knockout:CD45.1; and mirn23a/23b double knockout:CD45.1), spun down, and re-suspended in ice cold PBS to a final concentration of 107 cells/ml. Recipient mice were lethally X-ray irradiated (RS 2000 Biological Research Irradiator) 5–6 hours before transplant. A dose of 100ul cell suspension (106 cells) was retroorbitally injected. Reconstitution was monitored by analysis of peripheral blood. Mice were cheek bled at 6 and 12 weeks post-transplant. Blood was collected in Eppendorf tubes coated with EDTA (0.5M). Red blood cells were removed by ACK lysis. Nucleated peripheral blood was stained for flow cytometry with CD45.1 FITC, CD45.2 PE, B220 APC, CD11b APC-Cy7 antibody conjugates obtained from BioLegend (San Diego, CA, USA).
Gene Expression Analyses
Total RNA was isolated from in vitro cell lines using TRIzol (Thermo Fisher Scientific Life Sciences), according to the manufacturer’s protocol. RNA from primary lineage negative cells was isolated using miRNeasy kit from Qiagen (Germantown, MD, USA). For the gene array, the RT2 Apoptosis Profiler Array (Qiagen, Germantown, MD, USA) was used according to manufacturers protocol, with 1ug starting RNA material. For qPCR analysis, cDNA was reverse transcribed from 400 ng lin- primary cell RNA using TaqMan MicroRNA reverse transcription kit, according to the manufacturer’s protocol (Thermo Fisher Scientific Life Sciences). Quantitative analysis was performed using gene-specific TaqMan (Thermo Fisher Scientific Life Sciences) or SYBR Green (Thermo Fisher Scientific Life Sciences) reagents. All experiments were performed in triplicate using CFX96 C1000 system (Bio-Rad Laboratories, Hercules, CA, USA). Relative gene expression was calculated using the comparative threshold cycle method. GAPDH was used to normalize expression across different RNA preparations.
BrdU Assays
All mice in these studies were previously administered 3 doses of pIpC as described in previous methods section and were used for BrdU experiments 4 weeks after their final pIpC injection. Mice were injected with 2ug of BrdU (BD Biosciences, Billerica, MA) diluted in 200uL of sterile phosphate buffered saline by intraperitoneal injection 16 hours prior to analysis. Bone marrow cells were harvested from the femurs and tibias of these mice and cell fixation and permeabilization was done according to BD Pharmingen BrdU flow kit protocol (BD Biosciences, Billerica, MA). BrdU incorporation in HSPCs was evaluated by flow cytometry using a panel including Sca1 (D7)-FITC, Lineage cocktail- Biotin, Avidin-Texas Red, BrdU-APC, and c-Kit (2B8)-APC/Cy7.
Annexin V/7AAD Analysis
Primary bone marrow cells were harvested from the femurs and tibias of mice 4 weeks post deletion with pIpC. Staining for annexin V and 7AAD was done according to the FITC Annexin V apoptosis detection kit with 7-AAD (Biolegend, San Diego, CA). Analysis by flow cytometry was done using a combination of antibodies including Annexin V- FITC, Sca1- PE, Lineage cocktail-biotin, TR-Avidin, 7AAD, and c-Kit-APC/Cy7.
Statistical Analysis
Statistical data are presented as the mean +/− standard error of the mean (SEM). Differences between sample groups were determined by performing an unpaired student t-test. Analysis was performed using PRISM software version 6.0 (Graphpad software, La Jolla, CA, USA).
Results
Myeloid and lymphoid progenitor populations are altered in mirn23a−/− and mirn23a−/−mirn23b−/− mice
We previously observed that mirn23a−/− mice have increased CLP production and B cell development with a concomitant decrease in GMP (Granulocyte Monocyte Progenitor) production and myeloid cell development[18]. In this report we were interested in determining if compound loss of mirn23a and its paralog cluster mirn23b would result in a more severe hematopoietic phenotype. We first generated a mirn23b germline allele by crossing Mir23btm1MTm/Mmjax (Jackson Laboratories) to EIIa-CRE transgenic mice (Supplementary Figure 1)[28, 29]. Hematopoiesis was examined in 6 week old mirn23b−/− mice. No differences in bone marrow cellularity were observed between wildtype and mirn23b knockout mice (Supplementary Figure 2A). In contrast to mirn23a−/− mice, we observed no differences in bone marrow B cell and myeloid populations by flow cytometric analysis (Supplementary Figure 2B, C, and D). Similarly no significant differences in hematopoietic populations were observed in the spleen.
To investigate whether compound loss of mirn23a and mirn23b results in more severe defects in adult hematopoiesis than loss of mirn23a alone, we generated mirn23a−/−mirn23bfl/fl: Mx1-Cre mice. A conditional allele of mirn23b was used due to circumvent potential embryonic lethality of germline knockout double mice[26, 27]. A floxed allele of mirn23b was generated by crossing a mirn23b targeted mouse from Jackson Laboratory (Mir23btm1MTm/Mmjax) to an ACT-FLPe transgenic mouse (Supplementary Figure 1)[30]. Hematopoietic populations were assayed four weeks after mirn23b deletion with pIpC. The ability of pIpC to induce CRE mediated deletion of the floxed mirn23b allele was confirmed for by genomic DNA PCR (bone marrow, spleen, thymus) and at the RNA level by qRT-PCR (Bone marrow, Supplementary Figure 3A, 3B). The pIpC treated mirn23a−/−mirn23bf/f:MX1-CRE mice are referred to as mirn23a−/−mirn23b−/− throughout the manuscript.
