Abstract
Continual monitoring of secreted biomarkers from organ-on-a-chip models is desired to understand their responses to drug exposure in a noninvasive manner. To achieve this goal, analytical methods capable of monitoring trace amounts of secreted biomarkers are of particular interest. However, a majority of existing biosensing techniques suffer from limited sensitivity, selectivity, stability, and require large working volumes, especially when cell culture medium is involved, which usually contains a plethora of nonspecific binding proteins and interfering compounds. Hence, novel analytical platforms are needed to provide noninvasive, accurate information on the status of organoids at low working volumes. Here, we report a novel microfluidic aptamer-based electrochemical biosensing platform for monitoring damage to cardiac organoids. The system is scalable, low-cost, and compatible with microfluidic platforms easing its integration with microfluidic bioreactors. To create the creatine kinase (CK)-MB biosensor, the microelectrode was functionalized with aptamers that are specific to CK-MB biomarker secreted from a damaged cardiac tissue. Compared to antibody-based sensors, the proposed aptamer-based system was highly sensitive, selective, and stable. The performance of the sensors was assessed using a heart-on-a-chip system constructed from human embryonic stem cell-derived cardiomyocytes following exposure to a cardiotoxic drug, doxorubicin. The aptamer-based biosensor was capable of measuring trace amounts of CK-MB secreted by the cardiac organoids upon drug treatments in a dose-dependent manner, which was in agreement with the beating behavior and cell viability analyses. We believe that, our microfluidic electrochemical biosensor using aptamer-based capture mechanism will find widespread applications in integration with organ-on-a-chip platforms for in situ detection of biomarkers at low abundance and high sensitivity.
Graphical Abstract
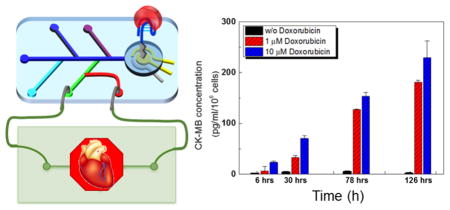
Drug development is a lengthy and costly process associated with high incidences of attrition.1,2 In addition, roughly 20% of drugs in the past four decades have been recalled because of cardiotoxicity and other cardiac safety issues.3,4 To solve this problem, human cell-based three-dimensional (3D) tissue/organ models that can precisely recapitulate the important architecture and functionality of their in vivo counterparts as viable platforms for drug screening prior to clinical trials have been developed.5,6 These physiologically relevant human organ models have shown several major advantages over conventional animal models such as more accurate prediction of human responses.6,7 Furthermore, recent developments in microfluidics have offered new approaches to fabricate biomimetic human organoid models that simulate both the biology and the physiological microenvironment of the human system, termed organs-on-chip.8–10 Despite recent advances in organs-on-chip systems, it is difficult to obtain information about secreted biomarkers that correlate with the behavior and status of these organoids during long-term culture. For example, it is well-known that the cardiac cells may remain viable yet tend to lose their metabolic activity within a few days.11 Hence, it is critical to develop methods to probe (disease) biomarker secretion by the cells during continual culture or drug screening processes. In addition, the small medium volumes in these organs-on-a-chip platforms mandate that the sensing systems should consume very little sample fluids, be able to be multiplexed (i.e., can monitor multiple biomarkers), are noninvasive to the organoids, and amenable to microfluidic devices allowing for seamless integration with organ constructs.
Measurement of secreted biomarkers by the organoids has been one of the approaches to assess the bioactivity and viability of the organoids in a noninvasive manner. Several detection methods have thus been utilized for label-free biosensing of secreted molecules such as those based on enzyme linked immunosorbent assays (ELISA)12 and surface plasmon resonance (SPR),13 among others.14 However, most of these existing techniques suffer from limited sensitivity and selectivity, especially when complex biological environments such as cell culture medium is involved, which usually contains a plethora of nonspecific proteins and interfering compounds but trace amounts of biomarkers of interest. The low abundance of secreted biomarkers is of critical concern particularly when it comes to disease markers such as those specific for cardiac damage or injury including creatine kinase (CK)-MB (<1 ng mL−1) and troponins.15,16 The current gold standard for such measurements relies on ELISA, which is typically insufficient in sensitivity in addition to the needs for off-chip operations that consume significant sample volumes. Therefore, a robust biosensor platform featuring high sensitivity and selectivity is an urgent need for probing cell behaviors in microphysiometry systems. To this end, electrochemical impedance spectroscopy (EIS) based sensor with high sensitivity and ultra low limit-of-detection (LOD) at low cost has numerous advantages compared to other biosensing techniques. Moreover, electrochemical biosensors are compatible with microfluidics technologies, allowing for facile integration with microfluidic organs-on-a-chip platforms for continual monitoring of (multiple) secreted biomolecules in a noninvasive manner while consuming only small volumes of media.17,18
To improve the selectivity and stability along with high sensitivity of a biosensor operating within complex biological environments such as cell culture medium, aptamers, which are nucleic acids with meticulously designed sequences, have been widely employed as the sensitive recognition elements of bio-sensors for the detection and quantification of various analytes such as nucleic acid, proteins, and cells.19–26 In comparison to antibodies, aptamers seldom aggregate, facilitating their uniform deposition over the sensing surfaces.27,28 In addition, they can easily be chemically modified with functional groups to provide them with tailored properties.29,30 Furthermore, the sensitivity and selectivity of aptamers are relatively stable with respect to disturbances such as changes in pH, temperature, and interferents over lengthy periods. Therefore, to measure low levels of soluble biomarkers, aptamers are excellent candidates as a class of receptors for use in an electrochemical biosensing platform compared to conventional antibodies, where higher sensitivity has been reported for aptamer-based electrochemical immunosensors.31
Here, we report the development of an aptamer-based electrochemical biosensor that can be integrated with a microfluidic platform for in-line detection of secreted protein biomarkers from an organ-on-a-chip device. In particular, the sensor was functionalized with aptamers specific to a cardiac injury biomarker in extremely low abundance, CK-MB. This aptamer-based electrochemical biosensor was analyzed for its sensitivity and compared with antibody-based immunosensors. Their performances were further assessed in conjunction with a microfluidic heart-on-a-chip system constructed from human embryonic stem cell-derived cardiomyocytes (ESC-CMs). The trace changes (<1 ng mL−1) in secreted CK-MB levels by the cardiac organoids upon drug insults in a dose-dependent manner were measured by the aptamer-based electrochemical immunosensors, and compared to other organoid evaluation methods including beating rates and cellular viability. The developed system is versatile and can be used to detect various cell-secreted biomarkers.