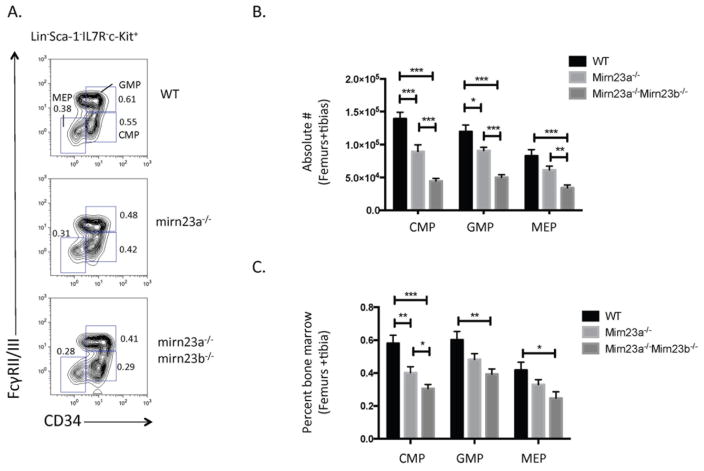
Evaluation of nucleated bone marrow cell numbers in wildtype, mirn23a−/−, and mirn23a−/−mirn23b−/− mice 4 weeks post treatment with pIpC revealed that mirn23a−/−mirn23b−/− mice have significantly decreased bone marrow cellularity compared to wildtype and mirn23a−/− mice, while no difference was observed between wildtype and mirn23a−/− cellularity (Supplementary Figure 3C). In 5–6 week old germline mirn23a−/− knockout mice, we previously observed that loss of mirn23a results in increased CLPs with decreased CMPs (Common Myeloid Progenitors) and downstream GMPs and MEPs (Megakaryocyte Erythroid Progenitors). To evaluate whether these populations were further altered in mirn23a−/−mirn23b−/− mice, we stained nucleated bone marrow cells from wildtype, mirn23a−/−, and mirn23a−/−mirn23b−/− mice with myeloid progenitor cell surface markers and analyzed for CMP, GMP, and MEP populations by flow cytometry (Figure 1A). This revealed that at the absolute cell number level (which accounts for differences in bone marrow cellularity), mirn23a−/− mice showed decreased CMPs, GMPs, and MEPs compared to WT mice, and mirn23a−/−mirn23b−/− showed an even further significant decrease between mirn23a−/− and mirn23a−/−mirn23b−/− populations (Figure 1B). By percent difference (which does not account for BM cellularity), the CMP is significantly reduced between WT, mirn23a−/−, and mirn23a−/−mirn23b−/− mice (Figure 1C). The GMP and MEP populations, while decreased between mirn23a−/− and mirn23a−/−mirn23b−/− populations, was not statistically significant.
Figure 1. Loss of or compound loss of mirn23a/mirn23b results in decreased production of bone marrow myeloid progenitors.
Bone marrow was isolated from the femurs and tibias of WT, mirn23a−/−, and mirn23a−/−mirn23bflox/flox (labeled mirn23a−/−mirn23b−/−) mice treated with pIpC for 4 weeks. A) Following ACK lysis, CMP, GMP, and MEP populations were identified by flow cytometry. Representative plots are shown. Numbers represent percent of total bone marrow population B) Differences in the absolute number (which includes differences in mouse cellularity) of CMP, GMP, and MEP populations were analyzed for statistical significance. C) Differences in percent CMPs (which excludes cellularity differences) were also analyzed for statistical significance. P-values were determined using an unpaired student’s t-test. WT (n=12), mirn23a−/− (N=12), and mirn23a−/−mirn23b−/− (n=11) animals were examined. *(p<0.05), ** (p<0.01), *** (p<0.001).
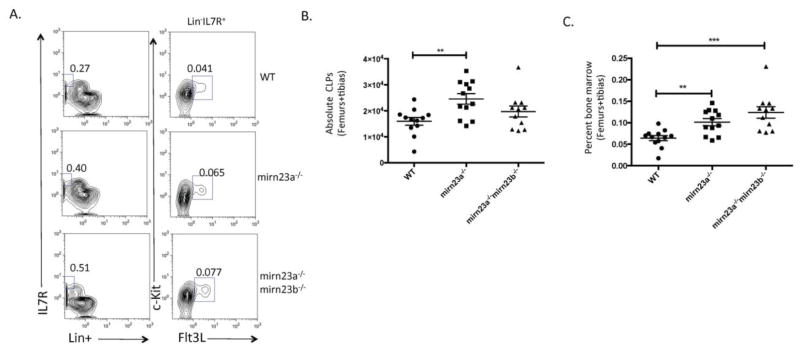
To see if the decreased CMPs were accompanied by increased CLPs, we stained nucleated bone marrow fractions from the three cohorts of mice and analyzed CLP populations by flow cytometry (Figure 2A). Absolute CLPs were significantly increased in mirn23a−/− mice compared to wildtype mice, while no significant differences were observed in the absolute number of mirn23a−/−mirn23b−/− CLPs between wildtype and mirn23a−/− mice (Figure 2B). However, at the percent level, mirn23a−/−mirn23b−/− CLPs are significantly increased compared to wildtype mice (Figure 2C). The percent of CLPs in mirn23a−/−mirn23b−/− mice is slightly increased compared mirn23a−/− mice, but this increase is not statistically significant (Figure 2C). Overall, these results show that compound loss of mirn23a/mirn23b miRNAs results in increased CLP production at the expense of myeloid progenitors.
Figure 2. Loss of or compound loss of mirn23a/mirn23b results in increased production of bone marrow common lymphoid progenitors.
Bone marrow was isolated from the femurs and tibias of WT, mirn23a−/−, and mirn23a−/−mirn23bflox/flox (labeled mirn23a−/−mirn23b−/−) mice treated with pIpC for 4 weeks. A) Following ACK lysis, CLP populations (Lin−IL7R+Flt3+c-Kit+) were analyzed from the nucleated bone marrow of WT, mirn23a−/−, and mirn23a−/−mirn23b−/− mice. Numbers represent percent of total bone marrow population. Representative plots are shown. B) Differences in absolute CLP populations were analyzed for statistical significance. C) Percent of CLPs (which excludes cellularity differences) were significantly increased in mirn23a−/− and mirn23a−/−mirn23b−/− mice compared to WT controls. P-values were determined using an unpaired student’s t-test. WT (n=12), mirn23a−/− (N=12), and mirn23a−/−mirn23b−/− (n=11) animals were examined. *(p<0.05), ** (p<0.01), *** (p<0.001).