RESULTS AND DISCUSSION
To monitor secreted CK-MB antigens, EIS measurements were utilized. A microelectrode set having a reference electrode (RE), a counter electrode (CE), and a working electrode (WE) was custom designed and fabricated (Figure 1a), for convenient integration with a microfluidic sensing platform. To achieve a stable biosensing system, gold (Au) was chosen as the electrode material since it has numerous advantages including favorable electron transfer kinetics, relatively good stability, and convenient covalent bonding with functional groups such as thiol-containing molecules (through the formation of Au-sulfur (S) bonds).32,33 The metal layers were deposited using an electron beam deposition process where a shadow mask was adopted for patterning. The Au CE and WE were deposited on top of a titanium (Ti) seed layer (20 nm), where a palladium (Pd) layer with a thickness of 20 nm was sandwiched in between two layers, to block the diffusion of Ti into the Au layer during the annealing procedure (300 °C for 6 h in a furnace).
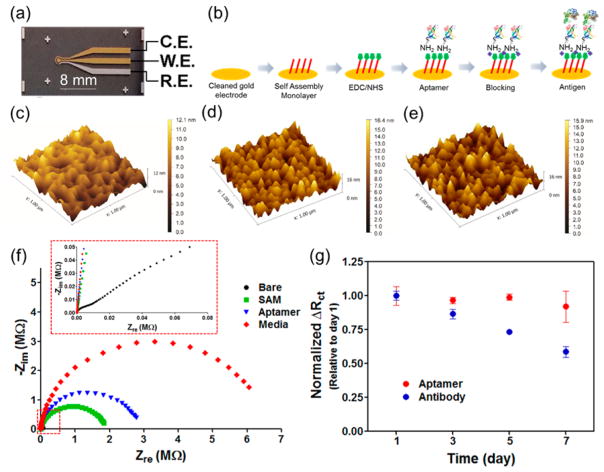
Figure 1.
(a) Photograph of the microfabricated electrode set. (b) Schematic diagram of immobilization steps of aptamers onto the microelectrode. AFM surface characterization of (c) bare Au WE electrode, (d) antibody-, and (e) aptamer-functionalized microelectrode. (f) EIS measurements after immobilization of aptamer and subsequent molecules onto bare microelectrode. (g) Normalized ΔRct over time showing the denaturation of antibody and aptamer biosensors. The aptamer and antibody-based biosensors were fabricated on the same day and stored in DPBS at 4 °C.
We then developed an on-chip process for attaching antibodies and aptamers onto the surface of the Au electrodes based on a combination of the Au–S bonding mechanism, the carbodiimide chemistry, and the streptavidin–biotin reactions.34 Figure 1b shows schematic illustration of the immobilization steps of aptamers onto the Au surface. In the first step, the Au surface was functionalized by immobilizing a self-assembling monolayer (SAM) from spacers containing thiol groups on one end and carboxyl groups on the other terminal. The creation of a covalent bond between SAM (exposed carboxyl groups) and amine-terminated aptamers was then achieved through the N-ethylcarbodiimide (EDC)/N-hydroxysuccinimide (NHS)-based carbodiimide chemistry. Functionalization of antibodies was similar, except for one additional step, that is, streptavidin-amine was subsequently attached to the SAM layer using carbodiimide chemistry followed by grafting of the biotinylated antibodies via the streptavidin–biotin interaction. These processes are simple and efficient, allowing for functionalization of the Au electrodes with antibodies/aptamers in a highly reproducible manner.
To investigate the variations on the Au electrode surface before and after functionalization with antibodies/aptamers, the atomic force microscopy (AFM) analysis was performed (Figure 1c–e). The mean surface roughness of the bare electrodes obtained from AFM measurements was 1.79 nm as shown in Figure 1c. The surface roughness of the electrodes functionalized with aptamers and antibodies was found to increase to 2.75 and 2.53 nm, respectively. The covalent immobilization method led to a relatively uniform distribution of the aptamers and antibodies on the surface of the electrodes (Figure 1d and e). Successful functionalization of each antibody/ aptamer layer on bare electrodes was confirmed by a subsequent increase in the impedance signal obtained after deposition of the each layer (Figure 1f and Figure S1). The Nyquist plot obtained from the bare electrodes was almost a straight line, which was characteristic of a diffusion-limited process (Figure 1f, inset). However, a significant semicircle curve appeared in the high frequency region after immobilization of the SAM layers on the electrode. The Rct increased to ~2 MΩ, indicating that electron 3−/4− and the electrode was strongly transfer between [Fe(CN)6] blocked by the SAM layers on the electrode.35 In addition, the Rct value obtained from the Au electrode after immobilization of the SAM was larger than the value obtained from the SAM functionalized Au electrodes reported previously.36,37 We believe that this discrepancy was due to the smaller surface area of our WE (~0.5 mm2) compared to that of previously reported Au disk electrodes (~8.0 mm2).36,38
Shelf Life and Selectivity of the Electrochemical Biosensors
The shelf life of a biosensor is an important factor during monitoring of biomarkers. To investigate the shelf life of the aptamer and antibody-based biosensors, the two devices were stored under moderately wet conditions at 4 °C since the biosensors will be exposed to prolonged wet conditions during real sample analysis. We prepared both biosensors on the same day and stored them for different time periods in dulbecco’s phosphate-buffered saline (DPBS) at 4 °C before they were used to measure CK-MB (1 ng mL−1). As shown in Figure 1g, the normalized ΔRct values obtained from the aptamer-based biosensors showed relatively long shelf life for up to 7 days of storage. In comparison, the normalized ΔRct values obtained from antibody-coated biosensors decreased dramatically (~60%) over the same period of time. These results indicate that the aptamer-based biosensors had a much longer shelf life compared to antibody-functionalized electrodes likely because of the denaturation of the protein-based antibodies. The improved shelf life of the aptamer biosensors also potentially enables their use in monitoring of biomarkers over extended periods of time.