Compound loss of mirn23a and mirn23b results in increased lymphoid development at the expense of myeloid development
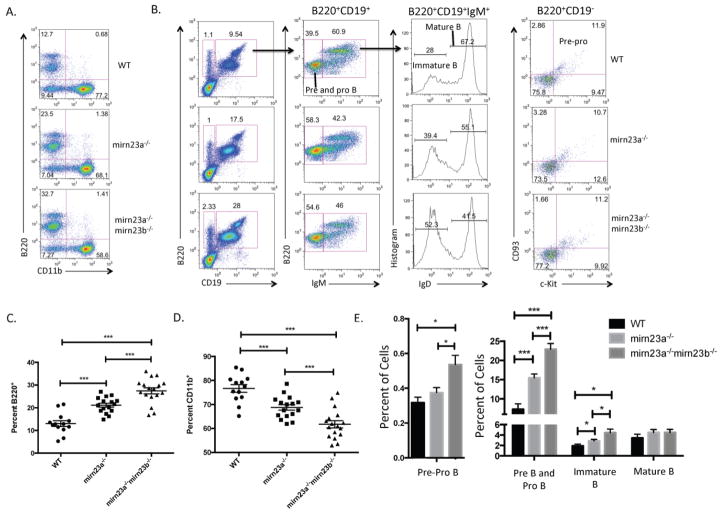
Since mirn23a−/−mirn23b−/− mice have an altered composition of CLP and CMP cells compared to wildtype and mirn23a−/− mice, we next sought to determine whether downstream B cell and myeloid cell populations were altered in these mice. To investigate this, we stained nucleated bone marrow cells for cell surface markers B220 (pan B cell marker) and CD11b (pan myeloid cell marker) (Figure 3A). We also evaluated B cell developmental populations by cell surface markers B220, CD19, c-Kit, CD93, IgM, and IgD (Figure 3B). These analyses revealed that the percent of B220+ B cells was significantly increased in mirn23a−/− mice compared to wildtype mice, and further increased in mirn23a−/−mirn23b−/− mice (Figure 3C). This increase in B220+ B cells was at the expense of CD11b+ myeloid cells, which were decreased in mirn23a−/− mice, and further decreased in mirn23a−/−mirn23b−/− mice (Figure 3D). The analysis of B cell progenitor populations revealed that all progenitor B cell populations are increased while mature/recirculating B cells are unchanged, indicating that there is no developmental block in B cell development in mirn23a−/− or mirn23a−/−mirn23b−/− mice (Figure 3E). Lastly, since we previously observed that an increase in B cell gene expression was responsible for increased B cell development in mirn23a−/− mice, we speculated that a further increase in B cell specific gene expression could enhance the lymphoid bias observed in double knockout mice. To test this, we isolated Lin- RNA and analyzed for B cell gene expression by qRTPCR. This analysis revealed significantly increased expression of lymphoid specific TFs Ebf1, Pax5, Ikzf1, and Mef2c in double knockout HSPCs (Supplementary Figure 5A).
Figure 3. Loss of mirn23a or compound loss of mirn23a/mirn23b results in increased B cell development at the expense of myeloid development.
A) Nucleated bone marrow was isolated from the femurs and tibias of WT, mirn23a−/−, and mirn23a−/−mirn23bflox/flox (labeled mirn23a−/−mirn23b−/−) mice treated with pIpC for 4 weeks. B cell (B220+) and myeloid cell (CD11b+) populations were analyzed by flow cytometry. WT (n=13), mirn23a−/− (N=16), and mirn23a−/−mirn23b−/− mice (N=17) were examined. Representative plots are shown. B) B cell populations were further characterized into different developmental states using cell surface markers B220, CD19, IgM, IgD, CD93, and c-Kit. WT (n=8), mirn23a−/− (N=13), and mirn23a−/−mirn23b−/− mice (N=12) were examined. Representative plots are shown. Numbers represent percent of the previously gated population. C) The percent of B220+ B cells was analyzed for statistical significance. D) The percent of CD11b+ myeloid cells were analyzed for statistical significance. E) B cell developmental stages were analyzed for statistical significance. *(p<0.05), ** (p<0.01), *** (p<0.001).
Splenic B cells are increased in mirn23a−/−mirn23b−/− mice
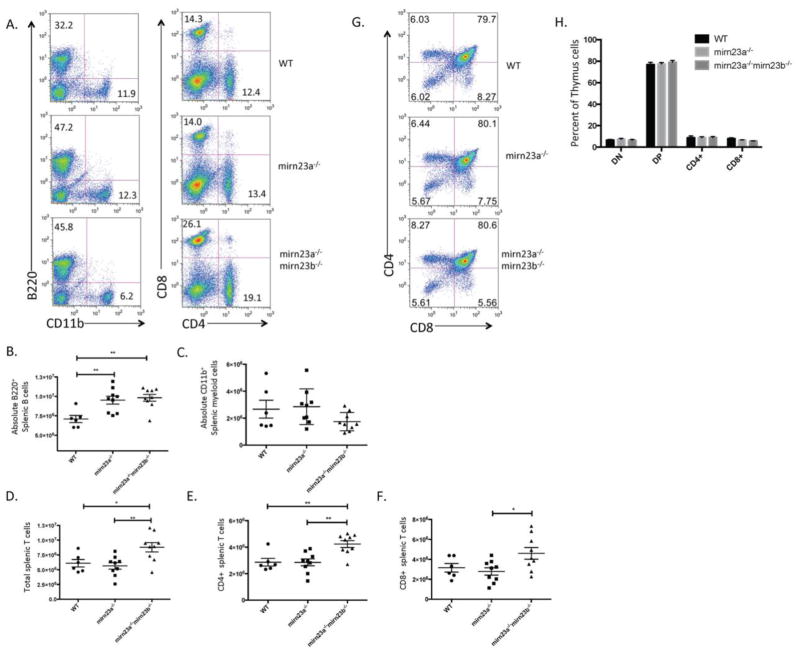
To investigate whether increased B cell development and decreased myeloid development in the bone marrow persisted into the periphery, we isolated spleens from wildtype, mirn23a−/−, and mirn23a−/−mirn23b−/− mice 4 weeks post pIpC and stained nucleated cell fractions for cell surface expression of B220 and CD11b (Figure 4A). We also investigated T cell numbers in the spleen by cell surface expression of CD4 and CD8 (Figure 4A). Unlike in the bone marrow, there was no difference in splenic cell numbers when comparing the three genotypes. Examination of B220+ populations in the spleen revealed that mirn23a−/− and mirn23a−/−mirn23b−/− mice have increased splenic B cell populations compared to wildtype mice, with no significant differences observed between mirn23a−/− and mirn23a−/−mirn23b−/− mice (Figure 4B). Surprisingly, no significant differences were observed between the numbers of CD11b+ myeloid cells between the three groups, although our variability within genotypes was high, potentially masking a difference (Figure 4C). The number of total T cells, CD4+ T cells, and CD8+ T cells was unchanged between wildtype and mirn23a−/− mice, consistent with the 5–6 week old mirn23a−/− mice previously described (Figures 4D–4F). However, analysis of mirn23a−/−mirn23b−/− T cell populations revealed that total T cells, CD4+ T cells, and CD8+ T cells were all slightly increased compared to wildtype and mirn23a−/− mice (Figure 4D–4F). To investigate whether this increased number of T cells was due to a developmental defect, we then isolated the thymus from the three cohorts and stained for CD4 and CD8 expression (Figure 4G). This revealed no significant differences in T cell development in the thymus, indicating that increased T cell numbers are not the result of increased T cell development in the thymus (Figure 4H). Overall, this work shows that the increased B cell development persists into the periphery of double knockout mice.