For applications in complex biological environments such as cell culture medium containing a large amount of proteins (bovine serum albumin (BSA)) and other small molecules, the aptamer-based based biosensor should be selective only toward CK-MB antigens. To prevent nonspecific binding of proteins onto the electrode surface of electrode, we used cell culture medium as the blocking agent instead of conventional blocking agents such as BSA. When we blocked the aptamer-functionalized electrode surface only by BSA, the impedance changed after incubation with the culture medium for human ESC-CMs even without the CK-MB antigen. This cell culture medium was composed of various materials such as BSA, L-glutamine, insulin, and vitamin A, etc. In particular, those components with smaller molecular weights, such as L-glutamine, insulin, and vitamin A might penetrate into the BSA blocking layer on aptamer-functionalized electrode. These incorporated molecules within the BSA blocking layer might induce an impedance change after incubation compared with the pristine BSA blocking layer. Therefore, we used the cell culture medium as the blocking agent and then optimized the incubation time to block the electrode after immobilization of aptamers using the cell culture medium, which did lead to a large increase in the impedance signal. However, the 30 min incubation was sufficient to block the surface of electrode since we observed that the incubation period exceeding 30 min did not significantly change the impedance of the electrodes anymore (data not shown). In an ideal setting, neither the medium itself (i.e., ions) nor the proteins and small molecules should interfere with CK-MB sensing. The selectivity of the aptamer-based biosensors was evaluated against other biomarkers, including 10 ng mL−1 albumin and glutathione S-transferase alpha (GST-α) that are well-known liver biomarkers. The results in Figure S2 demonstrated that, after exposure to these antigens, the impedance measured from the electrodes did not show significant changes, revealing that nonspecific binding did not occur during the incubation of the biosensor with albumin and GST-α antigens. However, subsequent addition of 10 ng mL−1 CK-MB was found to result in a significant increase in the impedance as observed in Figure S2. These data confirmed that the developed aptamer immunosensor demonstrated good selective detection for CK-MB, due to strong aptamer-target interaction.
Sensitivity of the Biosensors
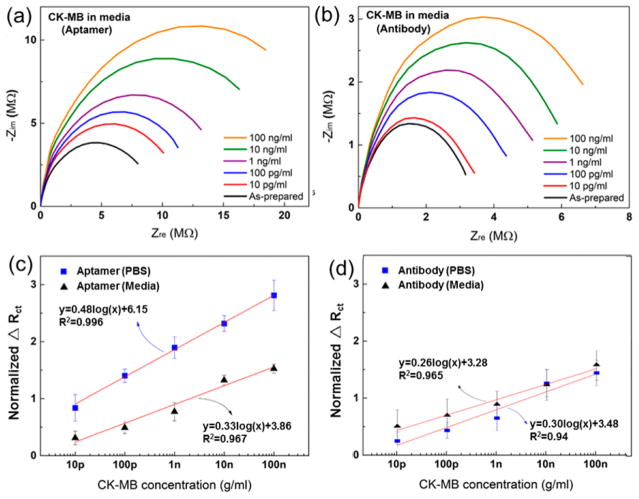
We next evaluated the sensitivity of both types of biosensors. After incubation of the biosensors with different concentrations of CK-MB antigens, the EIS measurements were performed at each antigen concentration (Figure 2a and b). To eliminate any possible individual variation of physical and electrochemical difference that may be present among the different electrodes, we determined normalized ΔRct via dividing the Rct of CK-MB (or any biomarker) by the Rct of media, which is termed the “as-prepared” signal, and was used as the baseline of the biosensors. The standard curves were next obtained by plotting normalized ΔRct at a range of concentrations (0.01–100 ng mL−1) in both DPBS and culture media. As shown in Figure 2c and d), the aptamer-based biosensor was found to be more sensitive than the antibody-based biosensor in measuring CK-MB levels in both DPBS and medium. The slope of the calibration curves for CK-MB antigens prepared in DPBS and medium measured by the aptamer-based biosensors were 0.48 and 0.33, respectively, while these values measured using antibody-based biosensors were 0.30 and 0.26, respectively. Therefore, the sensitivities of aptamer-based biosensors for CK-MB measurements were 1.58 and 1.26 times higher those of antibody-based biosensors in DPBS and culture medium, respectively. However, the reduced sensitivity for both aptamer- and antibody-based biosensors in the cell culture medium was likely associated with the interference of nonspecific protein (~99 mg mL−1 BSA) or small molecules surrounding the minute concentration of CK-MB antigen in the culture medium compared to the much less complex environment of DPBS. These results indicated that the aptamers coated onto the microelectrodes could be efficiently used to detect CK-MB in a complex microenvironment at a high sensitivity with a LOD of 2.4 pg mL−1 and a wide dynamic range (0.01–100 ng mL−1). This LOD obtained from the aptamer-based electrochemical biosensor for CK-MB detection is lower than those obtained by other similar EIS detection methods that used an Au electrode array (10 pg mL−1)39 or other methods including immunochemiluminometric assay (~1 ng mL−1),40,41 amperometric assay (0.11 μg mL−1),42 capillary immunoassay (0.6 ng mL−1),43 SERS-based ELISA (42.5pg mL−1),44 antibody-based SPR sensors (2.71 ng mL−1),45 and slightly lower than that of silicon nanowire field-effect transistor-based biosensor (SiNW-FET) (100 fg mL−1) because of field-effect amplification of the FETs.46 However, the fabrication of SiNW-FETs is complex and expensive, while our EIS detection method based on aptamers is sufficiently sensitive with a wide detection range for the cardiac biomarker, CK-MB. In addition, continual increase in impedance signals were observed in all cases (Figure 2c and d), suggesting that even high concentrations of up to 100 ng mL−1 of CK-MB did not result in the saturation of the electrodes. Therefore, after a minute amount of CK-MB is captured on the electrode surface, there is a potential to continue using these electrodes for multiple times before saturation.
Figure 2.
Nyquist curves for (a) aptamer and (b) antibody-based sensors measured in medium. Calibration curves for (c) aptamer- and (d) antibody-based sensors measured in DPBS and medium.
Overall, aptamer-based biosensors possessed several key advantages for measuring biomarkers in complex environments such as the culture medium, compared with antibody-based bio-sensors. The aptamer-based biosensors have ultrahigh sensitivity on the order of a few pg mL−1. Furthermore, the shelf life of aptamer-based biosensors was found to be at least 7 days, which may potentially be extended to several weeks and even months when frozen due to the stability of the aptamers. In comparison, antibody-based biosensors appeared to have a lower shelf life, which significantly limits their ease of use. It is also expected that aptamers can endure harsh conditions such as extreme pH, while antibodies will only operate under near physiological conditions.31,47,48 On the basis of these observations, aptamer-based biosensors can function as an excellent biosensing platform to monitor extremely low concentrations of CK-MB produced by heart-on-a-chip systems.
Heart-on-Chip and Cardiotoxicity Studies
Development of biomimetic heart-on-a-chip systems offers new insights to monitor cardiac functions for quantitative exploration of drug-induced cardiotoxicity.49,50 Noninvasive continuous measurement of cellular metabolic activity is needed to probe the activity of drugs on such models. Measurement of cardiac organoid functionality has conventionally been carried out by optical and electrophysiological approaches that observe the physical contraction of the cardiac organoids and detect the action potential of the cardiomyocytes.51,52 To date, there have only been a few reports in determining the functionality of heart-on-a-chip systems that have utilized biomolecular sensing.53,54 Moreover, cardiac injury/damage biomarkers, such as CK-MB, myosin, and troponins, are expressed at low levels by the cardiomyocytes upon exposure to toxic agents, which are highly diagnostic and predictive of cardiac damage.16 Hence, a detection platform that can be directly integrated with a heart-on-a-chip system for real time or near real time monitoring of secreted cardiac biomarkers for cardiotoxicity assessment is of great importance.
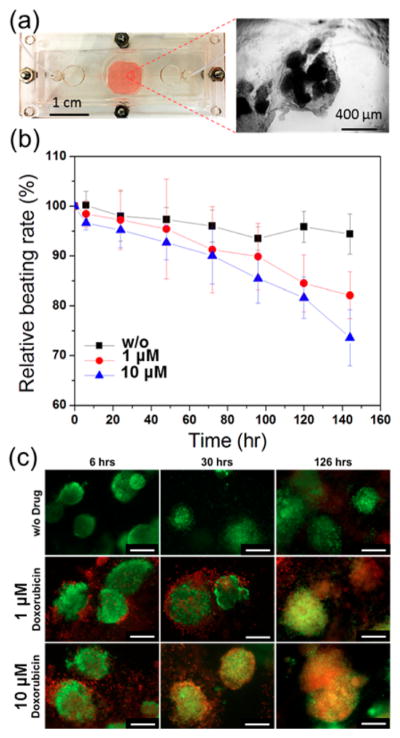
A cardiac bioreactor was constructed by modifying our recently published protocols (Figure 3a).55 Cardiac spheroids (~200 μm in diameter) derived from ESC-CMs were embedded inside a gelatin methacryloyl (GelMA) hydrogel, dispensed, and photo-cross-linked at the bottom of the bioreactor to generate the human cell-based heart-on-chip platform (Figure 3a, inset). The cardiac spheroids were successfully cultured for up to 5 days in the bioreactors without leakage or contamination. Doxorubicin (DOX), an anticancer drug, was used as the model drug to induce cardiac injury in the cardiac spheroids. The exposure of cardiomyocytes to DOX is expected to increase the levels of secreted CK-MB antigens by the cardiomyocytes and result in changes in their beating rates.56,57 Indeed, Figure 3(b) indicates that the relative beating rates of the cardiomyocytes in the heart-on-chips significantly decreased over time after the cells were exposed to DOX, resulting in reduced beating rates of 85% and 82% corresponding to drug concentrations of 1 and 10 μM, respectively. The viability of the cardiomyocytes was further analyzed using a live/dead assay. As shown in Figure 3c, while the cells in the control study (no drug) exhibited high cell viability at all time points. The cells that were treated with drugs had a decrease in their viability over a period of 5 days, especially in the group where 10 μM DOX was used. The changes in beating rates and the viability assessment of the heart-on-a-chip system confirmed the capability of DOX to induce cardiotoxicity in the in vitro cardiac model which was detected using microfluidic biosensors.
Figure 3.
(a) Photograph of the microfluidic bioreactor and the optical micrograph of the cardiac spheroids. (b) Measured normalized beating rates of cardiomyocytes over time after exposure to different concentrations (0, 1, and 10 μM) of DOX. (c) Live/dead assays of cardiomyocytes obtained after exposure of the cells to different concentrations of DOX up to 126 h. Live cells were stained green and dead cells were stained red. (Scale bar: 200 μm.)