Figure 4. Lymphoid development is increased in the periphery of mirn23a−/− and mirn23a−/−mirn23b−/− mice.
A) Nucleated splenic cells were isolated from the spleens of WT, mirn23a−/−, and mirn23a−/−mirn23b−/− mice and analyzed by flow cytometry for immune cell populations. WT (n=6), mirn23a−/− (N=9), and mirn23a−/−mirn23b−/− mice (N=9) were examined. B cell and myeloid populations were analyzed by B220 and CD11b expression respectively. T cell populations were analyzed by CD4 and CD8 expression. Representative plots are shown. Numbers represent percent of total nucleated splenic population. B–F) B cell, myeloid cell, and T cell populations were analyzed for statistically significant differences in the spleens of wildtype, mirn23a−/−, and mirn23a−/−mirn23b−/− mice. G) Thymus cells were isolated from WT, mirn23a−/−, and mirn23a−/−mirn23b−/− mice and analyzed for T cell development by cell surface markers CD4 and CD8. WT (n=4), mirn23a−/− (N=4), and mirn23a−/−mirn23b−/− mice (N=4) were examined. Representative plots are shown. H) No significant differences were observed in the thymus of mirn23a−/− or mirn23a−/−mirn23b−/− mice. P values were determined using unpaired students t-test. *(p<0.05), ** (p<0.01), *** (p<0.001).
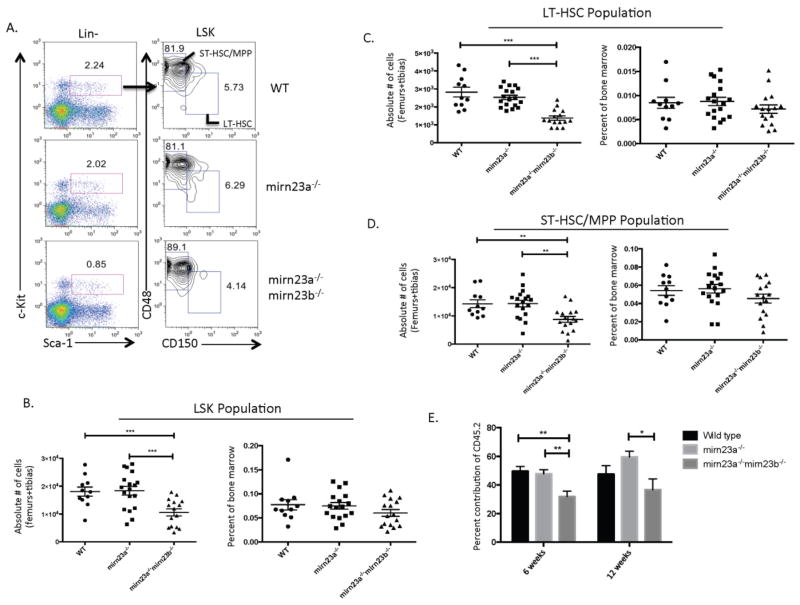
Deceased Mirn23a−/−mirn23b−/− bone marrow cellularity is accompanied by decreased HSPC populations
To investigate whether the decreased bone marrow cellularity in mirn23a−/−mirn23b−/− mice was accompanied by decreased HSPC (Hematopoietic Stem and Progenitor Cell) production, we evaluated LSK, ST-HSC/MPP and LT-HSC populations by flow cytometry (Figure 5A). This revealed that mirn23a−/−mirn23b−/− mice had decreased LSK, ST-HSC/MPP, and LT-HSC populations compared to both the wildtype and mirn23a−/− populations by overall number, while the percent of bone marrow that these populations represent was slightly decreased but not statistically significant (Figure 5B–5D).
Figure 5. Compound loss of mirn23a/mirn23b results in decreased stem cell production and bone marrow cellularity.
Bone marrow was isolated from the femurs and tibias of WT, mirn23a−/−, and mirn23a−/−mirn23bflox/flox (labeled mirn23a−/−mirn23b−/−) mice treated with pIpC for 4 weeks. A) Nucleated bone marrow was harvested and stained for LSK, LT-HSC (LSK CD150+CD48−), and ST-HSC/MPP (LSK CD150−CD48+) populations by flow cytometry. Representative plots are shown. Numbers represent percent of cells from the previously gated population. B) The total number of LSK cells is significantly decreased in mirn23a−/−mirn23b−/− mice compared to WT and mirn23a−/− mice. The percent of bone marrow cells that are LSK are unchanged C) The LT-HSC population is significantly decreased in mirn23a−/−mirn23b−/− mice compared to WT and mirn23a−/− mice. The percent LT-HSCs is unchanged. D) The ST-HSC/MPP population is significantly decreased in mirn23a−/−mirn23b−/− mice compared to WT and mirn23a−/− mice. The percent ST-HSC/MPP’s are unchanged. P-values were determined using unpaired students t-test. For bone marrow analysis, WT (n=11), mirn23a−/− (n=18), and mirn23a−/−mirn23b−/− (n=15) animals were examined. For spleen cellularity counts, WT (n=6), mirn23a−/− (n=9), and mirn23a−/−mirn23b−/− (n=9) animals were examined. E) Competitive transplant assays performed with an equal ratio of WT CD45.1+ and CD45.2+ (WT, mirn23a−/−, or mirn23a−/−mirn23b−/−) donor marrow coinjected into lethal irradiated mice. Contribution to peripheral blood of CD45.2+ cells is shown at 6 and 12-weeks post-transplant. At 6 weeks 15, 21, and 14 mice transplanted with WT, mirn23a−/−, and mirn23a−/−mirn23b−/− bone marrow were examined respectively. At 12 weeks n=10, n=7 and n=10 mice of the above groups were analyzed respectively. P values were determined using unpaired student t-test. * p<0.05, ** p<0.01, *** p<0.001.
To determine whether double knockout stem cells have a functional defect that contributes to the decreased population, we performed competitive transplant assays with WT, mirn23a−/−, or mirn23a−/−mirn23b−/− donor bone marrow transplanted into lethally irradiated congenic recipients. At 6 weeks, contribution of WT and mirn23a−/− donors was approximately ~50%, while the contribution from mirn23a−/−mirn23b−/− donors was significantly decreased (p<0.01 vs WT and mirn23a−/−) to ~32% (Figure 5E). The trend of decreased contribution to peripheral blood of double knockout cells was also observed at 12 weeks. However due to higher variability in contribution observed between recipient mice within experimental groups only the contribution from double knockout cells was decreased when compared to mirn23a−/− (p<0.05) cells. A less pronounced phenotype at 12 weeks potentially could also be due to repopulation by mirn23a−/−mirn23bf/f cells that did not undergo Cre mediated excision. These results demonstrate there is at least a short-term defect in repopulation by double knockout stem and progenitor cells. A few mice were examined for contribution of the CD45.2+ marrow to the myeloid (CD11b+) and B lymphoid lineages. Consistent with our previous observation with transplanted MPPs [20] we observed that mirn23a−/− and double knockout cells have increased B220+ B cell and decreased CD11b+ myeloid populations compared to wildtype cells (Supplemental Figure 4). We unfortunately were unable to examine a large enough cohort of mice 16 weeks post-transplant to make definitive conclusions regarding long-term reconstitution. A number of mice developed sever dermatitis before reaching the 16 weeks post-transplant that met our IACUC guideline for euthanasia. Together, these results show that mirn23a−/−mirn23b−/− mice have decreased HSPC populations in their bone marrow, and that double knockout cells have at least a short-term deficit in hematopoietic reconstitution.
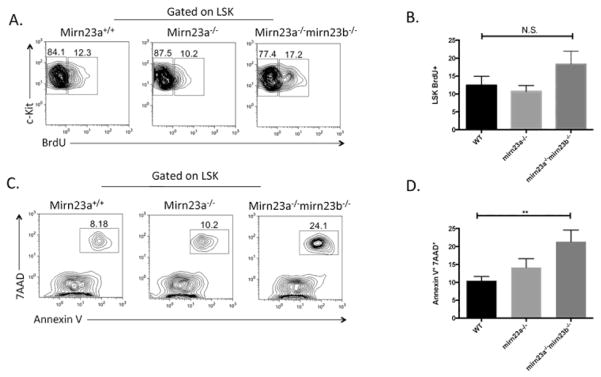
Apoptosis is increased in mirn23a−/−mirn23b−/− LSK cells
We next sought to determine if changes in HSPC proliferation and/or apoptosis contribute to the stem cell phenotypes observed at 4 weeks post deletion. To investigate whether any proliferation defects were present, we injected mice intraperitoneally with BrdU 16 hours prior to analysis and harvested bone marrow and evaluated by flow cytometry (Figure 6A). This revealed a slight 3–4% increase in LSK Brdu+ mirn23a−/−mirn23b−/− proliferation rates, but these were not statistically significant compared to WT or mirn23a−/− cells (Figure 6B).
Figure 6. Compound loss of mirn23a/mirn23b results in increased stem cell apoptosis.
Wildtype (n=9), mirn23a−/− (n=10), and mirn23a/mirn23b DKO mice (n=7) mice were treated with pIpC and analyzed 4 weeks post deletion for apoptosis and proliferation in their LSK populations. A) Mice were injected intraperitoneally with BrdU 16 hours prior to analysis. Representative BrdU plots are shown. B) No significant differences were observed for Brdu+ populations. C) Representative plots for Annexin V/7AAD staining are shown. D) Apoptosis was significantly increased in mirn23a/mirn23b deficient LSK populations compared to WT and mirn23a−/− mice. P values were determined using unpaired students t-test. * p<0.05, ** p<0.01, *** p<0.001.
To evaluate apoptosis in the stem and progenitor cell compartment of WT, mirn23a−/−, and mirn23a−/−mirn23b−/− mice, we harvested cells from pIpC treated mice 4 weeks after the final injection and stained cells for LSK (Lin-Sca1+Kit+) markers along with 7AAD and Annexin V (Figure 6C). This analysis revealed that mirn23a−/−mirn23b−/− cells had significantly increased Annexin V+ 7AAD+ populations compared to WT cells (Figure 6D). Mirn23a−/− cells had slightly increased Annexin V+ 7AAD+ populations compared to WT cells, but this phenotype was not significant.
To investigate genes that could be responsible for increased apoptosis in mirn23a−/−mirn23b−/− mice, we collected RNA from Lin- cells and assayed for known pro-apoptotic markers that are predicted or validated targets of the mirn23a/b clusters by qRT-PCR. This analysis revealed that Bim (Bcl2l11), Apaf1, and Caspase 9 (Casp9) were all increased in mirn23a−/− and mirn23a−/−mirn23b−/− cells, while Pten was unchanged (Supplemental Figure 5B). However, since we noticed similar levels of overexpression between mirn23a−/− and mirn23a−/−mirn23b−/−, changes in these genes alone are not likely responsible for the changes in apoptosis. To use a more comprehensive approach to identify genes involved with the apoptotic phenotype, we conducted apoptotic gene expression profile arrays between wildtype and mirn23a−/− cells, as well as between mirn23a−/− and mirn23a−/−mirn23b−/− cells (Supplementary Figure 5C, 5D). This analysis revealed 11 apoptotic genes upregulated >2 fold between wildtype and mirn23a−/− cells, and 9 apoptotic genes upregulated >2 fold between mirn23a−/− and mirn23a−/−mirn23b−/− cells. These results show that apoptosis is increased in mirn23a−/−mirn23b−/− mice compared to WT mice and that this phenotype may contribute to the stem cell defect observed 4 weeks post-mirn23b deletion.