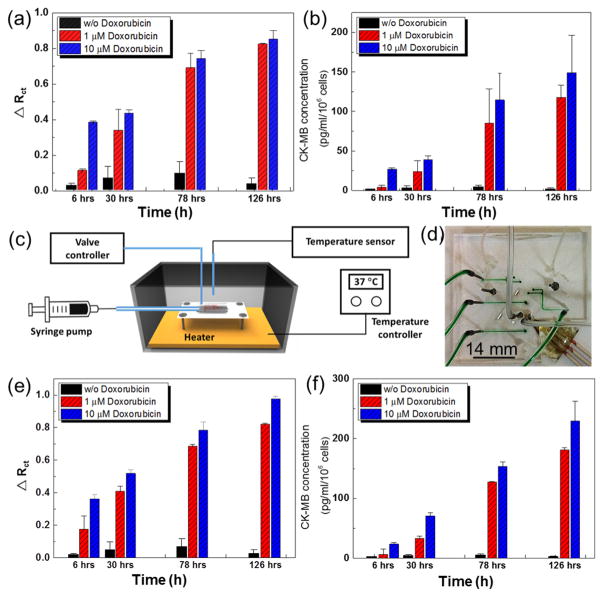
It is further expected that the amount of CK-MB in the media collected from the cardiac bioreactors should also increase over time as the cardiomyocytes undergo damage upon exposure to DOX. To evaluate the response of the cardiac organoids to DOX, the CK-MB biosensor was utilized to measure the CK-MB levels in bioreactors administered with 0, 1, and 10 μM DOX after 6, 30, 78, and 126 h. For these tests, the CK-MB specific aptamer-based biosensor was incubated with bioreactor samples for 30 min and then the EIS was measured (Figure 4a). The normalized ΔRct data obtained from the EIS measurements were correlated to the concentrations of CK-MB using a calibration curve (Figure 4b). The concentrations of secreted CK-MB antigens remained at negligible levels in cardiac bioreactors with no drug treatment throughout the entire period. However, administration of 1 μM and 10 μM DOX induced significant cell damage resulting in an increase in CK-MB levels in both dose- and time-dependent manner, clearly indicating the cytotoxic effect of DOX toward the cardiomyocytes. The concentrations of CK-MB in the media retrieved from the cardiac bioreactors after 30, 78, and 126 h were measured to be 24, 80, and 120 pg mL−1 cell−1, respectively, after 1 μM DOX treatment, while the group treated with a 10 μM DOX reached 45, 115, and 149 pg mL−1 cell−1 (Figure 4b). Comparing the beating behavior and cellular viability in Figure 3b and c, it can be concluded that the DOX treatment of cardiomyocytes influenced the cells in a dose-dependent manner where higher doses of DOX reduced the beating rates as well as the cellular viability. In addition, we have also observed that higher DOX concentrations resulted in elevated levels of CK-MB secretion from the cardiac organoids. This was expected since the CK-MB is known to be a myocyte injury biomarker.58
Figure 4.
(a) Normalized Rct values of CK-MB levels obtained using aptamer biosensors after the cardiac organoids were exposed to no-drug and 1 and 10 μM DOX for up to 126 h. (b) Calculated CK-MB concentrations obtained using the calibration curve. (c) Schematic illustration of the drug administration system connected to the microfluidic heart-on-a-chip bioreactor. (d) Photograph of the microfluidic bioreactor with an integrated microelectrode set. (e) Normalized Rct values of CK-MB levels obtained on-chip using aptamer-based sensors after the cardiac organoids were exposed to no-drug and 1 and 10 μM of DOX for up to 126 h. All the functionalization and detection steps were carried on chip. (f) CK-MB concentrations versus time obtained using the calibration curve.
After confirming that our aptamer-based biosensor could successfully measure trace amounts of CK-MB present in the media following cardiac damage caused by DOX, the sensor chip was further integrated with a heart-on-a-chip platform for in situ monitoring (Figure 4c). Installation of the microfluidic and sensing modules into the portable incubator allowed ease of use and automated handling of the integrated system. It also protected the vulnerable or sensitive components of the bio-sensor from potential damage or contamination. This chip was built from polydimethylsiloxane (PDMS) using soft lithography, which possessed a fluid loop for medium circulation and sampling, 2 inlets for electrolyte injection and washing, 1 chamber hosting the electrode, and 1 common outlet (Figure 4d). The microfluidic chip consisted of three layers including a fluidic layer, a valve layer, and a thin-membrane in between. Each channel was controlled by a pneumatic valve via pressurized N2 gas. To visually distinguish the valve layer from the microfluidic layer and prevent the diffusion of gas to the fluidic channel, the valve channels were filled with food dye (green color in Figure 4d). The medium from the cardiac bioreactor was sampled at predetermined time points for biomarkers measurements which were conducted later on by the microfluidic biosensor. The medium in the organoid modules was driven by a peristaltic pump, while the liquid handling of the reagents for electrochemical sensing was conducted by externally actuated pneumatic valves programmed to operate in an automated manner. This microfluidic chip required approximately 9 μL of sample volume. The concentrations of CK-MB levels secreted by the cardiac organoids upon treatment of different concentrations of DOX after 6, 30, 78, and 126 h were then calculated from ΔRct values (Figure 4e and f). Similar to off-line results, on-chip measurements (heart-on-a-chip platform) also revealed a similar trend, where higher levels of CK-MB antigens were detected, which corresponded to the use of higher concentrations of DOX and longer exposure times. These results clearly demonstrated that our aptamer-based electrochemical biosensor can be integrated with microfluidic organs-on-a-chip platforms and can be used for in situ, continual measurement of biomarkers until its saturation by cell-secreted biomarkers. We also demonstrated that trace amounts of biomarkers (pg mL−1) can be readily detected with high sensitivity. This is the first time that an integrated sensor was used to directly obtain cell-secreted biomarkers from a cardiac-organoid-on-a-chip system for rapid detection without any delays or off-chip processing. This critical feature of our aptamer-based biosensors can be conveniently expanded to other organ models where the detection of trace amounts of (damage) biomarkers is desired.
CONCLUSIONS
In this Article, we have developed a novel integrated label-free electrochemical biosensing platform combining an in-house designed Au microelectrode and a microfluidic system, to achieve fast and sensitive measurement of secreted biomarkers at low concentrations from a human cardiac-organoid-on-a-chip model. The use of aptamers as the antigen receptors was shown to significantly increase the sensitivity and shelf life of the biosensor in comparison to antibody-based biosensors. The biosensor functionalized with aptamers specific to cardiac damage biomarker CK-MB was then validated using samples taken from a human heart-on-a-chip model. Successful detection of trace amounts of CK-MB secreted by the cardiac organoids upon drug treatments was in a dose-dependent manner. The results obtained from biosensors were in agreement with the changes in the beating rates and cellular viability assessments. We believe that, our unique microfluidic electrochemical biosensor based on the aptamer-capturing mechanism can be conveniently extended to the measurement of a wide variety of other biomarkers of interest. With further development in multiplexing and optimization, these aptamer-based microfluidic electrochemical biosensors are expected to find widespread applications in monitoring organ-on-a-chip platforms for continual in situ detection of biomarkers at trace amounts.
EXPERIMENTAL METHODS
Preparation of Microelectrode
The diameter of the WE was 800 μm, the width of CE and RE were 150 μm, and gap between WE and other two electrodes was 200 μm, and the diameter of detection area was about 1500 μm. The glass substrate was diced to specific dimensions using a dicing saw machine. The diced glass chips were cleaned by acetone, isopropyl alcohol, and deionized water and dried at 120 °C in an oven. The shadow mask of 0.25 mm thickness with apertures was attached to the glass substrate. Twenty-nanometer-thick Ti, 20-nm-thick Pd, and 500-nm-thick Au were selectively deposited on the glass using the shadow mask by an e-beam evaporator to create the WE and CE. Then, the second shadow mask for RE patterning was attached to the Au-deposited glass substrate. In the same manner, 20-nm-thick Ti, 20-nm-thick Pd, and 500-nm-thick silver (Ag) were deposited on the glass substrate. Finally, the electrodes were annealed at 300 °C for 6 h in a furnace.
Electrochemical Measurements
Measurements of EIS spectrum was performed using an electrochemical workstation CHI660E (CH instrument, Inc.) with a conventional three electrode system. For the EIS technique, the initial potential was set to 0.1 V and the range of frequencies were scanned from 0.1 Hz to 100 kHz at 5 mV of amplitude. All measurements werecarried out in 50 mM K3Fe(CN)6 electrolyte solution.
Fabrication of Microfluidic Chips
The transparent polymer film was used as a photomask in a photolithography process on a layer of SU-8 photoresist, where the UV area created a positive relief of channel structure. This SU-8 structure was then used as master for casting PDMS channels by replica molding. To create the PDMS mold, the SU-8 master was covered with a liquid PDMS prepolymer in a mixture of 1:10 base polymer: curing agent and cured in the oven at 80 °C for 1 h. The cured PDMS mold was peeled off from the master. To prepare the thin membrane, PDMS was poured in a Petri dish and a film with a 40 μm of thickness was formed by spin coater. Then the PDMS layer was cured at 80 °C in an oven for 1 h. The three layers were bonded together after being treated with air plasma for 70 s where the microelectrodes were housed inside the fluidic channel. To connect the microfluidic chip to a syringe pump, metal connectors and tubes were inserted into the inlet and outlet ports. All the fluidic channels were initially filled with DPBS from the outlet to the inlet with a syringe pump at a flow rate of 1.0 mL h−1.
Immobilization Process for Aptamer and Antibody Biosensors
After cleaning the fabricated microelectrodes, DPBS and ethanol were flowed for 7 and 10 min, respectively. After that step, the SAM solution aimed to generate the thiol terminated SAM was flowed from a dedicated channel for 10 min and incubated in the reaction chamber for 1 h by using 11-mercaptoundecanoic acid (11-MUA). Then, the microelectrode in reaction chamber was rinsed with ethanol and DPBS subsequently for 10 and 7 min, respectively, to remove the unbound SAM molecules. The mixed solution of EDC (50 mM) and NHS (50 mM) was injected into the reaction channel and incubated for 15 min to start the activation of NHS ester, which was able to react with primary amine groups in aptamer or streptavidin by the amide bond. We obtained CK-MB 1A aptamer with a 5'Amine from OTCbiotech to specially capture CK-MB1, which had little-to-no cross-reactivity with CK-MB2. To immobilize the antibodies, 10 μM streptavidin solution was flowed for 7 min to the reaction chamber and incubated for 1 h after incubation of the NHS/EDC solution. Then, a 10 μM biotinylated antibody solution was flowed for 7 min into the reaction chamber and incubated for 1 h. This was followed by DPBS wash for 7 min, during which the unspecific sites were blocked by flowing and incubating cell culture media for 7 and 30 min, respectively. To measure CK-MB antigens, different concentrations of CK-MB biomarkers were flowed into the channel and incubated for 7 and 30 min, respectively.
Culturing Cardiospheres in the Microfluidic Bioreactor
Cryovials (Nalgene, Thermo Scientific) that worked as media reservoirs and bubble traps were connected between the outlet of the peristaltic pump and the inlet of the bioreactor. This system allowed sustainability of the bioreactor and enabled long-term media sample collection without the need to interfere with the bioreactor. Media consisting of RPMI (Sigma-Aldrich) and B27 supplement (Gibco, Life Technologies) were added. The bioreactor was perfused to allow for sufficient delivery/exchange of nutrients, oxygen, and waste.55 The spheroids mixed with 10% GelMA were cultured in the bioreactor operated with a peristaltic pump. Media was collected from the bioreactor (by changing the reservoir tube each time) at 6, 30, 78, and 126 h. In addition, the entire system was designed to remain bubble-free during the culture process to avoid potential interference. After including a bubble trap and testing for 6 days, there was no evidence of leakage or bubbles in the system indicating that it could be used for long-term culture of cells.