Discussion
To investigate the effect of compound deletion of paralogous microRNA clusters mirn23a and mirn23b on hematopoietic development, we generated a mirn23a−/−mirn23bfl/fl Mx1-Cre inducible knockout mouse and evaluated hematopoiesis at 4 weeks post deletion with pIpC. While deletion of mirn23a alone does not result in any obvious hematopoietic stem or progenitor cell (HSPC) defects, mirn23a/mirn23b double knockout mice show decreased bone marrow cellularity and stem cell populations. We observed that compound knockout cells are at a competitive disadvantage compared to wildtype or mirn23a single knockout cells in a transplant setting. Preliminary mechanistic studies revealed that apoptosis was increased in double knockout LSK populations, suggesting that this may be the mechanism leading to decreased HSPC populations. Lineage output in these mice is also affected by a gradient of miRNA expression that skews hematopoietic differentiation to the B cell lineage at the expense of the myeloid lineage. Deletion of mirn23a (which removes ~70% of total mirn23a/mirn23b miRNA expression) results in a ~2-fold increase in B cell development compared to wildtype mice, and complete deletion of mirn23a/mirn23b microRNAs in double knockout mice results in a further increase in B cell development, almost 3-fold up (by percent, not absolute number) compared to wildtype mice. We attempted to analyze long-term deletion of mirn23a/mirn23b in our model mouse, but by 8 weeks time, mice have regained some expression of miR-23b miRNAs and the stem cell/bone marrow cellularity defect is no longer observable, a drawback of the Mx1-Cre model that has been reported by others[31]. Repopulation by mirn23a−/−mirn23bf/f cells that did not undergo Cre mediated deletion may have also affected the robustness of the 12-week transplant data. For those experiments donor mice were treated to 6 rounds of pIpC injections as opposed to 3 but its still possible that some cells escaped deletion.
Molecular cues that govern HSC identity and function have been extensively studied but remain largely unknown. Most of our current knowledge is based on studies of signaling pathways and their downstream effectors. MiRNAs add an additional layer of complexity to HSC regulation since single miRNAs can bind several different mRNA transcripts while simultaneously being regulated by several independent genes themselves. Thus, elucidating a detailed molecular description of how mirn23a/mirn23b deficiency results in decreased stem cell populations will be difficult. However, several pathways and proteins involved in HSC maintenance have previously been shown or are predicted to be targeted by mirn23a/mirn23b miRNAs.
We have previously shown that miR-24 can downregulate pro-apoptotic proteins Bcl-2-like protein 11 (Bim) and Caspase 9 in hematopoietic cells[32]. Here we observe that Bim, Caspase 9 and Caspase 9 co-factor Apaf1 are all increased at the RNA level in mirn23a−/−mirn23b−/− hematopoietic cells. Bim has previously been shown to be expressed in HSPC compartments and has the potential to regulate their survival and transplant efficiency, as Bim deficient cells outcompete wildtype cells in a transplant setting [33]. Similarly loss of Caspase 9 or Apaf1 results in impaired function of hematopoietic stem cells in transplantation models[34]. Bim, Caspase 9 and Apf1 have all been shown to be direct targets of miRNAs coded for by the mirn23a/b clusters[35–38]. Precise regulation of these apoptotic regulators may be required for HSC homeostasis. In our mice, mirn23a/mirn23b deficiency appears to result in an increase in Bim, Caspase 9 and Apaf1 levels resulting in decreased hematopoiesis through increased cell death of stem and progenitor cells. However, since we observed increased expression in both the mirn23a−/− and mirn23a−/−mirn23b−/− mice, there are likely other contributing genes and pathways involved in the apoptotic phenotype. Gene expression profile arrays revealed ~15–20 potential target genes that could influence apoptosis as well, but the importance of these genes to the HSC deficit in double knockout mice is not yet known (Supplemental Figure 5C, 5D).
Mirn23a and mirn23b loss may lead to downregulation of the PI3K/AKT pathway in stem cell populations. Validated miR-24 target Tribbles 3 (Trib3) has the ability to negatively regulate AKT kinase activity [39, 40], and is expressed in the HSC [41]. We have observed that loss of mirn23a or knockdown of miR-24 results in increased Trib3 expression in hematopoietic cells. Similar to mirn23a/mirn23b DKO mice, deletion of AKT1 and AKT2 results in a decrease in LSK and LT-HSC populations in mice and additional studies have shown that attenuation of AKT is essential for maintaining HSC quiescence [42]. At this point, molecular regulation of the HSC by mirn23a/mirn23b miRNAs is speculative. Future studies will be aimed at delineating these complex molecular pathways.
These findings have important implications in our understanding of miRNAs. MiRNAs typically arise from gene duplication events during evolution, and thus, functional redundancy from miRNAs within an organism is much more prevalent than with other genes [43]. This is important when designing genetic knockout studies with miRNAs since removal of a single miRNA can be compensated for by a functionally or genetically redundant miRNA. In our model, deletion of the mirn23a cluster alone, which leaves about 30% total mirn23a/mirn23b expression, is enough to convey normal HSC maintenance with no phenotypic defects in any of the LSK cell compartments. However, when both miRNA clusters are lost, all compartments of the LSK are significantly decreased, resulting in a downstream cytopenia. This posits a model in which functional redundant miRNAs mask a phenotype previously not described. As attention shifts from overexpression studies to genetic knockouts with miRNAs, these paradigms of functional redundancy need to be considered during experimental design.
Lastly, these results have potentially important biomedical implications. Understanding the genetic mechanisms that govern HSC function and hematopoietic differentiation will be critical for the development of novel therapeutic agents to treat a wide variety of clinical hematological disorders such as cytopenia and leukemia/lymphoma. In addition to this, understanding the regulatory mechanisms that maintain normal HSC homeostasis will be critical to generate functional HSCs from iPS cells to be used in regenerative medicine. Targeted delivery of miRNA mimics or agonists to members of the mirn23a and mirn23b miRNA clusters may be helpful in these clinical pursuits.
Supplementary Material
The mirn23b miRNA cluster gene is embedded in an intron of the aminopeptidase O gene. A) Mir23btm1MTm/Mmjax (36298-JAX) were obtained from Jackson Laboratory (Bar Harbor, ME). These mice were used to generate a deletion allele and a floxed conditional allele of mirn23b. To generate a germline null and a floxed conditional allele of mirn23b, the original Mir23btm1MTm/Mmjax strain was mated to B) B6.FVB-Tg(EIIa-cre)C5379Lmgd/J mice (Jackson Laboratory 003724) and C) B6;SJL-Tg(ACTFLPe)9205Dym/J (Jackson Laboratory 003800) strains respectively. D) The conditional mirn23b allele can be converted to a deletion allele with the expression of CRE recombinase.
Nucleated cells were isolated from the bone marrow of WT (n=4) and mirn23b−/− mice (n=4) and analyzed by flow cytometry for stem cell and immune cell populations. A) Bone marrow cellularity is unchanged between WT and mirn23b−/− mice. B) B cell and myeloid cell populations are unchanged in mirn23b−/− mice. C) CLP populations are unchanged in mirn23b−/− mice. D) CMP, GMP, and MEP populations are unchanged in mirn23b−/− mice. E) LT-HSC and ST-HSC/MPP populations are unchanged in mirn23b−/− mice. F) Splenic immune cell populations are unchanged in mirn23b deficient mice. P values were determined using unpaired student t-test.
Evaluation of CRE mediated mirn23b excision in bone marrow cells 4 weeks post pIpC injection by A) Genomic DNA PCR and B) qRT-PCR analysis of miRs-23b, -27b and -24 expression. C) Bone marrow was isolated from the femurs and tibias of WT, mirn23a−/−, and mirn23a−/−mirn23bflox/flox (labeled mirn23a−/−mirn23b−/−) mice treated with pIpC for 4 weeks. Bone was isolated and following ACK lysis, total nucleated cells were counted to measure nucleated bone marrow cellularity.
Competitive transplant assays performed with an equal ratio of WT CD45.1+ and CD45.2+ (WT, mirn23a−/−, or mirn23a−/−mirn23b−/−) donor marrow coinjected into lethal irradiated mice. Contribution to peripheral blood of nucleated CD45.2+ cells to myeloid lineage (CD11b+) and B lymphoid (B220) as well as populations negative for CD11b and B220 (DN) are shown at 6 (A) and 12 (B)-weeks post-transplant. Three, three, and five mice transplanted with WT, mirn23a−/−, and mirn23a−/−mirn23b−/− bone marrow were examined respectively. P values were determined using unpaired student t-test. * p<0.05, ** p<0.01.
Lin- RNA was isolated from WT, mirn23a−/−, and mirn23a−/−mirn23b−/− mice 4 weeks after pIpC injection and used for subsequent gene expression analyses. A) Analysis of B cell lineage specific transcription factors in Lin- hematopoietic cells were analyzed by qRTPCR. B) qRT-PCR was performed on primary Lin- cells from mice 4 weeks post pIpC treatment. Pro-apoptotic genes Bcl2l11 (Bim), Apaf1, and Caspase 9 (Casp9) were all increased in mirn23a−/− and mirn23a/mirn23b DKO mice compared to WT lin- cells. P values were determined using unpaired students t-test. *(p<0.05), ** (p<0.01), *** (p<0.001). C–D) Apoptosis RT profiler arrays were carried out in WT vs mirn23a−/− and mirn23a−/− vs mirn23a−/−mirn23b−/− populations by qRTPCR. Differences in gene expression greater than 2-fold and 1.5-fold are shown for (C) WT vs mirn23a−/− and (D) mirn23a−/− vs mirn23a−/−mirn23b−/− cells.
Highlights.
Mirn23a/Mirn23b deficient mice exhibit decreased HSPCs and bone marrow cellularity
Loss of mirn23a/mirn23b in HSPC populations results in increased apoptosis
Mirn23a/mirn23b loss skews hematopoietic differentiation towards the B cell lineage
Mirn23a/mirn23b loss alters hematopoietic and apoptotic gene expression pathways
Acknowledgments
This work was supported by National Institute of Diabetes and Digestive and Kidney Disease Grant R01DK109051 (RD) and an IUSM Biomedical Enhancement Grant (RD). We would also like to thank the staff of the University of Notre Dame Freimann Life Sciences Center. We would also like to thank Charles Tessier and the Indiana University flow cytometry core. We would also like to acknowledge Sridhar Rao (Blood Research Institute, Milwaukee WI) for reading and commenting on the manuscript before submission.
Footnotes
Authorship
J.K., A.B., B.R., R.K., and R.D. were responsible for study conception and design. J.K., A.B., N.K., and R.B. were responsible for performing the experiments and data acquisition. J.K., N.K., R.B., R.K., and R.D. were responsible for data interpretation. The manuscript was prepared by J.K., A.B. and R.D.
Disclosures
The authors have no financial conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gottgens B. Regulatory network control of blood stem cells. Blood. 2015;125:2614–2620. doi: 10.1182/blood-2014-08-570226. [DOI] [PubMed] [Google Scholar]
- 2.Boulais PE, Frenette PS. Making sense of hematopoietic stem cell niches. Blood. 2015;125:2621–2629. doi: 10.1182/blood-2014-09-570192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley interdisciplinary reviews Systems biology and medicine. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Ema H. Mechanisms of self-renewal in hematopoietic stem cells. Int J Hematol. 2016;103:498–509. doi: 10.1007/s12185-015-1919-5. [DOI] [PubMed] [Google Scholar]
- 5.Sharma S, Gurudutta GU, Satija NK, et al. Stem cell c-KIT and HOXB4 genes: critical roles and mechanisms in self-renewal, proliferation, and differentiation. Stem Cells Dev. 2006;15:755–778. doi: 10.1089/scd.2006.15.755. [DOI] [PubMed] [Google Scholar]
- 6.Aggarwal R, Lu J, Pompili VJ, Das H. Hematopoietic stem cells: transcriptional regulation, ex vivo expansion and clinical application. Curr Mol Med. 2012;12:34–49. doi: 10.2174/156652412798376125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008;14:1036–1040. doi: 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 12.Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci U S A. 2009;106:238–243. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontana L, Pelosi E, Greco P, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 16.Gerloff D, Grundler R, Wurm AA, et al. NF-kappaB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 2015;29:535–547. doi: 10.1038/leu.2014.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurkewich JL, Bikorimana E, Nguyen T, et al. The mirn23a microRNA cluster antagonizes B cell development. J Leukoc Biol. 2016 doi: 10.1189/jlb.1HI0915-398RR. [DOI] [PubMed] [Google Scholar]
- 19.Kong KY, Owens KS, Rogers JH, et al. MIR-23A microRNA cluster inhibits B-cell development. Exp Hematol. 2010;38:629–640. e621. doi: 10.1016/j.exphem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurkewich JL, Hansen J, Klopfenstein N, et al. The miR-23a~27a~24-2 microRNA cluster buffers transcription and signaling pathways during hematopoiesis. PLoS Genet. 2017;13:e1006887. doi: 10.1371/journal.pgen.1006887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo S, Lu J, Schlanger R, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci U S A. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han YC, Park CY, Bhagat G, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu W, Dooley J, Chung SS, et al. miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood. 2015;125:2206–2216. doi: 10.1182/blood-2014-06-585273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lechman ER, Gentner B, van Galen P, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11:799–811. doi: 10.1016/j.stem.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song SJ, Ito K, Ala U, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell. 2013;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy L, Bikorimana E, Lapid D, Choi H, Nguyen T, Dahl R. MiR-24 is required for hematopoietic differentiation of mouse embryonic stem cells. PLoS Genet. 