Characterization of Cardiac Spheroids after DOX Injection (Cell Morphology, Beating Rates)
The cardiac organoids were continuously challenged with 0, 1, and 10 μM DOX added to the medium during perfusion culture. The beating rates of the cardiac spheroids were measured using an optical microscope (Zeiss Axio Observer D1) at desired time points for up to 140 h. The beating rates were normalized to those at time 0 of respective samples. Three bioreactors were analyzed for each drug condition.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge funding from the Defense Threat Reduction Agency (DTRA) under Space and Naval Warfare Systems Center Pacific (SSC PACIFIC) Contract No. N6601-13-C-2027. The authors also acknowledge funding form the Institute for Soldier Nanotechnology, National Institutes of Health (HL092836, EB012597, AR057837, HL099073), the Office of Naval Research Young Investigator award, ONR PECASE Award. T.K. and H.A. thank Scientific and Technological Research Council of Turkey (TUBITAK). H.A. also sincerely thank Eskisehir Osmangazi University and Prof. Dr. M. Bahaddin Acat. Y.S.Z. acknowledges the National Institutes of Health National Cancer Institute Pathway to Independence Award (1K99CA201603-01A1). The publication of this material does not constitute approval by the government of the findings or conclusions herein.
Footnotes
Author Contributions
S.R.S., Y.S.Z., and D.-J.K. contributed equally to this work.
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.6b02028.
Materials, fabrication of the bioreactor, human pluripotent stem cell culture and differentiation, characterization of functionalized microelectrodes, and live/dead assay (PDF)
References
- 1.Hughes JD, Blagg J, Price DA, Bailey S, Decrescenzo GA, Devraj RV, Ellsworth E, Fobian YM, Gibbs ME, Gilles RW, Greene N, Huang E, Krieger-Burke T, Loesel J, Wager T, Whiteley L, Zhang Y. Bioorg Med Chem Lett. 2008;18:4872–4875. doi: 10.1016/j.bmcl.2008.07.071. [DOI] [PubMed] [Google Scholar]
- 2.Kola I, Landis J. Nat Rev Drug Discovery. 2004;3:711–716. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 3.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. Nat Rev Drug Discovery. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 4.Schiller LR, Johnson DA. Am J Gastroenterol. 2008;103:815–819. doi: 10.1111/j.1572-0241.2008.01818.x. [DOI] [PubMed] [Google Scholar]
- 5.Bhise NS, Ribas J, Manoharan V, Zhang YS, Polini A, Massa S, Dokmeci MR, Khademhosseini A. J Controlled Release. 2014;190:82–93. doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang YS, Ribas J, Nadhman A, Aleman J, Selimovic S, Lesher-Perez SC, Wang T, Manoharan V, Shin SR, Damilano A, Annabi N, Dokmeci MR, Takayama S, Khademhosseini A. Lab Chip. 2015;15:3661–3669. doi: 10.1039/c5lc00666j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esch EW, Bahinski A, Huh D. Nat Rev Drug Discovery. 2015;14:248–260. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young EWK. Integr Biol. 2013;5:1096–1109. doi: 10.1039/c3ib40076j. [DOI] [PubMed] [Google Scholar]
- 9.Huh D, Torisawa Y-s, Hamilton GA, Kim HJ, Ingber DE. Lab Chip. 2012;12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 10.Lee E, Song HHG, Chen CS. Curr Opin Chem Eng. 2016;11:20–27. doi: 10.1016/j.coche.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serena E, Cimetta E, Zatti S, Zaglia T, Zagallo M, Keller G, Elvassore N. PLoS One. 2012;7:e48483. doi: 10.1371/journal.pone.0048483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Real-Fernandez F, Rossi G, Lolli F, Papini AM, Rovero P. MethodsX. 2015;2:141–144. doi: 10.1016/j.mex.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappi G, Spiga FM, Moncada Y, Ferretti A, Beyeler M, Bianchessi M, Decosterd L, Buclin T, Guiducci C. Anal Chem. 2015;87:5278–5285. doi: 10.1021/acs.analchem.5b00389. [DOI] [PubMed] [Google Scholar]
- 14.Sivanesan A, Izake EL, Agoston R, Ayoko GA, Sillence M. J Nanobiotechnol. 2015;13:43. doi: 10.1186/s12951-015-0102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerszten RE, Wang TJ. Nature. 2008;451:949–952. doi: 10.1038/nature06802. [DOI] [PubMed] [Google Scholar]
- 16.Casella M, Dello Russo A, Russo E, Al-Mohani G, Santangeli P, Riva S, Fassini G, Moltrasio M, Innocenti E, Colombo D, Bologna F, Izzo G, Gallinghouse JG, Di Biase L, Natale A, Tondo C. Cardiology Journal. 2014;21:516–523. doi: 10.5603/CJ.a2013.0153. [DOI] [PubMed] [Google Scholar]
- 17.Nwankire CE, Venkatanarayanan A, Glennon T, Keyes TE, Forster RJ, Ducree J. Biosens Bioelectron. 2015;68:382–389. doi: 10.1016/j.bios.2014.12.049. [DOI] [PubMed] [Google Scholar]
- 18.Magliulo M, Mallardi A, Gristina R, Ridi F, Sabbatini L, Cioffi N, Palazzo G, Torsi L. Anal Chem. 2013;85:3849–3857. doi: 10.1021/ac302702n. [DOI] [PubMed] [Google Scholar]
- 19.McCauley TG, Hamaguchi N, Stanton M. Anal Biochem. 2003;319:244–250. doi: 10.1016/s0003-2697(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 20.Tombelli S, Minunni M, Mascini M. Biomol Eng. 2007;24:191–200. doi: 10.1016/j.bioeng.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Kara P, de la Escosura-Muñiz A, Maltezda Costa M, Guix M, Ozsoz M, Merkoçi A. Biosens Bioelectron. 2010;26:1715–1718. doi: 10.1016/j.bios.2010.07.090. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Tuleouva N, Ramanculov E, Revzin A. Anal Chem. 2010;82:8131–8136. doi: 10.1021/ac101409t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai RY, Plaxco KW, Heeger AJ. Anal Chem. 2007;79:229–233. doi: 10.1021/ac061592s. [DOI] [PubMed] [Google Scholar]