2015;11:e1004959. doi: 10.1371/journal.pgen.1004959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, Yao N, Liu G, et al. Functional screen reveals essential roles of miR-27a/24 in differentiation of embryonic stem cells. EMBO J. 2015;34:361–378. doi: 10.15252/embj.201489957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakso M, Pichel JG, Gorman JR, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park CY, Jeker LT, Carver-Moore K, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez CI, Buchholz F, Galloway J, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 31.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen T, Rich A, Dahl R. MiR-24 promotes the survival of hematopoietic cells. PLoS One. 2013;8:e55406. doi: 10.1371/journal.pone.0055406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labi V, Bertele D, Woess C, et al. Haematopoietic stem cell survival and transplantation efficacy is limited by the BH3-only proteins Bim and Bmf. EMBO molecular medicine. 2013;5:122–136. doi: 10.1002/emmm.201201235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu EP, McLellan M, Ding L, et al. Caspase-9 is required for normal hematopoietic development and protection from alkylator-induced DNA damage in mice. Blood. 2014;124:3887–3895. doi: 10.1182/blood-2014-06-582551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, Xu J, Li L, et al. MicroRNA-23a/b and microRNA-27a/b suppress Apaf-1 protein and alleviate hypoxia-induced neuronal apoptosis. Cell Death Dis. 2014;5:e1132. doi: 10.1038/cddis.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lian S, Shi R, Bai T, et al. Anti-miRNA-23a oligonucleotide suppresses glioma cells growth by targeting apoptotic protease activating factor-1. Current pharmaceutical design. 2013;19:6382–6389. doi: 10.2174/13816128113199990509. [DOI] [PubMed] [Google Scholar]
- 37.Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, Srivastava D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J Exp Med. 2011;208:549–560. doi: 10.1084/jem.20101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker JC, Harland RM. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev. 2009;23:1046–1051. doi: 10.1101/gad.1777709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan MC, Hilyard AC, Wu C, et al. Molecular basis for antagonism between PDGF and the TGFbeta family of signalling pathways by control of miR-24 expression. EMBO J. 2010;29:559–573. doi: 10.1038/emboj.2009.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 41.de Graaf CA, Kauppi M, Baldwin T, et al. Regulation of hematopoietic stem cells by their mature progeny. Proc Natl Acad Sci U S A. 2010;107:21689–21694. doi: 10.1073/pnas.1016166108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warr MR, Pietras EM, Passegue E. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley interdisciplinary reviews Systems biology and medicine. 2011;3:681–701. doi: 10.1002/wsbm.145. [DOI] [PubMed] [Google Scholar]
- 43.Hertel J, Stadler PF. Hairpins in a Haystack: recognizing microRNA precursors in comparative genomics data. Bioinformatics. 2006;22:e197–202. doi: 10.1093/bioinformatics/btl257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The mirn23b miRNA cluster gene is embedded in an intron of the aminopeptidase O gene. A) Mir23btm1MTm/Mmjax (36298-JAX) were obtained from Jackson Laboratory (Bar Harbor, ME). These mice were used to generate a deletion allele and a floxed conditional allele of mirn23b. To generate a germline null and a floxed conditional allele of mirn23b, the original Mir23btm1MTm/Mmjax strain was mated to B) B6.FVB-Tg(EIIa-cre)C5379Lmgd/J mice (Jackson Laboratory 003724) and C) B6;SJL-Tg(ACTFLPe)9205Dym/J (Jackson Laboratory 003800) strains respectively. D) The conditional mirn23b allele can be converted to a deletion allele with the expression of CRE recombinase.
Nucleated cells were isolated from the bone marrow of WT (n=4) and mirn23b−/− mice (n=4) and analyzed by flow cytometry for stem cell and immune cell populations. A) Bone marrow cellularity is unchanged between WT and mirn23b−/− mice. B) B cell and myeloid cell populations are unchanged in mirn23b−/− mice. C) CLP populations are unchanged in mirn23b−/− mice. D) CMP, GMP, and MEP populations are unchanged in mirn23b−/− mice. E) LT-HSC and ST-HSC/MPP populations are unchanged in mirn23b−/− mice. F) Splenic immune cell populations are unchanged in mirn23b deficient mice. P values were determined using unpaired student t-test.
Evaluation of CRE mediated mirn23b excision in bone marrow cells 4 weeks post pIpC injection by A) Genomic DNA PCR and B) qRT-PCR analysis of miRs-23b, -27b and -24 expression. C) Bone marrow was isolated from the femurs and tibias of WT, mirn23a−/−, and mirn23a−/−mirn23bflox/flox (labeled mirn23a−/−mirn23b−/−) mice treated with pIpC for 4 weeks. Bone was isolated and following ACK lysis, total nucleated cells were counted to measure nucleated bone marrow cellularity.
Competitive transplant assays performed with an equal ratio of WT CD45.1+ and CD45.2+ (WT, mirn23a−/−, or mirn23a−/−mirn23b−/−) donor marrow coinjected into lethal irradiated mice. Contribution to peripheral blood of nucleated CD45.2+ cells to myeloid lineage (CD11b+) and B lymphoid (B220) as well as populations negative for CD11b and B220 (DN) are shown at 6 (A) and 12 (B)-weeks post-transplant. Three, three, and five mice transplanted with WT, mirn23a−/−, and mirn23a−/−mirn23b−/− bone marrow were examined respectively. P values were determined using unpaired student t-test. * p<0.05, ** p<0.01.
Lin- RNA was isolated from WT, mirn23a−/−, and mirn23a−/−mirn23b−/− mice 4 weeks after pIpC injection and used for subsequent gene expression analyses. A) Analysis of B cell lineage specific transcription factors in Lin- hematopoietic cells were analyzed by qRTPCR. B) qRT-PCR was performed on primary Lin- cells from mice 4 weeks post pIpC treatment. Pro-apoptotic genes Bcl2l11 (Bim), Apaf1, and Caspase 9 (Casp9) were all increased in mirn23a−/− and mirn23a/mirn23b DKO mice compared to WT lin- cells. P values were determined using unpaired students t-test. *(p<0.05), ** (p<0.01), *** (p<0.001). C–D) Apoptosis RT profiler arrays were carried out in WT vs mirn23a−/− and mirn23a−/− vs mirn23a−/−mirn23b−/− populations by qRTPCR. Differences in gene expression greater than 2-fold and 1.5-fold are shown for (C) WT vs mirn23a−/− and (D) mirn23a−/− vs mirn23a−/−mirn23b−/− cells.