- 24.Du L, Wu C, Peng H, Zou L, Zhao L, Huang L, Wang P. Sens Actuators, B. 2013;187:481–487. [Google Scholar]
- 25.Liss M, Petersen B, Wolf H, Prohaska E. Anal Chem. 2002;74:4488–4495. doi: 10.1021/ac011294p. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew Chem. 2005;117:5592–5595. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 27.Stoltenburg R, Reinemann C, Strehlitz B. Biomol Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Ruigrok VJB, Levisson M, Eppink MHM, Smidt H, van der Oost J. Biochem J. 2011;436:1–13. doi: 10.1042/BJ20101860. [DOI] [PubMed] [Google Scholar]
- 29.Burbulis I, Yamaguchi K, Yu R, Resnekov O, Brent R. Nat Methods. 2007;4:1011–1014. doi: 10.1038/nmeth1127. [DOI] [PubMed] [Google Scholar]
- 30.Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Mol Ther–Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayasena SD. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 32.Hakkinen H. Nat Chem. 2012;4:443–455. doi: 10.1038/nchem.1352. [DOI] [PubMed] [Google Scholar]
- 33.Xue Y, Li X, Li H, Zhang W. Nat Commun. 2014;5:4348. doi: 10.1038/ncomms5348. [DOI] [PubMed] [Google Scholar]
- 34.Rehak M, Snejdarkova M, Otto M. Biosens Bioelectron. 1994;9:337–341. doi: 10.1016/0956-5663(94)80033-2. [DOI] [PubMed] [Google Scholar]
- 35.Orazem ME, Tribollet B. Electrochemical Impedance Spectroscopy. Vol. 48 John Wiley & Sons; 2011. [Google Scholar]
- 36.Miodek A, Regan EM, Bhalla N, Hopkins NA, Goodchild SA, Estrela P. Sensors. 2015;15:25015–25032. doi: 10.3390/s151025015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan J, Ai J, Zhang Y, Geng X, Gao Q, Cheng Z. Sci Rep. 2016;6:19806. doi: 10.1038/srep19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Usiskin RE, Maruyama S, Kucharczyk CJ, Takeuchi I, Haile SM. J Mater Chem A. 2015;3:19330. [Google Scholar]
- 39.Yu X, Lv R, Ma Z, Liu Z, Hao Y, Li Q, Xu D. Analyst. 2006;131:745–750. doi: 10.1039/b517148b. [DOI] [PubMed] [Google Scholar]
- 40.Pasceri V, Patti G, Nusca A, Pristipino C, Richichi G, Di Sciascio G. Circulation. 2004;110:674–678. doi: 10.1161/01.CIR.0000137828.06205.87. [DOI] [PubMed] [Google Scholar]
- 41.Harris B, Nageh T, Marsden J, Thomas M, Sherwood R. Ann Clin Biochem. 2000;37:764–769. doi: 10.1258/0004563001900075. [DOI] [PubMed] [Google Scholar]
- 42.Moreira FTC, Dutra RAF, Noronha JP, Sales MGF. Biosens Bioelectron. 2014;56:217–222. doi: 10.1016/j.bios.2013.12.052. [DOI] [PubMed] [Google Scholar]
- 43.Torabi F, Mobini Far HR, Danielsson B, Khayyami M. Biosens Bioelectron. 2007;22:1218–1223. doi: 10.1016/j.bios.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 44.Chon H, Lee S, Yoon SY, Lee EK, Chang SI, Choo J. Chem Commun. 2014;50:1058–1060. doi: 10.1039/c3cc47850e. [DOI] [PubMed] [Google Scholar]
- 45.Kim D-H, Cho I-H, Park J-N, Paek S-H, Cho H-M, Paek S-H. Biosensors and Bioelectronics. 2016 doi: 10.1016/j.bios.2016.08.035. [DOI] [Google Scholar]
- 46.Zhang GJ, Chai KTC, Luo HZH, Huang JM, Tay IGK, Lim AEJ, Je M. Biosens Bioelectron. 2012;35:218–223. doi: 10.1016/j.bios.2012.02.052. [DOI] [PubMed] [Google Scholar]
- 47.Lee JO, So HM, Jeon EK, Chang H, Won K, Kim Y. Anal Bioanal Chem. 2008;390:1023–1032. doi: 10.1007/s00216-007-1643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Sullivan C. Anal Bioanal Chem. 2002;372:44–48. doi: 10.1007/s00216-001-1189-3. [DOI] [PubMed] [Google Scholar]
- 49.Zhang YS, Aleman J, Arneri A, Bersini S, Piraino F, Shin SR, Dokmeci MR, Khademhosseini A. Biomed Mater. 2015;10:034006. doi: 10.1088/1748-6041/10/3/034006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCain ML, Sheehy SP, Grosberg A, Goss JA, Parker KK. Proc Natl Acad Sci U S A. 2013;110:9770–9775. doi: 10.1073/pnas.1304913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim SB, Bae H, Cha JM, Moon SJ, Dokmeci MR, Cropek DM, Khademhosseini A. Lab Chip. 2011;11:1801–1807. doi: 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Masse S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M. Nat Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas CA, Jr, Springer PA, Loeb GE, Berwald-Netter Y, Okun LM. Exp Cell Res. 1972;74:61–66. doi: 10.1016/0014-4827(72)90481-8. [DOI] [PubMed] [Google Scholar]
- 54.Grosberg A, Alford PW, McCain ML, Parker KK. Lab Chip. 2011;11:4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, Lang Q, Zhang YS, Shin SR, Calzone G, Annabi N, Shupe T, Bishop C, Atala A, Dokmeci MR, Khademhosseini A. Biofabrication. 2016;8:014101. doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 56.Fredericks S, Collinson PO, Holt DW. Clin Chem. 2000;46:432–435. [PubMed] [Google Scholar]
- 57.Saad SY, Najjar TAO, Alashari M. J Biochem Mol Toxicol. 2004;18:78–86. doi: 10.1002/jbt.20012. [DOI] [PubMed] [Google Scholar]
- 58.Rosalki SB, Roberts R, Katus HA, Giannitsis E, Ladenson JH, Apple FS. Clin Chem. 2004;50:2205–2213. doi: 10.1373/clinchem.2004.041749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.